Abstract
RNA-binding proteins, and in particular TAR DNA-binding protein 43 (TDP43), are central to the pathogenesis of motor neuron diseases and related neurodegenerative disorders. Studies on human tissue have implicated several possible mechanisms of disease and experimental studies are now attempting to determine whether TDP43-mediated neurodegeneration results from a gain or a loss of function of the protein. In addition, the distinct possibility of pleotropic or combined effects — in which gains of toxic properties and losses of normal TDP43 functions act together — needs to be considered.
TAR DNA-binding protein 43 (TDP43) was identified 5 years ago as the major constituent of the proteinaceous inclusions that are characteristic of most forms of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD, now known as FTLD-TDP; see BOX 1)1. Since then, numerous groups have confirmed this finding2–10 and have shown that dominantly inherited genetic mutations within the gene that encodes TDP43, TAR DNA binding protein (TARDBP), are linked with ALS and FTLD-TDP phenotypes11. With over 600 studies published on TDP43 and its role in ALS and FTLD, there is now an emerging consensus that TDP43 protein is mechanistically linked to neurodegeneration.
Box 1. Nosology of frontotemporal lobar degeneration.
The classification of frontotemporal lobar degeneration (FTLD) has undergone a dramatic transformation over the past decade, mainly driven by remarkable advances in understanding the pathobiology of these disorders. A subset of FTLD cases were initially given the diagnosis of ‘dementia lacking distinctive histopathology’ (DLDH) after post-mortem examination because, although extensive neurodegeneration and reactive gliosis were seen throughout the CNS, no specific pathologic features were detected146. This was partially rectified when immunohistochemistry for ubiquitin revealed a unique pattern of ubiquitin-positive inclusions in the majority of these DLDH cases147. The use of ubiquitin immunohistochemistry resulted from knowledge that many intracellular inclusions that are associated with neurodegenerative diseases are ubiquitylated, including the inclusions formed by tau and α-synuclein. Thus, these enigmatic cases were given the diagnosis of ‘FTLD with ubiquitin-positive, tau and α-synuclein-negative inclusions’ (also known as FTLD-U). However, when these ubiquitin-positive inclusions were found to be TAR DNA-binding protein 43 (TDP43)-positive by immunohistochemistry and TDP43 was found to be the most sensitive and reliable marker of inclusions in cases such as these that also had a disease specific biochemical TDP43 signature, TDP43 was proposed to be the disease protein in FTLD-U1. Accordingly, FTLD cases with prominent TDP43 pathology were renamed FTLD-TDP and those with abundant tau pathology were renamed FTLD-Tau, and the small minority of cases characterized by RNA-binding protein FUS inclusions were designated FTLD-FUS84,148. Thus, FTLD-TDP, FTLD-Tau and FTLD-FUS represent 50%, 45% and 5% of all FTLD cases, respectively, at post-mortem examination. Finally, the finding that amyotrophic lateral sclerosis (ALS), the most common motor neuron disease, was often associated with FTLD-TDP prompted evaluation of ALS tissue, which revealed that the ubiquitylated skeins and round inclusions found in ALS were comprised of TDP43 protein in association with a disease-specific biochemical TDP43 signature similar to FTLD-TDP1. Furthermore, nearly all sporadic ALS cases and familial ALS cases with mutations of the gene that encodes TDP43 — TARDBP — are TDP43 proteinopathies. Thus, FTLD-TDP and ALS are two seemingly distinct neurodegenerative diseases based on their clinical manifestations, but they are both TDP43 proteinopathies and they therefore represent a spectrum of diseases ranging from relatively pure motor neuron degeneration in ALS to more prominent frontal and temporal neocortical involvement in FTLD-TDP, leading to a variable phenotype ranging from weakness and respiratory failure to psychiatric aberrations, dementia and eventual death.
TDP43 is a 414-amino-acid protein with two RNA recognition motifs (RNA-recognition motif 1 (RRM1) and RRM2) and a carboxy-terminal glycine-rich domain (FIG. 1a). Nearly all of the described ALS-associated TDP43 mutations are dominant missense mutations within the glycine-rich domain, suggesting that altering the function of this domain is sufficient to induce neurodegeneration11–15.
Figure 1. The genetics, pathology and biochemistry of TDP43 proteinopathies.
a | TAR DNA-binding protein 43 (TDP43) protein contains two RNA-recognition motifs (RNA-recognition motif 1 (RRM1) and RRM2), a carboxy-terminal glycine-rich domain, a bipartite nuclear localization signal (NLS) and a nuclear export signal (NES). Numerous mutations (shown by short vertical lines) have been linked to sporadic and familial forms of amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD). These are almost exclusively found within or immediately adjacent to the glycine-rich domain with the exception of an Asp169Gly mutation within exon 4 (the site at which TDP43 cleavage putatively occurs is shown by an arrow). Known TDP43 phosphorylation sites (Ser 379, Ser403 + Ser404, and Ser409 + Ser 410), when heavily phosphorylated, also contribute to the disease-specific TDP43 biochemical signature (these sites are indicated by asterisks)58. b | TDP43 immunohistochemistry of human FTLD brain reveals intracytoplasmic inclusions affecting neurons. Dystrophic neurites and glia also exhibit TDP43 inclusions (not shown). An image of the dentate gyrus stained with an anti-TDP43 antibody shows cells without inclusions with normal nuclear immunoreactivity, whereas inclusion-bearing cells show a loss of normal nuclear staining that leads to a presumptive loss of TDP43 function in the nucleus. TDP43 immunofluorescence (shown in green) of an intranuclear inclusion in FTLD-TDP43 is shown. TDP43 immunostaining using a phospho-specific anti-TDP43 antibody of human ALS spinal cord shows characteristic round inclusions (shown by brown immunohistochemistry) and skeins (shown by red immunofluorescence) in lower motor neurons. c | Biochemical analysis demonstrates the distinct biochemical disease-specific TDP43 signature characterized by the accumulation of sarkosyl-insoluble TDP43 species comprised of phosphorylated TDP43, ubiquitylated high molecular weight TDP43 and truncated C-terminal fragments.
The initial flurry of studies on TDP43 in ALS and FTLD-TDP described TDP43 pathology and biochemistry in human tissue (FIG.1b,c). In quick succession, TDP43 proteinopathies were documented in a wide range of other neurodegenerative diseases, including Alzheimer’s disease, other tauopathies and Lewy body disorders characterized by α-synuclein inclusions16,17. The extent of TDP43 pathology in these other diseases is limited in terms of both the amount and distribution of TDP43 compared with cases of primary ALS and FTLD-TDP. Thus, the formation of TDP43 pathology, although a primary event in FTLD-TDP and ALS, may be a secondary event in these other diseases, and it remains to be determined whether abnormal TDP43 exacerbates the extent of neurodegeneration in these patients.
Detailed reviews of the pathology of TDP43 proteinopathies are available elsewhere16,17. The major disease-specific findings in FTLD-TDP and ALS — as well as, to a lesser extent, other disorders in which TDP43 pathology is found — include abnormal ubiquitylation and phosphorylation of TDP43, the presence of sarko-syl-insoluble TDP43 inclusions, the presence of truncated 20–25-kDa TDP43 C-terminal fragments (CTFs; particularly in the cerebral cortex), mislocalization of TDP43 protein, and loss of normal nuclear TDP43 expression. Careful interpretation of each of these findings is warranted because it is unknown which of these observations are causally linked to neurodegeneration and which are secondary to the disease pathology or are epiphenomena.
A common theme of neurodegenerative diseases is the presence of insoluble aggregates that, if they are intracellular, are often abnormally phosphorylated and ubiquitylated. The unique features of TDP43 proteinopathies are the mislocalization of the cognate protein to the cytoplasm and the loss of its normal nuclear localization. Which of these features are mechanistically linked to neurodegeneration is currently unknown and so all of the observed features of neurodegenerative diseases with TDP43 pathology, referred to as TDP43 proteinopathies, need to be considered when formulating models of disease mechanisms.
Despite the progress towards describing the full spectrum of TDP43 pathology in human neurodegenerative diseases, the fundamental question of whether TDP43 dysfunction mediates neurodegeneration through gain of toxic function or a loss of normal function remains unanswered. To formulate a mechanistic model of TDP43-mediated neurodegeneration, we will review the normal functions of TDP43 and then examine the known abnormal features of TDP43 protein in the diseased human brain. We will use this as a framework for understanding mechanisms of disease in TDP43 proteinopathies. In doing so, we will highlight the most relevant human, animal and tissue culture findings in the context of the pathways that may be responsible for TDP43-mediated neurodegeneration in ALS, FTLD-TDP and related TDP43 proteinopathies. We aim to provide here an in-depth consideration of TDP43; however, it is important to note the growing number of proteins that are linked to ALS and FTLD, information on these proteins is available in other recent reviews18–20.
Normal TDP43 function
The best-studied functions of TDP43 are those concerning its regulation of RNA (FIG. 2). Evidence from converging lines of research suggests that TDP43 regulates RNA in a variety of ways. The RRM1 domain of TDP43 is critical for its binding to single-stranded RNA21–24. TDP43 preferentially binds UG repeats, but is also found to be associated with non-UG repeat sequences21,24–27. Indeed, studies using high-throughput sequencing of RNA isolated by crosslinking immunoprecipitation (HITS-CLIP) revealed that TDP43 binds to a large proportion of the transcriptome (>6,000 RNA species), preferentially localizing to introns (including deep intronic sites), 3′ untranslated regions (UTRs) and non-coding RNAs25,27. The list of TDP43 RNA targets was enriched for long transcripts, and transcripts related to synaptic activity or neuronal development25,27.
Figure 2. Normal functions of TDP43.
TAR DNA-binding protein 43 (TDP43) exhibits multiple normal biological functions, predominantly those that regulate RNA pathways. a | TDP43 is a component of heterogenous nuclear ribonucleoprotein (hnRNP) particles, which regulate splicing of pre-mRNA species. b | TDP43 also binds to mRNA sequences, particularly within the 3′ untranslated region, and affects mRNA stability and turnover. c | TDP43 is thought to play a part in mRNA trafficking, as TDP43 undergoes rapid nucleo–cytoplasmic shuttling and is localized within dendritic RNA granules. d | TDP43 is also a component of the Drosha complex, which functions to process primary microRNAs. e | TDP43 can act as a transcriptional repressor by binding to single stranded DNA (ssDNA) promoter sequences. f | TDP43 also colocalizes with stress granules that are thought to sequester and protect mRNAs under conditions of stress.
Consistent with its function as an RNA-binding protein, TDP43 associates with members of the heterogenous nuclear ribonucleoprotein (hnRNP) family of proteins that include hnRNP A2/B1, hnRNP A1, hnRNP C1/C2 and hnRNP A3 (REFS 28–30) (FIG. 2a). The interactions of TDP43 with hnRNPs are dependent on its C-terminal glycine-rich domain28,31. hnRNP complexes are known to regulate splicing, and TDP43 regulates the splicing of human cystic fibrosis transmembrane conductance regulator32, survival of motor neuron33, apolipoprotein A2 (APOA2)34 and serine/arginine-rich splicing factor 2 (SC35)35. Furthermore, transcriptome-wide analysis indicates that the splicing of many additional transcripts is probably mediated by TDP43 (REFS 25,27). In addition to its effects on RNA splicing, TDP43 also influences mRNA turnover: its expression regulates mRNA levels of cyclin-dependent kinase 6 (REF. 36), histone deacetylase 6 (REF. 37), Futsch (the Drosophila melanogaster homologue of microtubule-associated protein 1B (MAP1B))38 and low molecular weight neurofilament4 (FIG. 2b). The localization of TDP43 in RNA granules within neuronal processes also suggests that TDP43 is involved in RNA trafficking39,40 (FIG. 2c). A variety of stressors cause TDP43 to be redistributed from the nucleus into the cytoplasm, where it resides within stress granules, in which it presumably has a role in trafficking or stabilizing mRNAs30,39,41–48 (FIG. 2f). Finally, TDP43 is known to interact with the RNA-binding protein FUS (also known as translocated in liposarcoma (TLS)), another RNA-binding protein implicated in motor neuron disease and FTLD29,30.
Aside from its role in mRNA pathways, TDP43 is also thought to regulate microRNA (miRNA) biogenesis. TDP43 localizes to the perichromatin fibres in which miRNA biogenesis is thought to occur, and binds to the primary miRNA processing Drosha complex. Furthermore, knockdown of TDP43 alters the level of several miRNAs29,49–52 (FIG. 2d). TDP43 also interacts with DNA — in particular, single-stranded DNA — and was identified as a potential regulator of acrosomal protein SP10 (ACRVI; also known as SP10) gene expression during spermatogenesis24,53–56 (FIG. 2e). Indeed, TDP43 derives its name from its discovery in 1995, when it was shown to interact with the HIV-1 TAR DNA sequence motif22. Binding of TDP43 to these DNA sequences results in inhibition of transcription through unknown mechanisms.
Post-translational modification of TDP43
Like other proteins that form neurodegenerative disease-related intracellular inclusions, such as tau and α-synuclein, pathological TDP43 aggregates are ubiquitylated and phosphorylated1. Under normal circumstances, ubiquitylated and phosphorylated TDP43 is not readily detected in brain tissue. Thus, the very presence of these modified TDP43 species in ALS and FTLD-TDP is abnormal and seems to be disease specific; however, it is not clear whether these modifications lead to aggregate formation and/or neurotoxicity, or whether they represent a normal reaction to the presence of an intracellular aggregate and are therefore indirectly related to TDP43-mediated neurodegeneration. Interestingly, human tissue studies indicate that not all TDP43 inclusions are ubiquitin positive. In particular, so-called ‘pre-inclusions’ (granular, less dense cytoplasmic inclusions) are often not ubiquitin positive, suggesting that ubiquitylation is a relatively late phenomenon in the disease process2,4. By contrast, antibodies that specifically detect TDP43 that is phosphorylated at serine residues 409 and 410 seem to recognize most TDP43 inclusions as well as the truncated 25-kDa TPD43 CTFs57,58. These studies suggest that phosphorylation precedes ubiquitylation, but they do not address whether these post-translational modifications are mechanistically involved in TDP43-mediated neurodegeneration.
One important recent finding that may help to clarify the role of altered ubiquitylation (and consequently, of impaired protein degradation) in FTLD-TDP and ALS is the identification of rare dominant missense mutations within ubiquilin 2 (UBQLN2) that are associated with X-linked ALS59. UBQLN2 is the gene that encodes for UBQLN2 protein, which is thought to be involved in protein degradation. The disease-associated mutations were shown to cause impaired ubiquitin–proteasome degradation in cultured cells. Pathologic examination of CNS tissue from patients with UBQLN2 mutations showed that the CNS of these patients harbour TDP43 inclusions in addition to novel UBQLN2-positive inclusions, suggesting that perturbation of protein degradation pathways is mechanistically linked to the formation of inclusions, including TDP43 inclusions. These human studies serve as the backdrop for the experimental studies in cultured cells and animals reviewed below.
Ubiquitylation
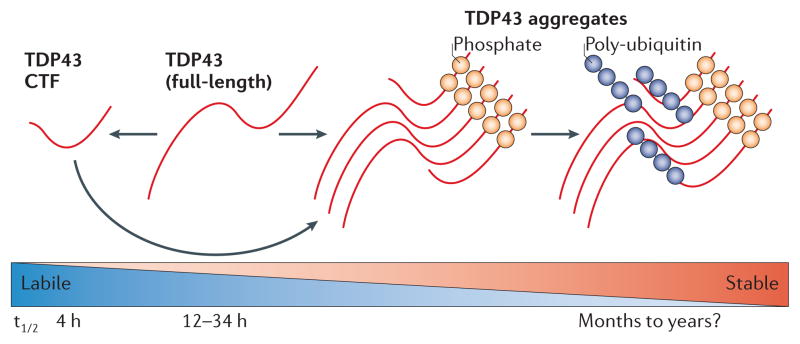
Cells are thought to dispose of TDP43 protein through the ubiquitin–proteasome system (UPS), consistent with the ubiquitylation of overexpressed TDP43 CTFs in a variety of cell lines and over-expressed full-length TDP43 in a variety of transgenic animal models60–70. Thus, disruption of the UPS might contribute to increased levels of ubiquitylated TDP43 in ALS and FTLD-TDP. Indeed, inhibition of the UPS leads to increased levels of phosphorylated TDP43 aggregates in cultured cells69. Full-length TDP43 has an exceptionally long half-life of 12–34 h in most cell types, whereas TDP43 CTFs have a half-life of only ~4 h29,71 (FIG. 3). As both full-length TDP43 and truncated TDP43 are degraded by the UPS65,69,70, the differences in their turnover rates may be due to other factors such as their differential interactions with the UPS, particularly as TDP43 CTFs lack the two nuclear localization signal (NLS) motifs and are therefore predominantly cytoplasmic65,69. Given the lability of TDP43 CTFs, the accumulation of cytoplasmic TDP43 CTFs in diseased brain and various cellular models is remarkable and may be a consequence of increased cleavage of full-length TDP43 or an impaired ability to degrade TDP43 CTFs.
Figure 3. TDP43 modification, stability and turnover.
Both proteosomal and autophagasomal degradation of TAR DNA-binding protein 43 (TDP43) protein has been described. We found that full-length TDP43 is a long-lived protein with a half-life of greater than 34 hours, although other studies have reported that it has a half-life ranging from 4 to 12 hours depending on the cell type29,71. Truncation of TDP43 results in the production of carboxy-terminal fragments (CTFs) that are rapidly translocated to the cytoplasm and degraded. TDP43 aggregates can form under various conditions in which CTFs and full-length TDP43 protein seem to co-aggregate, and TDP43 aggregation is tightly linked with the presence of phosphorylated TDP43. Ubiquitylation of TDP43 aggregates seems to occur late in the lifecycle of an inclusion. Given that TDP43 aggregates are resistant to degradation, different TDP43 isoforms and conformers exhibit different turnover rates, ranging from the labile soluble CTFs to stable insoluble aggregates. t1/2, half-life.
Autophagosome-mediated degradation — in which cytoplasmic proteins or organelles are targeted to membrane-bound vesicles that are in turn fused with lysosomes — may also be involved in TDP43 protein turnover72–76. Evidence for autophagic degradation of TDP43 includes known interactions of TDP43 with a variety of proteins that are involved in autophagy or related pathways, including the endosomal sorting complexes required for transport (ESCRTs)75, ubiquilin 1 (UBQLN)73, sequestosome 1 (REF. 74) and STAM-binding protein (STAMBP; also known as AMSH)77. ESCRTs are required to traffic proteins into multivesicular bodies, and depleting cells of ESCRT subunits inhibits autophagy and results in increased ubiquitylated, cytoplasmic TDP43 (REF. 75). Notably, mutations of charged multivesicular body protein 2B (CHMP2B), an ESCRT subunit, are linked to FLTD, although these rare cases do not exhibit TDP43 inclusions78. Thus, perturbation of autophagy-related pathways results in a clinical phenotype similar to FTLD-TDP, and this may reflect degeneration of similar neuronal populations. Finally, UBQLN is an adaptor protein that functions to deliver ubiquitylated proteins to the proteasome and also regulates autophagy. UBQLN overexpression in HeLa cell cultures results in increased numbers of cytoplasmic TDP43 aggregates that colocalize with both UBQLN and a marker of LC3-positive autophagasomes73. However, overexpression of UBQLN reduced survival in D. melanogaster and enhanced toxicity in HeLa cell cultures despite reducing TDP43 levels79.
These studies demonstrate a role for the UPS and autophagosome systems in TDP43 turnover, and the identification of dominant UBQLN2 mutations suggests that a primary alteration in protein degradation pathways can lead to TDP43-mediated neurodegeneration. However, there is little evidence that a protein degradation defect is primarily responsible for TDP43 dysfunction in sporadic and familial cases without UBQLN2 mutations.
Phosphorylation
TDP43 CTFs or cytoplasmic TDP43 are phosphorylated in a variety of cell lines that are engineered to overexpress TDP43 (REFS 65,67–70). Overexpression of full-length TDP43 in a variety of transgenic animal models also leads to the presence of phosphorylated TDP43 aggregates that are similar to those seen in ALS and FTLD-TDP60–64,66. Phosphorylated TDP43 proteins exhibit a longer half-life than non-phosphorylated proteins, suggesting that phosphorylation may inhibit UPS-mediated degradation and thus contribute to aggregate formation70. Alternatively, as TDP43 phosphorylation is associated with TDP43 insolublility, it remains possible that aggregation may render TDP43 CTFs less vulnerable to UPS-mediated degradation and that phosphorylation is merely a marker of aggregation (FIG. 3). Another study showed that mutating serine residues 409 and 410 of disease-associated mutant TDP43 to alanines (thereby mimicking non-phosphorylated TDP43) eliminated its neurotoxicity when overexpressed in a Caenorhabditis elegans model80. Although wild-type TDP43 was also moderately neurotoxic, introducing the same serine to alanine mutations in wild-type TDP43 did not mitigate neurotoxicity80.
In summary, phosphorylation seems to be a relatively early event in the onset and progression of ALS and FTLD-TDP. There is some evidence that TDP43 phosphorylation may result in differential degradation and/ or toxicity of the protein, but the precise role of TDP43 phosphorylation in mechanisms of disease remains unclear (FIG. 3). TDP43 phosphorylation is tightly correlated with TDP43 aggregation, and thus it is difficult to know whether phosphorylation is a cause or effect of aggregation. Indeed, phosphorylation is not an absolute requirement for TDP43 cleavage, aggregation or toxicity in cell culture systems81,82. Also, disease-associated TDP43 mutations have not been shown to alter TDP43 phosphorylation, and the effects of TDP43 phosphorylation on normal TDP43 function are unknown.
TDP43 insolubility and aggregation
The defining histopathology of FTLD-TDP and ALS is the presence of dense TDP43 aggregates in affected neurons and glia of the CNS1,83. The presence of abnormal proteinaceous aggregates is a common theme among most age-related neurodegenerative diseases, although in most cases whether the inclusions are harmful, protective or both at different stages of the disease process remains a matter of debate (FIG. 4). Indeed, the strong correlation between TDP43 aggregation and a host of other pathologic changes (such as nuclear clearance, ubiquitylation, phosphorylation and mislocalization) makes it difficult to definitively designate aggregation as the pathogenic event that culminates in disease.
Figure 4. Lifecycle of TDP43 pathology.
Normal neurons show robust intranuclear TAR DNA-binding protein 43 (TDP43) immunoreactivity (shown in red) with little cytoplasmic TDP43. So-called ‘pre-inclusions’ have been described, and these consist of granular cytoplasmic aggregates that are positive for phospho-TDP43 epitopes (p409 and p410) but that are often negative for ubiquitin. Neurons with pre-inclusions show characteristic loss of normal nuclear staining. Bona fide inclusions exhibit a variety of morphologies ranging from dense round inclusions and skeins in motor neurons to dystrophic neurites, cytoplasmic inclusions or intranuclear ‘cat eye’ inclusions in other neurons. Neuronophagia can rarely be seen in amyotrophic lateral sclerosis, and neuronophagic cell clusters have been reported to be associated with TDP43 inclusions96.
The lifecycle of TDP43 inclusions has been surmised based on both pathologic and experimental observations. The appearance of granular cytoplasmic TDP43 aggregates that are designated as pre-inclusions has been postulated to be an early event in the formation of TDP43 inclusions2–4,10. Notably, neurons that harbour pre-inclusions also exhibit the characteristic absence of normal nuclear TDP43 immunoreactivity. Mature aggregates in ALS are characteristically dense, round inclusions or filamentous skeins in the cytoplasm of affected motor neurons, whereas in FTLD-TDP they are typified by a variety of cytoplasmic, neuritic and intranuclear inclusions in a diverse group of forebrain neurons1,83–85. The importance of the diversity of morphologies is unknown.
Purified TDP43 protein is prone to aggregation in vitro86, consistent with the presence of TDP43 inclusions in many cellular and animal models of TDP43 proteinopathy60–66,68,69. In cell culture systems, only rare TDP43 aggregates are seen upon overexpression of full-length TDP43. However, overexpression of either cytoplasmically restricted TDP43 (restricted by mutating its NLS motifs) or truncated TDP43 CTFs yields numerous cytoplasmic inclusions65,68,69. Transducing recombinant pre-formed TDP43 aggregates into the cytoplasm of TDP43 overexpressing cells can also induce TDP43 aggregates56, presumably through a ‘seeding’ reaction, similar to that recently observed for tau and α-synuclein87–90. Consistent with these data is the recent identification of a C-terminal prion-like domain in TDP43 using a computational algorithm to identify prion-like domains91. This prion-like domain has been implicated in aggregation of TDP43 in cultured cells, and it is thought to confer biochemical properties associated with prions including aggregation, protease resistance and cell-to-cell propagation (but not necessarily infectivity)86,92–94.
The biochemical correlate of TDP43 inclusions is the accumulation of various sarkosyl-insoluble TDP43 species. A sizeable proportion of the 43-kDa TDP43 protein is normally sarkosyl-insoluble, and the amount of this normal TDP43 protein is not appreciably different in normal and diseased brain homogenates. However, other insoluble TDP43 species that are found in diseased brains include a phosphorylated 45-kDa species, a high molecular weight ubiquitylated species and CTFs of approximately 20–25 kDa1 (FIG. 1c).
Examination of patients with sporadic ALS who had the disease for an unusually long duration (10–20 years) showed that the number of TDP43 inclusions was low relative to standard cases of sporadic ALS95. The fact that the number of inclusion-bearing neurons is low in cases of ALS with long disease duration suggests that neurons that harbour TDP43 inclusions earlier in the disease process are prone to degeneration. Indeed, TDP43 aggregates are seen in the centre of all neuronophagic cell clusters in the anterior horn of the spinal cord in ALS96. The observation that there is selective neuronal degeneration of TDP43 inclusion-bearing cells suggests that TDP43 aggregates are tightly linked with neurodegeneration in a cell autonomous manner. However, these findings do not tell us whether TDP43 aggregates themselves are toxic, or whether they are a response to injury and therefore just a marker of another neurotoxic process.
Transgenic animals that express full-length TDP43 also exhibit TDP43-positive inclusions, although in general they are rare60–64,66. Quantification of TDP43-positive inclusions in transgenic mice revealed no correlation between the number of inclusions and neurodegeneration66. Thus, TDP43 aggregation itself does not seem to be an absolute requirement for TDP43-mediated neurodegeneration in vivo, although it is possible that TDP43 inclusions are sufficient to induce neuron death. Nevertheless, TDP43 inclusions have been postulated to result in a toxic gain of function by virtue of abnormal ‘activity’ conferred by altered conformation1. Finally, several — but not all — TDP43 transgenic mouse models exhibit cytoplasmic inclusions comprised of mitochondrial aggregates that are generally negative for TDP43 (REFS 60–64). Similar mitochondrial inclusions are not observed in human cases and their relevance to human disease is unknown. However, TDP43 toxicity in yeast is partly dependent on mitochondria and oxidative stress97, suggesting that independent of aggregation, one possible mechanism of neurodegeneration may be abnormal alterations in metabolic pathways.
The effect of TDP43 aggregation on neuronal viability remains unclear. Experimental models have demonstrated that TDP43 aggregates are not necessary for TDP43-mediated neurodegeneration, although this does not preclude the possibility that aggregates may still contribute to neuron dysfunction and death. In particular, the emerging concept of prion-like cell-to-cell transmission remains to be tested with regard to TDP43 proteinopathies, and more clarity on this concept for TDP43 proteinopathies could advance understanding of how TDP43 aggregation and/or fibrilization contribute to the anatomic spread and clinical progression of these diseases.
TDP43 truncation
As described above, the biochemical signature of FTLD-TDP includes the presence of truncated TDP43 protein, evident as 20–25-kDa species in sarkosyl-insoluble brain lysates1,98. Amino-terminal sequencing of brain-derived TDP43 fragments demonstrated that at least one of these fragments consists of a CTF beginning at Arg208 of TDP43 (REF. 65). However, there is considerable variability in the number of CTF bands from case to case. Interestingly, inclusions within the brain of ALS and FTLD-TDP patients are readily labelled with antibodies that recognize the C terminus of TDP43, but not with N-terminal TDP43 antibodies99. By contrast, spinal cord inclusions are labelled with both N- and C-terminal TDP43 antibodies, suggesting that they are comprised of full-length TDP43 (REF. 99). This regional heterogeneity in terms of CTF formation suggests that it may not be necessary for TDP43-mediated neurodegeneration. Another ~35-kDa fragment was found in lymphoblastoid cell lines derived from TDP43 mutation carriers13,100, but this species is low or absent in brain lysates1,57,58. Furthermore, in our experience, it is variably present in ALS and FTLD-TDP human brain homogenates and its presence does not correlate with disease status. These various fragments are presumably generated by proteolytic cleavage of full-length TDP43, although they could reflect splice variants or cryptic transcription start sites44. The apoptosis regulator caspase 3 has been proposed as the major protease responsible for generating the 25-kDa fragment in cultured cells44,81,82,101 (FIG. 5). As TDP43 contains three caspase 3 cleavage consensus sequences, and caspase 3 is able to cleave TDP43 into 25- and 35-kDa fragments both in vitro and in cultured cells, this suggestion is plausible101.
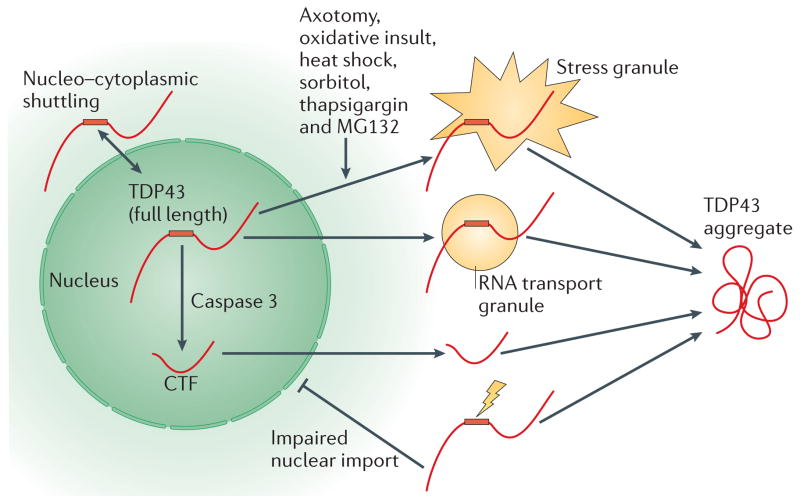
Figure 5. Subcellular localization of TDP43 protein.
TAR DNA-binding protein 43 (TDP43) shows a predominantly nuclear localization, although nucleo–cytoplasmic shuttling of TDP43 has been shown and low levels of cytoplasmic TDP43 can be demonstrated. TDP43 accumulation in the cytoplasm can be induced by a variety of cellular stressors that result in the formation of TDP43-positive stress granules. TDP43 protein has also been found in RNA transport granules within neuronal processes. TDP43 protein is thought to undergo proteolytic cleavage by caspase 3 to generate a carboxy-terminal fragment (CTF). As the CTF no longer contains the bipartite nuclear localization signal (NLS, shown by a red rectangle), CTFs translocate into the cytoplasm, where they may participate in aggregate formation. Experimentally generated mutations of the NLS and disease-associated mutations (shown by a yellow lightning bolt) have been found to increase the amount of cytoplasmic versus nuclear TDP43. MG132 is a proteasome inhibitor.
TDP43 contains two NLS motifs that lie between the N terminus and RRM1 (REF. 69). As TDP43 CTFs lack these motifs, they are predominantly cytoplasmic (FIG. 5). Furthermore, the C-terminal domain of TDP43 was found to be critical for aggregation in vitro86. Thus, TDP43 cleavage by caspase 3 or another protease may be one factor that accounts for the cytoplasmic localization and aggregation TDP43. Indeed, overexpression of truncated TDP43 results in cytoplasmic aggregates that are phosphorylated and ubiquitylated44,65,68,81,82,101,102. However, although expressing TDP43 CTFs consistently leads to cytoplasmic aggregates, de novo intranuclear cleavage of full-length TDP43 yields distinct results71; introducing a tobacco etch virus protease cleavage site within TDP43 induced cleavage of TDP43 within the nucleus, generating TDP43 CTFs that were efficiently translocated into the cytoplasm and rapidly degraded. In contrast to direct expression of TDP43 CTFs, these de novo CTFs aggregated only in the presence of a second event or ‘hit’ such as the introduction of natively misfolded CTFs71.
The presence of TDP43 CTFs has been evaluated in several rodent models overexpressing TDP43, many of which exhibit some level of lower molecular weight TDP43 fragments61–64,103. The first TDP43 transgenic mouse model was found to exhibit CTFs before onset of gait abnormalities or neurodegeneration, and levels of these CTFs increased with disease progression62. This initial report was confirmed by other groups that also reported a correlation between disease progression and the accumulation of TDP43 CTF61,63. However, several biochemical differences have been found between TDP43 CTFs in transgenic mice and human brains. 35-kDa fragments, when present, tend to be found at much higher levels in the brains of transgenic mice than in human brains61–63. TDP43 CTFs are highly soluble in mice extracts62–64,103, and partition into nuclear, as opposed to cytoplasmic, fractions63. Furthermore, not all TDP43 transgenic mice exhibit TDP43 CTFs despite having very similar neurodegenerative phenotypes, indicating that CTFs may not be absolutely necessary for TDP43-mediated neurodegeneration60,66. A transgenic mouse expressing TDP43 CTFs in the absence of full-length protein has yet to be described.
Few studies have addressed whether TDP43 CTFs confer a gain or loss of function. One group found that TDP43 CTFs impair neurite growth during differentiation of cultured rodent neurons, and that full-length TDP43 rescues this phenotype, suggesting that TDP43 CTFs act by a dominant-negative mechanism104. However, expression of full-length TDP43 in D. melanogaster was toxic to a variety of neuronal cell types, whereas expression of TDP43 CTFs did not result in neurotoxicity105,106. Importantly, TDP43-mediated toxicity in D. melanogaster and chick motor neurons required RNA binding activity106. As TDP43 CTFs are deficient in RNA binding, this suggests that TDP43-mediated neurodegeneration is not due to abnormal TDP43 cleavage but instead is due to a toxic gain of function of full-length TDP43. Clearly, additional mechanistic studies are required to determine the effect of TDP43 CTFs on normal TDP43 function.
TDP43 mislocalization
Although TDP43 is predominantly nuclear, interspecies heterokaryon assays have shown that it shuttles between nuclear and cytoplasmic compartments, which is consistent with studies that have been able to detect low levels of cytoplasmic TDP43 in normal cells21–23,69,107 (FIG. 2). A striking feature of TDP43 proteinopathies is the reversal of this partitioning such that most TDP43 proteins are mislocalized to the cytoplasm or neurites in the form of aggregates1. Although most TDP43 inclusions are extranuclear, some are also seen within the nucleus where they exhibit a characteristic ‘cat eye’ or lenticular morphology1,83–85 (FIG. 1b). TDP43 is normally localized to euchromatic regions of the nucleoplasm, including perichromatin fibrils and nuclear speckles where transcription and cotranscriptional splicing occurs49,108. Thus, although some TDP43 inclusions are nuclear, these can still be considered mislocalized. Four subtypes of FTLD-TDP have been described, and although intranuclear TDP43 inclusions can be seen in multiple subtypes, they are particularly prevalent in FTLD-TDP type D, the subtype that is associated with mutations in the gene encoding the valosin-containing protein (VCP)83–85. Little is known about why VCP mutations result in increased numbers of intranuclear inclusions.
TDP43 contains two NLS motifs between the N terminus and RRM1, and one nuclear export signal (NES) within RRM2, and these are the major cis elements regulating nuclear localization69,107 (FIG. 1a). The A90V ALS-associated mutation of TDP43 results in increased cytoplasmic partitioning, presumably because this mutation is located between the two NLS motifs109. Rare cases of FTLD-TDP have been linked to mutations in the 3′ UTR of sigma non-opioid intracellular receptor 1 (SIGMAR1) that increase SIGMAR1 expression and are thought to increase the ratio of cytoplasmic to nuclear TDP43 through unknown mechanisms110. A D. melanogaster model of mutant VCP-mediated neurodegeneration has shown that VCP and TDP43 interact genetically and that disease-associated VCP mutations result in a redistribution of TDP43 from the nucleus to the cytoplasm111. In addition to these genetic determinants, a variety of cellular stressors increase cytoplasmic TDP43 levels. For example, axotomy results in a time-dependent increase in cytosolic TDP43 expression41–43. Additionally, in response to a variety of other stressors, TDP43 or its 35-kDa CTFs redistribute to the cytoplasm and colocalize with stress granules30,39,44–47 (FIG. 5). Thus, both genetic and environmental factors seem to influence TDP43 localization. The fact that cytoplasmic TDP43 pathology is found to be secondary to a variety of genetic mutations, and the prevalence of cytoplasmic TDP43 pathology in a variety of neurodegenerative diseases other than ALS and FTLD-TDP suggest that there are a variety of stressors or insults that may lead to cytoplasmic localization and aggregation.
Several cell or animal model studies have attempted to address whether cytoplasmic TDP43 causes neurodegeneration. We, and others, have found that mutation of one or both NLS motifs renders TDP43 predominantly cytoplasmic, resulting in cytoplasmic aggregates both in cultured cells and transgenic mice66,67,69. These cytoplasmic aggregates were also shown to recruit non-mutated nuclear TDP43 into the cytoplasm of cultured cells69. Others have used automated time-lapse microscopy to show TDP43-mediated toxicity in primary rat neurons overexpressing fluorescently tagged TDP43. They found that the disease-associated A315T mutation resulted in increased neurotoxicity compared to the neurotoxicity in wild-type TDP43, and that neurotoxicity correlated with the amount of cytoplasmic TDP43 expression irrespective of inclusion bodies or even the presence of the A315T mutation112. Consistent with these findings, expression of human TDP43 in the developing D. melanogaster eye resulted in mild eye abnormalities (disruption of ommatidia and few necrotic patches) that were worsened by mutating the NLS motifs113. However, the same authors found that expressing human TDP43 in an adult fly using an inducible pan-neuronal system reduced lifespan to a greater extent than expressing mislocalized human TDP43 (that is, TDP43 with NLS or NES mutations)113. Finally, transgenic mice expressing TDP43 with an NLS mutation resulted in cytoplasmic TDP43 and rare cytoplasmic inclusions66. However, similar patterns of neurodegeneration were observed in mice expressing wild-type human TDP43, albeit to a lesser extent (due to lower TDP43 expression levels).
Thus, overexpression of both cytoplasmic and nuclear TDP43 seems to be toxic in animal models. Although cytoplasmic localization of TDP43 is not an absolute requisite for TDP43-mediated neurotoxicity, mislocalized TDP43 could still potentially contribute to neurodegeneration. In theory, TDP43 may retain its biological activity such as RNA binding but may do so in an abnormal location, resulting in a toxic gain of function. Although several cell culture models indicate that cytoplasmic TDP43 is toxic, animal models have yielded mixed results, and further work is required to determine the mechanistic importance of cytoplasmic versus nuclear TDP43.
Nuclear clearance and autoregulation
The most intriguing pathologic change that is associated with ALS and FTLD-TDP is the loss of normal nuclear TDP43 immunoreactivity in inclusion-bearing cells1. This nuclear clearance phenomenon seems to be a very early event in ALS and FTLD-TDP, occurring in cells that harbour pre-inclusions or ubiqutin-negative inclusions2–10. A distinction between mislocalization and nuclear clearance should be made, as the presence of an abnormally localized inclusion does not necessarily translate into the reduction of a normal protein in its normal location. For example, clearance or absence of normal huntingtin protein is not seen in the neurons bearing huntingtin inclusions that are associated with Huntington’s disease114. Presumably, the localization of TDP43 within specific nucleoplasmic domains is required for its normal function, and therefore nuclear clearance is expected to disrupt normal function. Remarkable progress has been made towards understanding the normal functions of TDP43, partly driven by the hypothesis that a loss of normal function may play a role in ALS and FTLD-TDP.
As described above, TDP43 exhibits a diverse array of normal functions. Thus, understanding which pathways may be important in terms of mediating neurotoxicity is difficult. Indeed, genome-wide analyses thus far have indicated that a large number of genes and transcripts are regulated by TDP43 expression making identification of disease-relevant pathways difficult25,27,60,66,115. However, one finding is of particular relevance to the phenomenon of autoregulation. As is the case for many RNA regulatory proteins, TDP43 downregulates its own mRNA transcript levels25,27,116 (FIG. 6a). Thus, overexpression of exogenous TDP43 leads to a decrease in endogenous TDP43 mRNA and protein in cultured cells and transgenic mice66,69,116. Furthermore, TDP43 protein was found to physically bind to its own mRNA transcript within the 3′ UTR, providing a mechanism of autoregulation25,27,116. Studies disagree with regard to what happens after TDP43 binds to its cognate mRNA (nonsense-mediated decay versus other mechanisms leading to exosome-mediated decay)25,116. Whether this autoregulatory mechanism contributes to nuclear clearance in affected human brain has not been definitively demonstrated. However, in transgenic mice overexpressing human TDP43 protein, endogenous TDP43 expression is reduced66. Perhaps over-abundant TDP43 protein in inclusion-bearing neurons can similarly reduce endogenous TDP43 expression leading to nuclear clearance. Furthermore, neurodegeneration in transgenic mice correlates better with the loss of normal endogenous TDP43 expression than with post-translational modification of TDP43, aggregation, truncation or subcellular localization, indicating that the dysregulation of normal activity is linked to neuron loss66.
Figure 6. Autoregulation of TDP43 and models of TDP43 toxicity.
a | Normal TAR DNA-binding protein 43 (TDP43) autoregulation. TDP43 expression is tightly regulated under normal conditions, and overexpression of exogenous TDP43 results in a reduction of endogenous TDP43 expression. This autoregulation of TDP43 expression by itself is mediated at the level of mRNA stability. Within the 3′ untranslated region (UTR) of TDP43 mRNA is a binding site for TDP43 protein that is critical for autoregulation. TDP43 binding to this 3′ UTR site promotes degradation of TDP43 mRNA, at least partly through the exosome. There are conflicting reports about the mechanism of autoregulation, in particular whether alternative splicing leading to nonsense-mediated decay (NMD) is the mechanism of TDP43 autoregulation. b | Loss of autoregulation. According to this model, the exposure of neurons to as-yet-unidentified stressors can lead to cytoplasmic mislocalization of TDP43 (1), and this is perhaps related to the stress granule response. Given TDP43’s propensity to aggregate, TDP43 forms phosphorylated pre-inclusions within the cytoplasm that sequester free TDP43 protein. This cytoplasmic sequestration leads to a loss of normal nuclear TDP43. If autoregulation occurs within the nucleus, a loss of TDP43 autoregulation ensues (2), which results in increased TDP43 mRNA and protein (3). This further exacerbates TDP43 aggregation (4). This vicious cycle leads to cell death (5) possibly through a variety of gain and loss of functions. c | Gain of autoregulation. In this model, neurons that are exposed to unknown stressors can undergo cytoplasmic mislocalization of TDP43 protein (1), and this is perhaps related to the stress granule response. Given the propensity of TDP43 to aggregate, TDP43 forms phosphorylated pre-inclusions within the cytoplasm which are resistant to degradation. If autoregulation occurs within the cytoplasm, an increase in cytoplasmic TDP43 may result in an increase in TDP43 autoregulation (2) that would decrease TDP43 mRNA and therefore decrease synthesis of new TDP43 protein (3). This reduction in TDP43 protein synthesis leads to a loss of normal nuclear TDP43 protein. Given the plethora of normal nuclear TDP43 functions, the absence of nuclear TDP43 is detrimental to neuronal viability, increasing the stress response (4) and leading to cell death (5). Again, this model allows for the possibly of a variety of gain and loss of functions that coordinately result in toxicity.
To more definitively demonstrate that a loss of TDP43 function results in neurodegeneration, multiple cellular and animal models have been generated in which TDP43 expression is reduced or eliminated. Knockdown of TDP43 in Neuro 2a cells inhibits neurite growth and diminishes cell viability, possibly through inactivation of RHO family GTPases117, whereas knockdown in HeLa or U2OS cells induces apoptosis through the CDK6–etino-blastoma pathway36. Knockout of TDP43 in embryonic stem cells inhibits proliferation115. Disrupting TDP43 in D. melanogaster has yielded conflicting results with one group reporting early lethality37, and others describing a semi-lethal phenotype with reduced survival into adulthood, reduced lifespan, locomotor defects, abnormal neuromuscular junctions and reduced dendritic branching118. Interestingly, in Danio rerio, both overexpression of human TDP43 and knockdown of zebrafish TARDBP resulted in the same swimming behaviour defects that are associated with short motor neuron axons that have premature and excessive branching119. Finally, several groups have generated TDP43 knockout mice, and in each instance all TDP43 knockout mice die during embryonic stages owing to defective outgrowth of the inner cell mass120–122. Even postnatal knockout of TDP43 using a tamoxifen-inducible knockout mouse model resulted in rapid lethality115. Although these mouse studies clearly demonstrate that TDP43 is required for development and even postnatal viability, they are unable to address whether a loss of TDP43 function is toxic to postmitotic neurons. This question awaits analysis of postnatal brain or spinal cord-specific knockout mice. However, taken together, the studies that are summarized above make it clear that TDP43 regulates a wide range of cellular processes and that the loss of normal TDP43 function is likely to be harmful. Thus, nuclear clearance of normal TDP43 in ALS and FTLD-TDP is likely to play an important role in TDP43 mediated neurodegeneration.
TDP43, and ALS and FTLD genetics
There has been a renaissance in the human genetics of ALS and FTLD. The list of genes that are associated with either ALS or FTLD include TARDBP12–15, FUS123, superoxide dismutase 1 (SOD1)124, UBLN2 (REF. 59), optineurin (OPTN)125, granulin (GRN)126,127, valosin containing protein (VCP)128, ataxin 2 (ATXN2)129, sigma non-opioid intracellular receptor 1 (SIGMAR1)110,130 and C9ORF72 (REFS 131,132). Despite the variety of genes that are implicated in ALS and FTLD, there is remarkable homogeneity in terms of downstream pathology: nearly every patient with ALS or FTLD that is linked to a genetic mutation exhibits TDP43 pathology. Thus, in the vast majority of cases, dysregulation of TDP43 is the common downstream mechanism of neurodegeneration. Exceptions to this rule are the absence of TDP43 pathology in cases that are linked to SOD1 or FUS mutations123,133. Although TDP43 inclusions are not observed in FUS mutant cases, FUS is an RNA-binding protein that is found in complex together with TDP43, and ALS-associated TARDBP mutations promote the interaction between TDP43 and FUS29,30. Thus, there is strong speculation that dysregulation of TDP43 and FUS may be mechanistically linked at the level of RNA processing. Indeed, FUS also exhibits a prion-like domain91,134 and forms inclusions in affected neurons123. Furthermore, in D. melanogaster models of neurodegeneration, two independent groups have found that TDP43 and FUS act together within a common genetic pathway135,136.
Moreover, it was recently shown that an intronic hexa-nucleotide repeat expansion of C9ORF72 can cause ALS-FTLD131,132. Nearly nothing is known about the protein product of C9ORF72. However, the mutation leads to differential splicing of the C9ORF72 transcript and the formation of nuclear RNA foci131. The presence of nuclear RNA foci is reminiscent of myotonic dystrophies that are linked to nucleotide repeat expansions in which abnormal RNA foci are thought to sequester RNA-binding proteins and alter the biogenesis of other cellular RNAs137. Similar RNA foci are observed in other nucleotide repeat disorders, and so the association between ALS and intermediate-length polyglutamine repeats in ATXN2 is intriguing (although RNA foci have not been reported in association with ATXN2 expansions)129. Notably, ATXN2 polyglutamine repeats that are associated with ALS are all interrupted by between one and three CAA codons within the CAG expansion138. Together, these findings suggest that toxic RNA species associated with an abnormal RNA primary structure may be one mechanism related to downstream dysregulation of TDP43 and RNA biogenesis.
In summary, several of the genetic causes and modulators of ALS and FTLD support the emerging theme of abnormal RNA processing as central to the molecular mechanisms of neurodegeneration in ALS and FTLD. The plethora of genes and proteins that are associated with TDP43 pathology provides support and plausibility for the notion that common neurodegeneration pathways that are linked to TDP43 and RNA mis-metabolism can be identified that underlie sporadic as well as familial ALS and FTLD including those autosomal and X-linked dominant forms of these disorders that are caused by distinctly different genetic mutations.
Models of TDP43-mediated neurodegeneration
Although enormous advances have been made in demonstrating that pathological TDP43 is linked to neurodegeneration in ALS and FTLD-TDP, there is still much progress to be made to understand how the normal functions of TDP43 relate to the mechanisms of TDP43-mediated neurodegeneration. We now have an idea of the key steps that occur from the onset of TDP43-mediated neurodegenerative disease through to the culmination of these events in dysfunction and death of affected neurons in ALS, FTLD-TDP and related TDP43 proteinopathies (FIGS 3,4). Advances in TDP43 research have been extremely rapid considering that 7 years elapsed following the discovery of amyloid-β as the building block of vascular amyloid in Alzheimer’s disease139 before mutations linked to Alzheimer’s disease were found in the amyloid-β precursor protein (APP) gene in 1991 (REFS 140–142). It took four more years to generate animal models of Alzheimer’s disease-like amyloid-β plaque pathology143,144, and more than 25 years later the mechanisms of amyloid-β-mediated neurodgeneration remain incompletely understood145.
As is often the case for disease processes, multiple pathways are likely to be involved, and the downstream consequences of TDP43 phosphorylation, aggregation, cleavage, mislocalization and clearance from the nucleus are unclear. Perhaps it is easiest to conjecture that the absence of normal nuclear TDP43 implies that there is a loss of normal TDP43 function. Given the plethora of normal nuclear TDP43 functions, this alone might lead to neurodegeneration. Alternatively, perhaps TDP43 retains its ability to bind RNA, and therefore its cytoplasmic localization acts as a peripheral sink, thereby implicating a toxic gain of function. Or maybe TDP43 CTFs act in a dominant negative fashion by occupying hnRNP binding sites without being able to bring along its usual RNA cargo.
In an attempt to synthesize the literature on the various potential mechanisms of neurodegeneration, we will consider the normal autoregulation of TDP43 (FIG. 6a) and two hypothetical models of TDP43-mediated neurodegeneration (FIG. 6b,c). The first model, which we term the ‘loss of autoregulation’ model, was proposed by two groups independently and is updated here25,116 (FIG. 6b). Although the initial insult is generally unknown (aside from cases that are linked to genetic mutations), one possible initiating event may be the presence of abnormal, toxic RNA species. Cellular stress is thought to cause a redistribution of TDP43 from the nucleus to the cytoplasm. This is akin to the formation of stress granules that has been observed experimentally, although there is currently little evidence showing an upregulation of stress granules in ALS and FTLD. Alternatively, RNA transport granules that are usually localized within peripheral neurites may be abnormally distributed. According to this model, this redistribution results in the aggregation of TDP43 into pre-inclusions that are phosphorylated, variably ubiquitylated and difficult to degrade. These cytoplasmic inclusions lead to further redistribution of normal TDP43 from the nucleus as protein is sequestered into inclusions, accounting for nuclear clearance. If TDP43 autoregulation occurs within the nucleus as suggested25, there would be a loss of TDP43 autoregulation and increased TDP43 expression, leading to increased aggregate formation. This would feed forward through a vicious cycle leading to cell death.
Alternatively, a ‘gain of autoregulation’ model can be imagined (FIG. 6c). According to this model, a stressor can lead to redistribution of TDP43 into the cytoplasm resulting in aggregation (similar to the loss of autoregulation model). If autoregulation of TDP43 mRNA is a cytoplasmic event and TDP43 aggregates retain the ability to bind RNA and therefore autoregulate its cognate RNA, then an increase in cytoplasmic TDP43 may lead to increased autoregulation, thereby decreasing TDP43 mRNA and decreasing newly synthesized TDP43 protein in affected cells. This decrease in synthesis may account for the nuclear clearance phenomenon (that is, aggregates represent the existing pool of TDP43 protein and not newly synthesized protein). Given the myriad of normal TDP43 functions, loss of normal nuclear TDP43 may further increase cellular stress, again resulting in a vicious cycle leading to cell death.
Conclusions
The exact mechanisms by which cell death occurs are not known. However, using the pathological and biochemical signature of abnormal TDP43 in human disease as a guide, many advances in our understanding of TDP43 biology have been made. We now speculate that many pathways may be involved in TDP43-mediated neurodegeneration, including gains of functions (mediated by aggregates or abnormal cytoplasmic function) and losses of functions (mediated by nuclear clearance, CTFs, RNA dysregulation or even UPS dysfunction). Indeed, the two proposed models highlight the possibility that gains and losses of function together may affect neuronal viability and are not mutually exclusive, highlighting the complexities of understanding neurodegenerative diseases, and in particular TDP43 proteinopathies. The fact that TDP43 pathology is found in nearly all cases of sporadic and familial ALS or FTLD (excluding ALS caused by SOD1 or FUS mutations and FTLD due to primary tauopathies) speaks to the centrality of TDP43 in the pathogenesis of these diseases. Furthermore, the mechanisms of TDP43-mediated neurodegeneration is likely to involve abnormal RNA processing, either in the form of abnormal splicing, differential RNA stability or the presence of toxic RNA species. The current challenges in understanding TDP43 proteinopathies are largely due to the extraordinary complexity of TDP43 functions, including the large number of mRNAs with which TDP43 interacts in potentially important pathways and networks linked to normal physiological or metabolic processes. Equally daunting is identifying the disease-relevant RNAs that are mechanistically linked to neurodegeneration, with which TDP43 interacts or regulates, so this remains an imposing challenge due to the vast number of RNAs targeted by TDP43 protein. Also, understanding how the diverse genetic (and presumably environmental) factors lead to downstream TDP43 pathology is a major obstacle to mechanistic understanding across the field of neurodegenerative diseases in general, and this is true for TDP43 proteinopathies as well. However, these challenges also represent compelling and largely tractable opportunities to identify shared molecules and pathways. There are also opportunities in understanding TDP43 proteinopathies within the context of neurodegenerative diseases in general, such as the role of prion-like cell-to-cell transmission in the progression and spread of protein misfolding diseases. Thus, despite the considerable progress in elucidating the mechanisms of TDP43-mediated neurodegeneration, much remains to be discovered and the opportunities to dissect disease-relevant gains and losses remains a dominant focus of TDP43 biology. However, given the amazing progress in understanding ALS and FTLD since the discovery of TDP43 as the disease protein in these disorders in 2006, there is a sense of optimism among scientists in this field. There is now belief that we have the ideas, the tools and the skills to advance our understanding of ALS, FTLD and related TDP43 proteinopathies to the point at which we can translate this understanding into improvements in the diagnosis and treatment of these disease, if these efforts were appropriately resourced.
Acknowledgments
We thank our many colleagues working on TDP43 in the Center for Neurodegenerative Disease Research (CNDR) and the Department of Neurology at the University of Pennsylvania for extensive collaborations that provided essential input at many stages of our research on TDP43 since 2006, including L. Kwong who provided immunoblot images. The studies from CNDR that are summarized here were supported by the National Institutes of Health (grants AG10124, AG17586, K08AG039510 and training grant T32 AG00255). V.M.-Y.L. is the John H. Ware III Chair of Alzheimer’s Research and J.Q.T. is the William Maul Measey-Truman G. Schnabel, Jr, MD Professor of Geriatric Medicine and Gerontology at the University of Pennsylvania. The authors would like to thank the families of their patients who made this research possible.
Glossary
- Dominant missense mutation
An alteration of a single nucleotide within a gene, resulting in a codon that encodes for an amino acid that is different from normal. Thus, a change in a single allele is sufficient to result in the mutation-associated phenotype
- Epiphenomena
Secondary processes or events that occur in parallel to a primary event and that may even be a result of the primary event. However, epiphenomena are in addition to the course of a disease and are not necessarily causally related to the primary mechanisms of disease
- High-throughput sequencing of RNA isolated by crosslinking immunoprecipitation
(HITS-CLIP). A method to identify the RNA-binding sites of a given protein in which protein–RNA interactions are stabilized by UV crosslinking. The protein of interest is immunoprecipitated, and the interacting RNA species are identified using high-throughput next-generation sequencing platforms
- RNA granules
Macromolecular structures in neurons enriched with RNA and RNA-binding proteins. They are thought to be involved in the preservation and transport of mRNA
- Stress granules
Dense cytosolic protein and RNA aggregations that appear under conditions of cellular stress. The RNA molecules are thought to be stalled translation pre-initiation complexes
- Multivesicular bodies
Endosomal intermediates in which small membrane vesicles are enclosed within a limiting membrane. The internal vesicles are thought to form by invagination and budding from the limiting membrane
- Adaptor protein
A protein that contributes to cellular function by recruiting other proteins to a complex. Such molecules often contain several protein–protein interaction domains
- Neuronophagia
The process in which dying neurons are cleared by phagocytic cells including microglia
- Interspecies heterokaryon assays
A test that is performed on cells that contain multiple genetically different nuclei from different species. The test is capable of demonstrating low levels of dynamic nucelo–cytoplasmic shuttling by measuring the transport of a nuclear protein from a donor nucleus to a receptor nucleus
Footnotes
Competing interests statement
The authors declare no competing financial interests.
FURTHER INFORMATION
Edward B. Lee’s homepage: http://www.med.upenn.edu/tnr/index.shtml
Virginia M.-Y. Lee and John Q. Trojanowski’s homepage: http://www.med.upenn.edu/cndr
ALL LINKS ARE ACTIVE IN THE ONLINE PDF
References
- 1.Neumann M, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. TDP43 protein is identified biochemically, immunohistochemically and by amino acid sequence analysis as the major component of proteinaceous ubiquitin-positive inclusions in FTLD and ALS. Pathologic TDP43 is found to be ubiquitylated, phosphorylated and cleaved, and is associated with nuclear clearance of normal TDP43. [DOI] [PubMed] [Google Scholar]
- 2.Giordana MT, et al. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol. 2010;20:351–360. doi: 10.1111/j.1750-3639.2009.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brandmeir NJ, et al. Severe subcortical TDP-43 pathology in sporadic frontotemporal lobar degeneration with motor neuron disease. Acta Neuropathol. 2008;115:123–131. doi: 10.1007/s00401-007-0315-5. [DOI] [PubMed] [Google Scholar]
- 4.Strong MJ, et al. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol Cell Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 5.Pamphlett R, Luquin N, McLean C, Jew SK, Adams L. TDP-43 neuropathology is similar in sporadic amyotrophic lateral sclerosis with or without TDP-43 mutations. Neuropathol Appl Neurobiol. 2009;35:222–225. doi: 10.1111/j.1365-2990.2008.00982.x. [DOI] [PubMed] [Google Scholar]
- 6.Dickson DW, Josephs KA, Amador-Ortiz C. TDP-43 in differential diagnosis of motor neuron disorders. Acta Neuropathol. 2007;114:71–79. doi: 10.1007/s00401-007-0234-5. [DOI] [PubMed] [Google Scholar]
- 7.Davidson Y, et al. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 2007;113:521–533. doi: 10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- 8.Cairns NJ, et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am J Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujita Y, Mizuno Y, Takatama M, Okamoto K. Anterior horn cells with abnormal TDP-43 immunoreactivities show fragmentation of the Golgi apparatus in ALS. J Neurol Sci. 2008;269:30–34. doi: 10.1016/j.jns.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 10.Mori F, et al. Maturation process of TDP-43-positive neuronal cytoplasmic inclusions in amyotrophic lateral sclerosis with and without dementia. Acta Neuropathol. 2008;116:193–203. doi: 10.1007/s00401-008-0396-9. [DOI] [PubMed] [Google Scholar]
- 11.Pesiridis GS, Lee VM, Trojanowski JQ. Mutations in TDP-43 link glycine-rich domain functions to amyotrophic lateral sclerosis. Hum Mol Genet. 2009;18:R156–R162. doi: 10.1093/hmg/ddp303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gitcho MA, et al. TDP-43 A315T mutation in familial motor neuron disease. Ann Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kabashi E, et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nature Genet. 2008;40:572–574. doi: 10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 14.Sreedharan J, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. The first of several reports identifying TARDBP missense mutations, confirming the role of TDP43 in both sporadic and familial ALS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Van Deerlin VM, et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mackenzie IR, Rademakers R, Neumann M. TDP-43 and FUS in amyotrophic lateral sclerosis and frontotemporal dementia. Lancet Neurol. 2010;9:995–1007. doi: 10.1016/S1474-4422(10)70195-2. [DOI] [PubMed] [Google Scholar]
- 17.Geser F, Lee VM, Trojanowski JQ. Amyotrophic lateral sclerosis and frontotemporal lobar degeneration: a spectrum of TDP-43 proteinopathies. Neuropathology. 2010;30:103–112. doi: 10.1111/j.1440-1789.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Da Cruz S, Cleveland DW. Understanding the role of TDP-43 and FUS/TLS in ALS and beyond. Curr Opin Neurobiol. 2011 Aug 1; doi: 10.1016/j.conb.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lagier-Tourenne C, Polymenidou M, Cleveland DW. TDP-43 and FUS/TLS: emerging roles in RNA processing and neurodegeneration. Hum Mol Genet. 2010;19:R46–R64. doi: 10.1093/hmg/ddq137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ward ME, Miller BL. Potential mechanisms of progranulin-deficient FTLD. J Mol Neurosci. 2011 Sept 3; doi: 10.1007/s12031-011-9622-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ayala YM, et al. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J Mol Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 22.Ou SH, Wu F, Harrich D, Garcia-Martinez LF, Gaynor RB. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang HY, Wang IF, Bose J, Shen CK. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 24.Buratti E, Baralle FE. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J Biol Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. Functional anlaysis of TDP43 protein as an RNA-binding protein that regulates alternative splicing of pre-mRNA. [DOI] [PubMed] [Google Scholar]
- 25.Polymenidou M, et al. Long pre-mRNA depletion and RNA missplicing contribute to neuronal vulnerability from loss of TDP-43. Nature Neurosci. 2011;14:459–468. doi: 10.1038/nn.2779. The identification of RNA molecules that physically interact with TDP43 protein using HITS-CLIP analysis. Binding of a large proportion of the transcriptome is observed, including binding of the TDP43 RNA itself, and this is mechanistically linked to autoregulation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buratti E, Brindisi A, Pagani F, Baralle FE. Nuclear factor TDP-43 binds to the polymorphic TG repeats in CFTR intron 8 and causes skipping of exon 9: a functional link with disease penetrance. Am J Hum Genet. 2004;74:1322–1325. doi: 10.1086/420978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tollervey JR, et al. Characterizing the RNA targets and position-dependent splicing regulation by TDP-43. Nature Neurosci. 2011;14:452–458. doi: 10.1038/nn.2778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buratti E, et al. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J Biol Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. A study of the interaction between the C terminus of TDP43 and other members of the hnRNP complex, including hnRNP A2/B1 and hnRNP A1. This paper also shows that TDP43 regulates splicing in the early stages of spliceosomal assembly. [DOI] [PubMed] [Google Scholar]
- 29.Ling SC, et al. ALS-associated mutations in TDP-43 increase its stability and promote complexes with FUS/TLS. Proc Natl Acad Sci USA. 2010;107:13318–13323. doi: 10.1073/pnas.1008227107. A proteomic analysis of proteins found in complex with TDP43, which includes the hnRNP proteins, FUS protein and components of the Drosha microprocessor complex. ALS-associated mutations increase the interaction between TDP43 and FUS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freibaum BD, Chitta RK, High AA, Taylor JP. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.D’Ambrogio A, et al. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buratti E, et al. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bose JK, Wang IF, Hung L, Tarn WY, Shen CK. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mercado PA, Ayala YM, Romano M, Buratti E, Baralle FE. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dreumont N, et al. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2010;38:1353–1366. doi: 10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ayala YM, Misteli T, Baralle FE. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc Natl Acad Sci USA. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fiesel FC, et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. EMBO J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Godena VK, et al. TDP-43 regulates Drosophila neuromuscular junctions growth by modulating Futsch/MAP1B levels and synaptic microtubules organization. PloS ONE. 2011;6:e17808. doi: 10.1371/journal.pone.0017808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang IF, Wu LS, Chang HY, Shen CK. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 40.Elvira G, et al. Characterization of an RNA granule from developing brain. Mol Cell Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Moisse K, et al. Cytosolic TDP-43 expression following axotomy is associated with caspase 3 activation in NFL−/− mice: support for a role for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1296:176–186. doi: 10.1016/j.brainres.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 42.Moisse K, et al. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1249:202–211. doi: 10.1016/j.brainres.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 43.Sato T, et al. Axonal ligation induces transient redistribution of TDP-43 in brainstem motor neurons. Neuroscience. 2009;164:1565–1578. doi: 10.1016/j.neuroscience.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 44.Nishimoto Y, et al. Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J Biol Chem. 2010;285:608–619. doi: 10.1074/jbc.M109.022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colombrita C, et al. TDP-43 is recruited to stress granules in conditions of oxidative insult. J Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 46.Volkening K, Leystra-Lantz C, Yang W, Jaffee H, Strong MJ. Tar DNA binding protein of 43 kDa (TDP-43), 14-3-3 proteins and copper/zinc superoxide dismutase (SOD1) interact to modulate NFL mRNA stability. Implications for altered RNA processing in amyotrophic lateral sclerosis (ALS) Brain Res. 2009;1305:168–182. doi: 10.1016/j.brainres.2009.09.105. [DOI] [PubMed] [Google Scholar]
- 47.Dewey CM, et al. TDP-43 is directed to stress granules by sorbitol, a novel physiological osmotic and oxidative stressor. Mol Cell Biol. 2011;31:1098–1108. doi: 10.1128/MCB.01279-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McDonald KK, et al. TAR DNA-binding protein 43 (TDP-43) regulates stress granule dynamics via differential regulation of G3BP and TIA-1. Hum Mol Genet. 2011;20:1400–10. doi: 10.1093/hmg/ddr021. [DOI] [PubMed] [Google Scholar]
- 49.Casafont I, Bengoechea R, Tapia O, Berciano MT, Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J Struct Biol. 2009;167:235–241. doi: 10.1016/j.jsb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Buratti E, et al. Nuclear factor TDP-43 can affect selected microRNA levels. FEBS J. 2010;277:2268–2281. doi: 10.1111/j.1742-4658.2010.07643.x. [DOI] [PubMed] [Google Scholar]
- 51.Fukuda T, et al. DEAD-box RNA helicase subunits of the Drosha complex are required for processing of rRNA and a subset of microRNAs. Nature Cell Biol. 2007;9:604–611. doi: 10.1038/ncb1577. [DOI] [PubMed] [Google Scholar]
- 52.Gregory RI, et al. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 53.Acharya KK, Govind CK, Shore AN, Stoler MH, Reddi P. P cis-requirement for the maintenance of round spermatid-specific transcription. Dev Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- 54.Kuo PH, Doudeva LG, Wang YT, Shen CK, Yuan HS. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Abhyankar MM, Urekar C, Reddi PP. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J Biol Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- 56.Furukawa Y, Kaneko K, Watanabe S, Yamanaka K, Nukina N. A seeding reaction recapitulates intracellular formation of Sarkosyl-insoluble transactivation response element (TAR) DNA-binding protein-43 inclusions. J Biol Chem. 2011;286:18664–18672. doi: 10.1074/jbc.M111.231209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Neumann M, et al. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hasegawa M, et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Deng HX, et al. Mutations in UBQLN2 cause dominant X-linked juvenile and adult-onset ALS and ALS/dementia. Nature. 2011;477:211–215. doi: 10.1038/nature10353. The identification of dominant mutations in UBQLN2 that are associated with X-linked juvenile and adult-onset ALS and ALS-dementia. The results implicate abnormal protein degradation pathways in the pathogenesis of motor neuron disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Shan X, Chiang PM, Price DL, Wong PC. Altered distributions of Gemini of coiled bodies and mitochondria in motor neurons of TDP-43 transgenic mice. Proc Natl Acad Sci USA. 2010;107:16325–16330. doi: 10.1073/pnas.1003459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stallings NR, Puttaparthi K, Luther CM, Burns DK, Elliott JL. Progressive motor weakness in transgenic mice expressing human TDP-43. Neurobiol Dis. 2010;40:404–414. doi: 10.1016/j.nbd.2010.06.017. [DOI] [PubMed] [Google Scholar]
- 62.Wegorzewska I, Bell S, Cairns NJ, Miller TM, Baloh RH. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. The first transgenic mouse overexpressing TDP43 is described. It demonstrates selective neurodegeneration and death, recapitulating many of the major features of ALS and FTLD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wils H, et al. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc Natl Acad Sci USA. 2010;107:3858–3863. doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Xu YF, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30:10851–10859. doi: 10.1523/JNEUROSCI.1630-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Igaz LM, et al. Expression of TDP-43 c-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J Biol Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. This paper shows that overexpression of disease-associated C-terminal TDP43 fragments leads to cytoplasmic aggregation. The aggregates are ubiquitylated and phosphorylated. These changes are associated with changes in RNA splicing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Igaz LM, et al. Dysregulation of the ALS-associated gene TDP-43 leads to neuronal death and degeneration in mice. J Clin Invest. 2011;121:726–738. doi: 10.1172/JCI44867. Overexpression of nuclear or cytoplasmic human TDP43 protein in transgenic mice results in selective neurodegeneration recapitulating many of the major features of FTLD and upper motor neuron disease. Transgene expression leads to downregulation of endogenous TDP43 mRNA and protein, and this autoregulation phenomenon is the best correlate of neurodegeneration. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nonaka T, et al. Phosphorylated and ubiquitinated TDP-43 pathological inclusions in ALS and FTLD-U are recapitulated in SH-SY5Y cells. FEBS Lett. 2009;583:394–400. doi: 10.1016/j.febslet.2008.12.031. [DOI] [PubMed] [Google Scholar]
- 68.Nonaka T, Kametani F, Arai T, Akiyama H, Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum Mol Genet. 2009;18:3353–3364. doi: 10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- 69.Winton MJ, et al. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J Biol Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. This paper shows that overexpression of TDP43 protein harbouring mutations of the NLS domain results in cytoplasmic TDP43 protein that accumulates as cytoplasmic aggregates, implicating abnormal TDP43 protein localization in the pathogenesis of ALS and FTLD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang YJ, et al. Phosphorylation regulates proteasomal-mediated degradation and solubility of TAR DNA binding protein-43 C-terminal fragments. Mol Neurodegener. 2010;5:33. doi: 10.1186/1750-1326-5-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pesiridis GS, Tripathy K, Tanik S, Trojanowski JQ, Lee VM. A “Two-hit” hypothesis for inclusion formation by carboxyl-terminal fragments of TDP-43 protein linked to RNA depletion and impaired microtubule-dependent transport. J Biol Chem. 2011;286:18845–18855. doi: 10.1074/jbc.M111.231118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Urushitani M, Sato T, Bamba H, Hisa Y, Tooyama I. Synergistic effect between proteasome and autophagosome in the clearance of polyubiquitinated TDP-43. J Neurosci Res. 2010;88:784–797. doi: 10.1002/jnr.22243. [DOI] [PubMed] [Google Scholar]
- 73.Kim SH, et al. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J Biol Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Brady OA, Meng P, Zheng Y, Mao Y, Hu F. Regulation of TDP-43 aggregation by phosphorylation and p62/SQSTM1. J Neurochem. 2011;116:248–259. doi: 10.1111/j.1471-4159.2010.07098.x. [DOI] [PubMed] [Google Scholar]
- 75.Filimonenko M, et al. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Wang X, et al. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 77.Suzuki S, et al. AMSH is required to degrade ubiquitinated proteins in the central nervous system. Biochem Biophys Res Commun. 2011;408:582–588. doi: 10.1016/j.bbrc.2011.04.065. [DOI] [PubMed] [Google Scholar]
- 78.Skibinski G, et al. Mutations in the endosomal ESCRTIII-complex subunit CHMP2B in frontotemporal dementia. Nature Genet. 2005;37:806–808. doi: 10.1038/ng1609. [DOI] [PubMed] [Google Scholar]
- 79.Hanson KA, Kim SH, Wassarman DA, Tibbetts RS. Ubiquilin modifies TDP-43 toxicity in a Drosophila model of amyotrophic lateral sclerosis (ALS) J Biol Chem. 2010;285:11068–11072. doi: 10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liachko NF, Guthrie CR, Kraemer BC. Phosphorylation promotes neurotoxicity in a Caenorhabditis elegans model of TDP-43 proteinopathy. J Neurosci. 2010;30:16208–16219. doi: 10.1523/JNEUROSCI.2911-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dormann D, et al. Proteolytic processing of TAR DNA binding protein-43 by caspases produces C-terminal fragments with disease defining properties independent of progranulin. J Neurochem. 2009;110:1082–1094. doi: 10.1111/j.1471-4159.2009.06211.x. [DOI] [PubMed] [Google Scholar]
- 82.Zhang YJ, et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc Natl Acad Sci USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sampathu DM, et al. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am J Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Mackenzie IR, et al. A harmonized classification system for FTLD-TDP pathology. Acta Neuropathol. 2011;122:111–113. doi: 10.1007/s00401-011-0845-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mackenzie IR, et al. Heterogeneity of ubiquitin pathology in frontotemporal lobar degeneration: classification and relation to clinical phenotype. Acta Neuropathol. 2006;112:539–549. doi: 10.1007/s00401-006-0138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson BS, et al. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J Biol Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Frost B, Ollesch J, Wille H, Diamond MI. Conformational diversity of wild-type Tau fibrils specified by templated conformation change. J Biol Chem. 2009;284:3546–3551. doi: 10.1074/jbc.M805627200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo JL, Lee VM. Seeding of normal Tau by pathological Tau conformers drives pathogenesis of Alzheimer-like tangles. J Biol Chem. 2011;286:15317–15331. doi: 10.1074/jbc.M110.209296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen C, et al. α-synuclein propagates from mouse brain to grafted dopaminergic neurons and seeds aggregation in cultured human cells. J Clin Invest. 2011;121:715–725. doi: 10.1172/JCI43366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Luk KC, et al. Exogenous α-synuclein fibrils seed the formation of Lewy body-like intracellular inclusions in cultured cells. Proc Natl Acad Sci USA. 2009;106:20051–20056. doi: 10.1073/pnas.0908005106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Cushman M, Johnson BS, King OD, Gitler AD, Shorter J. Prion-like disorders: blurring the divide between transmissibility and infectivity. J Cell Sci. 2010;123:1191–1201. doi: 10.1242/jcs.051672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gitler AD, Shorter J. RNA-binding proteins with prion-like domains in ALS and FTLD-U. Prion. 2011;5:179–187. doi: 10.4161/pri.5.3.17230. A review of recent characterization of prion-like domains within TDP43 and FUS, and the implications of such domains in the progression of disease. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Guo W, et al. An ALS-associated mutation affecting TDP-43 enhances protein aggregation, fibril formation and neurotoxicity. Nature Struct Mol Biol. 2011;18:822–830. doi: 10.1038/nsmb.2053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fuentealba RA, et al. Interaction with polyglutamine aggregates reveals a Q/N-rich domain in TDP-43. J Biol Chem. 2010;285:26304–26314. doi: 10.1074/jbc.M110.125039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nishihira Y, et al. Sporadic amyotrophic lateral sclerosis of long duration is associated with relatively mild TDP-43 pathology. Acta Neuropathol. 2009;117:45–53. doi: 10.1007/s00401-008-0443-6. [DOI] [PubMed] [Google Scholar]
- 96.Pamphlett R, Kum Jew S. TDP-43TDP43 inclusions do not protect motor neurons from sporadic ALS. Acta Neuropathol. 2008;116:221–222. doi: 10.1007/s00401-008-0392-0. [DOI] [PubMed] [Google Scholar]
- 97.Braun RJ, et al. Neurotoxic 43-kDa TAR DNA-binding protein (TDP-43) triggers mitochondrion-dependent programmed cell death in yeast. J Biol Chem. 2011;286:19958–19972. doi: 10.1074/jbc.M110.194852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Arai T, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 99.Igaz LM, et al. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am J Pathol. 2008;173:182–194. doi: 10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rutherford NJ, et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zhang YJ, et al. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Caccamo A, et al. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J Biol Chem. 2009;284:27416–27424. doi: 10.1074/jbc.M109.031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Tsai KJ, et al. Elevated expression of TDP-43 in the forebrain of mice is sufficient to cause neurological and pathological phenotypes mimicking FTLD-U. J Exp Med. 2010;207:1661–1673. doi: 10.1084/jem.20092164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yang C, et al. The C-terminal TDP-43 fragments have a high aggregation propensity and harm neurons by a dominant-negative mechanism. PloS ONE. 2010;5:e15878. doi: 10.1371/journal.pone.0015878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li Y, et al. A Drosophila model for TDP-43 proteinopathy. Proc Natl Acad Sci USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Voigt A, et al. TDP-43-mediated neuron loss in vivo requires RNA-binding activity. PloS ONE. 2010;5:e12247. doi: 10.1371/journal.pone.0012247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ayala YM, et al. Structural determinants of the cellular localization and shuttling of TDP-43. J Cell Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 108.Thorpe JR, Tang H, Atherton J, Cairns NJ. Fine structural analysis of the neuronal inclusions of frontotemporal lobar degeneration with TDP-43 proteinopathy. J Neural Transm. 2008;115:1661–1671. doi: 10.1007/s00702-008-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Winton MJ, et al. A90V TDP-43 variant results in the aberrant localization of TDP-43 in vitro. FEBS Lett. 2008;582:2252–2256. doi: 10.1016/j.febslet.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luty AA, et al. Sigma nonopioid intracellular receptor 1 mutations cause frontotemporal lobar degeneration-motor neuron disease. Ann Neurol. 2010;68:639–649. doi: 10.1002/ana.22274. [DOI] [PubMed] [Google Scholar]
- 111.Ritson GP, et al. TDP-43 mediates degeneration in a novel Drosophila model of disease caused by mutations in VCP/p97. J Neurosci. 2010;30:7729–7739. doi: 10.1523/JNEUROSCI.5894-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Barmada SJ, et al. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Miguel L, Frebourg T, Campion D, Lecourtois M. Both cytoplasmic and nuclear accumulations of the protein are neurotoxic in Drosophila models of TDP-43 proteinopathies. Neurobiol Dis. 2011;41:398–406. doi: 10.1016/j.nbd.2010.10.007. [DOI] [PubMed] [Google Scholar]
- 114.DiFiglia M, et al. Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science. 1997;277:1990–1993. doi: 10.1126/science.277.5334.1990. [DOI] [PubMed] [Google Scholar]
- 115.Chiang PM, et al. Deletion of TDP-43 down-regulates Tbc1d1, a gene linked to obesity, and alters body fat metabolism. Proc Natl Acad Sci USA. 2010;107:16320–16324. doi: 10.1073/pnas.1002176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ayala YM, et al. TDP-43 regulates its mRNA levels through a negative feedback loop. EMBO J. 2011;30:277–288. doi: 10.1038/emboj.2010.310. A molecular analysis of the mechanisms of TDP43 autoregulation, including identification of the 3′ UTR binding site and the role of exosome-mediated RNA decay. Nonsense-mediated decay is not implicated in this process. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Iguchi Y, et al. TDP-43 depletion induces neuronal cell damage through dysregulation of Rho family GTPases. J Biol Chem. 2009;284:22059–22066. doi: 10.1074/jbc.M109.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Feiguin F, et al. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 119.Kabashi E, et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum Mol Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 120.Sephton CF, et al. TDP-43 is a developmentally regulated protein essential for early embryonic development. J Biol Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wu LS, et al. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 122.Kraemer BC, et al. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010;119:409–419. doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Kwiatkowski TJ, Jr, et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009;323:1205–1208. doi: 10.1126/science.1166066. The identification of FUS mutations associated with familial ALS. As FUS is an RNA-binding protein, this discovery further supports the hypothesis that aberrant RNA processing is involved in the pathogenesis of ALS and FTLD. [DOI] [PubMed] [Google Scholar]
- 124.Deng HX, et al. Amyotrophic lateral sclerosis and structural defects in Cu, Zn superoxide dismutase. Science. 1993;261:1047–1051. doi: 10.1126/science.8351519. [DOI] [PubMed] [Google Scholar]
- 125.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 126.Baker M, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 127.Cruts M, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 128.Watts GD, et al. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nature Genet. 2004;36:377–381. doi: 10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 129.Elden AC, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. Based on data from a yeast genetic screen, this paper identifies intermediate-sized polyglutamine repeats as a risk factor for developing ALS in humans. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Al-Saif A, Al-Mohanna F, Bohlega S. A mutation in sigma-1 receptor causes juvenile amyotrophic lateral sclerosis. Ann Neurol. 2011 Aug 12; doi: 10.1002/ana.22534. [DOI] [PubMed] [Google Scholar]
- 131.Dejesus-Hernandez M, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. In this paper, a hexanucleotide repeat expansion in C9ORF72 is found to result in ALS, and this mutation accounts for the majority of familial ALS cases. The repeat expansion results in both altered splicing of the C9ORF72 transcript and the formation of nuclear C9ORF72 RNA foci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Renton AE. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. This study and reference 131 reported hexanucleotide repeat expansions in C9ORF72 that are pathogenic for ALS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mackenzie IR, et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. TDP43 pathology is seen in both sporadic and familial ALS cases, except in ALS cases that are associated with SOD1 mutations. This report suggests that cases that are associated with SOD1 mutations may involve a distinct disease process. [DOI] [PubMed] [Google Scholar]
- 134.Sun Z, et al. Molecular determinants and genetic modifiers of aggregation and toxicity for the ALS disease protein FUS/TLS. PLoS Biol. 2011;9:e1000614. doi: 10.1371/journal.pbio.1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Lanson NA, Jr, et al. A Drosophila model of FUS-related neurodegeneration reveals genetic interaction between FUS and TDP-43. Hum Mol Genet. 2011 Apr 12; doi: 10.1093/hmg/ddr150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Wang JW, Brent JR, Tomlinson A, Shneider NA, McCabe BD. The ALS-associated proteins FUS and TDP-43 function together to affect Drosophila locomotion and life span. J Clin Invest. 2011;121:4118–4126. doi: 10.1172/JCI57883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Wojciechowska M, Krzyzosiak WJ. Cellular toxicity of expanded RNA repeats: focus on RNA foci. Human Mol Genet. 2011;20:3811–3821. doi: 10.1093/hmg/ddr299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yu Z, et al. PolyQ repeat expansions in ATXN2 associated with ALS are CAA interrupted repeats. PloS ONE. 2011;6:e17951. doi: 10.1371/journal.pone.0017951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Glenner GG, Wong CW. Alzheimer’s disease: initial report of the purification and characterization of a novel cerebrovascular amyloid protein. Biochem Biophys Res Commun. 1984;120:885–890. doi: 10.1016/s0006-291x(84)80190-4. [DOI] [PubMed] [Google Scholar]
- 140.Goate A, et al. Segregation of a missense mutation in the amyloid precursor protein gene with familial Alzheimer’s disease. Nature. 1991;349:704–706. doi: 10.1038/349704a0. [DOI] [PubMed] [Google Scholar]
- 141.Chartier-Harlin MC, et al. Early-onset Alzheimer’s disease caused by mutations at codon 717 of the β-amyloid precursor protein gene. Nature. 1991;353:844–846. doi: 10.1038/353844a0. [DOI] [PubMed] [Google Scholar]
- 142.Murrell J, Farlow M, Ghetti B, Benson MD. A mutation in the amyloid precursor protein associated with hereditary Alzheimer’s disease. Science. 1991;254:97–99. doi: 10.1126/science.1925564. [DOI] [PubMed] [Google Scholar]
- 143.Hsiao K, et al. Correlative memory deficits, Aβ elevation, and amyloid plaques in transgenic mice. Science. 1996;274:99–102. doi: 10.1126/science.274.5284.99. [DOI] [PubMed] [Google Scholar]
- 144.Games D, et al. Alzheimer-type neuropathology in transgenic mice overexpressing V717F β-amyloid precursor protein. Nature. 1995;373:523–527. doi: 10.1038/373523a0. [DOI] [PubMed] [Google Scholar]
- 145.Holtzman DM, Morris JC, Goate AM. Alzheimer’s disease: the challenge of the second century. Sci Transl Med. 2011;3:77sr1. doi: 10.1126/scitranslmed.3002369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Knopman DS, Mastri AR, Frey WH, 2nd, Sung JH, Rustan T. Dementia lacking distinctive histologic features: a common non-Alzheimer degenerative dementia. Neurology. 1990;40:251–256. doi: 10.1212/wnl.40.2.251. [DOI] [PubMed] [Google Scholar]
- 147.Mackenzie IR, et al. Dementia lacking distinctive histology (DLDH) revisited. Acta Neuropathol. 2006;112:551–559. doi: 10.1007/s00401-006-0123-3. [DOI] [PubMed] [Google Scholar]
- 148.Mackenzie IR, et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 2010;119:1–4. doi: 10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]