Abstract
Lentiviral replication in its target cells affects a delicate balance between cellular co-factors required for virus propagation and immunoregulation for host defense. To better elucidate cellular proteins linked to viral infection we tested plasma from rhesus macaques infected with the simian immunodeficiency viral strain SIVsmm9, prior to, 10 days (acute) and 49 weeks (chronic) after viral infection. Changes in plasma protein content were measured by quantitative mass spectrometry by isobaric Tags for Absolute and Relative Quantitation (iTRAQ) methods. An 81 and 232% increase in SERPINA1 was seen during acute and chronic infection, respectively. Interestingly, gelsolin, vitamin D binding protein and histidine rich glycoprotein were decreased by 45% in acute conditions but returned to baseline during chronic infection. When compared to uninfected controls, a 48–103% increase in leucine rich alpha 2-glycoprotein, vitronectin and ceruloplasmin was observed during chronic viral infection. Observed changes in plasma proteins expression likely represent a compensatory host response to persistent viral infection.
Keywords: proteomics, plasma, biomarker, iTRAQ, monkey
Introduction
Host mechanisms for control of HIV infection remain incompletely understood after more than two decades of investigations 1. After initial infection, HIV-1 seroconversion occurs with a broad range of clinical manifestations or without symptoms that parallels robust unregulated viral replication 1, 2, followed by sustained low level chronic infection 3. The consequences of both phases of disease on innate immunity are regulated through alterations in cellular immunity. We postulate that changes in plasma factors reflects progression of disease and that they could serve to unveil relevant host immune responses to ongoing viral growth as biomarkers for disease progression 4.
Experiments analyzing viral-cell-host interactions from acquired human clinical samples are complicated, as viral infection per se cannot readily be separated from co-morbid conditions that commonly include illicit drug and alcohol use, opportunistic infections and environmental factors. This necessitates the use of animal models to evaluate host responses to viral infection. Simian immunodeficiency virus (SIV) infection of macaques is one most preferred model in studies of HIV-1 infection 5–7 as the model closely resembles lentiviral infection in its human host while assuring control of experimental infection conditions and precluding confounding factors 8.
Herein, isobaric Tags for Absolute and Relative Quantitation (iTRAQ) technology was used to assess plasma proteins that best reflect ongoing viral infection before and during the course of SIV infection 9–13. Plasma proteins were digested with trypsin and peptides were labeled with iTRAQ tags and recorded mass spectra of iTRAQ labeled peptides were used for reliable identification and quantitation. Additional information was obtained through manual examination of spectra to assess protein isoforms. The study provides a step towards unraveling for the associations of plasma proteins modified as a consequence of viral infection in a relevant animal model system for HIV-1 disease. Differential expression of proteins linked to control of innate immunity was found linked to host responses against ongoing viral infection.
Materials and Methods
Animal cohort, viral infection and sample collections
A cohort of 4 rhesus macaques (Indian strain) monkeys housed at the Yerkes National Primate Research Center (Atlanta, GA) was studied. For adaptation and stress reduction, animals were housed for 2 years prior to being exposed to experimental conditions. Blood was collected from the femoral vein by venipuncture into EDTA-containing vacutainers (BD Biosciences, Franklin Lakes, NJ) 7 weeks prior to SIV infection to establish baseline conditions. Plasma was obtained by collecting the upper fluid portion of centrifuged EDTA whole blood. Monkeys were infected with SIVsmm9 at an effective dose of 10,000 TCID50 by single intravenous inoculation. Plasma samples were obtained from SIV-infected rhesus macaques 10 days and 49 weeks after exposure, representing acute and chronic infection, respectively. Quantitation of viral infection was provided by mRNA quantification and reported values as log10 SIV mRNA copies per milliliter 14. Animal experiments were performed under IACUC approvals and following NIH guidelines. Archived samples frozen for several years at −80°C were used in this study. Expanded information regarding animal conditions, virology and sample collection is described in detail by Donahoe et al 14.
Sample preparation and immunodepletion
Prior to immunodepletion, proteases and infectious virus were inactivated in monkey plasma samples using 10 μL of 10% Triton X-100 and 50 μL of 20× Protease Inhibitor (Sigma-Aldrich Corp., St. Louis, MO) per mL of plasma. Samples were vortexed and incubated at room temperature for 15 min, then immediately placed on ice. To delipidate, the samples were centrifuged at 18,000 × g at 4°C for 15 min and collected the middle layer of the pre-cleared plasma. The standard manufacturer protocol for immunodepletion was followed using the ProteomeLab IgY-12 High Capacity Proteome Partitioning Kit (Beckman Coulter, Fullerton, CA) to remove the 12 most abundant proteins (albumin, IgG, fibrinogen, transferrin, IgA, IgM, haptoglobin, apolipoprotein A-I, apolipoprotein A-II, α1-antitrypsin, α1-acid glycoprotein and α2-macroglobulin). This kit contained pre-manufactured optimized buffers for sample loading, washing, eluting and regenerating, along with a HPLC affinity column LC10 (12.7 × 79.0 mm; capacity of 0.25 mL human plasma). Pre-cleared plasma (0.25 mL) was diluted with 0.375 mL of kit dilution buffer (10 mM Tris-HCl pH 7.4, 0.15 M NaCl) then filtered through 0.45 μm spin filters at 9000 × g for 1 min. Samples were injected onto the column at a flow rate of 0.5 mL/min. Flow-though fractions were collected and concentrated using VIVASPIN 15R (membrane 5000 MWCO, Sartorius BioLab Products, Cole-Parmer, Vernon Hills, IL) at 4000 × g for approximately 1.5 hr. The proteins bound to the column were eluted with elution buffer (0.1M glycine-HCl pH 2.5) then neutralized using 1 M Tris-HCl pH 8.0. The column was equilibrated with a solution of 10 mM Tris-HCl pH 7.4 and 0.15 M NaCl at a flow rate of 0.2 mL/min. Protein concentration was determined using Nanodrop ND-1000 (Thermo Fisher Scientific Inc., Waltham, MA) through the absorbance at 280 nm.
iTRAQ labeling
The standard Applied Biosystems Reagents Protocol for Plasma was followed for 4-plex iTRAQ labeling of 50 μg of plasma protein. The ABI iTRAQ reagents protocol for plasma called for acetone precipitation to remove buffers that might interfere with efficiency of iTRAQ labeling. However, we found that precipitation, with 200 proof ethanol, was most efficient for precipitation. In subsequent steps of trypsin digestion and iTRAQ labeling we used the following protocol. Ten volumes of cold 200 proof ethanol were added to each sample tube, vortexed and then placed in −20°C until precipitate formed. Following centrifugation at 13000 × g at 4°C for 15 min, the ethanol was decanted and 1 mL of cold 70% ethanol was added, vortexed and centrifuged at 13000 × g at 4°C for 5 min. Ethanol was decanted and the sample was dried in a speed vac for 10 min to remove any remaining ethanol. To each sample tube containing 50 μg of sample we added 25 μL of Sample Buffer – Plasma (ABI), 1 μL of denaturant and 2 μL of reducing reagent. Samples were vortexed for 20 sec, centrifuged to bring sample to bottom of tube and incubated at 60°C for 1 hr. In order to block cysteine residues 1 μL of freshly prepared 84 mM iodoacetamide (Sigma-Aldrich Corp, St. Louis, MO) was added. Samples were vortexed, centrifuged and incubated in the dark at room temperature for 30 min. After incubation, 10 μL of reconstituted trypsin (Trypsin, TPCK, ABI, 1 μg/μL) was added, vortexed and centrifuged, then incubated at 37°C overnight to digest each sample. Each iTRAQ reagent was brought to room temperature, dissolved in 70 μL of ethanol and the contents of one iTRAQ reagent tube was transferred to one sample tube. To ensure optimal efficiency of labeling, pH was maintained between 8.0–10.0. Samples were incubated at ambient room temperature for 1 hr, after which 100 μL of Milli-Q water was added to quench reaction for 30 min. The samples of each 4-plex experiment were pooled into an Eppendorf tube, frozen to −80°C and dried in a SpeedVac.
Peptide purification and strong cation exchange fractionation
Labeled samples were subjected to the next step of clean-up using a Waters Oasis MCX cartridge (Waters Corp, Milford, MA), a mixed-mode cation exchange cartridge packed with material containing both hydrophobic properties and negatively charged groups which. Use of Waters Oasis MCX cartridge was necessary to assure high sample quality and reproducible analysis. Briefly, 1 mL of 0.2% formic acid was added to each sample with pH 3.0 maintained. Each MCX cartridge was equilibrated by slowly passing 1 mL of 1:1 methanol:water across cartridge. Sample was applied at flow rate of 1 drop per second and cartridge was washed with 1 mL of 5% methanol, 0.1% formic acid followed by 1 mL of 100% methanol. The bound peptide was eluted with freshly prepared buffer (50 μL of 28% NH4OH, 950 μL of methanol), dried in speed vac and stored at −80°C until fractionated via strong cation exchange (SCX).
Strong cation exchange fractionation (SCX) was performed by using a liquid chromatography system (Beckman Coulter), which included an autosampler with a 500 μl injection loop, dual pump and a ultraviolet-visual (UV-Vis) detector set at 215 nm. The system was controlled using an IBM computer and 32 Karat 7.0 software (Beckman Coulter). The mobile phases consisted of mobile phase A (10 mM KH2PO4 + 25% acetonitrile, pH 2.7) and mobile phase B (10mM KH2PO4 + 25% acetonitrile + 500 mM KCl, pH 2.7). Each sample was resuspended in 290 μL of 0.1% formic acid and pH was maintained at less than or equal to 3.0. SCX fractionation of each iTRAQ labeled pooled sample was performed using a PolySULFOETHYL A column (100 × 2.1 mm, 5 μm, 3 Å; PolyLC, Columbia, MD). The flow rate was set at 0.100 mL/min. Samples were fractionated with a continuous salt gradient from 10 to 500 mM of KCl and collected as 12 fractions in 3 minute intervals between 43 min – 76 min (Fig. 2). After sample fractionation by SCX, the fractions were desalted with a RP-HPLC step gradient. Samples were loaded onto the C18 HPLC column (Jupiter 4 u Proteo 90A, 50 × 4.6 mm, 4 μm, Phenomenex, Torrance, CA) by using 95% mobile phase A (0.1% TFA) for 5 min and eluted using 50% mobile phase B (0.1% TFA in acetonitrile). Samples were dried in a speed vac and stored at −80°C until further processing.
Figure 2. Strong cation exchange fractionation.
Dotted line represents continuous gradient and corresponds to right y-axis. Solid line is a representative sample fractionation chromatogram and corresponds to left y-axis.
Tempo LC and MALDI-TOF/TOF
The Tempo LC MALDI robotic spotting system equipped with a C18 reversed phase capillary column (AB Sciex, Foster City, CA) was used to further fractionate peptides from SCX fractions, followed by data acquisition using 4800 MALDI TOF/TOF (AB Sciex) as previously published 15. Briefly, after using an in-house packed C18 column to separate fractions by HPLC gradient, LC fractions were spotted onto MALDI 1232-spot format plates, with a spotting interval of 24 seconds and applying 2.8 kV plate voltage. Data was acquired from LC MALDI spot fractions using 4800 MALDI TOF/TOF equipped with a 200 Hz repetition rate Nd:YAG laser. Spectrum from a total of 800 laser shots was accumulated for each TOF MS spectrum between 800 – 4000 m/z. Data dependent MS/MS mode was operated using CID gas (air) and 2 kV collision energy. Programmed laser stop conditions were employed for the accumulation of MS/MS spectra from 800 – 4000 laser shots.
Database searches
TOF MS and MS/MS spectra were analyzed using Protein Pilot v.2.0.1 software which utilizes the Paragon scoring algorithm 16. Search parameters included: iTRAQ 4plex (peptide labeled) sample type, iodoacetemide cys alkylation, trypsin digest, biological modifications ID focus and thorough search effort. The protein FASTA database used for searches was a concatenated “target-decoy” version of Macaca subset of the NCBI Ref Sequence (http://www.ncbi.nlm.nih.gov/RefSeq/) database (March, 2009; 44,047 protein entries) to which 110 ‘contaminant’ proteins were added (source: http://www.thegpm.org/crap/index.html). A total of 76,252 protein sequences comprised the target-decoy database.
Western blot assays
1-dimensional gel electrophoresis (1DE) was performed on 4 individual monkey plasma samples from 3 different time points: baseline, acute (10d) and chronic (49 wk) using NuPAGE gel system (Invitrogen Corp., Carlsbad, CA) with 4–12% gradient Bis-Tris gels under reducing conditions. For Western blot analyses, 1.5 μg of immunodepleted plasma protein were loaded per lane and the gel was transferred to PVDF membrane using Ready Gel™ Blotting sandwiches (Bio-Rad, Hercules, CA). The membranes were blocked using 10% skim milk in PBS with 2% Tween-20 (PBST). The following primary antibodies were used for protein validation: mouse anti-gelsolin MAb (BD Transduction Laboratories, San Jose, CA), rabbit anti-vitronectin PAb (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) and mouse anti-ceruloplasmin MAb (BD Transduction Laboratories). After incubation with the primary antibody, the membrane was incubated with the appropriate horseradish peroxidase conjugated goat anti-mouse (Jackson ImmunoResearch, West Grove, PA) or goat anti-rabbit (Jackson ImmunoResearch) secondary antibodies. SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) was used to detect a chemiluminescent signal recorded on Blue Lite X-ray film (ISCBioExpress, Kaysville, UT). Images were scanned into Adobe Photoshop Software and adjusted using “auto levels”. Then images were analyzed using ImageJ freeware available though NIH by inverting the image and a measurement box of exact same size was used for each band analysis. All Western blot analyses were performed in triplicate.
Statistical analysis
Protein/peptide abundance measures generated by iTRAQ were first processed using Protein Pilot v.2.0.1. The logarithm of the abundance measure was modeled as a function of animal, protein, peptide and experimental condition 17. Tag effects were not modeled as each sample from a single experimental condition was labeled with the same tag. Data for four experimental conditions were available: baseline, acute infection, chronic infection and a pooled sample.
All available abundance records (N=80,868) were normalized using an iterative backfitting procedure to remove the animal, protein and peptide effects (17 and SAS PROC itraqnorm provided by Douglas W. Mahoney, Mayo Clinic http://pubs.acs.org/doi/suppl/10.1021/pr700734f/suppl_file/pr700734f-file001.pdf, accessed 11/10/2009). Comparison of the distribution of the protein/peptide normalized (log) abundance measures by experimental condition were restricted to records with Protein Pilot assessed “confidence” of at least 50%; the analysis also excluded abundance measures from the ‘pooled samples’.
Results
Sample processing and iTRAQ labeling
The proteomic tests performed in this work are outlined in Figure 1. Plasma samples were immunodepleted from 12 most abundant proteins prior to subsequent proteomic profiling as previously published 18. Average protein yield from 250 μL of depleted sample was ~5%. The efficiency of immunodepletion of monkey proteins based on IgY12 human systems were verified by loading 2 μg of protein on a 4–12% Bis-Tris gel and performing 1DE (supplemental Fig. 1). Although 1DE showed efficient removal of most abundant proteins, mass spectrometry analysis still detected and quantitated peptides belonging to some of these proteins (data not shown). In support of these findings, Homo sapiens and Macaca mulatta sequence homology is high, however, there are likely non-overlapping epitopes in the pool of polyclonal antibodies, which may affect final efficiency of immunodepletion. We assume that if any given protein were not fully immunodepleted due to aforementioned reasons the resulting depletion would be proportional to all animals in study.
Figure 1.
Flow chart of experimental design.
After iTRAQ labeling, the three plasma samples from each monkey baseline 114, acute 115 and chronic 116 were combined with a pooled sample 117 (Fig. 1), however the pooled was not eventually used for further analysis. The complex mixtures of labeled peptides were fractionated by SCX chromatography (Fig. 2). We compared effectiveness of SCX fractionation by using salt step gradient versus continuous gradient. The continuous gradient generated more reproducible chromatography and thus more optimal fractionation (Fig. 2). When using MCX cartridges prior to SCX continuous gradient fractionation, we achieved increased sample load, optimal fractionation and approximately 7 times the amount of eluted peptide (data not shown), as compared to not using MCX cartridges prior to SCX step-gradient fractionation. As a result of enhanced fractionation, we obtained better MS/MS identification rate using the continuous gradient.
RP-nanoHPLC (2nd dimension) fractionation and MS analysis
Based on comparison of three mass spectrometry modes: QTrap4000, LTQOrbitrap in a PQD mode and 4800 MALDI-TOF/TOF, we choose the latter one as superior in protein identification and quantitation. SCX fractions were subjected to second dimension fractionation on RP-HPLC column with automatic spotting onto MALDI target plate using Tempo LC-MALDI TOF/TOF platform. Mass spectrometry data were acquired in a batch mode. Figure 3 is illustrates a representative sample fractionation chromatogram using Tempo LC, as well as MS and MS/MS spectra illustrating representative peptide fragmentation and quantitation data.
Figure 3. RP-HPLC and mass spectrometry identification and quantitation.
(A). Representative chromatogram for TempoLC (second dimension) fractionation; (B). M/z region of a precursor ion for ACEPGVDYVYK peptide (m.w. 1588.7942 Da); (C). Tandem mass fragmentation of ACEPGVDYVYK peptide; (D). M/z region showing iTRAQ reporter ions for ACEPGVDYVYK peptide quantitation.
Statistical analysis of differentially expressed proteins
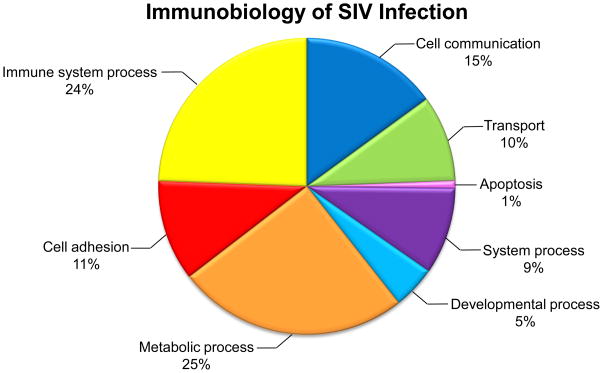
For each protein, the distributions of the normalized (log) abundance measures were compared using the Wilcoxon rank-sum test. The false discovery rate 19 was set at 5%. The relative protein abundance comparing experimental condition(j) to experimental condition(i) (where “j” and “i” represent any two experimental conditions to be compared) was estimated from normalized log(abundance) as: exp (mean{log(abundance)j } − mean{log(abundance)i }). Statistical analysis showed that levels of 64 proteins were differentially expressed and Figure 4 illustrates how these proteins are categorically involved in the immunobiology of SIV infection. Table 1 summarizes an abbreviated list of 30 proteins that were selected for further considerations based on their significant quantitative changes and potential role in SIV infection. A full list of proteins and peptides are included in Supplemental Table 1. Based on these data we have made two observations: first, despite immunodepletion of 12 most abundant proteins, differentially expressed and quantitated proteins represent medium to high abundant proteins and second, a majority of them represent proteins associated with inflammation generated by microbial infection.
Figure 4.
Pie chart illustrating biological process classification of 64 differentially expressed proteins identified in iTRAQ study of the immunobiology of SIV infection.
Table 1.
List of selected proteins differentially regulated in acute and chronic phase of SIV infected rhesus macaques.
| Protein | Baseline: acute ratioa* | Baseline: chronic ratiob* | Acute: chronic ratioc* | NCBI Accession # | Swiss- Prot | Molecular Wt | Postulated function |
|---|---|---|---|---|---|---|---|
| complement component 6 isoform 2; isoform 1 | NSC | 37% | NSC | 109077082 | P13671 | 104821 | forms pores in the plasma membrane of target cells |
| apolipoprotein A-IV | NSC | 52% | 73% | 109108832 | P06727 | 65037 | lipid transport |
| apolipoprotein B isoform 1&2 | −33% | 81% | 135% | 109102154 | P04114 | 515164 | carries cholesterol to tissues |
| apolipoprotein H isoform 2 (aka beta-2- glucoprotein 1 | −38% | NSC | 70% | 109116939 | P02749 | 38281 | involved in agglutination |
| ceruloplasmin (ferroxidase) isoform 3 | NSC | 48% | 46% | 109048809 | P00450 | 120486 | blue, copper-binding glycoprotein; ferroxidase activity oxidizing Fe2+ to Fe3+ without releasing radical oxygen species; iron transport across the cell membrane |
| complement component 3, partial | −27% | 20% | 64% | 109123141 | P01024 | 169789 | processing by C3 convertase is central reaction in classical and alternative complement pathways |
| complement component 4 binding protein, alpha chain precursor isoform 2 | NSC | 91% | 146% | 109018545 | 66598 | cleaved into 4a and 4b | |
| complement component 8, beta polypeptide | NSC | 43% | 41% | 109005039 | P07358 | 66770 | forms pores in the plasma membrane of target cells |
| complement component 9 | NSC | 43% | 38% | 109077053 | P02748 | 63168 | pore-forming subunit of the MAC |
| complement factor B isoform 1 | NSC | 50% | 33% | 109070536 | 124719 | cleaved into Ba and Bb; involved in regulation of immune reaction | |
| complement factor B isoform 3 | NSC | 30% | 34% | 109070540 | P00751 | 85406 | cleaved into Ba and Bb; involved in regulation of immune reaction |
| gelsolin precursor | −27% | NSC | 51% | 121116 | P06396 | 85698 | calcium-regulated, actin-binding protein regulating actin filament assembly and disassembly; inhibits apoptosis |
| histidine-rich glycoprotein | −45% | NSC | 95% | 109042262 | P04196 | 59326 | negative acute phase protein; immune complex clearance |
| pro-platelet basic protein (chemokine (C- X-C motif) ligand 7) | NSC | 188% | 393% | 109074506 | P02775 | 13849 | stimulates mitogenesis, synthesis of extracellular matrix, glucose metabolism and synthesis of plasminogen activator |
| serine (or cystine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 isoform 4 | 81% | 232% | 83% | 109084723 | P01009-2 | 46568 | serine protease inhibitor; C-terminal of cleaved SERPPINA1 may inhibit HIV-1 infection |
| serpin peptidase inhibitor, clade A, member 3 | NSC | 73% | 74% | 109084779 | 51875 | acute-phase protein produced in liver and induced during inflammation | |
| complement factor I precursor (C3B/C4B inactivator) | −32% | NSC | 73% | 109075363 | 47630 | regulates complement activation by cleaving cell-bound or fluid phase C3b and C4b | |
| beta globin | 1056% | 86% | −84% | 109107606 | P68871 | 15956 | oxygen transport from lung to peripheral tissues |
| complement component 5 | NSC | 51% | 75% | 109110418 | P01031 | 188305 | activation of C5 by a C5 convertase initiates the spontaneous assembly of the late complement components into the membrane attack complex |
| complement factor H isoform a precursor isoform 3 | −19% | 48% | 81% | 109019000 | P08603 | 139296 | cofactor in the inactivation of C3b by factor I; increases the rate of dissociation of the C3bBb complex (C3 convertase) and the (C3b)NBB complex (C5 convertase) in the alternative complement pathway |
| fetuin B isoform 3 | NSC | 64% | 60% | 109042265 | Q9UGM5 | 41941 | inhibitor of basic calcium phosphate precipitation |
| fibronectin 1 isoform 1 preproprotein | NSC | 73% | 42% | 109100908 | P02751 | 287731 | cell adhesion, cell motility, opsonization, wound healing, and maintenance of cell shape |
| hemopexin | NSC | 33% | 36% | 109107500 | P02790 | 51427 | binds and transports heme to the liver for breakdown and iron recovery |
| leucine-rich alpha-2- glycoprotein 1 | NSC | 103% | 82% | 109122991 | P02750 | 38201 | involved in protein- protein interaction, signal transduction, and cell adhesion and development |
| retinol-binding protein 4, plasma precursor | −37% | NSC | NSC | 109089986 | P02753 | 20774 | interacts with transthyretin, preventing its loss by filtration through the kidney glomeruli |
| transthyretin (prealbumin, amyloidosis type I) isoform 2; isoform 1 | −59% | NSC | 227% | 109121864 | P02766 | 15700 | thyroid hormone- binding protein |
| vitamin D-binding protein | −15% | NSC | 22% | 109074554 | P02774 | 52952 | transport Vitamin D, activate macrophages and osteoclasts, and enhance C5a chemotactic activity |
| plasminogen | −16% | 27% | 52% | 112807252 | P00747 | 90569 | activates urokinase- type plasminogen activator, collagenases and several complement zymogens, such as C1 and C5; cleaves fibrin, fibronectin, thrombospondin, laminin and von Willebrand factor |
| vitamin K- dependent protein S precursor | −39% | NSC | 108% | 113461945 | P07225 | 70333 | anticoagulant plasma protein |
| vitronectin | NSC | 65% | 62% | 109113730 | P04004 | 54360 | cell adhesion factor; inhibits membrane damaging effect of the terminal cytolytic complement pathway |
Values listed are statistically significant of p<0.05; NSC = no significant change
Values listed read percent change in acute infection when compared to baseline.
Values listed read percent change in chronic infection when compared to baseline.
Values listed read percent change in chronic infection when compared to acute infection.
Validation
For validation of our proteomic profiling results we utilized quantitative Western blot analysis. This method is the most straightforward orthogonal means and is based on specific antigen-antibody interaction. However, in some instances a limitation of Western blot validation exists in the lack of cross reactivity between antibodies raised against human proteins and monkey proteins, regardless of >90% sequence homology between human immunogen used for antibody production and identified monkey protein. Due to this limitation in detection, we have provided references linking selected iTRAQ identified proteins to HIV infection (Table 2).
Table 2.
List of select proteins with literature references to HIV-Protein relationships
| Protein | HIV-Protein Relationships | Reference | Protein Classificationa |
|---|---|---|---|
| complement component 3, partial | activated by HIV | 59–61 |
|
| histidine-rich glycoprotein (HRG) | modulated by HIV infection | 40, 62 |
|
| serine (or cystine) proteinase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 isoform 4 (SERPINA1) | inhibits formation of gp120; prevents HIV p55(gal) processing to p24; inhibits HIV-1 gp41 fusion peptide; increases STAT1 phosphorylation and inhibits HIV-long terminal repeat (LTR)-driven transcription; suppresses activation of the HIV-1-inducing transcription factor NF-{kappa}B | 29–32, 63 |
|
| leucine-rich alpha-2- glycoprotein 1 (LRG) | serum levels increase in HIV infection | 64 |
|
| plasminogen | uPA inhibits HIV infection; uPA/uPAR interaction inhibits HIV replication; severe HIV-1 associated dementia show increased levels of uPA (but not PAI-1) which are deminished by initiation of antiretroviral therapy | 43, 65, 66 |
|
Colored squares refer to biological processes as illustrated in Figure 4.
Three protein groups we chose to validate. One group is comprised of ceruloplasmin, gelsolin and vitronectin, all which were validated by Western blot analysis. The second group comprising of LRG, SERPINA1 and vitamin D binding protein the validation failed due to a lack of reactivity of existing antibodies. This is certainly an obstacle in data validation and other methods have to be employed such as Multiple Reaction Monitoring (MRM). The third group is represented by proteins of complement cascade which displays multiple bands representing various processed forms most notably complement C3. Some trypsin derived peptides may originate from either C3 or from C3b, iC3b, C3c, C3dg therefore representing total pool in the digest rather than reflecting differential expression of any particular form.
For validation using quantitative Western blot, ImageJ densitometry measurements were normalized between membranes, and averages and standard errors were calculated followed by a t-test. Fig. 5 shows validation results for three proteins: gelsolin, vitronectin and ceruloplasmin. For each protein we present results of statistical analysis (A) and results of quantitative Western blot (B). During acute infection gelsolin is down regulated while vitronectin and ceruloplasmin remain the same. All three proteins are up-regulated during chronic phase of infection. Both statistical analysis and Western blot validation indicated significant decrease of gelsolin during acute phase of infection, whereas vitronectin was not significantly changed during acute but significantly upregulated during chronic phase, which correlated these two methods of quantitation. Based on statistical analysis level of ceruloplasmin in plasma is not changed during acute phase and it is significantly elevated during chronic phase. However, Western blot validation showed small but significant decrease level of ceruloplasmin during acute phase and a significant increase during chronic.
Figure 5. Statistical analysis and Western blot validation of selected proteins.
(A). iTRAQ® ratios and significance based on statistical analysis. (B). Validation by quantitative Western blot of differentially regulated proteins. Timepoints marked with different letters indicate significant difference of pval<0.05 when compared to other timepoints. Standard error bars are given for Western blot validations.
This current study also shows that in instances when search algorithms recognize and differentiate between isoforms of one protein, manual verification of output data is necessary despite the required substantial effort. One example is ceruloplasmin, which has three entries in the NCBI database, while there is only one entry in UniProt and existence of isoforms is not listed. Protein Pilot software and subsequent statistical analysis listed two of these isoforms as separate proteins and as such were separately analyzed for quantitation. Because each isoform was not found in all samples, the resulting output may create false positive differences in protein expression, thus may influence the interpretation of biological changes in the biological system being investigated. However, manual data mining of identified peptides in ceruloplasmin isoforms 1 and 3 confirmed that there was only one peptide difference between each isoform and we concluded that these two isoforms should be treated as one protein (supplemental Figure 2). We validated ceruloplasmin under this assumption.
Discussion
Typically two quantitative proteomic platforms are used for profiling plasma: gel based 2-dimensional electrophoresis usually with difference gel electrophoresis (2DE DIGE) 20–22 and mass spectrometry based quantitation using isobaric Tags for Absolute and Relative Quantitation (iTRAQ) 23, 24. In our previous investigations of plasma samples we used 2DE DIGE platform which resulted in identification of previously not reported differential expression of proteins such as afamin and gelsolin 18, 25, 26. Differential expression of these proteins were further confirmed in similar analysis performed with SIV infected rhesus macaques 9, 27. While select proteins in the current study (gelsolin, ceruloplasmin, complement C3, vitronectin, for example) were previously documented being differentially expressed in human samples 18, 25, 26, the evaluation of these proteins in a broad range of interdisciplinary studies under distinct pathobiologic settings provides further validation for their importance in HIV-1 disease. Here we have expanded such studies by using the iTRAQ platform to quantitatively analyze differentially expressed plasma proteins during the course of SIV infection, with successful validation of selected statistically significant proteins.
Biomarkers for monitoring the progression of immunodeficiency virus infection are still limited to measurements of viral loads and T cell subtypes 4, 28. There are a handful of other cellular markers that correlate with systemic viral infection however they do not have adequate sensitivity and specificity to be used in clinical practice. Progression of disease over many years is a very complex process and includes distinct host responses occurring during the acute and chronic phases. Acute response to viral infection occurs within days to a few weeks after viral exposure and is characterized by rapid elevations in viral load and substantive decreases in the numbers and function of CD4+ T lymphocytes. Transition from acute to chronic phase is a complex process and one of reasons is that during acute infection, members of the complement cascade remain at the same level or decreased as prior to infection. Acute infection leads to humoral and cellular immune responses specific to the infecting viral strain that typically suppresses viral replication and partially restores CD4+ T cells. The resulting chronic viral disease stage becomes symptomatic for the host after several years when sufficient numbers of T cells are destroyed. SIV infection of rhesus macaques reflects this process and infected animals after initial acute phase enter chronic infection and eventually progress to terminal stage of AIDS 5–7. In this study we used this non-human primate model to investigate changes in plasma proteomes at three time points: before, during acute and chronic phases of infection to get an insight of organism’s response to viral infection in a very well controlled experiment.
Acute phase proteins (APPs) belong to a biochemically and functionally diverse group of proteins that are typically produced in liver as a response to inflammation. The observed changes in APPs likely represent host response to initial SIV infection. We observed that several positive acute phase proteins, including, serine proteinase inhibitors (SERPINS), complement, plasminogen and ceruloplasmin, as well as negative acute phase proteins such as transthyretin, retinol binding protein and histidine rich glycoprotein were affected by SIV infection.
SERPINS are a group of proteins that inhibit proteases and play key roles in controlling inflammation. Here we identify 2 SERPINS, A1 and A3 that are differentially regulated through the course of SIV infection. During the course of acute and chronic infection, SERPINA1 increased 81% and 232%, respectively, when compared to baseline levels. SERPINA1 inhibits the formation of gp120, as well as it inhibits HIV protease thus preventing HIV p55(gal) processing to p24, which are critical processes for HIV-1 morphogenesis 29, 30. Recently, it was demonstrated that a C-terminal portion of SERPINA1 inhibits HIV-1 infection by as much as 99%, while full length SERPINA1 only inhibited HIV-1 infection up to 70% 31, 32. These results provide supporting evidence that the host is unsuccessfully attempting to compensate for increased viral replication through many mechanisms and an increase in SERPINA1 might be one of them.
Leucine rich alpha 2-glycoprotein (LRG) expression has been shown to increase in various diseases 33, 34, however it’s physiological function has yet to be defined. Shirai et al propose LRG as a potential biomarker in certain inflammatory conditions as their data suggests that LRG is an up-regulated APP in response to stimulation of hepatocytes by proinflammatory cytokines 35. While we observed no significant change of LRG during acute infection, we observed a 103% increase during sustained chronic infection. Another study using iTRAQ platform also identified LRG as a novel inflammatory serum biomarker for autoimmune diseases 36. Our results, along with other published studies, provide evidence that while LRG may not be a biomarker for a specific disease, but a more universal marker for inflammation.
Histidine rich glycoprotein (HRG) is a negative APP 37–39 and has been documented to decrease in the acute states of AIDS 40. Our observation of a 45% decrease of HRG during acute infection, while rebounding to baseline levels in chronic infection supports these findings. HRG also interacts with several components of the complement system and assist in maintaining normal immune function, however, it’s full affect on the complement cascade is unclear 41.
We posit a relationship between plasminogen activity and vitronectin in affecting HIV-1 infection. The up-regulation of vitronectin during the chronic phase of SIV infection may indicate an effect on cell-associated viral transfers based on interactions with integrins and in cell adhesion as it has been shown that attachment of virus to its host cells is a critical early event for infection 42. It was previously shown that urokinase-type plasminogen activator (uPA) inhibits HIV-1 replication in macrophages and that this inhibition is directly linked to vitronectin-mediated cell adhesion 43. uPA activates plasminogen into the active serine protease plasmin 44 and both uPA and it’s receptor are expressed in immunocytes 45 and affect inflammatory processes 46. That plasminogen levels decreased without change in vitronectin during the acute phase of infection with both being upregulated during the chronic phase of infection, further supports the observation that plasminogen activator is dependent on vitronectin activity 43.
Other differentially expressed proteins identified in this study is comprised of several members of apolipoprotein family, hemopexin, vitamin D binding protein (VDBP), fibronectin, and vitamin K dependent Protein S, all which are also primarily made in the liver in response to inflammation. However, we observed changes in different directions e.g., no change from baseline to acute phase and increase as infection progressed to chronic phase or decrease during acute phase and restoration to pre-infection levels later in infection. These changes indicate that various components of organism’s response to viral infection are differentially regulated and defining a pattern will help us better understand the course of virus survival from immunesurveillance.
Interestingly most proteins of the complement cascade are down-regulated or not changed during acute phase however they are significantly up-regulated in chronic phase, which suggests that during initial infection, the complement cascade is suppressed by the virus and re-bounds as infection chronically progresses. The complement inflammatory cascade is part of the innate immune response bridging with acquired immunity by enhancing antibody responses and immunologic memory, lysing foreign cells and clearing immune complexes and apoptotic cells. Activation of the complement cascade is initiated through three pathways: classical, alternative and lectin. All pathways meet at the “hub” point, which is C3 and it, is cleaved to C3a and C3b triggering downstream reactions to produce anaphylatoxins C5b and C5a. The latter subsequently triggers formation of Membrane Attack Complex (MAC). In addition, C3 shows multiple pro-inflammatory functions involving histamine release from mast cells, smooth muscle contraction, increased vascular permeability 47, chemotaxis of dendritic cells and mast cells 48.
Complement C3 also modulates adaptive immunity at various levels, including cytokine release, T-cell proliferation, regulatory T cell development and B cell activation/differentiation 49–51. It has been reported that C3b and C3c can also control antigen proteolysis, which could be related to C3-mediated enhancement of antigen presentation, thus, aiding in the establishment of a specific adaptive immune response 52.
Complement components C5b, C6, C7, C8 and C9 create the MAC. Out of these five proteins we observed up-regulation of C6, C8 and C9, as well as C5, which is directly upstream of this complex. The host immune system may up-regulate C5 convertase to initiate formations of more MACs in response to viral infection, however we did not observe increased levels of C5b fragment which initiate formation of MAC. Because MAC is formed in a 1:1:1:1:1 ratio, up-regulation of C6 and C8 might be a confounding effect and seems unlikely that over-expression of these two elements will increase MAC formation. On the other hand the net level of any component circulating in blood might be affected by the fact that other cells outside of liver also produce some complement proteins.
Function and role of complement C4a has been mostly studies in the context of lupus erythromatosus and deficiency in either C4a or C4b is associated with autoimmunity. Recent study showed that patients with AIDS have persistent elevated levels of C4a which does not change with ART treatment while C4b levels remain unchanged 53. In our current approach using a monkey model, we report that levels of C4a increase 91% in chronic infection, which correlates with plasma samples from humans 18, 25, the biological consequence of this observation and previous reports that C4a, is strong inhibitor of blood monocytes chemotaxis at concentrations as low as 10(−16) mol/L 54 remains unclear.
Ceruloplasmin is a 132 kDa protein mostly synthesized in the liver, but also in the brain 55. It is a copper containing protein with ferroxidase activity oxidizing Fe+2 to less toxic Fe+3, which contributes to homeostasis. Defense against oxidant stress by scavenging superoxide radicals and oxidation of low density lipoproteins and its compensatory role to oxidative stress 55 has been postulated as beneficial in several pathological conditions including HIV infection 56. Unchanged (statistical analysis) or lower (Western blot analysis) levels of ceruloplasmin during acute phase of SIV infection may help the virus to escape from immune surveillance. Increased levels of ceruloplasmin (confirmed by statistical and Western blot analyses), which could be linked to increasing inflammation during late stages of disease, were also observed in sera of patients with HIV-associated dementia (HAD).
Gelsolin and its secretory form plasma gelsolin (pGELS) are distributed throughout body with body fluids. pGELS is not synthesized in liver and very little is known about regulation of its expression. Clinical studies have shown that pGELS is linked to a number of pathological conditions such as inflammation, cancer and amyloidosis 57. Increasing experimental evidence indicates that pGELS plays an important role in acute as well as chronic inflammatory responses although both mechanisms are different. For example in chronic rheumatoid arthritis there is crosstalk between cytoskeleton and immune system mediated by cytokines and transcriptional factors 58 involving pGELS. Spinardi and Witke concluded that modulation of pGELS expression might have beneficial effects such as inhibition of pGELS during acute inflammation and boosting pGELS in chronic inflammatory response 57. According to this interpretation, our observed down-regulation in gelsolin levels during acute phase may reflect host’s effort to combat viral infection.
Conclusions
Successful validation by Western blot of iTRAQ results supports its use for plasma proteomic analysis of SIV infection of rhesus macaques. SIV infection parallels the course of HIV disease in humans and as such provides valuable insight into the effect of the virus on the immune system. The results herein can be separated from other confounding factors such as co-infections, drug toxicity, multi-drug abuse, amongst others, which is very difficult to avoid when using samples from patients enrolled in clinical studies. Our data supports the notion that components of the complement cascade are differentially expressed throughout acute and chronic stages of infection and reflect the host’s response to microbial disease. Moreover, positive and negative acute phase proteins appears to best elucidate how the infected human host reacts, albeit unsuccessfully, to regulate ongoing SIV (HIV-1) infection.
Supplementary Material
Acknowledgments
The authors would like to thank University of Minnesota Center for Mass Spectrometry and Proteomics for assistance in data acquisition especially Drs. Thomas Krick and LeeAnn Higgins for in-depth discussion and consultation. We acknowledge NIDA grant DA 04498 which supported the monkey aspects of these studies that helps support all animal research at the Yerkes National Primate Research Center, located at Emory University in Atlanta, GA. This work was supported in part by the following grants: P20 RR15635, 1 P01NS043985-01, P20DA026146, 5R01NS36126, PO1NS31492, 2R01NS034239, P20RR15635, P30AI42845, P01MH64570, and 1 R01MH083516.
Footnotes
Supporting Information. Supporting information available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Pedersen C. Infection with human immunodeficiency virus type-1. Seroconversion chronic infection and the development of AIDS. Dan Med Bull. 1994;41(1):12–22. [PubMed] [Google Scholar]
- 2.Daar ES, Pilcher CD, Hecht FM. Clinical presentation and diagnosis of primary HIV-1 infection. Curr Opin HIV AIDS. 2008;3(1):10–5. doi: 10.1097/COH.0b013e3282f2e295. [DOI] [PubMed] [Google Scholar]
- 3.Ford ES, Puronen CE, Sereti I. Immunopathogenesis of asymptomatic chronic HIV Infection: the calm before the storm. Curr Opin HIV AIDS. 2009;4(3):206–14. doi: 10.1097/COH.0b013e328329c68c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mothe B, Ibarrondo J, Llano A, Brander C. Virological, immune and host genetics markers in the control of HIV infection. Dis Markers. 2009;27(3):105–20. doi: 10.3233/DMA-2009-0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roberts ES, Zandonatti MA, Watry DD, Madden LJ, Henriksen SJ, Taffe MA, Fox HS. Induction of pathogenic sets of genes in macrophages and neurons in NeuroAIDS. Am J Pathol. 2003;162(6):2041–57. doi: 10.1016/S0002-9440(10)64336-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox HS, Gold LH, Henriksen SJ, Bloom FE. Simian immunodeficiency virus: a model for neuroAIDS. Neurobiol Dis. 1997;4(3–4):265–74. doi: 10.1006/nbdi.1997.0159. [DOI] [PubMed] [Google Scholar]
- 7.Kapadia F, Vlahov D, Donahoe RM, Friedland G. The role of substance abuse in HIV disease progression: reconciling differences from laboratory and epidemiologic investigations. Clin Infect Dis. 2005;41(7):1027–34. doi: 10.1086/433175. [DOI] [PubMed] [Google Scholar]
- 8.Joag SV. Primate models of AIDS. Microbes Infect. 2000;2(2):223–9. doi: 10.1016/s1286-4579(00)00266-5. [DOI] [PubMed] [Google Scholar]
- 9.Pendyala G, Trauger SA, Siuzdak G, Fox HS. Quantitative plasma proteomic profiling identifies the vitamin E binding protein afamin as a potential pathogenic factor in SIV induced CNS disease. J Proteome Res. 9(1):352–8. doi: 10.1021/pr900685u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Auer J, Camoin L, Guillonneau F, Rigourd V, Chelbi ST, Leduc M, Laparre J, Mignot TM, Vaiman D. Serum profile in preeclampsia and intra-uterine growth restriction revealed by iTRAQ technology. J Proteomics. 73(5):1004–17. doi: 10.1016/j.jprot.2009.12.014. [DOI] [PubMed] [Google Scholar]
- 11.Kolla V, Jeno P, Moes S, Tercanli S, Lapaire O, Choolani M, Hahn S. Quantitative proteomics analysis of maternal plasma in Down syndrome pregnancies using isobaric tagging reagent (iTRAQ) J Biomed Biotechnol. 2010:952047. doi: 10.1155/2010/952047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee HJ, Na K, Choi EY, Kim KS, Kim H, Paik YK. Simple method for quantitative analysis of N-linked glycoproteins in hepatocellular carcinoma specimens. J Proteome Res. 9(1):308–18. doi: 10.1021/pr900649b. [DOI] [PubMed] [Google Scholar]
- 13.Latterich M, Abramovitz M, Leyland-Jones B. Proteomics: new technologies and clinical applications. Eur J Cancer. 2008;44(18):2737–41. doi: 10.1016/j.ejca.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 14.Donahoe RM, O’Neil SP, Marsteller FA, Novembre FJ, Anderson DC, Lankford-Turner P, McClure HH. Probable deceleration of progression of Simian AIDS affected by opiate dependency: studies with a rhesus macaque/SIVsmm9 model. J Acquir Immune Defic Syndr. 2009;50(3):241–9. doi: 10.1097/QAI.0b013e3181967354. [DOI] [PubMed] [Google Scholar]
- 15.Akkina SK, Zhang Y, Nelsestuen GL, Oetting WS, Ibrahlm HN. Temporal stability of the urinary proteome after kidney transplant: more sensitive than protein composition? J Proteome Res. 2009;8(1):94–103. doi: 10.1021/pr800646j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shilov IV, Seymour SL, Patel AA, Loboda A, Tang WH, Keating SP, Hunter CL, Nuwaysir LM, Schaeffer DA. The Paragon Algorithm, a next generation search engine that uses sequence temperature values and feature probabilities to identify peptides from tandem mass spectra. Mol Cell Proteomics. 2007;6(9):1638–55. doi: 10.1074/mcp.T600050-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Oberg AL, Mahoney DW, Eckel-Passow JE, Malone CJ, Wolfinger RD, Hill EG, Cooper LT, Onuma OK, Spiro C, Therneau TM, Bergen HR. 3rd, Statistical analysis of relative labeled mass spectrometry data from complex samples using ANOVA. J Proteome Res. 2008;7(1):225–33. doi: 10.1021/pr700734f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wiederin J, Rozek W, Duan F, Ciborowski P. Biomarkers of HIV-1 associated dementia: proteomic investigation of sera. Proteome Science. 2009;7:8–20. doi: 10.1186/1477-5956-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A. 2003;100(16):9440–5. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krogh M, Liu Y, Waldemarson S, Valastro B, James P. Analysis of DIGE data using a linear mixed model allowing for protein-specific dye effects. Proteomics. 2007;7(23):4235–44. doi: 10.1002/pmic.200700339. [DOI] [PubMed] [Google Scholar]
- 21.Tonge R, Shaw J, Middleton B, Rowlinson R, Rayner S, Young J, Pognan F, Hawkins E, Currie I, Davison M. Validation and development of fluorescence two-dimensional differential gel electrophoresis proteomics technology. Proteomics. 2001;1(3):377–96. doi: 10.1002/1615-9861(200103)1:3<377::AID-PROT377>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 22.Stephens A, Hannan NJ, Rainczuk A, Meehan K, Chen JI, Nicholls P, Rombauts L, Stanton PG, Robertson DM, Salamonsen L. Post-Translational Modifications and Protein-Specific Isoforms in Endometriosis Revealed by 2D DIGE. J Proteome Res. doi: 10.1021/pr901131p. [DOI] [PubMed] [Google Scholar]
- 23.Wu WW, Wang G, Baek SJ, Shen RF. Comparative study of three proteomic quantitative methods, DIGE, cICAT, and iTRAQ, using 2D gel- or LC-MALDI TOF/TOF. J Proteome Res. 2006;5(3):651–8. doi: 10.1021/pr050405o. [DOI] [PubMed] [Google Scholar]
- 24.Chong PK, Gan CS, Pham TK, Wright PC. Isobaric tags for relative and absolute quantitation (iTRAQ) reproducibility: Implication of multiple injections. J Proteome Res. 2006;5(5):1232–40. doi: 10.1021/pr060018u. [DOI] [PubMed] [Google Scholar]
- 25.Rozek W, Horning J, Anderson J, Ciborowski P. Sera Proteomic Biomarker Profiling in HIV-1 Infected Subjects with Cognitive Impairment. Proteomics - Clinical Applications. 2008;2:1498–1507. doi: 10.1002/prca.200780114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rozek W, Ricardo-Dukelow M, Holloway S, Gendelman HE, Wojna V, Melendez L, Ciborowski P. Cerebrospinal fluid proteomic profiling of HIV-1-infected patients with cognitive impairment. J Proteome Res. 2007 doi: 10.1021/pr070220c. in press. [DOI] [PubMed] [Google Scholar]
- 27.Pendyala G, Trauger SA, Kalisiak E, Ellis RJ, Siuzdak G, Fox HS. Cerebrospinal fluid proteomics reveals potential pathogenic changes in the brains of SIV-infected monkeys. J Proteome Res. 2009;8(5):2253–60. doi: 10.1021/pr800854t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciborowski P. Biomarkers of HIV-1-associated neurocognitive disorders: challenges of proteomic approaches. Biomarkers in Medicine. 2009;3(6):771–785. doi: 10.2217/bmm.09.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cordelier P, Strayer DS. Conditional expression of alpha1-antitrypsin delivered by recombinant SV40 vectors protects lymphocytes against HIV. Gene Ther. 2003;10(26):2153–6. doi: 10.1038/sj.gt.3302113. [DOI] [PubMed] [Google Scholar]
- 30.Cordelier P, Strayer DS. Mechanisms of alpha1-antitrypsin inhibition of cellular serine proteases and HIV-1 protease that are essential for HIV-1 morphogenesis. Biochim Biophys Acta. 2003;1638(3):197–207. doi: 10.1016/s0925-4439(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 31.Munch J, Standker L, Adermann K, Schulz A, Schindler M, Chinnadurai R, Pohlmann S, Chaipan C, Biet T, Peters T, Meyer B, Wilhelm D, Lu H, Jing W, Jiang S, Forssmann WG, Kirchhoff F. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell. 2007;129(2):263–75. doi: 10.1016/j.cell.2007.02.042. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro L, Pott GB, Ralston AH. Alpha-1-antitrypsin inhibits human immunodeficiency virus type 1. Faseb J. 2001;15(1):115–122. doi: 10.1096/fj.00-0311com. [DOI] [PubMed] [Google Scholar]
- 33.Wang H, Clouthier SG, Galchev V, Misek DE, Duffner U, Min CK, Zhao R, Tra J, Omenn GS, Ferrara JL, Hanash SM. Intact-protein-based High-resolution Three-dimensional Quantitative Analysis System for Proteome Profiling of Biological Fluids. Mol Cell Proteomics. 2005;4(5):618–625. doi: 10.1074/mcp.M400126-MCP200. [DOI] [PubMed] [Google Scholar]
- 34.Okano T, Kondo T, Kakisaka T, Fujii K, Yamada M, Kato H, Nishimura T, Gemma A, Kudoh S, Hirohashi S. Plasma proteomics of lung cancer by a linkage of multidimensional liquid chromatography and two-dimensional difference gel electrophoresis. Proteomics. 2006;6(13):3938–48. doi: 10.1002/pmic.200500883. [DOI] [PubMed] [Google Scholar]
- 35.Shirai R, Hirano F, Ohkura N, Ikeda K, Inoue S. Up-regulation of the expression of leucine-rich alpha(2)-glycoprotein in hepatocytes by the mediators of acute-phase response. Biochem Biophys Res Commun. 2009;382(4):776–9. doi: 10.1016/j.bbrc.2009.03.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Serada S, Fujimoto M, Ogata A, Terabe F, Hirano T, Iijima H, Shinzaki S, Nishikawa T, Ohkawara T, Iwahori K, Ohguro N, Kishimoto T, Naka T. iTRAQ-based proteomic identification of leucine-rich alpha-2 glycoprotein as a novel inflammatory biomarker in autoimmune diseases. Ann Rheum Dis. 69(4):770–4. doi: 10.1136/ard.2009.118919. [DOI] [PubMed] [Google Scholar]
- 37.Jones AL, Hulett MD, Parish CR. Histidine-rich glycoprotein: A novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol Cell Biol. 2005;83(2):106–18. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 38.Saigo K, Yoshida A, Ryo R, Yamaguchi N, Leung LL. Histidine-rich glycoprotein as a negative acute phase reactant. Am J Hematol. 1990;34(2):149–50. doi: 10.1002/ajh.2830340212. [DOI] [PubMed] [Google Scholar]
- 39.Jespersen J, Gram J, Bach E. A sequential study of plasma histidine-rich glycoprotein and plasminogen in patients with acute myocardial infarction and deep vein thrombosis. Thromb Haemost. 1984;51(1):99–102. [PubMed] [Google Scholar]
- 40.Morgan WT. Serum histidine-rich glycoprotein levels are decreased in acquired immune deficiency syndrome and by steroid therapy. Biochem Med Metab Biol. 1986;36(2):210–3. doi: 10.1016/0885-4505(86)90127-1. [DOI] [PubMed] [Google Scholar]
- 41.Manderson GA, Martin M, Onnerfjord P, Saxne T, Schmidtchen A, Mollnes TE, Heinegard D, Blom AM. Interactions of histidine-rich glycoprotein with immunoglobulins and proteins of the complement system. Mol Immunol. 2009;46(16):3388–98. doi: 10.1016/j.molimm.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 42.Lafrenie RM, Lee SF, Hewlett IK, Yamada KM, Dhawan S. Involvement of integrin alphavbeta3 in the pathogenesis of human immunodeficiency virus type 1 infection in monocytes. Virology. 2002;297(1):31–8. doi: 10.1006/viro.2002.1399. [DOI] [PubMed] [Google Scholar]
- 43.Elia C, Cassol E, Sidenius N, Blasi F, Castagna A, Poli G, Alfano M. Inhibition of HIV replication by the plasminogen activator is dependent on vitronectin-mediated cell adhesion. J Leukoc Biol. 2007;82(5):1212–20. doi: 10.1189/jlb.0407251. [DOI] [PubMed] [Google Scholar]
- 44.Dano K, Andreasen PA, Grondahl-Hansen J, Kristensen P, Nielsen LS, Skriver L. Plasminogen activators, tissue degradation, and cancer. Adv Cancer Res. 1985;44:139–266. doi: 10.1016/s0065-230x(08)60028-7. [DOI] [PubMed] [Google Scholar]
- 45.Blasi F, Vassalli JD, Dano K. Urokinase-type plasminogen activator: proenzyme, receptor, and inhibitors. J Cell Biol. 1987;104(4):801–4. doi: 10.1083/jcb.104.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chavakis T, Kanse SM, May AE, Preissner KT. Haemostatic factors occupy new territory: the role of the urokinase receptor system and kininogen in inflammation. Biochem Soc Trans. 2002;30(2):168–73. doi: 10.1042/. [DOI] [PubMed] [Google Scholar]
- 47.Stokol T, O’Donnell P, Xiao L, Knight S, Stavrakis G, Botto M, von Andrian UH, Mayadas TN. C1q governs deposition of circulating immune complexes and leukocyte Fcgamma receptors mediate subsequent neutrophil recruitment. J Exp Med. 2004;200(7):835–46. doi: 10.1084/jem.20040501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutzmer R, Lisewski M, Zwirner J, Mommert S, Diesel C, Wittmann M, Kapp A, Werfel T. Human monocyte-derived dendritic cells are chemoattracted to C3a after up-regulation of the C3a receptor with interferons. Immunology. 2004;111(4):435–43. doi: 10.1111/j.1365-2567.2004.01829.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Morgan BP, Marchbank KJ, Longhi MP, Harris CL, Gallimore AM. Complement: central to innate immunity and bridging to adaptive responses. Immunol Lett. 2005;97(2):171–9. doi: 10.1016/j.imlet.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 50.Kemper C, Atkinson JP. T-cell regulation: with complements from innate immunity. Nat Rev Immunol. 2007;7(1):9–18. doi: 10.1038/nri1994. [DOI] [PubMed] [Google Scholar]
- 51.Carroll MC. The complement system in regulation of adaptive immunity. Nat Immunol. 2004;5(10):981–6. doi: 10.1038/ni1113. [DOI] [PubMed] [Google Scholar]
- 52.Villiers CL, Cretin F, Lefebvre N, Marche PN, Villiers MB. A new role for complement C3: regulation of antigen processing through an inhibitory activity. Mol Immunol. 2008;45(13):3509–16. doi: 10.1016/j.molimm.2008.05.015. [DOI] [PubMed] [Google Scholar]
- 53.Stricker RB, Savely VR, Motanya NC, Giclas PC. Complement split products c3a and c4a in chronic lyme disease. Scand J Immunol. 2009;69(1):64–9. doi: 10.1111/j.1365-3083.2008.02191.x. [DOI] [PubMed] [Google Scholar]
- 54.Tsuruta T, Yamamoto T, Matsubara S, Nagasawa S, Tanase S, Tanaka J, Takagi K, Kambara T. Novel function of C4a anaphylatoxin. Release from monocytes of protein which inhibits monocyte chemotaxis. Am J Pathol. 1993;142(6):1848–57. [PMC free article] [PubMed] [Google Scholar]
- 55.Loeffler DA, LeWitt PA, Juneau PL, Sima AA, Nguyen HU, DeMaggio AJ, Brickman CM, Brewer GJ, Dick RD, Troyer MD, Kanaley L. Increased regional brain concentrations of ceruloplasmin in neurodegenerative disorders. Brain Res. 1996;738(2):265–74. doi: 10.1016/s0006-8993(96)00782-2. [DOI] [PubMed] [Google Scholar]
- 56.Sundaram M, Saghayam S, Priya B, Venkatesh KK, Balakrishnan P, Shankar EM, Murugavel KG, Solomon S, Kumarasamy N. Changes in antioxidant profile among HIV-infected individuals on generic highly active antiretroviral therapy in southern India. Int J Infect Dis. 2008;12(6):e61–6. doi: 10.1016/j.ijid.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 57.Spinardi L, Witke W. Gelsolin and diseases. Subcell Biochem. 2007;45:55–69. doi: 10.1007/978-1-4020-6191-2_3. [DOI] [PubMed] [Google Scholar]
- 58.Wagner M. Growth factor control of autophagy. Nat Cell Biol. 2005;7(3):212. doi: 10.1038/ncb0305-212. [DOI] [PubMed] [Google Scholar]
- 59.Bruder C, Hagleitner M, Darlington G, Mohsenipour I, Wurzner R, Hollmuller I, Stoiber H, Lass-Florl C, Dierich MP, Speth C. HIV-1 induces complement factor C3 synthesis in astrocytes and neurons by modulation of promoter activity. Mol Immunol. 2004;40(13):949–61. doi: 10.1016/j.molimm.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 60.Pendyala G, Fox HS. Proteomic and metabolomic strategies to investigate HIV-associated neurocognitive disorders. Genome Med. 2(3):22. doi: 10.1186/gm143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stoiber H, Soederholm A, Wilflingseder D, Gusenbauer S, Hildgartner A, Dierich MP. Complement and antibodies: a dangerous liaison in HIV infection? Vaccine. 2008;26 (Suppl 8):I79–85. doi: 10.1016/j.vaccine.2008.11.050. [DOI] [PubMed] [Google Scholar]
- 62.Rearden A, Hurford R, Luu N, Kieu E, Sandoval M, Perez-Liz G, Del Valle L, Powell H, Langford TD. Novel expression of PINCH in the central nervous system and its potential as a biomarker for human immunodeficiency virus-associated neurodegeneration. J Neurosci Res. 2008;86(11):2535–42. doi: 10.1002/jnr.21701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Congote LF. The C-terminal 26-residue peptide of serpin A1 is an inhibitor of HIV-1. Biochem Biophys Res Commun. 2006;343(2):617–22. doi: 10.1016/j.bbrc.2006.02.190. [DOI] [PubMed] [Google Scholar]
- 64.Weivoda S, Andersen JD, Skogen A, Schlievert PM, Fontana D, Schacker T, Tuite P, Dubinsky JM, Jemmerson R. ELISA for human serum leucine-rich alpha-2-glycoprotein-1 employing cytochrome c as the capturing ligand. J Immunol Methods. 2008;336(1):22–9. doi: 10.1016/j.jim.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Alfano M, Sidenius N, Blasi F, Poli G. The role of urokinase-type plasminogen activator (uPA)/uPA receptor in HIV-1 infection. J Leukoc Biol. 2003;74(5):750–6. doi: 10.1189/jlb.0403176. [DOI] [PubMed] [Google Scholar]
- 66.Sporer B, Koedel U, Popp B, Paul R, Pfister HW. Evaluation of cerebrospinal fluid uPA, PAI-1, and soluble uPAR levels in HIV-infected patients. J Neuroimmunol. 2005;163(1–2):190–4. doi: 10.1016/j.jneuroim.2005.02.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.