Abstract
Iron- and methane-cycling are important processes in wetlands with one connected to plant growth and the other to greenhouse gas emission, respectively. In contrast to acidic habitats, there is scarce information on the ecology of microbes oxidizing ferrous iron at circumneutral pH. The latter is mainly due to the lack of isolated representatives and molecular detection techniques. Recently, we developed PCR–DGGE and qPCR assays to detect and enumerate Gallionella-related neutrophilic iron-oxidizers (Ga-FeOB) enabling the assessment of controlling physical as well as biological factors in various ecosystems. In this study, we investigated the spatial distribution of Ga-FeOB in co-occurrence with methane-oxidizing bacteria (MOB) in a riparian wetland. Soil samples were collected at different spatial scales (ranging from meters to centimeters) representing a hydrological gradient. The diversity of Ga-FeOB was assessed using PCR–DGGE and the abundance of both Ga-FeOB and MOB by qPCR. Geostatistical methods were applied to visualize the spatial distribution of both groups. Spatial distribution as well as abundance of Ga-FeOB and MOB was clearly correlated to the hydrological gradient as expressed in moisture content of the soil. Ga-FeOB outnumbered the MOB subgroups suggesting their competitiveness or the prevalence of Fe2+ over CH4 oxidation in this floodplain.
Keywords: Gallionella, iron oxidation, methane oxidation, spatial patterns, geostatistics, riparian, wetland
Introduction
Wetland ecosystems are sites of intense biogeochemical cycling due to the interactions between the oxic surface and deeper anoxic soil layers making them highly productive ecosystems (Weiss et al., 2003; Gutknecht et al., 2006; Hartman et al., 2008; Burgin et al., 2011). Apart from being a crucial habitat for many animals, plants, and humans, wetlands are also the largest biological source of the greenhouse gas methane (Ringeval et al., 2011). Cycling of nutrients and emission of methane from wetlands is strongly intertwined due to the oxic–anoxic conditions in close proximity (Kogel-Knabner et al., 2010; Burgin et al., 2011) which is especially distinct for iron and methane (Laanbroek, 2010).
Oxic–anoxic interfaces in wetland soils are significantly extended by roots of wetland plants which are known to release oxygen from their roots, a phenomenon called “radial oxygen loss” (ROL; Armstrong, 1964; Bodelier et al., 2006; Laanbroek, 2010). Next to this, oxygen is also introduced into anoxic soils by other physical or biological processes (Doyle and Otte, 1997; Coci et al., 2005; Mermillod-Blondin and Lemoine, 2010) resulting in sharp redox gradients characteristic of wetland soils and sediments.
As one of the most important elements in biogeochemical redox reactions (Fortin and Langley, 2005), iron interacts with other elemental cycles, which in turn affect the growth and activities of plants and microorganisms. Iron oxides are widely distributed in wetland environments and will be reduced rapidly under anoxic conditions by anaerobic iron(III)-reducing bacteria, that couple the oxidation of organic carbon compounds to the reduction of iron (Fe3+) to ferrous iron (Fe2+; Lovley, 1997; Burgin et al., 2011). Ferrous iron can be oxidized chemically, however, in freshwater wetlands microbial activity accounts for a large part of the production of iron oxides under circumneutral pH conditions, despite the fact that chemical oxidation under these conditions proceeds much faster. This contradiction triggered studies detecting iron-oxidizing bacteria (FeOB) associated with iron oxides in samples from circumneutral environments (Emerson and Moyer, 1997; Weiss et al., 2003; Chan et al., 2009). Actual bacterial mediation of iron oxidation was also confirmed, further pointing to the important role of FeOB in the formation of iron oxides (Neubauer et al., 2002).
Despite the fact that a number of FeOB have been isolated and our understanding of the role of these microorganisms in iron oxidation has significantly progressed (Emerson et al., 2010) virtually nothing is known about the environmental factors affecting the diversity and distribution of FeOB in their natural habitats. Sundby et al. (2003) have proposed that seasonal changes of O2 concentration and pore water Fe2+ availability resulting from the growth and death of roots exert a major control on the precipitation of iron oxides. Neubauer et al. (2007) reported significant temporal changes in the iron-oxidizing community in microcosm studies with plants, which suggests that plant biomass and activity play a key role in rhizospheric Fe(II) oxidation. Changes in the Gallionella-related FeOB community composition have been observed to depend on season and hydrology in a tidal freshwater marsh (Wang et al., 2011), probably connected to the effects these parameter have on oxygen availability.
Next to chemical oxidation, there are quite a few biotic processes that consume oxygen, such as the oxidation of ammonium, methane, and sulfide, which are all products of anaerobic microbial processes. Thermodynamically, Fe(II) oxidation yields more energy than other chemolithotrophic reactions under oxygen-limiting conditions (Thauer et al., 1977), suggesting that sub-oxic conditions may be the niche occupied by FeOB in wetland soils. However, no information is available on the spatial distribution of niches suitable for FeOB as well as the competition for limiting amounts of oxygen between iron-oxidizing bacteria and other oxygen-consuming organisms.
A well characterized group of microbes also thriving at oxic–anoxic boundaries are aerobic methane-oxidizing bacteria (MOB), utilizing methane for carbon as well as energy source (Semrau et al., 2010). The aerobic MOB are represented in the phylum of the Proteobacteria and recently also MOB have been identified within the Verrucomicrobia (Op den Camp et al., 2009). Many environmental factors controlling the ecology of MOB have been studied and indentified (Conrad, 2007; Semrau et al., 2010; Bodelier, 2011). The proteobacterial MOB have traditionally been divided into type I and II, based on phylogeny, physiology, and morphology. On the basis of phylogeny, type I has been subdivided in three subtypes Ia, Ib, and Ic (Lüke and Frenzel, 2011). The obligatory aerobic metabolism of most MOB suggest that their environmental distribution and demands are similar to those for FeOB. For type II and Ia MOB different ecological strategies are emerging, type II being slow responders to substrate availability but persistent in the environment, whereas type Ia harbors species that respond and grow fast upon substrate availability (Steenbergh et al., 2010; Bodelier et al., 2012). Considering this, the vast ecological knowledge available about various environmental as well as isolated subtypes of MOB may be extrapolated to possible co-occurring FeOB. Besides using microbial co-occurrence, the distribution of FeOB according to spatial gradients of controlling environmental conditions is another way of obtaining ecological information. Since the soil is a highly heterogeneous environment, soil components, and properties may exhibit high spatial variations (Franklin and Mills, 2003; Baker et al., 2009; Ferreira et al., 2010). In particular, the perceived microbial community structure may depend on the spatial scale of observation, in the range of centimeters to a few hundred meters (Ettema and Wardle, 2002; Philippot et al., 2009b). Sampling of microbial communities in a spatial design followed by geostatistical analyses has rendered important ecological information on the functioning and distribution of ammonia oxidizers (Wessen et al., 2011), denitrifiers (Philippot et al., 2009b), methane oxidizers (Siljanen et al., 2011) as well as total microbial communities (Philippot et al., 2009a) by estimating the distribution in unsampled areas of experimental plots. However so far nothing is known about the spatial distribution of iron-oxidizing bacterial communities.
The aim of this paper is to assess the spatial distribution of the Gallionella-related FeOB (Ga-FeOB) and MOB communities and their interactions in an irregularly flooded riparian wetland. Collecting samples at different spatial scales representing a hydrological gradient allowed investigating the effect of flooding intensity. Special attention is paid to the influence of sampling scale and elevation on the distribution of both Ga-FeOB and MOB. Community structure of Ga-FeOB were measured by a previously designed nested PCR–DGGE and abundances of both Ga-FeOB and MOB by qPCR. Community was linked to environmental variables. Spatial distribution of bacterial groups was visualized by interpolated spatial maps. We hypothesized that the community structure of iron-oxidizing bacteria changes with sampling scale and elevation, having an effect on the presence of MOB or vice versa.
Materials and Methods
Site and sampling
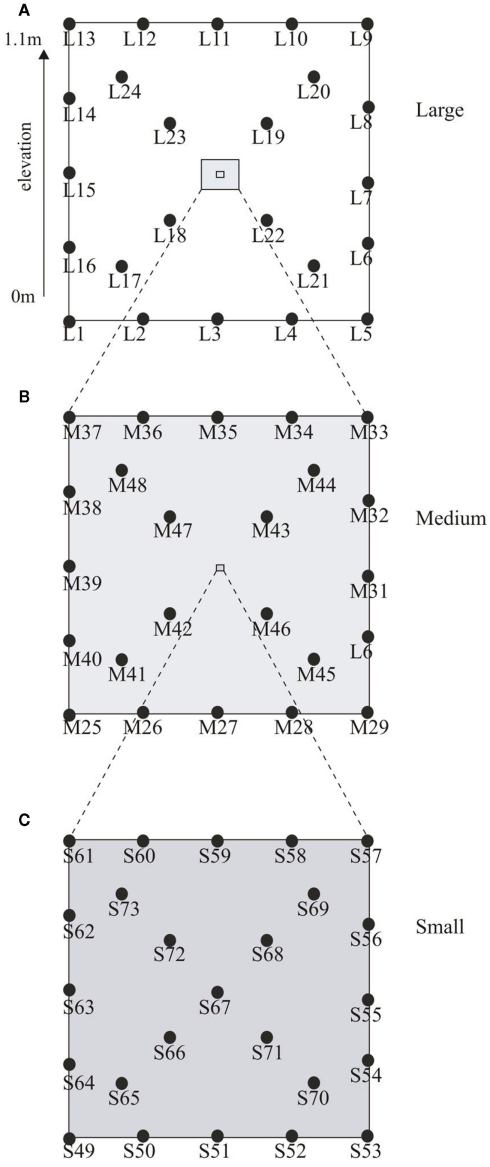
Soil samples were collected from the Ewijkse Waard (51°88′N, 5°73′E), a riparian floodplain along the River Waal (i.e., a tributary of the River Rhine) in the Netherlands, in November 2006. The soil properties have been described previously in detail (Kemnitz et al., 2004; Steenbergh et al., 2010; Bodelier et al., 2012). The sampling site is located on the embankment of a small oxbow lake which is connected to the river on one side but is separated from the river by an outflow barrier, ensuring that the oxbow lake does not become dry completely. This outflow barrier results in slow retreat of water from the sampling plot after flooding, which results in series of different flooding days within the plot; the highest part being flooded up to 2 weeks per year, while the lowest part can be flooded up to 150 days per year (Bodelier et al., 2012). To assess spatial dependence and heterogeneity a nested design was used as described by Franklin and Mills (2003). In total 73 soil cores (1.8 cm × 5 cm) were collected in a 10 m × 10 m plot (Figure 1A). The large-scale spatial level of sampling comprised 24 cores arranged evenly distributed along the sides and diagonals of the plot as depicted in Figure 1A. Nested around the intersection of the diagonals of the large plot, 23 samples were taken using the same distribution but now on a 1 m × 1 m grid (medium scale; Figure 1B). Nested around the same diagonal again 24 samples were obtained in a 20 cm × 20 cm grid (small scale; Figure 1C). The 10 m × 10 m plot was situated on a slope on the embankment of the oxbow lake resulting in an elevation difference of 1.1 m between the lowest and highest point of the plot. The samples taken at the three spatial scales were classified according to their elevation in the following elevation level classes, low (0–0.33 m), intermediate (0.34–0.66 m), and high (0.67–1.1 m). Soil water content (based on dry weight), soil density, total soil organic matter, and water filled pore space were analyzed using standard methods.
Figure 1.
Graphical representation of the sampling scheme. Depicted in (A) is the large-scale plot (10 m × 10 m) with an elevation from 0 to 1.1 m. Samples were taken at 2.5 m intervals along the sides and 2.35 m intervals along the diagonals. (B) Shows the medium plot (1 m × 1 m) with sampling interval of 25 cm along the sides and 23.5 cm along the diagonals while (C) depicts the small plot (20 cm × 20 cm) with samples intervals of 5.0 cm along the sides and 4.7 cm along the diagonals.
DNA extraction
DNA was extracted using a modification of the method described Yeates and Gillings (1998), based on the FastDNA spin kit for soil (MP Biomedicals, LLC, Solon, OH, USA), as described earlier (Pan et al., 2010). Soil (0.3 g) and 780 μl lysis buffer [200 mM NaPO4 pH 7.0; 1% (w/v) CTAB; 1.5 M NaCl; 2% (w/v) Polyvinylpyrrolidone K30; 5 mg ml−1 lysozyme (added directly before use)] was added into a multimix FastPrep tube and incubated at 37°C for 30 min. MT buffer (122 μl), provided with the kit, was added and tubes were shaken in the FastPrep instrument (MP, Biomedicals, LLC, Solon, OH, USA) for 30 s at 5.5 m s−1. Subsequently, samples were centrifuged for 15 min at 10000 rpm and 700 μl supernatant was collected. The pellet was re-extracted by adding lysis buffer (500 μl) and 50 μl MT buffer to the FastPrep tubes, shaken in the FastPrep instrument for 30 s at 5.5 m s−1 again followed by the transfer of the second 700 μl of supernatant into separate Eppendorf tubes. At this step, 2 μl × 700 μl supernatant was obtained from each sample. Five microliters of 10 mg ml−1 freshly made proteinase K was added to each tube. Tubes were incubated at 65°C for 30 min. Samples were extracted with phenol–chloroform–isoamyl alcohol (25:24:1), followed by a chloroform–isoamyl alcohol (24:1) extraction. One hundred twenty-five microliters of 7.5 M potassium acetate was added, samples were incubated on ice for 5 min, and then centrifuged at 10000 rpm for 10 min. Supernatants (2 × 700 μl per soil sample) were transferred to new tubes, 700 μl Binding Matrix was added, and tubes were mixed for 5 min on a rotator. Binding Matrix, with bound DNA, was pelleted by 1 min centrifugation at 10000 rpm. The supernatant was discarded and pellet was resuspended in 500 μl wash buffer. The resulting suspension was added into a Spin filter, and centrifuged for 1 min at 10000 rpm. The eluent was discarded and the pellet was washed again in 500 μl wash buffer. After discarding the second eluent, the Spinfilter was centrifuged for another 10 s to dry the pellet. The filter was taken into a new tube and 50 μl of TE pH 8.0 was added. The filter was incubated at room temperature for 1 min and centrifuged for 1 min. The filter was re-eluted in the same way with 50 μl of TE pH 8.0. The eluent collected in the catch tube contained the purified DNA. A total of 18 out of 73 samples (i.e., L1–5, M25, M31, M 46, M 49, S50, S52, S53, S56, S60, S63, S65, S67, and S72) did not give products during PCR procedures and were excluded from further analysis.
PCR–DGGE
A nested PCR–DGGE targeting Ga-FeOB was performed using a previously developed assay (Wang et al., 2009). Briefly, 1 μl of 50 ng μl−1 purified environmental DNA was used for the first step PCR with primer set 122F–998R. The PCR was run in a 50-μl reaction volume containing 5 μM of each primer, 2.5 mM of dNTPs, 1.5 mM MgCl2, 2 mM BSA, and 1.25 units Taq DNA polymerase. The PCR conditions included 1 cycle of 5 min at 94°C, followed by 30 cycles of 1 min at 94°C, 1 min at 55°C, and 1.5 min at 72°C, the final extension cycle was 10 min at 72°C. The PCR products were diluted 10 times and 2 μl of the dilution was used for a second-step PCR using universal primers set GC-F357 and 907R following a touch-down PCR cycle (Schäfer et al., 2001). Clones generated in a previous study (Wang et al., 2009), representing different phylogenetic taxa, were re-amplified using general bacterial DGGE primers GC-F357 and R907 (Muyzer et al., 1993), and used as markers. DGGE was performed with the Bio-Rad Protean II system as described previously (Muyzer et al., 1993). An 8% polyacrylamide gel with a vertical gradient of 30–60% of the denaturant was used to analyze the 550 bp PCR products. Equal volumes of the PCR products obtained after the second PCR reaction were loaded. The running conditions was 100 V at a constant temperature of 60°C in 23 L of 0.5 × TAE buffer (20 mM Tris acetate, 0.5 mM EDTA, pH 8.0) for 18 h. The DGGE gels were visualized using an UV transilluminator after ethidium bromide-staining. The representative bands were excised, purified, and sequenced. The sequences were checked and manually modified using Sequencher software (Gene Codes Corporation, City, USA) and aligned in the ARB program (Ludwig et al., 2004). A phylogenetic tree was constructed using the Neighbor Joining method. Sequences were deposited with GenBank under the following accession numbers JQ060106–JQ060114, representing DGGE bands G14–G37, respectively.
Real-time PCR
A recently developed real-time PCR assay (Wang et al., 2011) was used to quantify the Ga-FeOB bacteria in the samples collected. The primer set includes a degenerate forward primer 628F (GBMAGGCTAGAGTGTAGC) and a reverse primer 998R (CTCTGGAAACTTCCTGAC). DNA from soil samples was purified and diluted accurately to a concentration of 10 ng μl−1. The real-time PCR was performed in a 25-μl reaction volume containing 2.5 μl purified DNA and 22.5 μl SYBR® Green PCR Master Mix (Invitrogen). The concentration of each primer was 5 pmol μl−1. The PCR involved 45 cycles, with each cycle consisting of denaturation at 95°C for 20 s, annealing at 56°C for 20 s, and extension at 72°C for 45 s. Data acquisition was done at 82°C for 10 s, to avoid signal from primer dimer formation. The DNA copy number of Ga-FeOB in each sample was estimated by comparing the Ct value of each sample to those of the standard regression line, which was made by using reference clones (Wang et al., 2009, 2011). Three methanotrophic subgroups were quantified by pmoA-based quantitative PCR based on the assays described by Kolb et al. (2003). The type Ia, Ib, and II assays were carried out as previously described (Pan et al., 2010) with 25 μl reaction containing 12.5 μl 2 × SYBR green mix (AB gene, Epsom, UK), 2.5 μl of diluted DNA template and 0.8 mM each of primers. The samples were diluted accurately to 1 ng μl−1. The thermal cycle started with an initial denaturation at 95°C for 15 min, followed by 45 cycles of denaturation at 95°C for 20 s, annealing at 64°C for 20 s, and extension at 72°C for 45 s. Fluorescence was recorded at 84°C and DNA melting curve analysis was performed at temperatures ranging from 70 to 99°C. All assays (Ga-FeOB and MOB) were performed with a Rotor-Gene 6000 thermal cycling system (Corbett Research, Eight Mile Plains, QLD, Australia), where samples were added to aliquots of the master mixture using a CAS-1200 (Corbett Robotics Eight Mile Plains, QLD, Australia) liquid handling system. Every sample was performed in duplicate. Quantification analysis was performed by the Rotor-Gene software. A number of samples failed to reach the detection limit of the assays: Ga-FeOB (M25, M27, M31, M37, M48, S49–S59, S63, S64, S65, S67). Type Ia MOB (L2, S50, S52, S54–S59, S60, S61, S64, S65). Type Ib MOB (S52). Type II MOB (M45).
Data analysis
DGGE gels were analyzed using Phoretix gel analysis software (Phoretix International, Newcastle upon Tyne, UK). The number of bands of each lane was defined and a matrix of band intensity was created. Lanes were created manually, with a fixed width of 5% of the standard lane width. Each lane represents one sample. Background noise was subtracted by using the Rolling Ball algorithm with a radius of 50 pixels. Bands were detected automatically with a minimum slope of 100 and a noise reduction of 4. Then bands were assessed and corrected by eye, matched to a reference lane (Markers), and quantified. The relative abundance of each band was defined as the intensity ratio of each band to the total intensity of individual lanes.
Similarity between DGGE community profiles was analyzed using multivariate analyses in PRIMER 5 software (Plymouth Marine Laboratory, Plymouth, UK). Relative abundance data derived from the DGGE gels were used in non-metric Multidimensional Scaling (MDS) analyses. The input of MDS analyses were Bray–Curtis similarity matrices generated using log (x + 1) transformed relative abundances data. The MDS analyses results in a two-dimensional plot where the distance between samples indicates the similarity of these samples relative to other samples in the plot. The accuracy of the two-dimensional representation is indicated by the “Stress” value (Kruskal’s stress formula). Stress values <0.1 indicate a good ordination with no prospect of misleading interpretation. Stress values <0.2 still give a good two-dimensional representation where not too much reliance should be put on the detail. Theoretical aspects of the MDS analyses used are described by Clarke and Warwick (2001). One-way ANOSIM in PRIMER software test was used to compare samples between different scales (Clarke and Warwick, 1998). Data of the relative abundance of each band was log-transformed and analyzed using Statistica software package (Statistica version 9, StatSoft, Inc., Tulsa, USA), together with the qPCR results, and a number of defined environmental parameters. The distribution of bacterial abundance at different elevation levels and sampling scales was compared using ANOVA. Post hoc Tukey analysis was used to further compare the difference among samples. The relation between Ga-FeOB and MOB was done using non-parametric pair wise correlation in Statistica.
To model the spatial structures of the total abundance of Ga-FeOB (logarithm of the total 16s rRNA gene copy number), MOB (logarithm of the total pmoA gene copy number), and the moisture content (weight percentages) geostatistics were applied. These methods originate from mineralogy and soil science and are a common tool to identify and model spatial patterns (Legendre and Legendre, 1998). The basic principle is autocorrelation of spatial variability, i.e., close-set samples are more similar than those further apart (Ettema and Wardle, 2002). In a first step a variogram analysis was performed. In brief, semi-variances between samples were calculated and plotted against their spatial separation; the slope indicated whether a spatial structure is present or not (Ettema and Wardle, 2002). The Hawkins and Cressie’s modulus estimator was calculated to create variograms and maximum likelihood (ML) estimations with Matérn covariance function was fitted to semi-variance values (Cressie, 1993). The estimated model parameters have been used for kriging. This is a geostatistical method to visualize spatial structures of properties by spatial interpolation maps (Ribeiro and Diggle, 2001). Before kriging, goodness of fit of the model parameters has been tested by cross validation. To create spatial interpolation maps of moisture content, Ga-FeOB, and MOB abundances we used ordinary kriging applying global neighborhood. All geostatistical analyses were performed using the geostatistical data analysis software geoR as implemented in the statistical software R (Ribeiro and Diggle, 2001; R Development Core Team, 2010).
Results
Community structure of Gallionella-related FeOB
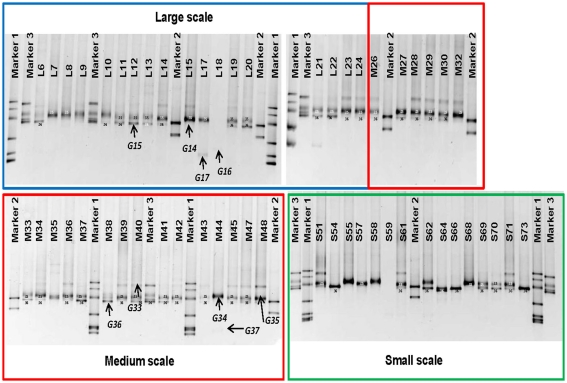
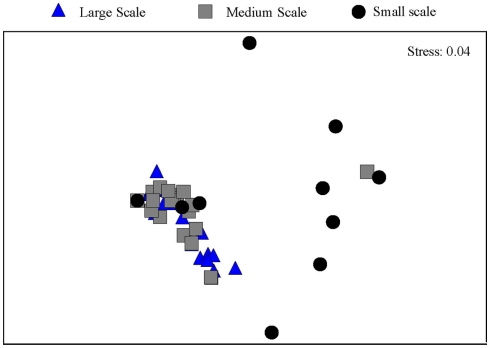
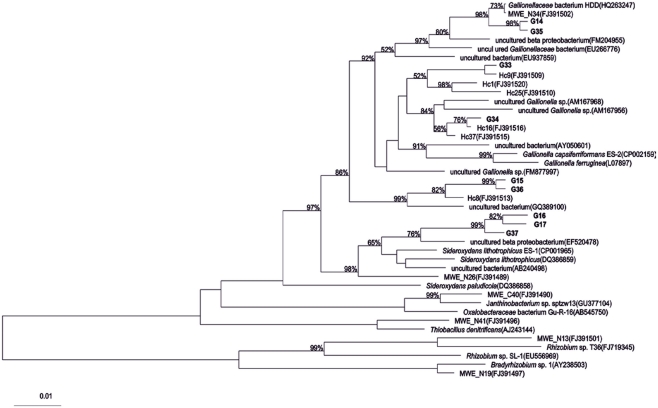
Between one and four bands in each sample were retrieved from the DGGE gels (Figure 2). There is a trend toward fewer bands in the samples collected at the small scale. Taking the relative intensities of the bands as input matrix for multivariate analyses using multidimensional scaling showed that the Ga-FeOB community in samples taken from large and medium scales differed from the samples collected at small scale, but not from each other (Figure 3; ANOSIM analyses, R = 0.42, large vs small, R = 0.43 medium vs small). A total of nine bands were retrieved from the DGGE gels and sequenced. They were distantly related to known cultures, with uncultured clones as most close relatives (Figure 4). The most dominant band (G35, G14, Figure 2) from the medium-scale and large-scale samples was closely related to a recently isolated neutrophilic Ga-FeOB strain HDD (Wang, 2011). The most closely isolated relative to Band G37, G16, and G17 is Sideroxydans lithotrophicus strain LD-1 (Weiss et al., 2007), a microaerophilic iron-oxidizing bacterium. Bands G33 and G34 were most closely related to uncultured Gallionella-related clones Hc9 and Hc16, respectively (Figure 4) isolated from a freshwater wetland (Wang et al., 2009). G15 and G 36 are most closely related to Gallionella-related clones (Hc8) obtained from the same wetland (Wang et al., 2009).
Figure 2.
DGGE patterns showing the distribution of iron-oxidizing bacteria at different sampling scales, which are indicated by colored boxes. Bands G14–G17 and G33–G37 were sequenced and shown in Figure 4. Sequenced clones obtained previously were used as markers. Marker1: MWE_C40(FJ391490), MWE_N41(FJ391496), MWE_N34(FJ391502), MWE_N13(FJ391501), MWE_N19(FJ391497), and MWE_N26(FJ391498), Marker2: Hc1(FJ391520), and Hc25(FJ391510), Marker3: Hc9(FJ391509), Hc37(FJ391515), Hc16 (FJ391516), and Hc8(FJ391513; Wang et al., 2009).
Figure 3.
Ordination by non-metric multidimensional scaling of log (x + 1) transformed relative abundance of the DGGE bands of the samples taken at different spatial scales.
Figure 4.
Phylogenetic tree of partial 16s rRNA gene sequences of DGGE bands (see Figure 2) obtained from different sampling scales. Bootstrap values based on 1000 replicate trees are shown near the nodes. The bar indicates 10% sequence difference. All sequences indicated with “Hc” or “MWE” are sequences derived from (Wang et al., 2009).
Effects of spatial scale and elevation level on the abundance and diversity of Gallionella-related FeOB and MOB
Spatial scale
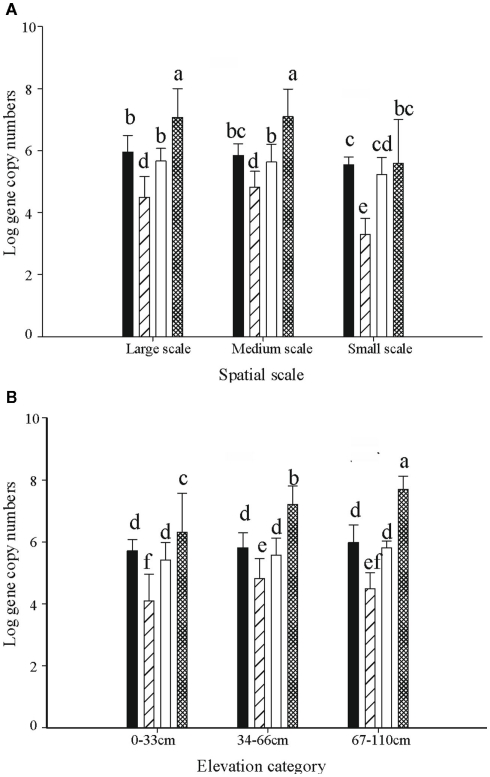
The abundance of Gallionella-related FeOB as determined by copy numbers of 16s rRNA genes, ranged from 1 × 103 to 3.9 × 108 per gram dry soil. Five out of 23 medium-scale samples did not yield product in the qPCR analyses, indicating that their 16s rRNA copy numbers were below the detection limit, and thus were excluded from the analyses. The highest copy number was observed in sample L20, taken in the high elevation part. The average copy numbers of the 16s rRNA gene from samples taken at large and medium scales, i.e., 4.6 × 107 and 3.7 × 107 copies per gram dry weight, respectively, were significantly higher than that of the small-scale samples (5.4 × 106 copies per gram dry weight; Figure 5A).
Figure 5.
The distribution of 16s rRNA copy numbers of Ga-FeOB and pmoA gene copy numbers of MOB (both expressed per gram of dry soil) at different spatial scales (A) and elevation levels (B). Indicated are means ± SD. Black bars depict type II, striped bars type Ia, and white bars type Ib MOB, while hatched bars show the Ga-FeOB. Different letters indicate significant differences (p < 0.01) between bars. For numbers of observations see footnote of Table 1.
One-way ANOVA test indicated significant differences of Gallionella-related iron-oxidizing bacteria as well as methane oxidizers across spatial scales (Table 1). Post hoc Tukey test indicated that the abundance of both Ga-FeOB and MOB in samples taken at small scale was significantly lower than those at large- and medium-scale plots (p < 0.05). No difference was observed between large- and medium-scale samples.
Table 1.
ANOVA analyses of effects of spatial factors and elevation on gene copy numbers of Ga-FeOB, type Ia, Ib, and II MOB.
| Factor | SS | DF | MS | F | p |
|---|---|---|---|---|---|
| SPATIAL | |||||
| Ga-FeOB | 2.197 | 2 | 1.098 | 6.82 | 0.0019 |
| Type II | 20.1750 | 2 | 10.0875 | 30.482 | 0.0000 |
| Type Ia | 2.617 | 2 | 1.308 | 4.941 | 0.0099 |
| Type Ib | 15.045 | 2 | 7.522 | 7.550 | 0.0014 |
| ELEVATION | |||||
| Ga-FeOB | 16.797 | 2 | 8.398 | 8.749 | 0.0006 |
| Type II | 0.615 | 2 | 0.307 | 1.670 | 0.1957 |
| Type Ia | 5.9770 | 2 | 2.9885 | 5.152 | 0.0087 |
| Type Ib | 1.361 | 2 | 0.680 | 2.400 | 0.0984 |
p Values in bold indicate statistically significant (p < 0.05) ANOVA F values and hence, significant effects of the variables on gene copy numbers. SS, sum of squares; DF, degrees of freedom; MS, mean square; F, ANOVA F statistic.
For factor “spatial”; FeOB (Large n = 24, Medium n = 18, Small n = 8); Type Ia MOB (Large n = 23, Medium n = 13, Small n = 11); Type Ib MOB (Large n = 24, Medium n = 23, Small n = 23); Type II MOB (Large n = 24, Medium n = 22, Small n = 23). For factor “elevation”; FeOB (Large n = 27, Medium n = 14, Small n = 10); Type Ia MOB (Large n = 33, Medium n = 17, Small n = 10); Type Ib MOB (Large n = 40, Medium n = 20, Small n = 10); Type II MOB (Large n = 42, Medium n = 20, Small n = 10).
At large- and medium-sampling scales, the total gene copy numbers of the different bacterial groups followed the sequence, Ga-FeOB > MOB type II > type 1b > type Ia (Figure 5A). Significant differences were detected between all the groups except for type II and type Ib MOB. In the small-scale sampling plot, the abundance of MOB type Ia was significantly lower than in the rest of the samples (Figure 5A).
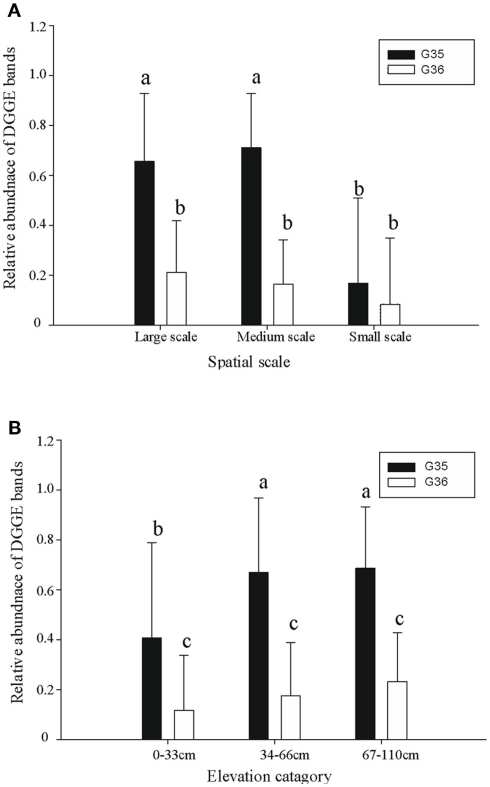
A diverse pattern of relative abundance of the bands retrieved from the DGGE gels was obtained. Among all the bands detected, band G35 and band G36 were the most abundant ones (Figure 2) off which G35 showed a similar trend to the average abundance detected using qPCR (Figure 6A). The relative abundance of band G35 dropped significantly at small-sampling scale as compared to large- and medium-scale samples whereas G36 did not. The rest of the bands did not show a clear trend given spatial level as a factor.
Figure 6.
The relative abundance of two dominant Ga-FeOB DGGE bands at different spatial scales (A) and elevation levels (B). Indicated are means ± SD. Different letters indicate significant differences (p < 0.01) between bars. For factor “spatial” number of observations is n = 18 (Large), n = 22 (Medium), n = 14 (Small) while for factor “elevation” this is n = 25 (Large), n = 19 (Medium), n = 10 (Small).
Elevation level
ANOVA analyses demonstrated that elevation significantly affected the numbers of FeOB and type I a MOB (Table 1). With the increase of elevation, the abundance of FeOB increased significantly (Figure 5B). Again in this case, the relative abundance of band G35 followed the same positive trend with increasing elevation which was not the case for G36 (Figure 6B). MOB showed a similar trend as the Ga-FeOB except for type Ia, which displayed maximum abundance in the intermediate elevation level (Figure 5B). There was a significant difference between the abundance of type Ia in samples taken from medium (33–46 cm) and low level (0–33 cm). No difference was observed among the abundance of other types of methane oxidizers along the elevation gradient.
The total 16s rRNA gene copy numbers detected for Ga-FeOB was much higher (10- to 100-fold) than the pmoA gene numbers of the different types of methane oxidizers at all elevation levels. The only reported 16s RNA copy numbers for iron-oxidizing genomes reported indicate two copies per genome (Fleming et al., 2011) while MOB generally have two to three copies of the pmoA gene (Kolb et al., 2003) supporting the absolute dominance of Ga-FeOB over MOB in terms of cell numbers. Overall, abundance of Ga-FeOB and MOB of all three sub-types were positively correlated (Table 2).
Table 2.
Spearman rank correlation table of measured environmental parameters and gene copy numbers of Ga-FeOB and type Ia, Ib, and II MOB.
| Elevation | Moisture (%) | Organic matter | WFPS | Type Ia | Type Ib | Type II | |
|---|---|---|---|---|---|---|---|
| Elevation | |||||||
| Moisture (%) | −0.54 | ||||||
| Organic matter | −0.08 | 0.21 | |||||
| WFPS | −0.28 | 0.14 | 0.03 | ||||
| Type Ia | 0.24 | −0.38 | −0.16 | −0.33 | |||
| Type Ib | 0.29 | −0.25 | −0.27 | −0.28 | 0.55 | ||
| Type II | 0.07 | −0.21 | −0.30 | −0.30 | 0.60 | 0.58 | |
| FeOB | 0.52 | −0.44 | −0.20 | 0.29 | 0.58 | 0.63 | 0.59 |
Values in bold are statistically different (p < 0.05).
WFPS, water filled pore space.
Spatial interpolation maps
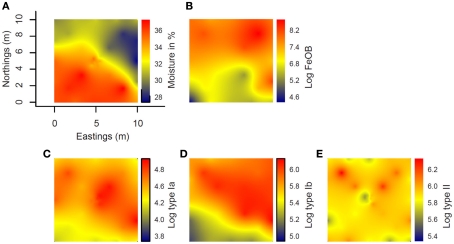
Interpolated maps revealed that Ga-FeOB and MOB abundances display different spatial patterns (Figures 7B–E). While a gradient was observed in Ga-FeOB and type I MOB abundances, type II MOB did not follow a gradient. A comparison between interpolated maps of soil moisture content and Ga-FeOB abundance data indicated a negative correlation between these two factors (Figures 7A,B). This was confirmed by Spearman correlation analyses using only the measured data (Table 2). Similarly, type I MOB abundances (Figures 7A,C,D) indicated a negative correlation with moisture content which was supported by Spearman correlation (Table 2). Ga-FeOB and type Ib showed also the highest spatial dependencies (Table 3). Type Ib displayed a very similar distribution as Ga-FeOB while type Ia abundance tends to be highest at intermediate elevations levels of the study site. Type II showed a distinctly deviating distribution (Figure 7E) which did also not correlate with moisture content (Table 2). Interestingly the lowest moisture content was observed not at the top of sampling plot, but to the side of the plot with the highest plant biomass (i.e., 0.56–1.12 m higher than the lowest point of the plot).
Figure 7.
Spatial interpolation maps (kriging maps) of the distribution at the study site of moisture content (A) and log-transformed gene copy number from Ga-FeOB specific qPCR (B), type Ia (C), type Ib (D), and type II methanotrophs specific qPCRs (E). Color bar to the right of each map indicates extrapolated values of either moisture content or log-transformed gene copy number values.
Table 3.
Parameters for geostatistical analysis of log-transformed qPCR data of FeOB and MOB taxa and percentage of moisture content.
| Parameter | Nugget | Sill | Spatial dependence | p-Range (m) p < 0.05 |
|---|---|---|---|---|
| Moisture | 1.856 | 6.514 | 0.71 | 5.38 |
| FeOB | None | 0.922 | 1.00 | 12.57 |
| Type Ia | 0.261 | 0.396 | 0.34 | 6.34 |
| Type Ib | 0.047 | 0.179 | 0.74 | 14.78 |
| Type II | 0.148 | 0.218 | 0.32 | 2.07 |
The nugget quantifies the amount of variability at distance of zero, the sill is the value where the variogram levels off, the spatial dependence indicates the proportion of variability of the data that was modeled, the practical range (p-range) equal to the distance at which 95% of the sill has been reached.
Discussion
The present study gives insights into the spatial patterns of microbes involved in two important geochemical processes along a hydrological gradient. Ga-FeOB outnumber MOB but their abundance is positively correlated to various subgroups of MOB, indicating that there are shared controlling environmental factors.
The strong increase in numbers of iron-oxidizing bacteria with elevation and the negative correlation with soil moisture content indicate that the hydrological gradient is one of the important environmental factors that control the distribution of the Ga-FeOB. However, the distribution of Ga-FeOB in the present study was opposite to the distribution of Ga-FeOB in a tidal freshwater marsh showing higher numbers in the regularly flooded, lower parts of the marsh compared to the higher parts (Wang et al., 2011). The strong relationship with Fe(III) availability and Ga-FeOB abundance in the study of Wang et al. (2011) pointed to iron as the primary controlling factor, a fact recently also observed in drinking water distribution systems (Li et al., 2010). Although we did not measure iron availability at time of sampling in the present study, an earlier study at the same site showed that total Fe in these soils is around 5 ppm (Conrad et al., 2008). This observation combined with the orders of magnitude higher numbers of Ga-FeOB found as compared to the tidal freshwater marsh, indicates that iron availability was not limiting at the floodplain site. As soil moisture content decreases with elevation (Figure 7A), the availability, and penetration depth of oxygen into the soil likely increases also, which in itself should favor bacterial iron oxidation. However, when soil oxygenation increases, more ferrous iron may be consumed by chemical iron oxidation. A decrease in soil moisture content may also lead to less iron reduction, which consequently slows down the redox turnover of iron (Blothe and Roden, 2009). Together, this would lead to low ferrous iron supply, a key factor for iron-oxidizing activity. Apparently, the conditions also in the upper parts of the floodplain are such that oxygen and ferrous iron are both available and that Ga-FeOB can compete with chemical oxidation.
All Ga-FeOB so far isolated are all lithotrophic obligatory aerobic bacteria which mainly are controlled by the availability of and O2 (Emerson et al., 2010). The dominant Ga-FeOB in this study (represented by band G35 and G36) display a clearly different distribution. G35 increases with elevation related to the lower water content and probably to the connected oxygen availability. The sequence is closely related to an isolate also derived from an irregular flooded riparian soil and tentatively called “Ferricurvibacter nieuwersluisensis”(Wang, 2011) which also grew in gradient tubes under microaerophilic conditions but also could tolerate exposure to higher amounts of oxygen (Wang, 2011). G36 did not display an elevation-related distribution in this irregular floodplain but is highly related to a sequence (Hc8 in Figure 2) which was the most dominant one in a tidal marsh, where it also correlated positively with iron availability (Wang, 2011). Although, we have no physiological data to support any firm conclusions regarding environmental control, it seems that within the Ga-FeOB there is habitat preference and niche differentiation likely related to the local regimes of O2 and or availability. Considering the limited number of isolates and lack of physiological knowledge of these isolates, it would be too speculative to discuss other environmental controls. However, considering the phylogenetic diversity in the proposed order Gallionellales (Emerson et al., 2010; Figure 4) it can be expected that behind the sequences there will be microbes with a broader metabolic repertoire and different oxygen tolerances than isolated so far, offering explanations for the environmental patterns observed in various environments.
The irregular flooded riparian floodplain studied and the tidal marsh investigated in a previous study (Wang et al., 2011) were very similar in terms of Ga-FeOB community composition but differed markedly in Ga-FeOB abundance Although the primers are targeting Gallionella-related FeOB only, the range of different sequence types or strains potentially detected by the primers used is much higher than detected in both studies (Wang et al., 2009) rendering it unlikely that the resemblance is due to a primer bias. This leads to the conclusion that physico-chemical conditions at the irregularly flooded site are better for performance of the dominant Ga-FeOB at that site. The alternative explanation would be that competition with other microbes and processes is more intense in the tidal freshwater marsh.
According to thermodynamics (Thauer et al., 1977; Sobolev and Roden, 2002), the oxidation processes should follow the order of ferrous iron > methane > sulfide > ammonium oxidation under conditions of oxygen limitation. It has been suggested by Neubauer et al. (2002) that neutrophilic Fe(II)-oxidizing bacteria may compete for limiting amounts of O2 in the rhizosphere and therefore influence other wetland biogeochemical cycles. van Bodegom et al. (2001b) also reported that iron oxidation was the most important oxidative process in a rice paddy rhizosphere and accounted initially for 97% of the consumed oxygen using a detailed mechanistic model (van Bodegom et al., 2001a). However, in an accompanying study these authors did not detect iron oxidizers by means of traditional cultivation-dependent most probable number counts and concluded that this oxidation was mainly chemical and that microbial iron oxidation was negligible (van Bodegom et al., 2001b). However, in our irregularly flooded site the Ga-FeOB outnumber the various groups of MOB yielding the first experimental evidence suggesting that on microbial population level microbial iron oxidation can prevail over methane consumption and that chemical iron oxidation may not be as dominant as assumed. We have to bear in mind though that this conclusion is based on DNA-based abundance assessment and not on measured processes.
The recent isolation and genomic description of a marine neutrophilic iron oxidizer provided evidence for the microaerophilic nature and high affinity for oxygen of FeOB, which would enable them to be active at lower oxygen concentrations than chemical oxidation and methane oxidizers can (Singer et al., 2011). However, there are no clues to the extent to which this information can be extrapolated to freshwater wetlands. The recent development of a cultivation-independent molecular detection assay allows for the reliably revealing of Ga-FeOB in environmental settings (Wang et al., 2009; Li et al., 2010) and the ability to relate their distribution to that of other microbes and chemical oxidation processes. The strong correlation we find in this study between the abundance of Ga-FeOB and some of the MOB indicates similar controlling factors, comparable competitive abilities or both. The scarce culture information of FeOB shows that neutrophilic obligate FeOB (Weiss et al., 2007) have similar growth rates as compared to methanotrophs (Dedysh et al., 2007; Belova et al., 2011). However, growth rates under environmental conditions can be quite different from optimal conditions in cultures (Mohanty et al., 2006; Steenbergh et al., 2010) and thus the interactions and competitive abilities of FeOB and MOB have to be verified under environmental conditions.
Alternative to the ability to compete with MOB, the dominance of FeOB over MOB may also be caused indirectly due to suppression of methanogenesis caused by substrate competition with iron reducers. Hence, sub-optimal methane supply would restrict the activities of MOB. The lag in methanogenic activity as measured in methane production assays on this site in an earlier study points into that direction (Kemnitz et al., 2004). Methane flux measurements executed on the same day as sampling was done in this study yielded values ranging from methane uptake (24 mg CH4 m−1/day) to net emission (180 mg CH4 m−1/day; data not shown). This highly variable emission, with negative values and values even in the range of rice fields does not allow for conclusive statements toward methane availability for MOB. However, earlier studies on this site demonstrated immediate in vitro methane consumption displaying rates in the range of high methane habitats (Steenbergh et al., 2010; Bodelier et al., 2012) suggesting continuous methane availability for MOB. It will require more detailed studies, monitoring soil iron (Fe2+, Fe3+) as well as methane availability in parallel to FeOB and MOB community assessments to obtain supporting evidence for “competitive ability” or the “substrate limitation” hypotheses.
In contrast to the distribution based on the measured points only, spatial interpolation maps of Ga-FeOB and MOB abundances reveal that Ga-FeOB and type Ib MOB display spatial autocorrelation which reflects the underlying hydrological gradient (i.e., combined effects of moisture content and elevation; Figure 7). Type Ia and II MOB displayed a distribution not spatially correlated to the hydrological gradient, with type Ia displaying highest abundances at intermediate moisture and type II showing a patchy distribution over the entire gradient. This differential distribution may be related to the different ecological “strategies” described for the various types of MOB (Steenbergh et al., 2010) with the fast-growing, responsive MOB (i.e., type Ia) abundant where substrates are available and type II occupying patches where survival of periods without substrate is required. A very similar spatial distribution of type II and type Ia MOB was found in a hydrological gradient in a littoral zone of a freshwater lake (Siljanen et al., 2011) supporting niche differentiation of MOB with respect to hydrology-associated environmental factors. In the study of Siljanen et al., type Ib also displayed a strong spatial autocorrelation with the hydrological gradient, but in contrast to the present study a positive relationship was found with moisture content. This can be explained by the fact that what is assigned as type Ib MOB contains uncultured as well as cultured representatives with quite different characteristics as might be concluded from their original habitats (Lüke et al., 2010; Semrau et al., 2010). Hence, interpreting the similarity between the distribution of type Ib and Ga-FeOB in our irregularly flooded riparian site as being groups that have similar ecological characteristics, would be rather speculative. The most likely explanation for the strong co-occurrence is the common dependence on oxic–anoxic interfaces and microaerophilic conditions, which Ga-FeOB require to compete successfully with chemical oxidation(Emerson et al., 2010) and to ensure the availability of O2, Fe2+, and methane. Additional support for common ecological niches comes from the fact that type Ib MOB are abundant members of the MOB community in rice field soils (Lüke et al., 2010; Ho et al., 2011), a habitat which is very similar to the studied floodplain soil (Conrad et al., 2008).
A striking phenomenon in our study was the clear deviating Ga-FeOB and MOB abundances at the small (i.e., cm) scale. For type Ia MOB and the FeOB, gene copies were below detection limit in a large number of samples (Typ Ia 11 out 23, FeOB 17 out 23), but also for type Ib as well as type II the abundances were generally significantly lower. Siljanen et al. (2011) observed much less spatial structure and variance at the cm scale within a littoral hydrological gradient than at a larger scale. Abundance is obviously affected by small scale differences, disconnected from the large-scale environmental gradient. The only measured variables that deviated in small scale plot are moisture and organic matter contents, which are both significantly higher in the small scale samples. The combination of these factors can put constraints on the availability of oxygen by a more intense competition with heterotrophs (van Bodegom et al., 2001b). Nevertheless, the lack of direct experimental evidence for competitive interactions between heterotrophs, Ga-FeOB, and MOB renders every explanation speculative. Fact is though that the observed local deviations in diversity and abundance of Ga-FeOB and MOB would have gone unnoticed in a more coarse sampling design along with the useful ecological information associated to it. Other authors have observed clear spatial patterns in response to agricultural management on meter scales (Philippot et al., 2009a,b; Wessen et al., 2011) using geostatistical approaches. It would be exciting to see whether the generalizations derived from these latter sampling schemes would hold when small scale assessments would have been executed.
Besides the variation between sampling scales there is also large variation in gene abundance over several orders of magnitude between samples from the same sampling scale, even when cm apart. This holds both true for Ga-FeOB as well as MOB. Hence, the possible solutions for the observed variability should come from factors affecting both groups of microbes. Considering the conflicting aspects of Fe(III) and methane availability (i.e., competition of methanogens and iron reducers), oxygen would be the most likely factor affecting both the distribution of Ga-FeOB and MOB. The sampled floodplain soil consists of heavy clay which has already limited air penetration that can even be worse when the soil is compacted. The latter has been show to influence methane oxidation in soil (Dalal et al., 2008; Gebert et al., 2011.). In this floodplain this can both be caused by the floodwater itself but also by trampling of the cows which are present in this area for nature management purposes. This would lead to a highly irregular pattern of compacted parts of the soil where possibilities for growth and activity for aerobes are very limited. The higher number of samples below the detection limit in the 20 cm × 20 cm plot may be simply because due to the size of cores taken, the whole surface area was samples and hence, the chance of hitting these “cold spots” is simply higher as compared to the large- and medium-scale samples.
Hence, heterogeneity of physico-chemical conditions and associated substrate availability may one part of the explanation. The physics of the soil may also influence biological factors like predation by protozoa (Ranjard and Richaume, 2001). The compaction may lead to a heterogenous pattern of soil space inaccessible for protozoa, leading to locally higher numbers of Ga-FeOB and MOB. However, since we have no detailed information on soil-macro and microstructure for the respective samples the explanations offered will remain speculative.
Conclusion
Gallionella-related neutrophilic iron-oxidizing bacteria are abundant throughout the studied riparian floodplain, their distribution reflecting the hydrological gradient. They outnumber various groups of methane oxidizers, demonstrating that FeOB are both able to compete for oxygen with methanotrophs as well as with chemical iron oxidation. The spatial distribution of Ga-FeOB resembles the distribution of type Ib MOB, whereas it deviates from those of type Ia and type II MOB pointing to the fact only one MOB subgroup has similar ecological strategies to Ga-FeOB. Abundances of Ga-FeOB as well as MOB differed significantly from larger scales when sampled on cm scale demonstrating common local limiting factors, which may indicate environmental boundaries for growth of Ga-FeOB and MOB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Foundation “De Ark” for permitting us to take samples at “Ewijkse Waard.” This study was part of the ESF-Eurodiversity program (ERAS-CT-2003-98049, 6th EU-framework program) and was financially supported by grants from the Netherlands Organization for Scientific Research (NWO; Grant no. 855.01.108) as by and by the Netherlands Darwin Center for Biogeology (grants 142.16.1031 and 142.16.1032). This publication is publication nr5218 of the Netherlands Institute of Ecology (NIOO-KNAW).
References
- Armstrong W. (1964). Oxygen diffusion from roots of some British bog plants. Nature 204, 801. 10.1038/204801a0 [DOI] [PubMed] [Google Scholar]
- Baker K. L., Langenheder S., Nicol G. W., Ricketts D., Killham K., Campbell C. D., Prosser J. I. (2009). Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biol. Biochem. 41, 2292–2298 10.1016/j.soilbio.2009.08.010 [DOI] [Google Scholar]
- Belova S. E., Baani M., Suzina N. E., Bodelier P. L. E., Liesack W., Dedysh S. N. (2011). Acetate utilization as a survival strategy of peat-inhabiting Methylocystis spp. Environ. Microbiol. Rep. 3, 36–46 10.1111/j.1758-2229.2010.00180.x [DOI] [PubMed] [Google Scholar]
- Blothe M., Roden E. E. (2009). Microbial iron redox cycling in a circumneutral-pH groundwater seep. Appl. Environ. Microbiol. 75, 468–473 10.1128/AEM.01742-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelier P. L. E. (2011). Interactions between nitrogenous fertilizers and methane cycling in wetland and upland soils. Curr. Opin. Environ. Sustain. 3, 379–388 10.1016/j.cosust.2011.06.002 [DOI] [Google Scholar]
- Bodelier P. L. E., Bar-Gilissen M. J., Meima-Franke M., Hordijk K. (2012). Structural and functional response of methane-consuming microbial communities to different flooding regimes in riparian soils. Ecol. Evol. 2, 106–127 10.1002/ece3.34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodelier P. L. E., Frenzel P., Drake H. L., Hurek T., Kusel K., Lovell C., Megonigal P., Reinhold-Hurek B., Sorrell B. (2006). “Ecological aspects of microbes and microbial communities inhabiting the rhizosphere of wetland plants,” in Wetlands and Natural Resource Management, eds. Verheven J. T. A., Beltman B., Bobbink R., Whigham D. F. (Berlin: Springer; ), 205–238 [Google Scholar]
- Burgin A. J., Yang W. H., Hamilton S. K., Silver W. L. (2011). Beyond carbon and nitrogen: how the microbial energy economy couples elemental cycles in diverse ecosystems. Front. Ecol. Environ. 9, 44–52 10.1890/090227 [DOI] [Google Scholar]
- Chan C. S., Fakra S. C., Edwards D. C., Emerson D., Banfield J. F. (2009). Iron oxyhydroxide mineralization on microbial extracellular polysaccharides. Geochim. Cosmochim. Acta 73, 3807–3818 10.1016/j.gca.2009.02.036 [DOI] [Google Scholar]
- Clarke K. R., Warwick R. M. (1998). Quantifying structural redundancy in ecological communities. Oecologia 113, 278–289 10.1007/s004420050379 [DOI] [PubMed] [Google Scholar]
- Clarke K. R., Warwick R. M. (2001). A further biodiversity index applicable to species lists: variation in taxonomic distinctness. Mar. Ecol. Prog. Ser. 216, 265–278 10.3354/meps216265 [DOI] [Google Scholar]
- Coci M., Riechmann D., Bodelier P. L. E., Stefani S., Zwart G., Laanbroek H. J. (2005). Effect of salinity on temporal and spatial dynamics of ammonia-oxidising bacteria from intertidal freshwater sediment. FEMS Microbiol. Ecol. 53, 359–368 10.1016/j.femsec.2005.01.016 [DOI] [PubMed] [Google Scholar]
- Conrad R. (2007). Microbial ecology of methanogens and methanotrophs. Adv. Agron. 96, 1–63 10.1016/S0065-2113(07)96005-8 [DOI] [Google Scholar]
- Conrad R., Klose M., Noll M., Kemnitz D., Bodelier P. L. E. (2008). Soil type links microbial colonization of rice roots to methane emission. Glob. Chang. Biol. 14, 657–669 10.1111/j.1365-2486.2007.01516.x [DOI] [Google Scholar]
- Cressie N. A. C. (1993). Statistics for Spatial Data. New York: John Wiley & Sons [Google Scholar]
- Dalal R. C., Allen D. E., Livesley S. J., Richards G. (2008). Magnitude and biophysical regulators of methane emission and consumption in the Australian agricultural, forest, and submerged landscapes: a review. Plant Soil 309, 43–76 10.1007/s11104-007-9446-7 [DOI] [Google Scholar]
- Dedysh S. N., Belova S. E., Bodelier P. L. E., Smirnova K. V., Khmelenina V. N., Chidthaisong A., Trotsenko Y. A., Liesack W., Dunfield P. F. (2007). Methylocystis heyeri sp nov., a novel type II methanotrophic bacterium possessing “signature” fatty acids of type I methanotrophs. Int. J. Syst. Evol. Microbiol. 57, 472–479 [DOI] [PubMed] [Google Scholar]
- Doyle M. O., Otte M. L. (1997). Organism-induced accumulation of iron, zinc and arsenic in wetland soils. Environ. Pollut. 96, 1–11 10.1016/S0269-7491(97)00014-6 [DOI] [PubMed] [Google Scholar]
- Emerson D., Fleming E. J., McBeth J. M. (2010). Iron-oxidizing bacteria: an environmental and genomic perspective. Annu. Rev. Microbiol. 64, 561–583 10.1146/annurev.micro.112408.134208 [DOI] [PubMed] [Google Scholar]
- Emerson D., Moyer C. (1997). Isolation and characterization of novel iron-oxidizing bacteria that grow at circumneutral pH. Appl. Environ. Microbiol. 63, 4784–4792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettema C. H., Wardle D. A. (2002). Spatial soil ecology. Trends Ecol. Evol. (Amst.) 17, 177–183 10.1016/S0169-5347(02)02496-5 [DOI] [Google Scholar]
- Ferreira T. O., Otero X. L., De Souza V. S., Vidal-Torrado P., Macias F., Firme L. P. (2010). Spatial patterns of soil attributes and components in a mangrove system in Southeast Brazil (Sao Paulo). J. Soils Sediments 10, 995–1006 10.1007/s11368-010-0224-4 [DOI] [Google Scholar]
- Fleming E. J., Langdon A. E., Martinez-Garcia M., Stepanauskas R., Poulton N. J., Masland E. D. P., Emerson D. (2011). What’s new is old: resolving the Identity of Leptothrix ochracea using single cell genomics, pyrosequencing and FISH. PLoS ONE 6, e17769. 10.1371/journal.pone.0017769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin D., Langley S. (2005). Formation and occurrence of biogenic iron-rich minerals. Earth Sci. Rev. 72, 1–19 10.1016/j.earscirev.2005.03.002 [DOI] [Google Scholar]
- Franklin R. B., Mills A. L. (2003). Multi-scale variation in spatial heterogeneity for microbial community structure in an eastern Virginia agricultural field. FEMS Microbiol. Ecol. 44, 335–346 10.1016/S0168-6496(03)00074-6 [DOI] [PubMed] [Google Scholar]
- Gebert J., Groengroeft A., Pfeiffer E. M. (2011). Relevance of soil physical properties for the microbial oxidation of methane in landfill covers. Soil Biol. Biochem. 43, 1759–1767 10.1016/j.soilbio.2010.07.004 [DOI] [Google Scholar]
- Gutknecht J. L. M., Goodman R. M., Balser T. C. (2006). Linking soil process and microbial ecology in freshwater wetland ecosystems. Plant Soil 289, 17–34 10.1007/s11104-006-9105-4 [DOI] [Google Scholar]
- Hartman W. H., Richardson C. J., Vilgalys R., Bruland G. L. (2008). Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc. Natl. Acad. Sci. U.S.A. 105, 17842–17847 10.1073/pnas.0709231105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A., Lüke C., Frenzel P. (2011). Recovery of methanotrophs from disturbance: population dynamics, evenness and functioning. ISME J. 5, 750–758 10.1038/ismej.2010.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemnitz D., Chin K. J., Bodelier P., Conrad R. (2004). Community analysis of methanogenic archaea within a riparian flooding gradient. Environ. Microbiol. 6, 449–461 10.1111/j.1462-2920.2004.00573.x [DOI] [PubMed] [Google Scholar]
- Kogel-Knabner I., Amelung W., Cao Z. H., Fiedler S., Frenzel P., Jahn R., Kalbitz K., Kolbl A., Schloter M. (2010). Biogeochemistry of paddy soils. Geoderma 157, 1–14 10.1016/j.geoderma.2010.03.009 [DOI] [Google Scholar]
- Kolb S., Knief C., Stubner S., Conrad R. (2003). Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69, 2423–2429 10.1128/AEM.69.5.2423-2429.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laanbroek H. J. (2010). Methane emission from natural wetlands: interplay between emergent macrophytes and soil microbial processes. A mini-review. Ann. Bot. 105, 141–153 10.1093/aob/mcp201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legendre P., Legendre L. (1998). Numerical Ecology. Amsterdam: Elsevier [Google Scholar]
- Li D., Li Z., Yu J., Cao N., Liu R., Yang M. (2010). Characterization of bacterial community structure in a drinking water distribution system during an occurrence of Red Water. Appl. Environ. Microbiol. 76, 7171–7180 10.1128/AEM.00707-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovley D. R. (1997). Microbial Fe(III) reduction in subsurface environments. FEMS Microbiol. Rev. 20, 305–313 10.1111/j.1574-6976.1997.tb00316.x [DOI] [Google Scholar]
- Ludwig W., Strunk O., Westram R., Richter L., Meier H., Yadhukumar, Buchner A., Lai T., Steppi S., Jobb G., Forster W., Brettske I., Gerber S., Ginhart A. W., Gross O., Grumann S., Hermann S., Jost R., Konig A., Liss T., Lussmann R., May M., Nonhoff B., Reichel B., Strehlow R., Stamatakis A., Stuckmann N., Vilbig A., Lenke M., Ludwig T., Bode A., Schleifer K. H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371 10.1093/nar/gkh293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke C., Frenzel P. (2011). Potential of pmoA amplicon pyrosequencing for methanotroph diversity studies. Appl. Environ. Microbiol. 77, 6305–6309 10.1128/AEM.05355-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lüke C., Krause S., Cavigiolo S., Greppi D., Lupotto E., Frenzel P. (2010). Biogeography of wetland rice methanotrophs. Environ. Microbiol. 12, 862–872 10.1111/j.1462-2920.2009.02131.x [DOI] [PubMed] [Google Scholar]
- Mermillod-Blondin F., Lemoine D. G. (2010). Ecosystem engineering by tubificid worms stimulates macrophyte growth in poorly oxygenated wetland sediments. Funct. Ecol. 24, 444–453 10.1111/j.1365-2435.2009.01624.x [DOI] [Google Scholar]
- Mohanty S. R., Bodelier P. L. E., Floris V., Conrad R. (2006). Differential effects of nitrogenous fertilizers on methane-consuming microbes in rice field and forest soils. Appl. Environ. Microbiol. 72, 1346–1354 10.1128/AEM.72.2.1346-1354.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyzer G., Dewaal E. C., Uitterlinden A. G. (1993). Profiling of complex microbial-populations by denaturing gradient gel-electrophoresis analysis of polymerase chain reaction-amplified genes-coding for 16s ribosomal-RNA. Appl. Environ. Microbiol. 59, 695–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer S. C., Emerson D., Megonigal J. P. (2002). Life at the energetic edge: kinetics of circumneutral iron oxidation by lithotrophic iron-oxidizing bacteria isolated from the wetland-plant rhizosphere. Appl. Environ. Microbiol. 68, 3988–3995 10.1128/AEM.68.8.3988-3995.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer S. C., Toledo-Duran G. E., Emerson D., Megonigal J. P. (2007). Returning to their roots: iron-oxidizing bacteria enhance short-term plaque formation in the wetland-plant rhizosphere. Geomicrobiol. J. 24, 65–73 10.1080/01490450601134309 [DOI] [Google Scholar]
- Op den Camp H. J. M., Islam T., Stott M. B., Harhangi H. R., Hynes A., Schouten S., Jetten M. S. M., Birkeland N. K., Pol A., Dunfield P. F. (2009). Environmental, genomic and taxonomic perspectives on methanotrophic Verrucomicrobia. Environ. Microbiol. Rep. 1, 293–306 10.1111/j.1758-2229.2009.00022.x [DOI] [PubMed] [Google Scholar]
- Pan Y., Bodrossy L., Frenzel P., Hestnes A., Krause S. B. M., Lüke C., Meima-Franke M., Siljanen H., Svenning M. M., Bodelier P. L. E. (2010). Impacts of inter- and intralaboratory variations on the reproducibility of microbial community analyses. Appl. Environ. Microbiol. 76, 7451–7458 10.1128/AEM.02185-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippot L., Bru D., Saby N. P. A., Cuhel J., Arrouays D., Simek M., Hallin S. (2009a). Spatial patterns of bacterial taxa in nature reflect ecological traits of deep branches of the 16S rRNA bacterial tree. Environ. Microbiol. 11, 3096–3104 10.1111/j.1462-2920.2009.01879.x [DOI] [PubMed] [Google Scholar]
- Philippot L., Cuhel J., Saby N. P. A., Cheneby D., Chronakova A., Bru D., Arrouays D., Martin-Laurent F., Simek M. (2009b). Mapping field-scale spatial patterns of size and activity of the denitrifier community. Environ. Microbiol. 11, 1518–1526 10.1111/j.1462-2920.2009.01879.x [DOI] [PubMed] [Google Scholar]
- Ranjard L., Richaume A. S. (2001). Quantitative and qualitative microscale distribution of bacteria in soil. Res. Microbiol. 152, 707–716 10.1016/S0923-2508(01)01251-7 [DOI] [PubMed] [Google Scholar]
- R Development Core Team. (2010). R: a Language and Environment for Statistical Computing. ver. 2.11.1. Available at: http://cran.r-project.org/
- Ribeiro J. R., Diggle P. J. (2001). geoR: a package for geostatistical analysis. R. J. 1, 1609–3631 [Google Scholar]
- Ringeval B., Friedlingstein P., Koven C., Ciais P., De Noblet-Ducoudre N., Decharme B., Cadule P. (2011). Climate-CH4 feedback from wetlands and its interaction with the climate-CO2 feedback. Biogeosciences 8, 2137–2157 10.5194/bg-8-2137-2011 [DOI] [Google Scholar]
- Schäfer H., Bernard L., Courties C., Lebaron P., Servais P., Pukall R., Stackebrandt E., Troussellier M., Guindulain T., Vives-Rego J., Muyzer G. (2001). Microbial community dynamics in Mediterranean nutrient-enriched seawater mesocosms: changes in the genetic diversity of bacterial populations. FEMS Microbiol. Ecol. 34, 243–253 10.1016/S0168-6496(00)00102-1 [DOI] [PubMed] [Google Scholar]
- Semrau J. D., Dispirito A. A., Yoon S. (2010). Methanotrophs and copper. FEMS Microbiol. Rev. 34, 496–531 [DOI] [PubMed] [Google Scholar]
- Siljanen H. M. P., Saari A., Krause S., Lensu A., Abell G. C. J., Bodrossy L., Bodelier P. L. E., Martikainen P. J. (2011). Hydrology is reflected in the functioning and community composition of methanotrophs in the littoral wetland of a boreal lake. FEMS Microbiol. Ecol. 75, 430–445 10.1111/j.1574-6941.2010.01015.x [DOI] [PubMed] [Google Scholar]
- Singer E., Emerson D., Webb E. A., Barco R. A., Kuenen J. G., Nelson W. C., Chan C. S., Comolli L. R., Ferriera S., Johnson J., Heidelberg J. F., Edwards K. J. (2011). Mariprofundus ferrooxydans PV-1 the first genome of a marine Fe(II) oxidizing Zetaproteobacterium. PLoS ONE 6, e25386. 10.1371/journal.pone.0025386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev D., Roden E. E. (2002). Evidence for rapid microscale bacterial redox cycling of iron in circumneutral environments. Antonie Van Leeuwenhoek 81, 587–597 10.1023/A:1020569908536 [DOI] [PubMed] [Google Scholar]
- Steenbergh A. K., Meima M. M., Kamst M., Bodelier P. L. E. (2010). Biphasic kinetics of a methanotrophic community is a combination of growth and increased activity per cell. FEMS Microbiol. Ecol. 71, 12–22 10.1111/j.1574-6941.2009.00782.x [DOI] [PubMed] [Google Scholar]
- Sundby B., Vale C., Caetano M., Luther G. W. (2003). Redox chemistry in the root zone of a salt marsh sediment in the Tagus Estuary, Portugal. Aquat. Geochem. 9, 257–271 10.1023/B:AQUA.0000022957.42522.9a [DOI] [Google Scholar]
- Thauer R. K., Jungermann K., Decker K. (1977). Energy conservation in chemotrophic anaerobic bacteria. Bacteriol. Rev. 41, 100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bodegom P., Goudriaan J., Leffelaar P. (2001a). A mechanistic model on methane oxidation in a rice rhizosphere. Biogeochemistry 55, 145–177 10.1023/A:1010640515283 [DOI] [Google Scholar]
- van Bodegom P., Stams F., Mollema L., Boeke S., Leffelaar P. (2001b). Methane oxidation and the competition for oxygen in the rice rhizosphere. Appl. Environ. Microbiol. 67, 3586–3597 10.1128/AEM.67.8.3586-3597.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. J. (2011). Ecology of Neutrophilic Iron-Oxidizing Bacteria in Wetland Soils. Ph.D. thesis, University of Utrecht, Utrecht [Google Scholar]
- Wang J. J., Muyzer G., Bodelier P. L. E., Laanbroek H. J. (2009). Diversity of iron oxidizers in wetland soils revealed by novel 16S rRNA primers targeting Gallionella-related bacteria. ISME J. 3, 715–725 10.1038/ismej.2009.37 [DOI] [PubMed] [Google Scholar]
- Wang J. J., Vollrath S., Behrends T., Bodelier P. L. E., Muyzer G., Meima-Franke M., Den Oudsten F., Van Cappellen P., Laanbroek H. J. (2011). Distribution and diversity of Gallionella-like neutrophilic iron oxidizers in a tidal freshwater marsh. Appl. Environ. Microbiol. 77, 2337–2344 10.1128/AEM.05008-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss J. V., Emerson D., Backer S. M., Megonigal J. P. (2003). Enumeration of Fe(II)-oxidizing and Fe(III)-reducing bacteria in the root zone of wetland plants: implications for a rhizosphere iron cycle. Biogeochemistry 64, 77–96 10.1023/A:1024953027726 [DOI] [Google Scholar]
- Weiss J. V., Rentz J. A., Plaia T., Neubauer S. C., Merrill-Floyd M., Lilburn T., Bradburne C., Megonigal J. P., Emerson D. (2007). Characterization of neutrophilic Fe(II)-oxidizing bacteria isolated from the rhizosphere of wetland plants and description of Ferritrophicum radicicola gen. nov sp nov., and Sideroxydans paludicola sp nov. Geomicrobiol. J. 24, 559–570 10.1080/01490450701670152 [DOI] [Google Scholar]
- Wessen E., Soderstrom M., Stenberg M., Bru D., Hellman M., Welsh A., Thomsen F., Klemedtson L., Philippot L., Hallin S. (2011). Spatial distribution of ammonia-oxidizing bacteria and archaea across a 44-hectare farm related to ecosystem functioning. ISME J. 5, 1213–1225 10.1038/ismej.2010.206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeates C., Gillings M. R. (1998). Rapid purification of DNA from soil for molecular biodiversity analysis. Lett. Appl. Microbiol. 27, 49–53 10.1046/j.1472-765X.1998.00383.x [DOI] [Google Scholar]