Abstract

Oxidative stress triggers DNA and lipid peroxidation, leading to the formation of electrophiles that react with DNA to form adducts. A product of this pathway, (3-(2′-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-α]purine-10(3H)-one), or M1dG, is mutagenic in bacterial and mammalian cells and is repaired by the nucleotide excision repair pathway. In vivo, M1dG is oxidized to a primary metabolite, (3-(2-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-α]purine-6,10(3H,5H)-dione, or 6-oxo-M1dG, which is excreted in urine, bile, and feces. We have developed a specific monoclonal antibody against 6-oxo-M1dG and have incorporated this antibody into a procedure for the immunoaffinity isolation of 6-oxo-M1dG from biological matrices. The purified analyte is quantified by LC-MS/MS using a stable isotope-labeled analogue ([15N5]-6-oxo-M1dG) as an internal standard. Healthy male Sprague–Dawley rats excreted 6-oxo-M1dG at a rate of 350–1893 fmol/kg·d in feces. This is the first report of the presence of the major metabolite of M1dG in rodents without exogenous introduction of M1dG.
Introduction
M1dG (3-(2′-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-α]purine-10(3H)-one) is one of many endogenous exocyclic lesions derived from the reaction of DNA with bifunctional electrophiles generated by lipid, protein, or DNA peroxidation. Being an endogenous constituent of human and rodent genomes,1−3 M1dG is mutagenic in bacterial and mammalian cells4,5 and leads to mispairing when replicated in vitro by multiple different DNA polymerases.6 M1dG is repaired by nucleotide excision repair.4
We have previously developed a monoclonal antibody against M1dG7 and incorporated the antibody into analytical schemes using immunoaffinity purification of M1dG followed by mass spectrometric detection.3,8 These studies indicated that adult humans excrete M1dG at a rate of 12 fmol/kg·d in urine.8 Subsequent investigations into the metabolism and elimination of M1dG revealed that administration of M1dG to rodents at doses from 8 mg/kg to 6 pg/kg leads to its rapid disappearance from plasma due to oxidation and formation of the single, stable metabolite, 6-oxo-M1dG (3-(2-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-α]purine-6,10(3H,5H)-dione).9−11 In both humans and rats, xanthine oxidase appears to be responsible for this oxidation, while aldehyde oxidase also plays a role in M1dG metabolism in humans.10,11 Because 6-oxo-M1dG is the sole metabolite of M1dG in rats, it appears to be an ideal surrogate for M1dG as an in vivo biomarker for oxidative stress.
All studies of M1dG metabolism in vivo have been conducted with exogenously administered deoxynucleoside. Thus, it is not known if 6-oxo-M1dG is actually present in unadulterated intact animals. In order to address this question, we developed a highly specific monoclonal antibody (mAb) against 6-oxo-M1dG and covalently linked the antibody to sepharose beads. The resultant antibody-sepharose matrix (the gel) was used for the immunoaffinity purification of 6-oxo-M1dG from urine and feces followed by LC-MS/MS quantification against the stable isotope-labeled internal standard, [15N5]-6-oxo-M1dG. Utilizing this method, we report for the first time that 6-oxo-M1dG is endogenously produced in rodents and is excreted in urine and feces.
Materials and Methods
Na2CO3, NaHCO3, NaCl, water, acetonitrile, mariculture keyhole limpet hemocyanin (mcKLH), HCl, acetic acid, and methanol were obtained from Thermo Fisher Scientific (Waltham, MA, USA). ABTS (diammonium salt), thimerosal, KH2PO4, Na2HPO4, KCl, 30% H2O2, Tween 20, bovine serum albumin (BSA), dimethylformamide (DMF), dimethylsulfoxide (DMSO), K2CO3, sodium periodate, sodium methoxide, NaBH4, hypoxanthine, xanthosine, xanthine, ethyl cis-3-iodoacrylate, ethyl cis-3-bromoacrylate aminopterin, ethanolamine, cyanogen bromide-activated sepharose beads, diethylene glycol, 2′-deoxyguanosine, sodium hydroxide, formic acid, sodium azide, glycine, and citric acid were obtained from Sigma-Aldrich (St. Louis, MO, USA). Horseradish peroxidase-conjugated goat antimouse IgG (H + L) [0.8 mg/mL] and IgG (Fc) were purchased from Jackson ImmunoResearch (West Grove, PA, USA). Phosphate buffered saline (PBS) was purchased from Invitrogen (Carlsbad, CA, USA). Chemicals were used as received from these commercial sources unless specified. Solvents were HPLC-grade purity or higher.
Synthesis of 6-oxo-M1Guo
The synthesis of 6-oxo-M1Guo was based on a previously described synthetic scheme11 with some modifications. Guanosine was dissolved in a minimum amount of a DMF/DMSO mixture (1:1, v/v). K2CO3 (1.5 equiv) was added to the solution and the reaction mixture was held at 60–65 °C for 18 h. Ethyl cis-3-iodoacrylate (0.25 equiv per h for 5 h) was added to the reaction mixture. Solvents were evaporated using a high-vacuum rotary evaporator. The residue was dissolved into a minimum amount of methanol, and sodium methoxide (1.5 equiv) was added (dropwise in a 0.5 M solution). 6-oxo-M1Guo was purified on Biotage SP1 flash chromatography system (Biotage, Uppsala, Sweden) using the following gradient; 1% to 5% acetonitrile in 30 column volumes followed by a 5% to 15% gradient in 10 column volumes. Separation was achieved on a 12 + M C18 column, and the purified compound showed a single sharp peak by reverse-phase HPLC. 1H NMR (600 MHz, DMSO-d6): δ 8.65 (d, 1H, H8, J = 7.8 Hz), 8.29 (s, 1H, H2), 6.24 (d, 1H, H7, J = 8.4 Hz), 5.82 (d, 1H, H1′), 5.48 (d, 1H, H2′–OH), 5.20 (d, 1H, H3′–OH), 5.03 (d, 1H, H5′–OH), 4.49 (d, 1H, H2′), 4.15 (d, 1H, H3′), 3.91 (d, 1H, H4′), 3.63–3.55 (m, 2H, H5′). The 1H NMR may be seen in Figure A of the Supporting Information.
Synthesis of 6-oxo-M1dG and Stably Labeled Analogue
6-oxo-M1dG and its stable isotope-labeled analogue, [15N5]-6-oxo-M1dG, were synthesized as previously described.12 Briefly, anhydrous dG (1.6 g, 6 mmol) and anhydrous K2CO3 (0.93 g, 6.75 mmol) were dissolved in anhydrous DMF (18 mL). One milliliter of a 0.55 M solution of ethyl cis-3-bromoacrylate in anhydrous DMF was added to the reaction every 15 min for 4 h. The reaction was stirred for an additional 2 h then cooled and filtered. The filtrate was evaporated under high vacuum and the residue was dissolved in water (4 mL) and 5% acetic acid (4 mL). The product was purified by reversed-phase (C18) medium pressure liquid chromatography with a Biotage SP1 apparatus (Biotage) using a prepacked 25 × 75 mm Biotage FLASH 25 + S cartridge (KP-C18-HS) and a FLASH 25 C18 samplet for sample loading. A pure final product, 6-oxo-M1dG (0.92 g, 50.6%), was obtained. [15N5]-6-oxo-M1dG was synthesized by substituting [15N5]-dG for dG (Cambridge Isotope Laboratories, Andover, MA, USA) and employing the scheme described above. The 1H NMR of 6-oxo-M1dG and its stably labeled analogue matched the spectrum previously reported for this compound.11 The purified products both showed only one peak by reverse-phase HPLC. Mass spectrometric analysis of 6-oxo-M1dG showed an [M + H]+ peak at m/z 320, while analysis of [15N5]-6-oxo-M1dG showed an [M + H]+ peak at m/z 325. The m/z = 325 corresponds to the 6-oxo-M1dG value plus 5 additional mass units from the 5 15N atoms incorporated into the [15N5]-dG starting material. A m/z 320 peak was not observed for [15N5]-6-oxo-M1dG, indicating that no unlabeled 6-oxo-M1dG was present in the internal standard.
Conjugation of 6-oxo-M1Guo to BSA and mcKLH
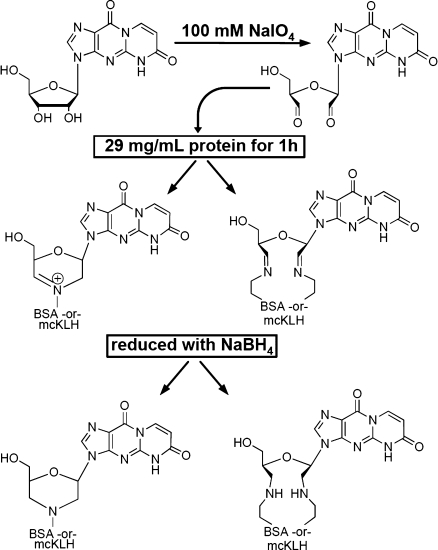
6-oxo-M1Guo (12 mg) was dissolved in 750 μL of 100 mM aqueous sodium periodate. Protein (20 mg of BSA or mcKLH) was reconstituted with 700 μL of PBS preadjusted to a pH of 9.5 with 5% K2CO3. The 6-oxo-M1Guo solution and the protein were combined and agitated. After 1 h, 45 μL of a 1 M diethylene glycol solution was added to the mixture (to quench excess oxidizing agent) followed by 700 μL of 0.45 M NaBH4 (aqueous). After another 12 h, the pH of the mixture was adjusted to ∼7.0 with 1.0 M formic acid. The mixture was kept at this pH for 1 h. Then, the pH was increased to 8.5 by the careful addition of 1 M aqueous ammonium hydroxide solution. This mixture was dialyzed against PBS buffer twice for 24 h. The sample was lyophilized and stored at 4 °C. Scheme 1 shows the conjugation reaction of 6-oxo-M1Guo with a lysine residue of the carrier protein. Figure B of the Supporting Information depicts the scheme for preparation of the conjugated protein.
Scheme 1. Conjugation Reaction between 6-oxo-M1Guo and the Carrier Protein (BSA or mcKLH).
Immunization and Hybridoma Preparation
Four BALB/cJ mice and four A/J mice (The Jackson Laboratory, Bar Harbor, ME, USA) were injected subcutaneously with 50 μg of 6-oxo-M1Guo-KLH and Freund’s complete adjuvant (primary boost). Four wk after the initial immunization, the mice were boosted subcutaneously (first boost) with the same dose of conjugate, but with the substitution of incomplete adjuvant (which was also used in subsequent boosts). Two wk after the first boost, the mice were tail bled and antibody titers were assessed by direct and competitive ELISA as described below. A second boost was administered 4 wk subsequent to the first, and after 2 wk, sera were again extracted and subjected to ELISA evaluation. A third boost was administered 25 wk after boost two, and the sera were collected and screened approximately 2 wk later. On the basis of the cumulative ELISA data, a single BALB/cJ mouse (BALB/cJ R) showing the most selective and concentrated anti-6-oxo-M1dG titer was chosen for a fourth and final boost given intraperitoneally and lacking adjuvant to prepare it for splenocyte extraction. Four days after the final boost, the mouse was sacrificed by cervical dislocation and the spleen harvested.
Splenocytes were isolated and subjected to polyethylene glycol-mediated fusion with both Sp/20 and NS1 murine myeloma cells (obtained from the University of Virginia and the University of Nebraska Medical Univeristy School of Dentistry, respectively) and allowed to recover for 24 h in liquid culture. The products of the fusion were evenly distributed into 24 96-well plates, with 12 plates corresponding to fusions of Sp/20 cells and splenocytes and 12 plates corresponding to fusions of NS1 cells and splenocytes. Cells were grown in Iscove’s Modified Dulbecco’s Medium with 20% Fetal Bovine Serum (Invitrogen). Hybridomas were selected by growing in the presence of aminopterin (5 × 10–8 M, Sigma) and HT supplement (1:100 dilution, Invitrogen) for 14 days with media and aminopterin replenishment every 3 days.
Supernatant was removed and screened for antigen-specific antibodies by ELISA, with 6-oxo-M1dGuo-BSA conjugate used as the antigen (described below). Positive hybridoma supernatants were rescreened in the presence of xanthine and 6-oxo-M1dG. Xanthine was employed as a competitor to identify antibodies not specific to 6-oxo-M1dG. Clones exhibiting anti-6-oxo-M1dG activity and specificity in addition to positive growth and productivity were selected and plated on 24-well plates. Of these, five clones were chosen based on activity, specificity, growth, and productivity as determined by direct and competitive ELISA analysis (described below) and then subsequently subjected to two rounds of subcloning. The clones were subsequently cryopreserved.
Note
Under Vanderbilt Institutional Care and Use Committee Protocol #M-07-109, the Vanderbilt Antibody and Protein Resource Core is permitted to use Freund’s complete adjuvant in the manner described above.
ELISA Procedure
The following solutions were prepared as follows and their use is described below: (a) carbonate-bicarbonate coating buffer (pH 9.6) was prepared from Na2CO3 (1.59 g/L), NaHCO3 (2.39 g/L), and thimerosal (0.10 g/L); (b) PBS-Tween (pH 7.4) was prepared from NaCl (8.00 g/L), KH2PO4 (0.20 g/L), Na2HPO4 (1.15 g/L), KCl (0.20 g/L), Tween 20 (1.00 mL/L), and thimerosal (0.10 g/L); (c) BSA (5.0 g) mixed in PBS-Tween (500 mL); (d) 1 nM ABTS solution in 70 mM citrate-phosphate buffer (pH 4.2) was prepared from citric acid (5.64 g/L), Na2HPO4 (5.84 g/L), and AzBTS-(NH4)2 (0.548 g).
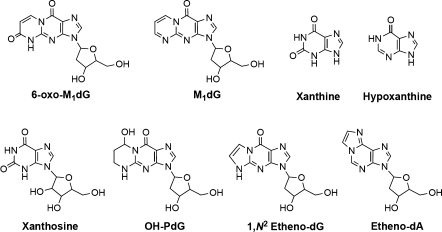
ELISA quality microtiter plates (Thermo Fisher Scientific; 96-well Immulon 2HB flat bottom microtiter plates or 384-well 4HB flat bottom plates) were coated with 50, 75, or 100 μL of a 10 μg/mL solution of 6-oxo-M1Guo-BSA antigen in carbonate-bicarbonate coating buffer (a) and incubated at 4 °C overnight. Plates were washed three times with 100 μL of PBS-Tween (b) using a Bio-Tek ELx405 automatic microplate washer (Winooski, VT, USA), then incubated with 100 μL of PBS-Tween (b) for 30 min at 37 °C. After 30 min, the PBS contents were discarded. Aliquots of murine serum dilutions, hybridoma supernatants, or purified antibody (depending on the stage of the antibody development process) were then incubated in the coated wells with or without the addition of varying concentrations of 6-oxo-M1dG and structural analogues (Figure 1) in a final volume of 100 μL PBS for 60 min at 37 °C. For wells that included 6-oxo-M1dG or structural analogues, the serum dilution used was 1:5000 in PBS-Tween (b), and the compounds depicted in Figure 1 were preincubated in the dilute murine serum for 45 min prior to the serum addition step. This allowed the nonspecific antibodies to bind the structural analogues, theoretically leaving behind only 6-oxo-M1dG-specific antibodies. Initial analysis revealed that xanthine was recognized nearly as well as 6-oxo-M1dG, which led to the use of this structural analogue in further competition studies.
Figure 1.
6-oxo-M1dG, M1dG, and structural analogues that were used as competitive antigens during ELISA analysis of murine sera, hybridomal supernatant, and purified antibodies.
The plates were then washed three times with PBS-Tween (b). Aliquots of diluted horseradish peroxidase-conjugated goat antimouse IgG Fc region-specific or IgG (H + L) secondary antibody (diluted 1:5000 in PBS-Tween/BSA) were added, and the plates were incubated at 37 °C for 60 min. After 60 min, the plates were washed three times and 100 μL of a freshly prepared ABTS solution (1.8 μL of H2O2 per 1 mL of ABTS) was immediately added to each well. To determine peroxidase activity, the absorbance at 414 nm was measured after 15 and 30 min for each well using a Bio-Tek Powerwave HT 340 plate reader with Gen5 software.
Antibody Purification and Isotype Determination
Selected clones were scaled up, inoculated into one-liter bioreactors (Wilson Wolf Manufacturing, New Brighton, MN, USA) and grown for 3–4 wk. Purification of mAb from the bioreactor supernatants was achieved by affinity chromatography on Protein-G sepharose (GE Healthcare, Piscataway, NJ, USA) followed by a final desalting step into PBS. Purified mAb were isotyped and subsequently quantified by SDS-PAGE electrophoresis followed by infrared Coomassie staining.
Linking Antibody to Sepharose Beads
Purified anti-6-oxo-M1dG mAb was loaded onto PD-10 columns (GE Healthcare) equilibrated with 25 mL coupling buffer (0.1 M NaHCO3, 0.5 M NaCl, pH 8.0 in water). The anti-6-oxo-M1dG mAb was eluted with 2.5 mL coupling buffer. Cyanogen bromide-activated sepharose beads (3.75 g) were prepared for conjugation by rapid swelling in 1 mM HCl followed by filtering, rinsing with 400 mL of 1 mM HCl and then 10 mL of coupling buffer. The sepharose beads were transferred to the antibody solution and gently rotated at 4 °C for about 20 h. The mixture was then filtered and washed with 500 mL of coupling buffer, and the residue was divided into four parts. Each quarter was placed in 14 mL of ethanolamine solution (1.0 M, pH 8.0) and gently rotated overnight at 4 °C. The sepharose beads were then filtered and washed with a low pH acetate buffer (0.1 M acetic acid, 0.1 M NaOH, and 0.5 M NaCl) followed by a 0.1 M Tris HCl buffer (0.5 M NaCl, pH 8.0). Each wash used 75 mL of buffer, and the wash cycle was repeated two more times. The resulting residue was suspended in 20 mL of 0.1 M Tris buffer with 0.5 M NaCl and 0.2% NaN3 (pH 8.0), and the final product (the gel) was stored at 4 °C.
6-oxo-M1dG Purification and Analysis
The gel was used to recover 6-oxo-M1dG and [15N5]- 6-oxo-M1dG from various fluids by the following general procedure. An aliquot of gel was prepared for use by rinsing with the following solutions: 50 mL of 0.1 M glycine (pH 2.7), 20 mL H20, 50 mL methanol, and 50 mL PBS. The gel was then resuspended in 3–5 mL of PBS. This cleaning procedure was necessary to remove any 6-oxo-M1dG that had bound to the mAb during preparation.
The pH of the fluid to be analyzed was adjusted to 7.0–8.0 with acetic acid or ammonium hydroxide. An aliquot of the washed and reconditioned gel was added to the sample, and the suspension was mixed gently for 1–4 h. The sample/gel suspension was filtered through a precleaned empty polypropylene SPE cartridge fitted with a polypropylene frit (Sigma-Aldrich). The filtered gel was washed with 10 mL of PBS, and the analytes were eluted with 2 mL of methanol. The eluant was dried under N2 gas, capped, and stored at −20 °C. The sample was reconstituted in 9:1 water/methanol (v/v) immediately prior to LC-MS/MS analysis.
Rat Urine and Fecal Sample Collection and Analysis
Male Sprague–Dawley rats were housed in metabolic cages. Urine and feces were collected at 24 h intervals and stored at −20 °C following collection. Urine samples were analyzed by thawing, adding 200–500 fmol of [15N5]-6-oxo-M1dG, and centrifuging the samples at 4000g for 25 min at 4 °C. An aliquot of supernatant was diluted 10–15-fold with PBS, and the pH of the final solution was adjusted to 7.5 ± 0.5 with either acetic acid or sodium hydroxide. The gel (prepared as described above) was introduced after pH adjustment, and the suspension was rotated gently end-over-end for 1–4 h. The suspension was filtered and the analyte eluted from the gel as described above.
Stored feces were thawed and weighed. The weighed sample was transferred to a clean vessel with 40 mL of PBS, and 200–500 fmol of [15N5]-6-oxo-M1dG were added. Dispersion of the sample was accomplished by rotating end-over-end for 2–3 h followed by sonication for 3–5 min. The suspension was then filtered through a mesh screen, with the collected solid material rinsed with 10 mL PBS. The filtrate was centrifuged at 4000g for 25 min at 4 °C. The supernatant was transferred to a clean vessel, diluted roughly 5 times with PBS, and the antibody gel (prepared as described above) was added. The suspension was rotated gently end-over-end for 1–4 h, filtered, and the analyte eluted from the gel as described above.
LC-MS/MS Analysis
Final LC-MS/MS analysis of gel-purified samples was accomplished on a Thermo Surveyor autosampler and MS pump in-line with a Thermo Quantum triple quadrupole mass spectrometer (Thermo Fisher Scientific). The mass spectrometer was equipped with an electrospray source and operated in positive ion mode. 6-oxo-M1dG and its internal standard, [15N5]-6-oxo-M1dG, were detected via selected reaction monitoring (SRM) with the following transitions, respectively; m/z 320 → 204 and 325 → 209. In both cases, the transition corresponds to cleavage of the glycosidic bond and loss of the deoxyribose moiety, with the positive charge remaining on the base.
The analytes were chromatographed on the following reverse-phase gradient system; 2% B to 15% B in 5 min, 15% B to 80% B in 1.5 min, followed by a 1 min hold at 80% B. Component A was water and component B was methanol/acetonitrile (3:1), and each component contained 0.1% acetic acid (v/v). The flow rate was 300 μL/min. The column used was a Phenomenex C18 (Phenomenex, Torrence, CA, USA; 10.0 × 0.2 cm, 3 μm) held at 35–40 °C. The column was equilibrated at the initial conditions for 3 min prior to each injection. Under this chromatographic regime, 6-oxo-M1dG eluted at approximately 5 min. 6-oxo-M1dG was quantitated by stable isotope dilution against [15N5]-6-oxo-M1dG. LC-MS/MS data was acquired and processed by Xcalibur software (Thermo Fisher Scientific).
Results
Selection and Purification of the Monoclonal Antibody
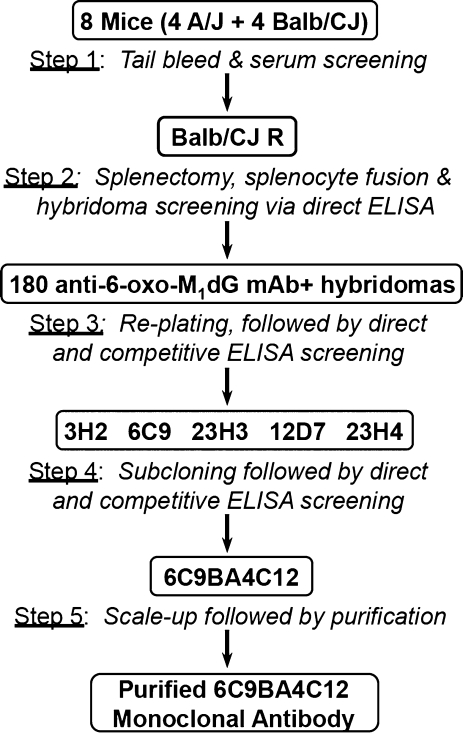
The overall scheme of the antibody generation and isolation process is depicted in Figure 2. Eight mice (four BALB/cJ and four A/J) were injected with 6-oxo-M1G conjugated to KLH as described in the Materials and Methods section. At 6, 10, and 35 wk after the initial innoculation, the mice were tail bled, and direct and competitive ELISA analyses of the sera were performed (Figure 2, step 1). All three bleeds from all 8 mice showed robust responses against the 6-oxo-M1G-BSA antigen, indicating that the immune systems of the mice were producing significant amounts of antibodies against the 6-oxo-M1G portion of the innoculant. Preinnoculation tail bleeds detected no antibodies against 6-oxo-M1G. Competitive ELISA screenings were executed to assess antibody specificity, with 6-oxo-M1dG and a series of structural analogues (Figure 1) serving as competing antigens.
Figure 2.
Schematic of monoclonal antibody generation and isolation procedure. Eight mice were inoculated with adduct/protein conjugate, followed by tail bleeding and testing for the presence of 6-oxo-M1dG antibodies using direct ELISA analysis (step 1). Splenocytes from a single mouse, Balb/CJ R, were fused with myeloma cells, plated on twenty-four 96-well plates, and screened using direct and competitive ELISA analyses (step 2). From this screen, 180 hybridoma cell lines showed anti-6-oxo-M1dG mAb production. These were replated and screened (step 3). Five parental cell lines were chosen for subcloning and further screening (step 4). On the basis of these final screenings, the cell line 6C9BA4C12 was chosen for scale-up and antibody purification (step 5).
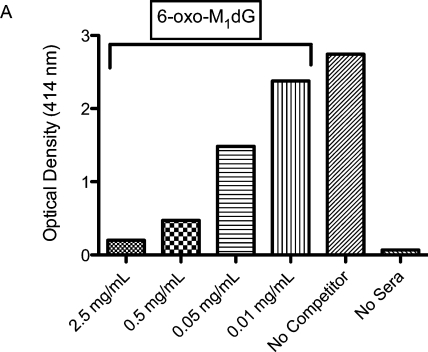
Figure 3 depicts a competitive ELISA analysis of the 35-week serum from mouse BALB/cJ R. This analysis employed 6-oxo-M1dG as the competing antigen and a robust concentration dependent decrease in optical density is observed, indicating the presence of anti-6-oxo-M1dG antibodies in the sera from this mouse. The results of similar analyses of the sera of all 8 inoculated mice can be seen in Supporting Information Figure C. Supporting Information Figure D displays the results from competitive ELISA analyses of the 35-week serum from all 8 inoculated mice where M1dG is employed as the competing antigen. Increasing amounts of M1dG do not result in a decrease in optical density, indicating that M1dG does not bind to the anti-6-oxo-M1dG antibodie(s) present in the sera.
Figure 3.
6-oxo-M1dG antibodies are present in the serum of mouse BALB/cJ R. The serum of mouse BALB/cJ R was subjected to competitive ELISA analysis with 6-oxo-M1dG used as the competitor. The displayed results represent the average of spectroscopic readings at 15 and 30 min postaddition of ABTS substrate.
On the basis of the ELISA analysis of murine sera, a single mouse (BALB/cJ R) was chosen for hybridoma production. Splenocytes from BALB/cJ R were fused with both Sp/20 and NS1 myeloma cells (Figure 2, step 2), and 1670 hybridomas were formed from 2304 fusions wells. ELISA analysis revealed that of the 1670 hybridomas, 180 showed production of anti-6-oxo-M1dG antibodies. These were grown in 24-well plates (Figure 2, step 3), and the supernatants were subjected to further ELISA screenings. On the basis of the screening data, five hybridomas were selected for subcloning and further screenings (Figure 2, step 4).
Figure 4 shows a representative ELISA analysis of the supernatants from two subclones (B and E2) of parental hybridoma cell line 6C9 in which xanthine and 6-oxo-M1dG are employed as competing antigens. The presence of unbound 6-oxo-M1dG decreases the optical density of both B and E2 supernatants, indicating the presence of anti-6-oxo-M1dG antibodies. For subclone E2, xanthine has a similar effect, demonstrating that E2’s mAb are not specific to 6-oxo-M1dG. However, xanthine has no effect on the optical density of subclone B supernatant, suggesting that mAb from subclone B are specific to 6-oxo-M1dG. On the basis of these data, subclone B was subjected to further subcloning but E2 was not. Supporting Information Figure E illustrates that cell line 6C9BA4, a subclone of hybridoma 6C9B, retained activity against 6-oxo-M1dG but did not react to xanthine.
Figure 4.
ELISA analysis of specificity of various subclonal antibodies. The specificity of antibodies produced by two hybridoma cell lines (B and E2) subcloned from the parental hybridoma cell line 6C9 were assessed using competitive ELISA analysis. Xanthine and 6-oxo-M1dG were used as competing antigens. The displayed results represent the average of spectroscopic readings of subclones E2 and B at 15 and 30 min postaddition of ABTS substrate.
On the basis of a review of the cumulative ELISA data, a final daughter cell line was selected from each parental hybridoma line, and these daughter cell lines were cryopreserved. The ELISA data indicated that antibodies from the cell line 6C9BA4C12 displayed the most promising expression and specificity. Thus, this cell line was subjected to large-scale expression and antibody purification. The final antibody was isotyped as IgG1-kappa.
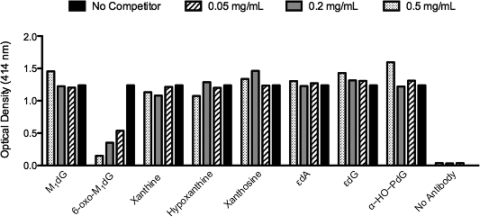
Figure 5 shows the ELISA responses of the mAb purified from 6C9BA4C12 in the presence of 6-oxo-M1dG and structural analogues. Only 6-oxo-M1dG attenuated the optical density, indicating a specificity of the purified mAb for 6-oxo-M1dG over close structural analogues.
Figure 5.
Specificity determination of antibodies produced from subclone 6C9BA4C12. Purified antibodies from the hybridomal subclone (6C9BA4C12) were screened for specificity for 6-oxo-M1dG in the presence of structural analogues using competitive ELISA analyses.
Performance of Covalently Linked mAb Gel and LC-MS/MS Analysis
The purified anti-6-oxo-M1dG mAb was covalently linked to sepharose beads as described above. The purification and analysis protocol outlined in Materials and Methods was used to quantify aliquots of 10 mM potassium phosphate (pH 8.0) spiked with varying amounts of 6-oxo-M1dG (0.25, 2.5, and 25 pmol) and 1.0 pmol of the internal standard, [15N5]-6-oxo-M1dG (n = 3 for each concentration of 6-oxo-M1dG). As shown in Table 1, the experimentally determined amount of 6-oxo-M1dG was within 12% of the known amount at each level. The average percent recovery of 6-oxo-M1dG ranged from 94% to 74%, while the internal standard was recovered at an average rate of 79%. Thus, the gel was able to bind 6-oxo-M1dG from an aqueous solution and released the analyte in the presence of methanol.
Table 1. Results of the Analysis of Buffer Spiked with Varying Amounts of 6-oxo-M1dG and 1.0 pmol of the Internal Standard [15N]-6-oxo-M1dGa.
| 6-oxo-M1dG per sample (pmol) | amount detected (pmol) | % accuracy | % recovery |
|---|---|---|---|
| 0.25 | 0.28 ± 0.02 | 11.7 ± 7.7% | 93.5 ± 7.2% |
| 2.5 | 2.73 ± 0.2 | 9.0 ± 6.0% | 82.2 ± 6.0% |
| 25.0 | 26.18 ± 0.6 | 4.7 ± 2.2% | 74.3 ± 2.5% |
The amount detected, % accuracy, and % recovery values are shown as the mean ± S.D. (n = 3 in all cases). The % recovery of the internal standard was 78.7 ± 3.4%.
The limit of detection (LOD) of the described LC-MS/MS system was established at approximately 10 fmol 6-oxo-M1dG on-column (Supporting Information Figure F, top) by analyzing a series of increasingly dilute 6-oxo-M1dG solutions. Given this LOD and the percent recovery observed from urine and fecal samples (approximately 30%), the limit of quantification of the assay was estimated to be 50 fmol per sample. Additionally, the LC-MS/MS system provided a linear response to 6-oxo-M1dG solutions over a 5000-fold concentration range and a single [15N5]-6-oxo-M1dG concentration (Supporting Information F, bottom).
These results indicate that the gel provided a sufficient level of recovery of the analyte and its internal standard as well as sufficient capacity for the expected levels of 6-oxo-M1dG present in rat urine and feces. They also demonstrate that the LC-MS/MS analysis of the purified sample provided sensitive and accurate quantification.
Analysis of Rat Urine and Feces for the Presence of 6-oxo-M1dG
Urine and feces collected from male Sprague–Dawley rats over 3 days were analyzed as described in the Materials and Methods section. Table 2 lists the results of the analysis. 6-oxo-M1dG was detected in urine from only one subject, animal 3, at a rate of 188 fmol/kg·d. However, 6-oxo-M1dG was eliminated in feces in all the animals at a rate of 350–1893 fmol/kg·d. Thus, it appears fecal elimination is the main route by which that 6-oxo-M1dG is excreted from the body.
Table 2. Amount of 6-oxo-M1dG Eliminated by Sprague–Dawley Rats via Urine and Fecesa.
| subject | urine | feces |
|---|---|---|
| animal 1 | <LOQ | 1893 ± 1963 |
| animal 2 | <LOQ | 350 ± 314 |
| animal 3 | 188 ± 62 | 534 ± 404 |
Values are given as the mean amount observed in fmol/kg·d ± S.D. (n = 3–5 for feces and 2 for urine).
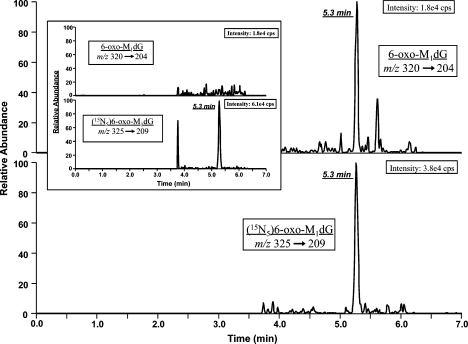
While the amounts of 6-oxo-M1dG are low, the assay provided a sufficient signal-to-noise ratio to permit accurate quantification. Figure 6 displays a sample chromatogram from a processed fecal sample. The upper trace (m/z 320 → 204) shows the analyte, while the bottom trace (m/z 325 → 209) represents the [15N5]-6-oxo-M1dG internal standard. Both compounds gave chromatographic peaks that were well above the background noise. The inset of Figure 6 is the internal standard alone after recovery from PBS. There is no peak in the 6-oxo-M1dG trace of the inset, which is illustrative of the fact that there was no isotopic impurity in the internal standard that could contribute to the 6-oxo-M1dG signal.
Figure 6.
6-oxo-M1dG is present in rat feces. A representative LC-MS/MS chromatogram of 6-oxo-M1dG and the internal standard [15N5]-6-oxo-M1dG isolated from rat feces is displayed. The inset represents a chromatogram of internal standard alone, purified from PBS.
Discussion
M1dG is an endogenous DNA adduct detectable in the genomic DNA of humans and rodents.2,3 Its presence is associated with peroxidative damage to cellular constituents, particularly polyunsaturated fatty acids, and DNA. Although many studies have shown that the lipid peroxidation product, malondialdehyde, can react in vitro with deoxyguanosine to form M1dG,1 recent studies in E. coli, with defined polyunsaturated fatty acid content in membrane phospholipids, indicate that this is not a major source of M1dG.13 Rather, the DNA peroxidation products, base propenals, appear to account for nearly all of the M1dG generated when bacteria are exposed to peroxynitrite (ONOO–).13 Thus, although M1dG appears to be a reliable marker of oxidative damage to cells, it primarily reflects oxidative damage to DNA.
M1dG is repaired by nucleotide excision repair4 and is excreted in urine at rather low levels in humans (12 ± 3.8 fmol/kg·d).8 A major contributor to the low levels of M1dG is its oxidative metabolism to 6-oxo-M1dG.9−11 This suggests 6-oxo-M1dG may be superior to M1dG as a noninvasive marker of oxidative damage. In the present manuscript, we describe the generation of a monoclonal antibody that exhibits sensitive and specific binding to 6-oxo-M1dG and its use in a quantitative assay suitable for analysis of urine and feces.
In the assay, the sepharose-bound mAb is used to isolate 6-oxo-M1dG from urine and feces. The purified sample is subjected to LC-MS/MS analysis where 6-oxo-M1dG is quantified via stable isotope dilution against the synthetic internal standard, [15N5]-6-oxo-M1dG, which is introduced to the sample prior to antibody purification.
The antibody gel shows excellent recovery of 6-oxo-M1dG from buffer solutions. The analyte is recovered at 74% or greater from 10 mM potassium phosphate over a 100-fold range of 6-oxo-M1dG, 0.25 pmol to 25.0 pmol. The internal standard was recovered at 79% (1.0 pmol) from these same solutions, and spiked buffer solutions were quantified accurately to ±12%. These data demonstrate that 100 μL of the antibody gel is able to bind at least 26 pmol of 6-oxo-M1dG from solution, and LC-MS/MS detection of the purified sample is quantitative.
The recovery of [15N5]-6-oxo-M1dG from saline solutions was greater than 78% but its recovery from urine and feces was only ∼29%. This suggests a matrix effect where other nucleosides, deoxynucleosides, and/or endogenous congeners compete with 6-oxo-M1dG for antibody binding sites. Although there was minimal cross-reactivity of the antibody with individual members of a panel of exocyclic adducts and purine oxidation products (Figures 1 and 5), it is possible that high levels of low-avidity nucleosides in urine and feces compete with the low levels of endogenous 6-oxo-M1dG for binding sites on the mAb.
6-oxo-M1dG was detected in the feces of each of three different Sprague–Dawley rats but was detected in the urine of only one of the rats (Table 2). The fecal levels of 6-oxo-M1dG (350–1893 fmol/kg·day) were significantly higher than the urinary levels (188 fmol/kg·day). This is consistent with previous findings that demonstrate M1dG is excreted mainly in urine, whereas 6-oxo-M1dG is excreted mainly in feces.10,11
Because 6-oxo-M1dG is present endogenously in rats, it suggests that its metabolic precursor, M1dG, is also produced endogenously in rats and metabolized to 6-oxo-M1dG. This idea is consistent with the finding that M1dG is present in rodent and human genomic DNA2,3 and with studies from this laboratory demonstrating that 6-oxo-M1dG is the sole metabolite of M1dG.10,11,14
It may be argued that endogenous 6-oxo-M1dG is generated in the nucleoside pool rather than arising from direct damage to DNA. However, this seems unlikely if one considers that base propenals are the principal precursors to M1dG.13 Base propenal formation is triggered by abstraction of the hydrogen from the C-4′ position of the deoxyribosyl unit of double-stranded DNA. The base propenal produced then reacts with a deoxyguanosine residue in duplex DNA to form M1dG. In comparison, the yields of base propenals are much lower when deoxynucleosides are oxidized instead of duplex DNA. Furthermore, M1dG formation as a reaction of base propenals and free deoxyguanosine is a bimolecular process. If base propenals are formed in duplex DNA, they are generated in close proximity to deoxyguanosine residues in the DNA duplex, which should maximize the probability of their reaction with a deoxyguanosine residue. This is not the case for base propenals generated in the nucleoside pool.
Another possible source of both 6-oxo-M1dG and M1dG is diet. One or both of these compounds may have been ingested, then excreted or metabolized. It was not possible to control for this possibility under the reported experimental regime, so future studies with animals fed nucleic acid-free diets will be required to test this hypothesis.
The ability to measure excreted 6-oxo-M1dG has the potential for clinical relevance in assessing the levels of oxidative DNA damage in humans. Since the basal levels of both deoxynucleoside adducts are low, they should provide high sensitivity to increases triggered by oxidative damage. Given that the analytical method described herein is applicable to feces and urine, the possibility exists for preclinical or clinical studies conducted in a noninvasive fashion.
Glossary
Abbreviations
- M1dG
3-(2′-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-α]purine-10(3H)-one)
- 6-oxo-M1dG
3-(2-deoxy-β-d-erythro-pentofuranosyl)-pyrimido[1,2-f]purine-6,10(3H,5H)-dione)
- ABTS
2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)
- equiv
equivalents
- mAb
monoclonal antibody
- mcKLH
mariculture keyhole limpet hemocyanin
- ELISA
Enzyme-Linked Immunosorbent Assay
- BSA
bovine serum albumin
- PBS
phosphate-buffered saline
- LC-MS/MS
liquid chromatography-tandem mass spectrometry
- SRM
selected reaction monitoring
Supporting Information Available
Proton NMR of 6-oxo-M1Guo; scheme of the generation of the 6-oxo-M1Guo/protein conjugate; sera reactivity with 6-oxo-M1dG; lack of reactivity of sera with M1dG; specificity of sera antigens for 6-oxo-M1dG; limit of detection and linearity of the LC-MS/MS system. This material is available free of charge via the Internet at http://pubs.acs.org.
This work was supported by NIH grant R37 CA087819 (to L.J.M.).
Author Contributions
⊥ These authors contributed equally to the manuscript.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Basu A. K.; O’Hara S. M.; Valladier P.; Stone K.; Mols O.; Marnett L. J. (1988) Identification of adducts formed by reaction of guanine nucleosides with malondialdehyde and structurally related aldehydes. Chem. Res. Toxicol. 1, 53–59. [DOI] [PubMed] [Google Scholar]
- Chaudhary A. K.; Nokubo M.; Reddy G. R.; Yeola S. N.; Morrow J. D.; Blair I. A.; Marnett L. J. (1994) Detection of endogenous malondialdehyde-deoxyguanosine adducts in human liver. Science 265, 1580–1582. [DOI] [PubMed] [Google Scholar]
- Rouzer C. A.; Chaudhary A. K.; Nokubo M.; Ferguson D. M.; Reddy G. R.; Blair I. A.; Marnett L. J. (1997) Analysis of the malondialdehyde-2′-deoxyguanosine adduct pyrimidopurinone in human leukocyte DNA by gas chromatography/electron capture/negative chemical ionization/mass spectrometry. Chem. Res. Toxicol. 10, 181–188. [DOI] [PubMed] [Google Scholar]
- Fink S. P.; Reddy G. R.; Marnett L. J. (1997) Mutagenicity in Escherichia coli of the major DNA adduct derived from the endogenous mutagen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 94, 8652–8657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanderVeen L. A.; Hashim M. F.; Shyr Y.; Marnett L. J. (2003) Induction of frameshift and base pair substitution mutations by the major DNA adduct of the endogenous carcinogen malondialdehyde. Proc. Natl. Acad. Sci. U.S.A. 100, 14247–14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddukuri L.; Eoff R. L.; Choi J. Y.; Rizzo C. J.; Guengerich F. P.; Marnett L. J. (2010) In vitro bypass of the major malondialdehyde- and base propenal-derived DNA adduct by human Y-family DNA polymerases kappa, iota, and Rev1. Biochemistry 49, 8415–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevilla C. L.; Mahle N. H.; Eliezer N.; Uzieblo A.; O’Hara S. M.; Nokubo M.; Miller R.; Rouzer C. A.; Marnett L. J. (1997) Development of monoclonal antibodies to the malondialdehyde-deoxyguanosine adduct, pyrimidopurinone. Chem. Res. Toxicol. 10, 172–180. [DOI] [PubMed] [Google Scholar]
- Hoberg A. M.; Otteneder M.; Marnett L. J.; Poulsen H. E. (2004) Measurement of the malondialdehyde-2′-deoxyguanosine adduct in human urine by immuno-extraction and liquid chromatography/atmospheric pressure chemical ionization tandem mass spectrometry. J. Mass Spectrom. 39, 38–42. [DOI] [PubMed] [Google Scholar]
- Knutson C. G.; Akingbade D.; Crews B. C.; Voehler M.; Stec D. F.; Marnett L. J. (2007) Metabolism in vitro and in vivo of the DNA base adduct, M1G. Chem. Res. Toxicol. 20, 550–557. [DOI] [PubMed] [Google Scholar]
- Knutson C. G.; Skipper P. L.; Liberman R. G.; Tannenbaum S. R.; Marnett L. J. (2008) Monitoring in vivo metabolism and elimination of the endogenous DNA adduct, M1dG {3-(2-deoxy-beta-d-erythro-pentofuranosyl)pyrimido[1,2-alpha]purin-10(3H)-one}, by accelerator mass spectrometry. Chem. Res. Toxicol. 21, 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otteneder M. B.; Knutson C. G.; Daniels J. S.; Hashim M.; Crews B. C.; Remmel R. P.; Wang H.; Rizzo C.; Marnett L. J. (2006) In vivo oxidative metabolism of a major peroxidation-derived DNA adduct, M1dG. Proc. Natl. Acad. Sci. U.S.A. 103, 6665–6669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szekely J.; Wang H.; Peplowski K. M.; Knutson C. G.; Marnett L. J.; Rizzo C. J. (2008) ″One-pot″ syntheses of malondialdehyde adducts of nucleosides. Nucleosides, Nucleotides Nucleic Acids 27, 103–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou X.; Taghizadeh K.; Dedon P. C. (2005) Chemical and biological evidence for base propenals as the major source of the endogenous M1dG adduct in cellular DNA. J. Biol. Chem. 280, 25377–25382. [DOI] [PubMed] [Google Scholar]
- Knutson C. G.; Wang H.; Rizzo C. J.; Marnett L. J. (2007) Metabolism and elimination of the endogenous DNA adduct, 3-(2-deoxy-beta-d-erythro-pentofuranosyl)-pyrimido[1,2-alpha]purine-10(3H)-one, in the rat. J. Biol. Chem. 282, 36257–36264. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.