Short abstract
A database of genes responsive to the c-Myc oncogenic transcription factor is described. The database Myc Target Gene prioritizes candidate target genes according to experimental evidence and clusters responsive genes into functional groups.
Abstract
We report a database of genes responsive to the Myc oncogenic transcription factor. The database Myc Target Gene prioritizes candidate target genes according to experimental evidence and clusters responsive genes into functional groups. We coupled the prioritization of target genes with phylogenetic sequence comparisons to predict c-Myc target binding sites, which are in turn validated by chromatin immunoprecipitation assays. This database is essential for the understanding of the genetic regulatory networks underlying the genesis of cancers.
Rationale
The MYC proto-oncogene encodes a prototypical oncogenic transcription factor that plays a central role in the genesis of many different human cancers [1,2]. MYC belongs to the family of myc genes that also includes Bmyc, MYCL and MYCN. First identified as the cellular homolog of the oncogenic retroviral v-myc oncogene, the MYC gene was then found to be altered by chromosomal translocation or amplification in human cancers. More recently, the deregulated expression of MYC in a wide variety of human cancers has become more apparent with the emergence of DNA microarray gene expression profiles.
The c-Myc transcription factor is a helix-loop-helix leucine zipper protein that dimerizes with an obligate partner, Max, to bind DNA sites, 5'-CACGTG-3', termed E-boxes. c-Myc also binds DNA sites that vary from this palindromic hexanucleotide canonical sequence [3]. DNA microarrays, as well as other methods for detecting differential gene expression, have led to the discovery of a vast new collection of Myc-responsive genes. These responsive genes are probably clustered into distinct transcriptomes that are cell-type and species specific, and can be further distinguished by whether they are directly or indirectly regulated by Myc. Only through the collective examination of the responsive genes will these transcriptomes become apparent. As such, we launched a Myc Target Gene database to begin to assemble and annotate individual Myc-responsive genes [1].
To date, most c-Myc target genes listed in the database have been identified through one or more differential expression screens including SAGE [4], DNA microarray [5] and subtractive hybridization [6]. Most of these putative targets exhibit no other evidence suggesting that they are directly responsive to c-Myc alone or to both c-Myc and other transcription factors which work coordinately with Myc. It has become a challenge to decipher which genetic targets, and thus which cellular pathways, are directly influenced by Myc in specific experimental systems. Various strategies have been utilized to provide additional evidence for direct Myc regulation of a small number of target genes. These include regulation by the Myc-estrogen receptor (MycER) chimeric protein (either in the presence or absence of cycloheximide), promoter reporter assays, expression following serum stimulation and correlation of expression with that of Myc in various cell systems. However, none of these approaches provide definitive evidence that a gene is a direct transcriptional target of Myc. One method, chromatin immunoprecipitation (ChIP), identifies genomic sequences that are bound by Myc in vivo, and provides substantial evidence that a gene is directly regulated by Myc [7].
Recently, we determined that bona fide c-Myc-binding sites that have been experimentally verified by ChIP, could be predicted from interspecies genomic sequence comparisons [8]. Most of the c-Myc targets we analyzed comprised one class of genes in which the canonical 5'-CACGTG-3' E-box, shown to be bound by c-Myc in vivo, is conserved between human and one or more rodent sequences. We sought to expand this analysis to genes for which there is a fair amount of evidence of direct regulation by Myc but which have not been analyzed by ChIP. The Myc Target Gene database [1] is the ideal starting point for this analysis. Here, we describe this database and the Myc Cancer Gene website [9]. Since this database allows for the prioritization of Myc-responsive genes according to the level of experimental evidence, we have applied the findings from the database to the identification of bona fide Myc target genes via phylogenetic sequence comparisons and the use of ChIP assays.
Myc Cancer Gene website and Target Gene database
The Myc website was designed to be a warehouse for information about Myc-responsive genes, altered MYC in human cancers and Myc protein-protein interactions. The website has an introduction to cancer genes for the lay public. The first of three databases is the Myc Target Gene database that includes all Myc-responsive genes reported in the literature. An international advisory board provides oversight of the content of the database and advises on references that may have been missed. In addition, the website provides a means for users to communicate with the advisory board and the website organizer. The database is updated quarterly unless a new, significant publication requires more immediate attention. The website also provides information and references on alterations of MYC genes in human cancers and links to a c-Myc protein-protein interaction database.
The Myc Target Gene database is searchable and provides an ability to prioritize the putative target genes according to the experimental evidence supporting the validity of the gene in question as an authentic target gene. The genes are listed with names given in the literature with priority given to official names in the LocusLink database, to which each entry is linked [10]. The 'description' of the gene is that given in LocusLink unless there is no official entry, in which case the best literature description is entered. Each gene, where possible, is characterized by the function of its product. The terms used are similar to those used for the Gene Ontology consortium [11]. However, we have simplified the functions and they are listed in a pull-down menu that facilitates the search function. Each gene is annotated as to whether it is upregulated (U) or downregulated (D) in response to Myc induction or overexpression. In cases where different studies reported different responses, for example upregulated in one study but downregulated in another, the entry of the gene is duplicated with the specific references cited at the end of the row of the table. References are provided as direct links to PubMed [12].
The level of experimental evidence for each gene is tabulated according to the technique used (M, microarray; D, differential cloning; S, SAGE; G, guess) as well as other specific experimental outcomes. An approach using the Myc-estrogen receptor hormone-binding domain allows investigators to determine whether a Myc-responsive gene behaves as a direct target gene [13]. A direct target is considered to be one for which c-Myc by itself is sufficient for its induction without an intermediate that requires new protein synthesis. In this system, the chimeric MycER protein is constitutively expressed and bound to the chaperone HSP90. Upon binding estrogenic compounds, such as 4-hydroxytamoxifen, the chimeric protein alters its conformation, disengages from the chaperone and translocates into the nucleus. The translocated MycER protein recognizes genomic targets and initiates transcription of target genes without requiring newly-synthesized proteins. On the basis of this concept, it is assumed that genes responding to ligand-stimulated MycER in the presence of cycloheximide are direct target genes. Two columns in the Myc database highlight the evidence, if any, from experiments with the inducible MycER system. The MycER system, however, would not identify a hypothetical class of target genes that requires both c-Myc and a target of c-Myc that encodes a transcription factor. This factor, in turn, cooperates with c-Myc in a feed-forward loop to induce the expression of yet another c-Myc target gene.
More fundamental observations are also annotated such as whether nuclear run-on studies were performed to support the hypothesis that a candidate gene is upregulated at the transcriptional level. Only a minority of the putative targets has been studied using nuclear run-on experiments. Another consideration is the time course of response to serum or growth stimulation (induction kinetics), which induces c-Myc expression followed rapidly by expression of c-Myc responsive and other genes. With the availability of the Rat1 fibroblast cell lines that lack both copies of c-myc through homologous recombination, Myc-responsive genes are characterized according to their expression in wild-type versus Myc null fibroblasts [14]. The database also notes whether a specific study surveyed different cell lines with varied Myc expression for the corresponding expression of a target gene in question. A column is dedicated to acknowledge target gene studies that used primary cells and vectors that force the expression of Myc in these non-immortalized, non-transformed cells.
Given that there is a wide range of experimental models, the database gives the cell type and species used in the collective studies for any specific gene. It is likely that patterns will emerge demonstrating a universal effect of Myc on a subset of genes independent of cell type. The patterns will also reveal tissue-specific effects of Myc when the database is further enriched.
Another feature of the database that is of fundamental importance is the annotation of the approaches taken to establish the interaction of Myc with the genomic locus of a target gene. The use of chromatin immunoprecipitation, reporter assays in transient transfection experiments, gel mobility shift assays and other techniques are shown in the database, when available. A number of columns are searchable for prioritization. For example, the database is able to prioritize according to whether genes have been validated by ChIP assays.
Features of Myc-responsive genes
The database (13 April 2003) contains 647 entries from a total of 117 references. Out of 647 entries 42 have three or more different references supporting them as Myc-responsive genes (Table 1). PTMA, ODC1, LDHA, NCL and NPM1 are highest on the list for the number of references. The vast majority of the entries represent only a more broad-based individual study using DNA microarray or SAGE. Comparison of SAGE data with the database reveals 61 out of 91 SAGE entries match with genes from other studies.
Table 1.
Myc-responsive genes reported in multiple systems
| Gene target | LocusLink ID number | Description | Regulation | Myc DNA binding |
| APEX | 328 | Endonuclease | U | C* |
| CAD | 790 | Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | U | C* |
| CCNA2 | 890 | Cyclin A2 | U | |
| CCND2 | 894 | CYCLIN D2 | U | C* |
| CCNE1 | 898 | CYCLIN E1 | U | |
| CDK4 | 1,019 | CDK4 Cyclin-dependent kinase 4 | U | G, C* |
| CDKN1A | 1026 | Cyclin-dependent kinase inhibitor 1A (p21, Cip1) | D | |
| CDKN2B | 1,030 | Cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) | D | G,C |
| CHC1 | 1104 | RCC1; chromosome condensation 1 | U | G |
| DDX18 | 8,886 | DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 18 (Myc-regulated), MrDb | U | |
| DUSP1 | 1,843 | Dual specificity phosphatase 1 | D | |
| EIF4E | 1,977 | Eukaryotic translation initiation factor 4E | U | G, C* |
| ENO1 | 2,023 | Enolase 1, (alpha) | U | C* |
| FASN | 2,194 | Fatty acid synthase | U | C* |
| FKBP4 | 2,288 | FK506 binding protein 4, 59 kDa | U | |
| FN1 | 2,335 | Fibronectin 1 | D | |
| GADD45A | 1,647 | Growth arrest and DNA-damage-inducible, alpha | D | |
| HSPA4 | 3,308 | Heat shock 70 kDa protein 4 | U | G |
| HSPCAL3 | 3,324 | Heat shock 90 kDa protein 1, alpha-like 3 | U | C* |
| HSPD1 | 3,329 | Heat shock 60 kDa protein 1 (chaperonin) | U | C* |
| HSPE1 | 3,336 | Heat shock 10 kDa protein 1 (chaperonin 10) | U | C* |
| LDHA | 3,939 | Lactate dehydrogenase A | U | G, C |
| MGST1 | 4,257 | Glutathione transferase. GST-1 | U | C* |
| MYC | 4,609 | v-myc myelocytomatosis viral oncogene homolog (avian) | D | |
| NCL | 4,691 | Nucleolin | U | C* |
| NME1 | 4,830 | Non-metastatic cells 1, protein (NM23A) expressed in | U | C* |
| NME2 | 4,831 | NM23-H2, non-metastatic cells 2, protein (NM23B) expressed in | U | C |
| NPM1 | 4,869 | Nucleophosmin, B23 | U | C |
| ODC1 | 4,953 | Ornithine decarboxylase 1 | U | G, C |
| PPAT | 5,471 | Phosphoribosyl pyrophosphate amidotransferase | U | C* |
| PTMA | 5,757 | Prothymosin, alpha (gene sequence 28) | U | C*, D, G |
| RPL23 | 9,349 | ribosomal protein L23 | U | C* |
| RPL3 | 6,122 | Ribosomal protein L3 | U | |
| RPL6 | 6,128 | Ribosomal protein L6 | U | |
| RPS15A | 6,210 | Ribosomal protein S15A | U | |
| SRM | 6,723 | Spermidine synthase | U | C* |
| TERT | 7,015 | Telomerase reverse transcriptase | U | G, C* |
| TFRC | 7,037 | Transferrin receptor (p90, CD71) | U | C* |
| THBS1 | 7,057 | Thrombospondin 1 | D | |
| TNFSF6 | 356 | Tumor necrosis factor (ligand) superfamily, member 6 | U | G |
| TP53 | 7,157 | Tumor protein p53 (Li-Fraumeni syndrome) | U | G |
| TPM1 | 7,168 | Tropomyosin 1 (alpha) | D |
Taken in aggregate, the Myc-responsive genes cluster into distinct functional groups. The largest group, with 120 entries, involves metabolism followed by protein synthesis with 82 entries. Twenty-five entries involve cell-cycle control and five are important for DNA replication. Fifteen, which are downregulated, involve cell adhesion. Interestingly, 24 entries involve chaperonins. This comprehensive view of Myc-responsive genes suggests that Myc affects gene expression globally. This view is compatible with a recent ChIP study suggesting that Myc may play a more general role in transcription [15].
Phylogenetic footprinting and ChIP validate direct Myc targets from the Myc Target Gene database
The Myc Target Gene database allows for the prioritization of putative target genes according to the level of experimental evidence that supports the candidacy of a specific gene as being a direct Myc target. While chromatin immunoprecipitation provides the best evidence for direct target genes, many of the genes identified by differential gene expression analysis have not been validated by ChIP. From this database, we sought to identify candidate-direct Myc target genes that exhibit positive data in at least four supportive experimental categories but, as yet, have not been validated by chromatin immunoprecipitation. From this list of 12 candidate genes, we identified, by phylogenetic sequence analysis, one or more conserved E-boxes in either the promoter or intron 1 of APEX1, BAX, DDX18, CDC25A, EIF4E, TOB3 and RCL. These seven genes were chosen for further analysis.
The cell system chosen for subsequent validation of these genes is the human B-cell line P493 that carries a tet-repressible exogenous MYC construct [16]. Untreated, exponentially growing P493 cells express roughly 30-fold higher MYC mRNA and protein levels than cells exposed to tetracycline for 72 hours (Figure 1a). As measured by quantitative real-time PCR, all candidate Myc target genes, with the exception of BAX, showed a significant (greater than 3.5-fold) elevation in mRNA levels in the untreated versus tet-treated B cells (Figure 1b). NPM1, which we previously identified as a direct Myc target, was used here as a positive control for both the gene expression and ChIP experiments [17].
Figure 1.
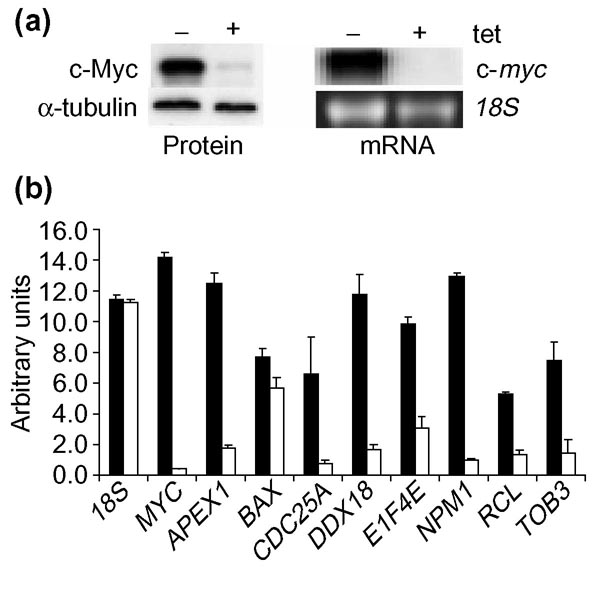
Expression of MYC and direct Myc target genes in treated (+ tet) and untreated (- tet) P493 cells. (a) Left panel reflects Myc protein expression by western blot analysis. Alpha-tubulin is shown as a loading control. (b) Real-time RT-PCR using SYBR Green to detect mRNA levels of indicated genes in both treated (white bars) and untreated (black bars) P493 cells. 18S RNA was used as an internal standard. MYC and NPM1 are shown as positive controls.
Figure 2 presents regulograms, graphical representations of the number of shared transcription factor binding sites in the context of high sequence conservation between two sequences, of six of the candidate genes plus NPM1. The regulograms are from the TraFaC database [18] and include the regions from each target gene that were analyzed by ChIP. The colored regions represent sequences that are at least 50% identical between human and mouse. All of these targets contain one or more conserved canonical Myc-binding sites in genomic regions exhibiting high levels of sequence identity. For instance, the entire APEX1 locus, which contains five exons and spans roughly 3 kb of sequence, is highly conserved. Within this locus, we identified three conserved canonical E-boxes, one in the promoter and two in exon 1 of the human gene.
Figure 2.
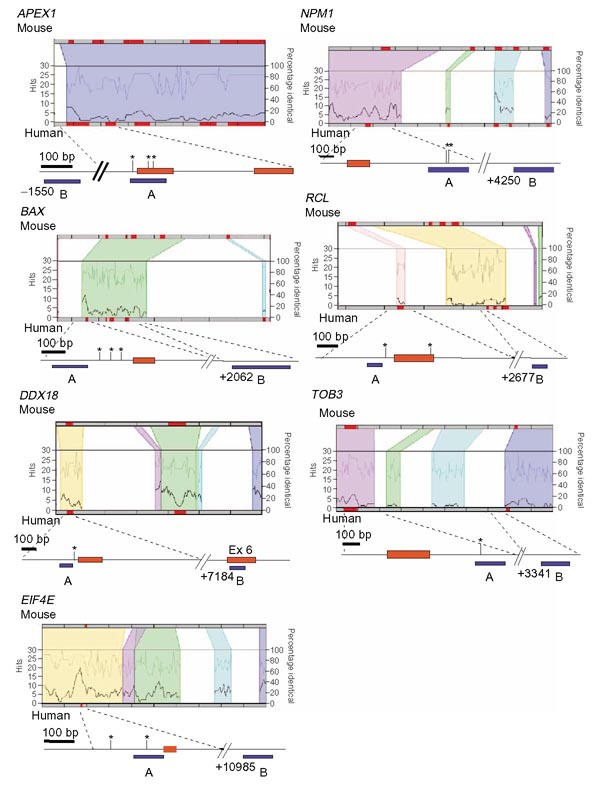
Regulograms of Myc targets analyzed by genomic sequence comparison and ChIP. Human and mouse sequences are aligned in each regulogram which depicts in colored regions the sequences with greater than 50% identity. Within these regions, the percentage of sequence identity is represented on the top line and the number of conserved transcription factor binding site 'hits' is represented by the bottom line. The genomic region analyzed by ChIP is mapped below each regulogram. Red boxes represent exons, with the first box always being exon 1. Vertical bars with asterisks indicate locations of conserved canonical E-boxes (5'-CACGTG-3'). Blue bars under each map represent fragments analyzed by ChIP. Ex, exon.
To determine whether c-Myc could directly bind conserved E-boxes, ChIP was performed with anti-Myc antibody, a control anti-hepatic growth factor (HGF) antibody, or no antibody using the human P493 B-cell line. PCR primers were designed to amplify two distinct regions, A and B, from each target locus. For each gene, fragment A contains or lies within 100 bp of the conserved binding sites whereas fragment B represents a negative control sequence up to 10 kb away from region A. It should be noted in Figure 3 that all predicted evolutionarily conserved E-box-containing DNA regions A were precipitated specifically with anti-Myc in the ChIP experiments. Glucokinase, which contains a promoter E-box but is not expressed in B cells, and all B fragments displayed only a background signal in the ChIP experiments. These findings provide proof-of-concept for the use of the database and phylogenetic footprinting and validate APEX1, BAX, DDX18, EIF4E, RCL and TOB3 as direct c-Myc target genes. Since we began these studies, two recent publications reporting genomic targets of c-Myc provide further evidence for the direct binding of c-Myc to APEX1, BAX, EIF4E and TOB3 [19,20]. Taken together, these studies support the strategy that we describe in this paper for the identification of candidate direct c-Myc target genes through the use of the Myc target gene database, phylogenetic footprinting and ChIP. The CDC25A gene does not contain an evolutionarily conserved canonical E-box and was investigated further as described below.
Figure 3.
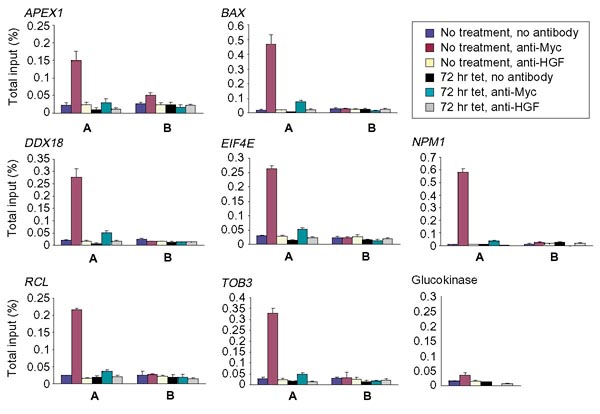
Chromatin immunoprecipitation of Myc targets in P493 cells. Each graph represents real-time PCR amplification of the A and B region of each gene using anti-Myc, anti-HGF and no antibody precipitated chromatin as template. Chromatin was precipitated from P493 cells that were either untreated or treated for 72 hours with tet. Bars represent the percentage of total input DNA for each ChIP sample.
It is notable that the DDX18 gene was originally identified as a c-Myc target by a chromatin immunoprecipitation screen with anti-Myc and anti-Max antibodies [21]. However, the initial precipitated cloned sequence was from a processed pseudogene segment corresponding to exon 6 of the functional gene. The identification of this pseudogene segment led to the identification of DDX18 as an authentic c-Myc target gene. This observation raises the important question of whether c-Myc is able to bind to E-boxes found in the pseudogene and exon 6 of the functional gene versus the phylogenetically-conserved E-box we mapped in the DDX18 promoter. To address this specifically, we examined the relative recovery of the DDX18 promoter versus exon 6 sequences in our ChIP experiments. As seen in Figure 3, no significant immunoprecipitated exon 6 sequences, represented by fragment B, were present in any of our ChIP samples. We surmise from these observations that c-Myc does not bind to the pseudogene sequence, but rather to the expressed DDX18 gene in the promoter region bearing a conserved Myc E-box.
Evidence for CDC25A as a c-Myc target was provided primarily by promoter-reporter assays, which do not reflect in situ binding of c-Myc to its target genomic sites [22]. Whilst we did not find a conserved canonical E-box (5'-CACGTG-3') we did find four non-canonical (5'-CATGTG-3') intron 1 E-boxes of which one is evolutionarily conserved (Figure 4). ChIP demonstrates that region A (conserved E-box) and region B (three E-boxes) were both significantly associated with c-Myc whereas the promoter region (Pr) and region C, corresponding to a region of intron 2, are not associated with c-Myc. Our ChIP findings contrast with the promoter-reporter assays, which showed high promoter-reporter activities for the intron 2 canonical Myc E-box region and lower activities for the intron 1 regions. Our findings do not support the association of c-Myc with the canonical intron 2 E-box. Furthermore, the intron 2 E-box lies within an Alu repeat element and it is unknown what, if any, role these sequences play in the regulation of gene expression. We surmise from these observations that reporter-promoter assays have grave limitations since they do not reflect the in situ status of the genomic sequences. Here, however, we provide the first evidence that conserved non-canonical E-boxes are bound by cMyc in vivo and may play a significant role in the regulation of direct Myc target genes.
Figure 4.
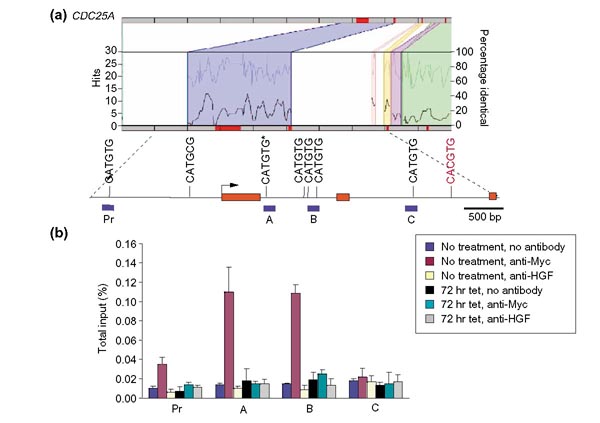
Regulogram and ChIP of CDC25A. (a) This represents the regulogram and genomic map for CDC25A. Exons 1 to 3 are depicted by the three red bars and the arrow indicates the transcriptional start site. All canonical and non-canonical Myc-binding sites are mapped onto this region. The single canonical site is illustrated in red and the conserved non-canonical 5'-CATGTG-3' site in intron 1 is indicated with an asterisks. Blue bars under map represent fragments analyzed by ChIP. (b) A graph of ChIP results from the four genomic regions indicated on the map is shown.
Our studies reported here combine a Myc-responsive gene database with phylogenetic footprinting and chromatin immunoprecipitation to validate a small set of candidate genes as direct c-Myc targets. We provide proof-of-concept that the use of phylogenetic footprinting is effective in predicting genomic Myc-binding sites. We found no evidence for the association of c-Myc to a canonical E-box found in a pseudogene, which has a corresponding authentic c-Myc target expressed gene, DDX18. Despite the elevated amounts of c-Myc protein in this B-cell model, we did not find promiscuous association of c-Myc to the DDX pseudogene or to other non-conserved canonical E-boxes found in the control B regions of NPM1 and CDC25A. The CDC25A gene was found to be associated with c-Myc through an evolutionarily conserved non-canonical (5'-CACATG-3') E-box indicating that broadening of the phylogenetic footprinting to non-canonical c-Myc-binding sites may be fruitful in the validation of direct Myc target genes by ChIP. It is notable that we have demonstrated previously another class (Class II) of genomic c-Myc-binding sites that may have drifted during evolution and hence are not conserved between species [8].
In conclusion, we provide proof-of-concept that a publicly-accessible database can be combined with sequence analysis and ChIP to begin to establish the transcriptional regulatory network underlying the ability of c-Myc to manifest its oncogenic properties in cancer cells. The future challenge is to exploit the database to identify transcription factors that cooperate with c-Myc to regulate subsets of functionally similar genes or functional transcriptomes (in the same metabolic pathway, for example) and transcriptional regulatory loops involved in c-Myc mediated phenotypes.
Materials and methods
Database data procurement
To initiate the Myc Target Gene database all publications reporting Myc-responsive genes were identified by PubMed [12] searches (keywords used: Myc, target gene, microarray, differential gene expression), from publication files kept by the website organizer and suggestions from the website advisory board. Information on each putative target gene was extracted from the publications using a worksheet that is also provided, when possible, to the authors of the publications for data verification. Information from the worksheets is entered into an Excel spreadsheet database that is designed for direct implementation as a searchable database on the website. For recent publications with extensive microarray data, data spreadsheets were requested from the authors and the gene entries were transferred to the database directly. Each gene entry was annotated according to the design of the Excel spreadsheet database, which is available upon request for non-profit uses. In some instances, authors have provided the organizer with preprints of their publications. To ensure that current and future publications on Myc target genes are included, abstracts of publications on Myc found in PubMed [12] are surveyed weekly by the website organizer and colleagues. In addition, the website provides a means for users to directly deposit unpublished data or to call attention to recently published results.
Phylogenetic comparisons
Human and rodent genomic sequences are available from the UCSC Genome Bioinformatics site [23] or TraFaC website [24,18]. The sequences analyzed included two kilobases upstream of the transcriptional start site through to the final exon for each target gene. The transcriptional start site and intron-exon boundaries were determined from the TraFaC alignments (regulograms) or by direct comparison of mRNA and genomic DNA. In order to find putative c-Myc-binding sites, sequences were searched using the nucleic acid motifs feature of the OMIGA software (Oxford Molecular Limited, Oxford, UK). The search parameters were user defined as the canonical (5'-CACGTG-3') or noncanonical (5'-CATGTG-3', 5'-CACGAG-3', 5'-CACGCG-3', 5'-CATGCG-3', 5'-CACGTTG-3') c-Myc-binding sites. The occurrence of putative c-Myc-binding sites in regions of high sequence identity were determined by examining the regulograms or the alignments produced from the dot plot sequence homology analysis of the OMIGA software.
Northern blot analysis
Total RNA was extracted from both tetracycline treated and untreated P493 cells using Trizol (Invitrogen, Carlsbad, CA). Ten micrograms RNA was run on a 1.2% agarose, 0.7 M formaldehyde denaturing gel before transferring to a nylon membrane overnight (Nytran, Schleicher and Schuell). Human myc cDNA plasmid fragment was isolated and labeled with 32P using a random prime labeling kit (Stratagene, La Jolla, CA). Hybridization signals were quantitated using a phosphorimager.
Western blot
Twenty micrograms total protein from cell lysates of both treated and untreated P493 cells were separated on a 10% SDS-polyacrylamide gel. Immunoblots were probed with the 9E10 monoclonal anti-Myc antibody and anti-α-tubulin (CP06, Oncogene Research Products, San Diego, CA).
Real-time RT-PCR
First-strand cDNA was synthesized from 2 μg total RNA using TaqMan reverse transcription reagents (Applied Biosystems, Branchburg, NJ). PCR primers were designed to amplify fragments that span intron/exon boundaries in the human cDNA sequences using the Primer Express software (Applied Biosystems). Real-time PCR was performed in the linear range using the SYBR Green Core Reagents kit on a 7700 Sequence Detection System (Applied Biosystems).
Chromatin immunoprecipitation (ChIP)
The cell line used for all ChIP analyses was the human B-cell line P493 carrying a conditional, tetracycline-regulated c-myc [16]. Cells were grown in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) and antibiotics. To repress c-myc expression, cells were exposed to 0.1 μg/ml tetracycline (tet) for 72 hours. Both untreated and tet-treated P493 cells were formaldehyde cross-linked and chromatin was precipitated as described previously [25]. The rabbit polyclonal c-Myc and HGF antibodies (anti-Myc sc-764 and anti-HGF sc-7949 [Santa Cruz Biotechnology, Inc, Santa Cruz, CA]) were each used to precipitate chromatin from 2 × 107 cells.
Real-time PCR
For real-time PCR, a SYBR green core reagents kit (Applied Biosystems) or the Failsafe Real-time PCR PreMix selection kit (Epicenter, Madison, WI) were utilized. Known quantities of total input DNA were used to generate a standard curve for determining the percentage of total input for each ChIP sample. All amplifications were carried out in the linear range of standard curves.
Acknowledgments
Acknowledgements
This work was supported in part by NIH grants CA51497, CA57341, LM07515, T32HL07525 and T32GM07819. We thank D Eick for P493 cells and J Hewitt and P Rusche for maintaining the Myc Target Gene database.
References
- The Myc Target Gene Database http://www.myccancergene.org/site/mycTargetDB.asp
- Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Huang J, Ma A, Kretzner L, Alt FW, Eisenman RN, Weintraub H. Binding of Myc proteins to canonical and noncanonical DNA sequences. Mol Cell Biol. 1993;13:5216–5224. doi: 10.1128/mcb.13.9.5216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Hermeking H. Characterization of the c-MYC-regulated transcriptome by SAGE: identification and analysis of c-MYC target genes. Proc Natl Acad Sci USA. 2002;99:6274–6279. doi: 10.1073/pnas.082005599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B, Shim H, Li Q, Wu C, Lee L, Maity A, Dang C. Identification of putative c-Myc responsive genes: characterization of rcl, a novel growth related gene. Mol Cell Biol. 1997;17:4967–4978. doi: 10.1128/mcb.17.9.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haggerty TJ, Zeller KI, Osthus RC, Wonsey DR, Dang CV. A strategy for identifying transcription factor binding sites reveals two classes of genomic c-Myc target sites. Proc Natl Acad Sci USA. 2003;100:5313–5318. doi: 10.1073/pnas.0931346100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myc Cancer Gene http://www.myccancergene.org/site/about.asp
- LocusLink http://www.ncbi.nlm.nih.gov/LocusLink/
- Gene Ontology Consortium http://www.geneontology.org/doc/index.expanded.shtml
- PubMed http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=PubMed
- Eilers M, Picard D, Yamamoto K, Bishop J. Chimaeras of myc oncoprotein and steroid receptors cause hormone-dependent transformation of cells. Nature. 1989;340:66–68. doi: 10.1038/340066a0. [DOI] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- Li Z, Van Calcar S, Qu C, Cavenee WK, Zhang MQ, Ren B. A global transcriptional regulatory role for c-Myc in Burkitt's lymphoma cells. Proc Natl Acad Sci USA. 2003;100:8164–8169. doi: 10.1073/pnas.1332764100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuhmacher M, Kohlhuber F, Holzel M, Kaiser C, Burtscher H, Jarsch M, Bornkamm GW, Laux G, Polack A, Weidle UH, et al. The transcriptional program of a human B cell line in response to Myc. Nucleic Acids Res. 2001;29:397–406. doi: 10.1093/nar/29.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller KI, Haggerty TJ, Barrett JF, Guo Q, Wonsey DR, Dang CV. Characterization of nucleophosmin (B23) as a Myc target by scanning chromatin immunoprecipitation. J Biol Chem. 2001;276:48285–48291. doi: 10.1074/jbc.M108506200. [DOI] [PubMed] [Google Scholar]
- UC/CHMC TraFaC Homology Server http://trafac.chmcc.org/trafac/index.jsp
- Fernandez PC, Frank SR, Wang L, Schroeder M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao DY, Watson JD, Yan PS, Barsyte-Lovejoy D, Khosravi F, Wong WW, Farnham PJ, Huang TH, Penn LZ. Analysis of Myc bound loci identified by CpG island arrays shows that Max is essential for Myc-dependent repression. Curr Biol. 2003;13:882–886. doi: 10.1016/S0960-9822(03)00297-5. [DOI] [PubMed] [Google Scholar]
- Grandori C, Mac J, Siebelt F, Ayer D, Eisenman R. Myc-Max heterodimers activate a DEAD box gene and interact with multiple E box-related sites in vivo. EMBO J. 1996;15:4344–4357. [PMC free article] [PubMed] [Google Scholar]
- Galaktionov K, Chen X, Beach D. Cdc25 cell cycle phosphatase as a target of c-myc. Nature. 1996;382:511–517. doi: 10.1038/382511a0. [DOI] [PubMed] [Google Scholar]
- UCSC Genome Bioinformatics http://genome.ucsc.edu/
- Jegga AG, Sherwood SP, Carman JW, Pinski AT, Phillips JL, Pestian JP, Aronow BJ. Detection and visualization of compositionally similar cis-regulatory element clusters in orthologous and coordinately controlled genes. Genome Res. 2002;12:1408–1417. doi: 10.1101/gr.255002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd KE, Wells J, Gutman J, Bartley SM, Farnham PJ. c-Myc target gene specificity is determined by a post-DNA binding mechanism. Proc Natl Acad Sci USA. 1998;95:13887–13892. doi: 10.1073/pnas.95.23.13887. [DOI] [PMC free article] [PubMed] [Google Scholar]


