Abstract
Fructose-bisphophate aldolase (FbaB), is an enzyme in glycolysis and gluconeogenesis in living organisms. The mutagenesis in a unique fbaB gene of Xanthomonas oryzae pv. oryzicola, the causal agent of rice bacterial leaf streak, led the pathogen not only unable to use pyruvate and malate for growth and delayed its growth when fructose was used as the sole carbon source, but also reduced extracellular polysaccharide (EPS) production and impaired bacterial virulence and growth in rice. Intriguingly, the fbaB promoter contains an imperfect PIP-box (plant-inducible promoter) (TTCGT-N9-TTCGT). The expression of fbaB was negatively regulated by a key hrp regulatory HrpG and HrpX cascade. Base substitution in the PIP-box altered the regulation of fbaB with the cascade. Furthermore, the expression of fbaB in X. oryzae pv. oryzicola RS105 strain was inducible in planta rather than in a nutrient-rich medium. Except other hrp-hrc-hpa genes, the expression of hrpG and hrpX was repressed and the transcripts of hrcC, hrpE and hpa3 were enhanced when fbaB was deleted. The mutation in hrcC, hrpE or hpa3 reduced the ability of the pathogen to acquire pyruvate and malate. In addition, bacterial virulence and growth in planta and EPS production in RΔfbaB mutant were completely restored to the wild-type level by the presence of fbaB in trans. This is the first report to demonstrate that carbohydrates, assimilated by X. oryzae pv. oryzicola, play critical roles in coordinating hrp gene expression through a yet unknown regulator.
Introduction
Carbohydrate nutrient acquisition is essential for bacterial pathogen growth to establish successful infections in host plants [1], [2], [3]. As in other living organisms, plant pathogenic bacteria carry out the catabolic process via the Emden-Meyerhof-Parnas (EMP) pathway of glycolysis, Entner–Doudoroff (ED), pentose phosphate pathway (PPP) and terminal oxidation mediated by the tricarboxylic acid (TCA) cycle to break down hexoses, like glucose, outside of their cells for energy and carbon molecules. Bacteria may also use gluconeogenesis to synthesize glucose from non-sugar C2 or C3 compounds or the intermediates of the TCA cycle when there is not sufficient hexoses in their immediate environment [4]. In Xanthomonas, ED, in conjunction with TCA, has been confirmed to be the predominant pathway for glucose catabolism, and a small portion (8 to 16%) of substrate glucose is routed into PPP [5], whereas the EMP pathway of glycolysis does not play a significant role in glucose catabolism, since Xanthomonas species, including rice bacterial leaf streak X. oryzae pv. oryzicola, lack an essential phosphofructokinase activity which converts fructose 6-phosphate to fructose-1,6-bisphosphate [5], [6], [7]. Moreover, little is known about the relationship of carbon metabolism to virulence.
The genome data of X. oryzae pv. oryzicola (http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=Xoc), X. oryzae pv. oryzae [8], [9], X. campestris pv. campestris [10], X. axonopodis pv. citri [11] and X. campestris pv. vesicatoria [12] show that xanthomonads possess essential genes for the EMP pathway of glycolysis, ED, PPP, gluconeogenesis and TCA cycle. Currently, great interests have been focused on whether or not and how the carbon metabolic pathways are involved in the virulence of plant pathogenic bacteria. For example, the glyceraldehyde-3-phosphate dehydrogenase (GAPDH), converting glyceraldehyde 3-phosphate to 1,3-bisphosphoglycerate, is required for ED, extracellular polysaccharide (EPS) production and full virulence of X. campestris pv. campestris [13]. The phosphogluconate dehydratase gene (edd) in ED is necessary for xanthan biosynthesis and the 6-phosphogluconate dehydrogenase gene (gndA) in PPP does not influence xanthan biosynthesis in X. oryzae pv. oryzae [6]. The malate: quinone oxidoreductase gene (mqo) in TCA cycle is required for the wild-type growth, disease symptom development and full virulence of Pseudomonas syringae pv. tomato DC3000 in Arabidopsis thaliana [14]. The phosphoenolpyruvate synthase gene (ppsA), converting pyruvate to phosphoenolpyruvate, is essential for gluconeogenesis, in planta growth, and full virulence of X. campestris pv. campestris [4]. However, little is known about other carbon metabolic factors.
Previous reports have confirmed that some carbohydrates and sulfur-containing amino acids have the ability to induce the expression of hrp genes in Gram-negative phytopathogenic bacteria [15], [16], [17], [18]. The hrp genes, normally within a 25–27 kb gene cluster in Xanthomonas species, encoding a type-III secretion system (T3SS), enable bacterial pathogens to trigger a rapid, localized, programmed hypersensitive response (HR) in nonhost plants and become pathogenic in hosts [18], [19], [20]. Expression of hrp genes is actually suppressed in nutrient-rich media but induced in planta and in apoplast-mimicking media, XVM2 containing sucrose and fructose for X. campestris pathovars or species [17], [18], [21], [22], [23]; XOM3 only containing xylose for X. oryzae pathovars [20], [24], [25], except inorganic salt(s), implying that some nutrients released from plant tissues, which are degraded for bacterial growth, may have effects on induction of hrp gene expression. For instance, the hrp expression in Ralstonia solanacearum is activated possibly by ubiquitous and non-diffusible molecules in the presence of pathogen-plant cell contact [15], [16], [26]. The above prompts us to assume that there are unknown correlations between carbon metabolism and the hrp system for bacterial pathogenesis in plants.
When Xanthomonas species interact with plants, some of the hrp gene products generate a pedestal-like T3S structure that traverses the two bacterial membranes [27], [28]. For example, a pilus-like secretion channel (HrpE), which is outside of HrcC [29], and also a translocon protein (HrpF) in the plant membrane [27], [30], [31], [32], [33]. As a whole, the T3S apparatus injects a number of effectors into the apoplast and cytosol of plant cells leading to disease in hosts or HR in non-hosts. Conceptually, expression of the hrp genes is controlled by two key regulatory genes, hrpG and hrpX, which are located outside of the hrp gene cluster [19]. HrpG is predicted to be an OmpR-type response regulator of a two-component signal transduction system and presumably perceives an environmental signal via an unknown sensor kinase [34], [35]. HrpX is an AraC-type of transcriptional activator [36] which forms a homodimer containing a helix-turn-helix domain which interacts with each TTCGC motif of the PIP-box (plant-inducible promoter) in the hrpB to hrpF promoter regions to activate transcription of hrp [26], [37], [38], [39] and T3S effector genes [34], [35], [40]. The PIP-box has been taken to identify novel HrpX regulons which possess a PIP-box upstream of a 30–32 base pairs followed by a conserved -10 box-like sequence, as TTCGB-N15–16-TTCGB- N30–32-YANNNT (B refers to the base C, G, or T but not A) in the promoter region [36], [41], [42], [43]. But few genes, like hrpF with an imperfect PIP-box (TTCGC-N8-TTCGT) or without following the -10 box-like motif in the promoter region, have been described as being expressed in a HrpX-dependent manner [38], [39]. Recently, the coordinated expression of Xanthomonas hrp-hrc-hpa expression is orchestrated by multiple two-component systems and transcriptional regulators such as Trh [44], Clp [45], Zur [40], LrpX [46], ColR/S [47], and PhoP/Q [48]. However, the expression of hrcC, hrpE and hpa3 genes is not obviously and completely controlled by these regulators, including HrpG and HrpX, in X. oryzae pv. oryzicola when the pathogen grows in hrp-inducing medium and in planta [20], implying that unknown regulator(s) may play roles in hrp gene expression.
To investigate uncertainty above, we screened our previous Tn5-tagged mutant library of X. oryzae pv. oryzicola [49] and got a mutant Mxoc0504 where the Tn5 was inserted in a unique gene fbaB. In this report, we present genetic evidence demonstrating that fbaB is required for gluconeogenesis, EPS production and the expression of hrp genes, as well as the full virulence of X. oryzae pv. oryzicola in rice.
Materials and Methods
Bacterial strains, culture media and growth conditions
Strains and plasmids used in this study are listed in Table 1. Escherichia coli strains were routinely grown in LB (Luria-Bertani) medium at 37°C [50]. X. oryzae pv. oryzicola strains were performed at 28°C in NA (1 g/L yest extract, 3 g/L beef extract, 5 g/L polypeptone, 10 g/L sucrose, 15 g/L agar), NB (NA without agar), NAN (NA without sucrose) or NAS (NA with 100 g/L sucrose), NY (NB without beef extract and sucrose), the non-carbohydrate minimal medium (NCM) (2 g/L (NH4)2SO4, 4 g/L K2HPO4, 6 g/L KH2PO4, 0.2 g/L MgSO4 · 7H2O) [4] or rice suepension cells [25] when required. Antibiotics were used when required at the following concentrations: kanamycin (Kan), 25 µg/ml; rifampicin (Rif), 50 µg/ml; ampicillin (Amp), 100 µg/ml and spectinomycin (Sp), 50 µg/ml.
Table 1. Strains and plasmids used in this study.
| Strains or plasmids | Relevant characteristicsa | Reference or source |
| E. coli | ||
| DH5α | F— Φ80dlacZ ΔM15Δ(lacZYA-argF)U169 endA1 deoR recA1 hsdR17(rK — mK +) phoA supE44 λ— thi-l gyrA96 relA1 | Clontech |
| S17-1λpir | recA,Thi, pro, hsdR−, M+, RP4: 2-Tc::Mu:KmTn7, λpir, Tpr, Smr | This lab |
| X. oryzae pv. oryzicola | ||
| RS105 | Wild type, the causal agent of bacterial leaf streak in rice, Rifr | This lab |
| RΔhrpG | A hrpG knock-out mutant of strain RS105, Rifr | [53] |
| RΔhrpX | A hrpX deletion mutant of strain RS105, Rifr | [53] |
| RΔhrcV | A hrcV deletion mutant of strain RS105, Rifr | [65] |
| RΔhrpE | A hrpE deletion mutant of strain RS105, Rifr | [65] |
| RΔhrcC | A hrcC deletion mutant of strain RS105, Rifr | [20] |
| RΔhpa3 | A hpa3 deletion mutant of strain RS105, Rifr | [20] |
| Mxoc0504 | A Xoryp_17640::Tn5 inserted mutant, Rifr, Kmr | This work |
| RΔfbaB | A fbaB deletion mutant of strain RS105, Rifr | This work |
| CRΔfbaB | RΔfbaB harboring pCfbaB, Rifr, Spr | This work |
| RS105(pfbaBaGUS) | The wild-type RS105 harboring pfbaBaGUS, Rifr, Spr | This work |
| RS105(pfbaBbGUS) | The wild-type RS105 harboring pfbaBbGUS, Rifr, Spr | This work |
| RS105(pfbaBcGUS) | The wild-type RS105 harboring pfbaBcGUS, Rifr, Spr | This work |
| RS105(pfbaBdGUS) | The wild-type RS105 harboring pfbaBdGUS, Rifr, Spr | This work |
| RΔhrpG(pfbaBaGUS) | RΔhrpG mutant harboring pfbaBaGUS, Rifr, Spr | This work |
| RΔhrpG(pfbaBbGUS) | RΔhrpG mutant harboring pfbaBbGUS, Rifr, Spr | This work |
| RΔhrpG(pfbaBcGUS) | RΔhrpG mutant harboring pfbaBcGUS, Rifr, Spr | This work |
| RΔhrpG(pfbaBdGUS) | RΔhrpG mutant harboring pfbaBdGUS, Rifr, Spr | This work |
| RΔhrpX(pfbaBaGUS) | RΔhrpX mutant harboring pfbaBaGUS, Rifr, Spr | This work |
| RΔhrpX(pfbaBbGUS) | RΔhrpX mutant harboring pfbaBbGUS, Rifr, Spr | This work |
| RΔhrpX(pfbaBcGUS) | RΔhrpX mutant harboring pfbaBcGUS, Rifr, Spr | This work |
| RΔhrpX(pfbaBdGUS) | RΔhrpX mutant harboring pfbaBdGUS, Rifr, Spr | This work |
| Plasmids | ||
| pMD18-T | pUC ori, cloning vector, Apr | Takara |
| pKMS1 | Suicide vector derivative from pK18mobGII, sacB+, Kmr | This lab |
| pHM1 | Spr or Smr IncW, Mob(p), Mob+, LacIP+, PK2 replicon, cosmid | This lab |
| pKΔfbaB | A 822 bp fusion cloned in pKMS1 for a 349 bp deletion in fbaB, Kmr | This work |
| pCfbaB | pHM1 expressing fbaB under its own promoter, Spr | This work |
| pfbaBaGUS | pHM1 expressing gusA under the fbaB promoter, Spr | This work |
| pfbaBbGUS | pHM1 expressing gusA under the site (b)-mutated promoter of fbaB, Spr | This work |
| pfbaBcGUS | pHM1 expressing gusA under the site (c)-mutated promoter of fbaB, Spr | This work |
| pfbaBdGUS | pHM1 expressing gusA under the site (d)-mutated promoter of fbaB, Spr | This work |
Apr = ampicillin resistance, Kmr = kanamycin resistance, Rifr = rifampicin resistance, Spr = spectinomycin.
DNA manipulation
DNA manipulation was performed following the standard procedures described by Sambrook [51]. The transconjugation between the X. oryzae pv. oryzicola and plasmids was performed as described by Turner [52]. Restriction enzymes and DNA ligases were performed in accordance with the manufacturer's instructions (Takara, Dalian, China). The PCR primers (Table S1) for gene targets in this report were purchased from Jinsite Biotechnology (http://www.croasia.net/company/jinsite_biotechnology_co.html). The genes cloned or amplified in this study were refered to the hrp clusters of X. oryzae pv. oryzicola RS105 strain (AF272885, AY875714) and the genome sequence of X. oryzae pv. oryzicola BLS256 strain (http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=Xoc).
Rice suspension cell cultures
Oryza sativa ssp. indica cv. Shanyou63, susceptible to X. oryzae pv. oryzicola RS105 strain, was used for callus induction. Seeds were dehulled and sterilized in 70% ethanol for 10 min and then in 50% commercial bleach with a few drops of Tween-20 for 30 min and then in 1% HgCl2 for 15 min. The sterilized seeds were washed 5 times with sterile distilled water and placed on N6 medium (10) with 2, 4-D (5 mg/L) for induction of rice callus at 28°C in the dark. The actively growing calli were selected and transferred to liquid N6 medium containing with 5 mg/L 2, 4-D and 1 mg/L kinetin (KT). The cells were maintained in the dark on a 7 day subculture schedule at a dilution of 1∶5 (inoculum: fresh medium). Generally, large amounts of rice suspension cells can be obtained after 4–5 week subculture and then dispersed or single round rice cells could be observed under the microscope.
Construction of a non-polar mutant in fbaB of X. oryzae pv. oryzicola
The non-polar mutant of fabB in X. oryzae pv. oryzicola RS105 strain was constructed by using homologous recombination as described by Jiang [53], using pKMS1 as a suicide vector. Two flanking fragments, left and right to fabB (Figure S1), were amplified using the genomic DNA of strain RS105 as the template and the primers fbaBI-F/fbaBI-R and fbaBII-F/fbaBII-R (Table S1), respectively, and then cloned into pMD18-T vectors (Takara, Dalian, China), respectively. After confirmed by sequencing, the two fragments were digested and cloned into the vector pKMS1 at BamHI and PstI sites, resulting in pKΔfbaB (Table 1). The plasmid pKΔfbaB was introduced into RS105 by electrotransformation, and then the electrotransformants were plated on NAN plates supplemented with kanamycin. The emerged colonies suggested that the first homologous crossover event occurred in the electron transformants in which the DNA of the deletion vector was integrated into either the left or the right border of fbaB in the recipient chromosome (Figure S1). The single colonies of the mutant produced by single homologous crossover event were then transferred to NBN broth to culture for 12 h at 28°C. Then the bacterial cell was plated on NAS plates. The single colonies emerged within 3–4 day were then picked up into NA and NA plus kanamycin plates, respectively. By contrast, these kanamycin sensitive colonies could be the mutants in which the second homologous crossover event occurred and then were confirmed by PCR amplification with the primer pair fbaBI-F/fbaBII-R (Table S1, Figure S1). Subsequently, Southern hybridization (DIG, Roche) was conducted to verify the deletion of the fbaB by using the left fragment as the probe (Figure S1). One of the confirmed mutants, RΔfbaB (Table 1), was used for further study.
Complementation of the fbaB mutant RΔfbaB
In order to complement the fbaB mutant RΔfbaB, a 1302 bp DNA fragment containing the entire fbaB gene (from 297 bp upstream of the start codon to the stop codon) was amplified by PCR using the total DNA of X. oryzae pv. oryzicola RS105 as the template and the primer pair fbaB-F/fbaB-R (Table S1). After being confirmed by sequencing, the amplified DNA fragment was cloned into pHM1 vector at HindIII and KpnI sites to create a recombinant plasmid pCfbaB (Table 1). Plasmid pCfbaB was then transferred into the RΔfbaB strain by electroporation. The transconjugants carrying pCfbaB were screened on NA plates with rifampicin and spectinomycin. A confirmed representative was verified by colony-PCR amplification for further study and named CRΔfbaB (Table 1).
HR and pathogenicity assays
HR and pathogenicity assays were performed as described [19]. Xanthomonas bacteria were grown in NB liquid with appropriate antibiotics at 28°C with shaking at 200 rpm for 16 h. The bacterial inocula were washed twice and resuspended in sterile water to 1×108 cfu/ml and used to infiltrate into tobacco leaves (Xanthi) for HR detection and into rice seedlings (cv. Shanyou63, susceptible to X. oryzae pv. oryzicola infection, two-week old) for water-soaking formation with needleless syringes, respectively, and to inoculate in adult rice plants (cv. Shanyou63, two-month old) by leaf-needling for lesion length measurement. All plants were grown and maintained in a greenhouse with 12-h day-night cycle illuminations with a fluorescent lamp and a constant temperature of 25°C with relative humidity at 75 to 80% [54]. Plant responses were scored at 24 h for HR, in 3 days for water-soaking symptoms, and in 14 days for lesion lengths after inoculation. Five leaves were inoculated for each independent experiment, and each treatment was repeated three times.
Determination of bacterial growth ability in planta and in minimal medium supplemented with different carbohydrates
Xanthomonas bacterial suspensions at 1×108 cfu/ml were infiltrated into the intercellular spaces of fully expanded leaves of rice (cv. Shanyou63, two-week old) with needleless syringes at three spots on each leaf. Three 0.8 cm diameter leaf discs were harvested with a cork borer from each infiltration area after infiltration. After being sterilized in 70% ethanol and 30% hypochlorite, the leaf discs were homogenized in 1 ml of distilled water. Diluted homogenates were plated on NA agar plates supplemented with appropriate antibiotics. The number of bacterial colonies on these plates was counted after incubation at 28°C until single colonies could be counted after 3 to 4 days. The number of bacterial CFU per square centimeter of leaf area was then estimated, and the standard deviation was calculated using colony counts from the three triplicate spots from each of the three samples per time point per inoculum. Experiments were repeated at least three times.
As to the detection of bacterial growth influenced by different carbohydrates, Xanthomonas bacteria were preincubated in 5 ml NB medium for 16–20 h at 28°C with shaking at 200 rpm until the OD600 value reached 0.6, and 2% of this culture was subcultured into 20 ml of the fresh NB for 16–18 h incubation. The bacterial cells were collected and washed twice, and resuspended to an optical density of 600 nm of 0.1 in 100 ml of the minimal medium NCM supplemented with different carbon source at 0.5%. For each time point, 200 ul of each culture was removed and determined by measuring OD600 against the medium blank. Data presented were from a representative experiment; the experiment was repeated independently three times.
Site-directed mutagenesis in the PIP-box motifs of fbaB by PCR amplification
There is a PIP-box sequence, TTCGT-N9-TTCGT, which is not typical to TTCGC-N15-TTCGC, following 30 base pair interval space before a -10 box-like motif CAGCAT in the fbaB promoter region. Base-substitution in the PIP-box sequence of the fbaB promoter region was performed via a PCR amplification strategy. Briefly, three substituted sequences, TTCGC-N9-TTCGT (fbaBb), TTCGT-N9-TTCGC (fbaBc) and TGATA-N9-TTCGT (fbaBd) within the fbaB promoter region were generated by using three primer pairs, fbaBa-F/fbaBb-R, fbaBa-F/fbaBc-R and fbaBa-F/fbaBd-R (Table S1), respectively, for PCR amplification with the genomic DNA of X. oryzae pv. oryzicola RS105 as the template. Then, these PCR products were cloned into pMD18-T vector and confirmed by sequencing for further study.
Construction of the fbaB reporter plasmids
To investigate whether the expression of fbaB is or not regulated by HrpG and HrpX, four fbaB reporter plasmids, pFbaBaGUS, pFbaBbGUS, pFbaBcGUS and pFbaBdGUS, were constructed by cloning the PIP-box promoter region and three mutated PIP-box promoters of the fbaB gene which were fused with the promoterless β-glucuronidase (gusA) gene into the broad-host-range cloning vector pHM1 (Table 1) at MCS (multiple cloning site). A 366 bp region upstream of the fbaB was amplified by PCR using the total DNA of the wild-type RS105 strain as the template and the primer pair FbaBPF/FbaBPR (Table S1). The amplified fragment of the wild-type promoter of fbaB, confirmed by sequencing, was fused with the promoterless gusA in the vector pHM1 at HindIII and EcoRI sites to create the recombinant plasmid pFbaBaGUS (Table 1). In contrast, the wild-type promoter was replaced by three site-directed substitutes in the PIP-box of the fbaB promoter, as mentioned above, and fused with the gusA gene in the vector pHM1 at HindIII and EcoRI sites, generating the recombinant plasmids pFbaBbGUS, pFabBcGUS and pFbaBdGUS (Table 1). The plasmids obtained were further confirmed by restriction analysis and sequencing.
Measurement of EPS production
EPS production was measured as previously described by Tang [55]. In brief, Xanthomonas bacteria were grown in 100 ml of NY medium supplemented with 2% (w/v) various sugars at 28°C with constant shaking at 200 rpm for 3 days. EPS was precipitated from the culture supernatant with ethanol, and dried to constant weight at 55°C, and weighed. Every experiment was repeated at least three times.
Semi-quantitative RT-PCR and Real-time quantitative RT-PCR
The expression of tested genes, including the reporter gusA, was assayed by semi-quantitative RT-PCR or real-time quantitative RT-PCR with corresponding primer pairs (Table S1). Xanthomonas bacteria were preincubated in 20 ml NB medium for 16–20 h, until the OD600 value reached 0.6, and 2% of this culture was subcultured into 20 ml of the fresh NB for 16–18 h incubation. The bacterial cells were collected and washed twice, and resuspended to an optical density of 600 nm of 2.0 by sterilized water. Then, 40 ul of bacterial suspension was inoculated into 1.5 ml of NB, NY medium or rice suspension cells incubating for 16 h at 25°C. As a template, total RNAs were extracted using the Trizol reagent (Takara, Dalian, China) according to the manufacturer's protocol. cDNA synthesis was conducted with AMV random primers (order no. D3801) provided by the manufacturer (Takara, Dalian, China). Before synthesis of the first-strand cDNA, total RNAs were digested with RNase-free DNase I (TaKaRa, Dalian, China) to remove potential traces of genomic DNAs. Semi-quantitative RT-PCR was performed on the ordinary PCR apparatus and the PCR program was as follows: step 1, 95°C for 3 min; step 2, 95°C for 20 s; step 3, 55°C for 30 s; step 4, 72°C for 40 s; 35 cycles from steps 2 to 4; and step 5, 72°C for 10 min. Real-time quantitative RT-PCR was performed on the Applied Biosystems 7500 real-time PCR System using SYBR Premix Ex Taq™ (Takara, Dalian, China), and the PCR thermal cycle condition was as following: denature at 95°C for 30 s and 41 cycles for 95°C, 5 s; 60°C, 34 s. The expression level of the 16S rRNA gene was used as an internal standard. The comparative-threshold method was used to calculate the relative mRNA level with respect to the corresponding transcript in cells cultured in NB or NY medium or rice suspension cells. All RT-PCRs were performed in triplicate.
Results
fbaB is required for full virulence and growth of X. oryzae pv. oryzicola in planta
The discovery of the fbaB gene as a virulence factor came from work aimed at the identification of genes involved in virulence alteration of X. oryzae pv. oryzicola RS105 strain in rice. The approach was to screen a Tn5-tagged mutant library of RS105 [20] for mutants that could impair virulence of the bacterium in rice. One mutant Mxoc0504 (Table 1), where the Tn5 transposon was inserted in an open-reading frame (ORF) of Xoryp_17640 at the 185 bp site (Figure S1), reduced virulence of X. oryzae pv. oryzicola RS105 in rice (data not shown). The genome location and genetic organization of the Tn5-insertion in Xoryp_17640 of X. oryzae pv. oryzicola RS105 suggests the presence of a transcriptionally active gene Xoryp_17640 (Figure S1). Fructose-bisphophate aldolase is an enzyme encoded by just one gene, which is highly conserved in Xanthomonas species (data not shown). It performs the reversible action of converting fructose-1,6-bisphosphate to dihydroxyacetone phosphate and glyceraldehyde 3-phosphate, which are involved in functional glycolytic and gluconeogenic pathways [1]. Thus, Xoryp_17640 is hereafter named as fbaB.
To faciliate the functional study of fbaB, a nonpolar fbaB mutant, named RΔfbaB (Table 1), was constructed by homologous suicide plasmid integration (Figure S1) (see Materials and Methods for detail). A complemented strain named CRΔfbaB was also constructed by introducing the recombinant plasmid pCfbaB, which carries the entire ORF fbaB with a 297 bp promoter region upstream of the start codon (Table 1), into the mutant RΔfbaB.
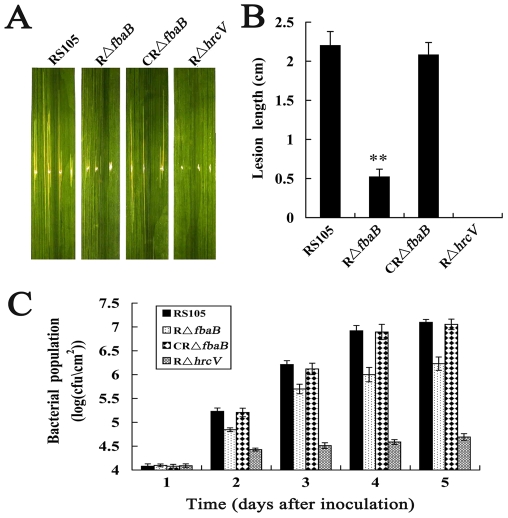
The virulence of the fbaB mutant RΔfbaB, the complemented strain CRΔfbaB and the wild-type RS105 was tested on the hybrid rice cultivar Shangyou63 by the leaf-needling inoculation method [4]. Although the mutant strain RΔfbaB still caused obvious bacterial leaf streak (BLS) symptoms, the symptoms were significantly less severe than that caused by the wild-type RS105 (Figure 1A). The mean lesion length caused by RΔfbaB was significantly reduced (P = 0.01, t test) by approximately 1.5 cm compared to the wild-type RS105, while the T3SS mutant RΔhrcV, used as a negative control, did not cause any BLS symptoms in rice (Figure 1A). The BLS lesion lengths caused by the complemented strain CRΔfbaB were however obiviously the same as those caused by the wild-type RS105 (Figure 1B). These results demonstrated that fbaB is required for full virulence of X. oryzae pv. oryzicola in rice.
Figure 1. fbaB is required for full virulence and growth of X. oryzae pv. oryzicola in planta.
(A) Symptoms caused by different X. oryzae pv. oryzicola strains suspended in water to OD600 = 0.3 (approximately 1×108 cfu/ml) on inoculated leaves of the host rice cv. Shanyou63 (susceptible cultivar) (2-month-old) by leaf-needling inoculation. Photographs were taken 14 days post-inoculation. RS105, the wild-type strain; RΔfbaB, the fbaB deletion mutant; CRΔfbaB, the complemented strain of RΔfbaB with the fbaB gene; RΔhrcV, a type III-deficient strain as a negative control. (B) Lesion lengths of rice bacterial leaf streak caused by different X. oryzae pv. oryzicola strains in rice. Values are the means ± standard deviations (SD) from three repeats, each with five leaves. The different symbol in each horizontal data column results from a paired, two-tailed Student t test relative to the wild-type. **, P = 0.01. (C) Bactrial growth capacity in inoculated leaves. Bacteria were recovered from the inoculated leaves every 24 hours in a period of 4 days post inoculation, and homogenized in sterile water. The homogenates were diluted and plated on NA plates with appropriate antibiotics. Bacterial CFU were counted after incubation at 28°C for 3 days. Data are the mean ± SD from three repeats.
In order to determine whether fbaB results in a decrease in the proliferation of X. oryzae pv. oryzicola in the host rice, we investigated the growth capacity of the fbaB mutant RΔfbaB, the complemented strain CRΔfbaB and the wild-type strain RS105 in planta. During the observation days, the bacterial number of the RΔfbaB mutant recovered from the infected rice leaves was significantly lower than that of the wild-type RS105 at each of the test points. The growth capacity of the RΔfbaB strain in planta was completely restored to the wild-type level by fbaB in trans (Figure 1C), whereas the T3SS mutant RΔhrcV did not grow more in inoculated rice tissues. These results indicated that the fbaB is required for growth of X. oryzae pv. oryzicola in planta.
fbaB is important in acquisition of fructose, pyruvate and malate for X. oryzae pv. oryzicola growth
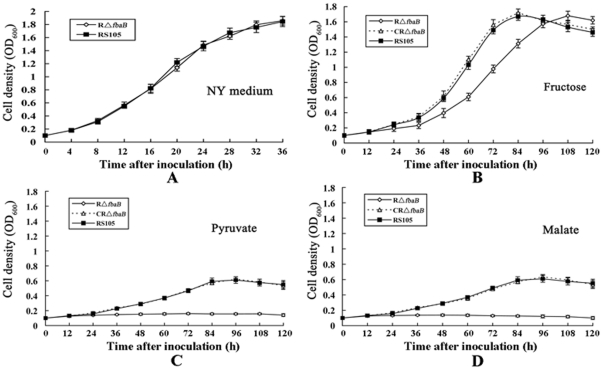
FbaB reversibly converts fructose-1,6-bisphosphate to dihydroxyacetone phosphate and glyceraldehyde 3-phosphate. This prompted us to investigate whether fbaB affects X. oryzae pv. oryzicola growth in a non-sugar NY medium (see Materials and Methods for detail). The result showed that the fbaB mutant RΔfbaB grew identically as the wild-type RS105 (Figure 2A), indicating that the RΔfbaB mutant was not auxotrophic. To further examine the effect of the fbaB gene on the ability of X. oryzae pv. oryzicola to utilize various carbon sources, the growth of the fbaB mutant RΔfbaB, the complemented strain CRΔfbaB and the wild-type RS105 were tested by using the liquid NCM (non-carbohydrate minimal medium) supplemented with glucose, sucrose, fructose, mannose, galactose, pyruvate and malate, respectively, as the sole carbon source. The growth of the RΔfbaB strain grew in a similar fashion to that of the wild-type RS105 when supplemented with glucose, sucrose, mannose or galactose (data not shown). However, the growth of RΔfbaB was significantly slower than that of the wild-type strain RS105 in liquid NCM supplemented with fructose as the sole carbon source, while the complemented strain CRΔfbaB with the fbaB gene restored the growth to the wild-type level (Figure 2B), suggesting that the mutation in fbaB diminishes the capability of X. oryzae pv. oryzicola to utilize fructose.
Figure 2. Growth curves of X. oryzae pv. oryzicola in sole carbon media.
RS105, the wild-type strain; RΔfbaB, the fbaB deletion mutant; CRΔfbaB, the complemented strain of RΔfbaB with the fbaB gene. The initial concentration of the tested strains was adjusted to OD600 of 0.1 with NCM supplemented with fructose, pyruvate or malate as the sole carbon source. Aliquots were taken in triplicate at intervals of 120 h after incubation at 28°C, and bacterial growth was determined by measuring OD600 against the medium blank. Values given are the means ± SD of triplicate measurements from a representative experiment; similar results were obtained in two other independent experiments.
Since pyruvate is the final product in glycolysis, and the initial carbohydrate for gluconeogenesis [1], the mutation in fbaB may presumably lead no gluconeogenesis. To seek this, we then investigated whether the mutagenesis in fbaB causes X. oryzae pv. oryzicola unable to utilize pyruvate or not. Indeed, the fbaB mutant RΔfbaB was unable to grow in NCM medium supplemented with pyruvate as the sole carbon source, whereas the complemented strain CRΔfbaB harboring the fbaB gene restored the ability to acquire pyruvate to the wild-type level (Figure 2C).
Pyruvate may be catalyzed by pyruvate carboxylase into oxaloacetate, or by the pyruvate dehydrogenase complex into acetyl-CoA which essentially flows into the TCA cycle. Malate is reversibly converted by malic enzyme into pyruvate for gluconeogenesis [1]. This promopted us to investigate whether or not the mutation in fbaB impairs the ability of X. oryzae pv. oryzicola to utilize malate for growth. The results showed that the fbaB mutant RΔfbaB was unable to grow in NCM supplemented with malate as the sole carbon source, while the complemented strain CRΔfbaB with fbaB was recovered to the wild-type level to use malate for growth (Figure 2D), implying that the conversion of malate into pyruvate can not flow through gluconeogenesis because of the mutation in fbaB.
Briefly, the above data indicate that fbaB of X. oryzae pv. oryzicola has limited influence on fructose utilization due to the presence of ED and PPP pathways when the downstream glycolysis is blocked, but plays important roles in gluconeogenesis when malate from TCA cycle is converted into pyruyate.
fbaB influences EPS production of X. oryzae pv. oryzicola
It has been demonstrated that EPS as a virulence factor plays an important role during bacterial infection [56], [57] and ED is necessary for xanthan biosynthesis and PPP does not influence xanthan biosynthesis in X. oryzae pv. oryzae [6]. In order to determine whether the mutation in fbaB has any effect on EPS production of X. oryzae pv. oryzicola, the EPS yields of the fbaB mutant RΔfbaB, the complemented strain CRΔfbaB and the wild-type RS105 were quantitatively measured after the strains grew in NY liquid medium supplemented with 2% of fructose, pyruvate and malate, respectively, for 3 days. Meanwhile, NY medium and NY medium added with 2% of glucose were used as the control. After the EPS of the tested strains was extracted from the cultures (see Materials and Methods for detail), we found that there were no significant (P = 0.01, t test) difference in EPS production among RΔfbaB, CRΔfbaB and RS105 when they grew in NY medium alone and NY medium containing 2% of glucose, respectively (Table 2). However, the EPS yield of RΔfbaB was significantly 30% less than that of the wild-type RS105 in NY plus 2% fructose medium, 80% less in NY plus pyruyate or malate (Table 2). By contrast, the fbaB gene restored EPS production of the mutant RΔfbaB to the wild-type level either in NY plus 2% fructose, or NY plus 2% pyruvate, or NY plus 2% malate (Table 2). The above results suggest that the mutation in fbaB leaves X. oryzae pv. oryzicola unable to sufficiently use fructose, and to completely uitilize pyruvate and malate. This provides evidence that the fbaB mutation on gluconeogenesis affects the ED pathway, resulting in less EPS production.
Table 2. EPS products in X. oryzae pv. oryzicola strains.
| Strainsa | EPS yield (g/100 ml)b | ||||
| NY | NY plus 2% glucose | NY plus 2% fructose | NY plus 2% pyruvate | NY plus 2% malate | |
| RΔfbaB | 0.05±0.009A | 0.88±0.082A | 0.52±0.012A | 0.12±0.012A | 0.10±0.009A |
| CRΔfbaB | 0.06±0.006A | 0.94±0.032A | 0.81±0.028B | 0.49±0.023B | 0.54±0.036B |
| RS105/pHM1 | 0.05±0.007A | 0.91±0.026A | 0.75±0.034B | 0.53±0.019B | 0.50±0.025B |
Strains were cultured in NY medium alone and supplemented with 2% various carbon sources.
Data presented are the means ± standard deviations of triplicate measurements from a representative experiment, and similar results were obtained in two other independent experiments. Different letters in each data column indicate significant differences (P = 0.01; t test).
fbaB is negatively regulated by HrpX and HrpG
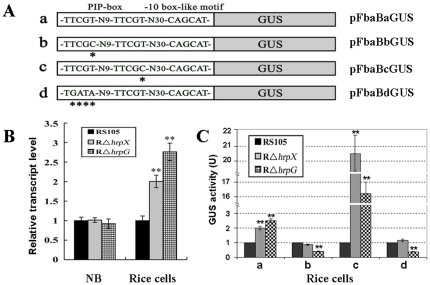
Previous reports have demonstrated that the PIP-box of HrpX regulons serves as a cis-regulated element in a HrpX-dependent manner [37], [38], [44]. Analysis of the promoter region of fbaB of X. oryzae pv. oryzicola BLS256 (Figure S1) by searching the existence of similar PIP-box sequence and by using a promoter-prediction software (http://www.fruitfly.org/seq_tools/promoter.html) revealed an imperfect PIP-box (TTCGT-N9-TTCGT) interval by 30 bp sequence with a -10 box-like motif (CAGCAT) upstream of the fbaB start codon (Figure 3A), suggesting that the expression of fbaB may be regulated by HrpX and HrpG, the latter controls the expression of hrpX [34], [35]. To investigate this, a real-time quantitative RT-PCR was employed to assay the action of the fbaB transcript with hrpX and hrpG. The fbaB relative transcript level displayed a significant increase (P = 0.01, t test) in the hrpX mutant RΔhrpX and the hrpG mutant RΔhrpG than that of the wild-type RS105 when the strains grew in rice suspension cells for 16 h. The expression of fbaB in RΔhrpG was higher than that in RΔhrpX, whereas there were no obvious difference of the fbaB expression among these three tested strain when they grew in NB medium (Figure 3B). These results demonstrate that the expression of fbaB is inducible in planta and repressed by HrpX and HrpG when the pathogen infects the host rice rather than in necrotrophic growth. The negative regulation of fbaB with HrpG and HrpX and the higher expression of fbaB in the hrpG mutant than in the hrpX mutant imply that other unkown factor(s) may involve in the regution of fbaB.
Figure 3. The expression of fabB in X. oryzae pv. oryzicola is negatively by hrpX and hrpG in rice suspension cells.
(A) Schematic map of the promoter region containing PIP-box and -10 box-like motif of fabB fused with a promoterless gusA gene. * stands for base substitutions. The constructs are listed on the right. (B) Expression analysis of fbaB by real-time quantitative RT-PCR. RNAs were isolated from cultures of the wild-type RS105, the hrpG deletion mutant RΔhrpG and the hrpX deletion mutant RΔhrpX strains which were grown in NB medium and rice suspension cells for 16 h, respectively. The relative mRNAs level of fbaB was calculated with respect to the level of the corresponding transcript in the wild-type RS105. (C) Effects of the mutated PIP-box on gusA transcript. The gusA transcript level by the wild-type PIP-box promoter (a) and three base-substituted PIP-box promoter (b, c, d) in the wild-type RS105, the hrpX mutant RΔhrpX and the hrpG mutant RΔhrpX were investigated, respectively. All the reporter strains above were cultured in rice suspension cells for 16 h and gusA transcript levels were then determined by real-time PCR. The transcript of gusA in the wild-type was taken as one unit. Data are the mean ± SD of triplicate measurements from a representative experiment; and similar results were obtained in two other independent experiments. The asterisks in each horizontal data column indicate significant differences at P = 0.01 by t test.
It has been demonstrated that HrpX regulates the expression of HrpX regulon genes by binding the PIP-box motif in promoter regions [36], [38], [39]; therefore, substitution of the fifth base in the motif TTCGC, or the complete mutation in the motif sequence itself, significantly alters promoter activity [26], [37], [38]. Fifth base substitution, TTCGB (B = T, C, or G) may increase or decrease the transcript level of a HrpX-regulated gene by up to 50% [38], [39]. In order to determine whether or not the two motifs of the imperfect PIP-box (TTCGT-N9-TTCGT) of fbaB are affected by HrpG and HrpX, the fifth base of the first or second TTCGT motif was substituted with a C to create TTCGC by site-direct substitutions, which generated pFbaBbGUS and pFbaBcGUS (Figure 3A), respectively. In addition, the first TTCGT motif was completely changed to a TGATA motif to produce pFbaBdGUS (Figure 3A) by using site-mutagenesis primers (Table S1). The GUS reporter strains (Table 1) were incubated in rice suspension cells for 16 h. The gusA transcript level was measured by real-time PCR (Figure 3C). Compared to pFbaBaGUS, pFbaBbGUS andpFbaBdGUS (Figure 3A) reduced the gusA transcript level in RΔhrpX (Figure 3C). This was similar to the gusA transcript level of pFbaBaGUS in the wild-type when compared to the hrpX mutant RΔhrpX (Figure 3C). However, the gusA transcript of pFbaBbGUS or pFbaBdGUS in the hrpG mutant RΔhrpG was significantly lower than that in the hrpX mutant or in the wild-type (Figure 3C). Intriguingly, the base substitution of the fifth residue in the right motif of the PIP-box (Figure 3A) significantly increased the gusA transcript of pFbaBcGUS in either RΔhrpX or RΔhrpG, compared with the wild-type (Figure 3C). The gusA expression of pFbaBcGUS in RΔhrpX was significantly higher (approximately 20-fold higher than that in the wild-type) than that in RΔhrpG (Figure 3C). The GUS activity assay also demonstrated the same results above (data not shown). The above evidence suggests that expression of fbaB is negatively regulated by HrpG and HrpX via the PIP-box promoter where a yet unknown factor might bind for the involved regulation.
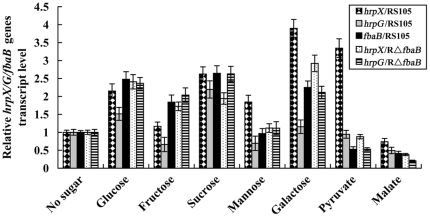
Various carbohydrates have different effects on expression of hrpG, hrpX and fbaB in X. oryzae pv. oryzicola
Environmental signals like carbon sources presenting in plants may serve as inducers or inhibitors of virulence-associated gene expression in plant bacteria [17], [21]. We sought to investigate transcript production of hrpG, hrpX and fbaB when X. oryzae pv. oryzicola is fed with different carbohydrates. The expression level of hrpG, hrpX and fbaB was measured by real-time PCR after the wild-type RS105 strain grew for 16 h in NY medium complemented with 0.5% of sucrose, galactose, glucose, mannose, fructose, pyruvate and malate, respectively. Using NY medium as the control, we found that, besides malate, the other six carbon sources enhanced the expression of hrpX. Fructose, mannose, galactose, pyruvate and malate had little effect on the expression of hrpG, while sucrose and glucose increased the transcript level of hrpG (Figure 4). The transcript level of fbaB was increased by sucrose, galactose, glucose and fructose rather than pyruvate and malate (Figure 4). Noticeably, fructose, mannose and malate repressed the expression of hrpG, pyruvate and malate inhibited the expression of fbaB, and malate suppressed the transcript of hrpX (Figure 4). The results above demonstrate that expression of certain genes involving the carbon metabolic pathways may be regulated by the two key hrp regulatory genes hrpG and hrpX, and in turn the expression of hrpG and hrpX may also be enhanced or repressed by the carbon sources or intermediates in the metabolic pathways.
Figure 4. Effects of different carbohydrates on the expression of hrpX, hrpG and fbaB in X. oryzae pv. oryzicola.
RNAs were isolated from cultures of the wild-type RS105 strain and the fbaB deletion mutant RΔfbaB grown in NY medium alone and NY supplemented with 0.5% of various carbohydrates for 16 h. The relative mRNAs levels of hrpX, hrpG and fbaB genes were calculated by real-time quantitative RT-PCR with respect to the level of the corresponding transcript in the wild-type RS105 cultured in NY medium alone. Data presented are the means ± SD of triplicate measurements from a representative experiment; similar results were obtained in two other independent experiments.
The negative regulation of fbaB with HrpG and HrpX prompted us to determine whether it is influenced by the carbohydrates tested in the previous section when fbaB is mutated. Using the same real-time PCR assay, we found that the relative mRNA level of hrpX was significantly reduced in the fbaB mutant than that in the wild-type RS105 strain when sucrose, mannose, galactose, pyruvate and malate were complemented in NY medium as the sole carbon sources, but increased when fructose was used. The expression of hrpG was significantly enhanced by glucose, fructose, sucrose, mannose and galactose, respectively, and repressed by pyruvate and malate (Figure 4). These results demonstrated that the mutation in fbaB may alter the expression of hrpG or hrpX when the sugars are used as the sole carbon source and the relationship of HrpX with HrpG may be influenced through unkown factor(s) affected by different carbonhydrates.
fbaB positively affects the expression of hrpG and hrpX, but negatively influences the transcripts of hrcC, hrpE and hpa3
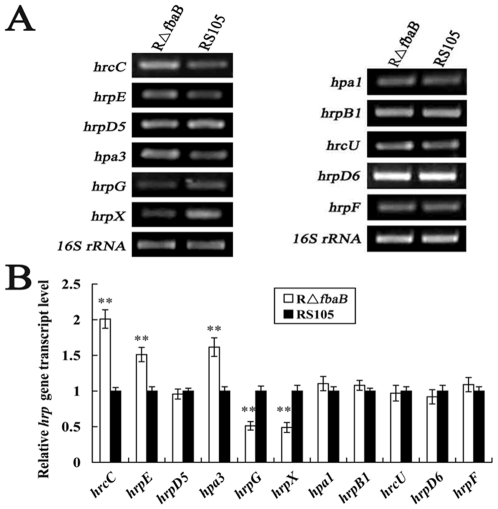
The data above demonstrated that expression of the key hrp regulatory genes, hrpG and hrpX, are induced or repressed when X. oryzae pv. oryzicola evidently uses sugars from plants as nutrient sources, implying that the expression of the hrp-hrc-hpa genes, which are regulated by HrpG and HrpX as reported [20], may be altered when fbaB is mutated. To investigate this, we employed a semi-quantitative (Figure 5A) and real-time RT-PCR (Figure 5B) with the specified primer pairs (Table S1) to evaluate the transcript production of representative hrp-hrc-hpa genes in the fbaB mutant RΔfbaB and the wild-type RS105 strain after incubation in rice suspension cells for 16 h, while the nutrient-rich medium NB was used as a synchronous control (data not shown). The results showed that: i) the expression of hrpG and hrpX in the fbaB mutant RΔfbaB was significantly (P = 0.01, t test) lower than that in the wild-type, implying that the dysfunction in glycolysis and gluconeogenesis by the mutation in fbaB represses the transcript of hrpG and hrpX; ii) the mRNA level of the tested genes, hrpD5, hrpD6, hpa1, hrpB1, hrcU and hrpF, in RΔfbaB was similar to that in the wild-type; iii) the transcriptional level of hrcC, hrpE and hpa3 (which was previously reported that their expression was not completely controlled by HrpG and HrpX in X. oryzae [24], [31]) in RΔfbaB was significantly higher than that in the wild-type (Figure 5), indicating that certain intermediates in glycolysis and gluconeogenesis derived from the aldol reaction [1] may influence the expression of hrcC, hrpE and hpa3. The above data suggest that the mutation in fbaB may alter the ability of X. oryzae pv. oryzicola to acquire carbon from its living niche which in turn represses the expression of hrpG and hrpX.
Figure 5. Influence of the mutation in fbaB on the expression of hrp-hrc-hpa genes of X. oryzae pv. oryzicola.
(A) Semi-quantitative RT-PCR analysis. RNAs were isolated from cultures of the wild-type RS105 strain and the fbaB mutant RΔfbaB grown in rice suspension cells for 16 h. The 16S rRNA gene of the pathogen is used as the standard internal control. The tested hrp-hrc-hpa genes were selected based on the reports [19], [20], [53] with the primer pairs (Table S1) and the sequence of the hrp clusters (AF272885, AY875714) was used as the reference. (B) Real-time quantitative RT-PCR analysis. The relative mRNA level of the tested hrp-hrc-hpa genes in the fbaB mutant RΔfbaB was calculated with respect to the level of the corresponding transcripts in the wild-type RS105 cultured in rice suspension cells for 16 h. Values given are the means ± SD of triplicate measurements from a representative experiment. The asterisks in each horizontal data column indicate significant differences. **, P = 0.01, t test. Experiment was repeated twice and yielded similar results.
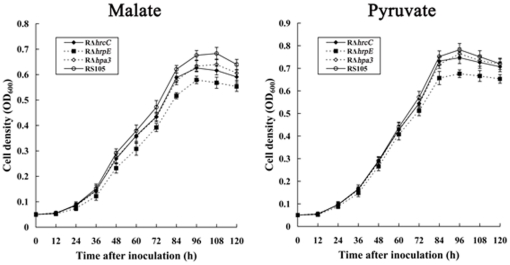
hrcC, hrpE and hpa3 are required in utilization of pyruvate and malate for X. oryzae pv. oryzicola
To verify our hypothesis that the hrcC, hrpE and hpa3 genes are involved in acquisition of pyruvate and malate, we tested the growth of the hrcC, hrpE and hpa3 mutants, RΔhrcC, RΔhrpE, and RΔhpa3 (Table 1), respectively, in NCM medium supplemented with sucrose, mannose, galactose, glucose, fructose, pyruvate and malate as the sole carbon sources, while the wild-type strain RS105 was used as the control. Indeed, the mutation in hrcC, hrpE or hpa3 reduced the growth of the pathogen when pyruvate and malate were used as the sole carbon source (Figure 6). By contrast, the growth of the hrpE mutant was affected much more than that of the hrcC or the hpa3 mutants by pyruvate and malate, respectively (Figure 6). These data suggests possible reasons why the expression of hrcC, hrpE and hpa3 was higher than other hrp-hrc-hpa genes when fbaB was mutated (Figure 5).
Figure 6. The mutation in hrcC, hrpE and hpa3 reduced the ability of X. oryzae pv. oryzicola to acquire pyruvate and malate.
RS105, the wild-type strain; RΔhrcC, the hrcC deletion mutant; RΔhrpE, the hrpE deletion mutant; RΔhpa3, the hpa3 deletion mutant. The initial concentration of the tested strains was adjusted to OD600 of 0.05 with NCM supplemented with pyruvate or malate as the sole carbon source. Aliquots were taken in triplicate at intervals of 120 h after incubation at 28°C, and bacterial growth was determined by measuring OD600 against the medium blank. Values given are the means ± SD of triplicate measurements from a representative result of other two similar independent experiments.
Discussion
In this study, we identified in X. oryzae pv. oryzicola RS105 strain a novel and unique virulence gene, fbaB, which encodes an fructose-bisphophate aldolase (FbaB), highly conserved in other Xanthomonads, and converts the intermediate fructose-1,6-bisphosphate to reversible dihydroxyacetone phosphate and glyceraldehyde 3-phosphate, which is essential for glycolysis and gluconeogenesis (Figure 7), and now has been shown to play a role in virulence. Genetic evidence presented here demonstrates that fbaB is required for X. oryzae pv.oryzicola to utilize fructose, pyruvate and malate so that the pathogen produces EPS and expands full virulence and growth in planta for adaptation. The mutation in fbaB does not make the pathogen auxotrophic and lethal. Interestingly, the expression of fbaB is negatively regulated by the HrpG and HrpX cascade via the imperfect PIP-box of fbaB possibly with an unknown regulator. The latter may presumably regulate the expression of hrcC, hrpE, and hpa3 and be influenced by the accumulation of pyruvate for the initiation of gluconeogenesis and malate from the TCA cycle (Figure 7). Intriguingly, the PIP-box spaced, by 30 base pairs with a -10 box-like motif is also highly conserved within the genome sequences of X. oryzae pv. oryzae PXO99A [9], KACC10331 [8], X. campestris pv. vesicatoria 85-10 strain [12], X. campestris pv. campestris 8004 [10], and X. axonopodis pv. citri 306 [11] (data not shown). This implies that the expression of fbaB homologues in other Xanthomonas species may be regulated by the same manner. However, whether fbaB of other Xanthomonas species plays a similar role as above in host-pathogen interactions needs to be further investigated.
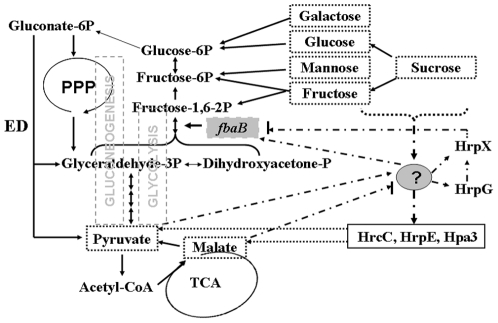
Figure 7. Working model of FbaB coordinating with hrp genes of X. oryzae pv. oryzicola in carbon metabolic pathoways.
The lined arrows from the carbohydrates in dashed-line boxes or the double lined arrows from the intermediates indicate carbon flows in glycolysis, gluconeogenesis, pentose phosphate pathway (PPP), entner–doudoroff (ED) and tricarboxylic acid (TCA) cycle pathways, respectively. The grey box displays fbaB encodes a fructose-bisphophate aldolase that converts vertically fructose-1,6-bisphosphate to dihydroxyacetone phosphate and glyceraldehyde 3-phosphate. The mutation in fbaB has no influence on ED and PPP pathways, but impairs glycolysis of the pathogen to use fructose and block gluconeogenesis to use pyruvate and malate. The expression of an unknown regulator in a cycled question mark may be enhanced by galactose, glucose, mannose, sucrose, fructose and pyruvate (as shown by dash-lined arrow) and repressed by malate (a dash-lined arrow with a stop bar). The unkown factor may differentially regulate the expression of hrpG or/and hrpX which down-regulate the expression of fbaB (a dash-lined arrow with a stop bar). The unknown regulator may also control the transcripts of hrcC, hrpE and hpa3 (other than other hrp-hrc-hpa genes) which are not completely regulated by HrpG and HrpX [20]. Being the components of the T3SS apparatus, HrcC, HrpE and Hpa3 may faciliate X. oryzae pv. oryzicola to utilize the intermediates, like pyruvate and malate, of the TCA cycle from plants.
X. oryzae pv. oryzicola is a nonvascular pathogen that enters through leaf stomata or wounds, and propagates and spreads in the intercellular spaces and the parenchyma apoplast to cause BLS in rice [19]. To reach the cell density for pathogenesis in plants, the pathogen has to be able to adapt to intercellular environments and also utilize available nutritional sources, especially carbohydrates, from the host plant. The mutation in fbaB does not affect the growth ability of X. oryzae pv. oryzicola when glucose, sucrose, mannose and galactose are used as the sole carbon source (data not shown), but impairs the growth rate when fructose is as the sole carbon (Figure 2), demonstrating that EMP pathway of glycolysis does not play an obvious role in sugar catabolism of X. oryzae pv. oryzicola. This is consistent with previous reports that Xanthomonads primarily employ ED, together with PPP, instead of EMP pathway of glycolysis to utilize glucose because of the lack of the phosphofructokinase (PFK) activity essential for a functional EMP, although a putative phosphofructokinase-encoding gene is annotated in the genomes of X. oryzae pv. oryzae, X. campestris pv. vesicatoria, and X. campestris pv. campestris [5], [6], [7], [13]. On the other hand, the mutagenesis in fbaB of X. oryzae pv. oryzicola results in the complete loss of the capability to grow when pyruvate and malate are used as the sole carbon source (Figure 2), implying that FbaB is essential for gluconeogenesis of X. oryzae pv. oryzicola (Figure 7). This is concordant with the fact that gluconeogenesis is indispensable for the pathogen to utilize pyruvate or the intermediates of the TCA cycle as the sole carbon sources in X. campestris pv. campestris [4], [13], [58]. Whether or not and how intermediates from the TCA cycle of host plants are acquired by the pathogen seems critically important in understanding mechanisms of plant-pathogen interactions.
Normally, UDP-glucose, UDP-galactose and dTDP-rhamnose are precursors or building blocks of EPS biosynthesis [7]. The galactosides UDP-glucose and UDP-galactose are synthesized from glucose-1-phosphate catalyzed from a precursor fructose-6-phosphate [7], [59]. Glucose-1-phosphate is converted from galactose, glucose or sucrose, and fructose-6-phosphate is metabolised from mannose, fructose or sucrose (Figure 7). The lack of phosphofructokinase (PFK) in xanthomonads [5], [6], [7], [13] may explain the reason that the mutation in fbaB of X. oryzae pv. oryzicola has no effects on EPS production when galactose, glucose, mannose and sucrose, rather than fructose, are used as the sole carbon source (data not shown). The mutagenesis in fbaB leads the complete loss of the ability to convert pyruvate and malate into glucose-1-phosphate via gluconeogenesis for EPS synthesis in X. oryzae pv. oryzicola, explaining the reason that the EPS production in the fbaB mutant is remarkably reduced when pyruvate and malate are used as the sole carbons (Table 2).
The hrp-encoded T3SS apparatus, together with other virulence factors, are often subject to be coordinated with the regulation of HrpG and HrpX which enables the pathogen to respond to environmental factors (such as pH and osmotic strength), plant signals (such as carbon sources, organic nitrogen, and phosphate), and catabolite repression that may be encountered during the infection [17], [18], [21], [23], [60]. This regulation is very complex and varies substantially between different Xanthomonas-plant pathosystems and in some cases even between closely related bacteria within the same pathosystem [61]. In this study, we found that different carbohydrates have different influences on expression of hrpG and hrpX. Sucrose, galactose, mannose, glucose, fructose and pyruvate significantly increase the expression of hrpX in X. oryzae pv. oryzicola when they used as the sole carbon source, while sucrose and glucose remarkably enhance the expression of hrpG (Figure 4). By contrast, the hrpX expression goes up while the hrpG expression decreases obviously when the pathogen grows on these carbon sources, respectively, and malate represses the expression of hrpG and hrpX (Figure 4), suggesting that other unknown factor(s) is (are) possibly involved differentially in regulation of the expression of hrpG and hrpX. This postulation conflicts with the concept that HrpG function as a positive activator upstream of HrpX in regulatory pathways of hrp gene expression. In fact, the expression of hrcT goes up when hrpG is mutated and there is no hrcT expression detected when hrpX is mutated in X. oryzae pv. oryzicola [20], supporting the above hypothesis.
The above findings also theoretically support the development of hrp-inducing media, XCV2 for X. campestris pv. vesicatoria [29], XOM2 for X. oryzae pv. oryzae [61], [62] and XOM3 for X. oryzae pv. oryzicola [25]. The major carbon source in plant leaf extract is sucrose, followed by glucose and fructose, and the dicarboxylic acid, malate, and the latter induces the secretion of extracellular enzymes and has a negative effect on the expression of the T3SS in X. campestris pv. campestris [63]. In P. syringae pv. phaseolicola, the expression of hrpAB, hrpC, and hrpD was reduced when citrate or succinate was added to fructose-or sucrose-containing medium [17]. Taken together, we assumed that hexoses from plant photosynthesis induce the expression of hrp genes and this action can be balanced by the intermediates from the TCA cycle of the plant pathogen. Thus, the disruption in carbon metabolic pathways reduces bacterial virulence in plants through alteration of the expression of global regulator genes, including hrpG and hrpX, in plant pathogenic bacteria. As we observed, the mutation in fbaB of X. oryzae pv. oryzicola leads the dysfunction of gluconeogenesis and the accumulation of intermediates, like malate, from the TCA cycle represses the expression of hrpG and hrpX (Figure 4 and 5B), enhances the bacterium to use the TCA intermediates by the help of HrcC, HrpE and Hpa3 (Figure 5B, 6 and 7), and may also affect the ability of the organism to obtain nutritents from the environment.
The interesting finding in this report is that the promoter region of fbaB of X. oryzae pv. oryzocola assembles the cis-element of PIP-box which is taken as the sequence of HrpX regulons (Figure 3A). The highly conserved PIP-box of the fbaB homologue in other typical Xanthomonas species (data not shown) suggests that the expression of fbaB may commonly be negatively regulated by HrpG and HrpX. Protein secretion assays demonstrated that FbaB is not secreted through the T2SS and T3SS (data not shown). In fact, the expression level of fbaB in the hrpG mutant is higher than than in the hrpX mutant (Figure 3B), suggesting yet unknown factor(s) may strongerly regulate the expression of HrpX than HrpG. This is consistent with the following findings. The base substitution in the fifth residue of the left motif TTCGT of the PIP-box significantly reduced the expression of fbaB when hrpG was mutated rather than in the hrpX mutant (Figure 3C). However, the substitution in the right motif TTCGT led the expression of fbaB to be 15–20 fold higher than the wild-type promoter when hrpG and hrpX are disrupted, respectively and the promoter activity in the hrpX mutant is significantly higher than that in the hrpG mutant (Figure 3C), implying that the alteration of the binding sites of the fbaB PIP-box promoter makes the expression of fbaB released from the regulation of HrpG and HrpX together with a yet unknown regulator (Figure 7). This unknown regulator may activate the expression of fbaB which is also regulated by the HrpG and HrpX cascade (Figure 7), or regulate the expression of HrpG by phosphorylation as speculated in R. solanacearum [64]. Unfortunately, our electrophoretic mobility shift assay (EMSA) showed that HrpX did not bind the fbaB promoter (data not shown), suggesting that HrpX, a transcriptional activator, may form a complex with a yet unknown factor to regulate the expression of fbaB. The expression of this unknown factor may be inhibited by intermediates, like malate, from the TCA cycle in X. oryzae pv. oryzicola, resulting in the lower expression of HrpG and HrpX when the block of gluconeogenesis is made by the fbaB mutation (Figure 3, 4, 5 and 6).
In addition, the expression of hrcC, hrpE and hpa3, other than other hrp-hrc-hpa genes, of X. oryzae pv. oryzicola is still activated when fbaB is mutated. Previously, we found that the mutation of hrpG and hrpX does not abolish the expression of hrcC, hrpE and hpa3 and postulated that a yet unknown factor may influence the expression of these genes [20], [53]. This speculation is in accordance with our hypothesis in this report that the hexoses from plant photosynthesis induce the expression of virulence-related genes, including hrp and fbaB genes, and the intermediates, like malate, from the TCA cycle of X. oryzae pv. oryzicola repress the expression of the unknown factor gene that will in turn, directly or indirectly, suppress the expression of HrpG and HrpX and increase the expression of hrcC, hrpE and hpa3 which is involved in nutrient acquirement of pyruvate and malate when X. oryzae pv. oryzicola contacts the host cells (Figure 5 and 6). Importantly, this report may also provide clues to investigate a yet unknown factor which presumably plays a central role in regulation between carbohydrate metabolism and the hrp system of X. oryzae pv. oryzicola mediated by pyruvate and malate (Figure 7).
Supporting Information
Schematic map and molecular analysis of fabB mutation in X. oryzae pv. oryzicola . The positions and orientations of Xoryp_17640, encoding FbaB, and other adjacent ORFs are shown by using the genome sequence of X. oryzae pv. oryzicola BLS256 strain as the reference (http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=Xoc). Arrows indicate locations and orientations of the ORFs or protein IDs, and lines indicate the intergenic sequences. ▾above ORF Xoryp_17640 presents the insertion site of a transposon Tn5 derivative in mutant Mxoc0504. A non-polar construction of a fabB deletion mutant was sketched (see Materials and methods for detail). The white box stands for a 349 bp deletion of fbaB. The fbaB mutant was verified by PCR with the primer pair upF/downR (Table S1) and by Southern hybridization with a 546 bp fragment of fbaB gene as the probe. Lane 1, the wild-type strain RS105; Lane2, the fabB mutant RΔfabB; Lane M, DL2000 or λ - EcoT14 DNA marker (TaKaRa, Dalian, China).
(TIFF)
Primers used in this study.
(DOC)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the State Key Basic Research and Development Project of China (2011CB16141), the National Natural Science Foundation of China (31071656 to Dr. Chen and 31000071 to Zou) and the Ph.D. Programs Foundation of Ministry of Education of China (20100073110045). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Eisenreich W, Dandekar T, Heesemann J, Goebel W. Carbon metabolism of intracellular bacterial pathogens and possible links to virulence. Nat Rev Microbiol. 2010;8:401–412. doi: 10.1038/nrmicro2351. [DOI] [PubMed] [Google Scholar]
- 2.Tamir-Ariel D, Navon N, Burdman S. Identification of genes in Xanthomonas campestris pv. vesicatoria induced during its interaction with tomato. J Bacteriol. 2007;189:6359–6371. doi: 10.1128/JB.00320-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang LF, Rong W, He CZ. Two Xanthomonas extracellular polygalacturonases, PghAxc and PghBxc, are regulated by type III secretion regulators HrpX and HrpG and are required for virulence. Mol Plant-Microbe Interact. 2008;21:555–563. doi: 10.1094/MPMI-21-5-0555. [DOI] [PubMed] [Google Scholar]
- 4.Tang DJ, He YQ, Feng JX, He BR, Jiang BL, et al. Xanthomonas campestris pv. campestris possesses a single gluconeogenic pathway that is required for virulence. J Bacteriol. 2005;187:6231–6237. doi: 10.1128/JB.187.17.6231-6237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zagallo AC, Wang CH. Comparative Glucose Catabolism of Xanthomonas Species. J Bacteriol. 1967;93:970–975. doi: 10.1128/jb.93.3.970-975.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim SY, Lee BM, Cho JY. Relationship between glucose catabolism and xanthan production in Xanthomonas oryzae pv. oryzae. Biotechnol Lett. 2010;32:527–531. doi: 10.1007/s10529-009-0193-0. [DOI] [PubMed] [Google Scholar]
- 7.Letisse F, Chevallereau P, Simon JL, Lindley N. The influence of metabolic network structures and energy requirements on xanthan gum yields. J Biotechnol. 2002;99:307–317. doi: 10.1016/s0168-1656(02)00221-3. [DOI] [PubMed] [Google Scholar]
- 8.Lee BM, Park YJ, Park DS, Kang HW, Kim JG, et al. The genome sequence of Xanthomonas oryzae pathovar oryzae KACC10331, the bacterial blight pathogen of rice. Nucleic Acids Res. 2005;33:577–586. doi: 10.1093/nar/gki206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salzberg SL, Sommer DD, Schatz MC, Phillippy AM, Rabinowicz PD, et al. Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A. BMC Genomics. 2008;9:204–221. doi: 10.1186/1471-2164-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Qian W, Jia Y, Ren SX, He YQ, Feng JX, et al. Comparative and functional genomic analyses of the pathogenicity of phytopathogen Xanthomonas campestris pv. campestris. Genome Res. 2005;15:757–767. doi: 10.1101/gr.3378705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Silva AC, Ferro JA, Reinach FC, Farah CS, Furlan LR, et al. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature. 2002;417:459–463. doi: 10.1038/417459a. [DOI] [PubMed] [Google Scholar]
- 12.Thieme F, Koebnik R, Bekel T, Berger C, Boch J, et al. Insights into genome plasticity and pathogenicity of the plant pathogenic bacterium Xanthomonas campestris pv. vesicatoria revealed by the complete genome sequence. J Bacteriol. 2005;187:7254–7266. doi: 10.1128/JB.187.21.7254-7266.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu GT, Xie JR, Chen L, Hu JR, An SQ, et al. Glyceraldehyde-3-phosphate dehydrogenase of Xanthomonas campestris pv. campestris is required for extracellular polysaccharide production and full virulence. Microbiology. 2009;155:1602–1612. doi: 10.1099/mic.0.023762-0. [DOI] [PubMed] [Google Scholar]
- 14.Mellgren EM, Kloek AP, Kunkel BN. Mqo, a tricarboxylic acid cycle enzyme, is required for virulence of Pseudomonas syringae pv. tomato strain DC3000 on Arabidopsis thaliana. J Bacteriol. 2009;191:3132–3141. doi: 10.1128/JB.01570-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aldon D, Brito B, Boucher C, Genin S. A bacterial sensor of plant cell contact controls the transcriptional induction of Ralstonia solanacearum pathogenicity genes. EMBO J. 2000;19:2304–2314. doi: 10.1093/emboj/19.10.2304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brito B, Aldon D, Barberis P, Boucher C, Genin S. A signal transfer system through three compartments transduces the plant cell contact-dependent signal controlling Ralstonia solanacearum hrp genes. Mol Plant-Microbe Interact. 2002;15:109–119. doi: 10.1094/MPMI.2002.15.2.109. [DOI] [PubMed] [Google Scholar]
- 17.Rahme LG, Mindrinos MN, Panopoulos NJ. Plant and environmental sensory signals control the expression of hrp genes in Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1992;174:3499–3507. doi: 10.1128/jb.174.11.3499-3507.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schulte R, Bonas U. Expression of the Xanthomonas campestris pv. vesicatoria hrp gene cluster, which determines pathogenicity and hypersensitivity on pepper and tomato, is plant inducible. J Bacteriol. 1992;174:815–823. doi: 10.1128/jb.174.3.815-823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zou LF, Wang XP, Xiang Y, Zhang B, Li YR, et al. Elucidation of the hrp Clusters of Xanthomonas oryzae pv. oryzicola That Control the Hypersensitive Response in Nonhost Tobacco and Pathogenicity in Susceptible Host Rice. Appl Environ Microbiol. 2006;72:6212–6224. doi: 10.1128/AEM.00511-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li YR, Zou HS, Che YZ, Cui YP, Guo W, et al. A novel regulatory role of HrpD6 in regulating hrp-hrc-hpa genes in Xanthomonas oryzae pv. oryzicola. Mol Plant-Microbe Interact. 2011 doi: 10.1094/MPMI-09-10-0205. (Accepted) [DOI] [PubMed] [Google Scholar]
- 21.Ankenbauer RG, Nester EW. Sugar-mediated induction of Agrobacterium tumefaciens virulence genes: structural specificity and activities of monosaccharides. J Bacteriol. 1990;172:6442–6446. doi: 10.1128/jb.172.11.6442-6446.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamir-Ariel D, Rosenberg T, Burdman S. The Xanthomonas campestris pv. vesicatoria citH gene is expressed early in the infection process of tomato and is positively regulated by the TctDE two-component regulatory system. Mol Plant Pathol. 2011;12:57–71. doi: 10.1111/j.1364-3703.2010.00652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tung SY, Kuo TT. Requirement for phosphoglucose isomerase of Xanthomonas campestris in pathogenesis of citrus canker. Appl Environ Microbiol. 1999;65:5564–5570. doi: 10.1128/aem.65.12.5564-5573.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Guo XX, Zou HS, Li YR, Zou LF, Chen GY. hrpD6 gene determines 27 Xanthomonas oryzae pv. oryzae to trigger hypersensitive response in tobacco and pathogenicity in rice. Acta Microbiologica Sinica. 2010;50:1155–1163. [PubMed] [Google Scholar]
- 25.Xiao YL, Li YR, Liu ZY, Xiang Y, Chen GY. Establishment of the hrp-inducing systems for the expression of the hrp genes of Xanthomonas oryzae pv. oryzicola. Acta Microbiologica Sinica. 2007;47:396–401. [PubMed] [Google Scholar]
- 26.Cunnac S, Boucher C, Genin S. Characterization of the cisacting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J Bacteriol. 2004;186:2309–2318. doi: 10.1128/JB.186.8.2309-2318.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He SY, Nomura K, Whittam TS. Type III protein secretion mechanism in mammalian and plant pathogens. Biochim Biophys Acta. 2004;1694:181–206. doi: 10.1016/j.bbamcr.2004.03.011. [DOI] [PubMed] [Google Scholar]
- 28.Hueck CJ. Type III protein secretion systems in bacterial pathogens of animals and 16 plants. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weber E, Koebnik R. Domain Structure of HrpE, the Hrp Pilus Subunit of 11 Xanthomonas campestris pv. vesicatoria. J Bacteriol. 2005;187:6175–6186. doi: 10.1128/JB.187.17.6175-6186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Büttner D, Bonas U. Port of entry-the type III secretion translocon. Trends in Microbiol. 2002a;10:186–192. doi: 10.1016/s0966-842x(02)02331-4. [DOI] [PubMed] [Google Scholar]
- 31.Büttner D, Bonas U. Getting across-bacterial type III effector proteins on their 16 way to the plant cell. EMBO J. 2002b;21:5313–5322. doi: 10.1093/emboj/cdf536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Büttner D, Nennstiel D, Klusener B, Bonus U. Functional analysis of HrpF, 21 a putative type III translocon protein from X. campestris pv. vesicatoria. J Bacteriol. 2002;184:2389–2398. doi: 10.1128/JB.184.9.2389-2398.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sugio A, Yang B, White FF. Characterization of the hrpF Pathogenicity 27 Peninsula of Xanthomonas oryzae pv. oryzae. Mol Plant-Microbe Interact. 2005;18:546–554. doi: 10.1094/MPMI-18-0546. [DOI] [PubMed] [Google Scholar]
- 34.Wengelnik K, Rossier O, Bonas U. Mutations in the regulatory gene hrpG of Xanthomonas campestris pv. vesicatoria result in constitutive expression of all hrp genes. J Bacteriol. 1999;181:6828–6831. doi: 10.1128/jb.181.21.6828-6831.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wengelnik K, Van den Ackerveken G, Bonas U. HrpG, a key hrp regulatory protein of Xanthomonas campestris pv. vesicatoria is homologous to two-component response regulators. Mol Plant-Microbe Interact. 1996;9:704–712. doi: 10.1094/mpmi-9-0704. [DOI] [PubMed] [Google Scholar]
- 36.Wengelnik K, Bonas U. HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J Bacteriol. 1996;178:3462–3469. doi: 10.1128/jb.178.12.3462-3469.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Furutani A, Nakayama T, Ochiai H, Kaku H, Kubo Y, et al. Identification of novel HrpXo regulons preceded by two cis-acting elements, a plant-inducible promoter box and a -10 box-like sequence, from the genome database of Xanthomonas oryzae pv. oryzae. FEMS Microbiol Lett. 2006;259:133–141. doi: 10.1111/j.1574-6968.2006.00265.x. [DOI] [PubMed] [Google Scholar]
- 38.Koebnik R, Krüger A, Thieme F, Urban A, Bonas U. Specific binding of the Xanthomonas campestris pv. vesicatoria AraC-type transcriptional activator HrpX to plant-inducible promoter boxes. J Bacteriol. 2006;188:7652–7660. doi: 10.1128/JB.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsuge S, Terashima S, Furutani A, Ochiai H, Oku T, et al. Effects on promoter activity of base substitutions in the cis-acting regulatory element of HrpXo regulons in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2005;187:2308–2314. doi: 10.1128/JB.187.7.2308-2314.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huang DL, Tang DJ, Liao Q, Li XQ, He YQ, et al. The Zur of Xanthomonas campestris is involved in hypersensitive response and positively regulates the expression of the hrp cluster via hrpX but not hrpG. Mol Plant-Microbe Interact. 2009;22:321–329. doi: 10.1094/MPMI-22-3-0321. [DOI] [PubMed] [Google Scholar]
- 41.Noël L, Thieme F, Nennstiel D, Bonas U. Two novel type III system-secreted proteins of Xanthomonas campestris pv. vesicatoria are encoded within the hrp pathogenicity island. J Bacteriol. 2002;184:1340–1348. doi: 10.1128/JB.184.5.1340-1348.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Oku T, Tanaka K, Iwamoto M, Inoue Y, Ochiai H, et al. Structural conservation of hrp gene cluster in Xanthomonas oryzae pv. oryzae. J Gen Plant Pathol. 2004;70:159–167. [Google Scholar]
- 43.Staslawicz BJ, Mudgett MB, Dangl JL, Galan JE. Common and contrasting themes of plant and animal diseases. Science. 2001;292:2285–2289. doi: 10.1126/science.1062013. [DOI] [PubMed] [Google Scholar]
- 44.Tsuge S, Nakayama T, Terashima S, Ochiai H, Furutani A, et al. Gene involved in transcriptional activation of the hrp regulatory gene hrpG in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2006;188:4158–4162. doi: 10.1128/JB.00006-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.He YW, Ng AY, Xu M, Lin K, Wang LH, et al. Xanthomonas campestris cell-cell communication involves a putative nucleotide receptor protein Clp and a hierarchical signalling network. Mol Microbiol. 2007;64:281–292. doi: 10.1111/j.1365-2958.2007.05670.x. [DOI] [PubMed] [Google Scholar]
- 46.Islam MR, Kabir MS, Hirata H, Tsuge S, Tsuyumu S. A leucine-rich protein, LrpX, is a new regulator of hrp genes in Xanthomonas oryzae pv. oryzae. J Gen Plant Pathol. 2009;75:66–71. [Google Scholar]
- 47.Zhang SS, He YQ, Xu LM, Chen BW, Jiang BL, et al. A putative colR (XC1049)-colS (XC1050) two-component signal transduction system in Xanthomonas campestris positively regulates hrpC and hrpE operons and is involved in virulence, the hypersensitive response and tolerance to various stresses. Res Microbiol. 2008;159:569–578. doi: 10.1016/j.resmic.2008.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Lee SW, Jeong KS, Han SW, Lee SE, Phee BK, et al. The Xanthomonas oryzae pv. oryzae PhoPQ two-component system is required for AvrXA21 activity, hrpG expression, and virulence. J Bacteriol. 2008;190:2183–2197. doi: 10.1128/JB.01406-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zou HS, Yuan L, Guo W, Li YR, Che YZ, et al. Construction of a Tn5-Tagged Mutant Library of Xanthomonas oryzae pv. oryzicola as An Invaluable Resource for Functional Genomics. Curr Microbiol. 2011;62:908–916. doi: 10.1007/s00284-010-9804-1. [DOI] [PubMed] [Google Scholar]
- 50.Miller JH. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 51.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 1989. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, U.S.A.
- 52.Turner PE. Phenotypic plasticity in bacterial plasmids. Genetics. 2004;167:9–20. doi: 10.1534/genetics.167.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang J, Zou HS, Li YR, Chen GY. Expression of the hrcC, hrpE and hpa3 genes is not regulated by the hrpG and hrpX genes in a rice pathogen Xanthomonas oryzae pv. oryzicola. Wei Sheng Wu Xue Bao. 2009;49:1018–1025. in Chinese. [PubMed] [Google Scholar]
- 54.Wang L, Makino S, Subedee A, Bogdanove AJ. Novel candidate virulence factors in rice pathogen Xanthomonas oryzae pv. oryzicola as revealed by mutational analysis. Appl Environ Microbiol. 2007;73:8023–8027. doi: 10.1128/AEM.01414-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang JL, Liu YN, Barber CE, Dow JM, Wootton JC, et al. Genetic and molecular analysis of a cluster of rpf genes involved in positive regulation of synthesis of extracellular enzymes and polysaccharide in Xanthomonas campestris pathovar campestris. Mol Gen Genet. 1991;226:409–417. doi: 10.1007/BF00260653. [DOI] [PubMed] [Google Scholar]
- 56.Denny TP. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Phytopathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 57.Yang W, Liu Y, Chen L, Gao T, Hu B, et al. Zinc uptake regulator (zur) gene involved in zinc homeostasis and virulence of Xanthomonas oryzae pv. oryzae in rice. Curr Microbiol. 2007;54:307–314. doi: 10.1007/s00284-006-0485-8. [DOI] [PubMed] [Google Scholar]
- 58.Say RF, Fuchs G. Fructose 1,6-bisphosphate aldolase/phosphatase may be an ancestral gluconeogenic enzyme. Nature. 2010;464:1077–1081. doi: 10.1038/nature08884. [DOI] [PubMed] [Google Scholar]
- 59.Hochster RM, Katznelson H. On the mechanism of glucose-6-phosphate oxidation in cell-free extracts of Xanthomonas phaseoli (XP8). Can J Biochem Physiol. 1958;36:669–689. [PubMed] [Google Scholar]
- 60.Winans SC. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J Bacteriol. 1990;172:2433–2438. doi: 10.1128/jb.172.5.2433-2438.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Seo YS, Sriariyanun M, Wang L, Pfeiff J, Phetsom J, et al. A two-genome microarray for the rice pathogens Xanthomonas oryzae pv. oryzsae and X. oryzae pv. oryzicola and its use in the discovery of a difference in their regulation of hrp genes. BMC Microbiol. 2008;8:99. doi: 10.1186/1471-2180-8-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Furutani A, Tsuge S, Ohnishi K, Hikichi Y, Oku T, et al. Evidence for HrpXo-dependent expression of type II secretory proteins in Xanthomonas oryzae pv. oryzae. J Bacteriol. 2004;186:1374–1380. doi: 10.1128/JB.186.5.1374-1380.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Watt TF, Vucur M, Baumgarth B, Watt SA, Niehaus K. Low molecular weight plant extract induces metabolic changes and the secretion of extracellular enzymes, but has a negative effect on the expression of the type-III secretion system in Xanthomonas campestris pv. campestris. J Biotechnol. 2009;140:59–67. doi: 10.1016/j.jbiotec.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 64.Yoshimochi T, Hikichi Y, Kiba A, Ohnishi K. The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signaling proteins. J Bacteriol. 2009;191:3424–3428. doi: 10.1128/JB.01113-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang YP, Zou LF, Zhou D, Chen GY. Key roles of hrpE gene of Xanthomonas oryzae pv. oryzicola in formation of Hrp pilus and pathogenicity in rice. Acta Phytopathologica Sinica. 2009;39:392–398. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Schematic map and molecular analysis of fabB mutation in X. oryzae pv. oryzicola . The positions and orientations of Xoryp_17640, encoding FbaB, and other adjacent ORFs are shown by using the genome sequence of X. oryzae pv. oryzicola BLS256 strain as the reference (http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=Xoc). Arrows indicate locations and orientations of the ORFs or protein IDs, and lines indicate the intergenic sequences. ▾above ORF Xoryp_17640 presents the insertion site of a transposon Tn5 derivative in mutant Mxoc0504. A non-polar construction of a fabB deletion mutant was sketched (see Materials and methods for detail). The white box stands for a 349 bp deletion of fbaB. The fbaB mutant was verified by PCR with the primer pair upF/downR (Table S1) and by Southern hybridization with a 546 bp fragment of fbaB gene as the probe. Lane 1, the wild-type strain RS105; Lane2, the fabB mutant RΔfabB; Lane M, DL2000 or λ - EcoT14 DNA marker (TaKaRa, Dalian, China).
(TIFF)
Primers used in this study.
(DOC)