Abstract
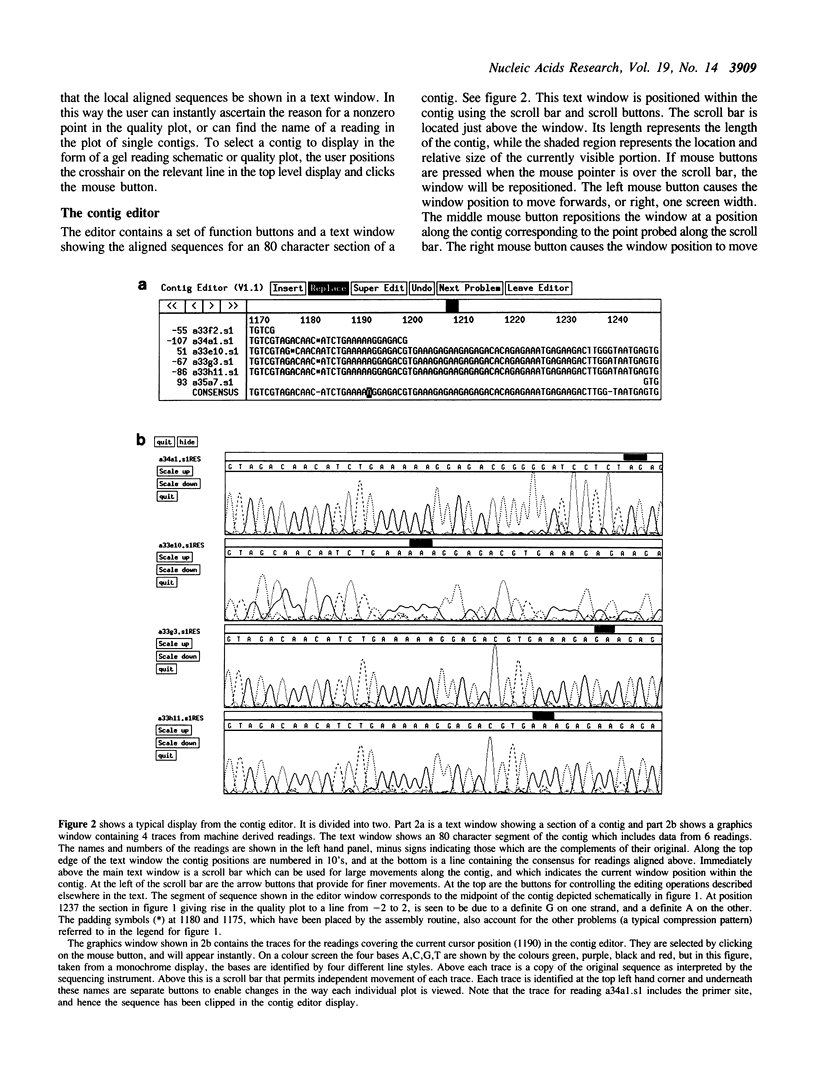
We describe a sequence assembly and editing program for managing large and small projects. It is being used to sequence complete cosmids and has substantially reduced the time taken to process the data. In addition to handling conventionally derived sequences it can use data obtained from Applied Biosystems,Inc. 373A and Pharmacia A.L.F. fluorescent sequencing machines. Readings are assembled automatically. All editing is performed using a mouse operated contig editor that displays aligned sequences and their traces together on the screen. The editor, which can be used on single contigs or for joining contigs, permits rapid movement along the aligned sequences. Insertions, deletions and replacements can be made in individual aligned readings and global changes can be made by editing the consensus. All changes are recorded. A click on a mouse button will display the traces covering the current cursor position, hence allowing quick resolution of problems. Another function automatically moves the cursor to the next unresolved character. The editor also provides facilities for annotating the sequences. Typical annotations include flagging the positions of primers used for walking, or for marking sites, such as compressions, that have caused problems during sequencing. Graphical displays aid the assessment of progress.
Full text
PDF



Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Chee M. S., Bankier A. T., Beck S., Bohni R., Brown C. M., Cerny R., Horsnell T., Hutchison C. A., 3rd, Kouzarides T., Martignetti J. A. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr Top Microbiol Immunol. 1990;154:125–169. doi: 10.1007/978-3-642-74980-3_6. [DOI] [PubMed] [Google Scholar]
- Johnston R. E., Mackenzie J. M., Jr, Dougherty W. G. Assembly of overlapping DNA sequences by a program written in BASIC for 64K CP/M and MS-DOS IBM-compatible microcomputers. Nucleic Acids Res. 1986 Jan 10;14(1):517–527. doi: 10.1093/nar/14.1.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola H., Söderlund H., Ukkonen E. SEQAID: a DNA sequence assembling program based on a mathematical model. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):307–321. doi: 10.1093/nar/12.1part1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts L. The worm project. Science. 1990 Jun 15;248(4961):1310–1313. doi: 10.1126/science.2356467. [DOI] [PubMed] [Google Scholar]
- Staden R. An improved sequence handling package that runs on the Apple Macintosh. Comput Appl Biosci. 1990 Oct;6(4):387–393. doi: 10.1093/bioinformatics/6.4.387. [DOI] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]



