Abstract
Oral Treponema species, most notably T. denticola, are implicated in the destructive effects of human periodontal disease. Progress in the molecular analysis of interactions between T. denticola and host proteins is reviewed here, with particular emphasis on the characterization of surface-expressed and secreted proteins of T. denticola involved in interactions with host cells, extracellular matrix components, and components of the innate immune system.
Keywords: spirochetes, virulence factors, periodontal disease
Approximately 30% of the US adult population has at least one periodontal site with demonstrable gingival recession and bone loss (1). In contrast to healthy gingival plaque in which the complex microbial community dominated by facultative Gram-positives, periodontal lesions harbors a microflora dominated by proteolytic Gram-negative anaerobes and spirochetes (2, 3). While a consortium of three organisms denoted as the ‘red complex’ (Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola) has the highest association with periodontal disease severity (4), spirochetes in particular are associated with severe and refractory periodontal conditions (2, 5–8). In periodontal lesions, spirochetes preferentially localize in the deepest part of the ‘pocket,’ specifically at the interface between the subgingival plaque and the epithelium (9). In advanced periodontitis, their ability to disrupt intercellular junctions likely contributes to invasion of underlying tissue (10–14). T. denticola, the most readily cultivable oral spirochete, is the model organism for studying both the unique biological features of these organisms and Treponema–host interactions in periodontal disease. Virulence-associated behaviors (15, 16) of T. denticola include adherence to host tissue and other microbes, motility, chemotaxis, tissue penetration, and release of immunomodulatory and cytopathic factors (reviewed in (17)). The numerical prevalence of spirochetes in disease, along with their high association with and location within diseased sites, strongly suggests that they play an important role in disease initiation and progression (4, 6, 9, 18).
Oral spirochetes, including T. denticola and over 50 other Treponema species, present unique challenges and opportunities to the field of molecular and cellular microbiology. Foremost among these challenges is the fact that at least 80% of oral Treponema spp. have never been cultured, and only T. denticola has been studied in detail. These nutritionally fastidious anaerobes possess features unique among bacteria, including cellular structure (19), motility apparatus (20), biosynthetic pathways (21), and outer membrane protein complexes (22, 23). As commensal residents and opportunistic pathogens of oral mucosal tissue, they offer a wide range of potential avenues for research into microbe–host interactions and signaling, microbial communities, microbial physiology, and molecular evolution. Thus, molecular level studies of oral spirochetes are timely and of high importance in understanding chronic bacterial infections such as periodontal disease.
Treponema denticola exists in a complex, multispecies biofilm environment in the gingival crevice. Numerous interbacterial interactions required for development and maintenance of the subgingival microbial community have been documented or proposed (24). These dynamic interactions comprise only part of the total of the environmental milieu in which these organisms have evolved. The oral microbiota live in a host mucosal environment consisting of several host cell types and extracellular matrix (ECM) components as potential substrates in addition to a fluid environment consisting of a complex and variable mix of saliva, gingival crevicular fluid, and serum components, including numerous antimicrobial components of both the innate and adaptive immune systems. As obligately host-associated organisms, oral spirochetes are extremely well adapted to survival in a eukaryotic host environment. This is reflected, as in many other host-associated microbes, in the relatively large number of T. denticola genes that can be clearly identified as having been acquired by horizontal gene transfer from an ancestral eukaryotic host (25–27). To understand the factors that allow commensal organisms to induce pathogenic responses under certain host environmental conditions, it is necessary to understand how they survive without causing disease. The focus of this review is on the interactions between T. denticola and host components that mediate both its persistence in the oral environment and its pathogenicity in periodontal disease. Primary attention will be given to interactions that are at least partially characterized and understood at the molecular level, and understudied areas will be pointed out where appropriate.
Research on oral spirochetes has progressed in recent years, driven partly by completion of the T. denticola genome sequence (25). Recent online release of the T. vincentii provisional annotated genome (http://www.ncbi.nlm.nih.gov/genomeprj/55865) and the unassembled genome sequence contigs of T. lecithinolyticum (28) have expanded the genomic resources for this group of oral microbes. Additionally, the Human Oral Microbiome Project is in the process of sequencing several other T. denticola strains (29). However, progress in molecular analysis of specific T. denticola behaviors has been considerably slowed by the limitations of currently available genetic systems for this organism, including extremely low transformation efficiency, few selectable markers (30), lack of reliable plasmid or other vectors for the most studied strain (31, 32), and lack of promoter-reporter systems. These significant technical issues, combined with the small number of researchers and the relatively low level of funding in this field, are continuing impediment to progress. This is reflected in the number of journal articles published on oral spirochetes relative to other periodontal pathogens. In 2010, approximately five times as many papers were published on P. gingivalis than were published on all oral spirochetes, including T. denticola, a ratio that has changed only slightly over the past 15 years.
Interactions with host cells
Exposure of cultured cells of various types to T. denticola (Fig. 1) results in monolayer detachment and proliferation inhibition (33–35), plasma membrane fibronectin (FN) degradation (36), membrane blebbing, decreased intercellular contact and cytoskeletal rearrangements (12, 35, 37), and loss of volume control (37). Most studies prior to the advent of molecular cloning and genome sequencing did not identify the specific T. denticola components responsible for the observed cellular responses. One example of a study that made some limited progress in this regard is contained in a series of reports by Shenker and coworkers on the antiproliferative effects of T. denticola on fibroblasts and monocytes (33, 38, 39). In these studies, certain protein fractions of T. denticola were identified as containing the active agents, but the identities of the proteins have not yet been determined.
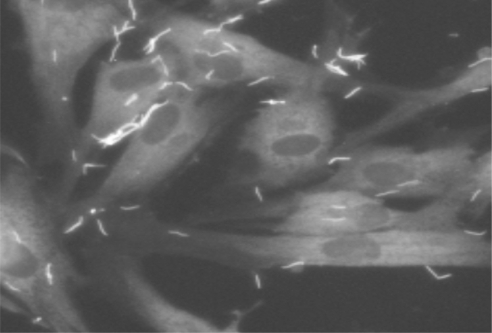
Fig. 1.
Immunofluorescence micrograph showing T. denticola adherence to periodontal ligament cells. Periodontal ligament cell monolayers were challenged with T. denticola 35405 for 2 hours, then washed extensively with PBS before staining. Periodontal ligament cells are stained with phalloidin. T. denticola is visualized with Alexa-fluor-labeled antiMsp antibodies.
Studies over the past 15 years utilizing purified proteins and isogenic T. denticola mutants have begun the process of identifying specific molecular pathways responsible for these rather complex cellular responses and, in some cases, have identified host proteins interacting with T. denticola effector proteins (Table 1 ). Two of the most prominent and widely studied outer membrane components of T. denticola are the major surface protein (Msp) and the PrtP protease complex (dentilisin, chymotrypsin-like protease (CTLP)) both of which have cytopathic effects in epithelial cells (40). Interestingly, although challenge with either Msp or dentilisin results in lysis of epithelial cells, Msp is hemolytic, whereas dentilisin is not (40), suggesting distinctly different mechanisms of interaction with different host receptor molecules.
Table 1.
Molecular analysis of T. denticola interactions with host components, using purified proteins and isogenic mutants
| Behavior | Protein(s) identified | Native activitya | Gene(s) | Recombinant activitya | Mutants | Mutant phenotypeb |
|---|---|---|---|---|---|---|
| Motility | n/d | n/d | fliFGH flgB, C, E, F | n/d | flgE tap1 | Non-motile (30, 140) |
| Chemotaxis | n/d | n/d | cheAWXY dmcA, dmcB | n/d | dmcA, dmcB, cheA | Non-motile, non-chemotactic (102, 141, 142) |
| Adherence to | ||||||
| Eukaryotic cells | dentilisin | + (40, 116) | n/d | |||
| ECM components | ||||||
| Fibronectin | Msp | + (22) | msp | + (67) | msp (143) | n/d |
| OppA | + (68) | oppA-F | n/d | oppA | ↓ binding (68) | |
| Fibrinogen | dentilisin | + (100) | n/d | prcA-prtP | n/d | |
| Msp | + (22) | msp | n/d | Msp | no change (143) | |
| Plasminogen | OppA | + (68) | oppA-F | n/d | oppA | ↓ binding (68) |
| Msp | n/d | msp | n/d | msp | ↓ binding (68) | |
| Laminin | Msp | + (22) | msp | + (67) | Msp | n/d |
| Hyaluronan | dentilisin (79) | n/d | msp | + (66) | n/d | |
| Keratin, collagen, heparin | msp | + (66) | n/d | |||
| Cytotoxicity | Msp | + (40) | msp | + (40) | n/d | |
| dentilisin | + (40) | prcB-prcA-prtP | n/d | |||
| Complement resistance (FH mediated) | FhbB | binds FH (115) | fhbB | + (123) | fhbB | ↓ FH binding; ↑serum sensitivity (124) |
| dentilisin | cleaves C3 (122) cleaves FH (115) | prtP | n/d | prtP | ↓ C3 cleavage (122); No FH cleavage (115) | |
| Abscess formation | dentilisin | n/d | prtP | n/d | prtP | ↓abscess formation (144) |
| dentipain | n/d | ideT | cleaves β-chain insulin (133) | ideT | ↓abscess formation (133) | |
| Iron acquisition | Hemin binding proteins | n/d | hbpA, hbpB | n/d | hbpA, hbpB | ↓ growth (145) |
Activity demonstrated [+, (citation)] or not reported [n/d].
Reported mutant phenotype (citation) or not reported [n/d].
Msp cytotoxicity is believed to be due to its demonstrated pore-forming activity in both planar lipid bilayers and in epithelial cell membranes (41, 42). This is consistent with the demonstrated lytic pore-forming activity of porins of other Gram-negative pathogens, including Neisseria spp. (43, 44). Interestingly, fibroblasts appear to be comparatively tolerant of Msp challenge, making them more tractable candidates for analysis of cellular responses. Studies by Ellen and coworkers have demonstrated that Msp disrupts the actin cytoskeleton of gingival fibroblasts, with specific effects, including disruption of actin stress fibers (45) and induction of de novo synthesis of subcortical actin filaments (35, 37, 46, 47). A specific peptide sequence within Msp has been shown to promote stress fiber formation (48, 49). Concurrently, and presumably related to this phenomenon, Msp blocks the ability of fibroblasts to regulate intracellular calcium gradients (50), which may contribute to the rather diverse range of morphological responses observed. The molecular mechanism of Msp-dependent calcium dysregulation, including identification of cellular Msp receptor(s), remains to be determined.
Dentilisin, a complex of three lipoproteins encoded by the prcB-prcA-prtP locus, has a wide range of cytopathic effects on whole cells and tissue. These effects are at least primarily due to its potent proteolytic activity of dentilisin, although as in other spirochetes, lipid moieties of the protease complex components may contribute to activation of innate immune responses (51). In particular, the ability of dentilisin activity to induce cell shrinkage and increased permeability of intercellular junctions have been shown to be a key factor in penetration and translocation of T. denticola through cell monolayers and ECM models (12, 13, 52). Further discussion of dentilisin's role in interaction with host proteins is found in subsequent sections dealing with interactions with specific host components.
T. denticola induces the production of various cytokines, including interleukin-1ß (IL-1ß), IL-6, IL-8, and TNF-α from various cell types (53–56). The specific T. denticola molecule(s) responsible have not yet been clearly identified. On the other hand, two of these studies (53, 55) also suggested that T. denticola hydrolyzes IL-8. Using wild-type and prtP mutant T. denticola strains, Miyamoto et al. showed that dentilisin activity was responsible for reduction of protein levels of inflammatory cytokines, including IL-1ß, IL-6, and TNF-α, detected in peripheral blood mononuclear cells following T. denticola challenge. This study highlights the difficulties encountered by investigators examining cellular responses to T. denticola. For this reason, it is important to include analyses of both protein expression and gene transcription when analyzing cellular responses that may be both induced by and subsequently modulated by proteolytic activity.
Interactions with ECM components
The ability to bind to or degrade ECM components is a hallmark of numerous mucosal pathogens. In addition to the direct tissue-destructive effects of ECM degradation, there is increasing evidence that degradation products of ECM components act as proinflammatory signals.
Fibronectin
Fibronectin is a dimeric, multidomain glycoprotein bound to the surface of eukaryotic cells and is a key component of the ECM. The molecular differences between the two major forms of FN (cellular FN and plasma FN) are due to differential mRNA splicing, and the two forms have distinct yet overlapping roles in formation and maintenance of ECM and in pathways of tissue homeostasis (reviewed in (57)). Numerous microbes, both commensal and pathogenic, exhibit FN-binding activity as a prominent behavior important for survival in the host environment. Molecular analysis of FN-binding proteins of several pathogens continues to be an important area of pathogenesis research over 30 years after the seminal discovery of FN binding to Staphylococcus aureus (58). Henderson et al. recently published an extensive review of FN function and the mechanisms and consequences of FN binding mediated by bacterial FN-binding proteins (59). Although the importance of T. denticola binding to FN has long been recognized, it has been studied rather less than the interaction between T. pallidum and FN. Due to the combination of the genetic similarities between T. pallidum and T. denticola and the complexities of working with T. pallidum, several investigators have studied T. denticola both in terms of periodontal pathogenicity and as a general model for Treponema–host interactions (60). Several key studies by the Ellen group beginning in the early 1990s established the role of FN in T. denticola interaction with host tissue. Dawson et al. characterized T. denticola binding to immobilized FN and proposed that one or more T. denticola FN-binding proteins were preferentially localized at the tips of spirochete cells (61, 62). In some strains, antiFN antibodies block T. denticola adherence to FN, further supporting the specificity of the binding interaction (63). Concurrent studies demonstrated the ability of T. denticola to induce degradation of cellular FN on gingival fibroblasts (36) and to induce cytoskeletal rearrangements that may be the result of FN-mediated signaling (37, 64, 65).
Although over 100 bacterial FN-binding proteins have been reported, the level of knowledge of these proteins and their interactions with FN vary widely. Detailed information on the structure of complexes formed between the bacterial proteins and FN is available in very few cases. In many instances, the only information available is that a protein binds FN in an in vitro assay, with or without information on kinetics, specificity, or FN target domain (extensively reviewed in (59)). It is rather common for bacteria to possess multiple distinct FN-binding proteins. Both experimental and genetic evidence suggests that T. denticola possesses at least two and many as nine or more proteins with FN-binding activity. Both Msp (22, 66, 67) and OppA (68) are reported to bind immobilized FN. Edwards et al. reported that Msp binds the N-terminal heparin I/fibrin I domain of FN (66) that is the most commonly reported target of bacterial FN-binding proteins. Although the specific domain targeted by OppA is not known, it is of interest that in ligand blotting experiments, both wild-type and Msp mutant T. denticola cells bind the same N-terminal FN domain (J.C. Fenno, unpublished observations).
Because T. denticola and T. pallidum survive in analogous mucosal environments and share many closely homologous proteins, it is reasonable to hypothesize that similar proteins might be involved in both microbes’ demonstrated FN-binding activity. Two of 10 potential adhesins predicted by genomic analysis of T. pallidum showed FN-binding activity when expressed in Escherichia coli (69). One of these (TP0155) shows 45% identity with T. denticola TDE2138, whereas the other (Tp0483) is not homologous with any T. denticola protein. Like TP0155, TDE2318, six other predicted T. denticola proteins contain an M23 peptidase domain. Bamford et al. recently reported recombinant expression and FN-binding activity of five of these seven T. denticola predicted FN-binding proteins. Although immunofluorescence microscopy suggested surface localization of at least one of the T. denticola proteins, three of the deduced protein sequences lack predicted signal peptide sequences and may not be secreted (70). It is not yet known whether or under what conditions all of the genes encoding these predicted FN-binding proteins are expressed. These intriguing results support continued research into the mechanisms of T. denticola-FN interactions.
One issue in research into bacterial–FN interactions is the distinction between plasma and cellular FN. Many studies of FN binding use plasma-derived FN (largely due to its availability and price) when modeling behaviors in which cellular FN may be more relevant. In the simplest analysis, FN plays important roles in blood clotting and wound healing, whereas cellular FN is incorporated into the ECM as a part of a fibrillar matrix (59) and is a target for bacterial adherence (59). However, interaction of T. denticola with fibroblasts and related cell types results in fragmentation and degradation of cellular FN (36, 71), and the resulting fragments may have modulatory effects on tissue homeostasis and inflammation (72–74). In vitro studies showed that purified dentilisin has the ability to degrade plasma FN (75) and T. denticola challenge resulted in FN degradation in fibroblasts (36), suggesting that dentilisin directly degrades cellular FN. However, Miao et al. reported that T. denticola dentilisin did not directly degrade cellular FN and that dentilisin induced MMP-2-dependent FN fragmentation in periodontal ligament cells. The MMP-2-dependent FN fragmentation pattern induced by dentilisin activity resembles the FN fragmentation pattern seen in gingival crevicular fluid from periodontal disease lesions. This suggests that the distinction between plasma and cellular FN may be very important in terms of how T. denticola and other oral microbes interact with this protein that is essentially ubiquitous in the subgingival environment.
Laminin and collagen
T. denticola is reported to bind laminin and collagen (62, 76, 77), and this has been proposed as a mechanism of adherence to cells and ECM in the oral cavity. T. denticola Msp (22, 66, 67) and OppA (68) bind laminin. Recombinant Msp binds type I collagen (66), and Msp blocks phagocytosis of collagen by fibroblasts (47). It is not clear whether this last phenomenon is due to Msp competing for collagen or to Msp disruption of fibroblast membrane function. Dentilisin activity has been reported to either directly degrade (75) or indirectly induce degradation (78) of these substrates. The few in vitro studies in the literature do not clearly establish the biological role of T. denticola binding and degradation of these substrates. Further studies are required to determine the significance of interactions of T. denticola with these ECM components in the subgingival environment.
Glycosoaminoglycans
Hyaluronan (HA) is a high molecular weight glycosoaminoglycan prevalent in ECM and connective tissue. The potential role of HA as a substrate for T. denticola adherence is supported by two studies employing distinctly different approaches. Edwards et al. demonstrated the ability of rMsp to bind to several immobilized substrates including HA (66). Haapasalo et al. reported T. denticola binding to immobilized HA and showed that HA blocked binding of T. denticola to HA-coated beads (79). Interestingly, inhibition of dentilisin activity also blocked HA binding by T. denticola cells, suggesting a role for dentilisin in HA binding. To date, these studies have not been followed up by analysis of isogenic mutants and purified native proteins.
Hyaluronidase-mediated degradation of HA increases the permeability of connective tissues and decreases the viscosity of body fluids (80). HA undergoes a high turnover rate, and HA degradation products act as an immune regulator, signaling through TLR2, TLR4, or both TLR2 and TLR4 in macrophages and dendritic cells (81). Hyaluronidases of pathogenic Staphylococcus, Streptococcus, and Clostridium spp. (among others) facilitate bacterial penetration through host tissues, and cell-bound HA serves as an antigenic disguise that prevents the recognition of bacteria by phagocytes (82). The ability to bind HA is an obvious prerequisite for HA degradation, and bacterial hyaluronidases generally also have HA-binding activity, but T. denticola interaction with HA has not yet been studied in detail. At least three species of cultivable oral spirochetes, including T. denticola produce a surface-expressed enzyme that hydrolyzes both HA and chondroitin sulfate (83). The 59-kDa enzyme purified from T. denticola (designated ‘HA/CSase’) hydrolyzed both of these ECM glycosoaminoglycans. Scott et al. suggested that this enzyme could have an active role in connective tissue degradation in periodontal disease (83). Analysis of the T. denticola genome suggests that this enzyme is encoded by TDE0471, predicted to encode a lipoprotein with sialidase activity. The relationship between this activity and the reported T. denticola HA binding activity is not known. The single study reporting HA binding also noted that there was no significant attachment of T. denticola to chondroitin-4-sulfate or dermatan sulfate (79).
Interactions with serum components
Nutrient acquisition
Routine growth of T. denticola requires serum supplementation of the complex medium formulation (84). Although a serum-free chemically defined medium has been reported, to date it has only been used in a single study of T. denticola fatty acid synthesis, primarily due to the technical challenge of its formulation (85). As an indication of the complexity of this medium, a commercial supplier has quoted a closely related formulation (OMIZ-P4 (86); ATCC Medium 2,131 (http://www.atcc.org)) at a price of well over US$1,000 per liter (J.C. Fenno, personal communication). Growth of T. denticola in a serum-containing medium is dependent on serum or albumin concentration (87). SDS-PAGE analysis of serum-containing culture medium during growth of T. denticola shows dentilisin-dependent degradation of albumin (J.C. Fenno, unpublished results).
Although T. denticola exhibits some chemotactic behavior toward glucose (88), it only weakly metabolizes glucose while preferentially deriving energy from amino acid fermentation (89). Consistent with its reliance on amino acids and peptides as primary nutrients, the T. denticola genome encodes eight ABC-type peptide uptake systems (25), presumably with a variety of peptide substrate specificities. To date, only one of these, encoded by OppA-F, has been characterized (68). OppA, the substrate binding protein of this system, is a 70-kDa outer membrane lipoprotein that binds FN, laminin, and plasminogen. The specific peptide substrate(s) taken up through OppA-F have not been determined, and an isogenic oppA mutant showed no defect in growth in culture. Several toxic peptide analogs specific for particular peptide uptake systems were tested and showed no difference in activity against wild-type and oppA mutant T. denticola (68). These results are most likely a reflection of the multiple redundant mechanisms of peptide acquisition in this organism.
Among cultivable oral organisms, T. denticola and members of the ‘black pigmented’ Bacteroidetes, including P. gingivalis produce the highest levels of hydrogen sulfide (H2S) (90), a cytotoxic compound found in elevated levels in periodontal lesions (91). Metabolism of glutathione by T. denticola produces H2S that is believed to play a role in the host tissue destruction seen in periodontitis (92). Recent work demonstrates that H2S induces apoptosis in oral epithelial cells (93) as well as gingival fibroblasts and periodontal ligament cells (94). In addition to the production of a toxic metabolite from a host-supplied nutrient (glutathione), the enzyme that catalyzes the final step in the glutathione metabolism pathway is cystalyisin, a cysteine desulfhydrase that was originally characterized due to its hemolytic activity (95).
Fibrinogen
The ability of bacterial pathogens to bind fibrinogen is linked to pathogenic behavior of several bacterial species, especially those such as staphylococci and streptococci involved in endocarditis (96). Fibrinogen is a plasma protein that is essential for wound healing and also plays a role in tissue homeostasis (reviewed in (97)). Several investigators have documented the ability of dentilisin activity to hydrolyze fibrinogen (23, 98–100) Bamford et al. reported purification of the native dentilisin complex (CTLP) by fibrinogen affinity chromatography (100). Although fibrinogen binding and degradation was generally interpreted as an example of T. denticola general proteolytic activity toward potential nutrients in the host environment, it was also recognized as a potential virulence behavior that could contribute to the dysregulation of tissue homeostasis characteristic of periodontal disease. Fibrinogen binding by the Msp in both native and recombinant forms has been reported in studies using T. denticola cells, native Msp, and recombinant Msp (22, 66, 76). Although no studies have reported the fibrinogen binding activity of msp-deficient T. denticola, results of recent work by Bamford et al. strongly imply that Msp is not the only fibrinogen-binding protein of T. denticola (70). This study used fibrinogen affinity chromatography to purify the native dentilisin protease complex from T. denticola extracts and subsequently demonstrated the hydrolytic activity of the purified protease toward fibrinogen. T. denticola exhibited saturable binding kinetics to soluble and immobilized fibrinogen. Because T. denticola thrives in periodontal lesions harboring damaged tissue, the ability to interact with fibrinogen may contribute both to colonization and environmental modulation by the spirochete.
Environmental signaling and chemotaxis
All microorganisms sense and respond to changes in their environment, especially for purposes of nutrient acquisition. Studies of chemotaxis pathways have progressed in T. denticola, both before and after publication of the genome. Studies by Shi and others characterized chemotaxis-related gene expression (101, 102) and chemotactic behavior toward potential nutrients (88) and the requirement for chemotaxis function for penetration of a model epithelium (103). These studies confirmed the expected important role of environmental sensing for this highly motile organism. The mechanism of environmental sensing typically involves one or more two-component systems consisting of a sensor kinase and a response regulator. A recent study characterized AtcS-AtcR, the first growth phase-regulated two-component system, identified in the T. denticola genome sequence (104). Expression of this system in T. denticola responds to environmental conditions, and likely functions to regulate expression of appropriate metabolic or biosynthetic pathways as nutrients are depleted. This and other signaling and regulatory systems are the subject of a recent review by Frederick et al. (105). In another study of potential importance to environmental responses, Gonzalez et al. (106) characterized the phosphoenolpyruvate-dependent phosphotransferase system of T. denticola. The authors reported that, because T. denticola lacks PTS permeases normally utilized by bacteria for the uptake of simple carbohydrates, it is likely that the Treponema phosphoryl-transfer chain has unique modes of signal detection and sensory transmission. The role of this phosphotransferase system in T. denticola metabolism or environmental sensing remains unknown.
Modulation of host immune response pathways
Modulation of host innate immune response pathways contributes to pathology of both chronic local infections and life-threatening systemic inflammatory conditions that may be induced or exacerbated by chronic bacterial challenge (107). The early literature on innate and acquired immune responses to oral spirochetes (reviewed in (108)) contains limited analyses of effects of specific antigens, and some apparently contradictory reports. More recent work strongly implicates T. denticola outer membrane protein, including the PrtP protease complex (dentilisin) and Msp in these processes (53, 109, 110). Cytopathic and immunomodulatory effects of these protein complexes are under study by several groups. PrtP (dentilisin) is one of only two known lipoproteins in the subtilase family, the other being SphB1, a subtilisin-autotransporter that catalyzes maturation of the virulence factor FhaB (filamentous hemagglutinin) at the surface of Bordetella pertussis (111). The T. denticola protease locus, including prcB, prcA, and prtP, is conserved in several oral Treponema spp. (112–114). Studies with T. denticola mutants implicate PrtP protease activity in modulation of complement regulatory protein Factor H activity (115), activation of MMP-2 (71), and disruption of intercellular junctions (13). Studies with wild-type strains suggest involvement of dentilisin in cell adherence and cytotoxicity (40, 116), FN degradation (12, 36), and modulation of IL-8 expression (53). Both protein complexes exhibit adhesin-like binding activities (22, 116–118), induce cytoskeletal rearrangements, (12, 46) and are cytotoxic to epithelial cells (12, 40). Msp (but not PrtP) is hemolytic (40) and disrupts intracellular calcium regulation (50). Msp disrupts cytoskeletal actin in both gingival fibroblasts and neutrophils (46, 119). PrtP protease activity degrades ECM components as well as IL-8 (53, 54), IL-1ß, IL-6, TNF-α (120), and other bioactive molecules involved in inflammatory vascular responses (98, 121).
Resistance to serum killing
The ability of periodontal organisms to survive the bactericidal activity present in serum and gingival crevicular fluid is suggestive of a specific mechanism of complement resistance. Although some evidence supports a role for the T. denticola surface protease dentilisin (CTLP) in serum resistance, due to its ability to hydrolyze α-chain of C3, producing iC3b (122), recent studies implicate a Factor H-binding lipoprotein in this behavior (115, 123). Because most spirochetal Factor H-binding proteins are acylated but otherwise relatively unconserved, this work utilized a genome- and proteomics-based approach to identify nine candidate genes encoding small lipoproteins containing coiled-coil domains (123). One of these (FhbB, encoded by TDE0108) bound Factor H to the cell surface, and the greatly increased susceptibility of an isogenic fhbB mutant to serum killing indicates that FhbB is primarily responsible for T. denticola complement resistance (124). Interestingly, the PrtP protease also plays a role in FhbB-mediated Factor H binding. Factor H bound to the T. denticola surface is cleaved by dentilisin activity to a size that closely approximates FHL-1 (115). However, as there was no difference in complement resistance between parent and prtP-deficient T. denticola strains (124), the precise role of dentilisin in interaction with the complement pathway remains to be determined.
Resistance to antimicrobial peptides
Antimicrobial peptides with a wide range of structure, charge, and amino acid content comprise a significant component of innate immunity. Several cell types associated with mucosal surfaces produce small (<6-kDa) cationic peptides, designated β-defensins (hBDs), that are active against a wide range of microbes. Antimicrobial activity of defensins is believed to be due to disruption of the bacterial cytoplasmic membrane, although the exact mechanism of action remains somewhat controversial (125). While defensin susceptibility of periodontal organisms varies with the particular defensin molecule, bacterial species and bacterial strain, Porphyromonas gingivalis, Aggregatibacter actinomycetemcomitans, and Fusobacterium nucleatum generally exhibit at least partial sensitivity to hBDs 1, 2, or 3 (126). In contrast, Brissette et al. reported that T. denticola is resistant to hBDs 2 and 3 (127), as are several oral Treponema spp. (128). The mechanism of resistance is not yet understood, but several mechanisms that contribute to resistance in other organisms have been ruled out, including dentilisin proteolytic activity, ABC efflux transporters, and competition with other host proteins for binding sites (129). Potential mechanisms of Treponema resistance to hBDs include proton motive force-driven efflux and reduced binding affinity due to absence of lipopolysaccharides (LPS) on the outer membrane of the cell. In any case, this striking insensitivity to killing effects of hBDs may contribute to the abundance of treponemes in periodontal disease lesions.
Degradation of immunoglobulins
The ability to hydrolyze immunoglobulins is a potential mechanism of evasion of adaptive immune responses. Although a number of microbial pathogens possess specific proteases with specific activity against immunoglobulins, T. denticola's ability to degrade immunoglobulins in vivo has been somewhat controversial. While dentilisin has demonstrated in vitro activity against IgG (23, 130), Hollmann and Van der Hoeven reported that intact T. denticola did not degrade IgA, IgG, or albumin (131). It is reasonable to question these results in light of the dentilisin studies and given the fact that serum albumin is a preferred nutrient source of T. denticola (87).
In a recent study, Ishihara et al. characterized IdeT, a T. denticola homolog of IdeS, that is an IgG-specific streptococcal protease (132). The C-terminal domain of the T. denticola protein (designated ‘dentipain’) is a protease that is cleaved and secreted into the extracellular environment. An isogenic mutant strain in which the dentipain domain was deleted was significantly less virulent in a mouse abscess model, suggesting that IdeT contributes to T. denticola virulence (133). Further studies are required to characterize the role of IdeT in interaction with immunoglobulins and other serum proteins in the subgingival environment.
Lipooligosaccharide (LOS) and lipoprotein effects
The T. denticola genome lacks an identifiable LPS synthesis pathway (25). Biochemical analysis of the T. denticola outer membrane revealed the presence of a novel glycolipid or lipooligosaccharide (LOS) that comprises the outer leaflet of the outer membrane bilayer (134). This LOS, although similar in overall structure and function to the lipopolysaccharide present on most Gram-negative organisms, has a distinctly different pattern of sugar molecules and lacks the lipid A component of a typical LPS and is also biochemically distinct from lipoteichoic acid. Classical LPS proinflammatory signaling is primarily through TLR4, with some involvement of TLR2 in many cases. However, in spirochetes, many of which lack LPS, lipoprotein-mediated signaling through TLR2 is a primary activator of inflammatory responses (51, 135). Rosen et al. reported activation of murine macrophages by both LOS and de-lipidated lipoproteins of T. denticola (56), but did not identify signaling pathways involved. Recently, the same group reported that LOS induced macrophage responses through a TLR4-MyD88 pathway, whereas the major outer membrane protein Msp induced innate immune responses through TLR2-MyD88 (136). It should be noted that Msp is not acylated, so the mechanism of activation may be distinct in this case. In closely related treponemes, T. maltophilum LOS activation of NF-κB was reported to be dependent on TLR2, while both TLR2 and TLR4 were involved in activation of innate immune responses to T. brennaborense LOS (137, 138). Other recent studies focused on downstream effects of likely TLR-mediated signaling events. Choi et al. reported that T. denticola LOS induced osteoclastogenesis and MMP-9 expression in cultured murine osteoblasts and suggested that these processes may be contributory mechanisms to bone loss associated with periodontal diseases (139). A similar study that focused on gingival fibroblasts reported significantly increased MCP-1, IL-8, PGE2, and MMP-3 in LOS-challenged fibroblasts.
Summary
Treponema denticola, the most readily cultivable oral spirochete, is the model organism for the study of spirochete–host interactions in periodontal disease. Research in recent years has greatly expanded our understanding of both the biology of spirochetes and the mechanisms utilized by these opportunistic commensal pathogens to both survive as members of the ‘normal’ oral flora as well as to participate in the induction of periodontal tissue destruction and thrive in the resulting environment. Numerous potentially pathogenic behaviors and potential virulence factors were identified for some of these behaviors, but it is rather striking that two multifunctional outer membrane protein complexes were reported to both directly and indirectly mediate cellular damage participate in many of the identified pathogenic behaviors of this organism. The surface-expressed PrtP lipoprotein protease complex (dentilisin) and the oligomeric Msp are among the most studied T. denticola virulence determinants. While this may overshadow the potential importance of other as yet poorly documented behaviors, it is evident that Msp and dentilisin are key players in T. denticola periodontal pathogenicity. Besides functioning in nutrient acquisition, dentilisin contributes to tissue penetration and cleaves several proteins involved in innate and adaptive immunity, likely favoring dysregulation of tissue homeostasis. Expression of dentilisin is required for native expression of the major surface protein (Msp) that binds FN, has cytotoxic pore-forming activity, and disrupts intracellular calcium responses. A deeper understanding of the roles of these and other T. denticola proteins in microbe–host interactions requires taking advantage of recent advances in spirochete molecular biology to analyze their cytopathic and immunomodulatory behaviors.
Acknowledgements
This work was supported by United States Public Health Service grant DE018221 (National Institute of Dental and Craniofacial Research Bethesda, MD, USA).
Conflict of interest and funding
There is no conflict of interest in the present study for the author.
References
- 1.Loesche WJ, Grossman NS. Periodontal disease as a specific, albeit chronic, infection: diagnosis and treatment. Clin Microbiol Rev. 2001;14:727–52. doi: 10.1128/CMR.14.4.727-752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–31. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- 3.Paster BJ, Boches SK, Galvin JL, Ericson RE, Lau CN, Levanos VA, et al. Bacterial diversity in human subgingival plaque. J Bacteriol. 2001;183:3770–83. doi: 10.1128/JB.183.12.3770-3783.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 5.Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729–39. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- 6.Loesche WJ, Syed SA, Schmidt E, Morrison EC. Bacterial profiles of subgingival plaques in periodontitis. J Periodontol. 1985;56:447–56. doi: 10.1902/jop.1985.56.8.447. [DOI] [PubMed] [Google Scholar]
- 7.Loesche WJ, Syed SA, Laughon BE, Stoll J. The bacteriology of acute necrotizing ulcerative gingivitis. J Periodontol. 1982;53:223–30. doi: 10.1902/jop.1982.53.4.223. [DOI] [PubMed] [Google Scholar]
- 8.Choi BK, Paster BJ, Dewhirst FE, Gobel UB. Diversity of cultivable and uncultivable oral spirochetes from a patient with severe destructive periodontitis. Infect Immun. 1994;62:1889–95. doi: 10.1128/iai.62.5.1889-1895.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellen RP, Galimanas VB. Spirochetes at the forefront of periodontal infections. Periodontol. 2005;38:13–32. doi: 10.1111/j.1600-0757.2005.00108.x. [DOI] [PubMed] [Google Scholar]
- 10.Frank RM. Bacterial penetration in the apical pocket wall of advanced human periodontitis. J Periodontal Res. 1980;15:563–73. doi: 10.1111/j.1600-0765.1980.tb00315.x. [DOI] [PubMed] [Google Scholar]
- 11.Saglie R, Newman MG, Carranza FA, Jr, Pattison GL. Bacterial invasion of gingiva in advanced periodontitis in humans. J Periodontol. 1982;53:217–22. doi: 10.1902/jop.1982.53.4.217. [DOI] [PubMed] [Google Scholar]
- 12.Uitto V-J, Pan YM, Leung WK, Larjava H, Ellen RP, Finlay BB, et al. Cytopathic effects of Treponema denticola chymotrypsin-like proteinase on migrating and stratified epithelial cells. Infect Immun. 1995;63:3401–10. doi: 10.1128/iai.63.9.3401-3410.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi B, Qi M, Kuramitsu HK. Role of dentilisin in Treponema denticola epithelial cell layer penetration. Res Microbiol. 2003;154:637–43. doi: 10.1016/j.resmic.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 14.Peters SR, Valdez M, Riviere G, Thomas DD. Adherence to and penetration through endothelial cells by oral treponemes. Oral Microbiol Immunol. 1999;14:379–83. doi: 10.1034/j.1399-302x.1999.140609.x. [DOI] [PubMed] [Google Scholar]
- 15.Salyers AA, Whitt DD. Bacterial pathogenesis: a molecular approach. Washington, DC: American Society for Microbiology; 1994. p. 30. [Google Scholar]
- 16.Hoepelman AI, Tuomanen EI. Consequences of microbial attachment: directing host cell functions with adhesins. Infect Immun. 1992;60:1729–33. doi: 10.1128/iai.60.5.1729-1733.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fenno JC, McBride BC. Virulence factors of oral treponemes. Anaerobe. 1998;4:1–17. doi: 10.1006/anae.1997.0131. [DOI] [PubMed] [Google Scholar]
- 18.Listgarten MA, Levin S. Positive correlation between the proportions of subgingival spirochetes and motile bacteria and susceptibility of human subjects to periodontal deterioration. J Clin Periodontol. 1981;8:122–38. doi: 10.1111/j.1600-051x.1981.tb02352.x. [DOI] [PubMed] [Google Scholar]
- 19.Holt SC. Anatomy and chemistry of spirochetes. Microbiol Rev. 1978;42:114–60. doi: 10.1128/mr.42.1.114-160.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Charon NW, Greenberg EP, Koopman MB, Limberger RJ. Spirochete chemotaxis, motility, and the structure of the spirochetal periplasmic flagella. Res Microbiol. 1992;143:597–603. doi: 10.1016/0923-2508(92)90117-7. [DOI] [PubMed] [Google Scholar]
- 21.Kent C, Gee P, Lee SY, Bian X, Fenno JC. A CDP-choline pathway for phosphatidylcholine biosynthesis in Treponema denticola . Mol Microbiol. 2004;51:471–81. doi: 10.1046/j.1365-2958.2003.03839.x. [DOI] [PubMed] [Google Scholar]
- 22.Haapasalo M, Müller K-H, Uitto V-J, Leung WK, McBride BC. Characterization, cloning, and binding properties of the major 53-kilodalton Treponema denticola surface antigen. Infect Immun. 1992;60:2058–65. doi: 10.1128/iai.60.5.2058-2065.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Uitto V-J, Grenier D, Chan EC, McBride BC. Isolation of a chymotrypsin-like enzyme from Treponema denticola . Infect Immun. 1988;56:2717–22. doi: 10.1128/iai.56.10.2717-2722.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolenbrander PE. Oral microbial communities: biofilms, interactions, and genetic systems. Annu Rev Microbiol. 2000;54:413–37. doi: 10.1146/annurev.micro.54.1.413. [DOI] [PubMed] [Google Scholar]
- 25.Seshadri R, Myers GS, Tettelin H, Eisen JA, Heidelberg JF, Dodson RJ, et al. Comparison of the genome of the oral pathogen Treponema denticola with other spirochete genomes. Proc Natl Acad Sci U S A. 2004;101:5646–51. doi: 10.1073/pnas.0307639101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown JR. Ancient horizontal gene transfer. Nat Rev Genet. 2003;4:121–32. doi: 10.1038/nrg1000. [DOI] [PubMed] [Google Scholar]
- 27.Duncan MJ. Oral microbiology and genomics. Periodontol 2000. 2005;38:63–71. doi: 10.1111/j.1600-0757.2005.00111.x. [DOI] [PubMed] [Google Scholar]
- 28.Chen T, Abbey K, Deng WJ, Cheng MC. The bioinformatics resource for oral pathogens. Nucleic Acids Res. 2005;33:W734–W40. doi: 10.1093/nar/gki361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dewhirst FE, Chen T, Izard T, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–17. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li H, Ruby J, Charon N, Kuramitsu H. Gene inactivation in the oral spirochete Treponema denticola: construction of an flgE mutant. J Bacteriol. 1996;178:3664–7. doi: 10.1128/jb.178.12.3664-3667.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slivienski-Gebhardt LL, Izard J, Samsonoff WA, Limberger RJ. Development of a novel chloramphenicol resistance expression plasmid used for genetic complementation of a fliG deletion mutant in Treponema denticola. Infect Immun. 2004;72:5493–7. doi: 10.1128/IAI.72.9.5493-5497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chi B, Chauhan S, Kuramitsu HK. Development of a system for expressing heterologous genes in the oral spirochete Treponema denticola and its use in expression of the Treponema pallidum flaA gene. Infect Immun. 1999;67:3653–6. doi: 10.1128/iai.67.7.3653-3656.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boehringer H, Taichman NS, Shenker BJ. Suppression of fibroblast proliferation by oral spirochetes. Infect Immun. 1984;45:155–9. doi: 10.1128/iai.45.1.155-159.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Reijntjens FM, Mikx FH, Wolters-Lutgerhorst JM, Maltha JC. Adherence of oral treponemes and their effect on morphological damage and detachment of epithelial cells in vitro. Infect Immun. 1986;51:642–7. doi: 10.1128/iai.51.2.642-647.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baehni PC, Song M, McCulloch M, Ellen RP. Treponema denticola induces actin rearrangement and detachment of human gingival fibroblasts. Infect Immun. 1992;60:3360–8. doi: 10.1128/iai.60.8.3360-3368.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ellen RP, Song M, McCulloch CA. Degradation of endogenous plasma membrane fibronectin concomitant with Treponema denticola 35405 adhesion to gingival fibroblasts. Infect Immun. 1994;62:3033–7. doi: 10.1128/iai.62.7.3033-3037.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.De Filippo AB, Ellen RP, McCulloch CA. Induction of cytoskeletal rearrangements and loss of volume regulation in epithelial cells by Treponema denticola . Arch Oral Biol. 1995;40:199–207. doi: 10.1016/0003-9969(95)98809-d. [DOI] [PubMed] [Google Scholar]
- 38.Lee W, Pankoski L, Zekavat A, Shenker BJ. Treponema denticola immunoinhibitory protein induces irreversible G1 arrest in activated human lymphocytes. Oral Microbiol Immunol. 2004;19:144–9. doi: 10.1111/j.0902-0055.2004.00129.x. [DOI] [PubMed] [Google Scholar]
- 39.Shenker BJ, Listgarten MA, Taichman NS. Suppression of human lymphocyte responses by oral spirochetes: a monocyte-dependent phenomenon. J Immunol. 1984;132:2039–45. [PubMed] [Google Scholar]
- 40.Fenno JC, Hannam PM, Leung WK, Tamura M, Uitto V.-J, McBride BC. Cytopathic effects of the major surface protein (Msp) and the chymotrypsin-like protease (CTLP) of Treponema denticola. Infect Immun. 1998;66:1869–77. doi: 10.1128/iai.66.5.1869-1877.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Egli C, Leung WK, Müller KH, Hancock RE, McBride BC. Pore-forming properties of the major 53-kilodalton surface antigen from the outer sheath of Treponema denticola . Infect Immun. 1993;61:1694–9. doi: 10.1128/iai.61.5.1694-1699.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mathers DA, Leung WK, Fenno JC, Hong Y, McBride BC. Major surface protein complex of Treponema denticola depolarizes and induces ion channels in HeLa cell membranes. Infect. Immun. 1996;64:2904–10. doi: 10.1128/iai.64.8.2904-2910.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ulmer JB, Burke CJ, Shi C, Friedman A, Donnelly JJ, Liu MA. Pore formation and mitogenicity in blood cells by the class 2 protein of Neisseria meningitidis . J Biol Chem. 1992;267:19266–71. [PubMed] [Google Scholar]
- 44.Massari P, King CA, MacLeod H, Wetzler LM. Improved purification of native meningococcal porin PorB and studies on its structure/function. Protein Expr Purif. 2005;44:136–46. doi: 10.1016/j.pep.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 45.Yang PF, Song M, Grove M, Ellen RP. Filamentous actin disruption and diminished inositol phosphate response in gingival fibroblasts caused by Treponema denticola. Infect Immun. 1998;66:696–702. doi: 10.1128/iai.66.2.696-702.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Amin M, Ho AC, Lin JY, Batista da Silva AP, Glogauer M, Ellen RP. Induction of de novo subcortical actin filament assembly by Treponema denticola major outer sheath protein. Infect Immun. 2004;72:3650–4. doi: 10.1128/IAI.72.6.3650-3654.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Batista da Silva AP, Lee W, Bajenova E, McCulloch CA, Ellen RP. The major outer sheath protein of Treponema denticola inhibits the binding step of collagen phagocytosis in fibroblasts. Cell Microbiol. 2004;6:485–98. doi: 10.1111/j.1462-5822.2004.00377.x. [DOI] [PubMed] [Google Scholar]
- 48.Amin M, Grove DA, Kapus A, Glogauer M, Ellen RP. An actin-stabilizing peptide conjugate deduced from the major outer sheath protein of the bacterium Treponema denticola. Cell Motil Cytoskeleton. 2007;64:662–74. doi: 10.1002/cm.20213. [DOI] [PubMed] [Google Scholar]
- 49.Amin M, Magnusson KE, Kapus A, Glogauer M, Ellen RP. Treponema denticola Msp-deduced peptide conjugate, P34BSA, promotes RhoA-dependent actin stress fiber formation independent of its internalization by fibroblasts. Cell Motil Cytoskeleton. 2008;65:406–21. doi: 10.1002/cm.20270. [DOI] [PubMed] [Google Scholar]
- 50.Wang Q, Ko KS, Kapus A, McCulloch CA, Ellen RP. A spirochete surface protein uncouples store-operated calcium channels in fibroblasts: a novel cytotoxic mechanism. J Biol Chem. 2001;276:23056–64. doi: 10.1074/jbc.M011735200. [DOI] [PubMed] [Google Scholar]
- 51.Hirschfeld M, Kirschning CJ, Schwandner R, Wesche H, Weis JH, Wooten RM, et al. Cutting edge: inflammatory signaling by Borrelia burgdorferi lipoproteins is mediated by toll-like receptor 2. J Immunol. 1999;163:2382–6. [PubMed] [Google Scholar]
- 52.Ellen RP, Ko KS, Lo KS, Grove DA, Ishihara K. Insertional inactivation of the prtP gene of Treponema denticola confirms dentilisin's disruption of epithelial junctions. J Mol Microbiol Biotechnol. 2000;2:581–6. [PubMed] [Google Scholar]
- 53.Asai Y, Jinno T, Ogawa T. Oral treponemes and their outer membrane extracts activate human gingival epithelial cells through toll-like receptor 2. Infect Immun. 2003;71:717–25. doi: 10.1128/IAI.71.2.717-725.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deng QD, Han Y, Xia X, Kuramitsu HK. Effects of the oral spirochete Treponema denticola on interleukin-8 expression from epithelial cells. Oral Microbiol Immunol. 2001;16:185–7. doi: 10.1034/j.1399-302x.2001.016003185.x. [DOI] [PubMed] [Google Scholar]
- 55.Nixon CS, Steffen MJ, Ebersole JL. Cytokine responses to Treponema pectinovorum and Treponema denticola in human gingival fibroblasts. Infect Immun. 2000;68:5284–92. doi: 10.1128/iai.68.9.5284-5292.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rosen G, Sela MN, Naor R, Halabi A, Barak V, Shapira L. Activation of murine macrophages by lipoprotein and lipooligosaccharide of Treponema denticola. Infect Immun. 1999;67:1180–6. doi: 10.1128/iai.67.3.1180-1186.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mao Y, Schwarzbauer JE. Fibronectin fibrillogenesis, a cell-mediated matrix assembly process. Matrix Biol. 2005;24:389–99. doi: 10.1016/j.matbio.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 58.Kuusela P. Fibronectin binds to Staphylococcus aureus. Nature (London) 1978;276:718–20. doi: 10.1038/276718a0. [DOI] [PubMed] [Google Scholar]
- 59.Henderson B, Nair S, Pallas J, Williams MA. Fibronectin: a multidomain host adhesin targeted by bacterial fibronectin-binding proteins. FEMS Microbiol Rev. 2011;35:147–200. doi: 10.1111/j.1574-6976.2010.00243.x. [DOI] [PubMed] [Google Scholar]
- 60.Ellen RP, Dawson JR, Yang PF. Treponema denticola as a model for polar adhesion and cytopathogenicity of spirochetes. Trends Microbiol. 1994;2:114–9. doi: 10.1016/0966-842x(94)90597-5. [DOI] [PubMed] [Google Scholar]
- 61.Dawson JR, Ellen RP. Clustering of fibronectin adhesins toward Treponema denticola tips upon contact with immobilized fibronectin. Infect Immun. 1994;62:2214–21. doi: 10.1128/iai.62.6.2214-2221.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dawson JR, Ellen RP. Tip-oriented adherence of Treponema denticola to fibronectin. Infect Immun. 1990;58:3924–8. doi: 10.1128/iai.58.12.3924-3928.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weinberg A, Holt Sc. Interaction of Treponema denticola TD-4, GM-1, and MS25 with human gingival fibroblasts. Infect Immun. 1990;58:1720–9. doi: 10.1128/iai.58.6.1720-1729.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ellen RP. Perturbation and exploitation of host cell cytoskeleton by periodontal pathogens. Microbes Infect. 1999;1:621–32. doi: 10.1016/s1286-4579(99)80062-8. [DOI] [PubMed] [Google Scholar]
- 65.Ko KS-C, Glogauer M, McCulloch CAG, Ellen RP. Treponema denticola outer membrane inhibits calcium flux in gingival fibroblasts. Infect Immun. 1998;66:703–9. doi: 10.1128/iai.66.2.703-709.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Edwards AM, Jenkinson HF, Woodward MJ, Dymock D. Binding properties and adhesion-mediating regions of the major sheath protein of Treponema denticola ATCC 35405. Infect Immun. 2005;73:2891–8. doi: 10.1128/IAI.73.5.2891-2898.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fenno JC, Müller K-H, McBride BC. Sequence analysis, expression and binding activity of recombinant major outer sheath protein (Msp) of Treponema denticola . J Bacteriol. 1996;178:2489–97. doi: 10.1128/jb.178.9.2489-2497.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fenno JC, Tamura M, Hannam PM, Wong GWK, Chan RA, McBride BC. Identification of a Treponema denticola OppA homologue that binds host proteins present in the subgingival environment. Infect Immun. 2000;68:1884–92. doi: 10.1128/iai.68.4.1884-1892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cameron CE, Brown EL, Kuroiwa JM, Schnapp LM, Brouwer NL. Treponema pallidum fibronectin-binding proteins. J Bacteriol. 2004;186:7019–22. doi: 10.1128/JB.186.20.7019-7022.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bamford CV, Francescutti T, Cameron CE, Jenkinson HF, Dymock D. Characterization of a novel family of fibronectin-binding proteins with M23 peptidase domains from Treponema denticola . Mol Oral Microbiol. 2010;25:369–83. doi: 10.1111/j.2041-1014.2010.00584.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miao D, Fenno JC, Timm JC, Joo NE, Kapila YL. Treponema denticola chymotrypsin-like protease (dentilisin) induces MMP-2-dependent fibronectin fragmentation in periodontal ligament cells. Infect Immun. 2011;79:806–11. doi: 10.1128/IAI.01001-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dobaczewski M, Gonzalez-Quesada C, Frangogiannis NG. The extracellular matrix as a modulator of the inflammatory and reparative response following myocardial infarction. J Mol Cell Cardiol. 2010;48:504–11. doi: 10.1016/j.yjmcc.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dai R, Iwama A, Wang S, Kapila YL. Disease-associated fibronectin matrix fragments trigger anoikis of human primary ligament cells: p53 and c-myc are suppressed. Apoptosis. 2005;10:503–12. doi: 10.1007/s10495-005-1880-5. [DOI] [PubMed] [Google Scholar]
- 74.Kapila YL, Kapila S, Johnson PW. Fibronectin and fibronectin fragments modulate the expression of proteinases and proteinase inhibitors in human periodontal ligament cells. Matrix Biol. 1996;15:251–61. doi: 10.1016/s0945-053x(96)90116-x. [DOI] [PubMed] [Google Scholar]
- 75.Grenier D, Uitto V-J, McBride BC. Cellular location of a Treponema denticola chymotrypsin-like protease and importance of the protease in migration through the basement membrane. Infect Immun. 1990;58:347–51. doi: 10.1128/iai.58.2.347-351.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haapasalo M, Singh U, McBride BC, Uitto V-J. Sulfhydryl-dependent attachment of Treponema denticola to laminin and other proteins. Infect Immun. 1991;59:4230–7. doi: 10.1128/iai.59.11.4230-4237.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Umemoto T, Namikawa I. Binding of host-associated treponeme proteins to collagens and laminin: a possible mechanism of spirochetal adherence to host tissues. Microbiol Immunol. 1994;38:655–63. doi: 10.1111/j.1348-0421.1994.tb01836.x. [DOI] [PubMed] [Google Scholar]
- 78.Sorsa T, Ingman T, Suomalainen K, Haapasalo M, Konttinen YT, Lindy O, et al. Identification of proteases from periodontopathogenic bacteria as activators of latent human neutrophil and fibroblast-type interstitial collagenases. Infect Immun. 1992;60:4491–5. doi: 10.1128/iai.60.11.4491-4495.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haapasalo M, Hannam P, McBride BC, Uitto V-J. Hyaluronan, a possible ligand mediating Treponema denticola binding to periodontal tissue. Oral Microbiol Immunol. 1996;11:156–60. doi: 10.1111/j.1399-302x.1996.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 80.Girish KS, Kemparaju K. The magic glue hyaluronan and its eraser hyaluronidase: a biological overview. Life Sci. 2007;80:1921–43. doi: 10.1016/j.lfs.2007.02.037. [DOI] [PubMed] [Google Scholar]
- 81.Jiang D, Liang J, Noble PW. Hyaluronan as an immune regulator in human diseases. Physiol Rev. 2011;91:221–64. doi: 10.1152/physrev.00052.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kreil G. Hyaluronidases–a group of neglected enzymes. Protein Sci. 1995;4:1666–9. doi: 10.1002/pro.5560040902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Scott D, Siboo R, Chan EC, Siboo R. An extracellular enzyme with hyaluronidase and chondroitinase activities from some oral anaerobic spirochaetes. Microbiology. 1996;142:2567–76. doi: 10.1099/00221287-142-9-2567. [DOI] [PubMed] [Google Scholar]
- 84.Fenno JC. Laboratory maintenance of Treponema denticola . Curr Protoc Microbiol . 2005 doi: 10.1002/9780471729259.mc12b01s00. 12B.1.1–12B.1.21. [DOI] [PubMed] [Google Scholar]
- 85.Wyss C. Fatty acids synthesized by oral treponemes in chemically defined media. FEMS Microbiol Lett. 2007;269:70–6. doi: 10.1111/j.1574-6968.2006.00609.x. [DOI] [PubMed] [Google Scholar]
- 86.Wyss C, Choi BK, Schupbach P, Guggenheim B, Gobel UB. Treponema maltophilum sp. nov., a small oral spirochete isolated from human periodontal lesions. Int J Syst Bacteriol. 1996;46:745–52. doi: 10.1099/00207713-46-3-745. [DOI] [PubMed] [Google Scholar]
- 87.Van Horn KG, Smibert RM. Albumin requirement of Treponema denticola and Treponema vincentii . Can J Microbiol. 1983;29:1141–8. doi: 10.1139/m83-175. [DOI] [PubMed] [Google Scholar]
- 88.Ruby JD, Lux R, Shi W, Charon NW, Dasanayake A. Effect of glucose on Treponema denticola cell behavior. Oral Microbiol Immunol. 2008;23:234–8. doi: 10.1111/j.1399-302X.2007.00417.x. [DOI] [PubMed] [Google Scholar]
- 89.Hespell RB, Canale-Parola E. Amino acid and glucose fermentation by Treponema denticola . Arch Mikrobiol. 1971;78:234–51. doi: 10.1007/BF00424897. [DOI] [PubMed] [Google Scholar]
- 90.Persson S, Edlund MB, Claesson R, Carlsson J. The formation of hydrogen sulfide and methyl mercaptan by oral bacteria. Oral Microbiol Immunol. 1990;5:195–201. doi: 10.1111/j.1399-302x.1990.tb00645.x. [DOI] [PubMed] [Google Scholar]
- 91.Gleissner C, Springborn I, Willershausen B. Evaluation of sulcular sulphide level monitoring using a portable sensor system. Eur J Med Res. 2002;7:491–501. [PubMed] [Google Scholar]
- 92.Chu L, Dong Z, Xu X, Cochran DL, Ebersole JL. Role of glutathione metabolism of Treponema denticola in bacterial growth and virulence expression. Infect Immun. 2002;70:1113–20. doi: 10.1128/IAI.70.3.1113-1120.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Murata T, Yaegaki K, Qian W, Herai M, Calenic B, Imai T, et al. Hydrogen sulfide induces apoptosis in epithelial cells derived from human gingiva. J Breath Res. 2008;2:017007. doi: 10.1088/1752-7155/2/1/017007. [DOI] [PubMed] [Google Scholar]
- 94.Zhang JH, Dong Z, Chu L. Hydrogen sulfide induces apoptosis in human periodontium cells. J Periodontal Res. 2010;45:71–8. doi: 10.1111/j.1600-0765.2009.01202.x. [DOI] [PubMed] [Google Scholar]
- 95.Chu L, Ebersole JL, Kurzban GP, Holt SC. Cystalysin, a 46-kilodalton cysteine desulfhydrase from Treponema denticola, with hemolytic and hemoxidative activities. Infect Immun. 1997;65:3231–8. doi: 10.1128/iai.65.8.3231-3238.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Moreillon P, Entenza JM, Francioli P, McDevitt D, Foster TJ, Francois P, et al. Role of Staphylococcus aureus coagulase and clumping factor in pathogenesis of experimental endocarditis. Infect Immun. 1995;63:4738–43. doi: 10.1128/iai.63.12.4738-4743.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Mosesson MW. Fibrinogen and fibrin structure and functions. J Thromb Haemost. 2005;3:1894–904. doi: 10.1111/j.1538-7836.2005.01365.x. [DOI] [PubMed] [Google Scholar]
- 98.Mäkinen PL, Mäkinen KK, Syed SA. Role of the chymotrypsin-like membrane-associated proteinase from Treponema denticola ATCC 35405 in inactivation of bioactive peptides. Infect Immun. 1995;63:3567–75. doi: 10.1128/iai.63.9.3567-3575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rosen G, Naor R, Kutner S, Sela MN. Characterization of fibrinolytic activities of Treponema denticola . Infect Immun. 1994;62:1749–54. doi: 10.1128/iai.62.5.1749-1754.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bamford CV, Fenno JC, Jenkinson HF, Dymock D. The chymotrypsin-like protease (CTLP) complex of Treponema denticola ATCC 35405 mediates fibrinogen adherence and degradation. Infect Immun. 2007;75:4364–72. doi: 10.1128/IAI.00258-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sim JH, Shi W, Lux R. Protein-protein interactions in the chemotaxis signalling pathway of Treponema denticola . Microbiology. 2005;151:1801–7. doi: 10.1099/mic.0.27622-0. [DOI] [PubMed] [Google Scholar]
- 102.Lux R, Sim JH, Tsai JP, Shi W. Construction and characterization of a cheA mutant of Treponema denticola . J Bacteriol. 2002;184:3130–4. doi: 10.1128/JB.184.11.3130-3134.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Lux R, Miller JN, Park NH, Shi W. Motility and chemotaxis in tissue penetration of oral epithelial cell layers by Treponema denticola . Infect Immun. 2001;69:6276–83. doi: 10.1128/IAI.69.10.6276-6283.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Frederick JR, Rogers EA, Marconi RT. Analysis of a growth-phase-regulated two-component regulatory system in the periodontal pathogen Treponema denticola . J Bacteriol. 2008;190:6162–9. doi: 10.1128/JB.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Frederick JR, Sarkar J, McDowell JV, Marconi RT. Molecular signaling mechanisms of the periopathogen, Treponema denticola . J Dent Res. 2011;90:1155–63. doi: 10.1177/0022034511402994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Gonzalez CF, Stonestrom AJ, Lorca GL, Saier MH., Jr Biochemical characterization of phosphoryl transfer involving HPr of the phosphoenolpyruvate-dependent phosphotransferase system in Treponema denticola, an organism that lacks PTS permeases. Biochem. 2005;44:598–608. doi: 10.1021/bi048412y. [DOI] [PubMed] [Google Scholar]
- 107.Henderson B, Poole S, Wilson M. Bacterial modulins: a novel class of virulence factors which cause host tissue pathology by inducing cytokine synthesis. Microbiol Rev. 1996;60:316–41. doi: 10.1128/mr.60.2.316-341.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ebersole JL. Systemic humoral immune responses in periodontal disease. Crit Rev Oral Biol Med. 1990;1:283–331. doi: 10.1177/10454411900010040601. [DOI] [PubMed] [Google Scholar]
- 109.Lee SH, Kim KK, Choi BK. Upregulation of intercellular adhesion molecule 1 and proinflammatory cytokines by the major surface proteins of Treponema maltophilum and Treponema lecithinolyticum, the phylogenetic group IV oral spirochetes associated with periodontitis and endodontic infections. Infect Immun. 2005;73:268–76. doi: 10.1128/IAI.73.1.268-276.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jun HK, Kang YM, Lee HR, Lee SH, Choi BK. Highly conserved surface proteins of oral spirochetes as adhesins and potent inducers of proinflammatory and osteoclastogenic factors. Infect Immun. 2008;76:2428–38. doi: 10.1128/IAI.01128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Coutte L, Willery E, Antoine R, Drobecq H, Locht C, Jacob-Dubuisson F. Surface anchoring of bacterial subtilisin important for maturation function. Mol Microbiol. 2003;49:529–39. doi: 10.1046/j.1365-2958.2003.03573.x. [DOI] [PubMed] [Google Scholar]
- 112.Correia FF, Plummer AR, Ellen RP, Wyss C, Boches SK, Galvin JL, et al. Two paralogous families of a two-gene subtilisin operon are widely distributed in oral treponemes. J Bacteriol. 2003;185:6860–9. doi: 10.1128/JB.185.23.6860-6869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Heuner K, Bergmann I, Heckenbach K, Gobel UB. Proteolytic activity among various oral Treponema species and cloning of a prtP-like gene of Treponema socranskii subsp. socranskii (1) . FEMS Microbiol Lett. 2001;201:169–76. doi: 10.1111/j.1574-6968.2001.tb10752.x. [DOI] [PubMed] [Google Scholar]
- 114.Klein EA, Dewhirst FE. Dentilisin is part of a conserved three-gene operon. J Dent Res. 2006;85(Spec Iss A):2126. www.dentalresearch.org. [Google Scholar]
- 115.McDowell JV, Huang B, Fenno JC, Marconi RT. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola . Infect Immun. 2009;77:1417–25. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Leung WK, Haapasalo M, Uitto V-J, Hannam PM, McBride BC. The surface proteinase of Treponema denticola may mediate attachment of the bacteria to epithelial cells. Anaerobe. 1996;2:39–46. [Google Scholar]
- 117.Weinberg A, Holt SC. Chemical and biological activities of a 64-kilodalton outer sheath protein from Treponema denticola strains. J Bacteriol. 1991;173:6935–47. doi: 10.1128/jb.173.21.6935-6947.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Hashimoto M, Ogawa S, Asai Y, Takai Y, Ogawa T. Binding of Porphyromonas gingivalis fimbriae to Treponema denticola dentilisin. FEMS Microbiol Lett. 2003;226:267–71. doi: 10.1016/S0378-1097(03)00615-3. [DOI] [PubMed] [Google Scholar]
- 119.Puthengady TB, Sun CX, Bajenova E, Ellen RP, Glogauer M. Modulation of human neutrophil functions in vitro by Treponema denticola major outer sheath protein. Infect Immun. 2006;74:1954–7. doi: 10.1128/IAI.74.3.1954-1957.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Miyamoto M, Ishihara K, Okuda K. The Treponema denticola surface protease Dentilisin degrades Interleukin-1{beta} (IL-1{beta}), IL-6, and Tumor Necrosis Factor Alpha. Infect Immun. 2006;74:2462–7. doi: 10.1128/IAI.74.4.2462-2467.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Mäkinen PL, Mäkinen KK, Syed SA. An endo-acting proline-specific oligopeptidase from Treponema denticola ATCC 35405: evidence of hydrolysis of human bioactive peptides. Infect Immun. 1994;62:4938–47. doi: 10.1128/iai.62.11.4938-4947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yamazaki T, Miyamoto M, Yamada S, Okuda K, Ishihara K. Surface protease of Treponema denticola hydrolyzes C3 and influences function of polymorphonuclear leukocytes. Microbes Infect. 2006;8:1758–63. doi: 10.1016/j.micinf.2006.02.013. [DOI] [PubMed] [Google Scholar]
- 123.McDowell JV, Frederick J, Stamm L, Marconi RT. Identification of the gene encoding the FhbB protein of Treponema denticola, a highly unique factor H-like protein 1 binding protein. Infect Immun. 2007;75:1050–4. doi: 10.1128/IAI.01458-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.McDowell JV, Frederick J, Goodman MP, Goetting-Minesky MP, Miller DP, Fenno JC, et al. Marconi. Identification of the primary mechanism of complement evasion by the periodontal pathogen, Treponema denticola . Mol Oral Microbiol. 2011;26:140–9. doi: 10.1111/j.2041-1014.2010.00598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Brogden KA. Antimicrobial peptides: pore formers or metabolic inhibitors in bacteria? Nat Rev Microbiol. 2005;3:238–50. doi: 10.1038/nrmicro1098. [DOI] [PubMed] [Google Scholar]
- 126.Komatsuzawa H, Ouhara K, Kawai T, Yamada S, Fujiwara T, Shiba H, et al. Susceptibility of periodontopathogenic and cariogenic bacteria to defensins and potential therapeutic use of defensins in oral diseases. Curr Pharm Des. 2007;13:3084–95. doi: 10.2174/138161207782110426. [DOI] [PubMed] [Google Scholar]
- 127.Brissette CA, Lukehart SA. Treponema denticola is resistant to human beta-defensins. Infect Immun. 2002;70:3982–4. doi: 10.1128/IAI.70.7.3982-3984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Brissette CA, Simonson LG, Lukehart SA. Resistance to human beta-defensins is common among oral treponemes. Oral Microbiol Immunol. 2004;19:403–7. doi: 10.1111/j.1399-302x.2004.00177.x. [DOI] [PubMed] [Google Scholar]
- 129.Brissette CA, Lukehart SA. Mechanisms of decreased susceptibility to beta-defensins by Treponema denticola . Infect Immun. 2007;75:2307–15. doi: 10.1128/IAI.01718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Grenier D, Mayrand D. Cleavage of human immunoglobulin G by Treponema denticola . Anaerobe. 2001;7:1–4. [Google Scholar]
- 131.Hollmann R, Van der Hoeven HJ. Inability of intact cells of Treponema denticola to degrade human serum proteins IgA, IgG and albumin. J Clin Periodontol. 1999;26:477–9. doi: 10.1034/j.1600-051x.1999.260711.x. [DOI] [PubMed] [Google Scholar]
- 132.Von Pawel-Rammingen U, Johansson BP, Bjorck L. IdeS, a novel streptococcal cysteine proteinase with unique specificity for immunoglobulin G. Embo J. 2002;21:1607–15. doi: 10.1093/emboj/21.7.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ishihara K, Wawrzonek K, Shaw LN, Inagaki S, Miyamoto M, Potempa J. Dentipain, a Streptococcus pyogenes IdeS protease homolog, is a novel virulence factor of Treponema denticola . Biol Chem. 2010;391:1047–55. doi: 10.1515/BC.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Schultz CP, Wolf V, Lange R, Mertens E, Wecke J, Naumann D, et al. Evidence for a new type of outer membrane lipid in oral spirochete Treponema denticola. Functioning permeation barrier without lipopolysaccharides. J Biol Chem. 1998;273:15661–6. doi: 10.1074/jbc.273.25.15661. [DOI] [PubMed] [Google Scholar]
- 135.Aliprantis AO, Yang RB, Mark MR, Suggett S, Devaux B, Radolf JD, et al. Cell activation and apoptosis by bacterial lipoproteins through toll-like receptor-2. Science. 1999;285:736–9. doi: 10.1126/science.285.5428.736. [DOI] [PubMed] [Google Scholar]
- 136.Nussbaum G, Ben-Adi S, Genzler T, Sela M, Rosen G. Involvement of Toll-like receptors 2 and 4 in the innate immune response to Treponema denticola and its outer sheath components. Infect Immun. 2009;77:3939–47. doi: 10.1128/IAI.00488-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Opitz B, Schroder NW, Spreitzer I, Michelsen KS, Kirschning CJ, Hallatschek W, et al. Toll-like receptor-2 mediates Treponema glycolipid and lipoteichoic acid-induced NF-kappaB translocation. J Biol Chem. 2001;276:22041–7. doi: 10.1074/jbc.M010481200. [DOI] [PubMed] [Google Scholar]
- 138.Schroder NW, Opitz B, Lamping N, Michelsen KS, Zahringer U, Gobel UB, et al. Involvement of lipopolysaccharide binding protein, CD14, and Toll-like receptors in the initiation of innate immune responses by Treponema glycolipids. J Immunol. 2000;165:2683–93. doi: 10.4049/jimmunol.165.5.2683. [DOI] [PubMed] [Google Scholar]
- 139.Choi BK, Lee HJ, Kang JH, Jeong GJ, Min CK, Yoo YJ. Induction of osteoclastogenesis and matrix metalloproteinase expression by the lipooligosaccharide of Treponema denticola . Infect Immun. 2003;71:226–33. doi: 10.1128/IAI.71.1.226-233.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Limberger RJ, Slivienski LL, Izard J, Samsonoff WA. Insertional inactivation of Treponema denticola tap1 results in a nonmotile mutant with elongated flagellar hooks. J Bacteriol. 1999;181:3743–50. doi: 10.1128/jb.181.12.3743-3750.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kataoka M, Li H, Arakawa S, Kuramitsu H. Characterization of a methyl-accepting chemotaxis gene, dmcA, from the oral spirochete Treponema denticola . Infect Immun. 1997;65:4011–6. doi: 10.1128/iai.65.10.4011-4016.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Li H, Arakawa S, Deng QD, Kuramitsu H. Characterization of a novel methyl-accepting chemotaxis gene, dmcB, from the oral spirochete Treponema denticola . Infect Immun. 1999;67:694–9. doi: 10.1128/iai.67.2.694-699.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fenno JC, Wong GWK, Hannam PM, McBride BC. Mutagenesis of outer membrane virulence determinants of the oral spirochete Treponema denticola . FEMS Microbiol Lett. 1998;163:209–15. doi: 10.1111/j.1574-6968.1998.tb13047.x. [DOI] [PubMed] [Google Scholar]
- 144.Ishihara K, Kuramitsu HK, Miura T, Okuda K. Dentilisin activity affects the organization of the outer sheath of Treponema denticola . J Bacteriol. 1998;180:3837–44. doi: 10.1128/jb.180.15.3837-3844.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Xu X, Kolodrubetz D. Construction and analysis of hemin binding protein mutants in the oral pathogen Treponema denticola . Res Microbiol. 2002;153:569–77. doi: 10.1016/s0923-2508(02)01370-0. [DOI] [PubMed] [Google Scholar]