Abstract
The deleterious and sometimes fatal outcomes of bacterial infectious diseases are the net result of the interactions between the pathogen and the host, and the genetically tractable fruit fly, Drosophila melanogaster, has emerged as a valuable tool for modeling the pathogen–host interactions of a wide variety of bacteria. These studies have revealed that there is a remarkable conservation of bacterial pathogenesis and host defence mechanisms between higher host organisms and Drosophila. This review presents an in-depth discussion of the Drosophila immune response, the Drosophila killing model, and the use of the model to examine bacterial–host interactions. The recent introduction of the Drosophila model into the oral microbiology field is discussed, specifically the use of the model to examine Porphyromonas gingivalis–host interactions, and finally the potential uses of this powerful model system to further elucidate oral bacterial-host interactions are addressed.
Keywords: Drosophila melanogaster, Porphyromonas gingivalis, Pathogen-host interactions, Periodontitis
Introduction
The use of invertebrate animal models has provided tremendous insight into the pathogen–host interactions of many human pathogens, and has revealed that many aspects of these interactions in higher host organisms are conserved in invertebrates. One of these animal models, the genetically tractable fruit fly, Drosophila melanogaster (Drosophila), has been well established as a model for studying the host–pathogen interactions of bacterial (1–9), fungal (10–17) and viral pathogens, and prior to the sequencing of the mosquito genome and the subsequent development of genetic tools, the malaria parasite (23–26); the model has also been used to probe the host defense response to infection.
This article will first provide an overview of the Drosophila model and address the advantages and drawbacks of the model for studying pathogen–host interactions. Next, the Drosophila immune response will be described in detail, and the various ways that the model has been used to study bacterial–host interactions will be addressed. Finally, the studies that used the Drosophila model to investigate Porphyromonas gingivalis–host interactions will be reviewed, followed by a discussion of the potential contributions that can be made using this model, toward a better understanding of oral bacteria–host interactions.
D. melanogaster as a model for studying host–pathogen interactions
Numerous studies have revealed that there is significant homology between the Drosophila immune response and the mammalian innate immune response (discussed in depth below). The absence of an adaptive immune response permits the study of the interactions between pathogens and the host's innate immune response in isolation. Drosophila are affordable to breed, are easy to handle, have a short generation time (10–14 days depending on the ambient temperature), have a clear endpoint (death), and can be used in quantities large enough to permit statistical analysis of the data. The Drosophila genome is fully sequenced (27–29), and numerous well-developed genetic tools are available for the manipulation and analysis of Drosophila responses [reviewed in (30, 31)]. Transposon mutagenesis has been used to successfully create loss-of-function mutants of at least 53% of Drosophila genes, with the ultimate goal of inactivating all genes (32, 33). Mutations are usually linked to visually identifiable markers, e.g. eye color or wing morphology, to allow for easy identification of mutant animals. Technologies for transgenic expression of genes in Drosophila are well developed (34–37) and have been enhanced by the development of expression systems like upstream activation sequence (UAS)/GAL4 (yeast transcription activator) (38, 39) that allow for temporally and spatially regulated expression; linking transgenes with reporter genes like lacZ (β galactosidase) or gfp (green fluorescent protein) allows gene expression to be monitored. Mutant and transgenic Drosophila lines are readily available at stock centers and have been extensively used to probe the interactions between pathogens and the Drosophila host. Drosophila loss-of-function immune response gene mutants have been used to examine the roles of the genes in the response to infection with various pathogens [11, (16, 19, 40–43) and Table 2], and transgenic Drosophila have been used to monitor the activation of immune response pathways upon infection (44, 45) and to examine the effects of transgenically expressed pathogen proteins on the host (21, 26, 46). Microarray and proteomic platforms as well as RNA interference (RNAi) lines and libraries have been developed and used to perform genome-wide analyses of Drosophila responses, in whole animals (41, 47–56) and in the well-established Drosophila cell culture lines, Schneider-2 (S2, embryonic-derived phagocytic cell) and malignant blood neoplasm (mbn-2) (56–59). As an estimated 50% of Drosophila genes have mammalian homologs (60), results from these and other studies are relevant to mammals. A comprehensive collection of Drosophila information can be found online at FlyBase (http://www.flybase.net) (61), including but not limited to gene annotation information, stock availability, images, references, and investigator contact information. Other databases containing Drosophila information include Berkeley Drosophila Genome Project (BDGP), Drosophila Interactions Database (DroID), and FlyView.
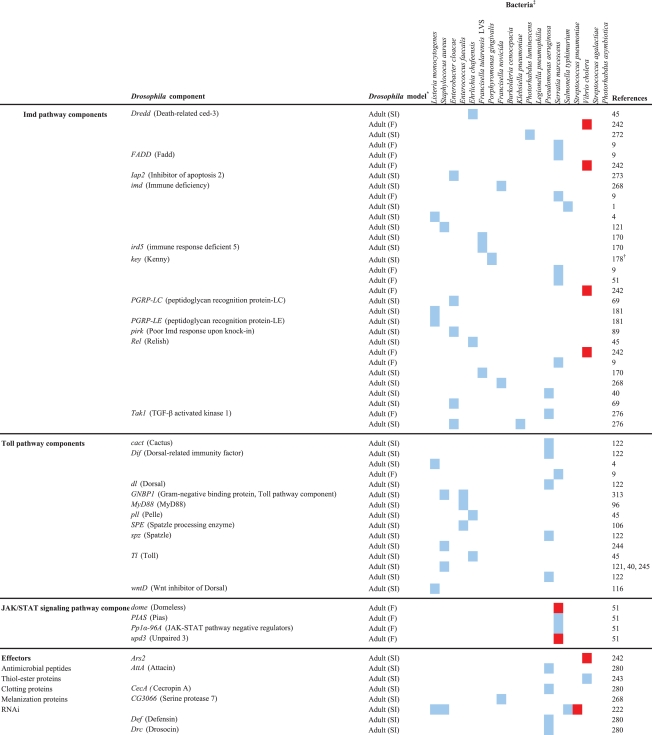
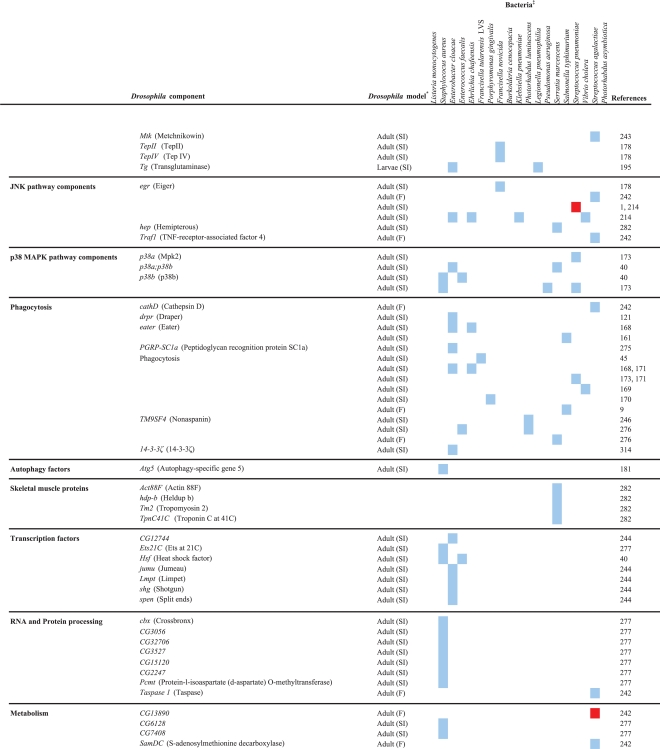
Table 2.
Drosophila components that are involved in the response to bacterial infection
 |
 |
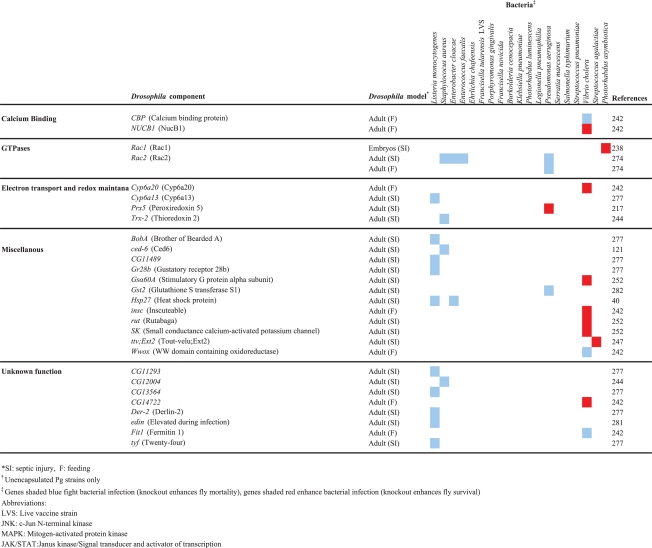 |
It is important to note that as Drosophila are not natural hosts for most of the human pathogens studied with this model and lack homologs for many mammalian immune response features, e.g. adaptive immunity, care should be taken when translating findings to mammals. Drosophila are usually reared at room temperature (25°C), and they can be incubated at temperatures of up to 30°C during infection experiments without affecting survival, although males become sterile at this temperature. Above 30°C, Drosophila physiological processes begin to deteriorate, and they are rapidly killed at 37°C (62), which is the optimal growth temperature for most human pathogens. In addition, some pathogen virulence genes are selectively expressed at 37°C, which could result in the absence of a virulence phenotype in the Drosophila model. Although the Drosophila hemolymph (blood) is not involved in respiration, it receives oxygen via the trachea, which likely makes it an inhospitable environment for obligate anaerobes. The Drosophila model may be more suitable for studying aerobic and facultatively anaerobic microbes; however, the model has been successfully used to examine the interactions between the obligately anaerobic oral bacterium P. gingivalis and the host. Even though P. gingivalis did not multiply in the flies, the bacterium viably persisted in them for up to 60 h after infection, likely aided by its high degree of aerotolerance (63).
In summary, the availability of a large number of well-developed tools for manipulating Drosophila genetics and analyzing Drosophila immune responses make it a powerful model for studying pathogen–host interactions, and numerous studies have convincingly demonstrated that strong correlation exists between microbial pathogenesis in mammals and in Drosophila.
The Drosophila immune system
Drosophila rely solely on an innate immune system to combat infecting microbes, and like mammals, they detect the presence of invading microbes using pattern recognition receptors, which recognize conserved microbial motifs and activate a response that is specific for the type of invading microbe [see (64) for a recent review].
Humoral immune response
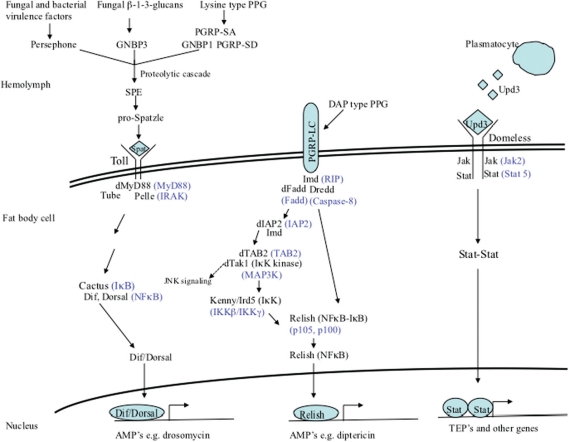
The Drosophila humoral response is mediated by three signaling pathways: the Imd (immune deficiency), Toll, and JAK/STAT (janus kinase/signal transducer and activator of transcription) pathways (Fig. 1). The hallmark of this response is the transient synthesis and release of antimicrobial peptides (AMPs) by the fat body – the major immune responsive tissue in Drosophila and the functional equivalent of the mammalian liver – directly into the hemolymph, where they accumulate to their effective concentrations. The Toll and Imd pathways are major regulators of Drosophila immune response genes and have been very well studied [see (65) for a recent in depth review].
Fig. 1.
Drosophila signaling pathways that regulate humoral responses. The Toll pathway regulates the expression of genes in response to the detection of Gram-positive type PPG or fungal β-1-3-glucans or activation by some fungal and bacterial proteases. The Imd pathway regulates the expression of genes in response to the detection of Gram-negative type PPG. The JAK/STAT pathway regulates the expression of genes in response to Upd3 signaling by plasmatocytes. Mammalian homologs of pathway components are shown in blue. Negative regulators of the pathways are not shown.
The Imd pathway
The Imd signaling pathway (Fig. 1) is homologous to the mammalian tumor necrosis factor receptor 1 signaling pathway, though they differ at the level of detection/activation. This pathway regulates both systemic and local (in response to natural infection) AMP expression, by fat body cells and gut epithelial cells, respectively, in response to primarily Gram-negative bacterial infections [(66–68) also see Table 2].
Activation and signal transduction
The pathway is activated by the direct detection of monomeric or polymeric meso-diaminopimelic (DAP)-type (Gram-negative type) peptidoglycan (PPG) by the transmembrane receptor, peptidoglycan recognition protein-LC (PGRP-LC) (69, 70). Binding of PPG by PGRP-LC leads to the intracellular recruitment of the adaptor molecule Imd (71–73) (receptor interacting protein, RIP, homolog), and the subsequent recruitment of Drosophila Fas-associated death domain protein (dFADD, FADD homolog) and the caspsase-8 homolog DREDD to the Imd/PGRP-LC complex (74). Drosophila inhibitor of apoptosis-2 (dIAP2) (75) and transforming-growth-factor-β activating kinase (dTAK1, IκB-kinase-kinase) (68) are also recruited, and dTAK1 becomes activated by TAK1-binding protein 2 (TAB2). dTAK1 phosphorylates the IκB-kinase complex (IKKβ/Ird5 and IKKγ/Kenny) (76), which in turn phosphorylates the dual domain nuclear factor kappa B (NFκB) family protein Relish (77–79). Relish is cleaved, possibly by DREDD, which separates the inhibitory ankyrin domain from the Rel-homology domain (80). The phosphorylated Rel domain then translocates into the nucleus where it activates the transcription of Imd regulated genes (81) (discussed below).
Downregulation
To prevent the deleterious effects of an unchecked inflammatory response, the Imd pathway is regulated on multiple levels. Several PGRPs function as N-acetylmuramoyl-l-alanine amidases that scavenge and degrade PPG, converting it into non-immunostimulatory fragments. These include PGRP-SC1, -SC2 (82), -SB1 (83), and –LB, which is itself upregulated by the pathway creating a negative feedback loop (84). PGRP-LF is a membrane-bound receptor that sequesters DAP-type PPG and prevents it from binding to PGRP-LC and activating Imd signaling (85). Two proteins Caspar (Fas-associated factor 1 homolog) and Dnr1 block DREDD mediated cleavage of Relish (86, 87). Pirk/Rudra/PIMS (PGRP-LC-interacting inhibitor of Imd signaling) functions in a negative feedback loop to disrupt Imd/PGRP-LC binding and inhibit Imd signaling (88–90), and the Ras/MAP kinase [mitogen-activated protein kinase (MAPK)] pathway negatively regulates Imd pathway activation by modulating the expression of Pirk/Rudra/PIMS (91). Finally, a ubiquitin-proteasome complex represses Imd signaling by degrading one of the pathway components, likely Relish (92).
The Toll pathway
The Toll signaling pathway (Fig. 1) is homologous to the mammalian Toll/IL-1 receptor-signaling pathway, although unlike the mammalian receptors Drosophila Toll does not directly recognize bacterial components. The pathway regulates systemic AMP expression by the cells of the fat body, primarily in response to fungal and Gram-positive bacterial infections [93–(97), also see Table 2].
Activation and signal transduction
The pathway is activated in response to the detection of lysine-type (Gram-positive type) PPG (LPPG) by circulating recognition molecules PGRP-SA (95) and Gram-negative binding protein 1 (GNBP1) (98), which function in a complex (99), and PGRP-SD (100). The Toll pathway is also activated in response to the detection of fungal cell wall β-1-3-glucans by GNBP3 or by the activation of the Drosophila protease Persephone by fungal- and bacterial-secreted virulence factors (101–104). A common proteolytic cascade is activated downstream of the recognition molecules (105) that culminates in the activation of Spatzle processing enzyme (SPE), which cleaves and activates the cytokine Spatzle (106). Activated Spatzle binds to the cell transmembrane receptor, Toll, and induces the receptor's dimerization, which recruits the adaptors, Drosophila myeloid differentiation factor 88 (dMyD88, homolog of mammalian MyD88), Tube (serine-threonine kinase), and Pelle (homolog of mammalian IL-1R associated kinase) intracellularly (96, 107). Subsequent to Pelle activation Cactus (inhibitor of kappa B, IκB) becomes phosphorylated and is then degraded, freeing the (NFκB) family proteins Dif and Dorsal to translocate into the nucleus and activate the expression of Toll-regulated genes (108–111). A functional redundancy exists between Dif and Dorsal in the control of immune response gene expression (as measured by drosomycin induction) in Drosophila larvae (112), but not in adults where Dif alone is sufficient for the induction of defensin and drosomycin (113), and the presence of either protein is sufficient for the induction of cecropin (94). Also, deformed epidermal autoregulatory growth factor 1 (Deaf1) is required downstream of Dorsal and Dif to activate genes (e.g. drosomycin) in response to fungal infections (114).
Downregulation
Like the Imd pathway, the Toll pathway is negatively regulated on multiple levels. Toll pathway activation induces the expression of Cactus (NFκB inhibitor) (115) and WntD (Wnt inhibitor of Dorsal) (116), creating negative feedback loops. Drosophila Ubc9 (dUbc9) also inhibits Toll pathway signaling at the level of Dif and Dorsal (117). Finally, a serine protease inhibitor (Spn1) acts upstream of SPE to downregulate Toll signaling in response to fungal infections, in a negative feedback manner (118).
Toll- and Imd-regulated genes
The Toll and Imd pathways regulate the expression of a subset of Drosophila genes that are induced upon septic injury (52, 53, 56). There is clearly some functional overlap and synergy in gene regulation between the pathways, as revealed by microarray- (53) and RNAi-analyses (119), the detection of Dif-Relish heterodimers in fly extracts (120), and the observation that Pseudomonas aeruginosa, Staphylococcus aureus, Listeria monocytogenes and Ehrlichia chafeensis activate both signaling pathways (4, 121–123). The most well characterized Drosophila immune effectors regulated by the pathways are the AMPs, of which there are seven classes. Cecropin (antibacterial) (124) and Defensin (antibacterial) (125) form ion channels in the cytoplasmic membrane (126, 127), diptericin (antibacterial) (128) and drosomycin (antifungal) (129, 130) act on the cytoplasmic membrane causing lysis (131, 132), Attacin (antibacterial) (133) interferes with outer membrane synthesis (133), Drosocin (antibacterial) (134) interferes with the activities of DnaK (135), and the mode of action of Metchnikowin (anti-bacterial and –fungal) (136) has not yet been determined. The Toll and Imd signaling pathways together regulate most of the AMPs, but diptericin and drosomysin are primarily regulated by Imd and Toll, respectively, and are, therefore, used to monitor pathway activation (53, 66, 137). Microarray analysis showed that 283 of 400 previously identified (52) Drosophila immune response genes are regulated by either one or both pathways. In addition to the AMPs and pathway regulators, the proteins encoded by these genes are involved in iron metabolism, opsonization, melanization, iron sequestration, coagulation, reactive oxygen species (ROS) production, Jun kinase (JNK) pathway signaling, stress response, and many proteases (53).
JAK/STAT pathway
The Drosophila JAK/STAT signaling pathway (Fig. 1) comprises the same components as the mammalian pathway, although they differ in the number of JAKs (one in Drosophila, four in mammals) and STATs (one in Drosophila, seven in mammals) they possess.
Activation and signal transduction
In response to septic injury, Drosophila plasmatocytes (macrophage like cells) secrete the cytokine, unpaired-3 (Upd3) (138, 139), which binds to the fat body cell transmembrane receptor, Domeless (homolog of mammalian class 1 cytokine receptor) (140, 141). Upd3 binding induces Domeless dimerization and the intracellular recruitment and phosphorylation of two Hopscotch (JAK) molecules, followed by two STAT92E (homolog of mammalian STAT5) molecules. Activated STAT92E molecules dimerize and translocate into the nucleus where they activate the expression of target genes (139).
Regulated genes
The JAK/STAT pathway regulates numerous physiological processes [reviewed in (142, 143) including immune responses; however, the contribution of this pathway to immune regulation has not been as well studied as the Toll and Imd pathways. In response to septic injury, the JAK/STAT pathway induces the expression of CG11501, which remains functionally uncharacterized (56) and, in combination with several other pathways (144), induces the expression of Turandots, which are stress-response proteins currently found only in Drosophila (145). The pathway is involved in the immune response in the Drosophila gut, where it helps maintain epithelial cell homeostasis, by regulating stem cell proliferation (51, 146). Activation of the JAK/STAT pathway in response to an intestinal Serratia marcescens infection negatively impacts Drosophila survival (51).
Downregulation
The JAK/STAT pathway is regulated on multiple levels like Imd and Toll. Drosophila protein inhibitor of activated STATs (dPIAS) inhibits STAT92E-dependent transcription by physically interacting with and blocking its DNA binding ability (147). Suppressor of cytokine signaling 36E (SOCS36E, mammalian soc5 homolog) expression is induced by JAK/STAT signaling and inhibits the pathway in a negative feedback loop (148, 149); a truncated form of STAT92E, which is transcribed from an alternate promoter, acts as a dominant negative regulator of JAK/STAT signaling (150); eye transformer, a receptor that is structurally related to Domeless, negatively regulates JAK/STAT signaling by interfering with STAT92E phosphorylation and activation (151).
Although the Drosophila antiviral immune response is not well characterized, a role for the Imd, Toll, and JAK-STAT pathways has been demonstrated (156–159).
Cellular immune response
The Drosophila cellular immune response is mediated by three sets of hemocytes: plasmatocytes, crystal cells, and lamellocytes, and while all three are found in larvae, only plasmatocytes are found in adults (160).
Plasmatocytes
These cells are similar to mammalian macrophages in that they express scavenger receptors (70, 161–165), secrete cytokines (139, 166), and are responsible for the phagocytosis of microbes (45, 167–170) and apoptotic cells (167, 171). Drosophila plasmatocytes also contribute to hemolymph coagulation by secreting the major clotting factor hemolectin (172), promote tolerance to intracellular bacterial infection (see below) (173), and utilize autophagy to control some intracellular pathogens (see below).
Phagocytosis
Phagocytosis involves several receptors many of which have mammalian homologs [see (174) for a recent in-depth review of Drosophila phagocytosis]. These include EGF repeat containing proteins Nimrod (162), Eater (161), and Draper (121), the scavenger receptors Croquemort (CD36 family), Peste (CD36 family) (165, 175), and Scavenger receptor CI (dSR-CI) (163), PGRP-LC (70), and Down syndrome cell adhesion molecule (Dscam, homolog of mammalian DSCAM) (164). Thus far, convincing in vivo evidence for the role of receptors in phagocytosis has only been obtained for Eater (161) and Nimrod (162). Phagocytosis may also be enhanced by the thiolester proteins (TEPs), which are members of the complement C3 (C3)/α2-macroglobulin (α2M) superfamily. The Drosophila genome encodes four Teps (I–IV) that are expressed at basal levels by fat body cells and larval hemocytes but strongly upregulated after a bacterial challenge (I, II, and IV in larvae and II and IV in adults); a fifth gene Tep V is believed to be a pseudogene (176). In vitro RNAi treatment of Drosophila S2 cells revealed that Teps II and III enhance the phagocytosis of Escherichia coli and S. aureus, respectively, likely by functioning as opsonins, while a sixth protein Tep VI (also known as macroglobulin complement related, Mcr) binds to and enhances the phagocytosis of Candida albicans (177). It has been suggested that Tep II may function as an α2M by virtue of its five splice isoforms that only differ in a domain that is similar to the bait region of α2Ms (176); however, this function has not been demonstrated. Although an in vivo role for the Teps (II and IV) in combating P. gingivalis infection has recently been demonstrated (178), Teps I–IV have been found to be dispensable for Drosophila survival after infection with a variety of other bacterial and fungal pathogens (179). RNAi studies have demonstrated that phagocytosis involves actin rearrangement (7, 177, 180), and phagocytosed microbes are taken up into a phagosome (57) and destroyed by mechanisms that are not yet clearly understood.
Autophagy
Autophagy is an evolutionarily conserved process by which cells envelope their cytoplasmic contents into double-membrane vesicles and, subsequently, deliver them to the lysosome for degradation. It is involved in the control of intracellular pathogen infections in mammals and was first reported as a Drosophila immune response by Yano et al., who observed that in vivo RNAi targeting an essential autophagy factor (Atg5) rendered Drosophila more susceptible to infection by the intracellular pathogen L. monocytogenes (181). They subsequently observed L. monocytogenes containing autophagosomes in Drosophila hemocytes using electron microscopy and identified the intracellular form of PGRP-LE (which detects DAP-type PPG) as the receptor responsible for the induction of autophagy. Autophagy is a part of the Drosophila antiviral immune response; however, the initiating receptor remains unidentified (182). Autophagy can also promote infection by pathogens such as the intracellular fungus Cryptococcus neoformans, which co-opts the autophagic system to maintain its lifestyle within Drosophila plasmatocytes (180).
Lamellocytes and crystal cells
Only present in Drosophila larvae, these cells are involved in encapsulation (lamellocytes) and melanization (crystal cells), immune responses that have no mammalian counterparts. Parasites that are too large to be phagocytosed, e.g. wasp eggs, are surrounded by lamellocytes and encapsulated, walling them off from the rest of the hemocoel (body cavity) (183, 184). The encapsulated particle is then destroyed by the local production of ROSs (185, 186), RNSs (187), and melanization, which is the synthesis and deposition of toxic melanin (188, 189). Melanization also occurs at the site of a cuticle breach and in Drosophila adults.
Coagulation
Coagulation is an immediate reaction that occurs at the site of a wound and prevents hemolymph loss, promotes wound healing, and acts as a secondary barrier to infection by immobilizing microbes (190, 191). Studies on hemolymph clots from Drosophila larvae revealed that the most abundant protein in the clot is hemolectin (homolog of mammalian clotting factor von Willebrand factor), which is secreted by plasmatocytes (172, 190, 192). Once the primary clot fibers have been formed, they are cross linked to each other and to other clot proteins like Fondue (193), by the enzyme transglutaminase (homolog of mammalian factor XIIIa) (194). Transglutaminase also accumulates on the surface of microbes and targets them to the clot, a role also observed for factor XIIIa in mammals (195). Drosophila clotting factor mutants have coagulation defects and are more sensitive to microbial infection (172, 190, 195).
Additional immune response features
MAPK pathways
Two MAPK pathways, the p38 MAPK pathway and the JNK pathway, contribute to the Drosophila immune response and are homologous to the mammalian pathways.
p38 MAPK pathway
p38 signaling in Drosophila has been shown to be involved in mediating stress responses (196–198) and immunity (40, 173, 199). Two MAPKs, p38a and p38b, are encoded by the Drosophila genome (196, 199), and they are activated in response to infection by the upstream MAPK-kinase (MKK) Licorne (homolog of mammalian MKK3) (199), which itself is activated by the MKK-kinase MEKK1 (homolog of mammalian MEKK4) (196, 198–200). Once activated, p38 can then regulate the activity of transcription factors, such as ATF2 and d-Jun to control gene expression (199, 201). p38 null Drosophila are more susceptible than wildtype animals to killing by bacterial (40, and Igboin et al., unpublished) and fungal pathogens (40). Also overexpression of p38b conferred increased survival to Drosophila infected with intracellular bacteria (e.g. Salmonella typhimurium) without a corresponding decrease in bacterial load, and this was due to a p38b-dependent sequestering of the bacteria within plasmatocyte phagosomes (173) (see below for a discussion of tolerance). p38 signaling is negatively regulated by Alphabet, a phosphatase that antagonizes MEKK1 phosphorylation (202).
JNK pathway
In Drosophila, immune activation of JNK pathway signaling can occur via the binding of the cytokine Eiger (mammalian tumor necrosis factor homolog) to its receptor Wengen (mammalian TNF receptor homolog) (203–205), also via the Imd pathway at the level of dTak1 (76), and mediates apoptosis, wound healing, morphogenesis, and immunity (206–208). Downstream of Eiger/Wengen binding, one or more JNK kinase kinases (JNKKK) are activated by currently poorly understood mechanisms. The JNKKK dTak1, a shared component with the Imd pathway (76), activates two JNKKs, Hemipterous (mammalian MKK7 homolog) and Mkk4 (mammalian MKK4 homolog) (209), which in turn activate Basket (mammalian JNK homolog) (199, 209, 210). Activated JNK can then turn on the expression of transcription factors like AP1 (211) and other target genes. JNK signaling regulates a subset of immune response genes induced after septic injury (56) and also regulates AMP gene expression by the Imd pathway, downstream of Relish (212, 213). It has been reported that Drosophila JNK signaling in response to infection can either be beneficial or detrimental, as evidenced by the observation that Eiger null mutants are more susceptible than wildtype animals to killing by some pathogens, e.g. P. gingivalis and S. aureus (178, 214), but are more resistant than wildtype animals to killing by some intracellular pathogens, e.g. S. typhimurium (1, 214). It has also been reported that Eiger null mutants are no more susceptible to Gram-positive cocci, including S. aureus, infections than wildtype animals (215). Several negative regulators of JNK signaling have been identified, including Alphabet, a phosphatase that antagonizes dTAK1 phosphorylation (202), Pva (receptor tyrosine kinase) and Peroxiredoxin (redox sensing enzyme), which antagonize dTAK1 phosphorylation of JNK (216, 217), Puc, a phosphatase that antagonizes JNK activity (218), and Relish, whose activation results in proteosomal degradation of dTAK1 (219).
Tolerance
Tolerance is a process by which plants and animals endure a microbial infection and is one of the more recently described immune responses of Drosophila. Tolerance differs from resistance, in which the host reduces the fitness of the pathogen, thereby reducing the pathogen load. Tolerance in Drosophila has only been observed in response to bacterial infections, and the hallmarks of this response are increased fly survival without a corresponding decrease in the bacterial load (1, 173, 220, 221) or an increase in bacterial load without a corresponding decrease in fly survival (222). As examples of the first scenario, S. typhimurium-infected eiger null Drosophila survived 3 days longer than wt animals even though their bacterial loads were similar (1), and Drosophila overexpressing p38b survived S. typhimurium, L. monocytogenes, and Legionella pneumophilia infections longer than wildtype animals without a reduction in bacterial load (173). The mechanism of tolerance to S. typhimurium was shown to be via the sequestration of the bacteria within Drosophila hemocytes. As an example of the second scenario, wildtype and CG3066 (signaling protease involved in melanization) null Drosophila were killed at the same rate by Burkholderia cepacia, even though up to 25 times more bacteria were present in the mutant animals (222).
Reactive intermediate production
As mentioned previously, ROSs, e.g. superoxide anion, and RNSs, e.g. nitric oxide, are produced in Drosophila larvae during the melanotic encapsulation of particles that are too large to be phagocytosed (185, 186, 223). They also play a role in the immune response in the Drosophila gut where they are induced in response to natural infection (i.e. ingestion) (224, 225).
Drosophila dual oxidase (dDuox) is present on gut epithelial cells and contains a nicotinamide-adenine-dinucleotide-phosphate (NADPH)-oxidase domain and a peroxidase domain. The NADPH oxidase domain generates superoxide anion, which dismutates to hydrogen peroxide (H2O2), and the peroxidase domain uses the H2O2 to produce hypochlorous acid (HOCl) (224). Excessive ROS production is prevented by immune responsive catalase, which converts the H2O2 to water and oxygen (226).
NO participates in signaling in the gut of Drosophila larvae, which upregulate nitric oxide synthase expression upon natural infection with some Gram-negative bacteria. NO contributes to the induction of the AMP, diptericin, in the gut and hemocytes of naturally infected Drosophila larvae, and pharmacological inhibition of NOS has been shown to impair the ability of Drosophila larvae to survive septic and natural infections (225). The availability of a NO synthase null Drosophila mutant should shed additional light on the role of NO in the immune response.
Iron sequestration
The Drosophila genome encodes conserved iron-binding proteins transferrin (227) and ferritin (228), which can limit iron availability to invading microbes. Transferrin, ferritin, and iron transporters are upregulated in response to microbial challenges, which suggests a role for iron sequestration in controlling microbial infections in Drosophila (49, 50, 52, 227, 229).
RNA interference (RNAi)
RNAi involves the sequence-specific degradation of mRNA and is involved in the Drosophila antiviral immune response (19, 22, 230, 231). The role of RNAi in the Drosophila immune response was first reported by Li et al., who observed the accumulation of Flockhouse virus (FHV)-specific small interfering RNAs (siRNAs) in infected S2 cells (232). Galiana-Arnoux et al. (233), Zambon et al. (234), and van Rij et al (230) demonstrated an in vivo role for RNAi in combating viral infections. Drosophila also possess dsRNA uptake machinery (235, 236), which is important for the systemic RNAi response to extracellularly released (by controlled export or from lysed host cells) viral RNA (237).
In summary, three distinct pathways, Imd, Toll, and JAK/STAT, mediate the Drosophila humoral response, and the main humoral effectors are the AMPs, which are synthesized and secreted by the fat body. Two MAPK signaling pathways, JNK and p38, also regulate Drosophila immune responses, and all five signaling pathways are homologous to the pathways in mammals. The cellular immune response in Drosophila adults is mediated by plasmatocytes, macrophage-like cells that detect the presence of invading microbes via scavenger receptors, and phagocytose and subsequently destroy said microbes. Phagocytosis may be aided by the Teps, C3/α2M family proteins that function as opsonins. Additional Drosophila immune defense strategies include the production of reactive intermediates (oxygen and nitrogen), RNAi (to combat viral pathogens), the sequestration of iron, autophagy (to combat intracellular pathogens), and endurance of infections.
Drosophila infection models
Drosophila cultured cell lines (S2 and mbn-2), embryos, larvae, and adults (male and female) have been used to study host–pathogen interactions; however, adults and cultured cells are the most widely used. Drosophila embryos, larvae, and adults offer the advantage of whole animal models to examine pathogen–host interactions, and in all three models, the readout for pathogen virulence is fly mortality. A Drosophila embryo model was developed to study Photorhabdus asymbiotica infection, as it offered the added advantage of allowing the interaction between the bacterium and actively migrating Drosophila hemocytes to be followed in vivo, and in real time, using confocal microscopy (238). Drosophila larvae and adults have been used to identify and characterize pathogen and host factors that promote or inhibit infection by pathogens like P. gingivalis (63, 178), Pseudomonas fluorescens (3, 239), P. aeruginosa (40, 122, 240), Vibrio cholera (241–243), S. aureus (244, 245), Klebsiella pneumonia (246), Streptococcus agalactiae (247), West Nile virus (19), Aspergillus fumigatus (13, 17), and Rhizopus oryzae (41). Drosophila adults have also been used to test the efficacy of antifungal (248–251) and antibacterial drugs (252–254) and other antibacterial therapies (255, 256). It is important to take into account the immunological differences between the models when choosing one. For example, Drosophila larvae possess all three types of hemocytes but adults possess only plasmatocytes. Also, larval hemocytes do not actively migrate to a site of infection but rather arrive there by direct capture from the open circulatory system, in which they are passively pumped around (257).
Drosophila S2- and mbn2-cell lines are used as models for plasmatocytes because they are immunocompetent, phagocytic, and express many plasmatocyte-specific scavenger receptors. They are very useful for studying the phagocytosis, trafficking, proliferation, and cell-to-cell spread of pathogens (2, 121, 177, 180, 258) and for large scale screening by RNAi to identify host factors that interact with pathogens (20, 51, 259–262).
Routes of infection
Three routes of infection have been used to introduce pathogens into Drosophila: injection/septic injury, ingestion/natural infection, and rolling (in fungal spores). The injection method introduces the pathogen directly into the Drosophila hemocoel and involves either pricking the body cavity (usually the thorax in adults) with a needle dipped into a concentrated culture or the injection of a precise dose of the pathogen using a nanoinjector. Ingestion is also referred to as natural infection and involves introducing the animals into vials containing filter disks soaked with media containing the bacterium of interest or vials containing a pregrown lawn of fungal spores. Some ingestion models require that the animals be starved prior to their introduction into food containing vials, which drives subsequently ingested food to the crop. The rolling method of infection has only been used with fungal pathogens and involves rolling the flies over a lawn of spores growing on a plate. The rolling and injection methods of infection require anesthetization, which is usually done with carbon dioxide, and the injection and rolling methods require subsequent transfer of the animals into vials with food. After infection, the animals are incubated at 25–30°C (embryos are covered with oil), and their survival is monitored. It is important to note that the route of infection affects the interaction of the pathogen with the host and can give different results. For example, the alb1 gene is involved in A. fumigatus virulence when the fungus is ingested by or introduced onto the cuticle of Drosophila but not when the fungus is injected into the animals (17).
Transgenic expression (non-infection based)
In situations where the Drosophila model is not permissive for infection, it has been possible to introduce pathogen virulence factors into the flies by transgenically expressing the virulence factors in the animals. Pathogen genes can be expressed ubiquitously (26) or in specific fly tissues such as the fat body (21), wings (263), and eyes (264) by placing the transgene under the control of tissue-specific promoters. This has facilitated the identification of host cell targets of Plasmodium sporozite proteins (26), anthrax lethal factor and edema factor (263, 265), pertussis toxin (266), HIV-1 viral protein U (21), and Epstein-Barr virus immediate-early protein BZLF1 (264) (Table 1).
Table 1.
Human bacterial pathogen genes that are involved in the infection of Drosophila
| Pathogen | Drosophila model (mode of infection)† | Pathogen genes involved in infection | References | |
|---|---|---|---|---|
| Promote infection | Inhibit infection | |||
| Bacillus anthracis | Adult (TE) | lef (lethal factor), cya (edema factor) | (263) | |
| Bordetella pertussis | Adult (TE) | ptx (pertussis toxin) | (266) | |
| Burkholderia cenocepacia | Adult (SI) | BCAL2831 (Two component regulatory system component) | cepR (Two component regulatory system component) | (267) |
| bscN (Type III secretion system ATP-binding protein) | ||||
| cepI (Two component regulatory system component) | ||||
| hldA (LPS biosynthesis) | ||||
| htrA (stress response protease) | ||||
| zmpA (zinc metalloprotease) | ||||
| zmpB (zinc metalloprotease) | ||||
| Francisella novicida | Adult (SI) | FTN_0649 (FAD-dependent 4Fe-4S ferrodoxin) | (268)* | |
| FTN_0869 (putative transglutaminase) | ||||
| FTN_0889 (putative transcriptional regulator) | ||||
| glpD (anaerobic glycerol-3-phosphate dehydrogenase) | ||||
| nadC (nicotinate-nucleotide pyrophosphorylase) | ||||
| OxyR (oxidative stress transcriptional regulator) | ||||
| pmrA response regulator | ||||
| udp (uridine phosphorylase) | ||||
| uvrA, uvrB, recB, ssb, mutM, ruvC (DNA repair) | ||||
| DNA repair: 9 | ||||
| Protein repair: 1 | ||||
| Transporter: 3 | ||||
| Other: 43 | ||||
| Adult (SI) | FPI genes: 14 | FTN_0119 | (271)* | |
| Other: 29 | pilA, pilB, pilM, fimT (Type IV pilus) | |||
| pckA ( Phosphoenolpyruvate carboxykinase) | ||||
| Adult (SI) | iglB, iglD (FPI proteins) | (269) | ||
| mglA (Virulence gene transcriptional regulator) | ||||
| Adult (SI) | Cell division: 1 | (58)* | ||
| DNA modification: 9 | ||||
| FPI: 3 | ||||
| Hypothetical: 24 | ||||
| Intergenic: 5 | ||||
| Metabolic: 41 | ||||
| Other: 13 | ||||
| Transcription/translation: 2 | ||||
| Transferases: 8 | ||||
| Transport: 28 | ||||
| Type IV Pili: 1 | ||||
| Unknown: 33 | ||||
| Francisella tularensis (LVS) | Adult (SI) | iglB, iglC, iglD (FPI proteins) | (170) | |
| mglA (Virulence gene transcriptional regulator) | ||||
| Klebsiella pneumoniae | Adult (SI) | TrpC (tryptophan biosynthesis) | (246) | |
| WaaQ (capsule and LPS biosynthesis) | ||||
| Listeria monocytogenes | Adult (SI) | actA (Actin polymerization) | (277) | |
| hly (Listeriolysin, hemolysin) | ||||
| Photorhabdus asymbiotica | Embryos (SI) | mcf1 (toxin) | (238) | |
| Porphyromonas gingivalis | Adult (SI) | capsular polysaccharide locus | (63) | |
| fimA (fimbriae) | ||||
| kgp (lysine specific protease) | ||||
| mfa (minor fimbriae) | ||||
| rgpA rgpB (arginine specific proteases) | ||||
| Pseudomonas aeruginosa | Adult (SI) | kerV (hypothetical methyltransferase) | (241) | |
| Adult (SI) | nrdD, nrdJ (Ribonucleotide reductases) | (301) | ||
| Adult (F) | lasl;rhll (Quorum sensing proteins) | qteE (Quorum sensing regulator) | (302) | |
| Adult (SI) | toxA (Exotoxin A) | (122) | ||
| Adult (SI) | iscR (Transcriptional regulator) | (303) | ||
| Adult (SI) | katA (Catalase A) | (303–305) | ||
| Adult (SI) | hcnC (Cyanide) | (279) | ||
| Adult (F) | exoS (Exotoxin) | (274) | ||
| Adult (SI) | OxyR (Oxidative stress transcriptional activator) | (306) | ||
| Adult (F) | relA (Nucleotide synthesis, stringent response) | (307) | ||
| Adult (F) | relA;spoT (Nucleotide synthesis, stringent response) | (308) | ||
| Adult (SI) | chpA, fimV, orf406, pilGHI, pilJ, pilK, pilL, orf2982 (Twitching motility) | (309) | ||
| Adult (SI) | dsbA (Disulfide isomerase) | (8, 122, 240) | ||
| Adult (F) | qscR | (5) | ||
| Adult (SI) | muxA (efflux pump) | (310) | ||
| pilA (Type IV pilus) | ||||
| Adult (SI) | exsA (Transcription activator, Type III secretory system) | (8) | ||
| exsD (Type III secretory apparatus component) | ||||
| pilV (Type IV pili, twitching motility) | ||||
| Adult (SI) | 33C7, 44B1 (Unknown) | (122) | ||
| gacA, mvfR (Two component regulatory system regulators, quorum sensing) | ||||
| mtrR, pstP (Transcriptional activators) | ||||
| phzB (Phenazine biosynthesis) | ||||
| plcS (Phospholipase C) | ||||
| pqsB (Hydroxy-alkylquinoline synthesis, quorum sensing) | ||||
| Adult (SI) | PA3001 (Oxidoreductase) | (311) | ||
| PA4489, PA5441 (Hypothetical, unknown function) | ||||
| pgk (Phosphoglycerate kinase) | ||||
| pgm (Phosphoglycerate mutase) | ||||
| pilI, cca (Twitching motility) | ||||
| pyrF (Orotidine decarboxylase) | ||||
| Adult (SI) | fabF1 (Beta-ketoacyl-acyl carrier protein synthase II) | (240) | ||
| flhB (Flagellar biosynthesis) | ||||
| opdO (Outer membrane porin) | ||||
| PA0272, prpR, hudR (transcription factors) | ||||
| PA0369, PA2077 (Hypotherical proteins) | ||||
| PA14_35740 (Transposase) | ||||
| PA2002 (Fatty acid transporter) | ||||
| pilF (Type IV pilus assembly protein) | ||||
| pvdI (Peptide synthesis) | ||||
| wspF (Methyl esterase) | ||||
| Pseudomonas fluorescens | larvae (F) | A4589 (Unknown function) | (3) | |
| gmd (LPS biosynthesis) | ||||
| gacA (Two component regulatory system protein) | (239) | |||
| Salmonella typhimurium | Adult (SI) | orgA (Salmonella pathogenicity island 1 component) | (1) | |
| phoP (two component regulatory system sensor) | ||||
| slrP (type III secretion effector) | ||||
| spiC, sseA, sseB, sseC,sseD (SPI2 translocation machinery) | ||||
| ssrA (Salmonella pathogenicity island 2 component) | ||||
| Serratia marcescens | Adult (SI) | wzm (LPS biosynthesis, O antigen) | (312) | |
| Streptococcus agalactiae | Adult (SI) | bca (alpha C protein, adhesin) | (247) | |
| clyE (cytolisin) | ||||
| cpsE (capsular polysaccharide) | ||||
| dltA (D-alanyl-lipoteichoic acid) | ||||
| Staphylococcus aureus | Adult (SI) | perR (Transcription regulator, oxidative stress and iron storage) | (254) | |
| pheP (Amino acid permease) | ||||
| Adult (SI) | atl (amidase–glucosaminidase, peptidoglycan degradation) | (245) | ||
| dltA (D-alanine ligase) | ||||
| mprF (lysylphosphatidylglycerol synthesis) | ||||
| SA0614 (Two component regulatory system component) | ||||
| SA0615 (Two component regulatory system component) | ||||
| ypfP (Glycolipid synthesis) | ||||
| Adult (SI) | itaS (lipoteichoic acid synthesis) | (121) | ||
| Vibrio cholera | Adult (F) | ctxB (Cholera toxin) | (241) | |
| kerV (hypothetical methyltransferase) | (241, 252) | |||
| Yersinia pseudotuberculosis | Adult (F) | kerV (hypothetical methyltransferase) | (241) | |
Studies where genome-wide screens were performed, and resulted in the identification of >20 genes. Genes that were further characterized are listed. If genes were functionally or otherwise classified, and the number of genes in each group was reported, we included the numbers in this table.
SI: septic injury, TE: transgenic expression (of pathogen virulence factors), F: feeding, R: rolling (in spores)
Abbreviations: LPS: lipopolysaccharide FPI: Francisella pathogenicity island LVS: Live vaccine strain
Drosophila–pathogen interactions
Drosophila as a model for analyzing bacterial virulence
Numerous studies have shown that bacterial virulence factors that are important for the successful infection of mammals are also involved in infecting Drosophila (1, 63, 170, 238, 240, 246, 247, 252, 254, 263, 267). Bacterial virulence factors that promote or inhibit infection in Drosophila, many of which are also involved in mammalian infection, are listed in Table 1. These include F. novicida oxidative stress transcriptional regulator, OxyR (268), L. monocytogenes actin polymerization protein, actA (4), and V. cholera toxin (252). P. aeruginosa has been extensively studied using the Drosophila model [see (64) for a recent review], and in the interest of brevity, this section of the review will address the use of the model to study Francisella tularensis virulence.
F. tularensis is a facultative intracellular pathogen that is the etiological agent of tularemia, a disease that affects humans and small vertebrates. Three subspecies, F. holartica, F. tularensis, and F. novicida, cause disease in humans and are vectored by blood-sucking insects. F. tularensis is able to infect and propagate in Drosophila mbn2 (170) and S2 cells (269) in vitro, killing the cells in the process. The bacterium also infects and kills adult flies in a dose-dependent manner (170, 269). Injection of GFP-expressing F. tularensis enabled the systemic spread of the bacteria and their localization to Drosophila hemocytes (about 55% of the bacteria were intracellular at the time of death) to be observed. Similar to infections in human cells, the bacteria are taken up into phagosomes in Drosophila S2 cells, which do not fuse with lysosomes, and from which the bacteria can escape within 30–60 min postinfection (269). The intracellular growth locus (igl) and macrophage growth locus (mgl) are involved in F. tularensis virulence in mice (270), and when several genes from these loci were mutated and the null mutants tested in Drosophila, they were found to be less virulent than the wildtype parental strain (170, 269).
Two hundred and forty-nine F. novicida transposon insertion mutants were individually evaluated in a Drosophila killing model, and 49 genes that were required for normal virulence in the flies were identified (Table 1) (271). The majority of the identified genes were novel F. novicida virulence factors; 43 mutants had attenuated killing, and the mutated loci in these strains included most of the Francisella pathogenicity island (FPI) genes; six mutants were hypervirulent in the model and included mutants in pilus assembly proteins, an outer membrane protein and a kinase. Most of the attenuated strains also had low proliferation abilities in mammalian macrophages (murine J774 cells), and many of them also demonstrated reduced cytotoxicity; the hypervirulent mutants were significantly more cytotoxic to the murine macrophages than the wildtype strains. The strong correlation between F. novicida mutant phenotypes in Drosophila and in mouse macrophages suggested that the survival of F. novicida-infected flies is a good indicator of the bacterium's pathogenesis in mammals.
A F. novicida transposon mutant library was screened to identify genes that were essential for growth and survival in Drosophila, and 149 negatively selected genes (56 confirmed, Table 1) were identified (268). Sixty of the genes had previously been identified as virulence factors in murine models. Seven of the negatively selected genes encoded proteins that are important for the bacterium's ability to resist oxidative damage, and the mutants showed increased sensitivity to reactive-oxygen producing agents in subsequent in vitro experiments. Screening the library in wt and Imd pathway-null Drosophila identified a subset of seven F. novicida genes that are required to resist Drosophila Imd-regulated defenses (genes that were negatively selected in wt flies but rescued in the mutant flies). Five of the seven mutants showed increased sensitivity to AMPs, modeled by polymyxin B, which are the main Imd-regulated immune effectors. The two mutants that showed no phenotype in the AMP killing assay were attenuated due to other Imd-dependent mechanisms, as similar levels of AMPs were induced in response to infection with these mutants as with the wt strain. Five genes, recB, pilA, pilB, pyrF, and manB, identified in this study as being essential for F. novicida growth and survival within the fly, were also identified by Ahlund et al. as being involved in F. tularensis virulence in Drosophila (271).
Another F. novicida transposon mutant library (3,050 alleles representing 1,448 genes) was first screened in cultured S2 cells to identify genes required for the intracellular proliferation of the bacterium, and then, a subset of the mutants was tested for their lethality in adult flies (Table 1) (58). Three hundred and ninety-four genes that when mutated resulted in a significant reduction in intracellular growth of the bacterium were identified, and 80 of the 168 subsequently tested mutants exhibited reduced lethality and proliferation in the adult animals. The mutated loci encoded FPI proteins and proteins involved in metabolism, type IV pili biogenesis, transport, and DNA modification. One hundred and thirty-five genes were also required for F. novicida replication in human macrophages. Genes that were newly identified as playing a role in F. novicida pathogenesis may not be identifiable by currently available mammalian models, or could be specifically required for the insect phase of the bacterium's life cycle. Four genes, minD, iglC, iglD, and FTN_0109, identified in this study as being impaired in growth and/or lethality in Drosophila, were also identified by Ahlund et al. as being involved in F. tularensis virulence in Drosophila (271). Seven genes, ruvC, glpD, mdaB, ilvE, kdpC, pilQ, and FTN_1014, identified in this study, were also identified by Moule et al. as being essential for F. novicida growth and survival within the fly (268).
The studies described above and referenced in Table 1 show that bacterial pathogens utilize many of the same virulence mechanisms to infect mammals and Drosophila, and that the Drosophila model is useful for identifying novel virulence factors.
Drosophila as a model for analyzing the host response to bacterial infection
The host response activated upon a bacterial infection can fight and resolve the infection or contribute to the pathology caused by the infection, and the vast array of Drosophila mutant and transgenic lines, and microarray, RNAi, and proteomics platforms has facilitated in-depth analyses of the host response to infection by a variety of pathogens. The results of studies in which several different Drosophila tools (immune response gene mutants, transgenic expression, whole genome microarray, and RNAi) were exploited to examine the host response to infection are discussed in this section of the review. Drosophila genes that are involved in the response to human bacterial pathogens are shown in Table 2.
Loss-of-function immune response gene mutants
A large collection of loss-of-function Drosophila mutants have been generated by numerous investigators in the field and are readily available for use to identify Drosophila immune response components that play a role during infection. The use of knockout mutants of pathway components demonstrated that the well-characterized Toll- and Imd-signaling pathways are involved in the response to a variety of bacterial infections (4, 45, 49, 122, 170, 272–275). For example, Drosophila Dif (NFκB)-null mutants were more susceptible to killing than wt animals by L. monocytogenes, demonstrating a role for the Toll pathway in the defense against the bacterium (4). Imd pathway mutants were more susceptible than wildtype animals to killing by F. tularensis LVS (170), and both Imd and Toll mutants were more susceptible to killing by Erlichia chaffeensis (45). Once a Drosophila–pathogen model has been established, mutants that are deficient in specific host processes can be used to examine the interaction between those processes and the microbe. For example, Phg1 (nonaspanin)-null Drosophila were found to be highly susceptible to Gram-negative (P. aeruginosa, E. cloacae, and K. pneumoniae) bacterial infections, and it was demonstrated that this enhanced susceptibility was due to the inefficient phagocytic abilities of the fly hemocytes (276).
A large library of 1,231 transposon insertion mutants representing 8% of the Drosophila genome was screened to identify host genes required to survive a L. monocytogenes septic injury infection (277). Eighteen Drosophila mutants with increased susceptibility to killing by the bacterium were identified (Table 2), and the mutated loci included those involved in ubiquitination, RNA processing, and transcription activation. A comparison of the growth rates of L. monocytogenes in sensitive mutants and wildtype animals identified two classes of mutants: those in which bacterial growth was elevated relative to wildtype and those in which the bacterial levels remained unchanged relative to wildtype. This suggested that the first class of mutants is immunocompromised, i.e. deficient in genes required to resist the L.monocytogenes infection, while the second class of mutants has reduced endurance-characterized by lowered survival without a corresponding increase in bacterial proliferation-to the bacterial infection. Surprisingly, no overlap was observed between the Drosophila genes identified in this study and genes identified in two previous RNAi screens, likely because the use of S2 cells for the RNAi screens limited the observations to the processes involved in the interaction between L.monocytogenes and just one type of cell, whereas the whole animal model allowed for a much wider variety of processes to be examined.
An even larger library of 6,200 Drosophila transposon mutants was screened for altered susceptibility to a V. cholera natural infection, and 16 mutants were identified that contained disruptions in genes encoding proteins with homology to conserved domain proteins or mammalian proteins of know function (242) (Table 2). Seven of the Drosophila mutants demonstrated enhanced resistance, while nine demonstrated lowered resistance relative to the wildtype strain. Interestingly, the Imd pathway was found to contribute to pathology under normal circumstances (i.e. pathway mutants were more resistant to infection). It was determined that the mechanism of Imd involvement in causing pathology was due in part to an enhancement of cholera toxin activity, as imd pathway mutants were more resistant than wildtype animals to killing by a cholera toxin null V. cholera mutant. A significantly higher number of apoptotic intestinal epithelial cells was observed in infected imd pathway mutants than in wildtype animals, which suggested that under normal circumstances programmed cell death- a defense mechanism against intracellular pathogens in eukaryotes- is repressed by the Imd pathway in response to V. cholera infection.
Transgenic Drosophila
As discussed above (Routes of infection), transgenic Drosophila expressing pathogen virulence factors have been used to identify host targets. Host genes that are absent from Drosophila have also been transgenically expressed in the flies to study their interactions with pathogens. For example, the paraoxonase (PON) family of enzymes can degrade the P. aeruginosa quorum sensing (QS) signaling molecule N-3-oxododecanoyl homoserine lactone, and although they are conserved in mammals and Caenorhabditis elegans, the enzymes are absent from Drosophila. A QS deficient P. aeruginosa strain, synthetic acyl-homoserine lactones (AHLs), and Drosophila overexpressing human PON1 were used to examine the effect of the enzyme on the signaling molecule and on P. aeruginosa virulence in vivo (278). Drosophila that were infected with the P. aeruginosa QS-deficient mutant and ingested a control sucrose solution survived significantly better that flies that ingested synthetic AHLs, showing that QS is important for P. aeruginosa virulence in vivo. When human PON1 was overexpressed in the Drosophila, the animals were protected from the lethality of a wildtype P. aeruginosa infection. When AHLs were administered to P. aeruginosa QS mutant-infected Drosophila, PON1 overexpressing Drosophila but not wildtype animals were protected from killing, which showed that the basis of the observed protection was the inactivation of the AHLs by PON1. Therefore, the manipulation of PON expression could be explored as a treatment for some infectious diseases. In another study, it was observed that Drosophila overexpressing rhodanase, a bovine enzyme that detoxifies cyanide, survived longer than wildtype animals when infected with cyanogenic P. aeruginosa strains (279).
Whole genome microarray analysis
Whole-genome transcriptional profiling of whole animals has been used to assess the Drosophila global immune response to various infections (280, 281). The transcriptional profiles of Drosophila septically infected with virulent (PA14) and avirulent (CF5) strains of P. aeruginosa were compared, in order to identify defence-specific genes (expression altered after CF5 infection) and pathogenesis-specific genes (expression altered after PA14 infection) (280). Based on these criteria, 213 defence-specific genes (133 upregulated, 80 downregulated) and 28 pathogenesis-specific genes (16 upregulated, 12 downregulated) were identified. Interestingly, while infection with strain CF5 significantly upregulated AMP gene expression, infection with strain PA14 did not. Microarray analysis of AMP transcript levels following infection with a PA14 avirulent isogenic mutant revealed similar levels of expression as CF5, suggesting that the ability to reduce Drosophila AMP expression is a virulence trait of PA14 and likely other virulent P. aeruginosa strains.
Although transcriptional profiling provides a large amount of data, a subset of genes can be selected for more in depth examination. For example, a follow-up study examined the defense-specific genes that were non-immunity related, in a bid to understand the susceptibility to infection that occurs following trauma (282). It was observed that skeletal muscle genes (SMGs) were upregulated upon CF5 infection but not upon PA14 infection, suggesting that PA14 may suppress SMG induction in response to infection. It was determined that the JNK pathway is involved in the regulation of SMGs, as the transcriptional profile of a JNK pathway hypomorph infected with CF5 showed almost no induction of SMGs. Also, bacterial counts were higher in the thoraces of the JNK hypomorph than the wildtype strain but similar in the abdomens, suggesting that the JNK pathway protects against P. aeruginosa infection by promoting local tissue reconstruction after an infection. Interestingly, this role for the JNK pathway in the local defense against P. aeruginosa in tissue was conserved in a mouse open wound trauma model.
RNAi analysis
RNAi has been used extensively to examine the Drosophila host response to microbial infection in recent years due to the establishment of RNAi lines (47), dsRNA libraries representing the majority of Drosophila genes (283, 284), and well-developed, relatively easy methodology for use with bacterial (259), fungal (59), and viral pathogens (285).
The generation of over 22,000 transgenic Drosophila RNAi lines representing 12,800 genes allow for the knock down of host genes in specific tissues of the whole animal (47). Thirteen thousand and fifty-three RNAi lines representing 78% of the genome were screened to identify host factors that affected Drosophila survival after an ingestion infection with S. marcescens (51). Eight hundred and eighty-five lines with altered survival were identified, of which 790 were susceptible (RNAi of the genes increased susceptibility to killing by S. marcescens), and 95 were resistant (RNAi of the genes decreased susceptibility to killing by S. marcescens). Subsequent RNAi screening of the genes in the Drosophila gut epithelium identified 166 genes that affected Drosophila survival, most notably JAK/STAT signaling pathway components. Further study of the role of JAK/STAT signaling revealed that the pathway negatively regulates Drosophila survival in response to a S. marcescens infection, due to a disruption of intestinal cell homeostasis.
Drosophila S2 cells readily take up dsRNA added to their culture medium, and use it to silence genes (286), and the Drosophila RNAi Screening Center developed an RNAi library representing 13,900 Drosophila genes (284). This library has been used to perform genome-wide screens in S2 cells that aimed to identify host factors involved in infection by several pathogens (20, 165, 260–262). A smaller targeted RNAi library representing 7,216 Drosophila genes that are conserved in metazoans (283) has been used to look specifically at host genes that may be relevant during human infection (177, 287, 288).
Drosophila as a model for examining polymicrobe–host interactions
Some human infectious diseases like periodontitis and cystic fibrosis are caused or exacerbated (in the case of CF) by polymicrobial communities; however, there is a dearth of in vivo models for studying the interactions among these microbes and between the microbes and the host. The polymicrobial community that colonizes the cyctic fibrosis airway results in persistent inflammation that ultimately destroys the lung, and P. aeruginosa is one of the primary CF pathogens. A Drosophila natural infection model was developed to examine the interaction between P. aeruginosa and other oropharyngeal (OF) species (including Streptococci, Neisseria, and Actinomyces) isolated from cystic fibrosis patients (289). A comparison of the survival of Drosophila infected with P. aeruginosa alone, each of the 40 OF species alone, and a combination of P. aeruginosa and the OF species identified three classes of microbes. The virulent class consisted of species that alone are able to kill Drosophila and enhanced the killing of Drosophila in combination with P. aeruginosa. The synergistic class represented species that alone are not pathogenic to Drosophila but, in combination with P. aeruginosa, significantly enhanced Drosophila killing. The avirulent group represented species that alone are not pathogenic to Drosophila and do not enhance the killing of Drosophila in combination with P. aeruginosa. A novel luminescence assay was used to monitor the expression of individual P. aeruginosa virulence genes in individual flies in real-time, during a coinfection with representatives of the synergistic class of OF species. P. aeruginosa virulence gene expression was altered in the presence of these OF species with half of the genes being upregulated, including several QS genes that are responsive to the interspecies signaling molecule autoinducer-2. This suggested that interspecies communication is important for modulating P. aeruginosa gene expression in a mixed microbial infection.
Using AMP (diptericin, cecropin, and drosomycin) expression as a readout for the immune response, three different host responses to a coinfection with P. aeruginosa and another OF species were observed: increased AMP expression as a result of the additive effect of both species, a suppression of AMP expression, and a synergistic activation of AMP expression to levels greater than would be achieved by the additive effect of both species. It was hypothesized that the hyperactivation of the immune response seen in the third instance could be detrimental to the animals. The results of this study demonstrate the complexity of polymicrobial- and polymicrobe–host-interactions and the power of the Drosophila model for deciphering these interactions.
Drosophila as a model for drug therapy and antibacterial screening
Drosophila provides the benefit of a whole animal context for drug and antibacterial testing, although the model is currently not as powerful as C. elegans for which high-throughput techniques have been developed (290). Drosophila can be used as an in vivo screen for antibiotics against bacteria like S. aureus (254) and V. cholera (252). Drosophila fed tetracycline or methicillin – which are used clinically to treat S. aureus infections – resisted a S. aureus infection that killed all flies fed a control sucrose solution (254). V. cholera infection in a Drosophila natural infection model mimics human cholera, and when the potassium ion channel blocker, clotrimazole, was co-administered with the bacterium the flies were resistant to the otherwise lethal infection (252).
Alternate antibacterial treatments such as phage therapy have been tested using Drosophila and have shown some promise. The lytic bacteriophage Caudovirales strains MPK1 and MPK6 were isolated based on their abilities to form plaques on lawns of P. aeruginosa (256). These phage also killed P. aeruginosa grown in broth culture, reducing the levels of the bacterium from 107 to 102 colony-forming units within 2 h. The efficacy of the phage against a P. aeruginosa infection in vivo was determined using a Drosophila septic injury model, and it was observed that when the phage were fed to the Drosophila, P. aeruginosa proliferation was inhibited and fly survival was enhanced. The phages were also protective against a P. aeruginosa infection in a mouse intraperitoneal model of infection.
D. melanogaster in oral microbiology
P. gingivalis
P. gingivalis is the first oral microbe that has been studied using the D. melanogaster model. More importantly, it is the first obligately anaerobic bacterium to be successfully studied using the model (63), despite the inhospitable oxygenated environment of the Drosophila hemolymph.
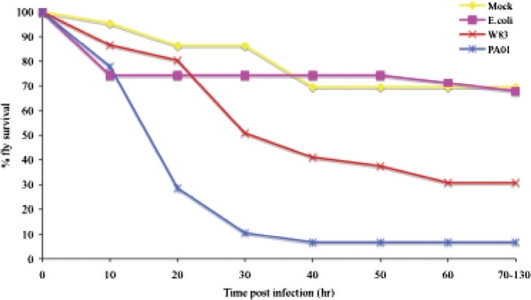
Using a septic injury route of infection, P. gingivalis was pathogenic to Drosophila, in a dose-dependent manner, with an intermediate level of pathogenicity between the non-pathogenic E. coil DH5-alpha and the highly pathogenic P. aeruginosa PA01 (Fig. 2). A comparison of clinically prevalent heteroduplex type strains of P. gingivalis revealed that all of the strains were virulent to some degree in Drosophila and that the highly disease-associated type strain, W83, was also the most pathogenic in the flies. P. gingivalis colony-forming unit levels did not increase in the Drosophila; however, the bacterium was able to persist in the flies up to 60 h postinfection. The relatively low temperature at which the infected flies were incubated (30°C), in addition to the oxygen rich environment of the hemolymph likely contribute to the inability of P. gingivalis to multiply in the host. However, P. gingivalis is aerotolerant and can survive exposure to air for up to 5 h without any loss in viability, which likely accounted for the bacterium's ability to persist in the Drosophila postinfection. P. gingivalis killing of Drosophila was not due to overt destruction of the fly tissues or a high bacterial burden as was observed with other pathogens, rather the observation that both live and heat-killed P. gingivalis effectively killed Drosophila suggested that the pathology may be due primarily to the host's own exaggerated immune response. Futher experiments are warranted to determine the exact cause of death of P. gingivalis-infected Drosophila, for example, in vivo RNAi could be used to dampen Drosophila immune responses and assess whether fly survival of P. gingivalis infection is enhanced as a result.
Fig. 2.
Survival curves of adult, female Drosophila infected with P. gingivalis and other bacterial species. E. coli DH5α (pink curve), P. aeruginosa strain PA01 (blue curve), P. gingivalis strain W83 (red curve), and vehicular control (yellow curve) (63).
Five P. gingivalis virulence gene mutants (arginine gingipains, lysine gingipain, major fimbriae, minor fimbriae, and capsule) that have been shown to be attenuated in rodent models of infection were also attenuated in the Drosophila model, demonstrating that P. gingivalis uses similar mechanisms of virulence in Drosophila and in mammals and that the bacterium's killing of Drosophila is multifactorial. It was hypothesized that as P. gingivalis spreads systemically (FITC-labeled bacteria were observed systemically using confocal microscopy), bacterial surface-associated components induce a systemic hyperactivation of the Drosophila immune response, which is not only bad for the bacterium but also harmful to the host.
To examine the Drosophila immune response to P. gingivalis infection (178), six immune response components were selected, and null mutants of these components were screened for altered susceptibility to P. gingvalis induced killing. Drosophila thiol-ester proteins, Tep II, Tep IV, and the JNK pathway ligand, Eiger (TNF homolog) were involved in the immune response against P. gingivalis infection, while the scavenger receptors Eater and Croquemort were dispensable for the response to P. gingivalis infection. Interestingly, the Imd pathway was initially found to be dispensable for the immune response against P. gingivalis, and because the strain that was used to infect the Drosophila (strain W83) is encapsulated, it was reasoned that the capsule may nullify any Imd pathway effects. This was supported by the observation that an unencapsulated P. gingivalis strain, strain 381, was significantly more pathogenic in Imd pathway-null flies than in wildtype flies. In a subsequent experiment, P. gingivalis strain W50UK, which is highly similar if not identical to W83, behaved like strain W83, while its isogenic capsule-null mutant, GPC, behaved like strain 381, when used to infect wildtype and Imd pathway-null flies, confirming that the bacterial capsule was responsible for the observed nullification of the Imd pathway.
To determine whether the P. gingivalis capsule prevented the activation of the Imd pathway by shielding the bacterium's peptidoglycan from detection by PGRP-LC, a dipt-LacZ reporter Drosophila strain was used to monitor activation of the pathway in response to W50UK and GPC infections. Beta galactosidase expression was detected in both cases, which demonstrated that the capsule of strain W50UK did not shield the bacterium's PPG from detection. To determine whether the P. gingivalis capsule could protect the bacterium from killing by Imd pathway regulated AMPs, the survival of strains W50UK and GPC after exposure to cecropin A and drosocin was compared. Strain GPC was significantly more susceptible to killing by the AMPs than W50UK, which showed that the P. gingivalis capsule mediates resistance to killing by Drosophila Imd-regulated AMPs. The results are also relevant to humans as the capsule also protected W50UK from killing by human beta-defensin-3. This was the first report to demonstrate that the P. gingivalis capsule is protective against host AMPs.
The future of the Drosophila model in the oral microbiology arena
Although the use of the Drosophila model for the study of oral microbiology is in its infancy, a wealth of information about oral bacteria–host interactions can be gleaned using this model. The major diseases of the oral cavity (periodontitis and caries) are polymicrobial in nature, and many putative periodontal (291–293) and caries (294, 295) pathogens have been identified and are yet to be characterized in terms of their interactions with the host. The introduction of the high-throughput Drosophila model will facilitate the screening of these potential pathogens, and the further characterization of species that are found to be virulent, as is being done with P. gingivalis. The Drosophila model is useful for studying P. gingivalis because this bacterium is aerotolerant, whereas a lot of oral bacteria will be killed by exposure to even a small amount of air and are, thus, unlikely to have virulent phenotypes in this model. Additional oral species have been screened for virulence using the Drosophila model, and a range of virulence has been observed (unpublished).
Further characterization of oral bacteria that are found to be pathogenic in the Drosophila model should involve screening for pathogen virulence factors, as well as host components that play a role in the response to infection. A large number of P. gingivalis mutants with in vitro phenotypes such as non-pigmentation, (296–299), enhanced biofilm forming ability, and loss of capsule synthesis (300) have been generated by transposon mutagenesis; however, likely due to the impracticality of using rodent models for large-scale screening, very few of these mutants have been tested for changes in virulence in vivo. The Drosophila model offers a high-throughput option to perform large-scale screening of these mutants to look for changes in virulence, which can then be further characterized. Also, the small size of Drosophila will make it relatively easy to profile its systemic response to infection, say with P. gingivalis, in a whole animal context, and could lead to the identification of novel host components that interact with the bacterium. Interesting candidates could be followed up on using null Drosophila mutants and/or in vivo RNAi.
The mixed microbial nature of periodontitis and caries warrants the study of polymicrobe interactions as well as polymicrobe–host interactions, and the work by Sibley et al (289) has laid the groundwork for the use of Drosophila to study these interactions in vivo. Studies are ongoing to examine the virulence of oral microbe mixtures comprised of P. gingivalis and other oral species, using Drosophila. The model could potentially be used to examine mixtures of microbes that are even more complex, possibly as complex as oral plaque.
Finally, Drosophila could potentially be used to test the efficacy of antimicrobials against oral bacteria in a whole-animal context; however, the model has not yet been developed for high-throughput drug testing.
It is important to keep in mind that Drosophila is not a natural host for oral microbes and that, due to anatomical and some host response – e.g. lack of an adaptive immune response – differences, the model is not directly comparable to humans. However, the large body of evidence obtained from studies in which the model was used to evaluate the host–microbe interplay has demonstrated that many mechanisms of microbial pathogenesis and the host response have been conserved between mammals and Drosophila. It is our belief that the addition of the Drosophila model to the repertoire of tools with which to study oral microbiology will facilitate infection research in this field considerably.
References
- 1.Brandt SM, Dionne MS, Khush RS, Pham LN, Vigdal TJ, Schneider DS. Secreted bacterial effectors and host-produced eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2004;2:e418. doi: 10.1371/journal.pbio.0020418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheng LW, Portnoy DA. Drosophila S2 cells: an alternative infection model for Listeria monocytogenes. Cell Microbiol. 2003;5:875–85. doi: 10.1046/j.1462-5822.2003.00327.x. [DOI] [PubMed] [Google Scholar]
- 3.de Lima Pimenta A, Di Martino P, Le Bouder E, Hulen C, Blight MA. In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microbes Infect. 2003;5:1177–87. doi: 10.1016/j.micinf.2003.09.002. [DOI] [PubMed] [Google Scholar]
- 4.Mansfield BE, Dionne MS, Schneider DS, Freitag NE. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell Microbiol. 2003;5:901–11. doi: 10.1046/j.1462-5822.2003.00329.x. [DOI] [PubMed] [Google Scholar]
- 5.Chugani SA, Whiteley M, Lee KM, D'Argenio D, Manoil C, Greenberg EP. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa . Proc Natl Acad Sci U S A. 2001;98:2752–7. doi: 10.1073/pnas.051624298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dionne MS, Ghori N, Schneider DS. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum . Infect Immun. 2003;71:3540–50. doi: 10.1128/IAI.71.6.3540-3550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elwell C, Engel JN. Drosophila melanogaster S2 cells: a model system to study Chlamydia interaction with host cells. Cell Microbiol. 2005;7:725–39. doi: 10.1111/j.1462-5822.2005.00508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fauvarque MO, Bergeret E, Chabert J, Dacheux D, Satre M, Attree I. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb Pathog. 2002;32:287–95. doi: 10.1006/mpat.2002.0504. [DOI] [PubMed] [Google Scholar]
- 9.Nehme NT, Liegeois S, Kele B, Giammarinaro P, Pradel E, Hoffmann JA, et al. A model of bacterial intestinal infections in Drosophila melanogaster . PLoS Pathog. 2007;3:e173. doi: 10.1371/journal.ppat.0030173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alarco AM, Marcil A, Chen J, Suter B, Thomas D, Whiteway M. Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J Immunol. 2004;172:5622–8. doi: 10.4049/jimmunol.172.9.5622. [DOI] [PubMed] [Google Scholar]
- 11.Apidianakis Y, Rahme LG, Heitman J, Ausubel FM, Calderwood SB, Mylonakis E. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot Cell. 2004;3:413–9. doi: 10.1128/EC.3.2.413-419.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bhabhra R, Miley MD, Mylonakis E, Boettner D, Fortwendel J, Panepinto JC, et al. Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun. 2004;72:4731–40. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chamilos G, Bignell EM, Schrettl M, Lewis RE, Leventakos K, May GS, et al. Exploring the concordance of Aspergillus fumigatus pathogenicity in mice and Toll-deficient flies. Med Mycol. 2010;48:506–10. doi: 10.3109/13693780903225813. [DOI] [PubMed] [Google Scholar]
- 14.Chamilos G, Lionakis MS, Lewis RE, Lopez-Ribot JL, Saville SP, Albert ND, et al. Drosophila melanogaster as a facile model for large-scale studies of virulence mechanisms and antifungal drug efficacy in Candida species. J Infect Dis. 2006;193:1014–22. doi: 10.1086/500950. [DOI] [PubMed] [Google Scholar]
- 15.Glittenberg MT, Silas S, Maccallum DM, Gow NA, Ligoxygakis P. Wild-type Drosophila melanogaster as an alternative model system for investigating the pathogenicity of Candida albicans . Dis Model Mech. 2011 Jul;4(4):504–14. doi: 10.1242/dmm.006619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamaris GA, Chamilos G, Lewis RE, Kontoyiannis DP. Virulence studies of Scedosporium and Fusarium species in Drosophila melanogaster . J Infect Dis. 2007;196:1860–4. doi: 10.1086/523765. [DOI] [PubMed] [Google Scholar]
- 17.Lionakis MS, Kontoyiannis DP. The growing promise of Toll-deficient Drosophila melanogaster as a model for studying Aspergillus pathogenesis and treatment. Virulence. 2010;1:488–99. doi: 10.4161/viru.1.6.13311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Battaglia PA, Zito S, Macchini A, Gigliani F. A Drosophila model of HIV-Tat-related pathogenicity. J Cell Sci. 2001;114:2787–94. doi: 10.1242/jcs.114.15.2787. [DOI] [PubMed] [Google Scholar]
- 19.Chotkowski HL, Ciota AT, Jia Y, Puig-Basagoiti F, Kramer LD, Shi PY, et al. West Nile virus infection of Drosophila melanogaster induces a protective RNAi response. Virology. 2008;377:197–206. doi: 10.1016/j.virol.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, et al. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890–3. doi: 10.1038/nature07151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Leulier F, Marchal C, Miletich I, Limbourg-Bouchon B, Benarous R, Lemaitre B. Directed expression of the HIV-1 accessory protein Vpu in Drosophila fat-body cells inhibits Toll-dependent immune responses. EMBO Rep. 2003;4:976–81. doi: 10.1038/sj.embor.embor936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukherjee S, Hanley KA. RNA interference modulates replication of dengue virus in Drosophila melanogaster cells. BMC Microbiol. 2010;10:127. doi: 10.1186/1471-2180-10-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schneider D, Shahabuddin M. Malaria parasite development in a Drosophila model. Science. 2000;288:2376–9. doi: 10.1126/science.288.5475.2376. [DOI] [PubMed] [Google Scholar]
- 24.Kotsyfakis M, Ehret-Sabatier L, Siden-Kiamos I, Mendoza J, Sinden RE, Louis C. Plasmodium berghei ookinetes bind to Anopheles gambiae and Drosophila melanogaster annexins. Mol Microbiol. 2005;57:171–9. doi: 10.1111/j.1365-2958.2005.04664.x. [DOI] [PubMed] [Google Scholar]
- 25.Brandt SM, Jaramillo-Gutierrez G, Kumar S, Barillas-Mury C, Schneider DS. Use of a Drosophila model to identify genes regulating Plasmodium growth in the mosquito. Genetics. 2008;180:1671–8. doi: 10.1534/genetics.108.089748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan J, Yang X, Mortin MA, Shahabuddin M. Malaria sporozoite antigen-directed genome-wide response in transgenic Drosophila. Genesis. 2009;47:196–203. doi: 10.1002/dvg.20483. [DOI] [PubMed] [Google Scholar]
- 27.Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, et al. The genome sequence of Drosophila melanogaster . Science. 2000;287:2185–95. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 28.Celniker SE, Wheeler DA, Kronmiller B, Carlson JW, Halpern A, Patel S, et al. Finishing a whole-genome shotgun: release 3 of the Drosophila melanogaster euchromatic genome sequence. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-12-research0079. RESEARCH0079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoskins RA, Carlson JW, Kennedy C, Acevedo D, Evans-Holm M, Frise E, et al. Sequence finishing and mapping of Drosophila melanogaster heterochromatin. Science. 2007;316:1625–8. doi: 10.1126/science.1139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Adams MD, Sekelsky JJ. From sequence to phenotype: reverse genetics in Drosophila melanogaster. Nat Rev Genet. 2002;3:189–98. doi: 10.1038/nrg752. [DOI] [PubMed] [Google Scholar]
- 31.Venken KJ, Bellen HJ. Emerging technologies for gene manipulation in Drosophila melanogaster . Nat Rev Genet. 2005;6:167–78. doi: 10.1038/nrg1553. [DOI] [PubMed] [Google Scholar]
- 32.Bellen HJ, Levis RW, Liao G, He Y, Carlson JW, Tsang G, et al. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics. 2004;167:761–81. doi: 10.1534/genetics.104.026427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, et al. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–7. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 34.Rubin GM, Spradling AC. Vectors for P element-mediated gene transfer in Drosophila . Nucleic Acids Res. 1983;11:6341–51. doi: 10.1093/nar/11.18.6341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Golic MM, Rong YS, Petersen RB, Lindquist SL, Golic KG. FLP-mediated DNA mobilization to specific target sites in Drosophila chromosomes. Nucleic Acids Res. 1997;25:3665–71. doi: 10.1093/nar/25.18.3665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Groth AC, Fish M, Nusse R, Calos MP. Construction of transgenic Drosophila by using the site-specific integrase from phage phiC31. Genetics. 2004;166:1775–82. doi: 10.1534/genetics.166.4.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Venken KJ, He Y, Hoskins RA, Bellen HJ. P[acman]: a BAC transgenic platform for targeted insertion of large DNA fragments in D. melanogaster. Science. 2006;314:1747–51. doi: 10.1126/science.1134426. [DOI] [PubMed] [Google Scholar]
- 38.Fischer JA, Giniger E, Maniatis T, Ptashne M. GAL4 activates transcription in Drosophila . Nature. 1988;332:853–6. doi: 10.1038/332853a0. [DOI] [PubMed] [Google Scholar]
- 39.Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–15. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, Xie C, Tian L, Hong L, Wu X, Han J. Participation of the p38 pathway in Drosophila host defense against pathogenic bacteria and fungi. Proc Natl Acad Sci U S A. 2010;107:20774–9. doi: 10.1073/pnas.1009223107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chamilos G, Lewis RE, Hu J, Xiao L, Zal T, Gilliet M, et al. Drosophila melanogaster as a model host to dissect the immunopathogenesis of zygomycosis. Proc Natl Acad Sci U S A. 2008;105:9367–72. doi: 10.1073/pnas.0709578105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levitin A, Marcil A, Tettweiler G, Laforest MJ, Oberholzer U, Alarco AM, et al. Drosophila melanogaster Thor and response to Candida albicans infection. Eukaryot Cell. 2007;6:658–63. doi: 10.1128/EC.00346-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mueller S, Gausson V, Vodovar N, Deddouche S, Troxler L, Perot J, et al. RNAi-mediated immunity provides strong protection against the negative-strand RNA vesicular stomatitis virus in Drosophila . Proc Natl Acad Sci U S A. 2010;107:19390–5. doi: 10.1073/pnas.1014378107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ferrandon D, Jung AC, Criqui M, Lemaitre B, Uttenweiler-Joseph S, Michaut L, et al. A drosomycin-GFP reporter transgene reveals a local immune response in Drosophila that is not dependent on the Toll pathway. EMBO J. 1998;17:1217–27. doi: 10.1093/emboj/17.5.1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Luce-Fedrow A, Von Ohlen T, Chapes SK. Ehrlichia chaffeensis infections in Drosophila melanogaster . Infect Immun. 2009;77:4815–26. doi: 10.1128/IAI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Avet-Rochex A, Bergeret E, Attree I, Meister M, Fauvarque MO. Suppression of Drosophila cellular immunity by directed expression of the ExoS toxin GAP domain of Pseudomonas aeruginosa . Cell Microbiol. 2005;7:799–810. doi: 10.1111/j.1462-5822.2005.00512.x. [DOI] [PubMed] [Google Scholar]
- 47.Dietzl G, Chen D, Schnorrer F, Su KC, Barinova Y, Fellner M, et al. A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila . Nature. 2007;448:151–6. doi: 10.1038/nature05954. [DOI] [PubMed] [Google Scholar]
- 48.Karlsson C, Korayem AM, Scherfer C, Loseva O, Dushay MS, Theopold U. Proteomic analysis of the Drosophila larval hemolymph clot. J Biol Chem. 2004;279:52033–41. doi: 10.1074/jbc.M408220200. [DOI] [PubMed] [Google Scholar]
- 49.Levy F, Rabel D, Charlet M, Bulet P, Hoffmann JA, Ehret-Sabatier L. Peptidomic and proteomic analyses of the systemic immune response of Drosophila . Biochimie. 2004;86:607–16. doi: 10.1016/j.biochi.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Vierstraete E, Verleyen P, Sas F, Van den Bergh G, De Loof A, Arckens L, et al. The instantly released Drosophila immune proteome is infection-specific. Biochem Biophys Res Commun. 2004;317:1052–60. doi: 10.1016/j.bbrc.2004.03.150. [DOI] [PubMed] [Google Scholar]
- 51.Cronin SJ, Nehme NT, Limmer S, Liegeois S, Pospisilik JA, Schramek D, et al. Genome-wide RNAi screen identifies genes involved in intestinal pathogenic bacterial infection. Science. 2009;325:340–3. doi: 10.1126/science.1173164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.De Gregorio E, Spellman PT, Rubin GM, Lemaitre B. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc Natl Acad Sci U S A. 2001;98:12590–5. doi: 10.1073/pnas.221458698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Gregorio E, Spellman PT, Tzou P, Rubin GM, Lemaitre B. The Toll and Imd pathways are the major regulators of the immune response in Drosophila . EMBO J. 2002;21:2568–79. doi: 10.1093/emboj/21.11.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart JM, et al. A genome-wide analysis of immune responses in Drosophila . Proc Natl Acad Sci U S A. 2001;98:15119–24. doi: 10.1073/pnas.261573998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irving P, Ubeda JM, Doucet D, Troxler L, Lagueux M, Zachary D, et al. New insights into Drosophila larval haemocyte functions through genome-wide analysis. Cell Microbiol. 2005;7:335–50. doi: 10.1111/j.1462-5822.2004.00462.x. [DOI] [PubMed] [Google Scholar]
- 56.Boutros M, Agaisse H, Perrimon N. Sequential activation of signaling pathways during innate immune responses in Drosophila . Dev Cell. 2002;3:711–22. doi: 10.1016/s1534-5807(02)00325-8. [DOI] [PubMed] [Google Scholar]
- 57.Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, et al. A systems biology analysis of the Drosophila phagosome. Nature. 2007;445:95–101. doi: 10.1038/nature05380. [DOI] [PubMed] [Google Scholar]
- 58.Asare R, Akimana C, Jones S, Abu Kwaik Y. Molecular bases of proliferation of Francisella tularensis in arthropod vectors. Environ Microbiol. 2010;12:2587–612. doi: 10.1111/j.1462-2920.2010.02230.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Phagocytosis of Candida albicans by RNAi-treated Drosophila S2 cells. Methods Mol Biol. 2009;470:347–58. doi: 10.1007/978-1-59745-204-5_24. [DOI] [PubMed] [Google Scholar]
- 60.Bernards A, Hariharan IK. Of flies and men—studying human disease in Drosophila . Curr Opin Genet Dev. 2001;11:274–8. doi: 10.1016/s0959-437x(00)00190-8. [DOI] [PubMed] [Google Scholar]
- 61.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, et al. FlyBase: enhancing Drosophila gene ontology annotations. Nucleic Acids Res. 2009;37:D555–9. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Krebs RA, Feder ME. Hsp70 and larval thermotolerance in Drosophila melanogaster: how much is enough and when is more too much? J Insect Physiol. 1998;44:1091–101. doi: 10.1016/s0022-1910(98)00059-6. [DOI] [PubMed] [Google Scholar]
- 63.Igboin CO, Moeschberger ML, Griffen AL, Leys EJ. Porphyromonas gingivalis virulence in a Drosophila melanogaster model. Infect Immun. 2011;79:439–48. doi: 10.1128/IAI.00784-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Limmer S, Quintin J, Hetru C, Ferrandon D. Virulence on the fly: Drosophila melanogaster as a model genetic organism to decipher host-pathogen interactions. Curr Drug Targets. 2011;12:978–99. doi: 10.2174/138945011795677818. [DOI] [PubMed] [Google Scholar]
- 65.Ganesan S, Aggarwal K, Paquette N, Silverman N. NF-kappaB/Rel proteins and the humoral immune responses of Drosophila melanogaster . Curr Top Microbiol Immunol. 2011;349:25–60. doi: 10.1007/82_2010_107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist gram-negative bacterial infection. EMBO Rep. 2000;1:353–8. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rutschmann S, Jung AC, Zhou R, Silverman N, Hoffmann JA, Ferrandon D. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat Immunol. 2000;1:342–7. doi: 10.1038/79801. [DOI] [PubMed] [Google Scholar]
- 68.Vidal S, Khush RS, Leulier F, Tzou P, Nakamura M, Lemaitre B. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NF-kappaB-dependent innate immune responses. Genes Dev. 2001;15:1900–12. doi: 10.1101/gad.203301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gottar M, Gobert V, Michel T, Belvin M, Duyk G, Hoffmann JA, et al. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature. 2002;416:640–4. doi: 10.1038/nature734. [DOI] [PubMed] [Google Scholar]
- 70.Ramet M, Manfruelli P, Pearson A, Mathey-Prevot B, Ezekowitz RA. Functional genomic analysis of phagocytosis and identification of a Drosophila receptor for E. coli . Nature. 2002;416:644–8. doi: 10.1038/nature735. [DOI] [PubMed] [Google Scholar]
- 71.Choe KM, Lee H, Anderson KV. Drosophila peptidoglycan recognition protein LC (PGRP-LC) acts as a signal-transducing innate immune receptor. Proc Natl Acad Sci U S A. 2005;102:1122–6. doi: 10.1073/pnas.0404952102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, et al. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci U S A. 1995;92:9465–9. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Georgel P, Naitza S, Kappler C, Ferrandon D, Zachary D, Swimmer C, et al. Drosophila immune deficiency (IMD) is a death domain protein that activates antibacterial defense and can promote apoptosis. Dev Cell. 2001;1:503–14. doi: 10.1016/s1534-5807(01)00059-4. [DOI] [PubMed] [Google Scholar]
- 74.Hu S, Yang X. dFADD, a novel death domain-containing adapter protein for the Drosophila caspase DREDD. J Biol Chem. 2000;275:30761–4. doi: 10.1074/jbc.C000341200. [DOI] [PubMed] [Google Scholar]
- 75.Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist gram-negative bacterial infection. Mol Cell Biol. 2006;26:7821–31. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Silverman N, Zhou R, Erlich RL, Hunter M, Bernstein E, Schneider D, et al. Immune activation of NF-kappaB and JNK requires Drosophila TAK1. J Biol Chem. 2003;278:48928–34. doi: 10.1074/jbc.M304802200. [DOI] [PubMed] [Google Scholar]
- 77.Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, et al. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila . Mol Cell. 1999;4:827–37. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- 78.Silverman N, Zhou R, Stoven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IkappaB kinase complex required for Relish cleavage and antibacterial immunity. Genes Dev. 2000;14:2461–71. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu Y, Wu LP, Anderson KV. The antibacterial arm of the Drosophila innate immune response requires an IkappaB kinase. Genes Dev. 2001;15:104–10. doi: 10.1101/gad.856901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, et al. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci U S A. 2003;100:5991–6. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Erturk-Hasdemir D, Broemer M, Leulier F, Lane WS, Paquette N, Hwang D, et al. Two roles for the Drosophila IKK complex in the activation of Relish and the induction of antimicrobial peptide genes. Proc Natl Acad Sci U S A. 2009;106:9779–84. doi: 10.1073/pnas.0812022106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2006;2:e14. doi: 10.1371/journal.ppat.0020014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mellroth P, Karlsson J, Steiner H. A scavenger function for a Drosophila peptidoglycan recognition protein. J Biol Chem. 2003;278:7059–64. doi: 10.1074/jbc.M208900200. [DOI] [PubMed] [Google Scholar]
- 84.Zaidman-Remy A, Herve M, Poidevin M, Pili-Floury S, Kim MS, Blanot D, et al. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity. 2006;24:463–73. doi: 10.1016/j.immuni.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 85.Maillet F, Bischoff V, Vignal C, Hoffmann J, Royet J. The Drosophila peptidoglycan recognition protein PGRP-LF blocks PGRP-LC and IMD/JNK pathway activation. Cell Host Microbe. 2008;3:293–303. doi: 10.1016/j.chom.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 86.Guntermann S, Primrose DA, Foley E. Dnr1-dependent regulation of the Drosophila immune deficiency signaling pathway. Dev Comp Immunol. 2009;33:127–34. doi: 10.1016/j.dci.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 87.Kim M, Lee JH, Lee SY, Kim E, Chung J. Caspar, a suppressor of antibacterial immunity in Drosophila . Proc Natl Acad Sci U S A. 2006;103:16358–63. doi: 10.1073/pnas.0603238103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Aggarwal K, Rus F, Vriesema-Magnuson C, Erturk-Hasdemir D, Paquette N, Silverman N. Rudra interrupts receptor signaling complexes to negatively regulate the IMD pathway. PLoS Pathog. 2008;4:e1000120. doi: 10.1371/journal.ppat.1000120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kleino A, Myllymaki H, Kallio J, Vanha-aho LM, Oksanen K, Ulvila J, et al. Pirk is a negative regulator of the Drosophila Imd pathway. J Immunol. 2008;180:5413–22. doi: 10.4049/jimmunol.180.8.5413. [DOI] [PubMed] [Google Scholar]
- 90.Lhocine N, Ribeiro PS, Buchon N, Wepf A, Wilson R, Tenev T, et al. PIMS modulates immune tolerance by negatively regulating Drosophila innate immune signaling. Cell Host Microbe. 2008;4:147–58. doi: 10.1016/j.chom.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 91.Ragab A, Buechling T, Gesellchen V, Spirohn K, Boettcher AL, Boutros M. Drosophila Ras/MAPK signalling regulates innate immune responses in immune and intestinal stem cells. EMBO J. 2011;30:1123–36. doi: 10.1038/emboj.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Khush RS, Cornwell WD, Uram JN, Lemaitre B. A ubiquitin-proteasome pathway represses the Drosophila immune deficiency signaling cascade. Curr Biol. 2002;12:1728–37. doi: 10.1016/s0960-9822(02)01214-9. [DOI] [PubMed] [Google Scholar]
- 93.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette Spatzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–83. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 94.Rutschmann S, Jung AC, Hetru C, Reichhart JM, Hoffmann JA, Ferrandon D. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila . Immunity. 2000;12:569–80. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- 95.Michel T, Reichhart JM, Hoffmann JA, Royet J. Drosophila Toll is activated by Gram-positive bacteria through a circulating peptidoglycan recognition protein. Nature. 2001;414:756–9. doi: 10.1038/414756a. [DOI] [PubMed] [Google Scholar]
- 96.Tauszig-Delamasure S, Bilak H, Capovilla M, Hoffmann JA, Imler JL. Drosophila MyD88 is required for the response to fungal and Gram-positive bacterial infections. Nat Immunol. 2002;3:91–7. doi: 10.1038/ni747. [DOI] [PubMed] [Google Scholar]
- 97.Rutschmann S, Kilinc A, Ferrandon D. Cutting edge: the toll pathway is required for resistance to gram-positive bacterial infections in Drosophila . J Immunol. 2002;168:1542–6. doi: 10.4049/jimmunol.168.4.1542. [DOI] [PubMed] [Google Scholar]
- 98.Gobert V, Gottar M, Matskevich AA, Rutschmann S, Royet J, Belvin M, et al. Dual activation of the Drosophila toll pathway by two pattern recognition receptors. Science. 2003;302:2126–30. doi: 10.1126/science.1085432. [DOI] [PubMed] [Google Scholar]
- 99.Wang L, Weber AN, Atilano ML, Filipe SR, Gay NJ, Ligoxygakis P. Sensing of Gram-positive bacteria in Drosophila: GNBP1 is needed to process and present peptidoglycan to PGRP-SA. EMBO J. 2006;25:5005–14. doi: 10.1038/sj.emboj.7601363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bischoff V, Vignal C, Boneca IG, Michel T, Hoffmann JA, Royet J. Function of the Drosophila pattern-recognition receptor PGRP-SD in the detection of Gram-positive bacteria. Nat Immunol. 2004;5:1175–80. doi: 10.1038/ni1123. [DOI] [PubMed] [Google Scholar]
- 101.Ligoxygakis P, Pelte N, Hoffmann JA, Reichhart JM. Activation of Drosophila Toll during fungal infection by a blood serine protease. Science. 2002;297:114–6. doi: 10.1126/science.1072391. [DOI] [PubMed] [Google Scholar]
- 102.Kim YS, Ryu JH, Han SJ, Choi KH, Nam KB, Jang IH, et al. Gram-negative bacteria-binding protein, a pattern recognition receptor for lipopolysaccharide and beta-1,3-glucan that mediates the signaling for the induction of innate immune genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:32721–7. doi: 10.1074/jbc.M003934200. [DOI] [PubMed] [Google Scholar]
- 103.Gottar M, Gobert V, Matskevich AA, Reichhart JM, Wang C, Butt TM, et al. Dual detection of fungal infections in Drosophila via recognition of glucans and sensing of virulence factors. Cell. 2006;127:1425–37. doi: 10.1016/j.cell.2006.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.El Chamy L, Leclerc V, Caldelari I, Reichhart JM. Sensing of danger signals and pathogen-associated molecular patterns defines binary signaling pathways upstream of Toll. Nat Immunol. 2008;9:1165–70. doi: 10.1038/ni.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kambris Z, Brun S, Jang IH, Nam HJ, Romeo Y, Takahashi K, et al. Drosophila immunity: a large-scale in vivo RNAi screen identifies five serine proteases required for Toll activation. Curr Biol. 2006;16:808–13. doi: 10.1016/j.cub.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 106.Jang IH, Chosa N, Kim SH, Nam HJ, Lemaitre B, Ochiai M, et al. A Spatzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10:45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 107.Sun H, Bristow BN, Qu G, Wasserman SA. A heterotrimeric death domain complex in Toll signaling. Proc Natl Acad Sci U S A. 2002;99:12871–6. doi: 10.1073/pnas.202396399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reichhart JM, Georgel P, Meister M, Lemaitre B, Kappler C, Hoffmann JA. Expression and nuclear translocation of the rel/NF-kappa B-related morphogen dorsal during the immune response of Drosophila . C R Acad Sci III. 1993;316:1218–24. [PubMed] [Google Scholar]
- 109.Ip YT, Reach M, Engstrom Y, Kadalayil L, Cai H, Gonzalez-Crespo S, et al. Dif, a dorsal-related gene that mediates an immune response in Drosophila . Cell. 1993;75:753–63. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- 110.Lemaitre B, Meister M, Govind S, Georgel P, Steward R, Reichhart JM, et al. Functional analysis and regulation of nuclear import of dorsal during the immune response in Drosophila . EMBO J. 1995;14:536–45. doi: 10.1002/j.1460-2075.1995.tb07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392:93–7. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]
- 112.Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 1999;18:3380–91. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-kappaB factor. Genes Dev. 1999;13:792–7. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Kuttenkeuler D, Pelte N, Ragab A, Gesellchen V, Schneider L, Blass C, et al. A large-scale RNAi screen identifies Deaf1 as a regulator of innate immune responses in Drosophila . J Innate Immun. 2010;2:181–94. doi: 10.1159/000248649. [DOI] [PubMed] [Google Scholar]
- 115.Nicolas E, Reichhart JM, Hoffmann JA, Lemaitre B. In vivo regulation of the IkappaB homologue cactus during the immune response of Drosophila . J Biol Chem. 1998;273:10463–9. doi: 10.1074/jbc.273.17.10463. [DOI] [PubMed] [Google Scholar]
- 116.Gordon MD, Dionne MS, Schneider DS, Nusse R. WntD is a feedback inhibitor of Dorsal/NF-kappaB in Drosophila development and immunity. Nature. 2005;437:746–9. doi: 10.1038/nature04073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Chiu H, Ring BC, Sorrentino RP, Kalamarz M, Garza D, Govind S. dUbc9 negatively regulates the Toll-NF-kappa B pathways in larval hematopoiesis and drosomycin activation in Drosophila . Dev Biol. 2005;288:60–72. doi: 10.1016/j.ydbio.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 118.Fullaondo A, Garcia-Sanchez S, Sanz-Parra A, Recio E, Lee SY, Gubb D. Spn1 regulates the GNBP3-dependent Toll-signalling pathway in Drosophila melanogaster . Mol Cell Biol. 2011 doi: 10.1128/MCB.01397-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tanji T, Hu X, Weber AN, Ip YT. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster . Mol Cell Biol. 2007;27:4578–88. doi: 10.1128/MCB.01814-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Tanji T, Yun EY, Ip YT. Heterodimers of NF-kappaB transcription factors DIF and Relish regulate antimicrobial peptide genes in Drosophila . Proc Natl Acad Sci U S A. 2010;107:14715–20. doi: 10.1073/pnas.1009473107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hashimoto Y, Tabuchi Y, Sakurai K, Kutsuna M, Kurokawa K, Awasaki T, et al. Identification of lipoteichoic acid as a ligand for draper in the phagocytosis of Staphylococcus aureus by Drosophila hemocytes. J Immunol. 2009;183:7451–60. doi: 10.4049/jimmunol.0901032. [DOI] [PubMed] [Google Scholar]
- 122.Lau GW, Goumnerov BC, Walendziewicz CL, Hewitson J, Xiao W, Mahajan-Miklos S, et al. The Drosophila melanogaster toll pathway participates in resistance to infection by the gram-negative human pathogen Pseudomonas aeruginosa . Infect Immun. 2003;71:4059–66. doi: 10.1128/IAI.71.7.4059-4066.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Luce-Fedrow A, Von Ohlen T, Boyle D, Ganta RR, Chapes SK. Use of Drosophila S2 cells as a model for studying Ehrlichia chaffeensis infections. Appl Environ Microbiol. 2008;74:1886–91. doi: 10.1128/AEM.02467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kylsten P, Samakovlis C, Hultmark D. The cecropin locus in Drosophila; a compact gene cluster involved in the response to infection. EMBO J. 1990;9:217–24. doi: 10.1002/j.1460-2075.1990.tb08098.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dimarcq JL, Hoffmann D, Meister M, Bulet P, Lanot R, Reichhart JM, et al. Characterization and transcriptional profiles of a Drosophila gene encoding an insect defensin. A study in insect immunity. Eur J Biochem. 1994;221:201–9. doi: 10.1111/j.1432-1033.1994.tb18730.x. [DOI] [PubMed] [Google Scholar]
- 126.Hultmark D, Steiner H, Rasmuson T, Boman HG. Insect immunity. Purification and properties of three inducible bactericidal proteins from hemolymph of immunized pupae of Hyalophora cecropia. Eur J Biochem. 1980;106:7–16. doi: 10.1111/j.1432-1033.1980.tb05991.x. [DOI] [PubMed] [Google Scholar]
- 127.Cociancich S, Ghazi A, Hetru C, Hoffmann JA, Letellier L. Insect defensin, an inducible antibacterial peptide, forms voltage-dependent channels in Micrococcus luteus . J Biol Chem. 1993;268:19239–45. [PubMed] [Google Scholar]
- 128.Wicker C, Reichhart JM, Hoffmann D, Hultmark D, Samakovlis C, Hoffmann JA. Insect immunity. Characterization of a Drosophila cDNA encoding a novel member of the diptericin family of immune peptides. J Biol Chem. 1990;265:22493–8. [PubMed] [Google Scholar]
- 129.Fehlbaum P, Bulet P, Michaut L, Lagueux M, Broekaert WF, Hetru C, et al. Insect immunity. Septic injury of Drosophila induces the synthesis of a potent antifungal peptide with sequence homology to plant antifungal peptides. J Biol Chem. 1994;269:33159–63. [PubMed] [Google Scholar]
- 130.Zhang ZT, Zhu SY. Drosomycin, an essential component of antifungal defence in Drosophila . Insect Mol Biol. 2009;18:549–56. doi: 10.1111/j.1365-2583.2009.00907.x. [DOI] [PubMed] [Google Scholar]
- 131.Keppi E, Pugsley P, Lambert J, Wicker C, Dimarcq JL, Hoffmann J, et al. Mode of action of diptericin A, a bactericidal peptide induced in the hemolymph of Phormia terranovae larvae. Arch Insect Biochem Physiol. 1989;10:229–39. [Google Scholar]
- 132.Tian C, Gao B, Rodriguez Mdel C, Lanz-Mendoza H, Ma B, Zhu S. Gene expression, antiparasitic activity, and functional evolution of the drosomycin family. Mol Immunol. 2008;45:3909–16. doi: 10.1016/j.molimm.2008.06.025. [DOI] [PubMed] [Google Scholar]
- 133.Asling B, Dushay MS, Hultmark D. Identification of early genes in the Drosophila immune response by PCR-based differential display: the Attacin A gene and the evolution of attacin-like proteins. Insect Biochem Mol Biol. 1995;25:511–8. doi: 10.1016/0965-1748(94)00091-c. [DOI] [PubMed] [Google Scholar]
- 134.Bulet P, Dimarcq JL, Hetru C, Lagueux M, Charlet M, Hegy G, et al. A novel inducible antibacterial peptide of Drosophila carries an O-glycosylated substitution. J Biol Chem. 1993;268:14893–7. [PubMed] [Google Scholar]
- 135.Otvos L, Jr O, I, Rogers ME, Consolvo PJ, Condie BA, Lovas S, et al. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry. 2000;39:14150–9. doi: 10.1021/bi0012843. [DOI] [PubMed] [Google Scholar]
- 136.Levashina EA, Ohresser S, Bulet P, Reichhart JM, Hetru C, Hoffmann JA. Metchnikowin, a novel immune-inducible proline-rich peptide from Drosophila with antibacterial and antifungal properties. Eur J Biochem. 1995;233:694–700. doi: 10.1111/j.1432-1033.1995.694_2.x. [DOI] [PubMed] [Google Scholar]
- 137.Romeo Y, Lemaitre B. Drosophila immunity: methods for monitoring the activity of Toll and Imd signaling pathways. Methods Mol Biol. 2008;415:379–94. doi: 10.1007/978-1-59745-570-1_22. [DOI] [PubMed] [Google Scholar]
- 138.Harrison DA, McCoon PE, Binari R, Gilman M, Perrimon N. Drosophila unpaired encodes a secreted protein that activates the JAK signaling pathway. Genes Dev. 1998;12:3252–63. doi: 10.1101/gad.12.20.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Agaisse H, Petersen UM, Boutros M, Mathey-Prevot B, Perrimon N. Signaling role of hemocytes in Drosophila JAK/STAT-dependent response to septic injury. Dev Cell. 2003;5:441–50. doi: 10.1016/s1534-5807(03)00244-2. [DOI] [PubMed] [Google Scholar]
- 140.Brown S, Hu N, Hombria JC. Identification of the first invertebrate interleukin JAK/STAT receptor, the Drosophila gene domeless. Curr Biol. 2001;11:1700–5. doi: 10.1016/s0960-9822(01)00524-3. [DOI] [PubMed] [Google Scholar]
- 141.Chen HW, Chen X, Oh SW, Marinissen MJ, Gutkind JS, Hou SX. mom identifies a receptor for the Drosophila JAK/STAT signal transduction pathway and encodes a protein distantly related to the mammalian cytokine receptor family. Genes Dev. 2002;16:388–98. doi: 10.1101/gad.955202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Agaisse H, Perrimon N. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol Rev. 2004;198:72–82. doi: 10.1111/j.0105-2896.2004.0133.x. [DOI] [PubMed] [Google Scholar]
- 143.Arbouzova NI, Zeidler MP. JAK/STAT signalling in Drosophila: insights into conserved regulatory and cellular functions. Development. 2006;133:2605–16. doi: 10.1242/dev.02411. [DOI] [PubMed] [Google Scholar]
- 144.Brun S, Vidal S, Spellman P, Takahashi K, Tricoire H, Lemaitre B. The MAPKKK Mekk1 regulates the expression of Turandot stress genes in response to septic injury in Drosophila . Genes Cells. 2006;11:397–407. doi: 10.1111/j.1365-2443.2006.00953.x. [DOI] [PubMed] [Google Scholar]
- 145.Ekengren S, Hultmark D. A family of Turandot-related genes in the humoral stress response of Drosophila . Biochem Biophys Res Commun. 2001;284:998–1003. doi: 10.1006/bbrc.2001.5067. [DOI] [PubMed] [Google Scholar]
- 146.Liu W, Singh SR, Hou SX. JAK-STAT is restrained by Notch to control cell proliferation of the Drosophila intestinal stem cells. J Cell Biochem. 2010;109:992–9. doi: 10.1002/jcb.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Betz A, Lampen N, Martinek S, Young MW, Darnell JE., Jr A Drosophila PIAS homologue negatively regulates stat92E. Proc Natl Acad Sci U S A. 2001;98:9563–8. doi: 10.1073/pnas.171302098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Karsten P, Hader S, Zeidler MP. Cloning and expression of Drosophila SOCS36E and its potential regulation by the JAK/STAT pathway. Mech Dev. 2002;117:343–6. doi: 10.1016/s0925-4773(02)00216-2. [DOI] [PubMed] [Google Scholar]
- 149.Callus BA, Mathey-Prevot B. SOCS36E, a novel Drosophila SOCS protein, suppresses JAK/STAT and EGF-R signalling in the imaginal wing disc. Oncogene. 2002;21:4812–21. doi: 10.1038/sj.onc.1205618. [DOI] [PubMed] [Google Scholar]
- 150.Henriksen MA, Betz A, Fuccillo MV, Darnell JE Jr. Negative regulation of STAT92E by an N-terminally truncated STAT protein derived from an alternative promoter site. Genes Dev. 2002;16:2379–89. doi: 10.1101/gad.1020702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kallio J, Myllymaki H, Gronholm J, Armstrong M, Vanha-aho LM, Makinen L, et al. Eye transformer is a negative regulator of Drosophila JAK/STAT signaling. FASEB J. 2010;24:4467–79. doi: 10.1096/fj.10-162784. [DOI] [PubMed] [Google Scholar]
- 152.Makki R, Meister M, Pennetier D, Ubeda JM, Braun A, Daburon V, et al. A short receptor downregulates JAK/STAT signalling to control the Drosophila cellular immune response. PLoS Biol. 2010;8:e1000441. doi: 10.1371/journal.pbio.1000441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Baeg GH, Zhou R, Perrimon N. Genome-wide RNAi analysis of JAK/STAT signaling components in Drosophila . Genes Dev. 2005;19:1861–70. doi: 10.1101/gad.1320705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Muller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871–5. doi: 10.1038/nature03869. [DOI] [PubMed] [Google Scholar]
- 155.Bach EA, Vincent S, Zeidler MP, Perrimon N. A sensitized genetic screen to identify novel regulators and components of the Drosophila janus kinase/signal transducer and activator of transcription pathway. Genetics. 2003;165:1149–66. doi: 10.1093/genetics/165.3.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Avadhanula V, Weasner BP, Hardy GG, Kumar JP, Hardy RW. A novel system for the launch of alphavirus RNA synthesis reveals a role for the Imd pathway in arthropod antiviral response. PLoS Pathog. 2009;5:e1000582. doi: 10.1371/journal.ppat.1000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Costa A, Jan E, Sarnow P, Schneider D. The Imd pathway is involved in antiviral immune responses in Drosophila . PLoS One. 2009;4:e7436. doi: 10.1371/journal.pone.0007436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zambon RA, Nandakumar M, Vakharia VN, Wu LP. The Toll pathway is important for an antiviral response in Drosophila . Proc Natl Acad Sci U S A. 2005;102:7257–62. doi: 10.1073/pnas.0409181102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dostert C, Jouanguy E, Irving P, Troxler L, Galiana-Arnoux D, Hetru C, et al. The Jak-STAT signaling pathway is required but not sufficient for the antiviral response of Drosophila . Nat Immunol. 2005;6:946–53. doi: 10.1038/ni1237. [DOI] [PubMed] [Google Scholar]
- 160.Lanot R, Zachary D, Holder F, Meister M. Postembryonic hematopoiesis in Drosophila . Dev Biol. 2001;230:243–57. doi: 10.1006/dbio.2000.0123. [DOI] [PubMed] [Google Scholar]
- 161.Kocks C, Cho JH, Nehme N, Ulvila J, Pearson AM, Meister M, et al. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila . Cell. 2005;123:335–46. doi: 10.1016/j.cell.2005.08.034. [DOI] [PubMed] [Google Scholar]
- 162.Kurucz E, Markus R, Zsamboki J, Folkl-Medzihradszky K, Darula Z, Vilmos P, et al. Nimrod, a putative phagocytosis receptor with EGF repeats in Drosophila plasmatocytes. Curr Biol. 2007;17:649–54. doi: 10.1016/j.cub.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 163.Ramet M, Pearson A, Manfruelli P, Li X, Koziel H, Gobel V, et al. Drosophila scavenger receptor CI is a pattern recognition receptor for bacteria. Immunity. 2001;15:1027–38. doi: 10.1016/s1074-7613(01)00249-7. [DOI] [PubMed] [Google Scholar]
- 164.Watson FL, Puttmann-Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, et al. Extensive diversity of Ig-superfamily proteins in the immune system of insects. Science. 2005;309:1874–8. doi: 10.1126/science.1116887. [DOI] [PubMed] [Google Scholar]
- 165.Philips JA, Rubin EJ, Perrimon N. Drosophila RNAi screen reveals CD36 family member required for mycobacterial infection. Science. 2005;309:1251–3. doi: 10.1126/science.1116006. [DOI] [PubMed] [Google Scholar]
- 166.Shia AK, Glittenberg M, Thompson G, Weber AN, Reichhart JM, Ligoxygakis P. Toll-dependent antimicrobial responses in Drosophila larval fat body require Spatzle secreted by haemocytes. J Cell Sci. 2009;122:4505–15. doi: 10.1242/jcs.049155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Charroux B, Royet J. Elimination of plasmatocytes by targeted apoptosis reveals their role in multiple aspects of the Drosophila immune response. Proc Natl Acad Sci U S A. 2009;106:9797–802. doi: 10.1073/pnas.0903971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Nehme NT, Quintin J, Cho JH, Lee J, Lafarge MC, Kocks C, et al. Relative roles of the cellular and humoral responses in the Drosophila host defense against three gram-positive bacterial infections. PLoS One. 2011;6:e14743. doi: 10.1371/journal.pone.0014743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Pham LN, Dionne MS, Shirasu-Hiza M, Schneider DS. A specific primed immune response in Drosophila is dependent on phagocytes. PLoS Pathog. 2007;3:e26. doi: 10.1371/journal.ppat.0030026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Vonkavaara M, Telepnev MV, Ryden P, Sjostedt A, Stoven S. Drosophila melanogaster as a model for elucidating the pathogenicity of Francisella tularensis . Cell Microbiol. 2008;10:1327–38. doi: 10.1111/j.1462-5822.2008.01129.x. [DOI] [PubMed] [Google Scholar]
- 171.Defaye A, Evans I, Crozatier M, Wood W, Lemaitre B, Leulier F. Genetic ablation of Drosophila phagocytes reveals their contribution to both development and resistance to bacterial infection. J Innate Immun. 2009;1:322–34. doi: 10.1159/000210264. [DOI] [PubMed] [Google Scholar]
- 172.Lesch C, Goto A, Lindgren M, Bidla G, Dushay MS, Theopold U. A role for Hemolectin in coagulation and immunity in Drosophila melanogaster . Dev Comp Immunol. 2007;31:1255–63. doi: 10.1016/j.dci.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 173.Shinzawa N, Nelson B, Aonuma H, Okado K, Fukumoto S, Miura M, et al. p38 MAPK-dependent phagocytic encapsulation confers infection tolerance in Drosophila . Cell Host Microbe. 2009;6:244–52. doi: 10.1016/j.chom.2009.07.010. [DOI] [PubMed] [Google Scholar]
- 174.Ulvila J, Vanha-Aho LM, Ramet M. Drosophila phagocytosis—still many unknowns under the surface. APMIS. 2011;119:651–62. doi: 10.1111/j.1600-0463.2011.02792.x. [DOI] [PubMed] [Google Scholar]
- 175.Franc NC, Dimarcq JL, Lagueux M, Hoffmann J, Ezekowitz RA. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity. 1996;4:431–43. doi: 10.1016/s1074-7613(00)80410-0. [DOI] [PubMed] [Google Scholar]
- 176.Lagueux M, Perrodou E, Levashina EA, Capovilla M, Hoffmann JA. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila . Proc Natl Acad Sci U S A. 2000;97:11427–32. doi: 10.1073/pnas.97.21.11427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Stroschein-Stevenson SL, Foley E, O'Farrell PH, Johnson AD. Identification of Drosophila gene products required for phagocytosis of Candida albicans . PLoS Biol. 2006;4:e4. doi: 10.1371/journal.pbio.0040004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Igboin CO, Tordoff KP, Moeschberger ML, Griffen AL, Leys EJ. Porphyromonas gingivalis-host interactions in a Drosophila melanogaster model. Infect Immun. 2011;79:449–58. doi: 10.1128/IAI.00785-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bou Aoun R, Hetru C, Troxler L, Doucet D, Ferrandon D, Matt N. Analysis of thioester-containing proteins during the innate immune response of Drosophila melanogaster . J Innate Immun. 2011;3:52–64. doi: 10.1159/000321554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Qin QM, Luo J, Lin X, Pei J, Li L, Ficht TA, et al. Functional analysis of host factors that mediate the intracellular lifestyle of Cryptococcus neoformans . PLoS Pathog. 2011;7:e1002078. doi: 10.1371/journal.ppat.1002078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Yano T, Mita S, Ohmori H, Oshima Y, Fujimoto Y, Ueda R, et al. Autophagic control of listeria through intracellular innate immune recognition in Drosophila . Nat Immunol. 2008;9:908–16. doi: 10.1038/ni.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Shelly S, Lukinova N, Bambina S, Berman A, Cherry S. Autophagy is an essential component of Drosophila immunity against vesicular stomatitis virus. Immunity. 2009;30:588–98. doi: 10.1016/j.immuni.2009.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Rizki TM, Rizki RM. Lamellocyte differentiation in Drosophila larvae parasitized by Leptopilina . Dev Comp Immunol. 1992;16:103–10. doi: 10.1016/0145-305x(92)90011-z. [DOI] [PubMed] [Google Scholar]
- 184.Nappi AJ, Silvers M. Cell surface changes associated with cellular immune reactions in Drosophila . Science. 1984;225:1166–8. doi: 10.1126/science.6433482. [DOI] [PubMed] [Google Scholar]
- 185.Nappi AJ, Vass E. Hydrogen peroxide production in immune-reactive Drosophila melanogaster . J Parasitol. 1998;84:1150–7. [PubMed] [Google Scholar]
- 186.Nappi AJ, Vass E, Frey F, Carton Y. Superoxide anion generation in Drosophila during melanotic encapsulation of parasites. Eur J Cell Biol. 1995;68:450–6. [PubMed] [Google Scholar]
- 187.Nappi AJ, Vass E, Frey F, Carton Y. Nitric oxide involvement in Drosophila immunity. Nitric Oxide. 2000;4:423–30. doi: 10.1006/niox.2000.0294. [DOI] [PubMed] [Google Scholar]
- 188.Ashida M. The prophenoloxidase cascade in insect immunity. Res Immunol. 1990;141:908–10. doi: 10.1016/0923-2494(90)90191-z. [DOI] [PubMed] [Google Scholar]
- 189.Nappi AJ, Vass E. Melanogenesis and the generation of cytotoxic molecules during insect cellular immune reactions. Pigment Cell Res. 1993;6:117–26. doi: 10.1111/j.1600-0749.1993.tb00590.x. [DOI] [PubMed] [Google Scholar]
- 190.Scherfer C, Karlsson C, Loseva O, Bidla G, Goto A, Havemann J, et al. Isolation and characterization of hemolymph clotting factors in Drosophila melanogaster by a pullout method. Curr Biol. 2004;14:625–9. doi: 10.1016/j.cub.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 191.Bidla G, Lindgren M, Theopold U, Dushay MS. Hemolymph coagulation and phenoloxidase in Drosophila larvae. Dev Comp Immunol. 2005;29:669–79. doi: 10.1016/j.dci.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 192.Goto A, Kumagai T, Kumagai C, Hirose J, Narita H, Mori H, et al. A Drosophila haemocyte-specific protein, hemolectin, similar to human von Willebrand factor. Biochem J. 2001;359:99–108. doi: 10.1042/0264-6021:3590099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Scherfer C, Qazi MR, Takahashi K, Ueda R, Dushay MS, Theopold U, et al. The Toll immune-regulated Drosophila protein Fondue is involved in hemolymph clotting and puparium formation. Dev Biol. 2006;295:156–63. doi: 10.1016/j.ydbio.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 194.Lindgren M, Riazi R, Lesch C, Wilhelmsson C, Theopold U, Dushay MS. Fondue and transglutaminase in the Drosophila larval clot. J Insect Physiol. 2008;54:586–92. doi: 10.1016/j.jinsphys.2007.12.008. [DOI] [PubMed] [Google Scholar]
- 195.Wang Z, Wilhelmsson C, Hyrsl P, Loof TG, Dobes P, Klupp M, et al. Pathogen entrapment by transglutaminase—a conserved early innate immune mechanism. PLoS Pathog. 2010;6:e1000763. doi: 10.1371/journal.ppat.1000763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Han SJ, Choi KY, Brey PT, Lee WJ. Molecular cloning and characterization of a Drosophila p38 mitogen-activated protein kinase. J Biol Chem. 1998;273:369–74. doi: 10.1074/jbc.273.1.369. [DOI] [PubMed] [Google Scholar]
- 197.Craig CR, Fink JL, Yagi Y, Ip YT, Cagan RL. A Drosophila p38 orthologue is required for environmental stress responses. EMBO Rep. 2004;5:1058–63. doi: 10.1038/sj.embor.7400282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Inoue H, Tateno M, Fujimura-Kamada K, Takaesu G, Adachi-Yamada T, Ninomiya-Tsuji J, et al. A Drosophila MAPKKK, D-MEKK1, mediates stress responses through activation of p38 MAPK. EMBO J. 2001;20:5421–30. doi: 10.1093/emboj/20.19.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Han ZS, Enslen H, Hu X, Meng X, Wu IH, Barrett T, et al. A conserved p38 mitogen-activated protein kinase pathway regulates Drosophila immunity gene expression. Mol Cell Biol. 1998;18:3527–39. doi: 10.1128/mcb.18.6.3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhuang ZH, Zhou Y, Yu MC, Silverman N, Ge BX. Regulation of Drosophila p38 activation by specific MAP2 kinase and MAP3 kinase in response to different stimuli. Cell Signal. 2006;18:441–8. doi: 10.1016/j.cellsig.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 201.Sano Y, Akimaru H, Okamura T, Nagao T, Okada M, Ishii S. Drosophila activating transcription factor-2 is involved in stress response via activation by p38, but not c-Jun NH(2)-terminal kinase. Mol Biol Cell. 2005;16:2934–46. doi: 10.1091/mbc.E04-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Baril C, Sahmi M, Ashton-Beaucage D, Stronach B, Therrien M. The PP2C Alphabet is a negative regulator of stress-activated protein kinase signaling in Drosophila . Genetics. 2009;181:567–79. doi: 10.1534/genetics.108.096461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Igaki T, Kanda H, Yamamoto-Goto Y, Kanuka H, Kuranaga E, Aigaki T, et al. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 2002;21:3009–18. doi: 10.1093/emboj/cdf306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kanda H, Igaki T, Kanuka H, Yagi T, Miura M. Wengen, a member of the Drosophila tumor necrosis factor receptor superfamily, is required for Eiger signaling. J Biol Chem. 2002;277:28372–5. doi: 10.1074/jbc.C200324200. [DOI] [PubMed] [Google Scholar]
- 205.Kauppila S, Maaty WS, Chen P, Tomar RS, Eby MT, Chapo J, et al. Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila . Oncogene. 2003;22:4860–7. doi: 10.1038/sj.onc.1206715. [DOI] [PubMed] [Google Scholar]
- 206.Moreno E, Yan M, Basler K. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr Biol. 2002;12:1263–8. doi: 10.1016/s0960-9822(02)00954-5. [DOI] [PubMed] [Google Scholar]
- 207.Sluss HK, Han Z, Barrett T, Goberdhan DC, Wilson C, Davis RJ, et al. A JNK signal transduction pathway that mediates morphogenesis and an immune response in Drosophila . Genes Dev. 1996;10:2745–58. doi: 10.1101/gad.10.21.2745. [DOI] [PubMed] [Google Scholar]
- 208.Bidla G, Dushay MS, Theopold U. Crystal cell rupture after injury in Drosophila requires the JNK pathway, small GTPases and the TNF homolog Eiger. J Cell Sci. 2007;120:1209–15. doi: 10.1242/jcs.03420. [DOI] [PubMed] [Google Scholar]
- 209.Chen W, White MA, Cobb MH. Stimulus-specific requirements for MAP3 kinases in activating the JNK pathway. J Biol Chem. 2002;277:49105–10. doi: 10.1074/jbc.M204934200. [DOI] [PubMed] [Google Scholar]
- 210.Baril C, Therrien M. Alphabet, a Ser/Thr phosphatase of the protein phosphatase 2C family, negatively regulates RAS/MAPK signaling in Drosophila . Dev Biol. 2006;294:232–45. doi: 10.1016/j.ydbio.2006.02.046. [DOI] [PubMed] [Google Scholar]
- 211.Kockel L, Homsy JG, Bohmann D. Drosophila AP-1: lessons from an invertebrate. Oncogene. 2001;20:2347–64. doi: 10.1038/sj.onc.1204300. [DOI] [PubMed] [Google Scholar]
- 212.Delaney JR, Stoven S, Uvell H, Anderson KV, Engstrom Y, Mlodzik M. Cooperative control of Drosophila immune responses by the JNK and NF-kappaB signaling pathways. EMBO J. 2006;25:3068–77. doi: 10.1038/sj.emboj.7601182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Kallio J, Leinonen A, Ulvila J, Valanne S, Ezekowitz RA, Ramet M. Functional analysis of immune response genes in Drosophila identifies JNK pathway as a regulator of antimicrobial peptide gene expression in S2 cells. Microbes Infect. 2005;7:811–9. doi: 10.1016/j.micinf.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 214.Schneider DS, Ayres JS, Brandt SM, Costa A, Dionne MS, Gordon MD, et al. Drosophila eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 2007;3:e41. doi: 10.1371/journal.ppat.0030041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Geuking P, Narasimamurthy R, Lemaitre B, Basler K, Leulier F. A non-redundant role for Drosophila Mkk4 and hemipterous/Mkk7 in TAK1-mediated activation of JNK. PLoS One. 2009;4:e7709. doi: 10.1371/journal.pone.0007709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bond D, Foley E. A quantitative RNAi screen for JNK modifiers identifies Pvr as a novel regulator of Drosophila immune signaling. PLoS Pathog. 2009;5:e1000655. doi: 10.1371/journal.ppat.1000655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Radyuk SN, Michalak K, Klichko VI, Benes J, Orr WC. Peroxiredoxin 5 modulates immune response in Drosophila . Biochim Biophys Acta. 2010;1800:1153–63. doi: 10.1016/j.bbagen.2010.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Martin-Blanco E, Gampel A, Ring J, Virdee K, Kirov N, Tolkovsky AM, et al. puckered encodes a phosphatase that mediates a feedback loop regulating JNK activity during dorsal closure in Drosophila . Genes Dev. 1998;12:557–70. doi: 10.1101/gad.12.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Park JM, Brady H, Ruocco MG, Sun H, Williams D, Lee SJ, et al. Targeting of TAK1 by the NF-kappa B protein Relish regulates the JNK-mediated immune response in Drosophila . Genes Dev. 2004;18:584–94. doi: 10.1101/gad.1168104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Corby-Harris V, Habel KE, Ali FG, Promislow DE. Alternative measures of response to Pseudomonas aeruginosa infection in Drosophila melanogaster . J Evol Biol. 2007;20:526–33. doi: 10.1111/j.1420-9101.2006.01267.x. [DOI] [PubMed] [Google Scholar]
- 221.Dionne MS, Pham LN, Shirasu-Hiza M, Schneider DS. Akt and FOXO dysregulation contribute to infection-induced wasting in Drosophila . Curr Biol. 2006;16:1977–85. doi: 10.1016/j.cub.2006.08.052. [DOI] [PubMed] [Google Scholar]
- 222.Ayres JS, Schneider DS. A signaling protease required for melanization in Drosophila affects resistance and tolerance of infections. PLoS Biol. 2008;6:2764–73. doi: 10.1371/journal.pbio.0060305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Tirouvanziam R, Davidson CJ, Lipsick JS, Herzenberg LA. Fluorescence-activated cell sorting (FACS) of Drosophila hemocytes reveals important functional similarities to mammalian leukocytes. Proc Natl Acad Sci U S A. 2004;101:2912–7. doi: 10.1073/pnas.0308734101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Ha EM, Oh CT, Bae YS, Lee WJ. A direct role for dual oxidase in Drosophila gut immunity. Science. 2005;310:847–50. doi: 10.1126/science.1117311. [DOI] [PubMed] [Google Scholar]
- 225.Foley E, O'Farrell PH. Nitric oxide contributes to induction of innate immune responses to gram-negative bacteria in Drosophila . Genes Dev. 2003;17:115–25. doi: 10.1101/gad.1018503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Ha EM, Oh CT, Ryu JH, Bae YS, Kang SW, Jang IH, et al. An antioxidant system required for host protection against gut infection in Drosophila . Dev Cell. 2005;8:125–32. doi: 10.1016/j.devcel.2004.11.007. [DOI] [PubMed] [Google Scholar]
- 227.Yoshiga T, Georgieva T, Dunkov BC, Harizanova N, Ralchev K, Law JH. Drosophila melanogaster transferrin. Cloning, deduced protein sequence, expression during the life cycle, gene localization and up-regulation on bacterial infection. Eur J Biochem. 1999;260:414–20. doi: 10.1046/j.1432-1327.1999.00173.x. [DOI] [PubMed] [Google Scholar]
- 228.Georgieva T, Dunkov BC, Dimov S, Ralchev K, Law JH. Drosophila melanogaster ferritin: cDNA encoding a light chain homologue, temporal and tissue specific expression of both subunit types. Insect Biochem Mol Biol. 2002;32:295–302. doi: 10.1016/s0965-1748(01)00090-x. [DOI] [PubMed] [Google Scholar]
- 229.Vierstraete E, Verleyen P, Baggerman G, D'Hertog W, Van den Bergh G, Arckens L, et al. A proteomic approach for the analysis of instantly released wound and immune proteins in Drosophila melanogaster hemolymph. Proc Natl Acad Sci U S A. 2004;101:470–5. doi: 10.1073/pnas.0304567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.van Rij RP, Saleh MC, Berry B, Foo C, Houk A, Antoniewski C, et al. The RNA silencing endonuclease Argonaute 2 mediates specific antiviral immunity in Drosophila melanogaster . Genes Dev. 2006;20:2985–95. doi: 10.1101/gad.1482006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Wang XH, Aliyari R, Li WX, Li HW, Kim K, Carthew R, et al. RNA interference directs innate immunity against viruses in adult Drosophila . Science. 2006;312:452–4. doi: 10.1126/science.1125694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Li H, Li WX, Ding SW. Induction and suppression of RNA silencing by an animal virus. Science. 2002;296:1319–21. doi: 10.1126/science.1070948. [DOI] [PubMed] [Google Scholar]
- 233.Galiana-Arnoux D, Dostert C, Schneemann A, Hoffmann JA, Imler JL. Essential function in vivo for Dicer-2 in host defense against RNA viruses in Drosophila . Nat Immunol. 2006;7:590–7. doi: 10.1038/ni1335. [DOI] [PubMed] [Google Scholar]
- 234.Zambon RA, Vakharia VN, Wu LP. RNAi is an antiviral immune response against a dsRNA virus in Drosophila melanogaster . Cell Microbiol. 2006;8:880–9. doi: 10.1111/j.1462-5822.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 235.Saleh MC, van Rij RP, Hekele A, Gillis A, Foley E, O'Farrell PH, et al. The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat Cell Biol. 2006;8:793–802. doi: 10.1038/ncbl439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Ulvila J, Parikka M, Kleino A, Sormunen R, Ezekowitz RA, Kocks C, et al. Double-stranded RNA is internalized by scavenger receptor-mediated endocytosis in Drosophila S2 cells. J Biol Chem. 2006;281:14370–5. doi: 10.1074/jbc.M513868200. [DOI] [PubMed] [Google Scholar]
- 237.Saleh MC, Tassetto M, van Rij RP, Goic B, Gausson V, Berry B, et al. Antiviral immunity in Drosophila requires systemic RNA interference spread. Nature. 2009;458:346–50. doi: 10.1038/nature07712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Vlisidou I, Dowling AJ, Evans IR, Waterfield N, ffrench-Constant RH, Wood W. Drosophila embryos as model systems for monitoring bacterial infection in real time. PLoS Pathog. 2009;5:e1000518. doi: 10.1371/journal.ppat.1000518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Olcott MH, Henkels MD, Rosen KL, Walker FL, Sneh B, Loper JE, et al. Lethality and developmental delay in Drosophila melanogaster larvae after ingestion of selected Pseudomonas fluorescens strains. PLoS One. 2010;5:e12504. doi: 10.1371/journal.pone.0012504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Kim SH, Park SY, Heo YJ, Cho YH. Drosophila melanogaster-based screening for multihost virulence factors of Pseudomonas aeruginosa PA14 and identification of a virulence-attenuating factor, HudA. Infect Immun. 2008;76:4152–62. doi: 10.1128/IAI.01637-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.An D, Apidianakis Y, Boechat AL, Baldini RL, Goumnerov BC, Rahme LG. The pathogenic properties of a novel and conserved gene product, KerV, in proteobacteria. PLoS One. 2009;4:e7167. doi: 10.1371/journal.pone.0007167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Berkey CD, Blow N, Watnick PI. Genetic analysis of Drosophila melanogaster susceptibility to intestinal Vibrio cholerae infection. Cell Microbiol. 2009;11:461–74. doi: 10.1111/j.1462-5822.2008.01267.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Park SY, Heo YJ, Kim KS, Cho YH. Drosophila melanogaster is susceptible to Vibrio cholerae infection. Mol Cells. 2005;20:409–15. [PubMed] [Google Scholar]
- 244.Jin LH, Shim J, Yoon JS, Kim B, Kim J, Kim-Ha J, et al. Identification and functional analysis of antifungal immune response genes in Drosophila . PLoS Pathog. 2008;4:e1000168. doi: 10.1371/journal.ppat.1000168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Tabuchi Y, Shiratsuchi A, Kurokawa K, Gong JH, Sekimizu K, Lee BL, et al. Inhibitory role for D-alanylation of wall teichoic acid in activation of insect Toll pathway by peptidoglycan of Staphylococcus aureus . J Immunol. 2010;185:2424–31. doi: 10.4049/jimmunol.1000625. [DOI] [PubMed] [Google Scholar]
- 246.Benghezal M, Fauvarque MO, Tournebize R, Froquet R, Marchetti A, Bergeret E, et al. Specific host genes required for the killing of Klebsiella bacteria by phagocytes. Cell Microbiol. 2006;8:139–48. doi: 10.1111/j.1462-5822.2005.00607.x. [DOI] [PubMed] [Google Scholar]
- 247.Baron MJ, Wong SL, Nybakken K, Carey VJ, Madoff LC. Host glycosaminoglycan confers susceptibility to bacterial infection in Drosophila melanogaster . Infect Immun. 2009;77:860–6. doi: 10.1128/IAI.00995-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Chamilos G, Lewis RE, Kontoyiannis DP. Lovastatin has significant activity against zygomycetes and interacts synergistically with voriconazole. Antimicrob Agents Chemother. 2006;50:96–103. doi: 10.1128/AAC.50.1.96-103.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Lamaris GA, Ben-Ami R, Lewis RE, Chamilos G, Samonis G, Kontoyiannis DP. Increased virulence of Zygomycetes organisms following exposure to voriconazole: a study involving fly and murine models of zygomycosis. J Infect Dis. 2009;199:1399–406. doi: 10.1086/597615. [DOI] [PubMed] [Google Scholar]
- 250.Lamaris GA, Ben-Ami R, Lewis RE, Kontoyiannis DP. Does pre-exposure of Aspergillus fumigatus to voriconazole or posaconazole in vitro affect its virulence and the in vivo activity of subsequent posaconazole or voriconazole, respectively? A study in a fly model of aspergillosis. J Antimicrob Chemother. 2008;62:539–42. doi: 10.1093/jac/dkn224. [DOI] [PubMed] [Google Scholar]
- 251.Lionakis MS, Lewis RE, May GS, Wiederhold NP, Albert ND, Halder G, et al. Toll-deficient Drosophila flies as a fast, high-throughput model for the study of antifungal drug efficacy against invasive aspergillosis and Aspergillus virulence. J Infect Dis. 2005;191:1188–95. doi: 10.1086/428587. [DOI] [PubMed] [Google Scholar]
- 252.Blow NS, Salomon RN, Garrity K, Reveillaud I, Kopin A, Jackson FR, et al. Vibrio cholerae infection of Drosophila melanogaster mimics the human disease cholera. PLoS Pathog. 2005;1:e8. doi: 10.1371/journal.ppat.0010008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Broderick KE, Feala J, McCulloch A, Paternostro G, Sharma VS, Pilz RB, et al. The nitric oxide scavenger cobinamide profoundly improves survival in a Drosophila melanogaster model of bacterial sepsis. FASEB J. 2006;20:1865–73. doi: 10.1096/fj.06-5780com. [DOI] [PubMed] [Google Scholar]
- 254.Needham AJ, Kibart M, Crossley H, Ingham PW, Foster SJ. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology. 2004;150:2347–55. doi: 10.1099/mic.0.27116-0. [DOI] [PubMed] [Google Scholar]
- 255.Estin ML, Stoltz DA, Zabner J. Paraoxonase 1, quorum sensing, and P. aeruginosa infection: a novel model. Adv Exp Med Biol. 2010;660:183–93. doi: 10.1007/978-1-60761-350-3_17. [DOI] [PubMed] [Google Scholar]
- 256.Heo YJ, Lee YR, Jung HH, Lee J, Ko G, Cho YH. Antibacterial efficacy of phages against Pseudomonas aeruginosa infections in mice and Drosophila melanogaster . Antimicrob Agents Chemother. 2009;53:2469–74. doi: 10.1128/AAC.01646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Babcock DT, Brock AR, Fish GS, Wang Y, Perrin L, Krasnow MA, et al. Circulating blood cells function as a surveillance system for damaged tissue in Drosophila larvae. Proc Natl Acad Sci U S A. 2008;105:10017–22. doi: 10.1073/pnas.0709951105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Serio AW, Jeng RL, Haglund CM, Reed SC, Welch MD. Defining a core set of actin cytoskeletal proteins critical for actin-based motility of Rickettsia . Cell Host Microbe. 2010;7:388–98. doi: 10.1016/j.chom.2010.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Agaisse H. Investigating the involvement of host factors involved in intracellular pathogen infection by RNAi in Drosophila cells. Methods Mol Biol. 2008;415:395–402. doi: 10.1007/978-1-59745-570-1_23. [DOI] [PubMed] [Google Scholar]
- 260.Akimana C, Al-Khodor S, Abu Kwaik Y. Host factors required for modulation of phagosome biogenesis and proliferation of Francisella tularensis within the cytosol. PLoS One. 2010;5:e11025. doi: 10.1371/journal.pone.0011025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Agaisse H, Burrack LS, Philips JA, Rubin EJ, Perrimon N, Higgins DE. Genome-wide RNAi screen for host factors required for intracellular bacterial infection. Science. 2005;309:1248–51. doi: 10.1126/science.1116008. [DOI] [PubMed] [Google Scholar]
- 262.Sessions OM, Barrows NJ, Souza-Neto JA, Robinson TJ, Hershey CL, Rodgers MA, et al. Discovery of insect and human dengue virus host factors. Nature. 2009;458:1047–50. doi: 10.1038/nature07967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Guichard A, Park JM, Cruz-Moreno B, Karin M, Bier E. Anthrax lethal factor and edema factor act on conserved targets in Drosophila . Proc Natl Acad Sci U S A. 2006;103:3244–9. doi: 10.1073/pnas.0510748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Adamson AL, Wright N, LaJeunesse DR. Modeling early Epstein-Barr virus infection in Drosophila melanogaster: the BZLF1 protein. Genetics. 2005;171:1125–35. doi: 10.1534/genetics.105.042572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Guichard A, McGillivray SM, Cruz-Moreno B, van Sorge NM, Nizet V, Bier E. Anthrax toxins cooperatively inhibit endocytic recycling by the Rab11/Sec15 exocyst. Nature. 2010;467:854–8. doi: 10.1038/nature09446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Fitch CL, de Sousa SM, O'Day PM, Neubert TA, Plantilla CM, Spencer M, et al. Pertussis toxin expression in Drosophila alters the visual response and blocks eating behaviour. Cell Signal. 1993;5:187–207. doi: 10.1016/0898-6568(93)90070-3. [DOI] [PubMed] [Google Scholar]
- 267.Castonguay-Vanier J, Vial L, Tremblay J, Deziel E. Drosophila melanogaster as a model host for the Burkholderia cepacia complex. PLoS One. 2010;5:e11467. doi: 10.1371/journal.pone.0011467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Moule MG, Monack DM, Schneider DS. Reciprocal analysis of Francisella novicida infections of a Drosophila melanogaster model reveal host-pathogen conflicts mediated by reactive oxygen and imd-regulated innate immune response. PLoS Pathog. 2010;6:e1001065. doi: 10.1371/journal.ppat.1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Santic M, Akimana C, Asare R, Kouokam JC, Atay S, Kwaik YA. Intracellular fate of Francisella tularensis within arthropod-derived cells. Environ Microbiol. 2009;11:1473–81. doi: 10.1111/j.1462-2920.2009.01875.x. [DOI] [PubMed] [Google Scholar]
- 270.Lauriano CM, Barker JR, Yoon SS, Nano FE, Arulanandam BP, Hassett DJ, et al. MglA regulates transcription of virulence factors necessary for Francisella tularensis intraamoebae and intramacrophage survival. Proc Natl Acad Sci U S A. 2004;101:4246–9. doi: 10.1073/pnas.0307690101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Ahlund MK, Ryden P, Sjostedt A, Stoven S. Directed screen of Francisella novicida virulence determinants using Drosophila melanogaster . Infect Immun. 2010;78:3118–28. doi: 10.1128/IAI.00146-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Aymeric JL, Givaudan A, Duvic B. Imd pathway is involved in the interaction of Drosophila melanogaster with the entomopathogenic bacteria, Xenorhabdus nematophila and Photorhabdus luminescens . Mol Immunol. 2010;47:2342–8. doi: 10.1016/j.molimm.2010.05.012. [DOI] [PubMed] [Google Scholar]
- 273.Valanne S, Kleino A, Myllymaki H, Vuoristo J, Ramet M. Iap2 is required for a sustained response in the Drosophila Imd pathway. Dev Comp Immunol. 2007;31:991–1001. doi: 10.1016/j.dci.2007.01.004. [DOI] [PubMed] [Google Scholar]
- 274.Avet-Rochex A, Perrin J, Bergeret E, Fauvarque MO. Rac2 is a major actor of Drosophila resistance to Pseudomonas aeruginosa acting in phagocytic cells. Genes Cells. 2007;12:1193–204. doi: 10.1111/j.1365-2443.2007.01121.x. [DOI] [PubMed] [Google Scholar]
- 275.Garver LS, Wu J, Wu LP. The peptidoglycan recognition protein PGRP-SC1a is essential for Toll signaling and phagocytosis of Staphylococcus aureus in Drosophila . Proc Natl Acad Sci U S A. 2006;103:660–5. doi: 10.1073/pnas.0506182103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bergeret E, Perrin J, Williams M, Grunwald D, Engel E, Thevenon D, et al. TM9SF4 is required for Drosophila cellular immunity via cell adhesion and phagocytosis. J Cell Sci. 2008;121:3325–34. doi: 10.1242/jcs.030163. [DOI] [PubMed] [Google Scholar]
- 277.Ayres JS, Freitag N, Schneider DS. Identification of Drosophila mutants altering defense of and endurance to Listeria monocytogenes infection. Genetics. 2008;178:1807–15. doi: 10.1534/genetics.107.083782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Stoltz DA, Ozer EA, Taft PJ, Barry M, Liu L, Kiss PJ, et al. Drosophila are protected from Pseudomonas aeruginosa lethality by transgenic expression of paraoxonase-1. J Clin Invest. 2008;118:3123–31. doi: 10.1172/JCI35147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Broderick KE, Chan A, Balasubramanian M, Feala J, Reed SL, Panda M, et al. Cyanide produced by human isolates of Pseudomonas aeruginosa contributes to lethality in Drosophila melanogaster . J Infect Dis. 2008;197:457–64. doi: 10.1086/525282. [DOI] [PubMed] [Google Scholar]
- 280.Apidianakis Y, Mindrinos MN, Xiao W, Lau GW, Baldini RL, Davis RW, et al. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc Natl Acad Sci U S A. 2005;102:2573–8. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Gordon MD, Ayres JS, Schneider DS, Nusse R. Pathogenesis of listeria-infected Drosophila wntD mutants is associated with elevated levels of the novel immunity gene edin. PLoS Pathog. 2008;4:e1000111. doi: 10.1371/journal.ppat.1000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Apidianakis Y, Mindrinos MN, Xiao W, Tegos GP, Papisov MI, Hamblin MR, et al. Involvement of skeletal muscle gene regulatory network in susceptibility to wound infection following trauma. PLoS One. 2007;2:e1356. doi: 10.1371/journal.pone.0001356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Foley E, O'Farrell PH. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2004;2:E203. doi: 10.1371/journal.pbio.0020203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, Ramadan N, et al. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 2006;34:D489–94. doi: 10.1093/nar/gkj114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Cherry S. RNAi screening for host factors involved in viral infection using Drosophila cells. Methods Mol Biol. 2011;721:375–82. doi: 10.1007/978-1-61779-037-9_23. [DOI] [PubMed] [Google Scholar]
- 286.Clemens JC, Worby CA, Simonson-Leff N, Muda M, Maehama T, Hemmings BA, et al. Use of double-stranded RNA interference in Drosophila cell lines to dissect signal transduction pathways. Proc Natl Acad Sci U S A. 2000;97:6499–503. doi: 10.1073/pnas.110149597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Cheng LW, Viala JP, Stuurman N, Wiedemann U, Vale RD, Portnoy DA. Use of RNA interference in Drosophila S2 cells to identify host pathways controlling compartmentalization of an intracellular pathogen. Proc Natl Acad Sci U S A. 2005;102:13646–51. doi: 10.1073/pnas.0506461102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Elwell CA, Ceesay A, Kim JH, Kalman D, Engel JN. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 2008;4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Sibley CD, Duan K, Fischer C, Parkins MD, Storey DG, Rabin HR, Surette MG. Discerning the complexity of community interactions using a Drosophila model of polymicrobial infections. PLoS Pathog. 2008;4:e1000184. doi: 10.1371/journal.ppat.1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Moy TI, Ball AR, Anklesaria Z, Casadei G, Lewis K, Ausubel FM. Identification of novel antimicrobials using a live-animal infection model. Proc Natl Acad Sci U S A. 2006;103:10414–9. doi: 10.1073/pnas.0604055103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–73. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–55. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Kumar PS, Griffen AL, Barton JA, Paster BJ, Moeschberger ML, Leys EJ. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–44. doi: 10.1177/154405910308200503. [DOI] [PubMed] [Google Scholar]
- 294.Gross EL, Leys EJ, Gasparovich SR, Firestone ND, Schwartzbaum JA, Janies DA, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–8. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Aas JA, Griffen AL, Dardis SR, Lee AM, Olsen I, Dewhirst FE, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–17. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Hoover CI, Yoshimura F. Transposon-induced pigment-deficient mutants of Porphyromonas gingivalis . FEMS Microbiol Lett. 1994;124:43–8. doi: 10.1111/j.1574-6968.1994.tb07259.x. [DOI] [PubMed] [Google Scholar]
- 297.Chen T, Dong H, Tang YP, Dallas MM, Malamy MH, Duncan MJ. Identification and cloning of genes from Porphyromonas gingivalis after mutagenesis with a modified Tn4400 transposon from Bacteroides fragilis . Infect Immun. 2000;68:420–3. doi: 10.1128/iai.68.1.420-423.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Shoji M, Ratnayake DB, Shi Y, Kadowaki T, Yamamoto K, Yoshimura F, et al. Construction and characterization of a nonpigmented mutant of Porphyromonas gingivalis: cell surface polysaccharide as an anchorage for gingipains. Microbiology. 2002;148:1183–91. doi: 10.1099/00221287-148-4-1183. [DOI] [PubMed] [Google Scholar]
- 299.Sato K, Kido N, Murakami Y, Hoover CI, Nakayama K, Yoshimura F. Lipopolysaccharide biosynthesis-related genes are required for colony pigmentation of Porphyromonas gingivalis . Microbiology. 2009;155:1282–93. doi: 10.1099/mic.0.025163-0. [DOI] [PubMed] [Google Scholar]
- 300.Davey ME, Duncan MJ. Enhanced biofilm formation and loss of capsule synthesis: deletion of a putative glycosyltransferase in Porphyromonas gingivalis . J Bacteriol. 2006;188:5510–23. doi: 10.1128/JB.01685-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Sjoberg BM, Torrents E. Shift in ribonucleotide reductase gene expression in Pseudomonas aeruginosa during infection. Infect Immun. 2011;79:2663–9. doi: 10.1128/IAI.01212-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Liang H, Duan J, Sibley CD, Surette MG, Duan K. Identification of mutants with altered phenazine production in Pseudomonas aeruginosa . J Med Microbiol. 2011;60:22–34. doi: 10.1099/jmm.0.022350-0. [DOI] [PubMed] [Google Scholar]
- 303.Kim SH, Lee BY, Lau GW, Cho YH. IscR modulates catalase A (KatA) activity, peroxide resistance and full virulence of Pseudomonas aeruginosa PA14. J Microbiol Biotechnol. 2009;19:1520–6. doi: 10.4014/jmb.0906.06028. [DOI] [PubMed] [Google Scholar]
- 304.Lee JS, Heo YJ, Lee JK, Cho YH. KatA, the major catalase, is critical for osmoprotection and virulence in Pseudomonas aeruginosa PA14. Infect Immun. 2005;73:4399–403. doi: 10.1128/IAI.73.7.4399-4403.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Shin DH, Choi YS, Cho YH. Unusual properties of catalase A (KatA) of Pseudomonas aeruginosa PA14 are associated with its biofilm peroxide resistance. J Bacteriol. 2008;190:2663–70. doi: 10.1128/JB.01580-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Lau GW, Britigan BE, Hassett DJ. Pseudomonas aeruginosa OxyR is required for full virulence in rodent and insect models of infection and for resistance to human neutrophils. Infect Immun. 2005;73:2550–3. doi: 10.1128/IAI.73.4.2550-2553.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Erickson DL, Lines JL, Pesci EC, Venturi V, Storey DG. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster . Infect Immun. 2004;72:5638–45. doi: 10.1128/IAI.72.10.5638-5645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Vogt SL, Green C, Stevens KM, Day B, Erickson DL, Woods DE, et al. The stringent response is essential for Pseudomonas aeruginosa virulence in the rat lung agar bead and Drosophila melanogaster feeding models of infection. Infect Immun. 2011;79:4094–104. doi: 10.1128/IAI.00193-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309.D'Argenio DA, Gallagher LA, Berg CA, Manoil C. Drosophila as a model host for Pseudomonas aeruginosa infection. J Bacteriol. 2001;183:1466–71. doi: 10.1128/JB.183.4.1466-1471.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Yang L, Chen L, Shen L, Surette M, Duan K. Inactivation of MuxABC-OpmB transporter system in Pseudomonas aeruginosa leads to increased ampicillin and carbenicillin resistance and decreased virulence. J Microbiol. 2011;49:107–14. doi: 10.1007/s12275-011-0186-2. [DOI] [PubMed] [Google Scholar]
- 311.Potvin E, Lehoux DE, Kukavica-Ibrulj I, Richard KL, Sanschagrin F, Lau GW, et al. In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ Microbiol. 2003;5:1294–308. doi: 10.1046/j.1462-2920.2003.00542.x. [DOI] [PubMed] [Google Scholar]
- 312.Kurz CL, Chauvet S, Andres E, Aurouze M, Vallet I, Michel GP, et al. Virulence factors of the human opportunistic pathogen Serratia marcescens identified by in vivo screening. EMBO J. 2003;22:1451–60. doi: 10.1093/emboj/cdg159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Pili-Floury S, Leulier F, Takahashi K, Saigo K, Samain E, Ueda R, et al. In vivo RNA interference analysis reveals an unexpected role for GNBP1 in the defense against Gram-positive bacterial infection in Drosophila adults. J Biol Chem. 2004;279:12848–53. doi: 10.1074/jbc.M313324200. [DOI] [PubMed] [Google Scholar]
- 314.Ulvila J, Vanha-Aho LM, Kleino A, Vaha-Makila M, Vuoksio M, Eskelinen S, et al. Cofilin regulator 14-3-3{zeta} is an evolutionarily conserved protein required for phagocytosis and microbial resistance. J Leukoc Biol. 2011;89:649–59. doi: 10.1189/jlb.0410195. [DOI] [PubMed] [Google Scholar]