Short abstract
An expressed sequence tag (EST) project on the adult tsetse midgut, the major organ system for establishment and early development of trypanosomes has been undertaken. The most notable block of genes upregulated in response to trypanosome challenge are a series of Toll and Imd genes and a series of genes involved in oxidative stress responses.
Abstract
Background
Tsetse flies transmit African trypanosomiasis leading to half a million cases annually. Trypanosomiasis in animals (nagana) remains a massive brake on African agricultural development. While trypanosome biology is widely studied, knowledge of tsetse flies is very limited, particularly at the molecular level. This is a serious impediment to investigations of tsetse-trypanosome interactions. We have undertaken an expressed sequence tag (EST) project on the adult tsetse midgut, the major organ system for establishment and early development of trypanosomes.
Results
A total of 21,427 ESTs were produced from the midgut of adult Glossina morsitans morsitans and grouped into 8,876 clusters or singletons potentially representing unique genes. Putative functions were ascribed to 4,035 of these by homology. Of these, a remarkable 3,884 had their most significant matches in the Drosophila protein database. We selected 68 genes with putative immune-related functions, macroarrayed them and determined their expression profiles following bacterial or trypanosome challenge. In both infections many genes are downregulated, suggesting a malaise response in the midgut. Trypanosome and bacterial challenge result in upregulation of different genes, suggesting that different recognition pathways are involved in the two responses. The most notable block of genes upregulated in response to trypanosome challenge are a series of Toll and Imd genes and a series of genes involved in oxidative stress responses.
Conclusions
The project increases the number of known Glossina genes by two orders of magnitude. Identification of putative immunity genes and their preliminary characterization provides a resource for the experimental dissection of tsetse-trypanosome interactions.
Background
The African trypanosomes that cause sleeping sickness in humans and nagana in livestock are cyclically transmitted by tsetse flies (Glossinidae). Tsetse flies are obligate blood feeders and ingest trypanosomes along with the blood meal from infected animals. In the fly, the trypanosomes undergo complex cycles of growth and development all of which occur within the lumen of the alimentary canal of the fly and, in the case of the brucei group trypanosomes, the salivary glands [1]. Tsetse flies are normally refractory to trypanosome infection with typically less than half the fly population becoming infected, even under ideal conditions in the laboratory. This is reflected in field infection rates which often fail to exceed 10% of the fly population. In addition, many of those that become infected fail to produce mature parasites and therefore never become infective and thus are incapable of transmitting the parasite. Many factors play a part in determining the success or failure of the infection and maturation process [2,3] with fly immunity factors of particular importance [4,5]. We are particularly interested in the immunological barriers to the initial establishment of trypanosome infection in the midgut [5]. In the longer term it will also be of interest to investigate the effects of fly immune mechanisms on the other stages of trypanosome maturation leading to the infective stage. However, the paucity of information on tsetse genes is currently a severe barrier to rapid progress in these areas. Here we report the sequencing of 21,427 expressed sequence tags (EST) from the midgut of adult Glossina morsitans morsitans, which were grouped in clusters potentially representing 8,876 unique genes. This increases the number of Glossina genes in public databases by two orders of magnitude. Putative functions for 3,884 were suggested by homology. Of these, 68 with putative immune-related functions were selected, macroarrayed and their transcriptional profiles were investigated following bacterial or trypanosome infection of the fly. All sequences generated in this project are available from the Sanger Glossina morsitans GeneDB database [6].
Results and discussion
The estimated complexity of the normalized library was 2.3 × 106. A total of 12,768 randomly selected clones were sequenced. Of these 1,128 had no insert and are not included in other figures. A total of 10,450 clones were sequenced from the 59 end, 10,977 were sequenced from the 39 end and 9,857 were sequenced from both ends. This yielded 21,427 ESTs (9,879,196 bp). Median EST size was 461 bp and 8,983 of the ESTs contained polyA tails. Overall, 3,761 59 and 39 read pairs from the same clone overlap, giving full sequence of a clone. The cDNA library insert size was tested by comparing the sequences to previously characterized Glossina genes (of which there are 19). Interestingly, 65 clones had higher than 95% homology to these genes across their length in both the forward and reverse direction. We estimate these clones range in size from 113 bp to 1,870 bp and we estimated the average insert size to be 990 bp. All the 39 sequences hitting the known Glossina genes had polyA tails but not always as long as the previously described sequences. The average distance of the 59 end start for the EST was 124 bp from the start of the cDNA. None of the clones contained full length cDNAs.
Clustering with Phrap (Phil Green, unpublished observations) produced 3,220 clusters with a median membership of 4.90 (range 2-135; 74.3% of the total EST). This left 5,656 singletons (25.7% of the total EST). The ESTs generated were 73.7% redundant which is relatively low considering that all clones were sequenced from both ends and hence there is an inherent level of redundancy caused by overlapping forward and reverse reads, which we estimate to be 10%. Preliminary sequencing of a library that had not been normalized demonstrated a redundancy of 87.5%, after just 181 reads. At a similar stage of sequencing the normalized library had a redundancy level of just 10%.
It is probable that the final midgut library contained ESTs representing transcripts from four sources: midguts exposed to trypanosomes from one to seven days; fat body from the same trypanosome-challenged flies; trypanosomes; and bacterial symbionts from the fly. Whilst the vast majority of mRNA used in library construction was of midgut origin the minor components would be expected to increase in representation during the normalization procedure. We found 356 ESTs that matched known T. brucei DNA sequences with a BLASTn score above 400 and these were eliminated. Tsetse flies also contain three bacterial symbionts [7]. Of these, Wigglesworthia and Sodalis have a strong presence in the midgut. We attempted to minimize the representation of these in the library by including a polyA mRNA purification step which acts to exclude bacterial sequences - most of which characteristically lack a polyA tail. In addition we analyzed the data specifically looking for bacterial sequences. Using BLASTX against Swall we identified 34 sequences with highest hits to bacterial sequences: identity varied between 46% and 96%. Of these 34 sequences, 20 were to the endosymbiont Wigglesworthia brevipalpis, the midgut endosymbiont found in another tsetse fly Glossina brevipalpis. The amino acid identity displayed by these 20 hits ranged from 52% to 91%. Given the very high abundance of Wigglesworthia in the midgut bacteriome it is clear that the polyA exclusion strategy was effective in minimizing the representation of bacterial sequences in the library. Of the remaining sequences, 3,884 sequences had matches in the Drosophila protein database, that is, 45% of the total number of clusters and singletons. We also found that 17.15% of the 59 alignments contained AUG in the alignment. Only 151 sequences had matches to proteins in Swall and not to Drosophila proteins. The high degree of similarity between the two species makes Glossina a very good comparative model for studying Drosophila and the Drosophila database an excellent resource for those studying Glossina. A description of the proportion of ESTs falling into different functional classes is given in Figure 1. The sequences have been submitted to GenBank: Accession numbers BX548257-BX569683.
Figure 1.

Classification of EST clusters using Gene Ontology TM. (a) Individual annotations were summarized by mapping to broad level terms from the 'biological process' ontology and (b) the 'molecular function'. ESTs which mapped to process unknown or molecular function unknown are not included in the charts.
Immune-related genes
Using BLASTX and Pfam we identified 78 homologs of genes with known or putative immunity-related functions. Although the insect midgut is known to be involved in the immune response [8-10] this is still a surprisingly high number of immune-related genes to find, particularly in the gut of an insect which feeds exclusively on (normally sterile) vertebrate blood. For example, an EST project centered on immune-competent, hemocyte-like cell lines from the malarial mosquito Anopheles gambiae [11] identified only 38 such clusters. Many explanations are possible for the comparatively high number found in Glossina. For example, the genes identified may not have immunity-related functions in the midgut. Alternatively, this high number may be a function of the presence of bacterial symbionts in the tsetse fly midgut [7] and the need to regulate their numbers. On the other hand, it may be a result of the low redundancy of the library and the comparatively large numbers of ESTs produced in this study.
Serine proteases and inhibitors
In this group, 15 genes have been identified. The presence of 11 putative proteinase inhibitors in a gut dedicated to the digestion of a high protein diet is surprising and suggests they have an important function. Serine protease genes possessing 'clip domains' are implicated in the activation of the prophenol oxidase cascade and other cascades associated with the immune response [12]. Four homologs of such serine proteases have been uncovered (Table 1). The function of such genes in a midgut environment remains to be determined. Many serine proteases involved in the immune response exist in fine balance with serine protease inhibitors to ensure that the impact of protease-activated cascades remains localized in time and space [13,14]. Serpins may have particular involvement in inactivating serine proteases with clip domains [13]. Some serpins are involved in the immune response. For example, Spn43Ac in Drosophila is involved in the regulation of Toll-mediated antifungal defense [15]. We have identified nine putative serpins, but it seems unlikely that these are involved in the regulation of complex insect-based cascades in the midgut. Instead, the large numbers of serpins found here may reflect the need to inactivate the complement and coagulation cascades in the blood meal - to protect the midgut epithelium and retain the meal in a physical state suitable for digestion, respectively. In support of the latter contention, Gmm-2766 is a homolog of Infestin which is reported in the NCBI protein database (AAK57342) as a novel thrombin inhibitor present in the midgut of the blood-sucking hemipteran Triatoma infestans. Serpins may also have an additional direct role in immunity as proteolytic enzymes are important virulence factors in many pathogens and proteinase inhibitors may have important roles in regulating disease [16].
Table 1.
Serine protease-related group of putative immune-related genes chosen for arraying
| Contig/EST | Length (bp) | E value | Homology | Putative function |
| Gmm-1598 * | 850 | 3e-41 | CG3066 (dm) | Serine carboxypeptidase |
| Tse112d02.q * | 473 | 4e-63 | Easter (dm) | Serine endopeptidase |
| Tse91h03 * | 576 | 1e-56 | CG3066 (dm) | Serine carboxypeptidase |
| Tse129g02 * | 476 | 3e-46 | PPAF (tm) | PPO activation |
| Tse105c10.q * | 557 | 4e-41 | TepIV (dm) | Complement c3/α2 macroglobulin |
| Tse48e12.q * | 553 | 7e-38 | TepIV (dm) | Complement c3/α2 macroglobulin |
| Gmm-3334 * | 1,454 | 1e-102 | Sp6 (dm) | Serpin |
| Gmm-3352 * | 2,529 | 1e-109 | Sp5 (dm) | Serpin |
| Gmm-2356 * | 962 | 4e-44 | CG9460 (dm) | Serpin |
| Gmm-2594 * | 570 | 2e-38 | CG9455 (dm) | Serpin |
| Gmm-0033 * | 452 | 1e-21 | Sp6 (dm) | Serpin |
| Tse72e04.q * | 436 | 5e-37 | Spn43Ab (dm) | Serpin |
| Gmm-2766 | 450 | 7e-06 | AAK57342.1 (ti) | Serpin |
| Tse72e08 * | 543 | 2e-23 | CG6680 (dm) | Serpin |
| Tse94c04.q * | 560 | 2e-52 | CG5392 (dm) | Serpin |
The contig (Gmm-) or clone identifier (Tse) is given for each gene. A 'q' following the clone identifier indicates sequencing was from the 39 end of the clone. The tables include the lowest BLASX E value (most significant similarity) together with putative function based on this homology. dm, Drosophila melanogaster; cb, Chrysomya bezziana; sp, Sarcophaga peregrina; ds, Drosophila simulans; tm, Tenebrio molitor; ti, Triatoma infestans; bm, Bombyx mori; ce, Caenorhabditis elegans; ag, Anopheles gambiae; gm, G. m. morsitans. *All genes with an Anopheles gambiae homolog at 1e-08 or less.
Two further components of this group are members of the complement C3/α2 macroglobulin superfamily, which are homologs of the TepIV gene in Drosophila. TepIV is strongly upregulated in response to immune challenge in adult Drosophila and it has been suggested that Tep genes in Drosophila may have complement-like properties [17]. Other suggested functions for members of this protein family are as proteinase inhibitors [16].
Adhesion proteins
In this group, 28 putative adhesion genes have been isolated (Table 2). We have eliminated homologs of enzymes involved in sugar metabolism which also contain chitin-binding domains. This leaves 14 molecules containing chitin-binding domains which may have an adhesive function and play a role in immunity. Tse36b05, Gmm-3093 and Gmm-2445 are homologs of a mucin, a peritrophic matrix constituent and a peritrophin, respectively, and may play a defensive barrier role in the midgut [18,19]. The cluster Gmm-1329 is a homolog of the Anopheles gambiae gene ICHIT which is immune inducible and highly expressed in the adult mosquito midgut [20]. Five other clusters are homologs of the Drosophila gene, Chit, which encodes a gene related to chitinase but which lacks catalytic activity. In Drosophila, the product encoded by Chit is an imaginal disk growth factor [21]. The function(s) of this group of genes in an adult insect is open to question, but may include an immune function given the immune responsiveness of other molecules carrying chitin-binding domains [20].
Table 2.
Adhesion molecules group of putative immune-related genes chosen for arraying
| Contig/EST | Length (bp) | E value | Homology | Putative function |
| Gmm-0398 | 539 | .002 | BRPCHIT (bm) | Chitin binding |
| Gmm-2843 | 1,061 | .003 | NP508588 (ce) | Chitin binding |
| Gmm-2445 * | 374 | 4e-23 | Peritrophin 15 (cb) | Chitin binding |
| Gmm-3093 | 1,046 | 2e-16 | CG7252 (dm) | Chitin binding |
| Gmm-3262 | 891 | 2e-06 | CG14125 (dm) | Chitin binding |
| Gmm-2386 * | 610 | 2e-31 | Chit (dm) | Chitin binding |
| Gmm-2904 * | 643 | 7e-65 | Chit (dm) | Chitin binding |
| Gmm-2945 * | 666 | 1e-62 | Chit (dm) | Chitin binding |
| Gmm-0048 * | 573 | 7e-67 | Chit (dm) | Chitin binding |
| Tse121d04 * | 618 | 3e-29 | Chit (dm) | Chitin binding |
| Gmm-2709 | 813 | 7e-13 | CG8473 (dm) | Chitin binding |
| Tse74h11 * | 584 | 9e-27 | CG9357 (dm) | Chitin binding |
| Tse98g05 * | 381 | 6e-29 | CG3044 (dm) | Chitin binding |
| Gmm-1329 * | 529 | 3e-08 | ICHIT (ag) | Chitin binding |
| Tse36b05 | 499 | 2e-25 | Hemomucin (dm) | Opsonization |
| Gmm-3156 * | 1,034 | 5e-67 | PGRP-LB (dm) | Binding bacteria |
| Tse70a12 * | 606 | 3e-28 | PGRP-LC (dm) | Binding bacteria |
| Tse81d11 * | 528 | 3e-27 | GNBP1 (dm) | Binding bacteria |
| Gmm-2608 * | 1,008 | 2e-16 | CG7228 (dm) | Scavenger receptor |
| Gmm-3328 * | 2,081 | 1e-139 | CG5750 (dm) | Scavenger receptor |
| Gmm-0072 * | 366 | 7e-23 | Crq (dm) | Scavenger receptor |
| Tse108b10.q * | 582 | 2e-65 | CG7228 (dm) | Scavenger receptor |
| Tse59b02.q * | 450 | 6e-63 | BEST:CK01577 (dm) | Scavenger receptor |
| Tse84d03.q | 472 | 1e-23 | CG2736 (dm) | Scavenger receptor |
| Tse29b09.q * | 488 | 6e-26 | Ldlr (dm) | Scavenger receptor |
| Gmm-2412 * | 666 | 3e-28 | LECA SARPE (sp) | C type lectin |
| Gmm-3236 * | 1,302 | 3e-89 | CG4115 (dm) | C type lectin |
| Tse33h03 * | 460 | 5e-62 | CG5335 (dm) | ConA type lectin |
See legend to Table 1.
Three homologs of pattern recognition proteins known to bind microbial surface molecules have been found. Two are homologs of peptidoglycan-binding proteins which are pattern recognition proteins involved in immune response pathways targeting bacterial infections [22-25]. The third is a homolog of a gram negative binding protein known to bind lipopolysaccharide and β 1-3 glucan from gram negative bacteria and fungi, respectively. The homolog is involved in the regulation of NF-κB-dependent antimicrobial peptide gene expression in Drosophila [26].
The sequencing uncovered seven homologs of scavenger receptor molecules which may have involvement in the immune response including a homolog of croquemort which is a macrophage receptor involved in the phagocytosis of apoptotic cells in Drosophila [27].
Of particular interest are three putative lectin genes. Two are homologs of c-type lectin superfamily members. One is from Drosophila and the other an immune responsive c-type lectin from the fleshfly Sarcophaga peregrina [28]. The third putative lectin (Tse33h03.q) is a member of the ConA-like lectin superfamily. Indirect evidence from sugar inhibition experiments suggests lectins play a role in determining the initial success of trypanosome infections in tsetse flies and stimulate the maturation of successful trypanosome infections [4,29]. Until now these genes have defied attempts to clone them, hindering precise analysis of their effects on trypanosomes. Further work on these lectins is underway.
Other putative immune-related genes
There are 35 genes in this group. An attacin gene, distinct from the previously reported AttA gene from Glossina [5], has been identified; attacins are important antimicrobial peptides in insects. It is interesting that other antimicrobial peptide genes, including ones already cloned and sequenced which are known to be expressed in the adult midgut [5], are notable by their absence from our EST list. Reactive oxygen species (ROS) play a role in insect immunity [30]; we identified 18 putative antioxidant genes consisting of five superoxide dismutases, three catalases, five peroxidases and five peroxiredoxins. Here we give details for the eight which were included in the macroarrays - we are carrying out further work on these antioxidant genes. This unusually high number of antioxidant genes may reflect the need of the fly to protect the midgut epithelium from oxygen radical attack on lipids caused by the abundance of heme molecules liberated from the digested blood meal [31]. Some of these antioxidant genes may also protect against ROS generated during immune responses. For example, Gmm-2058 is a homolog of Dpx4156, which is itself a homolog of mammalian and parasite genes believed to play a role in preventing oxidative damage by scavenging extracellular ROS released by immune effector cells [32,33].
Five clusters with homology to genes involved in iron metabolism were identified. Iron metabolism genes have been implicated in immunity [34,35]. The remaining 11 genes are homologs of genes involved in signaling pathways associated with the immune response. They include components of the well-studied antimicrobial peptide regulating pathways Toll and Imd [36]. They also include a homolog of the Drosophila Thor gene which has been implicated in post-transcriptional regulation of immune responses [37].
In the future, comparative studies of immune-related genes in different hematophagous species may be informative in the understanding of the relationships between insects and the parasites that they transmit. Publication of the full genome of A. gambiae makes this the benchmark species [38]. Of the 68 immunity-related genes used in the arrays, 54 had a homolog in the A. gambiae database at a BLASTX value of 1e-08 or less (Tables 1,2,3) suggesting that Glossina and Anopheles potentially show considerable overlap in the genes underpinning their immune systems. This suggests that future comparative studies of these species may help provide wide ranging insights into immune mechanisms in blood-sucking insects.
Table 3.
Miscellaneous group of putative immune-related genes chosen for arraying
| Contig/EST | Length (bp) | E value | Homology | Putative function |
| Gmm-0596 * | 560 | 7e-16 | Dorsal (dm) | Toll/signaling |
| Tse15b02 * | 403 | 3e-24 | ECSIT (dm) | Toll/signaling |
| Tse44a11.q | 571 | 2e-16 | Toll9 (dm) | Toll/signaling |
| Tse51g11 | 482 | 3e-23 | Relish (ds) | Imd/signaling |
| Tse37e05.q | 374 | 6e-14 | Relish (ds) | Imd/signaling |
| Gmm-1270 | 517 | 3e-27 | Imd (dm) | Imd/signaling |
| Gmm-3029 * | 1,073 | 8e-46 | Thor (dm) | Translational regulation |
| Gmm-2522 * | 972 | 2e-62 | P38b (dm) | MAP kinase |
| Tse131b02 * | 492 | 1e-32 | Mpk2 (dm) | MAP kinase |
| Tse41c10 * | 539 | 2e-61 | bsK (dm) | JNK cascade |
| Tse66g09.q * | 445 | 9e-49 | Stat92E (dm) | JAK-STAT cascade |
| Gmm-1963 | 561 | 1e-49 | Attacin (gm) | Antimicrobial |
| Gmm-2058 * | 953 | 4e-78 | Jafrac2 (dm) | Extracellular peroxidase |
| Gmm-0601 * | 458 | 1e-28 | Prx5037 (dm) | Extracellular peroxidase |
| Tse126a11 * | 453 | 9e-39 | Prx5037 (dm) | Extracellular peroxidase |
| Gmm-2087 * | 881 | 2e-98 | Prx6005 (dm) | Peroxidase |
| Gmm-2619 * | 957 | 1e-104 | Dpx2540 (dm) | Peroxidase |
| Gmm-3099 * | 824 | 1e-81 | Dpx4783 (dm) | Peroxidase |
| Gmm-1799 * | 585 | 2e-58 | Catalase (dm) | Oxidative stress |
| Gmm-2629 * | 950 | 1e-148 | Catalase (dm) | Oxidative stress |
| Gmm-1694 * | 460 | 6e-34 | Irp-1B (dm) | Iron metabolism |
| Gmm-0034 * | 474 | 8e-19 | Fer1HCH (dm) | Iron metabolism |
| Gmm-3307 * | 1,239 | 3e-88 | Fer2LCH (dm) | Iron metabolism |
| Gmm-3348 * | 1,747 | 2e-38 | Fer1HCH (dm) | Iron metabolism |
| Tse100h09 | 525 | 2e-53 | Transferrin (sp) | Iron metabolism |
See legend to Table 1.
Arrays
Both trypanosome and bacterial challenges lead to a significant number of downregulated genes (Figure 2) which may represent a malaise reaction of the gut to infection such as that recorded in Drosophila [39] and Manduca [40,41]. Only relatively small numbers of genes are upregulated in response to infection. However, based on their homology, many of the genes which are not upregulated probably do have an immune function - for example, Gmm-3156, Tse70a12 and Tse812d11 are all involved in bacterial binding.
Figure 2.
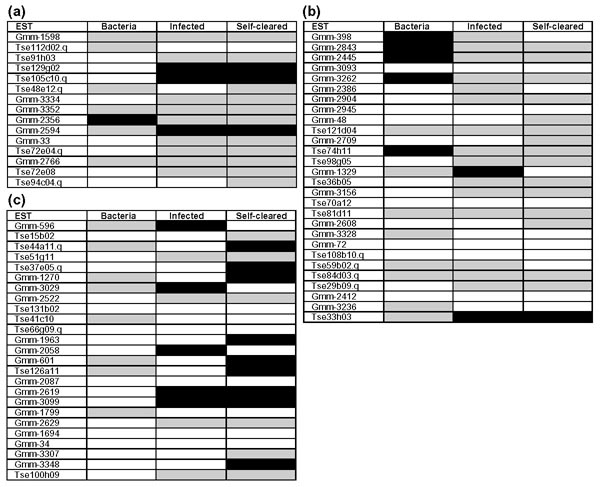
Expression profiles of putative immunity genes following bacteria or trypanosome challenge. Macroarrays were produced for the 68 putative immunity-related genes listed in Tables 1,2,3. The contig (Gmm-) or clone identifier (Tse) is given for each gene in the same order as Tables 1,2,3; so (a) contains the protease-related ESTs (Table 1), (b) contains the adhesion molecules (Table 2) and (c) contains the miscellaneous ESTs (Table 3). Bacteria column: one-day old, bacteria-fed flies divided by the average of one-day-old fed control flies; Infected column: trypanosome-infected flies divided by the average of an equivalent set of non-challenged control flies; Self cleared column: flies which have cleared a trypanosome infection divided by the average of an equivalent set of non-challenged control flies. For full details see Materials and methods. The following number of arrays (n) were screened: one-day old, fed control flies (n = 5); bacteria-fed flies (n = 4); non-trypanosome-challenged control flies (n = 5); self-cleared flies (n = 4); infected flies (n = 5). A black box indicates at least a doubling of expression levels in at least n-1 of the observations compared to the average of the control. A gray box indicates at least a halving of expression levels in at least n - 1 of the observations compared to the average of the control. A white box indicates no change in expression level.
Successful trypanosome infection results in considerable changes in the transcriptional profile (Figure 2). Comparing infected midguts to equivalent non-trypanosome challenged midguts, 10 genes are upregulated and 28 downregulated. Comparing midguts from tsetse which had self-cleared trypanosome infections to equivalent non-trypanosome challenged midguts, 13 genes were upregulated and 31 downregulated. Remarkably, six of the 10 genes upregulated and 25 of the 28 genes downregulated in infected flies show the same changes in self-cleared flies.
Two peroxidase homologs are upregulated in both self-cleared and infected flies. In addition two more peroxidase genes are upregulated in self-cleared flies. It is known that trypanosomes are particularly susceptible to ROS [42,43] and it is an interesting speculation that these genes may be upregulated to protect the fly against ROS which are generated during the tsetse immune response against trypanosomes (Zhengrong Hao and Serap Aksoy, unpublished observations). A homolog of the TepIV gene of D. melanogaster, a member of the larger c3/α2 macroglobulin family, is also upregulated in both trypanosome-infected and self-cleared flies. It has been suggested that invertebrate members of this family may have complement-like [17,44] or proteinase inhibitor functions [16]. Members of the Tep gene family of D. melanogaster are also upregulated on immune stimulation [17]. A serpin is upregulated in both self-cleared and infected flies and it is interesting to note that artificial proteinase inhibitors can have cytocidal effects on trypanosomes [45]. One of the three putative lectins, Tse33h03, is also upregulated in both self-cleared flies and infected flies. While the full length cDNA for the other two lectins described both have putative signal peptides and so are probably secreted from cells, this particular lectin is of the ConA superfamily and lacks a signal peptide and so is probably intracellular (M.J.L., unpublished observations). Therefore it is unlikely that this lectin has a direct interaction with trypanosomes.
Genes that are differently regulated between infected and self-cleared flies are clearly of particular interest. The peroxidase homologs are discussed above. Others upregulated in self-cleared flies but not in infected flies include homologs of members of both the Toll and Imd signaling pathways. This may suggest a role for these pathways in successful tsetse immune responses to trypanosomes. These findings are consistent with previous reports that immune peptides known to be regulated by these pathways in D. melanogaster are upregulated in response to trypanosome challenge [5]. Consistent with these findings, attacin, which can be regulated through both pathways in Drosophila [46], is upregulated in self-cleared flies.
The expression profile resulting from bacterial infection of the gut is quite distinct from that occurring in response to trypanosome infection (Figure 2). This suggests the possibility that different recognition pathways are involved in bacterial and trypanosome infections. Of particular note in these expression profiles is the upregulation of homologs of several genes implicated in binding chitin - only following bacterial infection. The consistent pattern of upregulation seen here adds support to the possibility of an immune function for chitin-binding molecules in insects [20]. The upregulation of genes involved in the Toll and Imd pathways and genes involved in the management of oxidative stress, uniquely during trypanosome infection, also merits further investigation.
The array data represent a first attempt to assess immune responses in the tsetse fly midgut. The ESTs reported here will permit more sophisticated approaches to be made in the future when it will be possible to determine the effect of the insect immune system on each of the developmental stages of the parasite. This will help us to understand why so few tsetse flies that have ingested trypanosomes go on to become infective.
The project has increased the number of Glossina genes in public databases by two orders of magnitude. Identification of the putative immunity genes provides a resource which should be of value in the experimental dissection of tsetse trypanosome interactions. For example, we are currently producing recombinant lectins to determine their impact on trypanosomes. The remarkably high proportion of the homologs found in the Drosophila database suggests that this rich resource will contribute much to our knowledge of tsetse biology. In particular, as annotation of the Drosophila genome proceeds it is likely that putative functions can be ascribed to many of the 4,485 ESTs for which no putative function has been ascribed.
Materials and methods
Flies and trypanosomes
Equal numbers of male and female flies of Glossina morsitans morsitans, originally established from puparia collected in Zimbabwe and maintained in Bristol University, were used for all experiments, and fed routinely on sterile horse blood unless otherwise stated. Flies were infected by adding approximately 106 trypanosomes of strain Trypanosoma brucei brucei TSW 196 per ml of blood to the first blood meal. For EST library construction a total of 191 trypanosome-challenged midguts (sectioned immediately anterior to the proventriculus and immediately anterior to the junction of midgut with the Malpighian tubules) were dissected from approximately equal numbers of adult male and female flies. The 191 midgut sample was obtained from approximately equal numbers of flies dissected daily from one to seven days after the infective blood meal. While attention was given to remove all fat body tissue during midgut dissections, minor contamination could not be ruled out since fat body tissue adheres strongly to the midgut in places.
For the array experiments self-cleared flies are defined as those which have had a trypanosome-infected meal but which have no microscopically detectable trypanosomes in the midgut when dissected six to nine days after the infective meal. Trypanosome infected flies are those which have trypanosomes in the midgut when dissected six to nine days after the infective meal.
Normalized library construction
A quantity of more than 2 μg of total RNA was extracted using RNAqueous™ (Ambion) according to the manufacturer's instructions. cDNA was cloned directionally into a phagemid vector (pT7T3-Pac). cDNA library normalization was performed according to 'method 4' of Bonaldo et al. [47]. This procedure is based on the hybridization of PCR-amplified cDNA inserts of a library with the library itself in the form of single-stranded circles. Following hybridization to a relatively low Cot of 5-10, the remaining single-stranded circles (normalized library) are purified over hydroxyapatite (HAP), converted to double-stranded circles by primer extension and electroporated into bacteria [47].
Sequencing
Each clone was sequenced using a T3 or T7 primer using ABI™ big dye terminator kits. The sequence was clipped for quality using Phred and vector using Cross Match (Phil Green, unpublished) and Svec_clip (Richard Mott, unpublished).
Sequence analysis
Each sequence was analyzed using BLASTX against Swall and Flybase proteins. Pfam [48] domains were identified using ESTwise (Ewan Birney, unpublished) and each Pfam domain was mapped to Interpro annotation. Contaminating T. brucei sequences were removed from the final set of clusters by screening them against all known T. brucei DNA sequences. Contaminating bacterial sequences from tsetse symbionts was a potential problem but the cDNA library construction procedures use a polyA purification step. This eliminated the vast majority of contaminating bacterial mRNA because they characteristically lack a polyA tail. In addition, all sequences were run against Swall. Those which returned the most significant hit to a bacterial sequence were recorded.
Gene Ontology (GO) [48] annotation was transferred to each sequence on the basis of BLASTX hits to Flybase proteins with a significance above E = 1 - 10 or, where there was a Pfam domain detected, the corresponding GO terms were transferred on the basis of Interpro to GO mapping.
Arrays
pT3T7-Pac plasmids containing the selected cDNA inserts were used as templates for PCR amplification of cDNA from the 68 selected putative immune-related genes. PCR products were purified using Qiagen miniprep columns and quantified by absorbance spectroscopy. PCR products were diluted to 15 ng/μl in 3 × SSC, 0.005% sarcosyl and quadrupley spotted onto each Hybond nylon membrane using a 96 pin, 1 μl slot pin replicator (V&P Scientific, Inc). Each membrane also contained three different internal controls: actin (Gmm-2387), GAPDH (Gmm-2682) and EIF (Gmm-1088) (all three of these Glossina genes were discovered in the course of this EST project).
mRNA for the generation of targets was purified from the midguts of flies with the physiological conditions listed below; each experiment was replicated for the number of times indicated. One-day-old flies, six hours after blood feeding (n = 5); One-day-old flies, six hours after feeding on blood containing heat-killed bacteria (0.5% volume:volume in blood each of Micrococcus luteus and Escherichia coli K12 RM148 grown to OD 0.5 at A600) (n = 5); Non trypanosome challenged six- to nine-day-old flies 48 hours after the last blood meal (n = 5); six- to nine-day-old flies, 48 hours after the last blood meal, which had been fed trypanosomes (see above) in the first blood meal, but which had self-cleared as determined by dissection (n = 4); six- to nine-day-old flies, 48 hours since the last blood meal, which had been fed trypanosomes in the first blood meal and which had become infected as determined by dissection (n = 5). In each replicate a separate array was hybridized with a separate target generated from a separate batch of experimental flies.
mRNA was extracted from midguts using Dynabeads and labeled with 32P using the Hotscribe First-Strand cDNA Labeling Kit (Amersham). Membranes were hybridized overnight in Ultrahyb solution (Ambion) and then washed for 2 × 5 mins in 2 × SSC, 0.1% SDS at 42°C followed by 2 × 30 mins in 0.1 × SSC, at 60°C. The membranes were scanned using a BioRad Molecular Imager FX. Image analysis was performed using ImaGene and GeneSight Lite software (BioDiscovery Inc, USA) according to the manufacturer's instructions. Background and negative control values were subtracted from each experimental value and any negative values resulting were set to zero. For normalization of total intensity on each array the mean of the internal control spots (GAPDH, actin and EIF) was averaged for each membrane separately. Values for the other spots on the membrane were then expressed relative to this average for the internal control spots. For each experimental comparison the average of the spots for each gene was calculated for all the array membranes forming the control in that pair (Figure 2) and those averages were then divided individually into the intensity reading for the corresponding genes on each of the experimental array membranes. A significant change in expression was deemed to have occurred if levels either consistently doubled or halved in at least n - 1 of the experimental array membranes. Median signal/background ratio was always >2.5.
Additional data files
The macroarray raw data (Additional data file 1) and a table showing the arrangement of spots on the arrays (Additional data file 2) are available with the online version of this article.
Supplementary Material
The macroarray raw data
A table showing the arrangement of spots on the arrays
Acknowledgments
Acknowledgements
This work was supported by a grant from the Wellcome Trust. Thanks to Jenny Berry and IAEA for providing tsetse flies.
References
- Aksoy S, Gibson WC, Lehane MJ. Perspectives on the interactions between tsetse and trypanosomes with implications for the control of trypanosomiasis. Adv Parasitol. 2003;53:1–84. doi: 10.1016/s0065-308x(03)53002-0. [DOI] [PubMed] [Google Scholar]
- Maudlin I, Welburn SC. Maturation of trypanosome infections in tsetse. Exp Parasitol. 1994;79:202–205. doi: 10.1006/expr.1994.1081. [DOI] [PubMed] [Google Scholar]
- Jordan AM. Trypanosomiasis control and African rural development. London: Longman;; 1986. [Google Scholar]
- Welburn SC, Maudlin I. Tsetse-typanosome interactions: rites of passage. Parasitol Today. 1999;15:399–403. doi: 10.1016/s0169-4758(99)01512-4. [DOI] [PubMed] [Google Scholar]
- Hao ZR, Kasumba I, Lehane MJ, Gibson WC, Kwon J, Aksoy S. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc Natl Acad Sci USA. 2001;98:12648–12653. doi: 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glossina morsitans GeneDB http://www2.genedb.org/genedb/glossina/index.jsp
- Aksoy S. Tsetse - a haven for microorganisms. Parasitol Today. 2000;16:114–118. doi: 10.1016/s0169-4758(99)01606-3. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Richman A, Muller HM, Kafatos FC. Molecular immune responses of the mosquito Anopheles gambiae to bacteria and malaria parasites. Proc Natl Acad Sci USA. 1997;94:11508–11513. doi: 10.1073/pnas.94.21.11508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehane MJ, Wu D, Lehane SM. Midgut-specific immune molecules are produced by the blood-sucking insect Stomoxys calcitrans. Proc Natl Acad Sci USA. 1997;94:11502–11507. doi: 10.1073/pnas.94.21.11502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzou P, Ohresser S, Ferrandon D, Capovilla M, Reichhart JM, Lemaitre B, Hoffmann JA, Imler JL. Tissue-specific inducible expression of antimicrobial peptide genes in Drosophila surface epithelia. Immunity. 2000;13:737–748. doi: 10.1016/s1074-7613(00)00072-8. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Casavant TL, Chang SR, Scheetz T, Roberts C, Donohue M, Schultz J, Benes V, Bork P, Ansorge W, et al. Anopheles gambiae pilot gene discovery project: identification of mosquito innate immunity genes from expressed sequence tags generated from immune-competent cell lines. Proc Natl Acad Sci USA. 2000;97:6619–6624. doi: 10.1073/pnas.97.12.6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang HB, Wang Y, Kanost MR. Pro-phenol oxidase activating proteinase from an insect, Manduca sexta: a bacteria-inducible protein similar to Drosophila easter. Proc Natl Acad Sci USA. 1998;95:12220–12225. doi: 10.1073/pnas.95.21.12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Kanost MR. The clip-domain family of serine proteinases in arthropods. Insect Biochem Mol Biol. 2000;30:95–105. doi: 10.1016/s0965-1748(99)00113-7. [DOI] [PubMed] [Google Scholar]
- Muta T, Iwanaga S. The role of hemolymph coagulation in innate immunity. Curr Opin Immunol. 1996;8:41–47. doi: 10.1016/s0952-7915(96)80103-8. [DOI] [PubMed] [Google Scholar]
- Levashina EA, Langley E, Green C, Gubb D, Ashburner M, Hoffmann JA, Reichhart JM. Constitutive activation of toll-mediated antifungal defense in serpin-deficient Drosophila. Science. 1999;285:1917–1919. doi: 10.1126/science.285.5435.1917. [DOI] [PubMed] [Google Scholar]
- Armstrong PB. The contribution of proteinase inhibitors to immune defense. Trends in Immunol. 2001;22:47–52. doi: 10.1016/s1471-4906(00)01803-2. [DOI] [PubMed] [Google Scholar]
- Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in Toll and JAK gain-of-function mutants of Drosophila. Proc Natl Acad Sci USA. 2000;97:11427–11432. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theopold U, Samakovlis C, Erdjument-Bromage H, Dillon N, Axelsson B, Schmidt O, Tempst P, Hultmark D. Helix pomatia lectin, an inducer of Drosophila immune response, binds to hemomucin, a novel surface mucin. J Biol Chem. 1996;271:12708–12715. doi: 10.1074/jbc.271.22.12708. [DOI] [PubMed] [Google Scholar]
- Lehane MJ. Peritrophic matrix structure and function. Annu Rev Entomol. 1997;42:525–550. doi: 10.1146/annurev.ento.42.1.525. [DOI] [PubMed] [Google Scholar]
- Dimopoulos G, Seeley D, Wolf A, Kafatos FC. Malaria infection of the mosquito Anopheles gambiae activates immune-responsive genes during critical transition stages of the parasite life cycle. EMBO J. 1998;17:6115–6123. doi: 10.1093/emboj/17.21.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura K, Shibata T, Saget O, Peel D, Bryant PJ. A new family of growth factors produced by the fat body and active on Drosophila imaginal disc cells. Development. 1999;126:211–219. doi: 10.1242/dev.126.2.211. [DOI] [PubMed] [Google Scholar]
- Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–759. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, Ferrandon D, Royet J. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–644. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RAB. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli. Nature. 2002;416:644–648. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- Choe KM, Werner T, Stoven S, Hultmark D, Anderson KV. Requirement for a peptidoglycan recognition protein (PGRP) in relish activation and antibacterial immune responses in Drosophila. Science. 2002;296:359–362. doi: 10.1126/science.1070216. [DOI] [PubMed] [Google Scholar]
- Kim YS, Han SJ, Ryu JH, Choi KH, Hong YS, Chung YH, Perrot S, Raibaud A, Brey PT, Lee WJ. Lipopolysaccharide-activated kinase, an essential component for the induction of the antimicrobial peptide genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:2071–2079. doi: 10.1074/jbc.275.3.2071. [DOI] [PubMed] [Google Scholar]
- Franc NC, Heitzler P, Ezekowitz RAB, White K. Requirement for croquemort in phagocytosis of apoptotic cells in Drosophila. Science. 1999;284:1991–1994. doi: 10.1126/science.284.5422.1991. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Komano H, Natori S. Expression of the lectin gene in Sarcophaga peregrina during normal development and under conditions where the defence mechanism is activated. J Insect Physiol. 1986;32:771–779. [Google Scholar]
- Maudlin I, Welburn SC. Lectin mediated establishment of midgut infections of Trypanosoma congolense and Trypanosoma brucei in Glossina morsitans. Trop Med Parasitol. 1987;38:167–170. [PubMed] [Google Scholar]
- Nappi AJ, Vass E, Frey F, Carton Y. Nitric oxide involvement in Drosophila immunity. Nitric Oxide. 2000;4:423–430. doi: 10.1006/niox.2000.0294. [DOI] [PubMed] [Google Scholar]
- Oliveira MF, Timm BL, Machado EA, Miranda K, Attias M, Silva JR, Dansa-Petretski M, de Oliveira MA, de Souza W, Pinhal NM, et al. On the pro-oxidant effects of haemozoin. FEBS Lett. 2002;512:139–144. doi: 10.1016/s0014-5793(02)02243-3. [DOI] [PubMed] [Google Scholar]
- Okado-Matsumoto A, Matsumoto A, Fujii J, Taniguchi N. Peroxiredoxin IV is a secretable protein with heparin-binding properties under reduced conditions. J Biochem. 2000;127:493–501. doi: 10.1093/oxfordjournals.jbchem.a022632. [DOI] [PubMed] [Google Scholar]
- Salazar-Calderon M, Martin-Alonso JM, de Eguino ADR, Casais R, Marin MS, Parra F. Fasciola hepatica: heterologous expression and functional characterization of a thioredoxin peroxidase. Exp Parasitol. 2000;95:63–70. doi: 10.1006/expr.2000.4495. [DOI] [PubMed] [Google Scholar]
- Weiss G, Wachter H, Fuchs D. Linkage of cell-mediated-immunity to iron-metabolism. Immunology Today. 1995;16:495–500. doi: 10.1016/0167-5699(95)80034-4. [DOI] [PubMed] [Google Scholar]
- Dunkov BC, Zhang DZ, Choumarov K, Winzerling JJ, Law JH. Isolation and characterization of mosquito ferritin and cloning of a cDNA that encodes one subunit. Arch Insect Biochem Physiol. 1995;29:293–307. doi: 10.1002/arch.940290307. [DOI] [PubMed] [Google Scholar]
- Levashina EA, Ohresser S, Lemaitre B, Imler JL. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J Mol Biol. 1998;278:515–527. doi: 10.1006/jmbi.1998.1705. [DOI] [PubMed] [Google Scholar]
- Bernal A, Kimbrell DA. Drosophila Thor participates in host immune defense and connects a translational regulator with innate immunity. Proc Natl Acad Sci USA. 2000;97:6019–6024. doi: 10.1073/pnas.100391597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. The genome sequence of the malaria mosquito Anopheles gambiae. Science. 2002;298:129–149. doi: 10.1126/science.1076181. [DOI] [PubMed] [Google Scholar]
- Daffre S, Kylsten P, Samakovlis C, Hultmark D. The lysozyme locus in Drosophila melanogaster - an expanded gene family adapted for expression in the digestive-tract. Mol Gen Genet. 1994;242:152–162. doi: 10.1007/BF00391008. [DOI] [PubMed] [Google Scholar]
- Dunn PE, Bohnert TJ, Russell V. Regulation of antibacterial protein-synthesis following infection and during metamorphosis of Manduca sexta. Ann NY Acad Sci. 1994;712:117–130. doi: 10.1111/j.1749-6632.1994.tb33567.x. [DOI] [PubMed] [Google Scholar]
- Russell V, Dunn PE. Antibacterial proteins in the midgut of Manduca sexta during metamorphosis. J Insect Physiol. 1996;42:65–71. doi: 10.1002/arch.940170202. [DOI] [PubMed] [Google Scholar]
- Gobert AP, Semballa S, Daulouede S, Lesthelle S, Taxile M, Veyret B, Vincendeau P. Murine macrophages use oxygen- and nitric oxide-dependent mechanisms to synthesize S-nitroso-albumin and to kill extracellular trypanosomes. Infect Immun. 1998;66:4068–4072. doi: 10.1128/iai.66.9.4068-4072.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlotra RK. Antioxidant defense mechanisms in parasitic protozoa. Crit Rev Microbiol. 1996;22:295–314. doi: 10.3109/10408419609105484. [DOI] [PubMed] [Google Scholar]
- Levashina EA, Moita LF, Blandin S, Vriend G, Lagueux M, Kafatos FC. Conserved role of a complement-like protein in phagocytosis revealed by dsRNA knockout in cultured cells of the mosquito, Anopheles gambiae. Cell. 2001;104:709–718. doi: 10.1016/s0092-8674(01)00267-7. [DOI] [PubMed] [Google Scholar]
- Bangs JD, Ransom DA, Nimick M, Christie G, Hooper NM. In vitro cytocidal effects on Trypanosoma brucei and inhibition of Leishmania major GP63 by peptidomimetic metalloprotease inhibitors. Mol Biochem Parasitol. 2001;114:111–117. doi: 10.1016/s0166-6851(01)00244-4. [DOI] [PubMed] [Google Scholar]
- Hedengren M, Borge K, Hultmark D. Expression and evolution of the Drosophila attacin/diptericin gene family. Biochem Biophys Res Commun. 2000;279:574–581. doi: 10.1006/bbrc.2000.3988. [DOI] [PubMed] [Google Scholar]
- Bonaldo MDF, Lennon G, Soares MB. Normalization and subtraction: two approaches to facilitate gene discovery. Genome Res. 1996;6:791–806. doi: 10.1101/gr.6.9.791. [DOI] [PubMed] [Google Scholar]
- Ashburner M. A biologist's view of the Drosophila genome annotation assessment project. Genome Res. 2000;10:391–393. doi: 10.1101/gr.10.4.391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The macroarray raw data
A table showing the arrangement of spots on the arrays


