Abstract
Understanding the structural organization of eukaryotic chromatin and its control of gene expression represents one of the most fundamental and open challenges in modern biology. Recent experimental advances have revealed important characteristics of chromatin in response to changes in external conditions and histone composition, such as the conformational complexity of linker DNA and histone tail domains upon compact folding of the fiber. In addition, modeling studies based on high-resolution nucleosome models have helped explain the conformational features of chromatin structural elements and their interactions in terms of chromatin fiber models. This minireview discusses recent progress and evidence supporting structural heterogeneity in chromatin fibers, reconciling apparently contradictory fiber models.
Keywords: Chromatin Histone Modification, Chromatin Structure, Epigenetics, Histones, Transcription Regulation, Chromatin Folding, Epigenetic Modifications, Higher Order Structure, Histone Tails
Introduction
The 3 billion DNA base pairs of the human genome are densely packed within eukaryotic chromatin, a nucleoprotein complex in which the DNA is wrapped around nucleosomes (see Fig. 1). Nucleosomes and higher order chromatin structures serve essential cellular functions, including condensation of meters-long genomic DNA by several orders of magnitude to enable its packaging into the micrometer-sized cell nucleus and regulation of DNA-directed processes, such as transcription, replication, recombination, and repair through local and dynamic unfolding of chromatin.
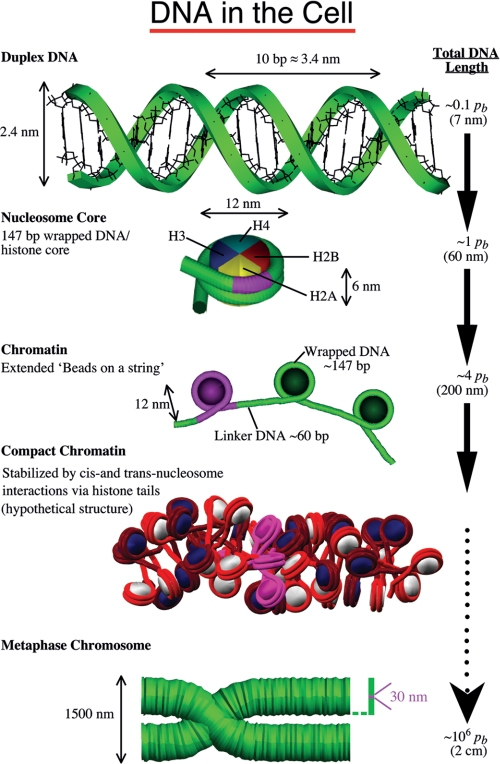
FIGURE 1.
Schematic view of the many levels of DNA folding in the cell. On length scales much smaller than the persistence length (pb), DNA can be considered straight. In eukaryotic cells, DNA wraps around a core of histone proteins to form the chromatin fiber. The fiber is shown in both the extended view and a hypothetical compact zigzag view (the “30-nm fiber”) deduced from a modeling study (29). Chromosomes are made up of a dense chromatin fiber, shown here in the metaphase stage. For reference, we highlight in pink in all the DNA/protein views the hierarchical organizational unit preceding it. The length scale on the right indicates the level of compaction involved.
Whether the higher order structure of nuclear chromatin is organized into a hierarchy of folding states (1) or non-hierarchical fractal geometry (2), previous landmark studies established the nucleosome as the repeating unit of chromatin (Figs. 1 and 2) (3). This basic unit consists of ∼200 bp of DNA, 147 bp of which are wound around the outside of a spool composed of core histones (two each of histones H2A, H2B, H3, and H4) to form the nucleosome core; the remainder (linker DNA) joins adjacent nucleosomes. At low salt, arrays of nucleosomes adopt an extended “beads-on-a-string” conformation (Fig. 1). In vitro, nucleosome arrays in solutions of physiological ionic strength form a compact higher order structure called the 30-nm diameter chromatin fiber. Determination of the detailed architecture of the chromatin fiber has occupied experimental and theoretical scientists for decades (4–8).
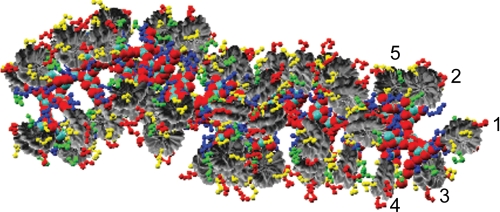
FIGURE 2.
Detailed view of the heteromorphic chromatin model shown in Fig. 1, with rendering of the core histone tails to show the complex inter- and intranucleosome interactions. The first five nucleosomes are marked to indicate the different interaction types. Histone tails are colored yellow (H2A), red (H2B), blue (H3), and green (H4).
In this minireview, we discuss recent progress in experimental analysis of chromatin folding, as well as specific molecular interactions and forces stabilizing compact chromatin. We include recent modeling studies that, consistent with experimental data, reveal a structural stability of internally heterogeneous chromatin fibers. These collective experimental and modeling approaches appear to reconcile contradictory chromatin fiber models by relating variable chromatin organization to functional roles.
Experimental Foundations
Nucleosome Core Structure
The nucleosome core can be reconstituted from ∼147-bp DNA fragments and histone octamers. Such particles have been crystallized and studied by x-ray diffraction (Fig. 3) (e.g. Refs. 9–14), revealing fine detail and differences from free DNA and providing solid anchors for modeling studies.
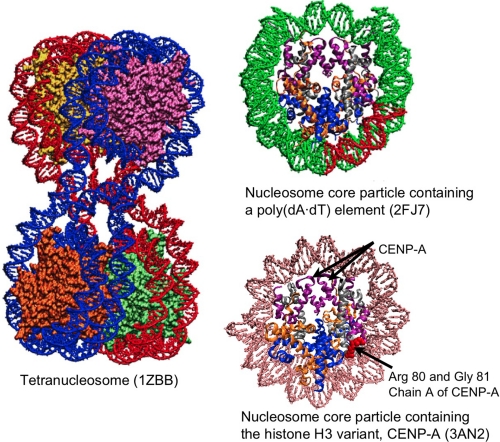
FIGURE 3.
Examples of crystal structures for the tetranucleosome and nucleosomes. Shown are the tetranucleosome complex (25), a nucleosome core particle containing a poly(dA·dT) tract (red) (12), and a human centromeric nucleosome containing the centromere-specific histone H3 variant CENP-A (18). In single nucleosome particles, the histone proteins are colored by type (i.e. H3, purple; H4, silver; H2A, orange; H2B, blue). Arrows point to residues present in CENP-A but not in the canonical H3 histone.
Each histone contains a well ordered domain responsible for the primary wrapping of DNA and tail regions, which make important points of contact between the protein and the DNA (see hypothetical model in Fig. 2). Specifically, an underwinding (10.2 versus ∼10. 5 bp/turn) of the nucleosome-bound DNA superhelix lines up neighboring DNA grooves to form a channel through which the H3 and H2B N-terminal tail domains pass. The tails likely play key roles in regulating biological processes, such as transcription, that require a conformational change in higher order chromatin structures. Unfortunately, the tail domains are typically poorly ordered in crystal structures of nucleosome cores due to the high salt concentrations required for crystallization. Because of this structural uncertainty (15), for modeling purposes, these domains are typically approximated as flexible polymers.
Solution studies revealed that the nucleosome core structure is highly dynamic and undergoes spontaneous and reversible unwrapping of DNA segments from the histone surface (16, 17); this flexibility provides accessibility and opportunity for interactions with DNA-binding proteins. In addition, it should be noted that inclusion of histone variants, e.g. centromeric histone H3 variant CENP-A (centromere protein A) (18, 19), may significantly alter both the extent and stability of nucleosome DNA wrapping.
Nucleosome Array Folding in Vitro: The 30-nm Fiber
Nucleosome arrays are characterized by variable linker DNA lengths ranging from 10 to 90 bp. Thus, the nucleosome repeat lengths (NRLs)2 vary from the shortest 155-bp NRL in fission yeast (20) to the longest NRL in echinoderm sperm (∼240 bp) (21). As mentioned above, arrays are further compacted in higher order chromosomal structures. In vitro, nucleosome arrays form a compact 30-nm chromatin fiber, whose exact structure (e.g. solenoid, zigzag, superbead, and others) has long been debated (Figs. 4 and 5) (3). Part of this structural uncertainty is due to variability in the native nucleosome arrays studied. A recent advance in understanding the internal organization of the chromatin fiber occurred with the construction of polynucleosome templates for precise positioning of histone octamers, initially from repeats of 5 S ribosomal DNA (22) and later from clone 601 DNA selected from random synthetic DNA sequences (23). Note that the strong positional sequences may create structural artifacts, so any generalizable conclusions require independent confirmation with native chromatin.
FIGURE 4.
Representative studies of chromatin structure providing foundations for the mesoscopic model of the 30-nm fiber. The models were selected among the many relevant works and are thus representative rather than complete. AFM, atomic force microscopy; SMFS, single-molecule force spectroscopy.
FIGURE 5.
Chromatin organization: ideal and deduced models for relaxed chromatin and stretched chromatin fibers. A–C, solenoid, zigzag, and heteromorphic models, respectively. A, ideal model (parallel and perpendicular views) for a 48-core 209-bp one-start solenoid fiber with six nucleosomes/turn in which DNA linkers (DNA segments shown in red connecting nucleosomes) are bent and neighboring nucleosomes (i±1 interactions (int.)) are in the closest contact. B, ideal two-start zigzag model (parallel and perpendicular views) for a 48-core 209-bp fiber in which DNA linkers are straight and i±2 nucleosomes are in the closest contact. C, heteromorphic architecture predicted by modeling and Monte Carlo simulations of 48-core 209-bp arrays with LH at room temperature (293 K), 0.15 m NaCl, and low concentration of magnesium ions and confirmed by cross-linking experiments (29). DNA linkers are shown in red, alternate nucleosomes are shown in white and blue, and LHs are shown as turquoise spheres. The view parallel to the fiber axis (left) and two enlarged nucleosome triplets are shown. Both straight and bent DNA linkers occur. In all views, connecting DNA linkers and DNA wrapped around the nucleosomes are colored in red; odd and even nucleosomes are white and blue, respectively; and LHs are shown in turquoise. The close-ups of trinucleosomes show both intra- and internucleosome interactions. The core histone tails are colored yellow (H2A), red (H2B), blue (H3), and green (H4). D, effect of NRL (173, 209, and 218 bp) and LH on the structure of the chromatin fiber as predicted from Monte Carlo simulations of 48-core arrays at 0.15 m monovalent ions (72). The center images also show the individual histone tail beads. Color coding is as described above. E, effect of various dynamic LH binding mechanisms on the chromatin unfolding mechanism for 24-core 209-bp fibers as revealed from stretching simulations mimicking single-molecule pulling experiments at monovalent salt conditions of 0.15 m (43). Panel 1, resulting force extension curves for fibers with one LH rigidly fixed to each core (blue curve) versus LHs that bind/unbind dynamically (average concentration of 0.8 LH/core; red curve) with added divalent cations. Dynamic LH binding/unbinding dramatically decreases the fiber stiffness and the forces needed for unfolding with respect to fibers with fixed LH, improving the agreement with experiments (33) significantly. pN, piconewtons. Panel 2, images representing unfolding intermediates at different forces along the dynamic LH curve in panel 1. Intermediates reveal “superbead-on-a-string” structures in which compact clusters coexist with extended fiber regions. Panel 3, effect of fast and slow dynamic LH binding/unbinding during chromatin fiber unfolding without divalent ions (43). The slow-rebinding LH molecules cause a more dramatic softening effect than a pool of fast-rebinding LH, whereas fast LH rebinding promotes formation of superbead-on-a-string conformations with compact clusters. Together, fast- and slow-binding LH pools provide facile fiber unfolding through heteromorphic superbead conformations.
Studies using 601 repeats have provided structural insights for oligonucleosomes with relatively short NRLs (167 and 177 bp) folded in the presence of divalent cations. For example, in cross-linking studies using arrays of 601 nucleosomes with 177-bp repeats, Richmond and co-workers (24) produced compelling evidence for a zigzag model in which nucleosome i is closest in space to nucleosomes i±2 (see contact patterns in Fig. 5, A–C). Zigzag folding was also evident in a subsequent crystallographic study of a tetranucleosome (25) with a 167-bp NRL.
However, Rhodes and co-workers (26, 27) produced strong evidence for the earlier interdigitated solenoid model (28) using electron microscopy of chromatin fibers reconstituted with longer NRLs (>177 bp) and with linker histone (LH) and divalent ions. More recently, cross-linking experiments combined with modeling have demonstrated an internal heterogeneity of compact 30-nm fibers (29). In particular, divalent ions appear to promote some bending of the linker DNAs, so compact structures for the chromatin fiber arise that combine mostly straight-linker (zigzag-like) fibers with a small percentage of bent linkers (Fig. 5C). Thus, bent-linker DNAs caused by divalent ions lead to a heteromorphic fiber form that combines features of both zigzag and solenoid models.
The heterogeneous nature of the 30-nm fiber is consistent with earlier electron microscopy (30) and atomic force microscopy (31) imaging. Other evidence for an irregular zigzag structure was provided by single-molecule experiments combined with a low-resolution model to interpret force/extension curves from pulling experiments by optical tweezers (32). However, recent single-molecule force microscopy studies (33) subjecting 25-nucleosome arrays with two linker lengths (NRLs of 167 and 197 bp) to forces up to 4 piconewtons suggested a fundamental one-start solenoid organization for the longer array. Clearly, many chromatin configurations are viable in ambient conditions. In fact, this multiplicity of conformations can be explained by the sensitivity of fiber architecture to a large number of internal and external factors (LH, NRL, ionic conditions, and variations in NRL from one core to the next; see next section). Indeed, such a dependence makes the chromatin fiber infinitely interesting and suitable for performing a rich array of functions in the cell.
Molecular Interactions and Factors Stabilizing Compact Chromatin
LHs Define the Path of Linker DNA
LHs play a crucial role in compacting 30-nm fibers (34, 35) by shielding the negatively charged linker DNA and thereby allowing close apposition of these segments in the folded fiber. Typical LHs have a tripartite structure, with extended positively charged C- and N-terminal domains protruding from a central globular domain. Mapping with micrococcal nuclease (29) or hydroxyl radicals (36) provided strong evidence for positioning of LH at the dyad axis with symmetric protection of ∼11 bp of each linker DNA. This symmetry and the ability of the globular domain alone to bring two linker DNA segments in close juxtaposition to form linker DNA stem motifs are consistent with energy-minimized models (37, 38).
Although the C- and N-terminal domains of LH are disordered in solution, modeling suggests that folding of the C terminus upon contacting the nucleosome contributes to the linker stem formation. On the basis of a striking homology between the C-terminal domain of LH and the HMG box fold motif, Bharath et al. (39) predicted that the folded C-terminal domain could induce formation of linker DNA stems by kinking inward and then diverging at the C-terminal domain. More recent analysis of experimental data and modeling studies have provided a nanoscale model of the LH-induced stem structure (38). Condensation consistent with folding of the C-terminal domain upon H1 binding to the nucleosome was recently confirmed by FRET experiments (40). These results are consistent with the LH C-terminal domain behaving as an intrinsically disordered domain (41).
Thus, LH refines the path of linker DNA within the chromatin fiber by forming rigid stems that reduce the separation angle of entering/exiting DNA (35, 42). Additionally, LH shields the electrostatic repulsion of the linker DNAs and promotes linker DNA bending. Still, recent single-molecule force studies (33) suggested a stabilizing rather than structure-determining effect of LH on higher order chromatin. This difference may reflect the intrinsic ability of LH to rearrange under the applied force and thus different organization of static and dynamic chromatin (Fig. 5E) (43).
Histone Tails Bridge Nucleosomes
The N-terminal domains of all core histones and the C-terminal domain of histone H2A lack a defined secondary structure when free in solution and are highly mobile when assembled into nucleosomes (15). These histone tails are highly positively charged but make only marginal contributions to individual nucleosome structure. Still, they are required for formation of higher order chromatin structures (44, 45), probably due to mediation of internucleosome interactions. A possible molecular mechanism of intrinsic nucleosome core interactions emerged from an x-ray analysis of nucleosome core crystals where the histone H4 N-terminal tail from one nucleosome contacts an acidic patch formed by histones H2A and H2B in a neighboring nucleosome (9). Evidence for H4 tail-H2A interactions was provided by cross-linking studies, suggesting both intra- and inter-array contacts (46, 47).
Recent cross-linking studies demonstrated that the same tail domains of core histones can alternate between intra- and inter-array interactions (47–49); we designate these as cis- and trans-interactions, respectively (see hypothetical model in Fig. 2). The inclusion of core histone variants (50) or mutations (51) altering the histone octamer surface can dispatch internucleosome interactions toward either cis-interactions and intra-array folding or trans-interactions and inter-array oligomerization.
Histone Post-translational Modifications Directly Alter Nucleosome Interactions and Chromatin Folding
Post-translational modifications (PTMs) of histone proteins have emerged as an important mechanism for modulating chromatin structure and function (52, 53). Although many of the histone PTMs affect chromatin structure and function by recruiting additional chromatin-remodeling or architectural factors consistent with the histone code hypothesis (54), histone acetylation and phosphorylation alter the protein charge and chemical properties of the amino acid side group and may directly affect internucleosome interaction and higher order folding.
For example, acetylation of Lys-16 in histone H4 was sufficient to destabilize salt-dependent folding of a nucleosome fiber (55). Surprisingly, charge alterations could not reproduce the action of Lys-16 acetylation; this suggests a highly specific interaction of the N-terminal domain with the histone octamer surface (56), perhaps dependent on formation of α-helical secondary structure by the histone H4 N-terminal tail (57). The other sites of histone N-terminal tail acetylation can also directly alter the folding and compaction in chromatin (58) and apparently act in a less specific manner in both cis- and trans-interactions (49). These modifications are likely functionally redundant as demonstrated by mutagenesis in vivo (59) and histone tail swapping experiments (60). This non-specificity is consistent with primarily electrostatic interactions of the N-terminal tails with other histones and DNA (61).
Besides acetylation, residue substitution experiments in which histone H4 was trimethylated at Lys-20 and probed by array sedimentation showed increased compaction of nucleosome arrays (11). Although the mechanism is unclear, the position of Lys-20 in the histone H4 tail (residues 16–20) may affect the internucleosome interaction with a surface of histone H2A. Moreover, ubiquitylation of histone H2B causes a notable disruption of a chromatin fiber structure through a mechanism distinct from histone acetylation (62).
Although the above studies were conducted on nucleosome arrays devoid of LH, other experiments have shown that histone PTMs can also override LH-stabilized fiber compaction. For example, citrullination of arginines in histone H3 and H4 N-terminal tails (63) and in vitro acetylation of chromatin fibers (64) or acetylation mimics within the H4 tail domain (49) override the strong stimulation of condensation of nucleosome arrays caused by LHs. Thus, histone acetylation may alter the secondary structure of the core histone tail domains in a way that does not simply abolish internucleosome interactions but actively interferes with other stabilizing chromatin interactions (65).
Effect of Nucleosome Positioning and NRL on Chromatin Compaction
Although NRL variations were initially proposed to alter chromatin fiber diameter proportionally to linker DNA length (21), more recent ultrastructural studies suggest a stepwise increase in the chromatin diameter from ∼33 nm for chromatin with 30–60-bp linkers to ∼42 nm for 70–90-bp linkers (26). The observed structural transitions were explained by topological variations of linker folding from the helical ribbon configuration to a crossed zigzag configuration (66), as well as by polymorphic chromatin models where the nucleosome linkers were tangentially oriented in the fiber and did not cross the fiber axis (67). For short NRLs typical of yeast and neuronal cells (167 bp), a smaller diameter of ∼21 nm with a zigzag morphology (68) was obtained, consistent with nucleosome arrangement in the tetranucleosome x-ray crystal structure (25).
Linker length variations were also predicted to have a strong effect on the chromatin higher order structure via altered internucleosome orientations (30, 31, 69, 70). However, experiments on nucleosomes with NRLs varied in a fixed pattern mimicking natural variations (207 ± 2) showed similar folding to uniform arrays of the same length (29). Consistent with these findings, modeling work has suggested that whereas short linkers are too rigid and long linkers are too flexible, medium values (in the range of 200–210 bp) can adopt variable conformations to optimize overall fiber compaction (Figs. 2 and 5D) (71, 72).
Modeling Approaches
Challenges and Approaches
As discussed in a recent modeling and simulation perspective (73), our computing power and algorithms have improved markedly over the past decade, rendering problems of greater scientific significance solvable with enhanced confidence and accuracy. Although all-atom simulations of nucleic acids have steadily increased in accuracy, scope, and length (e.g. microsecond simulations of solvated B-DNA dodecamer (73)), coarse-grained models are required to simulate macromolecular chromatin systems that are too large for atomic models and highly dynamic. Creation of such models is required to resolve key functional components of the molecular system while approximating others. Ultimately, modeling the various folded states of the chromatin fiber requires multiscale methods to bridge the resolution among different spatial and temporal scales.
As summarized in Fig. 4 (and in the indicated references), several groups have developed different models with various levels of complexity and involving different simulation techniques. In most of these models, nucleosome geometries are simplified by using a few key variables (e.g. nucleosome positions and charges and angular orientation between the wrapped and existing/entering DNA), water is treated implicitly, and configurations are generated by Monte Carlo sampling or via analytic formulations that sample various parameter ranges. Model validation is performed using available experimental data, such as salt-induced compaction of oligonucleosomes to reproduce experimental sedimentation coefficients and nucleosome packing ratios, diffusion behavior of oligonucleosomes, salt-dependent extension of histone tails measured by the tail-to-tail diameter of the core and radii of gyration for mononucleosomes, LH orientations, and internucleosome interaction patterns (see Ref. 61 for a summary of such model validation details and available experiments). Overall, such modeling is particularly useful for probing structural and energetic effects as a function of certain parameters or conditions like the ionic salt concentration, NRL, and the presence of LHs. Simulations can also suggest specific configurations to help interpret single-molecule pulling experiments by associating specific force versus extension data points with fiber conformations, as shown in Fig. 5.
Effect of Ionic Conditions, NRL, and LH on Fiber Architecture
As an example of these parameter dependences, Wong et al. (67) showed the dependence of fiber width on the linker DNA length and the orientation of LHs. Modeling of simplified coarse-grained nucleosome models by Rippe and co-workers (74) reinforced the large effect of the linker length and nucleosome twist angles on the extent of fiber compaction. Mesoscale modeling combined with experimental cross-linking studies that measured nucleosome interaction patters complemented by EM visualization (29) revealed a compaction pattern as LH and divalent ions are added: an open, disordered, zigzag organization for chromatin fibers without LH rearranges to form compact zigzag forms with LHs under monovalent ion conditions. With divalent ions, further compaction arises by bending a small portion of the linker DNAs to form a heteromorphic architecture (Fig. 5C).
In our model (61, 72), each nucleosome histone octamer (without protruding tails and with the wound DNA) is treated as an irregularly shaped electrostatic charged object with point charges parameterized to reproduce the atomistic electric field. Each flexible histone tail is modeled as a chain of spherical beads representing five amino acids each, and LHs are modeled using three beads representing the C-terminal, N-terminal, and globular domains (Fig. 5D). The linker DNA connecting the nucleosomes is treated using the wormlike chain elastic approximation for DNA. The total energy consists of bending, stretching, torsion, excluded volume, and electrostatic contributions (43, 61).
Studies investigating the folding of the chromatin fiber as a function of NRL, with and without LH, suggested that short-to-medium NRL fibers (173–209 bp) with LH condense into irregular zigzag structures and that solenoid features are viable only for longer NRLs (218–226 bp) (Fig. 5D) (71, 72). These studies suggest that medium NRL favors chromatin compaction throughout the cell cycle, unlike short and long NRL fibers: short NRL arrays fold into narrow fibers, whereas long NRLs do not easily lead to high packing ratios due to possible linker DNA bending. Furthermore, the histone tails influence fibers with medium NRL more easily. The small compaction effect of LH in short linker fibers is consistent with experiments (33, 68).
Recent modeling studies that mimic chromatin stretching experiments have proposed interesting roles for LH in fiber compaction under various dynamic mechanisms. A wealth of chromatin pulling experiments as summarized recently (75, 76) has emphasized the need for further mechanistic and structural interpretation of the force versus extension curves. For example, what factors stiffen the chromatin fiber, and what do the force versus extension curves imply regarding chromatin structure? Our studies focused on analyzing the stretching response of chromatin fibers as a function of the NRL and LH presence, including various binding mechanisms for LH. Indeed, LHs are known to be dynamic, with different binding ratios (77, 78). Because fiber resistance to stretching decreases markedly with dynamic compared with static LHs due to possible stem rearrangements in the former (Fig. 5E, panel 1), we have suggested that dynamic LH binding may be an essential mechanism to soften chromatin fibers and allow unfolding at typical forces corresponding to natural molecular motors (Fig. 5E) (43). Moreover, among the dynamic LHs, pools of fast- and slow-binding LHs may cooperatively induce fiber unfolding at low forces: lower binding affinity softens fibers due to stem destabilization, whereas higher binding affinity promotes superbead constructs that combine nucleosome clusters with stretched fiber regions. The combination may offer both flexibility and selective DNA exposure (Fig. 5E) (43). These results thus suggest how modeling can help identify the factors that stiffen or soften chromatin fibers, as well as propose conformations and pathways linked to experimental force versus extension curves. In particular, our conformations are consistent with an overall zigzag fiber arrangement (Fig. 5E) rather than solenoid interpretation (33).
Tertiary and Higher Order Structures, in Situ Structures, and Functional Connections
Higher Order Structures beyond the Chromatin Fiber
Folding of the nucleosome array into the 30-nm fiber is far from achieving the 5 orders of spatial compaction realized by the chromosomes near the end of the cell cycle (Fig. 1). Various looping, scaffolding, wrapping, and specific contacts with other proteins and possibly RNA have been suggested for this higher folding to occur.
In vitro, in the presence of divalent cations in excess of 2 mm, as well as in situ, in interphase eukaryotic cells, chromatin fibers self-interact to form tertiary chromatin structures (1, 6, 45). This type of chromatin compaction may account for the in situ chromatin fibers that are partially interdigitated (79) and in vitro chromatin structures with diameters of >30 nm (80, 81). In vitro, the formation of such structures is promoted by the same factors as the secondary structure: divalent cations, interactions between core histone N-terminal tails (46, 82), LHs (83), and heterochromatin architectural factors such as MENT, MeCP2, and Sir3 (84, 85). Modeling shows that longer nucleosome repeats typical of terminally differentiated cells promote lateral self-association of the chromatin fibers and nucleosome interdigitation (74).
Various ultrastructural studies, such as cryo-EM tomography (79), cryo-EM (86), electron spectroscopic imaging (87), and small-angle x-ray scattering (88, 89), have not yet provided a uniform view of chromatin organization in the interphase state. These techniques revealed 30-nm fibers in a number of cells with condensed chromatin, such as Echinodermata sperm, chicken erythrocytes, and mouse retina (79, 88, 90). However, chromatin in proliferating cells showed thicker (100-nm diameter) “chromonema fibers,” with none or few 30-nm structures detected (91–93). Even in the most condensed heterochromatic areas, no regular fibers of 30 nm in diameter were observed in the nuclei for interphase cells (94), thus raising the possibility that the 30-nm fiber is not a predominant structure in the interphase nucleus.
Dynamics of Higher Order Chromatin Folding
Although earlier structural models suggested a hierarchy of highly ordered static structures for eukaryotic chromatin, recent studies have revealed chromatin organization to be highly dynamic. This mobility includes the spontaneous reversible unfolding of the DNA segments from nucleosomes (16, 17) and oligonucleosome arrays (95), the transient association of chromatin architectural proteins like heterochromatin protein HP1 (96) with chromatin, and the accessibility of condensed chromatin inside metaphase chromosomes for extrachromosomal proteins (97). In particular, as discussed above in connection with Fig. 5E, LH is highly dynamic, binding to or unbinding from the nucleosomes rapidly (78, 98, 99), and LH binding dynamics have been suggested to affect fiber compaction (100). Modeling has suggested that fast- and slow-binding populations of LH (77) merge optimally in heterogeneous forms of fibers to make chromatin more amenable to molecular motors (Fig. 5E, panel 3) (43).
The dynamic nature of chromatin necessitates developing new structural and modeling approaches to capture transient structural intermediates of chromatin folding in vitro and in vivo and to relate them to existing nucleosome structures, energetically favorable molecular models, and physical maps and bioinformatic analyses of native chromatin structures. This convergence of experiments and modeling will undoubtedly help translate the linear organization of the nucleosomes and their interaction in three-dimensional space to the hierarchy of chromatin and DNA conformations associated with key regulatory processes via modulation of chromatin accessibility to nuclear proteins and mediation of epigenetic interactions.
Acknowledgments
We thank Rosana Collepardo-Guevara, Durba Roy, and Shereef Elmetwaly for assistance in preparing the figures.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 GM055164 (to T. S.) and R01 GM52426 (to J. H.). This work was also supported by National Science Foundation Grants MCB-0316771 (to T. S.) and MCB-1021681 (to S. G.) and Philip-Morris International (to T. S.).
- NRL
- nucleosome repeat length
- LH
- linker histone
- PTM
- post-translational modification.
REFERENCES
- 1. Woodcock C. L., Dimitrov S. (2001) Higher order structure of chromatin and chromosomes. Curr. Opin. Genet. Dev. 11, 130–135 [DOI] [PubMed] [Google Scholar]
- 2. Bancaud A., Huet S., Daigle N., Mozziconacci J., Beaudouin J., Ellenberg J. (2009) Molecular crowding affects diffusion and binding of nuclear proteins in heterochromatin and reveals the fractal organization of chromatin. EMBO J. 28, 3785–3798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. van Holde K. E. (1988) Chromatin, Springer-Verlag, New York [Google Scholar]
- 4. Felsenfeld G., Groudine M. (2003) Controlling the double helix. Nature 421, 448–453 [DOI] [PubMed] [Google Scholar]
- 5. Horn P. J., Peterson C. L. (2002) Molecular biology. Chromatin higher order folding–wrapping up transcription. Science 297, 1824–1827 [DOI] [PubMed] [Google Scholar]
- 6. Tremethick D. J. (2007) Higher order structures of chromatin: the elusive 30-nm fiber. Cell 128, 651–654 [DOI] [PubMed] [Google Scholar]
- 7. van Holde K., Zlatanova J. (2007) Chromatin fiber structure: where is the problem now? Semin. Cell Dev. Biol. 18, 651–658 [DOI] [PubMed] [Google Scholar]
- 8. Li G., Reinberg D. (2011) Chromatin higher order structures and gene regulation. Curr. Opin. Genet. Dev. 21, 175–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Luger K., Mäder A. W., Richmond R. K., Sargent D. F., Richmond T. J. (1997) Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature 389, 251–260 [DOI] [PubMed] [Google Scholar]
- 10. Davey C. A., Sargent D. F., Luger K., Maeder A. W., Richmond T. J. (2002) Solvent-mediated interactions in the structure of the nucleosome core particle at 1.9 Å resolution. J. Mol. Biol. 319, 1097–1113 [DOI] [PubMed] [Google Scholar]
- 11. Lu X., Simon M. D., Chodaparambil J. V., Hansen J. C., Shokat K. M., Luger K. (2008) The effect of H3K79 dimethylation and H4K20 trimethylation on nucleosome and chromatin structure. Nat. Struct. Mol. Biol. 15, 1122–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bao Y., White C. L., Luger K. (2006) Nucleosome core particles containing a poly(dA·dT) sequence element exhibit a locally distorted DNA structure. J. Mol. Biol. 361, 617–624 [DOI] [PubMed] [Google Scholar]
- 13. Iwasaki W., Tachiwana H., Kawaguchi K., Shibata T., Kagawa W., Kurumizaka H. (2011) Comprehensive structural analysis of mutant nucleosomes containing lysine to glutamine (KQ) substitutions in the H3 and H4 histone fold domains. Biochemistry 50, 7822–7832 [DOI] [PubMed] [Google Scholar]
- 14. Wu B., Dröge P., Davey C. A. (2008) Site selectivity of platinum anticancer therapeutics. Nat. Chem. Biol. 4, 110–112 [DOI] [PubMed] [Google Scholar]
- 15. Zheng C., Hayes J. J. (2003) Structures and interactions of the core histone tail domains. Biopolymers 68, 539–546 [DOI] [PubMed] [Google Scholar]
- 16. Li G., Levitus M., Bustamante C., Widom J. (2005) Rapid spontaneous accessibility of nucleosomal DNA. Nat. Struct. Mol. Biol. 12, 46–53 [DOI] [PubMed] [Google Scholar]
- 17. Tomschik M., Zheng H., van Holde K., Zlatanova J., Leuba S. H. (2005) Fast, long-range, reversible conformational fluctuations in nucleosomes revealed by single-pair fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. U.S.A. 102, 3278–3283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tachiwana H., Kagawa W., Shiga T., Osakabe A., Miya Y., Saito K., Hayashi-Takanaka Y., Oda T., Sato M., Park S. Y., Kimura H., Kurumizaka H. (2011) Crystal structure of the human centromeric nucleosome containing CENP-A. Nature 476, 232–235 [DOI] [PubMed] [Google Scholar]
- 19. Panchenko T., Sorensen T. C., Woodcock C. L., Kan Z. Y., Wood S., Resch M. G., Luger K., Englander S. W., Hansen J. C., Black B. E. (2011) Replacement of histone H3 with CENP-A directs global nucleosome array condensation and loosening of nucleosome superhelical termini. Proc. Natl. Acad. Sci. U.S.A. 108, 16588–16593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lantermann A. B., Straub T., Strålfors A., Yuan G. C., Ekwall K., Korber P. (2010) Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 17, 251–257 [DOI] [PubMed] [Google Scholar]
- 21. Athey B. D., Smith M. F., Rankert D. A., Williams S. P., Langmore J. P. (1990) The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: evidence in support of variable diameter models for chromatin. J. Cell Biol. 111, 795–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simpson R. T., Thoma F., Brubaker J. M. (1985) Chromatin reconstituted from tandemly repeated cloned DNA fragments and core histones: a model system for study of higher order structure. Cell 42, 799–808 [DOI] [PubMed] [Google Scholar]
- 23. Lowary P. T., Widom J. (1998) New DNA sequence rules for high-affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 [DOI] [PubMed] [Google Scholar]
- 24. Dorigo B., Schalch T., Kulangara A., Duda S., Schroeder R. R., Richmond T. J. (2004) Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science 306, 1571–1573 [DOI] [PubMed] [Google Scholar]
- 25. Schalch T., Duda S., Sargent D. F., Richmond T. J. (2005) X-ray structure of a tetranucleosome and its implications for the chromatin fiber. Nature 436, 138–141 [DOI] [PubMed] [Google Scholar]
- 26. Robinson P. J., Fairall L., Huynh V. A., Rhodes D. (2006) EM measurements define the dimensions of the “30-nm” chromatin fiber: evidence for a compact, interdigitated structure. Proc. Natl. Acad. Sci. U.S.A. 103, 6506–6511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Robinson P. J., Rhodes D. (2006) Structure of the “30-nm” chromatin fiber: a key role for the linker histone. Curr. Opin. Struct. Biol. 16, 336–343 [DOI] [PubMed] [Google Scholar]
- 28. Daban J. R., Bermúdez A. (1998) Interdigitated solenoid model for compact chromatin fibers. Biochemistry 37, 4299–4304 [DOI] [PubMed] [Google Scholar]
- 29. Grigoryev S. A., Arya G., Correll S., Woodcock C. L., Schlick T. (2009) Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc. Natl. Acad. Sci. U.S.A. 106, 13317–13322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Woodcock C. L., Grigoryev S. A., Horowitz R. A., Whitaker N. (1993) A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc. Natl. Acad. Sci. U.S.A. 90, 9021–9025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Leuba S. H., Yang G., Robert C., Samori B., van Holde K., Zlatanova J., Bustamante C. (1994) Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc. Natl. Acad. Sci. U.S.A. 91, 11621–11625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cui Y., Bustamante C. (2000) Pulling a single chromatin fiber reveals the forces that maintain its higher order structure. Proc. Natl. Acad. Sci. U.S.A. 97, 127–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kruithof M., Chien F. T., Routh A., Logie C., Rhodes D., van Noort J. (2009) Single-molecule force spectroscopy reveals a highly compliant helical folding for the 30-nm chromatin fiber. Nat. Struct. Mol. Biol. 16, 534–540 [DOI] [PubMed] [Google Scholar]
- 34. Thoma F., Koller T., Klug A. (1979) Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83, 403–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bednar J., Horowitz R. A., Grigoryev S. A., Carruthers L. M., Hansen J. C., Koster A. J., Woodcock C. L. (1998) Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher order folding and compaction of chromatin. Proc. Natl. Acad. Sci. U.S.A. 95, 14173–14178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Syed S. H., Goutte-Gattat D., Becker N., Meyer S., Shukla M. S., Hayes J. J., Everaers R., Angelov D., Bednar J., Dimitrov S. (2010) Single-base resolution mapping of H1-nucleosome interactions and three-dimensional organization of the nucleosome. Proc. Natl. Acad. Sci. U.S.A. 107, 9620–9625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fan L., Roberts V. A. (2006) Complex of linker histone H5 with the nucleosome and its implications for chromatin packing. Proc. Natl. Acad. Sci. U.S.A. 103, 8384–8389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Meyer S., Becker N. B., Syed S. H., Goutte-Gattat D., Shukla M. S., Hayes J. J., Angelov D., Bednar J., Dimitrov S., Everaers R. (2011) From crystal and NMR structures, footprints and cryo-electron micrographs to large and soft structures: nanoscale modeling of the nucleosomal stem. Nucleic Acids Res. 39, 9139–9154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bharath M. M., Chandra N. R., Rao M. R. (2003) Molecular modeling of the chromatosome particle. Nucleic Acids Res. 31, 4264–4274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Caterino T. L., Fang H., Hayes J. J. (2011) Nucleosome linker DNA contacts and induces specific folding of the intrinsically disordered H1 carboxyl-terminal domain. Mol. Cell. Biol. 31, 2341–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lu X., Hamkalo B., Parseghian M. H., Hansen J. C. (2009) Chromatin condensing functions of the linker histone C-terminal domain are mediated by specific amino acid composition and intrinsic protein disorder. Biochemistry 48, 164–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tóth K., Brun N., Langowski J. (2001) Trajectory of nucleosomal linker DNA studied by fluorescence resonance energy transfer. Biochemistry 40, 6921–6928 [DOI] [PubMed] [Google Scholar]
- 43. Collepardo-Guevara R., Schlick T. (2011) The effect of linker histone's nucleosome binding affinity on chromatin unfolding mechanisms. Biophys. J. 101, 1670–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Luger K., Richmond T. J. (1998) The histone tails of the nucleosome. Curr. Opin. Genet. Dev. 8, 140–146 [DOI] [PubMed] [Google Scholar]
- 45. Luger K., Hansen J. C. (2005) Nucleosome and chromatin fiber dynamics. Curr. Opin. Struct. Biol. 15, 188–196 [DOI] [PubMed] [Google Scholar]
- 46. Dorigo B., Schalch T., Bystricky K., Richmond T. J. (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J. Mol. Biol. 327, 85–96 [DOI] [PubMed] [Google Scholar]
- 47. Sinha D., Shogren-Knaak M. A. (2010) Role of direct interactions between the histone H4 tail and the H2A core in long-range nucleosome contacts. J. Biol. Chem. 285, 16572–16581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kan P. Y., Lu X., Hansen J. C., Hayes J. J. (2007) The H3 tail domain participates in multiple interactions during folding and self-association of nucleosome arrays. Mol. Cell. Biol. 27, 2084–2091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Kan P. Y., Caterino T. L., Hayes J. J. (2009) The H4 tail domain participates in intra- and internucleosome interactions with protein and DNA during folding and oligomerization of nucleosome arrays. Mol. Cell. Biol. 29, 538–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Zhou J., Fan J. Y., Rangasamy D., Tremethick D. J. (2007) The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat. Struct. Mol. Biol. 14, 1070–1076 [DOI] [PubMed] [Google Scholar]
- 51. Chodaparambil J. V., Barbera A. J., Lu X., Kaye K. M., Hansen J. C., Luger K. (2007) A charged and contoured surface on the nucleosome regulates chromatin compaction. Nat. Struct. Mol. Biol. 14, 1105–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 53. Wang G. G., Allis C. D., Chi P. (2007) Chromatin remodeling and cancer. Part I: covalent histone modifications. Trends Mol. Med. 13, 363–372 [DOI] [PubMed] [Google Scholar]
- 54. Jenuwein T., Allis C. D. (2001) Translating the histone code. Science 293, 1074–1080 [DOI] [PubMed] [Google Scholar]
- 55. Shogren-Knaak M., Ishii H., Sun J. M., Pazin M. J., Davie J. R., Peterson C. L. (2006) Histone H4K16 acetylation controls chromatin structure and protein interactions. Science 311, 844–847 [DOI] [PubMed] [Google Scholar]
- 56. Allahverdi A., Yang R., Korolev N., Fan Y., Davey C. A., Liu C. F., Nordenskiöld L. (2011) The effects of histone H4 tail acetylations on cation-induced chromatin folding and self-association. Nucleic Acids Res. 39, 1680–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yang D., Arya G. (2011) Structure and binding of the H4 histone tail and the effects of lysine 16 acetylation. Phys. Chem. Chem. Phys. 13, 2911–2921 [DOI] [PubMed] [Google Scholar]
- 58. Tse C., Sera T., Wolffe A. P., Hansen J. C. (1998) Disruption of higher order folding by core histone acetylation dramatically enhances transcription of nucleosomal arrays by RNA polymerase III. Mol. Cell. Biol. 18, 4629–4638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Dion M. F., Altschuler S. J., Wu L. F., Rando O. J. (2005) Genomic characterization reveals a simple histone H4 acetylation code. Proc. Natl. Acad. Sci. U.S.A. 102, 5501–5506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. McBryant S. J., Klonoski J., Sorensen T. C., Norskog S. S., Williams S., Resch M. G., Toombs J. A., 3rd, Hobdey S. E., Hansen J. C. (2009) Determinants of histone H4 N-terminal domain function during nucleosomal array oligomerization. Roles of amino acid sequence, domain length, and charge density. J. Biol. Chem. 284, 16716–16722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Arya G., Schlick T. (2009) A tale of tails: how histone tails mediate chromatin compaction in different salt and linker histone environments. J. Phys. Chem. A 113, 4045–4059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Fierz B., Chatterjee C., McGinty R. K., Bar-Dagan M., Raleigh D. P., Muir T. W. (2011) Histone H2B ubiquitylation disrupts local and higher order chromatin compaction. Nat. Chem. Biol. 7, 113–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wang Y., Li M., Stadler S., Correll S., Li P., Wang D., Hayama R., Leonelli L., Han H., Grigoryev S. A., Allis C. D., Coonrod S. A. (2009) Histone hypercitrullination mediates chromatin decondensation and neutrophil extracellular trap formation. J. Cell Biol. 184, 205–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Robinson P. J., An W., Routh A., Martino F., Chapman L., Roeder R. G., Rhodes D. (2008) 30-nm chromatin fiber decompaction requires both H4K16 acetylation and linker histone eviction. J. Mol. Biol. 381, 816–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wang X., Hayes J. J. (2007) Site-specific binding affinities within the H2B tail domain indicate specific effects of lysine acetylation. J. Biol. Chem. 282, 32867–32876 [DOI] [PubMed] [Google Scholar]
- 66. Wu C., Bassett A., Travers A. (2007) A variable topology for the 30-nm chromatin fiber. EMBO Rep. 8, 1129–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Wong H., Victor J. M., Mozziconacci J. (2007) An all-atom model of the chromatin fiber containing linker histones reveals a versatile structure tuned by the nucleosomal repeat length. PLoS ONE 2, e877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Routh A., Sandin S., Rhodes D. (2008) Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc. Natl. Acad. Sci. U.S.A. 105, 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Widom J. (1992) A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc. Natl. Acad. Sci. U.S.A. 89, 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Stehr R., Schöpflin R., Ettig R., Kepper N., Rippe K., Wedemann G. (2010) Exploring the conformational space of chromatin fibers and their stability by numerical dynamic phase diagrams. Biophys. J. 98, 1028–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Schlick T., Perisić O. (2009) Mesoscale simulations of two-nucleosome repeat length oligonucleosomes. Phys. Chem. Chem. Phys. 11, 10729–10737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Perišić O., Collepardo-Guevara R., Schlick T. (2010) Modeling studies of chromatin fiber structure as a function of DNA linker length. J. Mol. Biol. 403, 777–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T. E., 3rd, Laughton C. A., Orozco M. (2007) Refinement of the AMBER force field for nucleic acids: improving the description of α/γ conformers. Biophys. J. 92, 3817–3829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Stehr R., Kepper N., Rippe K., Wedemann G. (2008) The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys. J. 95, 3677–3691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chien F. T., van Noort J. (2009) 10 years of tension on chromatin: results from single-molecule force spectroscopy. Curr. Pharm. Biotechnol. 10, 474–485 [DOI] [PubMed] [Google Scholar]
- 76. Lavelle C., Victor J. M., Zlatanova J. (2010) Chromatin fiber dynamics under tension and torsion. Int. J. Mol. Sci. 11, 1557–1579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Carrero G., Crawford E., Hendzel M. J., de Vries G. (2004) Characterizing fluorescence recovery curves for nuclear proteins undergoing binding events. Bull. Math. Biol. 66, 1515–1545 [DOI] [PubMed] [Google Scholar]
- 78. Stasevich T. J., Mueller F., Brown D. T., McNally J. G. (2010) Dissecting the binding mechanism of the linker histone in live cells: an integrated FRAP analysis. EMBO J. 29, 1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Horowitz R. A., Agard D. A., Sedat J. W., Woodcock C. L. (1994) The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zigzag nucleosomal ribbon. J. Cell Biol. 125, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Widom J. (1986) Physicochemical studies of the folding of the 100 Å nucleosome filament into the 300 Å filament. Cation dependence. J. Mol. Biol. 190, 411–424 [DOI] [PubMed] [Google Scholar]
- 81. Grigoryev S. A., Bednar J., Woodcock C. L. (1999) MENT, a heterochromatin protein that mediates higher order chromatin folding, is a new serpin family member. J. Biol. Chem. 274, 5626–5636 [DOI] [PubMed] [Google Scholar]
- 82. Gordon F., Luger K., Hansen J. C. (2005) The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J. Biol. Chem. 280, 33701–33706 [DOI] [PubMed] [Google Scholar]
- 83. Carruthers L. M., Bednar J., Woodcock C. L., Hansen J. C. (1998) Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher order chromatin folding. Biochemistry 37, 14776–14787 [DOI] [PubMed] [Google Scholar]
- 84. McBryant S. J., Adams V. H., Hansen J. C. (2006) Chromatin architectural proteins. Chromosome Res. 14, 39–51 [DOI] [PubMed] [Google Scholar]
- 85. Grigoryev S. A., Bulynko Y. A., Popova E. Y. (2006) The end adjusts the means: heterochromatin remodeling during terminal cell differentiation. Chromosome Res. 14, 53–69 [DOI] [PubMed] [Google Scholar]
- 86. Eltsov M., Maclellan K. M., Maeshima K., Frangakis A. S., Dubochet J. (2008) Analysis of cryo-electron microscopy images does not support the existence of 30-nm chromatin fibers in mitotic chromosomes in situ. Proc. Natl. Acad. Sci. U.S.A. 105, 19732–19737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Fussner E., Djuric U., Strauss M., Hotta A., Perez-Iratxeta C., Lanner F., Dilworth F. J., Ellis J., Bazett-Jones D. P. (2011) Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 30, 1778–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Langmore J. P., Paulson J. R. (1983) Low-angle x-ray diffraction studies of chromatin structure in vivo and in isolated nuclei and metaphase chromosomes. J. Cell Biol. 96, 1120–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Paulson J. R., Langmore J. P. (1983) Low-angle x-ray diffraction studies of HeLa metaphase chromosomes: effects of histone phosphorylation and chromosome isolation procedure. J. Cell Biol. 96, 1132–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kizilyaprak C., Spehner D., Devys D., Schultz P. (2010) In vivo chromatin organization of mouse rod photoreceptors correlates with histone modifications. PLoS ONE 5, e11039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tumbar T., Sudlow G., Belmont A. S. (1999) Large-scale chromatin unfolding and remodeling induced by VP16 acidic activation domain. J. Cell Biol. 145, 1341–1354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kireeva N., Lakonishok M., Kireev I., Hirano T., Belmont A. S. (2004) Visualization of early chromosome condensation: a hierarchical folding, axial glue model of chromosome structure. J. Cell Biol. 166, 775–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Kireev I., Lakonishok M., Liu W., Joshi V. N., Powell R., Belmont A. S. (2008) In vivo immunogold labeling confirms large-scale chromatin folding motifs. Nat. Methods 5, 311–313 [DOI] [PubMed] [Google Scholar]
- 94. Fussner E., Ching R. W., Bazett-Jones D. P. (2011) Living without 30-nm chromatin fibers. Trends Biochem. Sci. 36, 1–6 [DOI] [PubMed] [Google Scholar]
- 95. Poirier M. G., Bussiek M., Langowski J., Widom J. (2008) Spontaneous access to DNA target sites in folded chromatin fibers. J. Mol. Biol. 379, 772–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Cheutin T., McNairn A. J., Jenuwein T., Gilbert D. M., Singh P. B., Misteli T. (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299, 721–725 [DOI] [PubMed] [Google Scholar]
- 97. Chen D., Dundr M., Wang C., Leung A., Lamond A., Misteli T., Huang S. (2005) Condensed mitotic chromatin is accessible to transcription factors and chromatin structural proteins. J. Cell Biol. 168, 41–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Misteli T., Gunjan A., Hock R., Bustin M., Brown D. T. (2000) Dynamic binding of histone H1 to chromatin in living cells. Nature 408, 877–881 [DOI] [PubMed] [Google Scholar]
- 99. Lever M. A., Th'ng J. P., Sun X., Hendzel M. J. (2000) Rapid exchange of histone H1.1 on chromatin in living human cells. Nature 408, 873–876 [DOI] [PubMed] [Google Scholar]
- 100. Brown D. T. (2003) Histone H1 and the dynamic regulation of chromatin function. Biochem. Cell Biol. 81, 221–227 [DOI] [PubMed] [Google Scholar]