Abstract
Synthetic biology aims to make biological engineering more scalable and predictable, lowering the cost and facilitating the translation of synthetic biological systems to practical applications. Increasingly sophisticated, rationally designed synthetic systems that are capable of complex functions pave the way to translational applications, including disease diagnostics and targeted therapeutics. Here, we provide an overview of recent developments in synthetic biology in the context of translational research and discuss challenges at the interface between synthetic biology and clinical medicine.
INTRODUCTION TO SYNBIO
Engineered, synthetic biological systems have important implications in medicine. However, the complexity of functions required to balance safety and therapeutic efficacy has limited our ability to build synthetic biological systems that can interface with, modulate, or reprogram natural systems within the human body. Synthetic biology is an emerging field that focuses on the development and application of engineering principles to the design, construction, and characterization of biological systems. One of the primary aims in this field is to make bioengineering more scalable and more predictable, thus reducing the cost and the development timeline of synthetic biological systems that could further disease prevention and treatment.
Early efforts in synthetic biology focused on the foundations of biological design and construction tools (1), including efforts to standardize the characterization, cataloging, and assembly of biological components (2). These tools have been used to build model systems that enable a greater understanding of natural biological systems or encode novel biological functions (3). More recently, focus in the field has shifted from building model systems to building biological systems that encode more complex behaviors (4, 5), such as producing fluorescent signals or initiating cellular apoptosis in response to disease biomarkers (6), identifying cancerous cells in a mixed culture (7), and modulating the growth of therapeutic cells in vivo (8) (Fig. 1).
Fig. 1.
Synthetic biology uses both natural and engineered biological components to construct genetic circuits that generate desired functional outputs in response to specified input signals. Synthetic biological systems have achieved various functions with translational potential, including initiating cell apoptosis in response to endogenous proteins such as β-catenin (6); discriminating cancer (HeLa) cells from other cell types (7); and controlling T cell proliferation in vivo using small-molecule drugs (8). PCMV, cytomegalovirus (CMV) promoter; PTRE, tetracycline-responsive promoter; PCAGop, CMV early enhancer element combined with chicken β-actin promoter followed by an intron with two LacO sites; PEF1α, elongation factor 1α promoter.
CREDIT: B. STRAUCH/SCIENCE TRANSLATIONAL MEDICINE
Although much of the activity in the field to date has focused on applications in bioprocessing and biosynthesis, potential applications in health and medicine are vast and compelling. The development of both bacterial and mammalian systems aimed at translational medicine applications, including disease treatment and prevention, has accelerated. Nevertheless, many challenges exist in moving state-of-the-art synthetic systems from the laboratory into human patients. In particular, we need to develop tools that can be implemented in physiological contexts and that demonstrate system robustness, functional precision, and host tolerability. In this Perspective, we highlight several promising engineered systems and discuss challenges that must be addressed before clinical use.
SYNTHETIC BACTERIAL SYSTEMS
Owing to the complexity of mammalian systems, it can be useful to first develop synthetic systems in simpler model organisms, such as yeast and bacteria, before transferring and reoptimizing in mammalian hosts. Furthermore, a central aim of synthetic biology is to facilitate the engineering of biology so that systems need not be constructed from scratch for each new application. Synthetic biologists have begun to construct integrated systems for translational applications by piecing together an increasingly sophisticated collection of biological “parts” with diverse functions. An early example explored the concept of targeted cancer treatment (9). Capitalizing on the hypoxic nature of tumor microenvironments and the fact that many bacterial species home in on tumors, researchers have constructed genetic circuits with sensors that detect hypoxia and bacterial cell density (9). In one circuit, a promoter that responds to formate dehydrogenase—an enzyme activated by anaerobic growth—was used for hypoxia detection. In another circuit, a sensing component detected high bacterial cell density through lux quorum-sensing (from the bacterium Vibrio fischeri). These sensors were coupled to the inv gene that encodes invasin (from Yersinia pseudotuberculosis), which initiates bacterial adhesion and invasion of mammalian cells. Engineered Escherichia coli that expressed this entire synthetic system were shown to effectively invade cultured mammalian cells under hypoxic conditions that mimicked the tumor microenvironment (9).
The power of synthetic biology to facilitate the development of novel systems with therapeutic potential is further illustrated by the International Genetically Engineered Machines (iGEM) competition, in which undergraduate students build functional genetic systems from standardized, interchangeable biological parts from the BioBricks Parts Registry (10). Several projects focused on engineering probiotics have been pursued in recent years, including the production of β-galactosidase to treat lactose intolerance (http://2008.igem.org/Team:Caltech); ratiometric modulation of T cell populations to address inflammatory bowel disease (http://2009.igem.org/Team:Stanford); and production of the toxic protein Tse2 to eliminate pathogenic bacteria in the gut (http://2010.igem.org/Team:Washington). In one example, E. coli were engineered to produce pyruvate oxidase under the control of the quorum-sensing transcriptional activator LuxR, which allowed the bacteria to generate cytotoxic amounts of hydrogen peroxide in the presence of other, invasive bacterial populations (http://2008.igem.org/Team:Caltech). This example provides preliminary support for the development of probiotic bacterial strains that can battle bacterial infections. Although these student projects are still in their infancy, and demonstrations have largely been confined to bacterial cultures, the concepts being explored feed directly into human therapeutics and translational applications. Indeed, researchers have demonstrated the feasibility of engineering commensal bacterial strains, such as Nissle 1917, to produce quorum-sensing proteins that interrupt cholera infection (11) or to secrete proteins that induce insulin production by mouse intestinal cells (12).
In addition to using engineered bacteria as therapeutic agents, synthetic systems have been constructed to interface with bacteria in vivo. Researchers have engineered bacteriophages to overexpress proteins that repress gene repair and disrupt oxidative stress response pathways in bacteria in mice, thus enhancing the effect of antibiotic therapy and improving survival when challenged with lethal doses of E. coli (13). Although challenges such as immunogenicity, toxin release, and development of phage resistance must be addressed before synthetic bacterial systems become available for human application, these early efforts highlight the potential of synthetic biology to generate integrated systems that can deliver therapeutic outputs in response to physiological cues or in combination with existing treatment strategies.
SYNTHETIC MAMMALIAN SYSTEMS
Moving toward higher-level organisms, synthetic biologists have constructed several systems to control mammalian cell behavior in response to either exogenous or endogenous input signals (14). Most existing inducible promoter systems require inducer molecules that are toxic to cells and therefore cannot be administered at sufficiently high concentrations to human subjects. In response, improvements in foundational technologies, such as DNA synthesis, have led to the de novo construction of heterologous small-molecule-responsive transgene expression systems. For example, the regulation of transgene expression in bioreactors as well as in animal models has been reported, using synthetic promoter systems that respond to the plant-derived metabolite phloretin and the vitamin biotin (15, 16). Molecules such as phloretin and biotin are ideal inducer molecules owing to their low toxicity and commercial availability, which should reduce regulatory hurdles toward their clinical use.
Another class of control systems that respond to exogenous inputs stems from the field of optogenetics, which has provided a powerful set of light-responsive tools for interrogating natural biological systems (17). Optogenetics technology has been incorporated into synthetic systems that exert spatiotemporal control over cell signaling (18). In these systems, photoreceptive phytochrome B and its binding partner were used to mediate the localization and activation of signaling molecules, such as guanine nucleotide exchange factors and their cognate G proteins. Precise and reversible control of protein localization and mammalian cell shape and motility can be achieved with light. A recent study reported a light-controlled synthetic regulatory circuit capable of attenuating glycemic excursions in type 2 diabetic mice through the use of either implanted fiber optics or direct illumination (19), suggesting that optogenetics can be used for cell manipulation in vivo in response to exogenous stimulation.
Synthetic systems that interface with endogenous input signals have also been demonstrated. In one example, researchers engineered a sensor circuit that responded to increasing uric acid levels. This circuit overexpressed a mammalian-codon-optimized gene derived from Aspergillus flavus that encodes for the uricase mUox, which converts urate—a toxic metabolic end-product—to allantoin, for easy excretion (20). This synthetic feedback system enabled cell-intrinsic control of urate homeostasis in human cell culture and mediated the reduction of pathologic urate levels in mice implanted with mammalian cells harboring this sensor circuit.
RNA controllers that regulate mammalian gene expression through protein-responsive modulation of alternative splicing were recently described (6). RNA aptamer sequences that specifically bind to endogenous proteins, such as β-catenin and nuclear factor κB (NF-κB), were integrated into key intronic locations near or flanking an alternatively spliced exon from the SMN1 mini-gene and then expressed in human embryonic kidney cells. Ligand binding to the RNA aptamer modulated inclusion of the alternatively spliced exon, which harbored a stop codon, regulating the expression of a downstream target gene. Incorporation of green-fluorescent protein (GFP) as the target gene provided a diagnostic “signal” (6). Alternatively, the inclusion of a functional target—the suicide gene thymidine kinase—generated therapeutic outputs, including cell death in response to β-catenin (Fig. 1).
Another recent study demonstrated a synthetic logic circuit capable of distinguishing human cervical cancer (HeLa) cells from other cell types by detecting the miRNA expression profile in each cell (Fig. 1) (7). The multilayered circuit incorporated constitutive and inducible promoters, transcriptional activator and repressor proteins, and binding sites for a set of endogenous miRNAs known to be uniquely highly or poorly expressed in HeLa cells relative to healthy cell types profiled in the MicroRNA Atlas (21). A fluorescent output signal was produced only in the presence of the correct combination of high and low levels of the selected miRNAs, allowing the identification of HeLa cells in a heterogenous cell population (Fig. 1) (7). This method opens doors to new cancer diagnostics and further illustrates the translational potential of synthetic systems that can interface with endogenous input signals.
The application of RNA controllers to disease treatment has been explored in the context of T cell immunotherapy. To achieve controlled T cell persistence, which is critical to the therapeutic efficacy of tumor-targeting T cells in cancer patients, a ribozyme-based system was demonstrated to respond to small-molecule drugs in a rapid, reversible, and dose-dependent manner and to effectively regulate human T cell proliferation in vivo (Fig. 1) (8). This control system coupled existing biological parts, including ligand-responsive ribozyme switches (22), to therapeutically relevant target genes that encode for T cell growth-promoting cytokines interleukin-2 (IL-2) and IL-15 to enable drug-responsive control of the timing and extent of T cell proliferation in vivo. Such a system, if applied to humans, could increase the precision and efficacy of cellular immunotherapy. Furthermore, the incorporation of the suicide gene thymidine kinase provided an effective means of treatment termination through the controlled ablation of engineered T cells, increasing the safety profile of cell-based therapeutics.
A common feature of the diverse synthetic mammalian systems discussed above is that they are composed of biological parts (such as promoters, RNA switches, and target genes) that have been rationally selected and systematically assembled, so that efficient optimization and modification of the systems are possible because the function of and interaction among the components are well understood. These studies highlight the adaptability of synthetic biological systems and the importance of engineering principles, such as component modularity and transportability across organisms, to the translation of synthetic systems in clinical applications.
BUILDING A FUTURE FOR CLINICAL SYNBIO
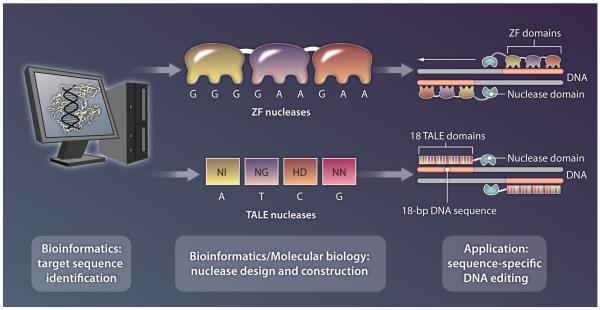
A general challenge facing synthetic biology is efficient integration into the appropriate cellular host without compromising the activity and integrity of either the synthetic system or its host. Commonly used laboratory techniques, such as transient transfection and probability-driven, non-site-specific integration of plasmid DNA into the host genome, are not suitable for clinical applications because of low integration efficiency, short-term performance consistency, and potential for off-target effects. Efforts to develop new genome-editing technologies have generated various methods for the rational design of zinc finger (ZF) nucleases that are capable of site-specific gene insertion, deletion, and disruption (Fig. 2) (23-26). More recently, transcription activator-like effectors (TALEs) have been coupled to nucleases so as to generate a novel class of genome-editing tools that can be reliably engineered for site-specific genomic manipulation (Fig. 2) (27). Notably, DNA assembly concepts and techniques developed in synthetic biology have been applied to the modular construction of TALE nucleases, allowing the efficient assembly of long, repetitive sequences that are otherwise difficult to construct (28, 29).
Fig. 2.
Emerging genetic engineering technologies enable site-specific integration of synthetic elements. ZFs and TALEs can be rationally designed to bind specific DNA sequences that have been identified through bioinformatics as targets of interest. Several computational algorithms have been developed to aid in the design of ZFs (23-26). TALEs follow well-defined rules such that specific amino acid pairs (NI, NG, HD, and NN) in the TALE sequence define the respective DNA base (A, T, C, and G) to be recognized. When coupled to nucleases, pairs of ZFs and TALEs bind to DNA to enable nuclease domain dimerization, site-specific cleavage of double-stranded DNA, and precise insertion of transgenic elements into target chromosomal locations.
CREDIT: B. STRAUCH/SCIENCE TRANSLATIONAL MEDICINE
Combining the targeting capability of ZF nucleases with the gene delivery efficiency of viral vectors, researchers have also engineered retroviral vectors containing ZF domains (30). The resulting vectors exhibit selective integration properties and eliminate the preference for genomic integration in transcriptional start sites, lowering the probability of insertional mutagenesis and oncogenesis—an important consideration for eventual use in humans. Furthermore, genetic manipulation techniques, such as DNA shuffling, have been applied to the generation of chimeric, adeno-associated viral vectors with enhanced transduction efficiencies and lowered immunogenicity, which would be suitable for gene therapy in patients (31).
As synthetic biologists make progress toward bringing engineered systems into the clinic, a number of unique challenges presented by in-human applications must be addressed. The most important of these requirements is system robustness since performance consistency and long-term stability will be demanded of any engineered system introduced into a human patient. In addition, safety requirements will necessitate stringent thresholds for off-target and toxicity effects. Lastly, practical challenges surrounding system implementation in primary human cells—such as upper bounds on the size of genetic programs that can be delivered by viral vectors or difficulties in stably expressing multiple protein components—will present serious limitations to current design practices in synthetic biology. The vast majority of systems that have been built to date comprise multiple synthetic proteins derived from diverse sources, including bacteria. It is difficult, if not impossible, to deliver systems with large genetic footprints into primary human cells by using clinically approved vectors and to sustain stable expression of all system components. In addition, systems comprising heterologous proteins are likely to elicit nonspecific immune responses in human hosts, thus rendering the system toxic to the patient.
These challenges compel synthetic biologists to develop innovative systems for clinical application. Customizable control systems that do not require heterologous protein components have already been demonstrated (6, 22, 32). In addition, researchers are focusing on developing expanded toolkits that allow for combinatorial control strategies within compact genetic footprints to support the design of more sophisticated and tightly controlled functions (33, 34). Lastly, collaborations between clinicians and synthetic biologists must be encouraged and cultivated to address the immediate needs and challenges of in-human application.
Synthetic biology is still a maturing field with many obstacles to overcome before it can fully realize its clinical potential. The synthetic systems highlighted in this Perspective have begun to address important medical challenges, such as the development of cancer-targeted therapeutics and diagnostics. Continued innovation promises to improve the efficacy and safety of next-generation treatment strategies. Synthetic biology is adding to the existing biological engineering toolbox, and successful translation of these technologies into the clinic will have a transformative impact on patient health, including cell-based therapies, regenerative medicine, and tissue and organ replacement.
Acknowledgments
Funding: C.D.S. is supported by funds from the National Institutes of Health, National Science Foundation, and Defense Advanced Research Projects Agency. Y.Y.C. is supported by the Harvard University Society of Fellows.
Footnotes
Competing interests: The authors declare no competing interests.
Citation: Y. Y. Chen, C. D. Smolke, From DNA to targeted therapeutics: Bringing synthetic biology to the clinic. Sci. Transl. Med. 3, 106ps43 (2011).
References
- 1.Endy D. Foundations for engineering biology. Nature. 2005;438:449–453. doi: 10.1038/nature04342. doi:10.1038/nature04342 Medline. [DOI] [PubMed] [Google Scholar]
- 2.Canton B, Labno A, Endy D. Refinement and standardization of synthetic biological parts and devices. Nat. Biotechnol. 2008;26:787–793. doi: 10.1038/nbt1413. doi:10.1038/nbt1413 Medline. [DOI] [PubMed] [Google Scholar]
- 3.Andrianantoandro E, Basu S, Karig DK, Weiss R. Synthetic biology: New engineering rules for an emerging discipline. Mol. Syst. Biol. 2006;2:2006. doi: 10.1038/msb4100073. 0028. doi:10.1038/msb4100073 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Haynes KA, Silver PA. Eukaryotic systems broaden the scope of synthetic biology. J. Cell Biol. 2009;187:589–596. doi: 10.1083/jcb.200908138. doi:10.1083/jcb.200908138 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Purnick PE, Weiss R. The second wave of synthetic biology: From modules to systems. Nat. Rev. Mol. Cell Biol. 2009;10:410–422. doi: 10.1038/nrm2698. doi:10.1038/nrm2698 Medline. [DOI] [PubMed] [Google Scholar]
- 6.Culler SJ, Hoff KG, Smolke CD. Reprogramming cellular behavior with RNA controllers responsive to endogenous proteins. Science. 2010;330:1251–1255. doi: 10.1126/science.1192128. doi:10.1126/science.1192128 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xie Z, Wroblewska L, Prochazka L, Weiss R, Benenson Y. Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science. 2011;333:1307–1311. doi: 10.1126/science.1205527. doi:10.1126/science.1205527 Medline. [DOI] [PubMed] [Google Scholar]
- 8.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl. Acad. Sci. U.S.A. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. doi:10.1073/pnas.1001721107 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J. Mol. Biol. 2006;355:619–627. doi: 10.1016/j.jmb.2005.10.076. doi:10.1016/j.jmb.2005.10.076 Medline. [DOI] [PubMed] [Google Scholar]
- 10.Smolke CD. Building outside of the box: iGEM and the BioBricks Foundation. Nat. Biotechnol. 2009;27:1099–1102. doi: 10.1038/nbt1209-1099. doi:10.1038/nbt1209-1099 Medline. [DOI] [PubMed] [Google Scholar]
- 11.Duan F, March JC. Interrupting Vibrio cholerae infection of human epithelial cells with engineered commensal bacterial signaling. Biotechnol. Bioeng. 2008;101:128–134. doi: 10.1002/bit.21897. doi:10.1002/bit.21897 Medline. [DOI] [PubMed] [Google Scholar]
- 12.Duan F, Curtis KL, March JC. Secretion of insulinotropic proteins by commensal bacteria: rewiring the gut to treat diabetes. Appl. Environ. Microbiol. 2008;74:7437–7438. doi: 10.1128/AEM.01019-08. doi:10.1128/AEM.01019-08 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4629–4634. doi: 10.1073/pnas.0800442106. doi:10.1073/pnas.0800442106 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weber W, Fussenegger M. Molecular diversity—The toolbox for synthetic gene switches and networks. Curr. Opin. Chem. Biol. 2011;15:414–420. doi: 10.1016/j.cbpa.2011.03.003. doi:10.1016/j.cbpa.2011.03.003 Medline. [DOI] [PubMed] [Google Scholar]
- 15.Gitzinger M, Kemmer C, El-Baba MD, Weber W, Fussenegger M. Controlling transgene expression in subcutaneous implants using a skin lotion containing the apple metabolite phloretin. Proc. Natl. Acad. Sci. U.S.A. 2009;106:10638–10643. doi: 10.1073/pnas.0901501106. doi:10.1073/pnas.0901501106 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber W, Lienhart C, Baba MD, Fussenegger M. A biotin-triggered genetic switch in mammalian cells and mice. Metab. Eng. 2009;11:117–124. doi: 10.1016/j.ymben.2008.12.001. doi:10.1016/j.ymben.2008.12.001 Medline. [DOI] [PubMed] [Google Scholar]
- 17.Deisseroth K, Feng G, Majewska AK, Miesenböck G, Ting A, Schnitzer MJ. Next-generation optical technologies for illuminating genetically targeted brain circuits. J. Neurosci. 2006;26:10380–10386. doi: 10.1523/JNEUROSCI.3863-06.2006. doi:10.1523/JNEUROSCI.3863-06.2006 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Levskaya A, Weiner OD, Lim WA, Voigt CA. Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature. 2009;461:997–1001. doi: 10.1038/nature08446. doi:10.1038/nature08446 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ye H, Daoud-El Baba M, Peng RW, Fussenegger M. A synthetic optogenetic transcription device enhances blood-glucose homeostasis in mice. Science. 2011;332:1565–1568. doi: 10.1126/science.1203535. doi:10.1126/science.1203535 Medline. [DOI] [PubMed] [Google Scholar]
- 20.Kemmer C, Gitzinger M, Daoud-El Baba M, Djonov V, Stelling J, Fussenegger M. Self-sufficient control of urate homeostasis in mice by a synthetic circuit. Nat. Biotechnol. 2010;28:355–360. doi: 10.1038/nbt.1617. doi:10.1038/nbt.1617 Medline. [DOI] [PubMed] [Google Scholar]
- 21.Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, Schermer B, Bissels U, Inman J, Phan Q, Chien M, Weir DB, Choksi R, De Vita G, Frezzetti D, Trompeter HI, Hornung V, Teng G, Hartmann G, Palkovits M, Di Lauro R, Wernet P, Macino G, Rogler CE, Nagle JW, Ju J, Papavasiliou FN, Benzing T, Lichter P, Tam W, Brownstein MJ, Bosio A, Borkhardt A, Russo JJ, Sander C, Zavolan M, Tuschl T. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. doi:10.1016/j.cell.2007.04.040 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. doi:10.1073/pnas.0703961104 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wright DA, Thibodeau-Beganny S, Sander JD, Winfrey RJ, Hirsh AS, Eichtinger M, Fu F, Porteus MH, Dobbs D, Voytas DF, Joung JK. Standardized reagents and protocols for engineering zinc finger nucleases by modular assembly. Nat. Protoc. 2006;1:1637–1652. doi: 10.1038/nprot.2006.259. doi:10.1038/nprot.2006.259 Medline. [DOI] [PubMed] [Google Scholar]
- 24.Maeder ML, Thibodeau-Beganny S, Sander JD, Voytas DF, Joung JK. Oligomerized pool engineering (OPEN): An ‘open-source” protocol for making customized zinc-finger arrays. Nat. Protoc. 2009;4:1471–1501. doi: 10.1038/nprot.2009.98. doi:10.1038/nprot.2009.98 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, Pierick CJ, Hoffman E, Maeder ML, Khayter C, Reyon D, Dobbs D, Langenau DM, Stupar RM, Giraldez AJ, Voytas DF, Peterson RT, Yeh JR, Joung JK. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA) Nat. Methods. 2011;8:67–69. doi: 10.1038/nmeth.1542. doi:10.1038/nmeth.1542 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sander JD, Maeder ML, Reyon D, Voytas DF, Joung JK, Dobbs D. ZiFiT (Zinc Finger Targeter): An updated zinc finger engineering tool. Nucleic Acids Res. 2010;38:W462–W468. doi: 10.1093/nar/gkq319. Web Server issue. doi:10.1093/nar/gkq319 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T, Huang S, Jiang WZ, Wright D, Spalding MH, Weeks DP, Yang B. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011;39:359–372. doi: 10.1093/nar/gkq704. doi:10.1093/nar/gkq704 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang F, Cong L, Lodato S, Kosuri S, Church GM, Arlotta P. Efficient construction of sequence-specific TAL effectors for modulating mammalian transcription. Nat. Biotechnol. 2011;29:149–153. doi: 10.1038/nbt.1775. doi:10.1038/nbt.1775 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li T, Huang S, Zhao X, Wright DA, Carpenter S, Spalding MH, Weeks DP, Yang B. Modularly assembled designer TAL effector nucleases for targeted gene knockout and gene replacement in eukaryotes. Nucleic Acids Res. 2011;39:6315–6325. doi: 10.1093/nar/gkr188. doi:10.1093/nar/gkr188 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lim KI, Klimczak R, Yu JH, Schaffer DV. Specific insertions of zinc finger domains into Gag-Pol yield engineered retroviral vectors with selective integration properties. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12475–12480. doi: 10.1073/pnas.1001402107. doi:10.1073/pnas.1001402107 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grimm D, Lee JS, Wang L, Desai T, Akache B, Storm TA, Kay MA. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J. Virol. 2008;82:5887–5911. doi: 10.1128/JVI.00254-08. doi:10.1128/JVI.00254-08 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Beisel CL, Chen YY, Culler SJ, Hoff KG, Smolke CD. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 2011;39:2981–2994. doi: 10.1093/nar/gkq954. doi:10.1093/nar/gkq954 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Win MN, Smolke CD. Higher-order cellular information processing with synthetic RNA devices. Science. 2008;322:456–460. doi: 10.1126/science.1160311. doi:10.1126/science.1160311 Medline. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deans TL, Cantor CR, Collins JJ. A tunable genetic switch based on RNAi and repressor proteins for regulating gene expression in mammalian cells. Cell. 2007;130:363–372. doi: 10.1016/j.cell.2007.05.045. doi:10.1016/j.cell.2007.05.045 Medline. [DOI] [PubMed] [Google Scholar]