Summary
Budding of COPI-coated vesicles from Golgi membranes requires an Arf-family G protein and the coatomer complex recruited from cytosol. Arf is also required with coatomer-related clathrin adaptor complexes to bud vesicles from the trans-Golgi network and endosomal compartments. To understand the structural basis for Arf-dependent recruitment of a vesicular coat to the membrane, we determined the structure of Arf1 bound to the γζ-COP subcomplex of coatomer. Structure-guided biochemical analysis reveals that a second Arf1-GTP molecule binds to βδ-COP at a site common to the γ- and β-COP subunits. The Arf1-binding sites on coatomer are spatially related to PtdIns4,5P2-binding sites on the endocytic AP2 complex, providing evidence that the orientation of membrane binding is general for this class of vesicular coat proteins. A bivalent GTP-dependent binding mode has implications for the dynamics of coatomer interaction with the Golgi and for the selection of cargo molecules.
Introduction
The transport of proteins and lipids between intracellular compartments is mediated largely via transport vesicles, and the underlying mechanisms of vesicle budding and fusion are conserved among eukaryotes. Vesicles are formed through the action of cytosolic coat protein complexes (COPs) that assemble on a membrane surface, capture cargo molecules, and polymerize into spherical cages to sculpt the membrane into a bud (Bonifacino and Glick, 2004). Cells contain a variety of COPs: COPII vesicles transport newly-synthesized proteins from the endoplasmic reticulum (ER) to the Golgi complex; COPI- or coatomer-coated vesicles are involved in retrograde traffic from the Golgi; and the clathrin polymeric coat combines with various adaptor complexes (AP1–4) to mediate budding from the trans-Golgi network, endosomal compartments as well as from the plasma membrane (Lee et al., 2004).
The assembly of coat proteins is initiated by the activation of an Arf-family G protein on the membrane. Thus, Arf1 regulates the formation of COPI vesicles and of clathrin-coated vesicles that contain the adaptor protein (AP) complexes AP1, AP3 and AP4 (Boehm et al., 2001; Serafini et al., 1991; Stamnes and Rothman, 1993). Likewise, COPII-vesicle budding involves Sar1 G-protein, an Arf homolog (Barlowe et al., 1994). By contrast, the AP2 clathrin adaptor for endocytic vesicle formation does not utilize an Arf-family G protein in this way; instead, AP2 binds to plasma membrane via the headgroups of phosphatidyl inositol-4,5-bisphosphate (PtdIns4,5P2) lipid molecules (Jackson et al., 2010).
COPI vesicle formation on the Golgi apparatus involves the stepwise recruitment to membranes of Arf1 and coatomer, a 550 kDa protein complex of seven COPs—α-, β-, β′-, γ-, δ-, ε-, and ζ-COP (Waters et al., 1991). First, Arf1-GDP is activated by a Golgi-localized exchange factor (Peyroche et al., 1996) to replace GDP with GTP, triggering a conformational change in Arf1 whereby the myristoylated and amphipathic N-terminal α helix is displaced from a surface groove of the G-protein and embedded in the bilayer (Amor et al., 1994; Antonny et al., 1997; Goldberg, 1998). Next, Arf1-GTP recruits coatomer through a direct, GTP-dependent interaction (Serafini et al., 1991). Membrane-associated coatomer then binds to cargo molecules and self-assembles to form a polyhedral cage that molds the membrane into a COPI-coated bud (Bremser et al., 1999).
This general reaction scheme for Arf1-dependent coat formation is probably conserved among the classes of clathrin vesicles that employ AP1, AP3 and AP4 tetrameric adaptors (see, for example, Boehm et al. [2001]). Indeed, the tetrameric βδ/γζ-COP complex of coatomer is related to APs in the sequence of its subunits and presumably in its overall architecture. A distinctive feature of the COPI system is that βδ/γζ-COP remains bound to the cage-forming αβ′ε-COP complex in cytosol, and the heptameric particle is recruited en bloc to Golgi membranes (Hara-Kuge et al., 1994), unlike the stepwise accretion of clathrin/AP and COPII (Sec23/24 and Sec13/31 components).
The architecture of AP2 and the molecular basis for its recruitment to membranes via PtdIns4,5P2 have been explored in considerable detail, and this body of work serves as a framework for exploring coat recruitment to membranes (Collins et al., 2002; Jackson et al., 2010). A PtdIns4,5P2-binding site on one of the large subunits (α-AP or β2-AP) is the primary point of attachment of AP2 tetramer to the membrane. Next, an extensive conformational change ‘opens’ the AP2 tetramer to afford a multivalent interaction with membrane via additional PtdIns4,5P2 binding sites (possibly four in total, including sites on β2- and μ2-adaptin subunits) and via cytoplasmic transport signals on transmembrane cargo proteins (Jackson et al., 2010).
Far less is known about the molecular basis for Arf-dependent assembly of coatomer and AP coat proteins. In this study, we combine structural and biochemical analyses of βδ/γζ-COP and Arf1 to provide a model for GTP-dependent recruitment of coatomer to membrane. The crystal structure of γζ-COP bound to Arf1 and a non-hydrolyzable GTP analog explains how GTP binding initiates coat recruitment, by defining the GTP-mediated changes in Arf1 that facilitate interaction with the coat protein. Strikingly, we find that βδ/γζ-COP interacts with two molecules of Arf1-GTP, which bind to quasi-equivalent sites on the large β-COP and γ-COP subunits. On the basis of this analysis, we propose a model for a bivalent GTP-dependent interaction of coatomer with membranes.
Results and Discussion
Experimental Approach
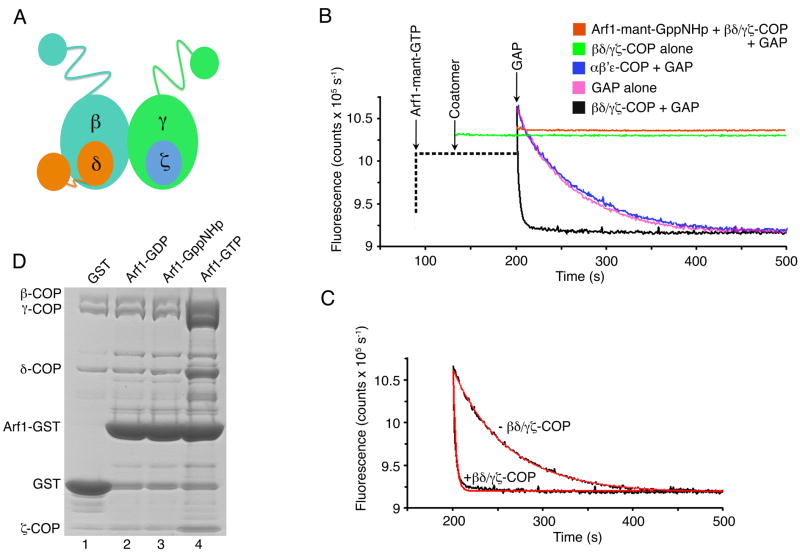
Interactions between Arf1-GTP and subunits of the βδ/γζ-COP complex were reported in biochemical cross-linking studies and via yeast two-hybrid analysis (Eugster et al., 2000; Sun et al., 2007). Likewise, interactions were described between Arf1 and subunits of AP4, involving the α-solenoid domain of ε-adaptin (equivalent to γ-COP) and the C-terminal domain of μ-adaptin, which is equivalent to δ-COP (Boehm et al. [2001], and see Figure 1A for an overview of βδ/γζ-COP and AP architecture). To map Arf1-binding sites to discrete domains of βδ/γζ-COP, we prepared the tetrameric, dimeric (βδ-COP and γζ-COP) and various truncated forms of the mammalian proteins by coexpression in baculovirus-infected insect cells (see Experimental Procedures). We then tested these proteins for an interaction with Arf1 using two distinct assays. The first assay used the truncated soluble form of human Arf1 (lacking the membrane-anchoring residues 1–17 [Paris et al., 1997]; hereafter referred to as human Arf1) bound to the fluorescent GTP analog mant-GTP (3′-O-[N-methyl-anthraniloyl]-GTP).
Figure 1. Assays for Coatomer Interaction with Arf1.
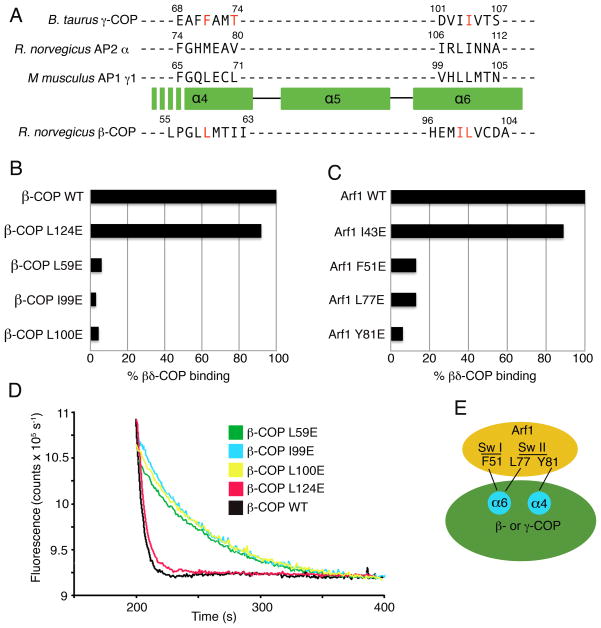
(A) Schematic diagram of βδ/γζ-COP, the cargo-binding tetramer of the COPI coat, based on the expected close similarity to AP adaptor complexes. Small (30 kDa) appendage domains on β- and γ-COP are joined to the α-solenoid (also called trunk) domains via flexible linkers. β- and γ-COP are related in sequence and structure. Likewise ζ-COP is related to the N-terminal domain of δ-COP, but the C-terminal domain of δ-COP (equivalent to the C-terminal domain of the μ2-AP subunit of AP2) breaks the —symmetrical relationship of γζ-COP and βδ-COP.
(B) The fluorescence GTPase assay. The graph shows time courses for four representative reactions. The fluorescence output of a solution of 0.5 μM soluble (lacking membrane-anchor residues 1–17) human Arf1-mant-GTP or Arf1-mant-GppNHp was monitored upon addition of various forms of coatomer, at a fixed concentration of GAP (40 μM human GAP1 catalytic core, residues 1–136). The left-hand portion of the curves, drawn as a dotted line, indicates the order of addition of the various protein components. A control experiment (orange curve) shows Arf1-mant-GppNHp in the presence of GAP and 0.1 μM βδ/γζ-COP; there is no fluorescence decrease with this non-hydrolyzable substrate. The experimental curve in magenta shows GTP hydrolysis on Arf1-mant-GTP caused by GAP alone. The curve in black shows rapid GTP hydrolysis on Arf1-mant-GTP catalyzed by GAP plus 0.1 μM βδ/γζ-COP. The blue curve shows that the cage-forming αβ′ε-COP complex (0.1 μM) has no effect on GAP-catalyzed GTP hydrolysis. Finally, the green curve shows that in the absence of GAP, βδ/γζ-COP alone has no affect on GTP hydrolysis.
(C) The graph shows least-squares fits to first order exponentials (red lines), using data (black lines) from (B). In these experimental conditions (see Experimental Procedures), the rate constant is 0.016 s−1 in the absence of βδ/γζ-COP, and 0.32 s−1 in the presence of 0.1 μM βδ/γζ-COP.
(D) Binding and elution (pull-down) assay to test the interaction between coatomer and yeast Arf1. GST or GST-Arf1 (residues 18–181) protein complexed with GDP, GppNHp or GTP was immobilized on glutathione sepharose beads. The beads were incubated with purified βδ/γζ-COP and then washed. (Total bound proteins are shown in Figure S1). GAP protein was added to trigger GTP hydrolysis and elution of βδ/γζ-COP. Eluted protein was analyzed by SDS-PAGE and Coomassie blue staining. The Arf1-GST bands released in the elution step represent only ~2.5% of the bead-bound Arf1-GST. See also Figure S1.
Figures 1B and 1C show an initial set of experiments in which the fluorescence output of a 0.5 μM solution of human Arf1-mant-GTP was continuously monitored (λ = 438 nm) as COPI proteins and Arf-GTPase activating protein (GAP; the catalytic core of human GAP1, residues 1–136) were added. This fluorometric assay monitors a decrease in fluorescence due to GTP hydrolysis, because Arf1-mant-GDP has a lower fluorescence output than Arf1-mant-GTP (see, for example, Ahmadian et al. [1997] for details of the mant fluorescence assay with Ras). Since the rate of GAP-dependent GTP hydrolysis is increased by coatomer (Goldberg, 1999), the assay reports on the interaction between Arf1 and COPI. In an initial experiment, the presence of GAP alone with the fluorescent Arf1 substrate caused a slow decrease in fluorescence (magenta curve in Figure 1B). The addition of 0.1 μM full-length βδ/γζ-COP to the solution containing GAP and Arf1-mant-GTP accelerated GTP hydrolysis 20-fold (black curve in Figure 1B). βδ/γζ-COP alone did not catalyze GTP hydrolysis (green curve); rather it synergized with GAP to accelerate GTP hydrolysis on Arf1. To confirm that the assay monitors GTP hydrolysis, we bound human Arf1 to the non-hydrolyzable form of the fluorescent nucleotide, mant-GppNHp, and found that the addition of βδ/γζ-COP and GAP caused no change in fluorescence (orange curve in Figure 1B). Finally, the addition of αβ′ε-COP had no effect on GTP hydrolysis; it did not accelerate hydrolysis beyond the GTPase rate due to GAP alone (blue curve in Figure 1B).
Figure 1C shows exponential fits to two of the datasets, indicating that under these experimental conditions the addition of 0.1 μM βδ/γζ-COP increases the GTPase rate about 20-fold (acceleration of the GTPase rate is several hundred-fold at the highest coatomer concentrations). We note in passing that it is currently unclear whether coatomer-stimulated GTP hydrolysis has a physiological role, in the context of full-length myristoylated Arf1, intact GAP1 protein, and Golgi membranes (Kliouchnikov et al., 2009). The purpose here is to utilize the coatomer stimulation of GTP hydrolysis as a sensitive and straightforward test for coatomer interaction with Arf1.
Figure 1D outlines the second, complementary assay for Arf1-GTP interaction with coatomer. We expressed yeast Arf1 (as before, this is the soluble truncated form of the G protein lacking residues 1–17) fused to the N terminus of glutathione S-transferase (GST), then replaced the bound nucleotide on Arf1 with GDP, GTP or GppNHp, and immobilized the fusion protein (or GST alone) on glutathione sepharose beads. We probed for a GTP-dependent interaction with βδ/γζ-COP in a two-stage procedure. First, purified βδ/γζ-COP was incubated with immobilized GST-Arf1 and the beads were washed to remove all unbound protein (protein bound to the beads at this stage is shown in Figure S1). Second, catalytic amounts of GAP protein were added to trigger GTP hydrolysis on bead-bound Arf1-GTP, thereby releasing coat proteins (see Experimental Procedures). As shown in Figure 1D, appreciable amounts of βδ/γζ-COP were eluted only from Arf1-GTP (lane 4). Although βδ/γζ-COP bound to Arf1-GppNHp in the first stage of the assay (Figure S1, lane 3), the coat protein was not eluted in the second stage because GTP hydrolysis is blocked by the non-hydrolyzable bond of GppNHp. (Hereafter we refer to this assay as the pull-down assay.)
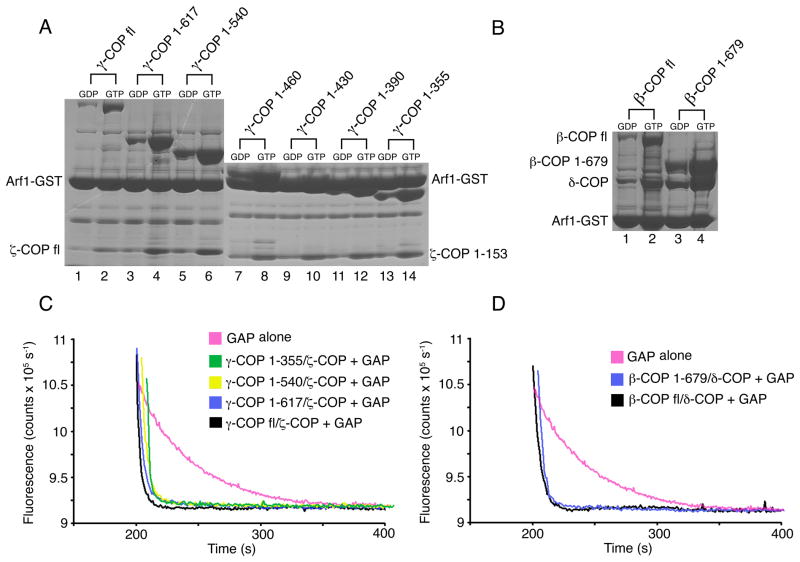
Identification of a Core γζ-COP Complex that Binds to Arf1-GTP
To map the Arf1-binding site on the βδ/γζ-COP complex we focused first on full-length and truncated forms of γζ-COP (Figure 2). Full-length γζ-COP bound to Arf1-GTP in the pull-down assay (Figure 2A, lane 2), and it retained the ability to accelerate GTP hydrolysis in the fluorescence assay (Figure 2C, black curve). Likewise, a form of γζ-COP lacking the C-terminal appendage domain of γ-COP (γ-COP 1–617) interacted with Arf1-GTP (Figure 2A, lane 4; Figure 2C, blue curve).
Figure 2. Identification of Arf1-binding sites on βδ/γζ-COP.
(A) GTP-dependent binding of Arf1 to γζ-COP dimers. The pull-down assay was utilized to test the Arf1-binding capacity of truncated forms of γ-COP. In the experiments, γ-COP was present in a dimer with full-length ζ-COP (lanes 1–6) or truncated (1–153) ζ-COP (lanes 7–14). All of the dimeric proteins, from the longest to the shortest forms (left to right), bind to Arf1.
(B) GTP-dependent binding of Arf1 to βδ-COP dimers. Arf1 binds to full-length βδ-COP (lane 2) and to a βδ-COP dimer lacking the β-COP appendage domain (lane 4).
(C) Various forms of γζ-COP, including the minimal γζ-COP dimer (γ-COP 1–355 and ζ-COP 1–153) identified in (A), are fully active in the fluorescence assay—that is, all synergize with GAP to catalyze GTP hydrolysis on Arf1. For clarity, the curves have been offset incrementally along the x-axis.
(D) The βδ-COP dimer can also synergize with GAP to catalyze GTP hydrolysis on Arf1. Curves have been offset incrementally along the x-axis.
Next, we prepared a series of γζ-COP truncated proteins that lack increasingly large portions of the C-terminus of the α-solenoid domain of γ-COP, together with just the structured core region of ζ-COP (residues 1–153). All the γ-COP proteins, including the shortest form (γ-COP 1–355) retain a dimer interaction with ζ-COP, as expected from knowledge of the related ασ-adaptin interaction within the AP2 crystal structure (Collins et al., 2002). As shown in Figure 2A, all the γζ-COP proteins interacted with Arf1. Figure 2C shows a subset of these complexes tested in the fluorescence assay, indicating that the shortest form of γ-COP, 1–355, retains the ability to accelerate GTP hydrolysis on Arf1-GTP.
The 1–355 polypeptide was the shortest form of γ-COP that remained stable during purification of the dimeric protein; shorter polypeptides probably compromise the stability of the α-solenoid C terminus in the vicinity of the binding site for ζ-COP. We conclude that the dimeric protein comprising γ-COP 1–355 and ζ-COP 1–153 constitutes a core complex that binds to Arf1-GTP, and it should be amenable to structural analysis.
A Second Binding Site for Arf1 on the βδ/γζ-COP Complex
The stoichiometry of the coatomer:Arf1 complex is unknown. When we tested the full-length βδ-COP dimer in the pull-down assay we were surprised to observe that it also bound to Arf1-GTP (Figure 2B, lane 2), and it stimulated GTP hydrolysis in the fluorescence assay (Figure 2D, black curve). A truncated form of βδ-COP lacking the β-COP appendage domain behaved similarly (Figures 2B and 2D). We conclude that coatomer interacts with a second molecule of Arf1-GTP. Rather than defining the Arf1-binding site on βδ-COP by truncation analysis, we mapped the site more precisely by structure-guided mutagenesis based on the crystal structure of γζ-COP bound to Arf1-GppNHp (see below).
Structure Determination of γζ-COP Bound to Arf1
Initial crystallization experiments focused on complexes of truncated bovine γζ-COP and human Arf1, but the various crystal forms we obtained were all of poor crystallographic quality. Instead, the complex comprising γζ-COP core (bovine γ-COP 1–355 plus ζ-COP 1–153) and S. cerevisiae Arf1 (residues 18–181) complexed with GppNHp crystallized in space group P42212 with two copies of the γζ-COP-Arf1 complex in the asymmetric unit, and the crystals diffracted X-rays to at least 2.9 Å resolution. The structure was determined by the single-wavelength anomalous diffraction method using selenium as the anomalous scatter, and refined to 2.9 Å resolution (see Experimental Procedures and Table 1).
Table 1.
Data Collection and Refinement Statistics
| Crystal | Native | Selenomethionine |
|---|---|---|
| Space group | P42212 | P42212 |
| Cell parameters (Å) | a=b=163.579, c=145.188 | a=b=163.827, c=144.928 |
| Data processing: | ||
| Wavelength (Å) | 1.075 | 0.9791 |
| Resolution (Å) | 50–2.9 | 50–3.2 |
| Measured reflections | 591546 | 780725 |
| Unique reflections | 83327 | 61351 |
| Rmerge (%)a | 6.1 (43.2) | 11.5 (47.6) |
| I/σ | 32.1 (4.8) | 28.6 (5.99) |
| Completeness (%) | 99.9 (100) | 99.9 (100) |
| Redundancy | 7.1 (7.1) | 12.7 (12.9) |
| Refinement statistics: | ||
| Data range (Å) | 35–2.9 | |
| Reflections | 83278 | |
| Non-hydrogen atoms | 8978 | |
| R.m.s. Δ bonds (A)b | 0.009 | |
| R.m.s. Δ angles (°)b | 1.29 | |
| R-factor (%)c | 20.6 | |
| Rfree (%)c, d | 26.0 | |
Highest resolution shell is shown in parenthesis.
Rmerge = 100 × ΣhΣi | Ii(h) − <I(h)> |/Σh<I(h)>, where Ii(h) is the ith measurement and <I(h)> is the weighted mean of all measurement of I(h) for Miller indices h.
Root-mean-squared deviation (r.m.s. Δ) from target geometry.
R-factor = 100 × Σ|FP − FP(calc)|/Σ FP.
Rfree was calculated with 5% of the data.
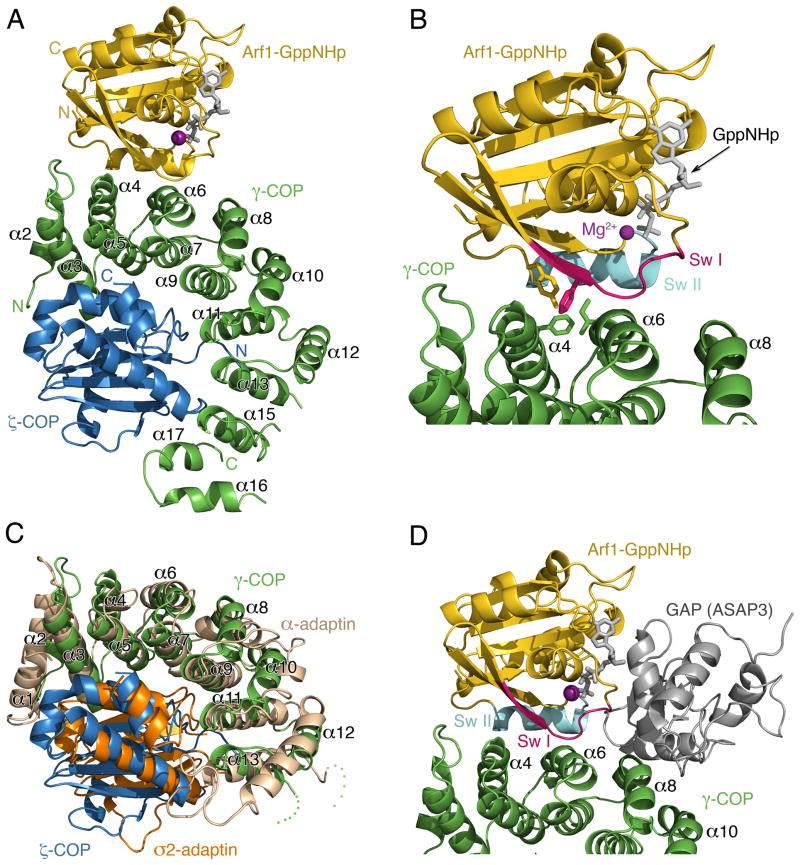
Overall Architecture of γζ-COP Bound to Arf1-GppNHp
The molecular model of γζ-COP bound to Arf1-GppNHp is presented in Figure 3. In the structure, the first ~315 residues of γ-COP adopt an α-solenoid fold (a curved, right-handed superhelix of α helices) comprising fifteen α helices in total, and the α-solenoid domain binds as an arc around the ζ-COP subunit (Figure 3A). ζ-COP closely resembles the longin-fold σ-adaptin subunit of the AP2 adaptor complex (Figure 3C). We have numbered the α helices of the α-solenoid fold according to the scheme introduced by Collins et al. (2002) for AP2. This can be assigned straightforwardly because there is a relatively close overlap (Figure 3C) of our crystal structure with the corresponding region of the AP2 complex (γ-COP is related to the α-adaptin subunit of AP2). The two structures are similar despite their negligible sequence homology (18% sequence identity between the ~350 amino acids of the α-solenoid regions).
Figure 3. Crystal Structure of γζ-COP Complexed with Arf1-GppNHp.
(A) Ribbon diagram with γ-COP colored green, ζ-COP blue and Arf1 gold. The α helices of the γ-COP α-solenoid domain are labeled according to the scheme introduced by Collins et al. (2002) for the AP2 adaptor complex (γ-COP is related to the α-adaptin subunit of AP2).
(B) Close up view in the same orientation as (A). The switch I and II elements of the G protein are indicated (red and cyan, respectively), and the side chains of several key interfacial residues are included (a more detailed analysis of interfacial residues is presented in Figure 4).
(C) Structural overlap of γζ-COP and the corresponding region of the ασ-adaptin dimer of AP2 (Collins et al., 2002). γ- and ζ-COP are colored green and blue as before; σ2-adaptin is orange and α-adaptin is light brown.
(D) A composite molecular model that includes the GAP catalytic domain of ASAP3, taken from the crystal structure of ASAP3 bound to Arf6 (Ismail et al., 2010). The model was created by superimposing the Arf6 and Arf1 structures. The picture is oriented as in (A).
Arf1 contacts residues of helices α4 and α6 on the outer surface of the γ-COP arc (Figure 3A and 3B). Thus, the G protein makes no contacts with ζ-COP, but it is apparent that ζ-COP is required in the dimer to stabilize γ-COP in its interaction with Arf1.
GTP-Dependent Contacts at the Interface of γζ-COP and Arf1
In the crystal structure there is clear electron density for bound GppNHp, and yeast Arf1 closely resembles the conformation of the isolated human Arf1-GppNHp protein (Goldberg, 1998). The GTP conformation is highly distinct from that adopted by Arf1-GDP, in which the switch I and II regions adopt altered conformations and the N-terminal membrane anchor’ is retracted into a surface groove of the protein (Amor et al., 1994). The finding that Arf1 adopts a canonical GTP-like conformation in complex with γζ-COP is important, as we can surmise that GTP binding to Arf1 will have two conformational consequences—the membrane anchor is released for interaction with the bilayer and the affinity for γζ-COP is increased—the net effect of which is membrane recruitment of coatomer. The molecular mechanism for Arf conformational switching leading to anchor release is understood in considerable detail (Amor et al., 1994; Beraud-Dufour et al., 1999; Goldberg, 1998), and our study now defines the GTP-triggered changes to Arf1 switch regions that facilitate interaction with coatomer.
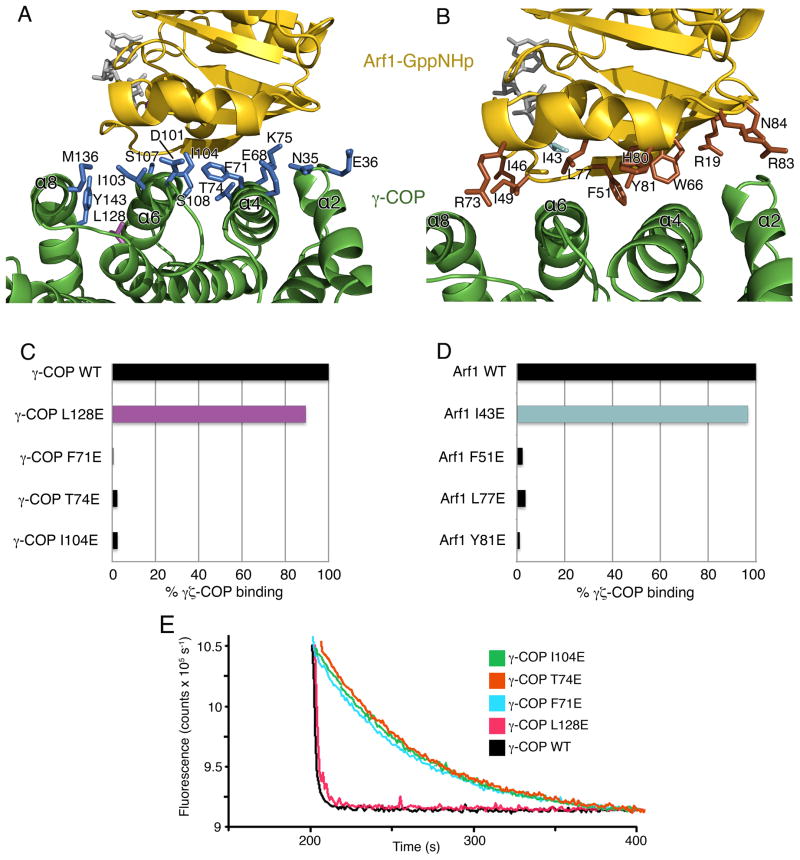
At the interface of Arf1-GppNHp and γζ-COP a set of predominantly hydrophobic side chains contribute to interactions (Figure 4). These are supplied mainly by helices α4 and α6 of γ-COP (Figure 4A) and the switch I and II elements of Arf1 (Figure 4B). There are just two hydrogen-bonding interactions at the center of the interface: residue Lys75 of γ-COP binds to the carbonyl oxygen atom of Arf1 residue His 80, possibly interacting with the helix dipole of the switch 2 α helix; and the side chain groups of γ-COP residue Thr74 and Arf1 residue Tyr 81 are hydrogen bonded. The central region of the interface involves a cluster of hydrophobic side chains that is likely to be especially important for protein-protein binding: namely, Arf1 residues F51, L77 and Y81, and γ-COP residues F71, T74 and I104. We mutated these residues individually to glutamic acid (to cause localized but significant changes) and tested the effect on the protein-protein interaction. The goals of this analysis were to analyze the interaction of Arf1 with γζ-COP, and also to pinpoint residue changes that we could exploit in subsequent efforts to map the interface of βδ-COP and Arf1. The six γ-COP and Arf1 mutations target surface-exposed side chains, and all the mutant proteins were soluble and well behaved during purification. We tested the γ-COP mutations (changes were made to full-length γ-COP and then coexpressed as γζ-COP dimers) in the pull-down assay and found that binding to Arf1-GTP was essentially abolished for all three mutant proteins (Figure 4C). As a control experiment, we generated an additional mutation, L128E of γ-COP, in this case targeting a surface residue distal to the protein-protein interface, and found that the mutant γζ-COP dimer retains approximately 90% of the binding capacity of wild-type protein (Figure 4C). To corroborate the results, we tested the mutant proteins in the fluorescence assay (Figure 4E). As expected, the control mutation L128E stimulated GTP hydrolysis almost as potently as wild-type γζ-COP, whereas F71E, T74E and I104E had no effect, and did not stimulate GTP hydrolysis beyond the rate due to GAP alone.
Figure 4. Interacting Surfaces of γζ-COP and Arf1.
(A) Close-up view showing the interface of γ-COP (green) and Arf1 (gold). Side-chain groups from γ-COP that reside at the interface (within 3.5Å of Arf1) are drawn in blue. One γ-COP residue not at the interface, Leu128, is colored purple; this residue was chosen as a control mutation for experiments shown in (C) and (E). The picture is rotated approximately 180° around a vertical axis relative to Figure 3A.
(B) Close-up view in a similar orientation to (A), with side-chain groups from Arf1 colored brown. In the Arf1 mutagenesis experiments, residue I43 (colored cyan) was chosen as a control mutation since it is distant from the protein-protein interface.
(C) Bar graph shows the effects of mutating γ-COP interfacial residues on the interaction between γζ-COP and Arf1, as measured by the pull-down assay. Mutations were introduced into full-length γ-COP. Binding is expressed as a percentage of the binding of wild type γζ-COP. The control γ-COP mutant, L128E, corresponds to the side chain colored purple in (A).
(D) Bar graph shows the effects of mutations in S. cerevisiae Arf1 on the interaction with full-length γζ-COP. Note that the interfacial residues chosen for mutagenesis—F51, L77 and Y81—are identical in human Arf1. The Arf1 control mutant I43E (cyan) is a valine residue in the human Arf1 sequence.
(E) The ability of γ-COP mutants to synergize with GAP in GTP hydrolysis was measured using the fluorescence assay. The effect of the γ-COP mutations is essentially the same as in the pull-down assay (C). Curves have been offset incrementally along the x-axis.
Experiments with the yeast Arf1 mutations gave a consistent set of results (Figure 4D). The three interface mutations, F51E, L77E and Y81E, caused almost complete loss of binding to γζ-COP in the pull-down assay; by contrast, a control mutant, I43E, that lies outside the protein-protein interface, bound to γζ-COP almost as tightly as wild-type Arf1-GTP.
In choosing Arf1 amino-acid residues for mutagenesis that could disrupt the interaction with coatomer we avoided sites that are implicated in the interaction with GAP protein, according to the crystal structure of the catalytic core of the GAP ASAP3 complexed with Arf6 (Ismail et al., 2010). This statement is of course redundant, in that the observed synergy of coatomer and GAP in catalyzing GTP hydrolysis almost certainly involves a tripartite complex of Arf1-GAP-coatomer with non-overlapping binding sites on Arf1-GTP for GAP and coatomer. But it is worth emphasizing the rarity of this arrangement—most Ras-superfamily G proteins bind their GAPs and effectors at overlapping binding sites so that effector stimulation of GTP hydrolysis is impossible (Vetter and Wittinghofer, 2001). In Figure 3D we show a composite model of γζ-COP and Arf1 plus the GAP catalytic domain of ASAP3 (taken from Ismail et al. [2010]). The GAP molecule binds over the GTPase active site and inserts an arginine ‘finger’ residue that bonds with the nucleotide to assist catalysis (Ismail et al., 2010). Importantly, there is no steric overlap of the GAP and γ-COP polypeptides in the composite model, although the two molecules converge closely in the vicinity of the switch II element (colored cyan in Figure 3D) near to the GTPase active site.
At present, we can only speculate on the mechanism by which coatomer stimulates GTP hydrolysis in the tripartite complex. There are two general possibilities: contacts between coatomer and GAP could enhance GAP affinity for Arf1-GTP; alternatively, coatomer may make a more direct catalytic contribution through interactions with switch II that assist in the proper positioning of the substrate water molecule (Kliouchnikov et al., 2009; Luo and Randazzo, 2008). Resolution of this issue will require further biochemical analyses together with a crystal structure of the tripartite complex.
Location of the Arf1-Binding Site on βδ-COP
The large γ-COP and β-COP subunits of coatomer are evolutionarily related, and will likely have similar overall structures based on knowledge of the corresponding subunits in the AP1 and AP2 crystals (Collins et al., 2002; Heldwein et al., 2004). Thus, the simplest model that explains our biochemical findings is that Arf1-GTP binds to βδ-COP and γζ-COP at quasi-equivalent sites on the large subunits. To test this, we targeted key residues on the α4 and α6 helices of β-COP for mutagenesis, based on a sequence alignment of the large subunits. Since γ-COP and β-COP have very low sequence homology (14% identity in the α-solenoid region of the mammalian proteins), we aligned the sequences in two steps (Figure 5A). First, we superimposed the crystal structures of γ-COP, α-AP and γ1-AP to yield a structure-based alignment of the α4–α6 region, and then used this to assist the sequence alignment of β-COP.
Figure 5. Mapping the Arf1-binding site on βδ-COP.
(A) Sequence- and structure-based alignment of the large subunits of COPI and AP complexes, in the vicinity of the α4- and α6-helices of the α-solenoid domain. The three upper sequences are aligned based on a structural overlap of γ-COP (this study), α2-adaptin (from the crystal structure of AP2 determined by Collins et al. [2002]) and γ1-adaptin (from the AP1 crystal structure determined by Heldwein et al. [2004]). The sequence of β-COP is aligned to the others based on sequence homology including the conservation of helical repeats of the α-solenoid fold. The residues of γ-COP and β-COP selected for mutagenesis are colored red. The location of β-COP residue Pro56 implies that the α4 helix will start downstream of this residue in β-COP; consistent with this, the key interface residues of γ-COP are located toward the C-terminal end of the α4 helix.
(B) Bar graph shows the effects of β-COP mutations on the interaction between βδ-COP and Arf1, as measured by the pull-down assay. Mutations were introduced into full-length β-COP protein. The β-COP residue L124 was selected as a site distant from the interface with Arf1; this residue corresponds to the mutation L128E on γ-COP (see Figure 4A).
(C) Bar graph shows the effects of mutations in S. cerevisiae Arf1 on the interaction with full-length βδ-COP. This is the same set of mutations that were used to test γζ-COP interactions in Figure 4D.
(D) The ability of β-COP mutants to synergize with GAP in GTP hydrolysis was measured using the fluorescence assay. The effect of the β-COP mutations is essentially the same as in the pull-down assay (B). Curves have been offset incrementally along the x-axis.
(E) Summary of the mapping experiments. Arf1 probably binds in a similar manner to γ-COP and β-COP, whereby the Arf1 switch regions interact with residues of the α4- and α6-helices of the α-solenoid domain.
On this basis, we selected three putative binding-site residues for mutagenesis—L59E, I99E and L100E, and these were incorporated into full-length β-COP. In the pull-down assay, all three mutant βδ-COP dimers lost 95% or more of the binding capacity to Arf1-GTP relative to wild-type βδ-COP (Figure 5B). By contrast a control mutation L124E, introduced at a distal site on β-COP, retained almost full binding capacity. Concordant results were obtained with the fluorescence assay; the control mutant, L124E, stimulated GTP hydrolysis almost as potently as wild-type βδ-COP, whereas L59E, I99E and L100E mutant proteins had no significant effect on the GAP-catalyzed GTPase reaction (Figure 5D).
Finally, the yeast Arf1 mutants that severely impaired interactions with γζ-COP were tested against full-length, wild-type βδ-COP protein (Figure 5C). Arf1 mutants F51E, L77E and Y81E retained at most 13% binding capacity to βδ-COP relative to wild-type Arf1, whereas the control Arf1 mutant I43E retained 90% binding capacity to βδ-COP.
In summary, these results strongly support a model in which Arf1-GTP molecules bind in a similar fashion to the β-COP and γ-COP subunits of coatomer. Although the atomic details will differ somewhat (considering the very low sequence homology of the two proteins), we predict a general binding mode for β-COP and γ-COP with Arf1-GTP involving hydrophobic residues contributed by the α4 and α6 helices that interact with a circumscribed surface region of the switch I and II elements of Arf1 (summarized in the schematic Figure 5E).
A Model for Arf1-Dependent Recruitment of Coatomer to Membranes
The finding that Arf1-GTP binds to both γζ-COP and βδ-COP is striking, but our data do not preclude the existence of additional Arf1-binding sites on coatomer. To address this issue, we introduced the Arf1 binding-site mutations, γ-COP I104E and β-COP L100E, into βδ/γζ-COP complexes that contained full-length versions of all four subunits, and then tested for binding to Arf1-GTP. As shown in Figure 6B, when we mutated the Arf1-binding site on γ-COP and βCOP individually, the interaction of βδ/γζ-COP with Arf1-GTP was reduced but not abolished. By contrast, the double mutation reduced the interaction to background levels. When we repeated the experiments using the fluorescence assay (Figure 6C) we obtained very similar results. The single-site mutants accelerated GTP hydrolysis at an intermediate rate, whereas the double-mutant form of βδ/γζ-COP had no effect on GTP hydrolysis; it did not accelerate hydrolysis beyond the GTPase rate due to GAP alone (Figure 6C, green curve).
Figure 6. Model for Membrane Recruitment of Coatomer.
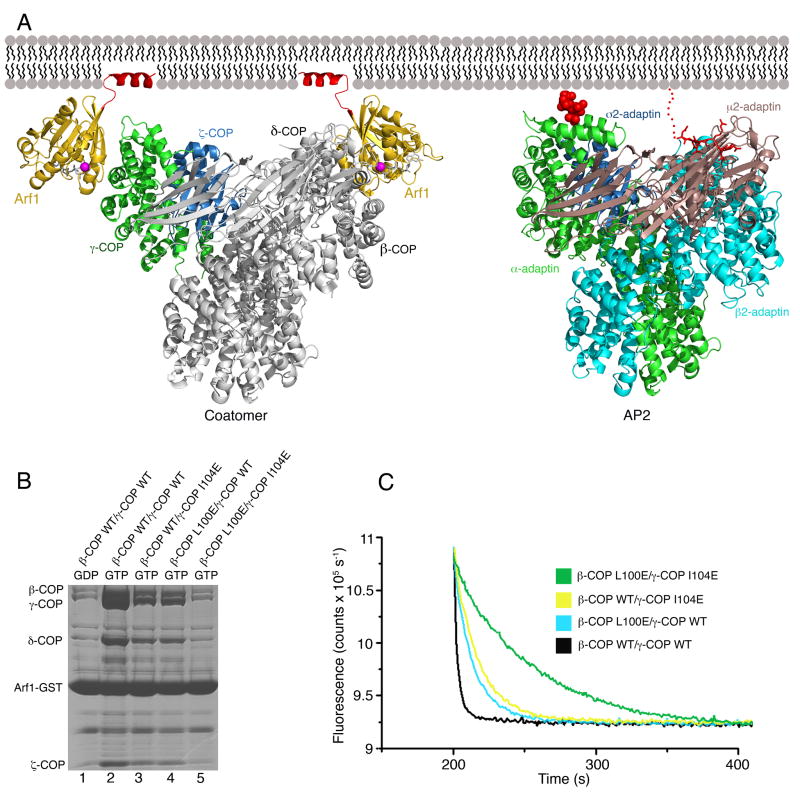
(A) Left panel shows a composite model of βδ/γζ-COP bound to membrane via two molecules of Arf1-GTP. The γζ-COP/Arf1 crystal structure is colored (following the scheme used in Figure 3) as is the second molecule of Arf1. The remainder of βδ/γζ-COP, in grey, is modeled based on homology with the AP2 adaptor complex; specifically, we used the crystal structure of the open conformation of AP2 (Jackson et al., 2010). The N-terminal amphipathic α helices of Arf1 (colored red) are modeled in their expected locations as membrane anchors. The right panel shows the structure of the open form of AP2 (as described by Jackson et al. [2010]) modeled in an interaction with membrane via a PtdIns4,5P2 headgroup (van der Waals spheres colored red) bound to the primary site on α-AP and a YxxΦ cargo sorting motif (stick representation colored red) that interacts with the μ2-AP subunit. For clarity, we have drawn AP2 in the same orientation as βδ/γζ-COP; this required only a slight shift of the membrane-bound orientation proposed by Jackson et al. (2010). See also Figure S2.
(B) Effects of mutations in the Arf1-binding sites of full-length βδ/γζ-COP complex, measured using the pull-down assay. Single mutations reduce but do not abolish Arf1 interaction (lanes 3 and 4), whereas a double mutation binds to Arf1 at background levels (compare lane 5 to lane 1).
(C) The effects of single and double mutations in the βδ/γζ-COP complex on GTP hydrolysis in the fluorescence assay.
Taken together with the finding that αβ′ε-COP does not bind to Arf1, these results imply that heptameric coatomer has a total of two binding sites for Arf1-GTP. On this basis, we built a composite model of βδ/γζ-COP in a bivalent, GTP-dependent interaction with membrane (Figure 6A, left panel). The model combines the γζ-COP/Arf1 crystal structure and the AP2 structure in its open conformation (Jackson et al., 2010). Since the γζ-COP and ασ-AP complexes overlap closely (Figure 3C), we simply extended the α-solenoid domain of γ-COP based on the α-AP structure. We modeled the second molecule of Arf1-GTP bound to β-COP, basing β-COP on β2-AP, and with the Arf1 molecule contacting the α4 and α6 helices in the same manner as observed in the γζ-COP-Arf1 crystals.
The presence of not one but two molecules of Arf1-GTP strongly constrains the possible orientations of the complex relative to the membrane surface. The membrane anchor of Arf1 (residues 1–13) is joined to the G-protein core by a very short—3 or 4 residue long—linker region (Liu et al., 2010), so we modeled the Arf1 molecules in close proximity to the membrane and in a symmetrical arrangement (i.e. an approximate two-fold axis normal to the membrane plane). The two molecules of Arf1 are approximately 110 Å apart in the model. Importantly, the membrane-bound orientation of the βδ/γζ-COP model is similar to that proposed by Jackson et al. (2010) for the open form of the AP2 tetramer, which adheres to membrane via PtdIns4,5P2 molecules (one PtdIns4,5P2 molecule is drawn in Figure 6A, right panel, bound to the primary binding site on α-AP). The conserved binding mode arises because the Arf1-binding sites on coatomer are spatially related to the PtdIns4,5P2-binding sites on AP2. More specifically, the membrane-proximal elements of the Arf1 molecule bound to γ-COP are in the vicinity of the primary PtdIns4,5P2 molecule bound to α-AP (Figure 6A); and the membrane-proximal elements of the Arf1 molecule bound to β-COP are in the vicinity of the PtdIns4,5P2 molecule at β2–AP (not shown in Figure 6A; see Jackson et al. [2010]). These observations suggest that the orientation of membrane binding is general for the COPI and AP class of coat proteins. Future studies should address the idea of a broadly applicable binding mode, and also whether AP1, AP3 and AP4 interactions with membrane involve one or two Arf1-GTP molecules adhered to the α4 and α6 helices of the large AP subunits.
The Sec23/24 complex of the COPII coat bears no resemblance to the COPI/AP coat proteins, despite being functionally equivalent. Sec23/24 binds to the Arf homolog Sar1, and in a Sec23/24-Sar1 model for membrane binding, Sar1 is oriented towards membrane in a similar manner to Arf1 in the βδ/γζ-COP complex (Figure S2). The Sec23 and Sec24 subunits are evolutionarily related, but only Sec23 binds to Sar1-GTP; in yeast Sec24, the equivalent site has been adapted to interact with the t-SNARE protein Sed5 (Mossessova et al., 2003).
This highlights the key question as to why coatomer binds to two molecules of Arf1-GTP. Studies of the endocytic AP2 adaptor suggest a possible answer (Jackson et al., 2010). AP2 undergoes a striking open-to-closed (also referred to as open-to-locked) conformational transition. In cytosol, AP2 adopts the closed form with low affinity for PtdIns4,5P2 and for cargo sorting signals (Collins et al., 2002). The shift to the open form is driven by binding to PtdIns4,5P2 and to cargo signals, and these binding events synergize such that AP2 binds stably and preferentially to cargo- and PtdIns4,5P2-enriched membranes (Jackson et al., 2010). Given the close structural and functional homology among the tetrameric adaptors, AP1–4 and COPI, it seems likely that related open-to-closed conformational transitions will operate to couple membrane binding to cargo packaging for all these coats. Jackson et al. (2010) have suggested that Arf1-GTP will act in a similar fashion to PtdIns4,5P2 to drive protein conformation toward the open state upon binding to AP1, AP3, AP4 and COPI; indeed, synergistic binding of cargo and Arf1-GTP has been reported in the case of AP1 (Lee et al., 2008). This proposal is supported by the close spatial relationship of the major PtdIns4,5P2 and Arf1-GTP binding sites. We suggest the following working model for coatomer binding to membrane, by analogy with the AP2 system (Jackson et al., 2010). First, the interaction of one molecule of membrane-bound Arf1-GTP with coatomer (via the site on γζ-COP) is the primary event that recruits a closed form of coatomer to membrane. Next, a second molecule of Arf1-GTP binds to coatomer at the βδ-COP site and thereby shifts the conformational equilibrium toward the open form of coatomer; this process synergizes with binding of cargo transport signals, which also serve to stabilize the open conformation (i.e. as observed for AP1 by Lee et al. [2008]). Our biochemical data do not address whether or to what extent Arf1 binding may elicit such conformational changes. Future studies on the large (~280 kDa) βδ/γζ-COP complex—involving a thorough dissection of βδ-COP—should address whether Arf1 binding to coatomer triggers AP2-like changes to the solenoid core of the tetramer and to the C-terminal domain of δ-COP (Jackson et al., 2010), to expose binding sites for cargo transport signals. To date, no transport signals have been identified that interact with βδ/γζ-COP at sites equivalent to the binding sites for YxxΦ and [ED]xxxL[LI] signals on AP2. Perhaps Arf1-GTP binding to βδ/γζ-COP will be needed to properly detect and dissect these cargo/coat interactions.
Finally, fluorescence microscopy studies reveal that the binding of coatomer and Arf1 to Golgi membranes is a dynamic process involving rapid membrane binding and release driven by cycles of GTP loading and hydrolysis on Arf1 (Presley et al., 2002). This continuous activity occurs on a faster timescale than vesicle budding and would seem, on the face of it, to mediate against the budding reaction. Intriguingly, there is evidence that GTP hydrolysis on Arf1 is required for efficient cargo packaging into COPI-coated vesicles (Nickel, 1998). According to our model for coatomer recruitment, a closed form of coatomer bound to membrane via a single molecule of Arf1-GTP, and devoid of cargo interactions, will be susceptible to membrane release upon GTP hydrolysis. Whereas the open form, induced by synergy of cargo and Arf1 binding, will be more resistant to dissociation by virtue of the bivalent Arf1 interaction (the lower probability of simultaneous loss of both Arf1 molecules via GTP hydrolysis). Moreover, depending on the competition of two reactions—loss of Arf1-GTP through hydrolysis versus rebinding of the second Arf1-GTP molecule to coatomer—the open, cargo-associated form of coatomer may constitute a long-lived (kinetically stable [Presley et al., 2002]) complex that commits to vesicle formation.
Experimental Procedures
Protein Production
Expression plasmids for full-length and truncated forms of R. norvegicus β-COP and B.taurus γ-COP were constructed using the pFastBac HTB vector (Invitrogen), whereas plasmids for full-length B. taurus δ-COP and for full-length and truncated (residues 1–153) B. taurus ζ-COP were constructed with pFastBac1 vector. Baculoviruses were generated by infecting Sf9 cells with recombinant bacmids prepared using DH10Bac cells, following the Bac-to-Bac protocol (Invitrogen). We used the type I isoforms of both γ-COP and ζ-COP.
The various dimeric and tetrameric coatomer complexes were produced by co-expression of the proteins in Hi-5 cells infected with the corresponding baculoviruses. Insect cells were harvested by centrifugation 48 hours post-infection, lysed by sonication, and lysate was clarified at 100,000 × g for 1 hour. Proteins were purified by Ni2+-affinity chromatography followed by ion-exchange chromatography (HiTrap Q HP column, GE Healthcare). Purified proteins were concentrated and stored at −80° C.
Site-directed mutagenesis of β-COP and γ-COP was carried out using the Phusion kit (New England Biolabs), and mutant proteins were produced in the same manner as wild-type proteins.
For pull-down assays, S. cerevisiae Arf1 (residues 18–181) was cloned into a modified pET28b vector (Novagen) with an N-terminal His6-smt3 moiety and a C-terminal GST tag. Bacterial cells were lysed and protein purified on a Ni2+-affinity column. The His6-smt3 tag was removed with Ulp1 protease and protein re-purified on a Ni2+-affinity column. Mutant proteins were generated with the Phusion kit and prepared in the same manner as wild-type protein.
For crystallographic studies, γζ-COP (comprising B. taurus γ-COP residues 1–355 and ζ-COP residues 1–153) was expressed in insect cells and purified as before, but with an additional gel filtration step (Superdex 200 column). γζ-COP complex incorporating selenomethionine was produced in Hi-5 cells grown in Sf-900 II medium supplemented with 50 mg/L selenomethionine and 50 mg/L L-cystine. S. cerevisiae Arf1 (residues 18–181) was cloned into pET28b, expressed and purified via Ni2+-affinity chromatography. The hexa-histidine tag was removed with thrombin, and the bound nucleotide was replaced with GppNHp (guanosine-5′-[(β,γ)-imido]triphosphate) as described (Bi et al., 2007). Selenomethionione-substituted Arf1 was expressed in M9 minimal media supplemented with selenomethionine.
Coatomer Binding and Elution (Pull-Down) Assay
To test binding of coatomer to Arf1, 200 μg of S.cerevisae Arf1-GST (residues 18–181, with bound GTP, GDP or GppNHp) was incubated with a 20 μl slurry of glutathione sepharose 4B beads (GE Healthcare) for 30 min at 22° C, and beads were then washed with binding buffer (25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM DTT, 5 mM MgCl2 and 0.2 % Triton X-100). The slurry was incubated with coatomer for 15 min at 22° C in a total volume of 100 μl binding buffer. Beads were washed once with 100 μl binding buffer, and coatomer was eluted from the beads by adding GTPase-activating protein (GAP) to trigger GTP hydrolysis. Specifically, H. sapiens GAP1 catalytic core (residues 1–136) was added to 1 μg/ml final concentration, and the mixture incubated for 5 min at 22° C. Supernatant was then analyzed by SDS-PAGE and Coomassie blue staining. For the bar graphs shown in Figures 4 and 5, protein bands were measured by densitometry, corrected for background, and binding of mutant coatomer forms was expressed as a percentage of binding of wild-type protein.
GTPase Fluorescence Assay
H. sapiens Arf1 (residues 18–181) was bound to the fluorescent nucleotide derivatives mant-GTP (3′-O-[N-methyl-anthraniloyl]-GTP) and mant-GppNHp (Jena Bioscience) as described (Bi et al., 2007). The yield for Arf1-mant-GTP was 90% (that is, 10 % of Arf1 remained bound to GTP), and the yield for Arf1-mant-GppNHp was 58%, according to absorption measurements at 280 and 355 nm. Unbound fluorescent nucleotides were removed by a final gel filtration step, and contaminating free mant-nucleotide was estimated as less than 1 part in 2000 for the Arf1-mant-GTP preparation.
The decrease in fluorescence, due to hydrolysis of mant-GTP on Arf1, was monitored on a fluorimeter (Fluoromax-4, Horiba Jobin-Yvon). All reactions were carried out at 25° C in 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM DTT and 5 mM MgCl2, using an excitation wavelength of 360 nm and emission measured at 438 nm. GTP hydrolysis was initiated by addition of H. sapiens GAP1 catalytic core (residues 1–136) to a reaction mixture containing Arf1-mant-GTP plus coatomer. Reaction rates were determined from exponential fits to the data. Where necessary, curves were corrected for drift in fluorescence signal. In all experiments the drift was at least two orders of magnitude slower than GTPase rates, so this correction has an insignificant effect on the measured GTPase rate constants.
By titrating coatomer against constant concentrations of Arf1•GAP, we estimate the affinity of coatomer for Arf1, in the presence of GAP, to be approximately Kd = 1 μM (data not shown).
Crystallization and SAD Structure Determination
For crystallization purposes, the B. taurus γζ-COP core complex was mixed with yeast Arf1-GppNHp at a 1:1.2 molar ratio, in 25 mM Tris-HCl pH 7.5, 150 mM NaCl, 5 mM DTT and 5 mM MgCl2. Crystals of the complex grew by the hanging-drop method over well solutions containing 9% PEG-400 and 100 mM MES-NaOH pH 6.5; hanging drops contained 2 μl of 30 mg/ml protein solution plus 2 μl of well solution. The crystals grew in space group P42212 with two copies of the complex in the asymmetric unit. For data collection, crystals were transferred into 25% PEG-400 and 100 mM MES-NaOH pH 6.5, and frozen at −80° C.
X-ray diffraction data were collected at beamline X-29 of the National Synchrotron Light Source (NSLS). An X-ray wavelength of 0.979 Å was used, corresponding to the peak of the Se fluorescence output of the irradiated crystal, and the structure was determined by the single-wavelength anomalous scattering (SAD) method. X-ray data were processed with the program HKL2000 (Otwinowski and Minor, 1997), and thirty-six selenium sites were identified (out of forty six) using the program Phenix (Adams et al., 2010). The resulting electron density map was improved by density modification, including non-crystallographic averaging, with the program RESOLVE (Terwilliger and Berendzen, 1999). Model building and refinement were carried out with the programs Coot (Emsley et al., 2010) and Phenix, respectively. The final model refined to an R factor of 20.6% (Rfree = 26.0%) for native data between 35 and 2.9 Å resolution (Table 1). The final protein model comprises 8618 non-hydrogen atoms; no water molecules are included in the model. There are no outliers in a Ramachandran plot of the final refined structure. The following residues have been omitted from both ncs-related copies of γζ-COP/Arf1-GppNHp due to weak electron density: residues 177–181 of Arf1; residues 1–19, 240–256, 277–287, and 312–355 of γ-COP; and residues 1–6, and 147–153 of ζ-COP. Additionally, residues 20–23 were omitted from one copy of γ-COP.
Supplementary Material
Acknowledgments
We thank staff at beamline X-29 of the NSLS for access to synchrotron facilities. We thank Changwook Lee and Romina Mancusso for assistance with protein studies.
Footnotes
Protein Data Bank Accession Number
Atomic coordinates and structure factors have been deposited in the Protein Data Bank under the PDB code 3TJZ.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadian MR, Hoffman U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- Amor JC, Harrison DH, Kahn RA, Ringe D. Structure of the human ADP-ribosylation factor 1 complexed with GDP. Nature. 1994;372:704–708. doi: 10.1038/372704a0. [DOI] [PubMed] [Google Scholar]
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Beraud-Dufour S, Paris S, Chabre M, Antonny B. Dual interaction of ADP ribosylation factor 1 with Sec7 domain and with lipid membranes during catalysis of guanine nucleotide exchange. J Biol Chem. 1999;274:37629–37636. doi: 10.1074/jbc.274.53.37629. [DOI] [PubMed] [Google Scholar]
- Bi X, Mancias JD, Goldberg J. Insights into COPII coat nucleation from the structure of Sec23 Sar1 complexed with the active fragment of Sec31. Dev Cell. 2007;13:635–645. doi: 10.1016/j.devcel.2007.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm M, Aguilar RC, Bonifacino JS. Functional and physical interactions of the adaptor protein complex AP-4 with ADP-ribosylation factors (ARFs) EMBO J. 2001;20:6265–6276. doi: 10.1093/emboj/20.22.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Glick BS. The mechanisms of vesicle budding and fusion. Cell. 2004;116:153–166. doi: 10.1016/s0092-8674(03)01079-1. [DOI] [PubMed] [Google Scholar]
- Bremser M, Nickel W, Schweikert M, Ravazzola M, Amherdt M, Hughes CA, Sollner TH, Rothman JE, Wieland FT. Coupling of coat assembly and vesicle budding to packaging of putative cargo receptors. Cell. 1999;96:495–506. doi: 10.1016/s0092-8674(00)80654-6. [DOI] [PubMed] [Google Scholar]
- Collins BM, McCoy AJ, Kent HM, Evans PR, Owen DJ. Molecular architecture and functional model of the endocytic AP2 complex. Cell. 2002;109:523–535. doi: 10.1016/s0092-8674(02)00735-3. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eugster A, Frigerio G, Dale M, Duden R. COPI domains required for coatomer integrity, and novel interactions with ARF and ARF-GAP. EMBO J. 2000;19:3905–3917. doi: 10.1093/emboj/19.15.3905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J. Structural basis for activation of ARF GTPase: mechanisms of guanine nucleotide exchange and GTP-myristoyl switching. Cell. 1998;95:237–248. doi: 10.1016/s0092-8674(00)81754-7. [DOI] [PubMed] [Google Scholar]
- Goldberg J. Structural and functional analysis of the ARF1-ARFGAP complex reveals a role for coatomer in GTP hydrolysis. Cell. 1999;96:893–902. doi: 10.1016/s0092-8674(00)80598-x. [DOI] [PubMed] [Google Scholar]
- Hara-Kuge S, Kuge O, Orci L, Amherdt M, Ravazzola M, Wieland FT, Rothman JE. En bloc incorporation of coatomer subunits during the assembly of COPI-coated vesicles. J Cell Biol. 1994;124:883–892. doi: 10.1083/jcb.124.6.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heldwein EE, Macia E, Wang J, Yin HL, Kirchhausen T, Harrison SC. Crystal structure of the clathrin adaptor protein 1 core. Proc Natl Acad Sci USA. 2004;101:14108–14113. doi: 10.1073/pnas.0406102101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail SA, Vetter IR, Sot B, Wittinghofer A. The Structure of an Arf-ArfGAP Complex Reveals a Ca2+ Regulatory Mechanism. Cell. 2010;141:812–821. doi: 10.1016/j.cell.2010.03.051. [DOI] [PubMed] [Google Scholar]
- Jackson LP, Kelly BT, McCoy AJ, Gaffry T, James LC, Collins BM, Honing S, Evans PR, Owen DJ. A large-scale conformational change couples membrane recruitment to cargo binding in the AP2 clathrin adaptor complex. Cell. 2010;141:1220–1229. doi: 10.1016/j.cell.2010.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliouchnikov L, Bigay J, Mesmin B, Parnis A, Rawet M, Goldfeder N, Antonny B, Cassel D. Discrete determinants in ArfGAP2/3 conferring Golgi localization and regulation by the COPI coat. Mol Biol Cell. 2009;20:859–869. doi: 10.1091/mbc.E08-10-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, Doray B, Govero J, Kornfeld S. Binding of cargo sorting signals to AP-1 enhances association with ADP ribosylation factor 1-GTP. J Cell Biol. 2008;180:467–472. doi: 10.1083/jcb.200709037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Liu Y, Kahn RA, Prestegard JH. Dynamic structure of membrane-anchored Arf-GTP. Nat Struct Mol Biol. 2010;2010:876–881. doi: 10.1038/nsmb.1853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo R, Randazzo PA. Kinetic analysis of Arf GAP1 indicates a regulatory role for coatomer. J Biol Chem. 2008;283:21965. doi: 10.1074/jbc.M802268200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:483–495. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Nickel W, Malsam J, Gorgas K, Ravazzola M, Jenne N, Helms JB, Wieland FT. Uptake by COPI-coated vesicles of both anterograde and retrograde cargo is inhibited by GTPγS in vitro. J Cell Sci. 1998;111:3081–3090. doi: 10.1242/jcs.111.20.3081. [DOI] [PubMed] [Google Scholar]
- Otwinowski W, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Paris S, Beraud-Dufour S, Robineau S, Bigay J, Antonny B, Chabre M, Chardin P. Role of protein-phospholipid interactions in the activation of ARF1 by the guanine nucleotide exchange factor Arno. J Biol Chem. 1997;272:22221–22226. doi: 10.1074/jbc.272.35.22221. [DOI] [PubMed] [Google Scholar]
- Peyroche A, Paris S, Jackson CL. Nucleotide exchange on ARF mediated by yeast Gea1 protein. Nature. 1996;384:479–481. doi: 10.1038/384479a0. [DOI] [PubMed] [Google Scholar]
- Presley JF, Ward TH, Pfeifer AC, Siggia ED, Phair RD, Lippincott-Schwartz J. Dissection of COPI and Arf1 dynamics in vivo and role in Golgi membrane transport. Nature. 2002;417:187–193. doi: 10.1038/417187a. [DOI] [PubMed] [Google Scholar]
- Serafini T, Orci L, Amherdt M, Brunner M, Kahn RA, Rothman JE. ADP-ribosylation factor is a subunit of the coat of Golgi-derived COP-coated vesicles: a novel role for a GTP-binding protein. Cell. 1991;67:239–253. doi: 10.1016/0092-8674(91)90176-y. [DOI] [PubMed] [Google Scholar]
- Stamnes MA, Rothman JE. The binding of AP-1 clathrin adaptor particles to Golgi membranes requires ADP-ribosylation factor, a small GTP-binding protein. Cell. 1993;73:999–1005. doi: 10.1016/0092-8674(93)90277-w. [DOI] [PubMed] [Google Scholar]
- Sun Z, Anderl F, Frohlich K, Zhao L, Hanke S, Brugger B, Wieland F, Bethune J. Multiple and stepwise interactions between coatomer and ADP-ribosylation factor-1 (Arf1)-GTP. Traffic. 2007;8:582–593. doi: 10.1111/j.1600-0854.2007.00554.x. [DOI] [PubMed] [Google Scholar]
- Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–1304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Waters MG, Serafini T, Rothman JE. ‘Coatomer’: a cytosolic protein complex containing subunits of non- clathrin-coated Golgi transport vesicles. Nature. 1991;349:248–251. doi: 10.1038/349248a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.