Summary
The Notch pathway is a crucial cell-fate regulator in the developing heart. Attention in the past centered on Notch function in cardiomyocytes. However, recent advances demonstrate that region-specific endocardial Notch activity orchestrates the patterning and morphogenesis of cardiac chambers and valves through regulatory interaction with multiple myocardial and neural crest signals. Notch also regulates cardiomyocyte proliferation and differentiation during ventricular chamber development and is required for coronary vessel specification. Here, we review these data and highlight disease connections, including evidence that Notch-Hey-Bmp2 interplay impacts adult heart valve disease and that Notch contributes to cardiac arrhythmia and preexcitation syndromes.
Introduction
Notch is an evolutionarily conserved signaling pathway that regulates cell-fate specification, differentiation, and patterning. Mutations of Notch ligands and receptors are implicated in numerous congenital and acquired human diseases (reviewed in Garg, 2006; Chabriat et al., 2009). Notch acts locally, specifying individual fates among a group of equivalent neighboring cells or directing a field of cells towards a given developmental fate. Notch activity is crucial in organs with complex architecture, such as the heart, that requires the coordinated development of multiple parts. Functional studies in vertebrates demonstrate that Notch is required for cardiovascular development (reviewed in Kokubo et al., 2005; High and Epstein, 2008; Niessen and Karsan, 2008; MacGrogan et al., 2010) and its importance for this process in humans is demonstrated by the fact that mutations in several Notch signaling elements underlie congenital cardiovascular disease (High and Epstein, 2008; Macgrogan et al., 2011). Despite the knowledge gained in the past years, the molecular mechanisms and cellular processes that Notch regulates in the developing cardiovascular system remain only partially understood.
This review focuses on recent advances in the field of Notch signaling in cardiac development, and discusses them in the context of the “developmental logic of Notch function” (Artavanis-Tsakonas and Muskavitch, 2010) and its role as a cell-fate regulatory and patterning signal. We begin by describing the elements of the Notch pathway and their expression during mouse cardiac development. We then discuss how Notch signaling patterns the embryonic endocardium, enabling region-specific differentiation and critical interactions of the endocardium (or its derived mesenchyme) with other cardiac tissues (cardiac neural crest, myocardium), so that specialized structures (cardiac valves and chambers) are generated. We also discuss the importance of Notch in cell fate specification during coronary vessel formation and the implication of Notch in cardiac pathologies, including aortic valve disease, cardiomyopathy and abnormalities of cardiac conduction.
The Notch signaling pathway
Notch proteins are single-pass transmembrane receptors with large extracellular regions (NECD) composed of 29–36 tandem epidermal growth factor like repeats, a shorter membrane-spanning portion, and an intracellular domain (NICD), which contains, among other motifs, a transcriptional activation domain. When released from the cell membrane, NICD can function as a transcription factor (Kopan, 2002). Four Notch proteins (Notch1–Notch4) have been identified in mammals (Kopan and Ilagan, 2009; Figure 1). Notch proteins are processed in the Golgi by proteolytic cleavage by a furin-like convertase (Logeat et al., 1998; S1 cleavage site, Figure 1). Modified Notch is then targeted to the cell surface as a heterodimer held together by non-covalent interactions. Once in the membrane, the NECD is available to interact with membrane-bound ligands of the Delta or Serrate/Jagged families expressed by neighboring cells, and cell-cell contact is required for signaling (Figure 1). Productive ligand-receptor interaction depends on the activity of E3 ubiquitin ligases such as mind bomb-1, which in the signaling cell ubiquitylates the ligand and promotes its endocytosis (Itoh et al., 2003). This event facilitates S2 cleavage of the receptor (Figure 1). The remaining Notch fragment becomes susceptible to cleavage by γ-secretase (at the S3 site), resulting in release of NICD, which translocates to the nucleus of the receiving cell (Kopan and Ilagan, 2009). In the nucleus, NICD binds directly to the transcription factor RBPJK/CSL/Su(H) (Jarriault et al., 1995; Figure 1). NICD–RBPJK binding displaces corepressors that repress target genes in the absence of Notch signaling, and allows recruitment of the transcriptional co-activator Mastermind-like (MAML). Formation of the RBPJK-NICD-MAML ternary complex leads to direct transcriptional activation of target genes, including those encoding basic-helix-loop-helix (bHLH) repressor transcription factors of the Hes and Hey (HESR) families (Iso et al., 2003). The spectrum of immediate Notch targets is large and several genes may be activated in parallel, including those encoding repressor transcription factors like Snail1 (Timmerman et al., 2004; Sahlgren et al., 2008) or p21 (Rangarajan et al., 2001), or c-Myc, with a context-dependent activator or repressor function (Weng et al., 2006). Other genes involved in a variety of functions are also regulated by Notch (reviewed in Andersson et al., 2011).
Figure 1. The Notch pathway.
Membrane-bound Notch ligands (Dll1,3,4 and Jag1,2) are characterized by a Delta/Serrate/Lag2 (DSL) motif (yellow) located in the extracellular domain. The Notch receptor (Notch1-4) is processed at the S1 site by a furin protease, sugar-modified by Fringe in the Golgi, and inserted into the membrane as a heterodimer with a large extracellular domain (NECD). Ligand-receptor interaction leads to two consecutive cleavage events (at S2 and S3 sites), respectively carried out by an ADAM protease and presenilin, which release the Notch intracellular domain (NICD). NICD translocates to the nucleus and forms a transcriptional activation complex after binding to Mastermind (MAML) and CSL/RBPJk/Su(H). This ternary complex activates the transcription of a set of target genes including Hes, Hey and others.
Cardiac development and Notch activity: a general map
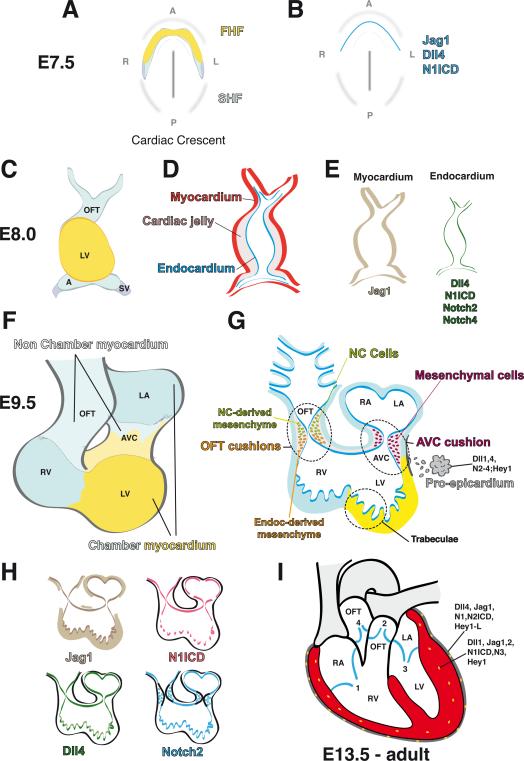
Cardiac morphogenesis in the mouse begins at around E7.5 when, as the embryo folds, a subset of mesodermal cells progressively distributes antero-laterally to form the cardiac crescent at ~8.0 (Figure 2A). This crescent contains two populations of precardiac cells, the first and second heart fields (FHF and SHF, Figure 2A), that include progenitors of the first cardiac tissues, the myocardium and endocardium. At this stage, N1ICD (Del Monte et al., 2007) and the Notch ligands Dll4 (Duarte et al., 2004) and Jag1 (our unpublished data) are detected in the primitive E7.5 endocardium (Figure 2B) suggesting active Notch signaling and early endocardial specification.
Figure 2. Cardiac development and Notch pathway expression in the mouse.
(A) E7.5, cardiac crescent. Progenitor cell populations of the first and second heart fields (FHF, yellow; SHF, light blue) have fused in the midline of the embryo. (B) Jag1, Dll4 and N1ICD are expressed in the developing endocardial tubes (blue). (C) E8.0, heart tube stage. The approximate contributions of FHF and SHF are shown. (D) The heart tube has an outer myocardial layer and an inner endocardial endothelium separated by cardiac jelly (gray). (E) Jag1 is expressed in the myocardium while Dll4, N1ICD, Notch2 and Notch4 are expressed in the endocardium. (F) E9.5, looped heart. AVC formation separates the atria from the ventricles. The approximate contributions of FHF and SHF are shown. (G) Valve primordia are formed between E9.5–10.5. The diagram shows the contributions of endocardium-derived mesenchyme to the AVC and of endocardium-derived plus neural crest (NC) mesenchyme to the OFT. Ventricular chamber development begins with trabeculae formation in the left and right ventricles. At this stage, the proepicardium arises in the inflow region of the heart and expresses Dll1,4; N2–4 and Hey1. (H) In the E9.5–10.5 heart, Jag1 is expressed in chamber myocardium and endocardium; Dll4 and N1ICD are expressed in valve and atrial endocardium and at the base of trabecular endocardium. Notch2 is expressed in chamber endocardium and valve mesenchyme. (I) E13.5-adult heart. The four chambers and four valves are shown (1, tricuspid valve; 2, pulmonary valve; 3, mitral valve; 4, aortic valve). The epicardium covering the heart and expressing various Notch elements is shown in gray and the coronary vessels in yellow. Expression of Notch elements in epicardium and coronary arteries is indicated. In all panels the ventral aspect of the heart is shown (anterior to the top). A, atria; AVC, atrio-ventricular canal; LA, left atrium, LV, left ventricle; OFT, outflow tract; RV, right ventricle.
As development proceeds, the bilateral cardiac primordia coalesce and fuse at the embryonic midline to form a single primary heart tube at ~E8.0 (Figure 2C). This primitive heart consists of two layers, an inner endocardium and outer myocardium, separated by an extracellular matrix (ECM) termed cardiac jelly (Figure 2D). At this stage, Jag1 expression is maintained in the endocardium and is turned on in the primitive myocardium (Loomes et al., 1999), while restricted endocardial expression is displayed by Dll4, Notch2 (Timmerman et al., 2004), N1ICD (Del Monte et al., 2007), and Notch4 (Uyttendaele et al., 1996; Figure 2E).
The heart tube grows by incorporating SHF cells into the anterior (arterial) and posterior (venous) poles. The heart tube simultaneously initiates a characteristic R-loop, or rightward bend, and the initial anterior-posterior polarity transforms into a right-left patterning (Figure 2F). In the E9.5 mouse heart, N1ICD expression is relatively uniform in prospective valve endocardium, namely the endocardium lining the atrio-ventricular canal (AVC) and the outflow tract (OFT, Figure 2G,H). In contrast, N1ICD expression in the ventricles is restricted to the endocardium at the base of the developing trabeculae (Del Monte et al., 2007), similar to Dll4 (Grego-Bessa et al., 2007 and Figure 2G, H). Jag1 is strongly expressed in AVC endocardium and myocardium (Loomes et al., 1999), while strong Notch2 expression is detected in chamber endocardium and transformed mesenchymal cells (Loomes et al., 2002; Figure 2G, H).
At E9.5, a third cardiac tissue, the epicardium, develops through the addition of cells originating from a transient structure called the proepicardium, which arises from splanchnic mesoderm posterior to the inflow region of the heart and expresses Dll1, 4, Jag1, Notch2–4 and Hey1 (Del Monte et al., 2011; Grieskamp et al., 2011 and Figure 2G). Epicardial cells, which express Dll4, Jag1, Notch1, N2ICD, Notch3 and Hey1-L (Del Monte et al., 2011; Grieskamp et al., 2011), migrate and cover the entire myocardial surface by E11.0. Epicardial cells then undergo epithelial-mesenchymal transformation (EMT) to generate subepicardial mesenchyme, expressing Dll4, Jag1, N1ICD and N2ICD (Del Monte et al., 2011 and Figure 2G), which differentiates into various lineages, including coronary smooth muscle cells and cardiac interstitial cells (Mikawa and Gourdie, 1996; Christoffels et al., 2009).
Epicardium, myocardium and endocardium are intimately associated with one another and their interactions result in a highly coordinated pattern of proliferation and differentiation in the developing heart. In the mature four-chambered heart, the endocardium lines the lumen of the cardiac chambers and contributes to the atrio-ventricular (AV) valves and to a portion of the OFT (semilunar) valves; the myocardium forms the contractile tissue; and the epicardium contributes to the smooth muscle cells of the coronary vasculature (Figure 2I), the subepicardial mesenchyme, part of the AV valves, and the cardiac fibroblasts. Several Notch elements are expressed in dynamic patterns at these later stages and in the adult heart, suggesting that Notch plays complex and reiterated roles in heart morphogenesis and homeostasis.
Endocardial patterning and lateral induction
Notch patterns the early endocardium, which expresses many Notch elements and whose interaction with the myocardium is critical for cardiac development. Studies in mice show that Notch loss-of-function (RBPJk or Notch1 mutants) impairs expression of endocardial markers of chamber and valve tissue. Thus, while early markers of endothelium (CD31/Pecam-1) and myocardium (MLC2v, Irx4) are expressed, transcription of Irx5 and other endocardial markers is abrogated in Notch targeted mutants, indicating that endocardial patterning is lost at a time when myocardial patterning is unaffected (Timmerman et al., 2004; Grego-Bessa et al., 2007; Figure 3A). Gain-of-function experiments, in contrast, show that ectopic N1ICD expression in endocardium throughout the heart extends the uniform N1ICD distribution observed in prospective valve tissue thus expanding valve patterning to ventricular endocardium (Luna-Zurita et al., 2010).
Figure 3. Signals in cardiac patterning.
(A) Top: Wild type. Notch1 activity (red) is found in the endocardium of the AVC, OFT, LA and basal region of trabeculae. Hey1, Hey2 and HeyL are differentially expressed in chamber and AVC endocardium. Myocardial Bmp2 expression (blue) is confined to the AVC and OFT myocardium by Hey1 (green) and Hey2 (brown), expressed in the chambers. Middle: Systemic Notch loss of function (Notch LOF) in RBPJk mutants leads to Hey down-regulation and ectopic Bmp2 expression in the endocardium. Endocardial AVC and chamber patterning is lost. Notch inactivation does not affect myocardial patterning or the myocardial expression domains of Hey1, Hey2 or Bmp2. Bottom: Notch gain-of-function (Notch GOF) by ectopic N1ICD expression in endocardium and myocardium (Nkx2.5-Cre; RosaN1ICD). Expression of Hey1, 2 and HeyL expands throughout the endocardium. Hey1 is expressed throughout the myocardium (green), repressing Bmp2 in the AVC. Valve patterning is lost in the myocardium and expands throughout the endocardium. (B) Pathway establishing chamber vs. valve domains in the developing heart. Cardiogenic signals activate Tbx20 expression, which drives Hey1 and Hey2 and represses Tbx2 expression in chamber myocardium. Hey1 and Hey2 confine Bmp2 expression to prospective valve tissue. Tbx2 and 3 are activated by Bmp2 and restricted to valve territory by Tbx20 and Hey. Forced Tbx2 expression represses Hey genes (dashed lines). Color codes as in (A); abbreviations as in Figure 2.
A unique feature of Notch activity in the heart is that, unlike the situation in the CNS and other tissues, Notch inactivation in the embryonic endocardium results in decreased Dll4 ligand expression (Timmerman et al., 2004; Grego-Bessa et al., 2007), while Notch activation expands Dll4 expression throughout this tissue (Rutenberg et al., 2006; Watanabe et al., 2006; Luna-Zurita et al., 2010). This implies the existence of a Notch-dependent positive feedback loop regulating Dll4 expression in the embryonic endocardium. Furthermore, it suggests that Notch in the endocardium functions by a lateral induction mechanism whereby endocardial cells behave as a developmental field, and are specified together (and not individually) as valve or chamber endocardium. These observations also apply to the vascular endothelium, as the effect of Notch manipulation on Dll4 expression is the same (Luna-Zurita et al., 2010). In the CNS, individual Dll1-expressing cells become neuroblasts and via lateral inhibition, inhibit this fate in neighboring cells that down-regulate ligand expression via a negative feedback loop (Chitnis et al., 1995; de la Pompa et al., 1997).
Establishing the valve territory
The AVC is located between the prospective atrial and ventricular chamber regions (Figure 2F) and is a source of signals promoting formation of the endocardial cushions, precursors of the cardiac valves (Person et al., 2005; Aanhaanen et al., 2011). Bmp2 is an essential patterning signal in AVC myocardium where it activates Tbx2/3 expression (Yamada et al., 2000). Tbx2 is responsible for maintaining a primitive myocardium phenotype in the AVC and throughout the inner curvature of the heart (Christoffels et al., 2004). Cardiac-specific deletion of Bmp2 causes a failure of AVC specification (Ma et al., 2005), and mice lacking Tbx2 show defective AVC patterning and ectopic expression of chamber-specific genes (Harrelson et al., 2004; Aanhaanen et al., 2009). These studies establish a Bmp2-Tbx2/3 regulatory axis specifying AVC myocardium (Figure 3B; Yamada et al., 2000; Ma et al., 2005).
Ectopic expression experiments (Rutenberg et al., 2006; Watanabe et al., 2006; Luna-Zurita et al., 2010) have suggested a critical role for Notch in confining Bmp2 expression to the AVC myocardium (Figure 3A,B). Ectopic N1ICD expression in the myocardium activates the Notch target Hey1 throughout this tissue, which in turn represses Bmp2 and Tbx2 in AVC, reflecting partial (Watanabe et al., 2006) or total (Luna-Zurita et al., 2010) expansion of chamber territory at the expense of the AVC (Figure 3A,B). Tbx20, which represses Tbx2 (Singh et al., 2009), also impacts this pathway and is required for chamber-specific gene expression and differentiation upstream of Hey (Stennard et al., 2005; Kokubo et al., 2007; Singh et al., 2009). The functional requirement for Tbx20 extends to the adult heart, where it regulates ion channel expression (Shen et al., 2011). This regulatory interplay between Tbx20, Tbx2 and Hey transcription factors defines two developmental domains (chambers and valves) in the developing myocardium (Figure 3B).
Although the results from N1ICD gain-of-function experiments in myocardium have been informative (Rutenberg et al., 2006; Watanabe et al., 2006; Luna-Zurita et al., 2010), Notch1 is not normally expressed in the early and mid-gestation myocardium, and inactivation of Notch signaling in RBPJk-targeted mutants does not affect myocardial Hey1, Hey2 or Bmp2 expression (Timmerman et al., 2004; Luna-Zurita et al., 2010). In the endocardium, however, Notch1 does function via Hey genes to repress Bmp2 (Luna-Zurita et al., 2010; Figure 3A,B). Ectopic N1ICD in the endocardium activates Hey1, Hey2 and HeyL (Luna-Zurita et al., 2010) and RBPJk mutant embryos show strongly reduced endocardial Hey gene expression and Bmp2 activation.
Ventricular preexcitation and arrhythmias
At late stages of cardiac development, AVC myocardium partially regresses, although some derivatives contribute to the muscular support of the AV valves, the conduction system including the AV node, and muscular components of the left ventricle. Recent studies suggest that alterations in Notch signaling can modulate AVC myocardial remodeling, resulting in relatively common forms of structural heart disease associated with potentially lethal conduction defects (Aanhaanen et al., 2011; Akazawa and Komuro, 2011; Rentschler et al., 2011).
Normally, the electrical activities of the atria and ventricles are insulated from one another by the non-conducting annulus fibrosus, and impulses generated in the atria can travel to the ventricles only via the relatively slow-conducting AV node. However, in Wolff-Parkinson-White (WPW) syndrome and related disorders, additional muscular pathways can electrically couple atria and ventricles, allowing rapid AV conduction devoid of the physiologic protection provided by the AV node against ventricular tachycardia induced by rapid atrial activity. This leaves affected patients at risk of life-threatening arrhythmias. When NI1CD is expressed in developing mouse myocardium (Mlc2v-Cre driver), all resulting mice develop ventricular pre-excitation that mimics WPW (Rentschler et al., 2011). Interestingly, a related phenotype is seen in mice lacking the AVC patterning transcription factor Tbx2 (Aanhaanen et al., 2011).
These results suggest that ventricular preexcitation can arise from deficient patterning of the AVC-derived myocardium, whereby persistence of AVC myocardium results in strands of myocardial tissue that electrically connect atria and ventricles, explaining the etiology of cardiac pre-excitation syndromes such as WPW. However, details of the underlying developmental mechanisms remain to be explored. For example, although AVC patterning occurs early in heart development (Harrelson et al., 2004; Aanhaanen et al., 2009), Mlc2v-Cre;NICD mice do not appear to have early AVC patterning defects (Rentschler et al., 2011). Persistence of AVC myocardial strands is a late embryonic event, and ventricular pre-excitation is not evident until post-natal life. Furthermore, Tbx2/3 expression is unaffected in Mlc2v-Cre;NICD mice, suggesting that Notch activation does not cause WPW as a result of Tbx2/3 repression. The relationship between Notch and Tbx2/3 at late stages of AVC remodeling remains to be defined, and the mechanisms by which Notch can promote functional bypass tract formation (and whether it does so in humans) will require further research.
Signal integration in cardiac valve development
Cardiac valve formation begins at around E9.5 in the mouse, when myocardial signals from AVC and OFT regions instruct adjacent endocardial cells to undergo EMT. During this process, endocardial cells lose their endothelial cobblestone-like morphology and become migratory mesenchyme, invading the underlying extracellular matrix to form the cushion mesenchyme (reviewed in Hinton and Yutzey, 2011).
In addition to its patterning role in the myocardium, Bmp2 is a crucial EMT-inducing signal that via Alk3 and Alk6 receptors activation in the endocardium, triggers EMT and cushion formation (Gaussin et al., 2002; Ma et al., 2005; Figure 4).
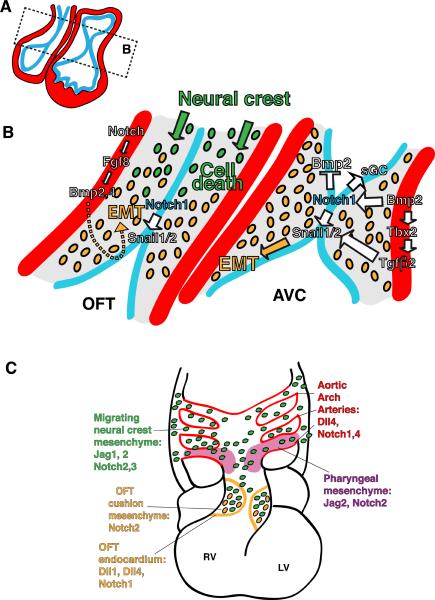
Figure 4. Tissue interactions during cardiac EMT and valve morphogenesis.
(A) Diagram of an E10 looped heart. (B) Detail of the boxed area in (A) highlighting the OFT and AVC endocardial cushions where EMT takes place and valves form. Myocardium, red; endocardium, blue; extracellular matrix, gray. Signals (arrows) from endocardium and myocardium converge to trigger EMT. Migratory NC mesenchyme invades the OFT region. Its interaction with endocardial mesenchyme is crucial for valve morphogenesis. (C) Diagram of an E12.5 heart showing the color-coded expression of Notch elements during OFT and AAA remodeling. NC migrate from the dorsal neural tube around the AAA and toward the OFT cushions. They will also contribute to the SMC of the AAA.
The endocardium is not passive during EMT, but is the source of crucial EMT-inducing signals, including Notch, whose elements are expressed in AVC and OFT endocardium prior to valve formation (Del Monte et al., 2007; Figure 2G,H). Targeted inactivation of RBPJk or Notch1 results in severely hypoplastic endocardial cushions due to impaired EMT (Timmerman et al., 2004). In E9.5 RBPJk mutants, AVC endocardial cells display features of activated pre-migratory cells but fail to detach from one another and invade the cardiac jelly. RBPJk mutants also show defective transforming growth factor beta 2 (Tgfβ2) transcription in endocardium and myocardium, suggesting a non-cell autonomous effect of Notch signaling on Tgfβ2 expression (Timmerman et al., 2004). N1ICD-RBPJK activates the EMT driver Snail1, whose expression is severely reduced in RBPJk mutant AVC and OFT endocardium (Timmerman et al., 2004). Loss of Snail1 expression prevents down-regulation of cadherin-mediated endocardial cell adhesion, and EMT is blocked. Wild-type explants of AVC and OFT tissue are able to undergo EMT when explanted onto a 3D-collagen gel (Markwald et al., 1996). However, explants from Notch1 or RBPJk mutants, or wild-type explants treated with a Notch-signaling inhibitor, do not undergo EMT, indicating that Notch is crucial for this process (Timmerman et al., 2004).
The importance of endocardial Notch signaling for EMT and its role as a signal integrator was demonstrated by Notch1 gain of function studies (Luna-Zurita et al., 2010). Endothelial- and endocardial-specific N1ICD expression in the mouse embryo leads to ectopic expression of Snail1,2 and other EMT-specific genes. In collagen gel explant assays, ventricular endocardial cells from transgenic embryos unexpectedly transform and behave like AVC endocardium, migrating over the gel surface and expressing Snail1,2, Tgfβ2, Periostin and other mesenchyme markers, although they do not invade the gel matrix. This partial, non-invasive EMT behavior becomes fully invasive upon addition of BMP2 to the explant medium, suggesting that EMT is indeed a two-phase process, involving independent regulation of epithelial cell detachment and matrix invasion. Notch1 signaling is attenuated after myocardial Bmp2 deletion, and BMP2-induced ventricular EMT is reduced by Notch inhibition, suggesting that Bmp2 acts upstream of Notch in EMT (Luna-Zurita et al., 2010).
Experiments with endothelial cells have suggested a mechanism of myocardial Bmp2 and endocardial Notch1 signal integration during EMT: Bmp2-Alk3/6 signaling inhibits glycogen synthase kinase-3β (Gsk3β) activity, which otherwise would phosphorylate (and inhibit) Snail1. These data suggest that in the prospective valve tissue, endocardial Notch1 promotes Snail1 expression, while myocardium-derived Bmp2 activates its endocardial receptors, triggering Snail1 expression and its nuclear stabilization via Gsk3β inhibition (Luna-Zurita et al., 2010). Thus, acquisition of the invasive phenotype by valve mesenchyme involves Notch1- and Bmp2-induced Snail1 expression and Bmp2-mediated Snail1 nuclear accumulation, leading to sustained expression of downstream mesenchymal genes.
A recent report uncovered a Notch-dependent autocrine loop in the AVC that activates expression of the soluble guanylyl cyclase (sGC) heterodimer, the nitric oxide (NO) receptor (Chang et al., 2011). Notch-RBPJK signals also promote NO production via induction of the activin-A and PI3-kinase/Akt pathway. The resulting increased NO signaling contributes to AVC EMT. This finding is intriguing since it suggests a link between Notch, NO and valve disease (Figure 4A,B; see below).
Outflow tract development and septation
The OFT is the arterial pole of the heart where the right and left ventricles empty into the pulmonary trunk and the aorta, respectively. OFT development involves coordinated interactions between endocardium, endocardial-derived mesenchyme, cardiac and smooth muscle progenitors from the SHF, and cardiac neural crest (NC) (Hutson and Kirby, 2007; Epstein, 2010). At midgestation, the single vessel emerging from the looped heart, the truncus arteriosus, is remodeled under the influence of invading NC and septates to form two vessels, the aorta and the pulmonary trunk. At the same time, the OFT rotates and aligns with the ventricles. If OFT alignment/rotation is disturbed, the result is double-outlet right ventricle (DORV), in which aorta and pulmonary trunk arise from the right ventricle, an over-riding aorta, or a related mal-alignment syndrome. Failure of OFT septation results in persistent truncus arteriosus (PTA), or, if partial septation occurs, an aorto-pulmonary window.
At E12.5, Notch elements are broadly expressed in the developing OFT (Loomes et al., 2002; High et al., 2007). Jag1 is expressed in NC-derived cells surrounding the aortic arch arteries (AAA); Jag2 in the pharyngeal region, including cells around the AAA; Dll1 only in OFT endocardium, and Dll4 in the endothelium of AAA. Notch1 is expressed in the endothelium lining the AAA and in the OFT endocardium; Notch2 is expressed throughout the pharyngeal mesenchyme and NC- derived cells surrounding the AAA, similarly to Notch3; Notch4 is expressed in endothelial cells of the AAA (Figure 4C). Notch inhibition in cardiac NC derivatives was achieved by breeding mice expressing dominant-negative MAML (DNMAML) with NC-specific Cre drivers (Pax3 and Wnt1), and results in aortic arch patterning defects, pulmonary artery stenosis and ventricular septal defects. Notch is also essential for differentiation of cardiac NC precursors into smooth muscle cells although it does not affect their proliferation or migration into the OFT (High et al., 2007).
The cardiac progenitor driver Islet1-Cre and the SHF-specific driver Mef2c-AHF-Cre were bred with DNMAML or Jag1flox to abrogate Notch signaling in cardiac progenitors, including those of the SHF. These mice show complex OFT and AAA defects including DORV and PTA, indicating that Jag1 signaling is essential in SHF derivatives (High et al., 2009). At E10.5, Isl1-Cre;DNMAML embryos show hypoplasia of the right ventricle and OFT cushions, and the NC-derived mesenchyme normally found within the OFT cushions is missing (High et al., 2009). Pharyngeal explants containing distal OFT cultured with a Notch inhibitor show reduced Fgf8 expression. Bmp4, a Fgf8 effector in OFT development (Park et al., 2008), is down-regulated in OFT cushions and myocardium of Islet1-Cre;DNMAML and Islet1-Cre;Jag1flox/flox mutants (High et al., 2009). OFT explant assays show that EMT is severely impaired in Islet1-Cre;DNMAML mutants. This defect can be rescued by adding Fgf8 (High et al., 2009). These results suggest that Notch regulates Fgf8 and Bmp4 expression in SHF myocardium, thereby inducing EMT in adjacent endothelial cells (Figure 4B). The regulatory mechanism and the specific Notch elements, besides Jag1, involved in OFT valve development remain to be studied. In addition, detailed analysis of Islet1-Cre expression in pharyngeal mesoderm and neural crest, and the functions of Notch targets such as Hes1 (Rochais et al., 2009) in this process will need to be pursued in the future.
Semilunar valve morphogenesis
During valve development, thinning and elongation of the valve primordia leads to leaflet maturation, while the ECM is restructured into a highly stratified and VIC-dense mature matrix (reviewed in Hinton and Yutzey, 2011). A specific feature of OFT (semilunar) valves morphogenesis is the infiltration by migratory NC cells (Jiang et al., 2000; de Lange et al., 2004; Jain et al., 2010). As previously discussed, Notch inhibition in SHF results in complex OFT and AAA abnormalities reminiscent of those caused by NC defects (High et al., 2009). Pax3 is a marker of premigratory NC, and its inactivation causes defects in many NC derivatives. Recent studies demonstrate that cardiac defects resulting from loss of Pax3 include dysmorphic, thickened and functionally incompetent semilunar valves (Jain et al., 2011). Interestingly, Notch inhibition in the SHF, which causes secondary NC abnormalities, also affects semilunar valve development. Thus, Islet1- or Mef2c-AHFCre;DNMAML mice show abnormal NC patterning in the OFT, indicated by the reduced expression of postmigratory NC markers (Jain et al., 2011). These findings suggest that Notch mediates signaling between cardiac progenitors, including those of the SHF, and migrating NC. This crosstalk probably occurs before these cell populations have invaded the heart proper, while they are in apposition at ~E9.5 in pharyngeal tissues (Jain et al., 2011). Interestingly, at E17.5, the semilunar valve leaflets of Mef2c-AHF-Cre; DNMAML embryos are 2.2 times bigger than those of wild-type littermates and contain an excess of ECM. In utero assessment of cardiac function by echocardiography shows moderate-to-severe aortic insufficiency. These data demonstrate that Notch inhibition in SHF progenitors leads to abnormal OFT valve morphogenesis and function (Jain et al., 2011). NC is known to contribute to OFT valve mesenchyme as early as E10.5, but these cells largely disappear as development proceeds, so that by E17.5 few NC derivatives remain (Jiang et al., 2000; de Lange et al., 2004). Pax3-deficient NC mutants and SHF Notch mutants with defective NC patterning have hypocellular OFT cushions at early stages, probably caused by reduced numbers of NC mesenchyme derivatives (High et al., 2009). However, by late gestation, these mutants have hypercellular semilunar valves (Jain et al., 2011). Semilunar valve morphogenesis is normally associated with a gradual decrease in mesenchymal cell proliferation and increased apoptosis (Hinton and Yutzey, 2011). Mef2c-AHF-Cre or Islet-Cre; DNMAML embryos show deficient apoptosis (Jain et al., 2011).
These data suggest that Notch mediates interactions between NC- and endocardium-derived mesenchymal cells, triggering the apoptosis that contributes to OFT valve remodeling (Jain et al., 2011; Figure 4B). This illustrates some of the fundamental developmental processes that are governed by Notch during OFT valve morphogenesis, including proliferation, differentiation and apoptosis.
Cardiac valve disease: recapitulation of an embryonic regulatory circuit
Abnormalities of valve remodeling are associated with development of bicuspid aortic valves (BAV), a common adult cardiac disease affecting 1–2% of the population, which is associated with important morbidity and mortality (Tzemos et al., 2008). BAV is often undetected in childhood, but affected adults are prone to aortic valve calcification, regurgitation and overall dysfunction (Friedman et al., 2008). Calcific aortic valve disease (CAVD) is the most common form of acquired valve disease in Western societies (Goldbarg et al., 2007). Aside from BAV, the most important factor associated with CAVD is age. As the obstructed flow and improper valve function progresses, the left ventricle has to increase its workload, which can lead to cardiac hypertrophy, dilatation, and heart failure. Examination of diseased valves reveals lipid and calcium deposition in the leaflets, endothelial cell activation, and inflammation (Otto, 2009).
NOTCH1 mutations occur in familial, non-syndromic, autosomal dominant CAVD associated with BAV (Garg et al., 2005). Two types of mutation were found in this study. One family has a premature stop codon in the NOTCH1 extracellular domain while in another family, a frameshift mutation results in gross alteration of the carboxy-terminus of the extracellular domain, with the addition of 74 incorrect amino acids. NOTCH1 haploinsufficiency was suggested to be the mechanism underlying this CAVD (Garg et al., 2005).Garg and coworkers showed that HEY1 and HEY2 repress the osteogenic master regulator RUNX2 through a physical interaction, suggesting that RUNX2 derepression may occur as a consequence of NOTCH1 mutation in these CAVD patients (Garg et al., 2005). Supporting an anti-osteogenic function, Notch1 was shown to repress valvular Bmp2 expression in adult murine aortic valve leaflets, and derepression of Bmp2 following Notch1 inhibition caused the formation of calcific nodules in valve interstitial cell cultures (Nigam and Srivastava, 2009). Recent work showed that Sox9, a transcription factor downstream of Notch during valve development (Luna-Zurita et al., 2010) is also Notch-responsive in the adult valve (Acharya et al., 2011), and its down-regulation may contribute to valve calcification (Peacock et al., 2010).
Heterozygous Notch1 mutant mice do not show BAV but develop signs of aortic valve calcification when fed a western diet, although unlike human patients, they do not develop stenosis (Nigam and Srivastava, 2009). In similar experiments, heterozygous RBPJk or Notch1 mice were fed a high-fat diet supplemented with vitamin D (Nus et al., 2011). In this setting, only the heterozygous RBPJk mice develop CAVD with significant hemodynamic disturbance. Pathological and molecular analysis reveals macrophage infiltration, enhanced collagen deposition, pro-osteogenic protein expression and calcification.
Notch inhibition in porcine VICs downregulates Hey1, activates osteogenic marker expression and increases calcified nodule formation (Nus et al., 2011). Despite the disparities between the humans and mice, the diet-dependent mouse genetic model of valve disease may prove very useful for studying the molecular mechanism of CAVD and the potential role of inflammation. The regulatory axis (Notch-Hey-Bmp2) identified in the embryo is also at work in the adult, and may be subverted in congenital or acquired disease, leading to valve calcification and stenosis. From the perspective of canonical Notch function, this regulatory axis suggests that the role of Notch in adult valve homeostasis includes the repression of osteoblast cell fate in VICs.
On a final note, an involvement of Notch-NO pathway crosstalk in BAV formation and valve disease is suggested by the link between Notch and NO signaling during valve development (Chang et al., 2011). This potential link is made even more intriguing by the fact that mice deficient for endothelial NO synthase have BAV (Lee et al., 2000) as do humans with NOTCH1 heterozygous mutations (Garg et al., 2005).
Ventricular trabeculation
During ventricular chamber development the myocardium differentiates into two distinct layers: an outer compact zone and an inner trabecular zone. Trabeculae are sheets of cardiomyocytes forming muscular ridges lined by endocardial cells (Sedmera et al., 2000; Moorman and Christoffels, 2003; Figure 5). Trabeculae are found in all vertebrates and are a landmark in the initiation of ventricular chamber formation (Moorman and Christoffels, 2003). Trabecular cardiomyocytes and the overlying endocardium are in close association, and communication between them regulates cardiomyocyte proliferation and differentiation. An early function of trabeculae is to increase myocardial mass in the absence of coronary circulation (Sedmera et al., 2000). As development proceeds, trabeculae are remodeled and compressed, resulting in increased thickness of the compact zone myocardium. Failure of trabeculae formation is embryonic lethal and failure to remodel causes cardiomyopathy (Oechslin and Jenni, 2011).
Figure 5. Notch and ventricular trabeculation.
The inner surface (endocardium) of the ventricular cavity is shown at the top, and the outer surface (epicardium) at the bottom. Note the apposition of endocardium and myocardium at the base of developing trabeculae, where active Dll4-Notch1 signaling (strong red nuclei) predominates and decays towards the apex (pink-white nuclei). Notch signals to the trabecular myocardium to promote cardiomyocyte proliferation and differentiation (yellow and blue arrows). Modified from (Grego-Bessa et al., 2007).
Embryos lacking Notch1 or RBPJk, systemically or in the endocardium, show defective trabeculation. Molecular analysis of these mutants reveals that expression of endocardial and myocardial trabecular differentiation markers is impaired, and ventricular cardiomyocyte proliferation is inhibited. These findings imply that endocardial Notch1 signaling is required for proliferation and differentiation of trabecular myocardium (Grego-Bessa et al., 2007).
At least three signaling pathways essential for trabeculation are affected in Notch mutants: Bmp10, Nrg1/ErbB and EphrinB2/EphB4. Bmp10 is expressed in trabecular myocardium where it sustains proliferation (Chen et al., 2004). Standard or endothelial-specific Notch inactivation markedly reduces Bmp10 activity and trabecular myocytes proliferation. Phenotypic rescue of Notch mutant embryos upon incubation in Bmp10-conditioned medium, suggests that in trabecular myocardium Notch modulates proliferation via Bmp10 (Grego-Bessa et al., 2007).
Endocardial Nrg1 is crucial for trabeculation (Meyer and Birchmeier, 1995) and signals to ErbB2–4 receptors, expressed on myocardium (Lee et al., 1995). Nrg1 activity is reduced in Notch mutants, and addition of Nrg1 to cultured Notch mutant embryos rescue their cardiomyocyte differentiation defect (Grego-Bessa et al., 2007).
The EphrinB2 ligand and EphB4 receptor signaling system are also required for trabeculation (Wang et al., 1998). Endocardial EphrinB2/EphB4 expression and activity is impaired in Notch mutants and EphrinB2 is a direct transcriptional target of N1ICD/RBPJk (Grego-Bessa et al., 2007). Additional studies indicate that Nrg1 transcription is reduced in EphrinB2 mutants while Bmp10 mutants show normal EphrinB2 and Nrg1 expression, suggesting that EphrinB2 acts upstream of Nrg1 and that both molecules may act independently of Bmp10 during trabeculation.
These data allowed us to suggest a model (Figure 5) in which Notch-mediated endocardium-myocardium interaction promotes transition of primitive myocardium to trabecular myocardium (EphrinB2- and Nrg1-dependent) and trabecular cardiomyocyte proliferation is sustained by Bmp10 (Grego-Bessa et al., 2007).
Epicardial development and cardiac repair?
Initial analysis of the role of Notch in the epicardium and its derivatives has been described in two recent reports (Grieskamp et al., 2011; Del Monte et al., 2011). In the first one, Tbx18-Cre; RBPJkflox/flox mutants have been characterized (Grieskamp et al., 2011). These mice show abnormal coronary vessels and histology reveals dilation of the lumens of myocardially-(arterial) and subepicardiallly- (venous) located vessels. Marker analysis shows that large coronary arteries and veins are correctly specified. Moreover, the reduction of intramyocardial capillaries suggests that coronary arteries and veins are expanded in the mutants at the expense of capillaries (Grieskamp et al., 2011). The authors conclude that the differentiation potential of mutant EPDCs is compromised, since they are unable to differentiate into smooth muscle cells (SMCs), suggesting that RBPJK-dependent Notch signaling is required for perivascular cell differentiation into SMCs (Grieskamp et al., 2011).
In the second report, Notch function in epicardial development was studied with the driver line mWt1/IRES/GFP-Cre (Wt1Cre)(Del Monte et al., 2011). Wt1Cre;Notch1flox/flox embryos show body and pericardial hemorrhages at E13.5 and hearts with severely reduced and disorganized coronary vascular plexi. Histology reveals that coronary vessels are particularly rare in the compact myocardium zone where coronary arteries form, whereas the area occupied by subepicardial vessels (coronary veins) is expanded. Coronary SMC marker expression is reduced, as is the expression of EphrinB2, confirming the arterial nature of the affected vessels (Del Monte et al., 2011).
These two reports indicate that Notch plays a key role in the specification and differentiation of cell fates during epicardium and coronary vessel development. The impaired coronary artery development in Wt1-Cre;Notchflox/flox mice (Del Monte et al., 2011) might be primarily caused by a loss of coronary artery progenitor specification. In Tbx18-Cre; RBPJkflox/flox mice, coronary artery fate specification is unaffected but SMC differentiation is impaired (Grieskamp et al., 2011). Notch has been previously shown to regulate SMC differentiation in the systemic vasculature (High et al., 2007; Feng et al., 2010), suggesting that this might be a general role for Notch.
Recently, the epicardium has been considered as a potential source for treating the damaged heart (Vieira and Riley, 2011). The identification of a Notch-positive sub-epicardial progenitor cell population expressing cardiac-related growth factors (Russell et al., 2010), raised hopes about their role in cardiac repair that have yet to be demonstrated.
Conclusions and perspectives
NOTCH pathway mutations in humans result in very specific cardiovascular developmental defects and syndromes. In this review we have described studies that have defined the roles of individual Notch elements during AVC and OFT formation and morphogenesis, and in the development of the valves, chambers, conduction system and coronary vasculature. These findings have important implications for understanding the etiology of congenital cardiac defects that impact neonatal, pediatric and adult populations.
The available data on Notch function in the heart indicate that: (1) Endocardial Notch, acting by lateral induction, regulates cell fate specification and tissue patterning to define chamber versus non-chamber (valve) domains; (2) Notch also regulates cell fate specification in epicardium, coronary vessels, and in the cardiac conduction system; (3) endocardial Notch and myocardial Bmp2 signaling converge on Snail1 to activate EMT, leading to valve primordium formation; (5) endocardial Notch activity modulates various myocardial signals (Tgfβ2, Bmp10) required for valve and trabecular development, suggesting that Notch has a non-cell autonomous effect in these distant cell populations; (7) During OFT valve morphogenesis Notch is required for myocardial Fgf8 and Bmp4 expression and EMT, and mediates the communication between endocardium- and NC-derived mesenchyme that results in the mature valve; (8) Notch influences cellular proliferation, differentiation and apoptosis during these processes; (9) The Notch-Hey-Bmp2 regulatory interplay in the embryonic valves extends to the adult, and its disruption causes aortic valve disease.
Despite important advances in recent years, the detailed tableau of Notch-mediated regulation of cardiac development has been only partially exposed. Future studies will need to decipher in detail the mechanisms of activation of the different tissue-specific Notch elements, both in space and time, as well as to relate these to clinical phenotypes. These challenges will require sophisticated genetic tools and animal models, combined with new expression and functional assays and imaging technologies. We believe that increasing our knowledge of how Notch regulates cardiac development will also help us to ascertain its potential for cardiac repair.
ACKNOWLEDGMENTS
We thank J. M. Pérez-Pomares for helpful discussions and critical reading of the MS and G. de Luxán and L. Luna-Zurita for generating the figures. S. Bartlett (CNIC) provided English editing. We apologize to colleagues for omissions due to space limitations. J.L.dlP is funded by the Spanish Ministry of Science and Innovation grants SAF2010-17555, RD06/0014/0038 (RECAVA), RD06/0010/1013 (TERCEL) and FP7-ITN 215761 (NotchIT). J.A.E. is supported by NIH U01 HL100405, R01 HL095634, the American Heart Association Jon Holden DeHaan Myogenesis Center, the WW Smith Endowed Chair and the Spanish Fund for Cardiovascular Regenerative Medicine. The CNIC is supported by the MICINN and the Pro-CNIC Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aanhaanen WT, Brons JF, Dominguez JN, Rana MS, Norden J, Airik R, Wakker V, de Gier-de Vries C, Brown NA, Kispert A, et al. The Tbx2+ primary myocardium of the atrioventricular canal forms the atrioventricular node and the base of the left ventricle. Circ Res. 2009;104:1267–1274. doi: 10.1161/CIRCRESAHA.108.192450. [DOI] [PubMed] [Google Scholar]
- Aanhaanen WT, Moorman AF, Christoffels VM. Origin and development of the atrioventricular myocardial lineage: insight into the development of accessory pathways. Birth Defects Res A Clin Mol Teratol. 2011;91:565–577. doi: 10.1002/bdra.20826. [DOI] [PubMed] [Google Scholar]
- Acharya A, Hans CP, Koenig SN, Nichols HA, Galindo CL, Garner HR, Merrill WH, Hinton RB, Garg V. Inhibitory role of notch1 in calcific aortic valve disease. PLoS One. 2011;6:e27743. doi: 10.1371/journal.pone.0027743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akazawa H, Komuro I. Navigational error in the heart leads to premature ventricular excitation. J Clin Invest. 2011;121:513–516. doi: 10.1172/JCI46038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson ER, Sandberg R, Lendahl U. Notch signaling: simplicity in design, versatility in function. Development. 2011;138:3593–3612. doi: 10.1242/dev.063610. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Muskavitch MA. Notch: the past, the present, and the future. Curr Top Dev Biol. 2010;92:1–29. doi: 10.1016/S0070-2153(10)92001-2. [DOI] [PubMed] [Google Scholar]
- Chabriat H, Joutel A, Dichgans M, Tournier-Lasserve E, Bousser MG. Cadasil. Lancet Neurol. 2009;8:643–653. doi: 10.1016/S1474-4422(09)70127-9. [DOI] [PubMed] [Google Scholar]
- Chang AC, Fu Y, Garside VC, Niessen K, Chang L, Fuller M, Setiadi A, Smrz J, Kyle A, Minchinton A, et al. Notch Initiates the Endothelial-to-Mesenchymal Transition in the Atrioventricular Canal through Autocrine Activation of Soluble Guanylyl Cyclase. Dev Cell. 2011;21:288–300. doi: 10.1016/j.devcel.2011.06.022. [DOI] [PubMed] [Google Scholar]
- Chen H, Shi S, Acosta L, Li W, Lu J, Bao S, Chen Z, Yang Z, Schneider MD, Chien KR, et al. BMP10 is essential for maintaining cardiac growth during murine cardiogenesis. Development. 2004;131:2219–2231. doi: 10.1242/dev.01094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Grieskamp T, Norden J, Mommersteeg MT, Rudat C, Kispert A. Tbx18 and the fate of epicardial progenitors. Nature. 2009;458:E8–9. doi: 10.1038/nature07916. discussion E9-10. [DOI] [PubMed] [Google Scholar]
- Christoffels VM, Hoogaars WM, Tessari A, Clout DE, Moorman AF, Campione M. T-box transcription factor Tbx2 represses differentiation and formation of the cardiac chambers. Dev Dyn. 2004;229:763–770. doi: 10.1002/dvdy.10487. [DOI] [PubMed] [Google Scholar]
- de la Pompa JL, Wakeham A, Correia KM, Samper E, Brown S, Aguilera RJ, Nakano T, Honjo T, Mak TW, Rossant J, et al. Conservation of the Notch signalling pathway in mammalian neurogenesis. Development. 1997;124:1139–1148. doi: 10.1242/dev.124.6.1139. [DOI] [PubMed] [Google Scholar]
- de Lange FJ, Moorman AF, Anderson RH, Manner J, Soufan AT, de Gier-de Vries C, Schneider MD, Webb S, van den Hoff MJ, Christoffels VM. Lineage and morphogenetic analysis of the cardiac valves. Circ Res. 2004;95:645–654. doi: 10.1161/01.RES.0000141429.13560.cb. [DOI] [PubMed] [Google Scholar]
- Del Monte G, Grego-Bessa J, Gonzalez-Rajal A, Bolos V, De La Pompa JL. Monitoring Notch1 activity in development: Evidence for a feedback regulatory loop. Dev Dyn. 2007;236:2594–2614. doi: 10.1002/dvdy.21246. [DOI] [PubMed] [Google Scholar]
- Del Monte G, Casanova JC, Guadix JA, Macgrogan D, Burch JB, Perez-Pomares JM, de la Pompa JL. Differential Notch Signaling in the Epicardium Is Required for Cardiac Inflow Development and Coronary Vessel Morphogenesis. Circ Res. 2011;108:824–836. doi: 10.1161/CIRCRESAHA.110.229062. [DOI] [PubMed] [Google Scholar]
- Duarte A, Hirashima M, Benedito R, Trindade A, Diniz P, Bekman E, Costa L, Henrique D, Rossant J. Dosage-sensitive requirement for mouse Dll4 in artery development. Genes Dev. 2004;18:2474–2478. doi: 10.1101/gad.1239004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein JA. Franklin H. Epstein Lecture. Cardiac development and implications for heart disease. N Engl J Med. 2010;363:1638–1647. doi: 10.1056/NEJMra1003941. [DOI] [PubMed] [Google Scholar]
- Feng X, Krebs LT, Gridley T. Patent ductus arteriosus in mice with smooth muscle-specific Jag1 deletion. Development. 2010;137:4191–4199. doi: 10.1242/dev.052043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman T, Mani A, Elefteriades JA. Bicuspid aortic valve: clinical approach and scientific review of a common clinical entity. Expert Rev Cardiovasc Ther. 2008;6:235–248. doi: 10.1586/14779072.6.2.235. [DOI] [PubMed] [Google Scholar]
- Garg V. Molecular genetics of aortic valve disease. Curr Opin Cardiol. 2006;21:180–184. doi: 10.1097/01.hco.0000221578.18254.70. [DOI] [PubMed] [Google Scholar]
- Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- Gaussin V, Van De Putte T, Mishina Y, Hanks MC, Zwijsen A, Huylebroeck D, Behringer RR, Schneider MD. Endocardial cushion and myocardial defects after cardiac myocyte-specific conditional deletion of the bone morphogenetic protein receptor ALK3. Proc Natl Acad Sci U S A. 2002;99:2878–2883. doi: 10.1073/pnas.042390499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldbarg SH, Elmariah S, Miller MA, Fuster V. Insights into degenerative aortic valve disease. J Am Coll Cardiol. 2007;50:1205–1213. doi: 10.1016/j.jacc.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Grego-Bessa J, Luna-Zurita L, del Monte G, Bolos V, Melgar P, Arandilla A, Garratt AN, Zang H, Mukouyama YS, Chen H, et al. Notch signaling is essential for ventricular chamber development. Dev Cell. 2007;12:415–429. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieskamp T, Rudat C, Ludtke TH, Norden J, Kispert A. Notch signaling regulates smooth muscle differentiation of epicardium-derived cells. Circ Res. 2011;108:813–823. doi: 10.1161/CIRCRESAHA.110.228809. [DOI] [PubMed] [Google Scholar]
- Harrelson Z, Kelly RG, Goldin SN, Gibson-Brown JJ, Bollag RJ, Silver LM, Papaioannou VE. Tbx2 is essential for patterning the atrioventricular canal and for morphogenesis of the outflow tract during heart development. Development. 2004;131:5041–5052. doi: 10.1242/dev.01378. [DOI] [PubMed] [Google Scholar]
- High FA, Zhang M, Proweller A, Tu L, Parmacek MS, Pear WS, Epstein JA. An essential role for Notch in neural crest during cardiovascular development and smooth muscle differentiation. J Clin Invest. 2007;117:353–363. doi: 10.1172/JCI30070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- High FA, Epstein JA. The multifaceted role of Notch in cardiac development and disease. Nat Rev Genet. 2008;9:49–61. doi: 10.1038/nrg2279. [DOI] [PubMed] [Google Scholar]
- High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. J Clin Invest. 2009;119:1986–1996. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinton RB, Yutzey KE. Heart valve structure and function in development and disease. Annu Rev Physiol. 2011;73:29–46. doi: 10.1146/annurev-physiol-012110-142145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutson MR, Kirby ML. Model systems for the study of heart development and disease. Cardiac neural crest and conotruncal malformations. Semin Cell Dev Biol. 2007;18:101–110. doi: 10.1016/j.semcdb.2006.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iso T, Kedes L, Hamamori Y. HES and HERP families: multiple effectors of the Notch signaling pathway. J Cell Physiol. 2003;194:237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jain R, Engleka KA, Rentschler SL, Manderfield LJ, Li L, Yuan L, Epstein JA. Cardiac neural crest orchestrates remodeling and functional maturation of mouse semilunar valves. J Clin Invest. 2011;121:422–430. doi: 10.1172/JCI44244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R, Rentschler S, Epstein JA. Notch and cardiac outflow tract development. Ann N Y Acad Sci. 2010;1188:184–190. doi: 10.1111/j.1749-6632.2009.05099.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarriault S, Brou C, Logeat F, Schroeter EH, Kopan R, Israel A. Signalling downstream of activated mammalian Notch [see comments] Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- Jiang X, Rowitch DH, Soriano P, McMahon AP, Sucov HM. Fate of the mammalian cardiac neural crest. Development. 2000;127:1607–1616. doi: 10.1242/dev.127.8.1607. [DOI] [PubMed] [Google Scholar]
- Kokubo H, Miyagawa-Tomita S, Johnson RL. Hesr, a Mediator of the Notch Signaling, Functions in Heart and Vessel Development. Trends Cardiovasc Med. 2005;15:190–194. doi: 10.1016/j.tcm.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Kokubo H, Tomita-Miyagawa S, Hamada Y, Saga Y. Hesr1 and Hesr2 regulate atrioventricular boundary formation in the developing heart through the repression of Tbx2. Development. 2007;134:747–755. doi: 10.1242/dev.02777. [DOI] [PubMed] [Google Scholar]
- Kopan R. Notch: a membrane-bound transcription factor. J Cell Sci. 2002;115:1095–1097. doi: 10.1242/jcs.115.6.1095. [DOI] [PubMed] [Google Scholar]
- Kopan R, Ilagan MX. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee KF, Simon H, Chen H, Bates B, Hung MC, Hauser C. Requirement for neuregulin receptor erbB2 in neural and cardiac development. Nature. 1995;378:394–398. doi: 10.1038/378394a0. [DOI] [PubMed] [Google Scholar]
- Lee TC, Zhao YD, Courtman DW, Stewart DJ. Abnormal aortic valve development in mice lacking endothelial nitric oxide synthase. Circulation. 2000;101:2345–2348. doi: 10.1161/01.cir.101.20.2345. [DOI] [PubMed] [Google Scholar]
- Logeat F, Bessia C, Brou C, LeBail O, Jarriault S, Seidah NG, Israel A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc Natl Acad Sci U S A. 1998;95:8108–8112. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loomes KM, Taichman DB, Glover CL, Williams PT, Markowitz JE, Piccoli DA, Baldwin HS, Oakey RJ. Characterization of Notch receptor expression in the developing mammalian heart and liver. Am J Med Genet. 2002;112:181–189. doi: 10.1002/ajmg.10592. [DOI] [PubMed] [Google Scholar]
- Loomes KM, Underkoffler LA, Morabito J, Gottlieb S, Piccoli DA, Spinner NB, Baldwin HS, Oakey RJ. The expression of Jagged1 in the developing mammalian heart correlates with cardiovascular disease in Alagille syndrome. Hum Mol Genet. 1999;8:2443–2449. doi: 10.1093/hmg/8.13.2443. [DOI] [PubMed] [Google Scholar]
- Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J Clin Invest. 2010;120:3493–3507. doi: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu MF, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Macgrogan D, Luna-Zurita L, de la Pompa JL. Notch signaling in cardiac valve development and disease. Birth Defects Res A Clin Mol Teratol. 2011;91:449–459. doi: 10.1002/bdra.20815. [DOI] [PubMed] [Google Scholar]
- MacGrogan D, Nus M, de la Pompa JL. Notch signaling in cardiac development and disease. Curr Top Dev Biol. 2010;92:333–365. doi: 10.1016/S0070-2153(10)92011-5. [DOI] [PubMed] [Google Scholar]
- Markwald R, Eisenberg C, Eisenberg L, Trusk T, Sugi Y. Epithelial-mesenchymal transformations in early avian heart development. Acta Anat. 1996;156:173–186. doi: 10.1159/000147845. [DOI] [PubMed] [Google Scholar]
- Meyer D, Birchmeier C. Multiple essential functions of neuregulin in development. Nature. 1995;378:386–390. doi: 10.1038/378386a0. [DOI] [PubMed] [Google Scholar]
- Mikawa T, Gourdie RG. Pericardial mesoderm generates a population of coronary smooth muscle cells migrating into the heart along with ingrowth of the epicardial organ. Dev Biol. 1996;174:221–232. doi: 10.1006/dbio.1996.0068. [DOI] [PubMed] [Google Scholar]
- Moorman AF, Christoffels VM. Cardiac chamber formation: development, genes, and evolution. Physiol Rev. 2003;83:1223–1267. doi: 10.1152/physrev.00006.2003. [DOI] [PubMed] [Google Scholar]
- Niessen K, Karsan A. Notch signaling in cardiac development. Circ Res. 2008;102:1169–1181. doi: 10.1161/CIRCRESAHA.108.174318. [DOI] [PubMed] [Google Scholar]
- Nigam V, Srivastava D. Notch1 represses osteogenic pathways in aortic valve cells. J Mol Cell Cardiol. 2009;47:828–834. doi: 10.1016/j.yjmcc.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nus M, Macgrogan D, Martinez-Poveda B, Benito Y, Casanova JC, Fernandez-Aviles F, Bermejo J, de la Pompa JL. Diet-Induced Aortic Valve Disease in Mice Haploinsufficient for the Notch Pathway Effector RBPJK/CSL. Arterioscler Thromb Vasc Biol. 2011;31:1580–1588. doi: 10.1161/ATVBAHA.111.227561. [DOI] [PubMed] [Google Scholar]
- Oechslin E, Jenni R. Left ventricular non-compaction revisited: a distinct phenotype with genetic heterogeneity? Eur Heart J. 2011;32:1446–1456. doi: 10.1093/eurheartj/ehq508. [DOI] [PubMed] [Google Scholar]
- Otto CM. Calcific aortic valve disease: outflow obstruction is the end stage of a systemic disease process. Eur Heart J. 2009;30:1940–1942. doi: 10.1093/eurheartj/ehp175. [DOI] [PubMed] [Google Scholar]
- Park EJ, Watanabe Y, Smyth G, Miyagawa-Tomita S, Meyers E, Klingensmith J, Camenisch T, Buckingham M, Moon AM. An FGF autocrine loop initiated in second heart field mesoderm regulates morphogenesis at the arterial pole of the heart. Development. 2008;135:3599–3610. doi: 10.1242/dev.025437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock JD, Levay AK, Gillaspie DB, Tao G, Lincoln J. Reduced sox9 function promotes heart valve calcification phenotypes in vivo. Circ Res. 2010;106:712–719. doi: 10.1161/CIRCRESAHA.109.213702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Person AD, Klewer SE, Runyan RB. Cell biology of cardiac cushion development. Int Rev Cytol. 2005;243:287–335. doi: 10.1016/S0074-7696(05)43005-3. [DOI] [PubMed] [Google Scholar]
- Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. Embo J. 2001;20:3427–3436. doi: 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentschler S, Harris BS, Kuznekoff L, Jain R, Manderfield L, Lu MM, Morley GE, Patel VV, Epstein JA. Notch signaling regulates murine atrioventricular conduction and the formation of accessory pathways. J Clin Invest. 2011;121:525–533. doi: 10.1172/JCI44470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochais F, Dandonneau M, Mesbah K, Jarry T, Mattei MG, Kelly RG. Hes1 is expressed in the second heart field and is required for outflow tract development. PLoS One. 2009;4:e6267. doi: 10.1371/journal.pone.0006267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell JL, Goetsch SC, Gaiano NR, Hill JA, Olson EN, Schneider JW. A dynamic notch injury response activates epicardium and contributes to fibrosis repair. Circ Res. 2010;108:51–59. doi: 10.1161/CIRCRESAHA.110.233262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Developmental patterning of the cardiac atrioventricular canal by Notch and Hairy-related transcription factors. Development. 2006;133:4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlgren C, Gustafsson MV, Jin S, Poellinger L, Lendahl U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc Natl Acad Sci U S A. 2008;105:6392–6397. doi: 10.1073/pnas.0802047105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedmera D, Pexieder T, Hu N, Clark EB. Developmental changes in the myocardial architecture of the chick. Anat Rec. 1997;248:421–432. doi: 10.1002/(SICI)1097-0185(199707)248:3<421::AID-AR15>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Sedmera D, Pexieder T, Vuillemin M, Thompson RP, Anderson RH. Developmental patterning of the myocardium. Anat Rec. 2000;258:319–337. doi: 10.1002/(SICI)1097-0185(20000401)258:4<319::AID-AR1>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- Shen T, Aneas I, Sakabe N, Dirschinger RJ, Wang G, Smemo S, Westlund JM, Cheng H, Dalton N, Gu Y, et al. Tbx20 regulates a genetic program essential to adult mouse cardiomyocyte function. J Clin Invest. 2011;121:4640–4654. doi: 10.1172/JCI59472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Horsthuis T, Farin HF, Grieskamp T, Norden J, Petry M, Wakker V, Moorman AF, Christoffels VM, Kispert A. Tbx20 interacts with smads to confine tbx2 expression to the atrioventricular canal. Circ Res. 2009;105:442–452. doi: 10.1161/CIRCRESAHA.109.196063. [DOI] [PubMed] [Google Scholar]
- Stennard FA, Costa MW, Lai D, Biben C, Furtado MB, Solloway MJ, McCulley DJ, Leimena C, Preis JI, Dunwoodie SL, et al. Murine T-box transcription factor Tbx20 acts as a repressor during heart development, and is essential for adult heart integrity, function and adaptation. Development. 2005;132:2451–2462. doi: 10.1242/dev.01799. [DOI] [PubMed] [Google Scholar]
- Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, et al. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzemos N, Therrien J, Yip J, Thanassoulis G, Tremblay S, Jamorski MT, Webb GD, Siu SC. Outcomes in adults with bicuspid aortic valves. Jama. 2008;300:1317–1325. doi: 10.1001/jama.300.11.1317. [DOI] [PubMed] [Google Scholar]
- Uyttendaele H, Marazzi G, Wu G, Yan Q, Sassoon D, Kitajewski J. Notch4/int-3, a mammary proto-oncogene, is an endothelial cell-specific mammalian Notch gene. Development. 1996;122:2251–2259. doi: 10.1242/dev.122.7.2251. [DOI] [PubMed] [Google Scholar]
- Vieira JM, Riley PR. Epicardium-derived cells: a new source of regenerative capacity. Heart. 2011;97:15–19. doi: 10.1136/hrt.2010.193292. [DOI] [PubMed] [Google Scholar]
- Wang HU, Chen ZF, Anderson DJ. Molecular distinction and angiogenic interaction between embryonic arteries and veins revealed by ephrin-B2 and its receptor Eph-B4. Cell. 1998;93:741–753. doi: 10.1016/s0092-8674(00)81436-1. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Kokubo H, Miyagawa-Tomita S, Endo M, Igarashi K, Aisaki KI, Kanno J, Saga Y. Activation of Notch1 signaling in cardiogenic mesoderm induces abnormal heart morphogenesis in mouse. Development. 2006;133:1625–1634. doi: 10.1242/dev.02344. [DOI] [PubMed] [Google Scholar]
- Weng AP, Millholland JM, Yashiro-Ohtani Y, Arcangeli ML, Lau A, Wai C, Del Bianco C, Rodriguez CG, Sai H, Tobias J, et al. c-Myc is an important direct target of Notch1 in T-cell acute lymphoblastic leukemia/lymphoma. Genes Dev. 2006;20:2096–2109. doi: 10.1101/gad.1450406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M, Revelli JP, Eichele G, Barron M, Schwartz RJ. Expression of chick Tbx-2, Tbx-3, and Tbx-5 genes during early heart development: evidence for BMP2 induction of Tbx2. Dev Biol. 2000;228:95–105. doi: 10.1006/dbio.2000.9927. [DOI] [PubMed] [Google Scholar]