Abstract
The role of circadian proteins in regulating whole body metabolism and bone turnover has been studied in detail and has led to the discovery of an elemental system for timekeeping involving the core genes Clock, Bmal1, Per, and Cry. Nocturnin, a peripheral circadian-regulated gene has been shown to play a very important role in regulating adipogenesis by deadenylation of key mRNAs and intra-cytoplasmic transport of PPARγ. The role that it plays in osteogenesis has previously not been studied in detail. In this report we examined in vitro and in vivo osteogenesis in the presence and absence of Nocturnin and show that loss of Nocturnin enhances bone formation and can rescue Rosiglitazone induced bone loss in mice. The circadian rhythm of Nocturnin is likely to be an essential element of marrow stromal cell fate.
Keywords: Nocturnin, rosiglitazone, PPARγ
Introduction
Factors that control mesenchymal stem cell fate are of great importance since these can influence the formation and differentiation of various tissues. In vertebrates, adipocytes and osteoblasts which form fat and bone respectively, originate from multipotent mesenchymal stem cells.(1) Any imbalance favoring either of these processes can lead to an array of disorders, one of which is age related osteoporosis.(2) Recent studies have shown that circadian-regulated clocks are important for modulating genes involved in metabolic processes and osteogenesis.(3) The clock genes Bmal1, Clock, Period(Per) and Cryptochrome (Cry) which form the central core of the circadian process, are critical regulators of metabolic homeostasis.(4) Moreover, the Per and Cry genes also impact bone turnover. For example, deletion of Per and Cry genes independently in mice leads to increased bone mass by affecting different arms of skeletal homeostasis, Per affects osteoblastogenesis, whereas Cry is thought to affect osteoclastogenesis.(5) At the molecular level, these genes interact with CREB, a transcription factor that controls sympathetic mediated bone formation.(6)
Nocturnin (Noc) is a circadian clock-controlled gene that is downstream of the adipogenic transcriptional factor PPARγ. It is not a part of the central core clock complex, but is transcriptionally regulated by central clock genes. It is also induced in response to serum shock, and generally upregulated in murine tissues in the early evening during feeding.(7,8,9) NOCTURNIN (NOC) belongs to the exonuclease–endonuclease phosphatase super family of deadenylases and regulates gene expression post-transcriptionaly,(10) utilizing its deadenylase properties. In addition, NOC has been shown to colocalize with PPARγ at the perinuclear region and help its translocation into the nucleus, resulting in an increase in adipogenic differentiation. The loss of this protein leads to a decrease in the nuclear localization of PPARγ resulting in altered nuclear transcription.(11) The pattern of NOC expression in the cytoplasm of cells shows strong localization to one area of the cytoplasm along with the perinuclear area. These lines of data strongly point to a role for NOC in adipogenesis, but Noc is also expressed in a variety of other tissues including early multipotent stem cells. For example, Noc −/− mice at 12 weeks of age show enhanced skeletal mass and increased osteoblastogenesis. (11) In this study, we examined the mechanisms responsible for Noc actions on the skeleton and asked whether this was related to control of adipogenesis through PPARγ or by directing bone marrow stromal cell lineage specification. We found that Noc overexpression leads to the suppression of a number of osteoblast transcriptional factors and markers like ATF4, Osteocalcin, and Osterix. We also observed that there was a slight decrease in the expression of the cholesterol metabolism enzyme CYP51. Furthermore, when Noc was over-expressed in preosteoblastic cells, the cells were deficient in forming an osteoblastic phenotype, whereas shRNAi-mediated knockdown of Noc led to an increase in osteoblast bone nodule formation.
Materials and Methods
Mouse strains and cells used
The mice used in this study were described in Refs. 11 and 12 and are on a C57BL/6J background. MC3T3E1 C4 preosteoblastic cells were used for overexpression of NOC in these cells and the vectors and generation of these cells were described in an earlier paper. (11)
Skeletal staining, tissue collection and histology
For skeletal staining the protocol was modified from Mcleod et al. (13) Bones were collected from different age groups, fixed in 4% paraformaldehdye, followed by decalcification in 14% EDTA solution, then processed for hematoxylin and eosin (H&E) staining.
Osteoblast differentiation media and alkaline phosphatase staining
MC3T3E1 C4 cells for differentiation assays were grown in osteoblast culture media containing 10% FBS α-MEM with β-glycerophosphate and ascorbic acid. Fresh media was added every other day. After the required time in culture, cells were processed for alkaline phosphatase staining using a kit purchased from Sigma.
Real time quantitative PCR
Primers used for studying gene expression profiles for mNoc, mATF4, mOsteocalcin, mOsterix, and mCyp51 will be provided upon request. The methodology for performing the real time PCR was described previously in Ref. 11, and the method used for analyzing the data was based on the method described by Livak and Schmittgen.(14)
Western blotting
Protein lysates were prepared as described in Ref. 15 and processed on SDS PAGE gels for Western blotting. The antibodies used in this study are anti-Flag monoclonal (Sigma-Aldrich, St. Louis, MO), Nocturnin (20), anti-Actin (Santa Cruz Biotechnology, Santa Cruz, CA)
MicroCT and rosiglitazone treatment
Four groups of animals, two negative control groups (Noc+/+) and two experimental groups (Noc −/−) were put on diet treatment at 12 weeks of age. The two groups of each genotype were put on control diet and diet with rosiglitazone (20 mg/kg/d for 8 weeks; research diets). The bones from the 20-week-old animals were collected and processed for microCT. All experiments were done in accordance with the rules set by the IACUC of the Maine Medical Center. The animals were housed at the Maine Medical Center Research Institute (Scarborough, ME).
Results
Osteoblast gene expression in the presence of Nocturnin
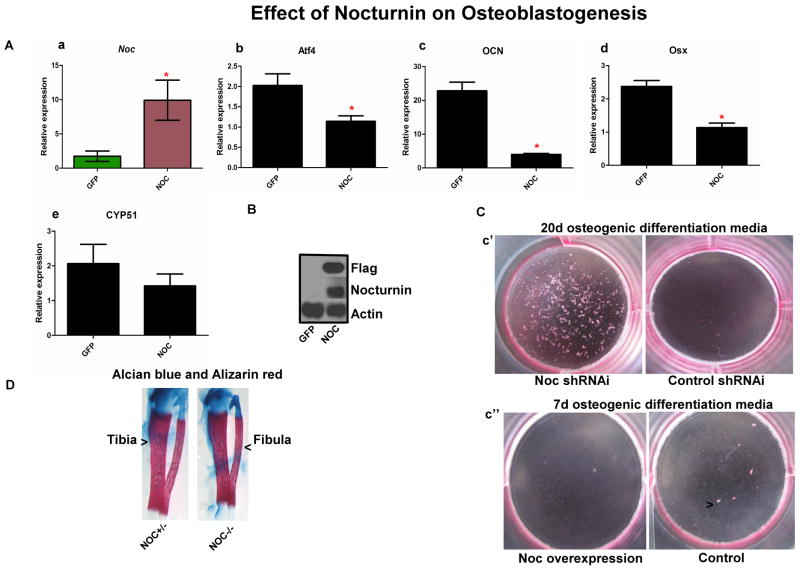
To study if Nocturnin regulates osteoblast gene expression we used MC3T3E1 preosteoblastic cells overexpressing Flag tagged NOC which has been described previously.(9) The overexpression of NOC in these cells did not lead to a major change in gene expression at the 1d time point (data not shown). But osteoblastic markers in a long term differentiation assay did differ, including Runx2, which we previously had shown was down regulated by Noc overexpression.(11) In the current study, we observed that the major osteoblast specific transcriptional factors, Osterix (Osx) and ATF4 (Atf4), showed a significantly lower expression in the presence of Noc. Moreover, the terminal differentiation marker, Osteocalcin (Ocn), in these cells was also significantly decreased. Additionally, recent studies have shown that the control of steriodogenic enzymes like Lanosterol 14 alpha-demethylase (CYP51) and cytochrome p450 family 11 subfamily A polypeptide 1 (CYP11a1) can be transcriptionally controlled by Runx2 and are expressed in osteoprogenitors cells. (16) As the expression of CYP51 is diurnal in the liver, (17) this prompted us to look at the expression of this gene in the Nocturnin overexpressing cells. Like the other osteoblast transcription factors we observed a decrease in the expression of this gene. (see Fig.1A).
Figure 1.
(A) Effect of Nocturnin on osteoblastogenesis. This figure shows effect of overexpression of (a) Noc on osteoblast gene marker expression as can be observed, there is a significant suppression of osteoblastogeneis, which is evident from the decrease in (b) ATF4 and (c) Osteocalci ; (d) Osterix we also saw a trend towards suppression in (e) CYP51 when Noc is overexpressed, in MC3T3E1 cells compared to GFP cells. (B) The increase in Noc is confirmed by the Flag tag Western blot along with a NOC protein levels in cells, Actin was used as the loading control. (C) (c’)The long term (20 days with change in media every two days) osteoblast differentiation assay on cells that have Noc knocked down show that there is a significant increase in nodule formation compared to control cells. (c’’) Osteoblast differentiation assay for 7 days showed decreased alkaline phosphatase staining in Noc overexpressed cells compared to control cells (n = 3, * indicates P < 0.05). (D) Alizarin red and Alcian blue staining showing the tibial and fibula bones from Noc control +/− and Noc null.
To further study the cellular phenotype in vitro, we used alkaline phosphatase staining of GFP and Noc overexpressing cells that were in osteogenic media for 7days and observed a decrease in alkaline phosphatase staining in the Noc overexpressing cells (see Fig 1Cc’’). Next we silenced Noc in MC3T3E1 cells and observed that the loss of NOC leads to an increase in nodule formation at day 20 of differentiation when cells were plated in osteogenic media (Fig 1C).
To study the early development of the long bones in the Noc−/− animals we used hind limbs from Noc−/− and compared them to Noc−/+ controls and stained the bones using alizarin red and alcian blue. The overt morphology of the tibia and femur was not different between mutant and controls Figure 1D and histological preparations showed no major observable difference in one week old femoral sections in the trabecular or cortical bone (data not shown).
Effect of rosiglitazone treatments on wild-type and Nocturnin null mice in vivo
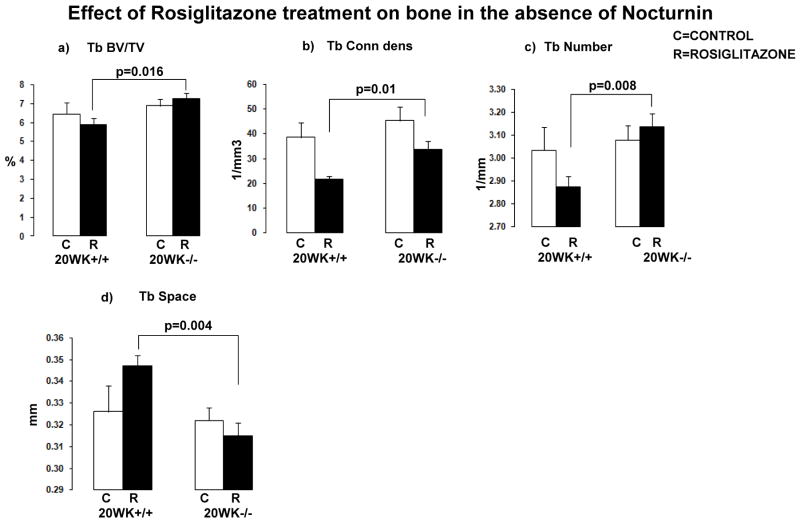
Nocturnin was previously identified as a gene that was induced by rosiglitazone treatment.(9) Deletion of Nocturnin in mice revealed an increase in bone mass at 12 weeks of age.(11) In wild type C57BL/6J mice, treatment with rosiglitazone leads to loss of bone mass and an increase in total body weight.(18) To test if the effect of rosiglitazone is mediated through an increase in expression of Nocturnin, we fed wild-type mice and Nocturnin null mice (both on a C57BL/6J background) a control diet and a rosiglitazone diet for an 8 week period from 12 weeks of age to 20 weeks of age. Skeletal parameters were measured using microCT. In Noc−/− mice, there was protection from trabecular bone loss compared to wild-type controls treated with rosiglitazone (Fig. 2).
Figure 2.
Effect of rosiglitazone treatment on bone in the absence of Nocturnin. Graphical representation of the data from mice that were on control diet and rosiglitazone diet. (a) Trabecular BV/TV, (b) trabecular connectivity density, (c) trabecular number, and (d) trabecular space. MicroCT evaluation of femurs from mice that were on rosiglitazone diet and compared to mice on control diet showed that there are increased trabecular parameters in the Noc null mice, suggesting that there is a protection effect of rosiglitazone treatment on bone in the absence of Noc. (n = 4 animals per group, P-values are indicated on the chart)
Discussion
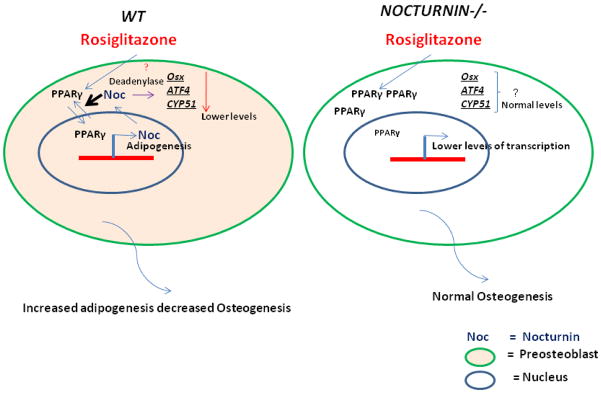
Previously we had shown that deletion of Nocturnin globally leads to an increase in bone mass in 12-week-old mice.(11) In the current study, we analyzed the potential mechanisms involved in this process both in vitro and in vivo by administering the PPARγ agonist rosiglitazone (Fig. 3). In the latter case, unlike wild type mice, Noc−/− mice were protected against rosiglitazone induced trabecular bone loss. To investigate the pathogenesis of this process we studied the effect of Nocturnin overexpression in pre-osteoblastic cells and saw suppression in transcript levels of two critical osteoblast transcriptional factors, ATF4 and Osterix. Yet the mechanism through which Nocturnin achieves this is not clear, since the major function of this protein is to act as a deadenylase in the cytoplasm where it can potentially bind to 3’UTRs of mRNA and control the stability of the transcript. Previously, we had shown that NOC does not enter the nucleus but can target IGF1 mRNA for degradation in the bone, thereby controlling some aspects of osteogenesis. (19) And we also showed that Runx2, the earliest osteoblast transcriptional factor was downregulated in the absence of Noc.(11,17) Taken together these lines of evidence suggest that Noc may work through an intermediary to regulate osteogenic differentiation possibly through its effects on PPARγ.
Figure 3.
Model showing the effect rosiglitazone treatment has on bone in the presence and absence of Nocturnin. The model illustrates the salient points of the paper: in a wild-type (WT) pre-osteoblast in the presence of rosiglitazone, Nocturnin helps PPARγ translocate in to the nucleus where it can help facilitate adipogenesis potentially over osteogenesis; and in the cytoplasm, Nocturnin can potentially bind to the mRNA’s of ATF4, Osterix, and CYP51 and negatively regulate them using its deadenylase function. In the absence of Nocturnin (Nocturnin−/−) there is less of PPARγ in the nucleus, along with loss of deadenylase regulation of the osteoblast marker transcripts, which results in protection from bone loss on rosiglitazone treatment.
Recent published data show that Runx2 regulates a number of steroidogenic enzymes like CYP51 and CYP11a1, both highly expressed in osteoprogenitors. Interestingly, the expression of CYP51 in the liver, where Noc is also highly expressed, has been shown to be diurnal.(17) Most recently, Nocturnin has been shown to regulate cholesterol transport in the gut. (20) And, importantly, CYP51 can modulate bone development since deletion of this enzyme in mice leads to several bone defects. (21) In our studies, CYP51 transcripts in Nocturnin overexpressing MC3T3E1 cells were reduced suggesting that Noc may impact Runx2 downstream genes very early in the differentiation process and this regulation is temporally determined.
In sum, we have shown that Noc, a circadian regulated deadenylase also plays an important role in MSC fate and, ultimately, in skeletal homeostasis. Noc (gene name Ccrn4l) evolved in vertebrate systems from the yeast gene Ccr4, which encodes a multifunctional protein that is both a cytoplasmic deadenylase and a transcriptional cofactor that is regulated by substrate availability. Clearly, during evolution, NOC’s function was altered, yet still remains in mammals as a critical factor regulating adipogenesis and osteogenesis. Our current data adds to the growing body of literature suggesting that timekeeping is an essential homeostatic process for bone turnover.
Acknowledgments
CJR was supported by NIH Grants AR045433 and AR54604.
Footnotes
Conflicts of interest
The authors declare no conflicts of interests
References
- 1.Phinney DG, Prockop DJ. Concise Review: Mesenchymal Stem/Multipotent Stromal Cells: The State of Transdifferentiation and Modes of Tissue Repair—Current Views. STEM CELLS. 2007;25:2896–2902. doi: 10.1634/stemcells.2007-0637. [DOI] [PubMed] [Google Scholar]
- 2.Nuttall ME, Gimble JM. Controlling the balance between osteoblastogenesis and adipogenesis and the consequent therapeutic implications. Current Opinion in Pharmacology. 2004;4:290–294. doi: 10.1016/j.coph.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 3.Fu L, Patel MS, et al. The Molecular Clock Mediates Leptin-Regulated Bone Formation. Cell. 2005;122(5):803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 4.Froy O. Metabolism and Circadian Rhythms—Implications for Obesity. Endocrine Reviews. 2010;31:1–24. doi: 10.1210/er.2009-0014. [DOI] [PubMed] [Google Scholar]
- 5.Maronde E, Schilling AF, et al. The Clock Genes Period 2 and Cryptochrome 2 Differentially Balance Bone Formation. PLoS ONE. 2010;5(7):e11527. doi: 10.1371/journal.pone.0011527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu L, et al. The Molecular Clock Mediates Leptin-Regulated Bone Formation. Cell. 2005;122:803–815. doi: 10.1016/j.cell.2005.06.028. [DOI] [PubMed] [Google Scholar]
- 7.Green CB, Besharse JC. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proceedings of the National Academy of Sciences. 1996;93(25):14884–14888. doi: 10.1073/pnas.93.25.14884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garbarino-Pico E, et al. Immediate early response of the circadian polyA ribonuclease nocturnin to two extracellular stimuli. RNA. 2007;13:745–755. doi: 10.1261/rna.286507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kawai M, Green CB, et al. Nocturnin: a circadian target of Pparg-induced adipogenesis. Annals of the New York Academy of Sciences. 2010;1192(1):131–138. doi: 10.1111/j.1749-6632.2009.05221.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9(4):337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 11.Kawai M, Green CB, et al. A circadian-regulated gene, Nocturnin, promotes adipogenesis by stimulating PPAR-γ nuclear translocation. Proceedings of the National Academy of Sciences. 2010;107(23):10508–10513. doi: 10.1073/pnas.1000788107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Green CB, Douris N, et al. Loss of Nocturnin, a circadian deadenylase, confers resistance to hepatic steatosis and diet-induced obesity. Proceedings of the National Academy of Sciences. 2007;104(23):9888–9893. doi: 10.1073/pnas.0702448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McLeod MJ. Differential staining of cartilage and bone in whole mouse fetuses by alcian blue and alizarin red S. Teratology. 1980;22(3):299–301. doi: 10.1002/tera.1420220306. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2-[Delta][Delta]CT Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Guntur AR, Reinhold MI, et al. Conditional ablation of Pten in osteoprogenitors stimulates FGF signaling. Development. 2011;138(7):1433–1444. doi: 10.1242/dev.058016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teplyuk NM, Zhang Y, et al. The Osteogenic Transcription Factor Runx2 Controls Genes Involved in Sterol/Steroid Metabolism, Including Cyp11a1 in Osteoblasts. Molecular Endocrinology. 2009;23(6):849–861. doi: 10.1210/me.2008-0270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Acimovic J, Fink M, et al. CREM modulates the circadian expression of CYP51, HMGCR and cholesterogenesis in the liver. Biochemical and Biophysical Research Communications. 2008;376(1):206–210. doi: 10.1016/j.bbrc.2008.08.126. [DOI] [PubMed] [Google Scholar]
- 18.Rzonca SO, Suva LJ, et al. Bone Is a Target for the Antidiabetic Compound Rosiglitazone. Endocrinology. 2004;145(1):401–406. doi: 10.1210/en.2003-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kawai M, Delany AM, et al. Nocturnin Suppresses Igf1 Expression in Bone by Targeting the 3′ Untranslated Region of Igf1 mRNA. Endocrinology. 2010;151(10):4861–4870. doi: 10.1210/en.2010-0407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Douris N, et al. Nocturnin Regulates Circadian Trafficking of Dietary Lipid in Intestinal Enterocytes. Current biology. 2011 doi: 10.1016/j.cub.2011.07.018. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keber R, Motaln H, et al. Mouse knockout of the cholesterogenic cytochrome P450 lanosterol 14α-Demethylase (CYP51) resembles Antley-Bixler syndrome. Journal of Biological Chemistry. 2011 doi: 10.1074/jbc.M111.253245. [DOI] [PMC free article] [PubMed] [Google Scholar]