Abstract
Background
Limited or no epidemiological information has been reported for rabies viruses (RABVs) isolated from livestock in the northeastern Brazilian states of Paraíba (PB) and Pernambuco (PE). The aim of this study was to clarify the molecular epidemiology of RABVs circulating in livestock, especially cattle, in these areas between 2003 and 2009.
Findings
Phylogenetic analysis based on 890 nt of the nucleoprotein (N) gene revealed that the 52 livestock-derived RABV isolates characterized here belonged to a single lineage. These isolates clustered with a vampire bat-related RABV lineage previously identified in other states in Brazil; within PB and PE, this lineage was divided between the previously characterized main lineage and a novel sub-lineage.
Conclusions
The occurrences of livestock rabies in PB and PE originated from vampire bat RABVs, and the causative RABV lineage has been circulating in this area of northeastern Brazil for at least 7 years. This distribution pattern may correlate to that of a vampire bat population isolated by geographic barriers.
Background
Rabies is a fatal infectious disease that causes encephalomyelitis. In Brazil, various rabies viruses (RABVs) have been isolated from numerous animal species, including dogs, foxes, cats, and cattle, as well as from hematophagous, insectivorous, and frugivorous bats. Vampire bats, particularly Desmodus rotundus, are an important rabies vector in Latin America. Transmission from vampire bats to humans has been reported, primarily in the Amazon regions of Brazil and Peru, and a large number of cases of cattle rabies transmitted by vampire bats also have been reported in Brazil [1]. Since the introduction of a regional elimination program, the incidence of human and canine rabies in Latin America has fallen by 90% over the past 20 years. However, northeastern Brazil remains a "hotspot" for human rabies because of circulation of the virus among the dog population [2]. Carnieli Jr. et al. reported the molecular characterization and epidemiology of RABVs isolated from canids in northeastern Brazil [3-5], and Shoji et al. reported the genetic and phylogenetic characterization of RABVs isolated from wild fox, insectivorous bats, and livestock in Paraíba (PB) [6]. In Olinda, a city in Pernambuco (PE), 7,062 patients underwent prophylactic antirabies treatment between 2002 and 2006 [7]. Molecular and geographic analyses of livestock rabies in central and southeast Brazil revealed that RABVs isolated from livestock were related to the virus found in vampire bat populations, and this epidemiological pattern was maintained over time and space in these areas [8-11]. However, little or no information on the molecular epidemiology of RABVs has been reported for the virus isolated from livestock in PB and PE. The aim of the present study was to analyse the molecular epidemiology of RABVs circulating among livestock, especially cattle, in these areas between 2003 and 2009.
Results
The sequences of 890 nt PCR products, corresponding to nucleotides 89-978 of the Pasteur vaccine (PV) strain, were determined for all 52 RABV isolates. Among the 52 isolates, the nucleotide and amino acid sequence identities were 97.7-100% and 97.9-100%, respectively.
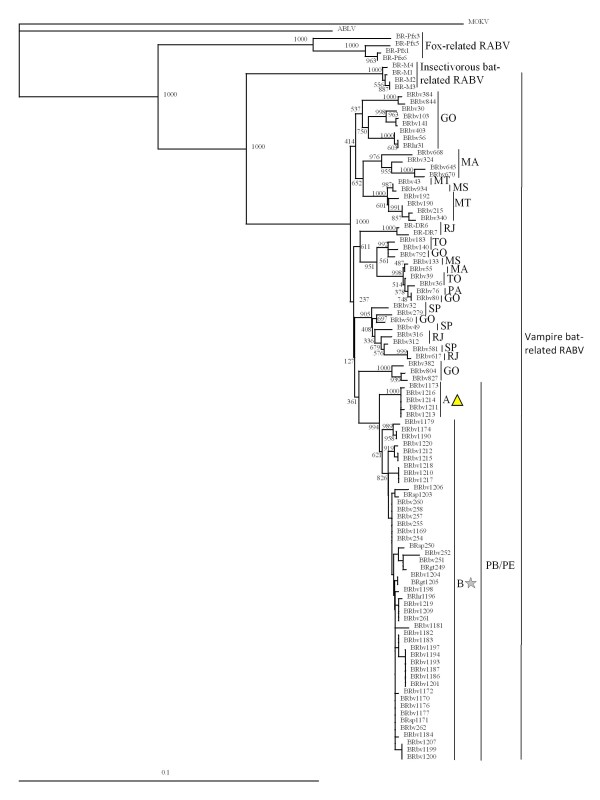
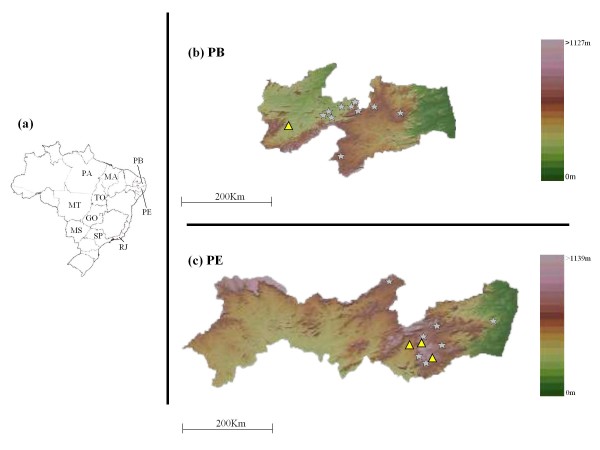
Phylogenetic analysis based on the sequences of 890 nt of the N gene revealed that the 52 RABV isolates included in this study clustered with a vampire bat-related RABV lineage; these isolates did not cluster with the dog-, fox-, or insectivorous bat-related RABVs (Figure 1). Comparison with RABVs isolated from other states in Brazil indicated that these 52 RABV isolates belonged to a single lineage; furthermore, this lineage was divided between a previously characterized main lineage (B) and a novel sub-lineage (A) consisting of several isolates located in PE. Geographical plotting showed that RABV isolates of the novel sub-lineage were derived from neighbouring areas (Figure 2). The topographical distributions of isolates from lineages A and B could not be distinguished in the areas covered by this study.
Figure 1.
A phylogenetic tree based on the nucleotide sequences of the N gene. A phylogenetic tree based on the nucleotide sequences of 890 nt (bases 89-978) of the N gene was constructed using the method devised by Saitou and Nei [12]; the bootstrap probabilities of each node were calculated using 1,000 replicates. The designations BRbv, BRsp, BRgt, BRhr, BR-Pfx, and BR-DR indicate samples from Brazilian cattle, sheep, goats, horses, foxes, and vampire bats, respectively. MOKV and ABLV denote the Mokola virus and Australian bat lyssavirus, respectively. State abbreviations are as follows: PB, Paraíba; PE, Pernambuco; GO, Goiás; SP, São Paulo; RJ, Rio de Janeiro; MT, Mato Grosso; TO, Tocantins; MA, Maranhão; PA, Pará; MS, Mato Grosso do Sul. The triangle and star symbols represent the new sub-lineage (group A) and the previously reported lineage (group B), respectively.
Figure 2.
Geographic distribution of rabies virus isolates in the Brazilian states of Paraíba and Pernambuco. (a) Map of Brazil indicating location of states pertinent to this study. State abbreviations are as follows: PB, Paraíba; PE Pernambuco; GO, Goiás; SP, São Paulo; RJ, Rio de Janeiro; MT, Mato Grosso; TO, Tocantins; MA, Maranhão; PA, Pará; MS, Mato Grosso do Sul. (b, c) Detailed geographic distribution of livestock isolates classified as genetic variants in the states of PB (b) and PE (c). The two symbols (triangle and star) correspond to the new sub-lineage and previously reported lineage, respectively, as used in Figure 1. Samples for which the geographic origin and the genetic variant are identical are illustrated using the same symbol. Brazilian maps were obtained from Brasil em Relevo - Embrapa Monitoramento por Satélite http://www.relevobr.cnpm.embrapa.br/.
Discussion
Segments of the N gene from the 52 RABV isolates, collected from PB and PE between 2003 and 2009, were sequenced and phylogenetically analysed. This N gene segment displayed greater than 97.7% nucleotide and amino acid sequence identity among these 52 RABVs. This correlation implies that the lineage has been maintained during transmission in PB and PE.
The phylogenetic analysis described here indicates that all of the livestock RABVs in PB and PE were derived from vampire bat rabies. Furthermore, the present study reveals that there are two RABV lineages that are separate from other regional Brazilian vampire bat-related RABV lineages. The first, sub-lineage A, consists of BRbv1173 (collected in PB in 2004) and another seven RABV isolates collected in PE in 2008-2009; the second, sub-lineage B, consists of isolates collected in PB in 2003-2009 and in PE in 2007-2009. Comparison among RABV isolates from multiple northern states of Brazil revealed that lineages from PB and PE were distinct from those obtained from the Maranhão (MA) state. Because of geographical barriers (mountains and rivers) between PB/PE and MA, it would be difficult for vampire bats (the prime vector for RABV) to move freely between PB/PE and MA. Geographical mapping (Figure 2) demonstrates that sub-lineage A was located mainly in high-altitude areas of PE, while sub-lineage B was widely distributed and present in both PE and PB. Thus, these two lineages seem to correlate with geographic factors and/or vampire bat populations, but do not appear to correlate with the year of isolation. Using a 203 nt segment (bases 109 to 311) of the N gene, Kobayashi et al. [8,9] reported the existence of at least 24 RABV genetic variants among vampire bat-transmitted cases of rabies in cattle in Brazil; the distribution of several of these RABV genetic variants was found to be delimited by geographic boundaries, including mountain ranges and rivers. Our analysis (data not shown) using the same 203 nt segment reveals that the 52 RABV isolates (from PB and PE) of the present study correlate with the same PB lineage described by Kobayashi et al. These results indicate that livestock rabies has been transmitted by vampire bats in PB and PE during this study period. Furthermore, this RABV lineage seems to have been circulating in this area for at least 7 years, with transmission affected by geographic factors and resulting in dispersion of vampire bats among regional populations.
Previous reports have shown human exposure to vampire bat-transmitted rabies in northeastern Brazil, including PE [1,7]. The present phylogenetic analysis suggests that rabies epidemics that occurred in cattle in PB and PE were transmitted by vampire bats. These RABV isolates comprised a lineage independent from that of other Brazilian isolates, with the distinction reflecting isolation from neighbouring regions by geographic factors. Thus, the vampire bat-derived rabies in this area represents an endemic disease, suggesting that the regional control of vampire bat rabies in this area may be a workable model for local elimination of human and livestock rabies.
Conclusions
The present study indicates that occurrences of livestock rabies in PB and PE were caused by vampire bat RABVs, and that this RABV lineage has been circulating in this area of northeastern Brazil for at least 7 years. This pattern of distribution may correlate to that of a vampire bat population isolated by geographic barriers.
Methods
The 52 RABV isolates used in this study were obtained from cattle (46), sheep (3), goats (2), and horse (1), and were collected in PB and PE between 2003 and 2009 (Table 1). Brain specimens from these livestock were diagnosed as RABV-positive by an immunofluorescent antibody test and a mouse inoculation test. These study procedures were implemented in accordance with the Institutional Guidelines for Animal Experiments at the Campina Grande University under the permission (number 129/2009) of the Committee for Experimental Animals of this College. Viral RNA was extracted from the brains of livestock using the QIAamp Viral RNA Mini Kit (Qiagen, Hilden, Germany). Nucleoprotein (N) gene sequences from the Brazilian RABV isolates were amplified using RT-PCR with primers JW12 (5'-ATGTAACACCYCTACAATG-3') (position: 55-73 of PV) and N8 (5'-AGTTTCTTCAGCCATCTC-3') (position: 1585-1568 of PV), followed by hemi-nested PCR with primer pairs as follows. Primer pair A consisted of JW12 and P2 (5'-CCCATATAACATCCAACAAAGTG-3') (position: 1029-1007 of PV), and generated a 975-nt amplicon. Primer pair B consisted of P1 (5'-CTACAATGGATGCCGACAAGA-3') (position: 66-86 of PV) and N8, and generated a 1,520 nt amplicon. The nucleotide sequences of RABVs from Brazilian foxes, livestock, and hematophagous and insectivorous bats were obtained from GenBank (Table 1). Cycle sequencing, nucleotide and amino acid sequence alignments, and phylogenetic analyses were performed as previously described [13,14]. The geographic origins of the RABV isolates sequenced from the Brazilian livestock were plotted at the municipal level of the respective federal states using MapInfo Professional GIS software (ver. 8.0, MapInfo Japan K.K., Tokyo, Japan). Brazilian maps were obtained from Brasil em Relevo - Embrapa Monitoramento por Satélite [15].
Table 1.
Brazilian rabies virus isolates used in this study
| Sample*1 | Species | Location | State*2 | Year | Lineage of PB and PE*3 | Accession No. | Reference |
|---|---|---|---|---|---|---|---|
| BR-DR6 | Desmodus rotundus | Laje de Muriae | RJ | 1998 | AB297633 | [16] | |
| BR-DR7 | Desmodus rotundus | Itaperuna | RJ | 1997 | AB297634 | [16] | |
| BR-M1(BR-Pbt1) | Molossus sp. | Patos | PB | AB206414 | [6] | ||
| BR-M2(BR-Pbt2) | Molossus sp. | Patos | PB | AB206415 | [6] | ||
| BR-M3(BR-Pbt3) | Molossus sp. | Patos | PB | AB206416 | [6] | ||
| BR-M4(BR-Pbt4) | Molossus sp. | Patos | PB | AB206417 | [6] | ||
| BR-Pfx1 | Fox | Patos | PB | 2002 | AB362483 | [13] | |
| BR-Pfx3 | Fox | Patos | PB | 2001 | AB206409 | [6] | |
| BR-Pfx5 | Fox | Patos | PB | 2002 | AB206411 | [6] | |
| BR-Pfx6 | Fox | Patos | PB | 2002 | AB207884 | [6] | |
| BRbv30 | Cattle | Morrinhos | GO | 1999 | AB083803 | [17] | |
| BRbv32 | Cattle | Sao Roque | SP | 1994 | AB083805 | [17] | |
| BRbv36 | Cattle | Nova Olinda | TO | 1998 | AB083809 | [17] | |
| BRbv39 | Cattle | Colinas | TO | 1999 | AB083811 | [17] | |
| BRbv43 | Cattle | Alto Taquari | MT | 1999 | AB083813 | [17] | |
| BRbv49 | Cattle | Piraju | SP | 1989 | AB083817 | [17] | |
| BRbv50 | Cattle | Corumbaiba | GO | 1999 | AB083818 | [17] | |
| BRbv55 | Cattle | Montes Altos | MA | 1998 | AB675602*4 | [8] | |
| BRbv56 | Cattle | Iporá | GO | 1998 | AB675603*4 | [8] | |
| BRbv76 | Cattle | Xinguará | PA | 2002 | AB675604*4 | [8] | |
| BRbv80 | Cattle | Ipameri | GO | 2001 | AB675605*4 | [8] | |
| BRbv103 | Cattle | Nova Crixás | GO | 2001 | AB675606*4 | [8] | |
| BRbv133 | Cattle | Xambioa | TO | 2000 | AB675607*4 | [8] | |
| BRbv140 | Cattle | Natividade | TO | 2000 | AB675608*4 | [8] | |
| BRbv141 | Cattle | Nova Crixas | GO | 2000 | AB675609*4 | [8] | |
| BRbv183 | Cattle | Natividade | TO | 2001 | AB675610*4 | [8] | |
| BRbv190 | Cattle | Pocone | MT | 2002 | AB675611*4 | [9] | |
| BRbv192 | Cattle | Nobres | MT | 2002 | AB675612*4 | [8] | |
| BRbv215 | Cattle | Rosario Oeste | MT | 2002 | AB675613*4 | [9] | |
| BRbv251(BR-Pbv1) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206423 | [6] |
| BRbv252(BR-Pbv2) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206424 | [6] |
| BRbv254(BR-Pbv3) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206425 | [6] |
| BRbv255(BR-Pbv5) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206426 | [6] |
| BRbv257(BR-Pbv7) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206427 | [6] |
| BRbv258(BR-Pbv8) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206428 | [6] |
| BRbv260(BR-Pbv10) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206429 | [6] |
| BRbv261(BR-Pbv11) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206430 | [6] |
| BRbv262(BR-Pbv12) | Cattle | Patos | PB | 2003 | PB/PE-B | AB206431 | [6] |
| BRbv279 | Cattle | Pirapozinho | SP | 2002 | AB675614*4 | [9] | |
| BRbv312 | Cattle | Paulo de Frontin | RJ | 1987 | AB675615 | This study | |
| BRbv316 | Cattle | Miguel Pereina | RJ | 2000 | AB675616*4 | [9] | |
| BRbv324 | Cattle | Itapecuru Mirim | MA | 2004 | AB675617*4 | [9] | |
| BRbv340 | Cattle | Nossa Senhora do Livramento | MT | 2004 | AB675618*4 | [9] | |
| BRbv382 | Cattle | Orizona | GO | 2002 | AB675619*4 | [9] | |
| BRbv384 | Cattle | Nova América | GO | 2002 | AB675620*4 | [9] | |
| BRbv403 | Cattle | Piranhas | GO | 2002 | AB675621*4 | [9] | |
| BRbv581 | Cattle | Tambaú | SP | 2003 | AB675622*4 | [9] | |
| BRbv617 | Cattle | Rio Claro | RJ | 2004 | AB675623*4 | [9] | |
| BRbv645 | Cattle | Capinzal do Norte | MA | 2004 | AB675624*4 | [9] | |
| BRbv668 | Cattle | Santo Antônio dos Lopes | MA | 2005 | AB675625*4 | [9] | |
| BRbv670 | Cattle | Godofredo Viana | MA | 2005 | AB675626*4 | [9] | |
| BRbv792 | Cattle | Morrinhos | GO | 2002 | AB675627*4 | [9] | |
| BRbv804 | Cattle | Pilar de Goiás | GO | 2005 | AB675628*4 | [9] | |
| BRbv827 | Cattle | Cocalzinho de Goiás | GO | 2005 | AB675629*4 | [9] | |
| BRbv844 | Cattle | Itapaci | GO | 2006 | AB675630*4 | [9] | |
| BRbv934 | Cattle | Bandeirantes | MS | 2005 | AB675631*4 | [9] | |
| BRbv1169 | Cattle | Patos | PB | 2004 | PB/PE-B | AB623080 | This study |
| BRbv1170 | Cattle | Santa Terezinha | PB | 2004 | PB/PE-B | AB623081 | This study |
| BRbv1172 | Cattle | São José do Bonfim | PB | 2004 | PB/PE-B | AB623082 | This study |
| BRbv1173 | Cattle | Itaporanga | PB | 2004 | PB/PE-A | AB623083 | This study |
| BRbv1174 | Cattle | São Vicente do Seridó | PB | 2004 | PB/PE-B | AB623084 | This study |
| BRbv1176 | Cattle | Patos | PB | 2005 | PB/PE-B | AB623085 | This study |
| BRbv1177 | Cattle | Santa Luzia | PB | 2005 | PB/PE-B | AB623086 | This study |
| BRbv1179 | Cattle | Areal | PB | 2006 | PB/PE-B | AB623087 | This study |
| BRbv1181 | Cattle | Monteiro | PB | 2006 | PB/PE-B | AB623089 | This study |
| BRbv1182 | Cattle | Junco do Seridó | PB | 2006 | PB/PE-B | AB623090 | This study |
| BRbv1183 | Cattle | São José do Sabugi | PB | 2006 | PB/PE-B | AB623091 | This study |
| BRbv1184 | Cattle | Santa Luzia | PB | 2006 | PB/PE-B | AB623092 | This study |
| BRbv1186 | Cattle | Patos | PB | 2007 | PB/PE-B | AB623093 | This study |
| BRbv1187 | Cattle | Patos | PB | 2007 | PB/PE-B | AB623094 | This study |
| BRbv1190 | Cattle | Areal | PB | 2007 | PB/PE-B | AB623096 | This study |
| BRbv1193 | Cattle | Patos | PB | 2007 | PB/PE-B | AB623097 | This study |
| BRbv1194 | Cattle | São José do Bonfim | PB | 2007 | PB/PE-B | AB623098 | This study |
| BRbv1197 | Cattle | São José do Bonfim | PB | 2007 | PB/PE-B | AB623099 | This study |
| BRbv1198 | Cattle | Brejinho | PE | 2007 | PB/PE-B | AB623106 | This study |
| BRbv1199 | Cattle | Patos | PB | 2008 | PB/PE-B | AB623100 | This study |
| BRbv1200 | Cattle | Patos | PB | 2008 | PB/PE-B | AB623101 | This study |
| BRbv1201 | Cattle | Patos | PB | 2008 | PB/PE-B | AB623102 | This study |
| BRbv1204 | Cattle | Patos | PB | 2008 | PB/PE-B | AB623103 | This study |
| BRbv1206 | Cattle | Patos | PB | 2008 | PB/PE-B | AB623104 | This study |
| BRbv1207 | Cattle | Patos | PB | 2009 | PB/PE-B | AB623105 | This study |
| BRbv1209 | Cattle | Vitória de Santo Antão | PE | 2008 | PB/PE-B | AB623107 | This study |
| BRbv1210 | Cattle | Venturosa | PE | 2008 | PB/PE-A | AB623108 | This study |
| BRbv1211 | Cattle | Pedra | PE | 2008 | PB/PE-A | AB623109 | This study |
| BRbv1212 | Cattle | Venturosa | PE | 2008 | PB/PE-A | AB623110 | This study |
| BRbv1213 | Cattle | Garanhuns | PE | 2008 | PB/PE-A | AB623111 | This study |
| BRbv1214 | Cattle | Venturosa | PE | 2008 | PB/PE-A | AB623112 | This study |
| BRbv1215 | Cattle | Paranatama | PE | 2008 | PB/PE-B | AB623113 | This study |
| BRbv1216 | Cattle | Venturosa | PE | 2008 | PB/PE-A | AB623114 | This study |
| BRbv1217 | Cattle | Belo Jardim | PE | 2008 | PB/PE-B | AB623115 | This study |
| BRbv1218 | Cattle | Belo Jardim | PE | 2009 | PB/PE-B | AB623116 | This study |
| BRbv1219 | Cattle | Lajedo | PE | 2009 | PB/PE-B | AB623117 | This study |
| BRbv1220 | Cattle | Garanhuns | PE | 2009 | PB/PE-A | AB623118 | This study |
| BRgt249(BR-Pgt1) | Goat | Patos | PB | 2003 | PB/PE-B | AB206437 | [6] |
| BRgt1205 | Goat | São Mamede | PB | 2008 | PB/PE-B | AB623077 | This study |
| BRhr31 | Horse | Ipora | GO | 1998 | AB083804 | [17] | |
| BRhr1196 | Horse | Patos | PB | 2007 | PB/PE-B | AB623076 | This study |
| BRsp250(BR-Psp1) | Sheep | Patos | PB | 2003 | PB/PE-B | AB206438 | [6] |
| BRsp1171 | Sheep | Santa Terezinha | PB | 2004 | PB/PE-B | AB623078 | This study |
| BRsp1203 | Sheep | Patos | PB | 2008 | PB/PE-B | AB623079 | This study |
*1 Names in parentheses are designations from Shoji et al. (2006)
*2 State abbreviations are as follows: PB Paraíba, PE Pernambuco, GO Goiás, SP São Paulo, RJ Rio de Janeiro, MT Mato Grosso, TO Tocantins, MA Maranhão, PA Pará, MS Mato Grosso do Sul
*3 Lineages are based on the phylogenetic tree of Figure 1
*4 We determined extra sequences in this study
Competing interests
The authors declare that they have no competing interests.
Authors' contributions
NM performed the molecular genetic studies and edited the manuscript; HK performed the RT-PCR and sequencing; TI, JABA, MLCRS, FHI, and TS participated in the study design, management, and coordination, and assisted in drafting the manuscript. All authors read and approved the final manuscript.
Contributor Information
Nobuyuki Mochizuki, Email: mochizuki.nobuyuki@gmail.com.
Hiroyuki Kawasaki, Email: hkat@i-revo.ne.jp.
Maria LCR Silva, Email: luacristiny@yahoo.com.br.
José AB Afonso, Email: afonsojab@oi.com.br.
Takuya Itou, Email: itou.takuya@nihon-u.ac.jp.
Fumio H Ito, Email: fumio@usp.br.
Takeo Sakai, Email: sakai.takeo@nihon-u.ac.jp.
Acknowledgements
This work was supported in part by a Grant-in-Aid for the Academic Frontier Project for Private Universities from the Ministry of Education, Culture, Sports, Science, and Technology of Japan.
References
- Schneider MC, Romijn PC, Uieda W, Tamayo H, da Silva DF, Belotto A, da Silva JB, Leanes LF. Rabies transmitted by vampire bats to humans: an emerging zoonotic disease in Latin America? Rev Panam Salud Publica. 2009;25(3):260–269. doi: 10.1590/s1020-49892009000300010. [DOI] [PubMed] [Google Scholar]
- Schneider MC, Aguilera XP, Barbosa da Silva J Junior, Ault SK, Najera P, Martinez J, Requejo R, Nicholls RS, Yadon Z, Silva JC. et al. Elimination of neglected diseases in latin america and the Caribbean: a mapping of selected diseases. PLoS Negl Trop Dis. 2011;5(2):e964. doi: 10.1371/journal.pntd.0000964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnieli P Jr, Brandão PE, Carrieri ML, Castilho JG, Macedo CI, Machado LM, Rangel N, de Carvalho RC, de Carvalho VA, Montebello L. et al. Molecular epidemiology of rabies virus strains isolated from wild canids in Northeastern Brazil. Virus Res. 2006;120(1-2):113–120. doi: 10.1016/j.virusres.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Carnieli P Jr, Castilho JG, Fahl Wde O, Veras NM, Carrieri ML, Kotait I. Molecular characterization of rabies virus isolates from dogs and crab-eating foxes in Northeastern Brazil. Virus Res. 2009;141(1):81–89. doi: 10.1016/j.virusres.2008.12.015. [DOI] [PubMed] [Google Scholar]
- Carnieli P Jr, Fahl Wde O, Castilho JG, Oliveira Rde N, Macedo CI, Durymanova E, Jorge RS, Morato RG, Spindola RO, Machado LM. et al. Characterization of rabies virus isolated from canids and identification of the main wild canid host in Northeastern Brazil. Virus Res. 2008;131(1):33–46. doi: 10.1016/j.virusres.2007.08.007. [DOI] [PubMed] [Google Scholar]
- Shoji Y, Kobayashi Y, Sato G, Gomes AA, Itou T, Ito FH, Sakai T. Genetic and phylogenetic characterization of rabies virus isolates from wildlife and livestock in Paraiba, Brazil. Acta Virol. 2006;50(1):33–37. [PubMed] [Google Scholar]
- Dantas-Torres F, Oliveira-Filho EF. Human exposure to potential rabies virus transmitters in Olinda, State of Pernambuco, between 2002 and 2006. Rev Soc Bras Med Trop. 2007;40(6):617–621. doi: 10.1590/s0037-86822007000600003. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Ogawa A, Sato G, Sato T, Itou T, Samara SI, Carvalho AA, Nociti DP, Ito FH, Sakai T. Geographical distribution of vampire bat-related cattle rabies in Brazil. J Vet Med Sci. 2006;68:1097–1100. doi: 10.1292/jvms.68.1097. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Sato G, Mochizuki N, Hirano S, Itou T, Carvalho AA, Albas A, Santos HP, Ito FH, Sakai T. Molecular and geographic analyses of vampire bat-transmitted cattle rabies in central Brazil. BMC Vet Res. 2008;4:44. doi: 10.1186/1746-6148-4-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnieli P Jr, Castilho JG, Fahl Wde O, Veras NM, Timenetsky Mdo C. Genetic characterization of Rabies virus isolated from cattle between 1997 and 2002 in an epizootic area in the state of Sao Paulo, Brazil. Virus Res. 2009;144(1-2):215–224. doi: 10.1016/j.virusres.2009.05.001. [DOI] [PubMed] [Google Scholar]
- Macedo CI, Carnieli P Junior, Fahl Wde O, Lima JY, Oliveira Rde N, Achkar SM, Castilho JG, Carrieri ML, Kotait I. Genetic characterization of rabies virus isolated from bovines and equines between 2007 and 2008, in the States of Sao Paulo and Minas Gerais. Rev Soc Bras Med Trop. 2010;43(2):116–120. doi: 10.1590/S0037-86822010000200002. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4(4):406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Kobayashi Y, Sato G, Hirano S, Itou T, Ito FH, Sakai T. Determination and Molecular Analysis of the Complete Genome Sequence of Two Wild-Type Rabies Viruses Isolated from a Haematophagous Bat and a Frugivorous Bat in Brazil. J Vet Med Sci. 2011;73(6):759–766. doi: 10.1292/jvms.10-0238. [DOI] [PubMed] [Google Scholar]
- Mochizuki N, Kobayashi Y, Sato G, Itou T, Gomes AA, Ito FH, Sakai T. Complete genome analysis of a rabies virus isolate from Brazilian wild fox. Arch Virol. 2009;154(9):1475–1488. doi: 10.1007/s00705-009-0475-9. [DOI] [PubMed] [Google Scholar]
- Brasil em Relevo Monitoramento por Satélite. http://www.relevobr.cnpm.embrapa.br/
- Kobayashi Y, Sato G, Kato M, Itou T, Cunha EM, Silva MV, Mota CS, Ito FH, Sakai T. Genetic diversity of bat rabies viruses in Brazil. Arch Virol. 2007;152(11):1995–2004. doi: 10.1007/s00705-007-1033-y. [DOI] [PubMed] [Google Scholar]
- Ito M, Itou T, Shoji Y, Sakai T, Ito FH, Arai YT, Takasaki T, Kurane I. Discrimination between dog-related and vampire bat-related rabies viruses in Brazil by strain-specific reverse transcriptase-polymerase chain reaction and restriction fragment length polymorphism analysis. J Clin Virol. 2003;26(3):317–330. doi: 10.1016/S1386-6532(02)00048-3. [DOI] [PubMed] [Google Scholar]