Background: Human islet amyloid polypeptide (hIAPP) fibrils of unknown structure are formed in type 2 diabetes.
Results: A hIAPP fibril structure was derived from EPR data, electron microscopy, and computer modeling.
Conclusion: The fibril is a left-handed helix that contains hIAPP monomers in a staggered conformation.
Significance: The results provide the basis for therapeutic prevention of fibril formation and growth.
Keywords: Amyloid, Computer Modeling, Electron Paramagnetic Resonance (EPR), Protein Structure, Structural Biology, Fibril, Helix, Human Islet Amyloid Polypeptide
Abstract
Misfolding and amyloid fibril formation by human islet amyloid polypeptide (hIAPP) are thought to be important in the pathogenesis of type 2 diabetes, but the structures of the misfolded forms remain poorly understood. Here we developed an approach that combines site-directed spin labeling with continuous wave and pulsed EPR to investigate local secondary structure and to determine the relative orientation of the secondary structure elements with respect to each other. These data indicated that individual hIAPP molecules take up a hairpin fold within the fibril. This fold contains two β-strands that are much farther apart than expected from previous models. Atomistic structural models were obtained using computational refinement with EPR data as constraints. The resulting family of structures exhibited a left-handed helical twist, in agreement with the twisted morphology observed by electron microscopy. The fibril protofilaments contain stacked hIAPP monomers that form opposing β-sheets that twist around each other. The two β-strands of the monomer adopt out-of-plane positions and are staggered by about three peptide layers (∼15 Å). These results provide a mechanism for hIAPP fibril formation and could explain the remarkable stability of the fibrils. Thus, the structural model serves as a starting point for understanding and preventing hIAPP misfolding.
Introduction
Alzheimer disease, Parkinson disease, type 2 diabetes mellitus, and spongiform encephalopathies are human diseases that involve misfolding and deposition of proteins (1). In type 2 diabetes, amyloid deposits found in the pancreata of 95% of patients are primarily composed of the 37-residue human islet amyloid polypeptide (hIAPP)3 (2). hIAPP is co-secreted with insulin by β-cells in the pancreatic islets of Langerhans (3–5) and is thought to play a role in carbohydrate metabolism (6–8) and satiety (9). hIAPP is monomeric in its physiological state but aggregated in the disease state. Evidence from cell and animal studies indicates a link between hIAPP misfolding and pancreatic β-cell dysfunction (10–13).
In disease, hIAPP undergoes a multistep misfolding process in which the monomer changes into various oligomeric forms and ultimately forms fibrils. A detailed molecular and mechanistic understanding of this process could facilitate therapeutic intervention. However, the three-dimensional structure of hIAPP fibrils or other misfolded forms remains elusive. FT-IR spectroscopy and circular dichroism show that the fibrils contain β-sheet structure (14, 15), and x-ray and electron diffraction indicate that the cross-β-strands are 4.7 Å apart and perpendicular to the fibril axis (16). Site-directed spin labeling and electron paramagnetic resonance (EPR) of hIAPP fibrils show these strands are arranged in a parallel, in-register structure (17). According to transmission electron microscopy (EM) and atomic force microscopy, hIAPP fibrils can have striated ribbon and twisted fibril morphologies (18).
Luca et al. (19) used solid-state NMR and electron microscopy to propose a model with residues 18–27 of hIAPP as a bend between two β-strands. A related, albeit slightly different, model has been suggested based upon the x-ray crystal structures of hIAPP fragments (20). Additionally, a number of theoretical models have been proposed for hIAPP fibrils, with one such model suggesting that hIAPP adopts a planar S-shaped fold with three β-strands (residues 8–16, 20–27, and 30–37) stacked in register (21). To obtain more detailed structural insight into the architecture of hIAPP fibrils, we adapted an EPR-based approach previously developed to determine the structure of membrane-bound α-synuclein (22). The EPR data are consistent with a structure in which individual hIAPP monomers within the fibril contain two β-strands with an interstrand separation that is greater than predicted in previous models. Computational refinement based on EPR results indicated that the two strands have an out-of plane stagger of about 15 Å. Stacking of multiple monomers results in a left-handed helical twist consistent with that observed for the fibril by electron microscopy. The model provides a basis for understanding the formation, extension, and stability of the hIAPP fibril.
EXPERIMENTAL PROCEDURES
Chemicals and Peptides
Hexafluoroisopropanol was obtained from Sigma-Aldrich. Synthetic wild-type hIAPP was obtained from Bachem Bioscience Inc. (King of Prussia, PA). The spin label, 1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate (MTSL), was obtained from Toronto Research Chemicals (Toronto, Ontario, Canada). Single- and double-cysteine mutants of hIAPP were purchased from Biomer Technology (Pleasanton, CA), with the native cysteines at positions 2 and 7 substituted by alanine (17). Peptides arrived 95% purified and lyophilized and were stored at −80 °C until use.
Spin Labeling and hIAPP Fibril Formation
Purified peptides were reacted with >5 molar excess MTSL for ∼1 h at room temperature. Unreacted MTSL was removed using a Toyopearl cation exchange column. The spin-labeled peptide was desalted using a C18 reverse phase SpinColumn (Harvard Apparatus, Holliston, MA) and eluted with 100% hexafluoroisopropanol. Peptide concentrations were calculated by UV absorbance at 280 nm in 6 m guanidine HCl using an extinction coefficient of 1400 m−1 cm−1. Labeled peptides were stored at −80 °C until use.
Stock solutions of spin-labeled hIAPP and wild-type hIAPP were lyophilized and then reconstituted with deionized water containing 0.5% acetic acid. hIAPP fibrils were formed in a spin-diluted state (17) by mixing stock solutions of spin-labeled hIAPP with unlabeled hIAPP (1:4 ratio) in 1.5-ml Eppendorf tubes. For fibril formation, aliquots of stock hIAPP were added to 10 mm phosphate buffer with 100 mm NaCl (pH 7.4) in a volume of 100 μl to give a concentration of 100 μm. Wild-type fibrils were grown for 1 week and then sonicated on ice at 1-min intervals using a tip sonicator for 10 min. The sonicated seeds were added to each spin-labeled sample at 10 weight by weight to encourage growth of amyloid fibrils. Samples were incubated at room temperature for 3–7 days to allow fibril formation.
Electron Microscopy
To examine fibril growth, 10 μl of fibril samples were adsorbed onto carbon- and Formvar-coated copper grids and negatively stained with 2% (w/v) uranyl acetate solution for 5 min. The stained grids were photographed using a JEOL JEM-1400 electron microscope at 80–100 kV. For unilateral metal shadowing experiments, specimens were freeze-dried on a carbon film and a 0.7-nm layer of tantalum/tungsten was deposited from an elevation angle of 30° in a BAL-TEC BAF 060 freeze-fracture machine (Leica Microsystems, Buffalo Grove, IL). The metal layer was stabilized by a uniform 5-nm carbon layer. Micrographs were recorded on a Philips CM120 microscope (FEI, Hillsboro, OR).
EPR Spectroscopy
Fibril samples were pipetted into a glass capillary (0.6-mm inner diameter, 0.84-mm outer diameter, VitroCom, Mountain Lakes, NJ) that was sealed at one end. The sample was centrifuged, and supernatant was removed to eliminate smaller aggregates and free spin labels. EPR spectra of fibrils were recorded at room temperature on a Bruker (Billerica, MA) EMX spectrometer with an HS resonator at 12-milliwatt incident microwave power and a scan range of 150 gauss. Mobility was calculated using the inverse of the central line width: the distance between the peak of the central line and the trough of the third line.
Pulsed EPR
Intramolecular distances were measured using four-pulse DEER experiments. Fibril samples were prepared by diluting 3% spin-labeled hIAPP (two spin labels per peptide) with unlabeled hIAPP and seeding with 10.0 wt% sonicated wild-type fibrils. Samples (300 μl) were incubated at room temperature for 3–7 days to allow fibril formation and then ultracentrifuged to remove supernatant containing smaller aggregates. The fibril pellet was collected into a quartz capillary (1.5-mm inner diameter, 1.8-mm outer diameter) and flash-frozen before DEER data were acquired at 78 K. Four-pulse DEER experiments were performed using a Bruker ELEXSYS E580 X-band pulse EPR spectrometer fitted with a 3-mm split ring (MS-3) resonator, a continuous flow helium cryostat (CF935), and a temperature controller (ITC503S, all Oxford Instruments). Data were fit using Gaussian distributions (23) in the DEERAnalysis2008 package (24).
Model Building
Computational refinement of the structure of the hIAPP fibril was performed using distances from DEER experiments (covering residues 12–36 of hIAPP) and data from EM and continuous wave EPR. The model included 101 copies of residues 12–36 of the 37-amino acid hIAPP peptide. The initial model was constructed as shown in supplemental Fig. S1. Each peptide was broken into two fragments (residues 12–26 and 27–36). The two fragments were each constructed as β-strands perpendicular to the fibril axis at a distance of 30 Å apart and with a 5 Å space between adjacent strands (supplemental Fig. S1, A and B). Seventeen spin labels were added to every fourth peptides at residues 12, 13, 14, 15, 16, 17, 18, 19, 24, 27, 28, 29, 30, 31, 32, 35, and 36, beginning from the first peptide (supplemental Fig. S1C). This starting model was based on CD and FT-IR data showing that hIAPP fibrils consist of β-sheets (19, 25); experimental data from fiber diffraction suggesting that the β-strands are perpendicular to the fibril axis and that each β-strand is about 5 Å apart from the neighboring strand (16); and site-directed spin labeling data suggesting that the β-strands are parallel and in-register (17). The starting structure was generated by the in-house programs FIBRIL and PRONOX (26), which were used to produce input files for AMBER10 (27).
Simulated Annealing Molecular Dynamics (SAMD)
SAMD calculations were performed in AMBER10 (27). After a brief minimization (3000 steps) of the starting structure, 10 cycles of SAMD were performed for 160 ps/cycle. Each SAMD cycle included a heating phase from 0 to 1200 K in 4 ps; maintenance of the temperature at 1200 K for a further 6 ps (during the first 10 ps, the force constants for the constraints were increased from 0.1 to 10.0); and then cooling to 0 K over 150 ps, with stepwise adjustments of the TAUTP parameter. SAMD was performed with a time step of 0.001 ps, a distance-dependent dielectric of 4, and a cut-off of 10.0 Å. The two fragments of each peptide were free to move in the SAMD calculations, under the constraints described below. Full details of the parameters and input files used for these calculations will be provided on request.
The constraints used in the calculations are shown in supplemental Table S1. DEER distances were implemented with ranges of ±2, ±4, or ±8 (supplemental Table S2) to reflect the width of the experimental distribution qualitatively. The “missing” peptide bond between residues 26 and 27 in each peptide was constrained to distances <5 Å. The distance from the first to the last peptide was limited to 468–481 Å to prevent the fibril from bending.
Several sets of calculations were performed with inclusion of constraints of the helical pitch of the fibril, based on the data from EM, with the goal of producing structures with pitches ranging from 200 to 4000 Å. The pitch constraint was implemented based on the assumption of an ∼5 Å interpeptide distance (as defined in the starting structure) and constraint of torsional angles between the β-strands of peptides i and i+10 in the fibril (supplemental Table S1). Thus, for example, a pitch of 500 Å is equivalent to a full helical turn over 100 peptides; therefore, the required torsional angle is −36° (for a left-handed helix) between the β-strands of peptides i and i+10. The equation used to generate the torsional constraints for a given pitch and details of the implementation of the constraints are given in a footnote to supplemental Table S1. SAMD calculations were also performed with no constraint of the pitch.
Structures were collected at the end of each SAMD cycle, giving a total of 10 structures. Each of these was energy-minimized (500 steps) to connect the missing peptide bonds and replace the spin labels with the original amino acids. Structural analysis was performed using AMBER10 (27) and PROCHECK (28) for checking the integrity of the structure. The chirality and pitch of the fibril helix, the distance between the two β-sheets, and the stagger of each peptide were analyzed using the in-house program FIBRIL. Distances between label pairs in the modeling structures were calculated and compared with the experimental DEER distances.
RESULTS
EM Analysis of Fibril Morphology
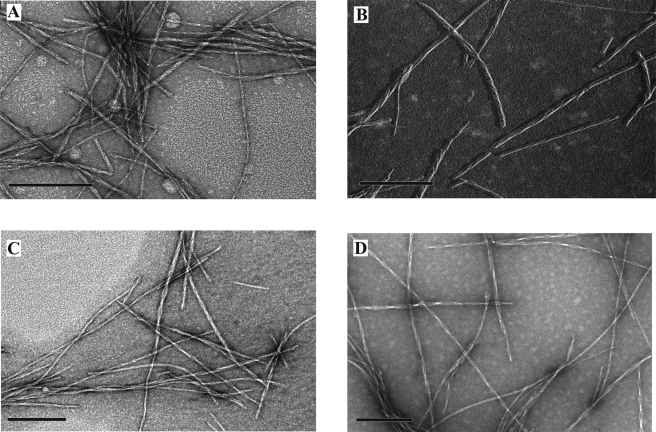
Because hIAPP fibrils can be polymorphic, we first developed a protocol involving multiple rounds of seeding that resulted in a relatively homogeneous fibril preparation. This approach yielded wild-type hIAPP fibrils with a predominantly (∼95%) twisted morphology (Fig. 1A) and a left-handed coil according to unilateral shadowing EM (Fig. 1B). These fibrils were used as seeds to induce formation of spin-labeled fibrils, which were grown from a mixture of labeled peptide and an excess of wild-type peptide (75% for singly labeled and 97% for doubly labeled peptides). According to negative stain electron microscopy, the resultant spin-labeled fibrils had the same morphology as those of the wild-type fibrils (Fig. 1, C and D).
FIGURE 1.
Electron microscope images of hIAPP fibrils showing twisted morphology. A, negatively stained wild-type hIAPP fibrils. B, unilateral metal shadowing of wild-type hIAPP fibrils showing a left-handed twist (metal was deposited from the right; deposited metal is shown in white, and shadows are black). C, negatively stained singly labeled (13R1) hIAPP fibrils. D, negatively stained double-labeled (13R1/18R1) hIAPP fibrils. Bar: 200 nm.
Local Mobility and Secondary Structure from Continuous Wave EPR Spectroscopy
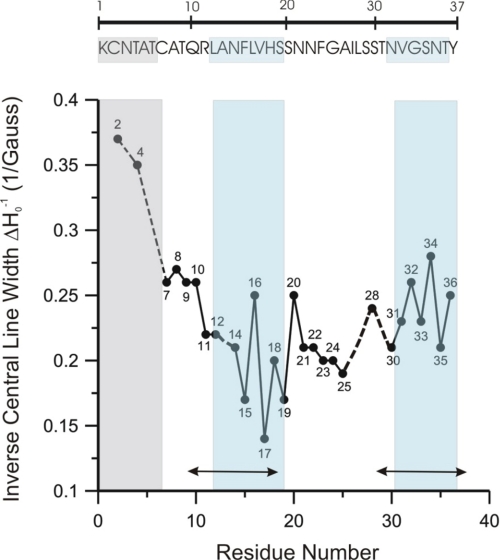
Next, 28 sets of hIAPP fibrils containing singly labeled peptides were analyzed by continuous wave EPR spectroscopy to obtain site-specific mobility and secondary structure information. The EPR spectra obtained under these conditions (supplemental Fig. S2) are highly sensitive to the label motion, which can be quantified using the inverse central line width (ΔHo−1), a commonly used semiquantitative mobility parameter. We found that residues 2 and 4 have high ΔHo−1 values, indicating that this N-terminal region is disordered in hIAPP fibrils. Residues 11–36 have generally low mobility values. Despite the general immobilization, enough contrast remains to reveal two regions (14–19 and 31–36) in which ΔHo−1 alternates with a periodicity of 2. Such a periodicity is commonly observed for β-sheet structures in which one face is more buried than the other. Because residue 13 was not included in the present study, it was not possible to monitor the periodicity starting from residue 12. However, based upon distance measurements presented below, residues 12 and 13 are included in one of the two β-strand regions (Fig. 2, blue). Residues 12–19 and 31–36 are also located within regions previously assigned to be in β-strand conformations according to solid-state NMR (Fig. 2). Residues 7–10 appear to be in a transition region in which the mobility is between that of the extreme N terminus and the most immobilized regions. No clear periodicity could be observed for the intrastrand region, which has previously been suggested to contain little or no β-sheet structure in hIAPP fibrils (19, 20). The solid-state NMR study investigated hIAPP fibrils with different morphology, but the assignment of secondary structure is in remarkably good agreement with our data, suggesting that regions of secondary structure may be similar in fibrils of different morphology.
FIGURE 2.
Residue mobilities of hIAPP fibrils containing 25% R1, as calculated from the inverse of central line width (ΔHo−1) of EPR spectra shown in supplemental Fig. S2. No mobility data were obtained for residue 13, but residue 12 was included in the strand region based upon the results from the four-pulse DEER distance measurements. The double-headed arrows indicate regions proposed to be in a β-sheet conformation based on solid-state NMR (19). The top panel shows the amino acid sequence of hIAPP 1–37. The boxed areas correspond to the residues outlined in the mobility graph below.
Intramolecular Distances from Four-pulse DEER Experiments
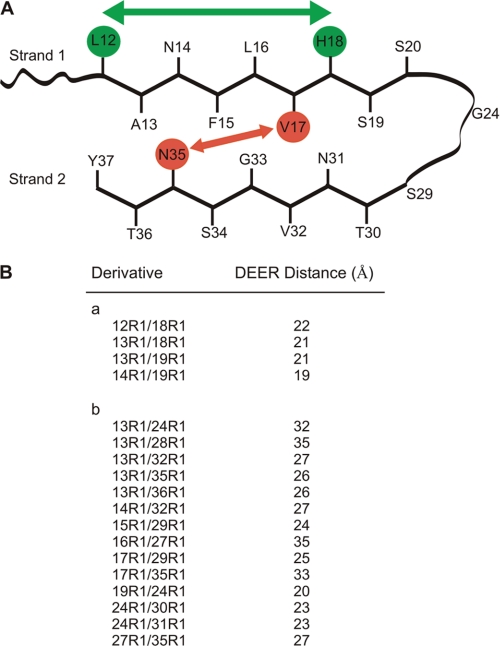
The EPR mobility data are consistent with the two-stranded, horseshoe-like model (Fig. 3) that has frequently been suggested for hIAPP and peptides in other amyloid fibrils (29–33). To test this model, intramolecular interlabel distances were determined in fibrils containing 18 different doubly labeled hIAPP peptides mixed with a 97% excess of wild-type hIAPP. None of these spectra displayed strong spin-spin interactions, which are typically seen for spin labels ≤15 Å apart (supplemental Fig. S3). To increase the range of the EPR distance measurements, four-pulse DEER experiments were performed (supplemental Fig. S4). First, we measured distances between residues expected to be in a β-sheet region (Fig. 3, supplemental Fig. S4A). Because a β-strand typically extends by ∼3.5 Å/amino acid, two spin labels (referred to here as R1) introduced at the i and i+6 positions should be ∼21 Å apart. The DEER measurements (Fig. 3B) are in excellent agreement with this prediction because the distances for the 12R1/18R1 and 13R1/19R1 pairs are 22 and 21 Å, respectively (Fig. 3A). The labels for the 13R1/18R1 and 14R1/19R1 pairs are spaced 5 amino acids apart, but the labels are facing in opposite directions and are predicted to be ∼20 Å apart. Again, the experimental distances support the presence of a β-strand for residues 12–19.
FIGURE 3.
Schematic model of hIAPP in protofilament and four-pulse DEER distance measurements. A, hIAPP with two β-strands regions connected by a loop. Placement of “inward” and “outward” oriented side chains is based on mobility measurements by continuous wave EPR (supplemental Fig. S3). Intrastrand (green) and interstrand (red) distances are indicated schematically. B, intramolecular DEER distances measured in doubly labeled hIAPP peptides within fibrils. Distances are from Gaussian fits of dipolar evolution data (supplemental Fig. S4).
Next, we measured 14 additional distances to investigate the spatial relationship between the two β-strands and other regions of the molecule (Fig. 3, supplemental Fig. S4B). Most of these distances (Fig. 3B) are inconsistent with the model presented in Fig. 3A. This is particularly apparent for the inter-β-strand distances (13R1/32R1; 13R1/35R1; 13R1/36R1; 14R1/32R1; 17R1/35R1), which range from 26 to 33 Å and are ∼20 Å longer than expected. Similarly, a number of distances between strand 1 and the non-β-strand region (20–30) are longer than those in the model in Fig. 3A (13R1/24R1; 13R1/28R1; 17R1/29R1).
The DEER data clearly reveal long distances between the strands, and the distributions for interstrand distances are wider than those for residues that are within the same strand (supplemental Fig. S4). This could indicate some structural heterogeneity within the fibrils, with somewhat variable orientations between the strands. However, it should also be noted that the accuracy with which the distance distributions can be determined is higher for the smaller distances because these require only short data acquisition times in four-pulse DEER experiments.
Structural Models
To generate structural models, we performed SAMD calculations using the EPR data as constraints. The calculations included a stack of 101 peptides with every fourth peptide being spin-labeled (supplemental Fig. S1), and a family of 10 structural models was generated. Remarkably, all of these models spontaneously exhibited left-handed helical structures that resemble the overall twisted morphology seen in the EM images. The mean pitch of the helix was 237.0 ± 13.5 Å (supplemental Table S3), which corresponds to a complete helical turn over about 50 peptides. This pitch is at the lower end of the range of pitches seen by EM. This could be due to the calculations including only a single stack of peptides, whereas the EM images (Fig. 1) show protofilaments wrapped around each other to form fibrils with variable pitches of 250 to about 1000 Å. Therefore, further SAMD calculations were performed (supplemental Tables S4–S7) to determine whether the DEER distances could also be consistent with these longer pitches. Calculations in which the pitch was nominally constrained to 250 Å (supplemental Table S4), 500 Å (supplemental Table S5), and 1000 Å (supplemental Table S6), respectively, gave structures with average pitches of 242.7 ± 4.4, 448.3 ± 10.6, and 833.0 ± 42.3 Å, respectively (supplemental Fig. S5). With stronger constraints, we were able to generate helical structures (all left-handed) with much larger pitches (supplemental Table S7).
One of the best established geometrical parameters for hIAPP fibrils is the distance between the β-sheets, which has been determined to be about 10 Å by electron diffraction (16). Calculation of this “sheet-to-sheet” distance for the derived models gave values of 8.8 ± 0.3, 10.8 ± 0.2, and 12.6 ± 0.4 Å for mean helical pitches of 242.7, 448.3, and 833.0 Å, respectively (supplemental Tables S4–S6), but much higher sheet-to-sheet distances for larger pitches (supplemental Table S7). DEER data were also inconsistent with helical pitches >1000 Å. For example, the mean 13R1/35R1 distance in models with pitches ≤1000 Å was consistent with the DEER distance, but this consistency was lost in models with higher helical pitches (supplemental Fig. S6). The 13R1/35R1 distance spans the two β-sheets and should be sensitive to the sheet-to-sheet distance. Therefore, we suggest that these results support an upper limit for the helical pitch of close to 1000 Å, about the same as that seen in the EM images.
A structural model with a pitch of about 500 Å is shown in Fig. 4. The peptide conformation has a horseshoe shape (Fig. 4A), but with the important distinction that one β-strand is displaced with respect to the plane of the second β-strand (Fig. 4B). This displacement can be seen by observing the location of a single peptide in the fibril (Fig. 4, C and D), in which the two β-strands within each peptide have a stagger of about ∼15 Å, causing one β-strand in peptide i to be adjacent to the opposite β-strand in peptide i+3. The resulting helical twist produced by this arrangement of peptides is shown in Fig. 4E.
FIGURE 4.
Model of hIAPP fibril structure. A, a typical hIAPP peptide incorporated in the fibril, viewed along the fibril axis. B, the same peptide viewed along an axis orthogonal to the fibril axis, showing the stagger of the two β-strands of the peptide. C, a section of the structural model showing the stagger of the peptides (shown as blue, green, and orange ribbons; the relationship of blue-colored peptides is indicated). D, rotation of the structural model with the axes of β-strands orthogonal to the fibril. E, a structural model containing 101 peptides showing a turn of the left-handed helix of ∼90° between the centers of the red boxes. The helix has an average pitch of 440 Å.
DISCUSSION
In this study, we investigated the structure of hIAPP fibrils using electron microscopy, electron paramagnetic resonance, and computational refinement. The EPR data are consistent with a peptide conformation with two β-strands forming two opposing β-sheets that wrap around one another, but the long interstrand distances are inconsistent with previous models. Our computational refinement revealed that the two strands in each peptide are staggered by about 15 Å, and this staggered relationship leads to a left-handed twisted morphology within the protofilament. This stagger is not present in the model of striated ribbon fibrils (19) and may represent the underlying structural difference between those two morphologies. An interesting consequence of the twisted arrangement with staggered β-strands is the significant exposure of a hydrophobic surface at the fibril ends. These “sticky ends” are likely to promote the capture of further monomeric hIAPP and thus may facilitate fibril elongation.
A detailed analysis of the geometry of the derived models indicated an upper limit for the helical pitch of about 1000 Å. Comparison of structures with different pitches revealed only small differences in peptide conformation and slight changes in the angle between adjacent β-strands, but these differences resulted in significant differences in the helical pitch. The general consistency between the structures derived at an atomic level and the macroscopic EM images is remarkable, particularly because only a single stack of peptides was used in the computational refinement. Our findings provide a direct connection between the macroscopic EM images and a structural model at the molecular level.
The finding that minor structural changes at the monomer level can lead to significant variations in the long-range twist could explain why fibrils of hIAPP and other amyloid proteins can vary significantly in twist along the fibril axis (16, 18). Subtle structural changes might occur during fibril growth, and such changes could be sufficient to result in a significant alteration in fibril pitch. We note that our structural models only consider a single stack of peptides and that additional restrictions may occur from the wrapping of multiple structures around each other within fibrils or perhaps also within protofilaments (19). The twisted arrangement of the protofilaments and the out-of-plane stagger of each monomer also suggest a reason for the particular stability of hIAPP fibrils. This arrangement results in the absence of a discrete plane of hydrogen bonds orthogonal to the fibril axis, and thus, there is no plane that would be susceptible to fracture.
Although a small stagger has been inferred from mutagenesis data (30) and solid-state NMR results (32) for amyloid-beta fibrils, the present results differ significantly from all previously proposed fibril structures. The difference arises from the ability to determine long-range distances, which are essential for defining the overall structure and defining the stagger between the two β-strands. These distances complement those from solid-state NMR, which is effective for determining distances <10 Å. The refined family of structures represents the first example in which structural investigation at the atomistic level is able to reproduce the overall morphology observed by EM. In our previous studies, we have combined continuous wave and pulsed EPR with computational refinement to investigate structures of membrane proteins (22, 34). The results of the current study demonstrate that this approach is also applicable to amyloid protein misfolding. The approach should be applicable to studying the structures of fibrils from other amyloid proteins, as well as those of the various oligomeric misfolded forms of these proteins.
Supplementary Material
Acknowledgment
Computation for the work described in this study was supported by the University of Southern California Center for High-performance Computing and Communications.
This work was supported, in whole or in part, by National Institutes of Health Grant AG027936 (to R. L.).
This article was selected as a Paper of the Week.

This article contains supplemental Tables S1–S7 and Figs. S1–S6.
- hIAPP
- human islet amyloid polypeptide
- MTSL
- 1-oxyl-2,2,5,5-tetramethyl-Δ3-pyrroline-3-methyl methanethiosulfonate
- DEER
- double electron-electron resonance
- SAMD
- simulated annealing molecular dynamics.
REFERENCES
- 1. Stefani M. (2004) Protein misfolding and aggregation: new examples in medicine and biology of the dark side of the protein world. Biochim. Biophys. Acta 1739, 5–25 [DOI] [PubMed] [Google Scholar]
- 2. Clark A., Lewis C. E., Willis A. C., Cooper G. J., Morris J. F., Reid K. B., Turner R. C. (1987) Islet amyloid formed from diabetes-associated peptide may be pathogenic in type-2 diabetes. Lancet 330, 231–234 [DOI] [PubMed] [Google Scholar]
- 3. Clark A., de Koning E. J., Hattersley A. T., Hansen B. C., Yajnik C. S., Poulton J. (1995) Pancreatic pathology in non-insulin-dependent diabetes (NIDDM). Diabetes Res. Clin. Pract. 28, S39–47 [DOI] [PubMed] [Google Scholar]
- 4. Westermark P., Wernstedt C., Wilander E., Hayden D. W., O'Brien T. D., Johnson K. H. (1987) Amyloid fibrils in human insulinoma and islets of Langerhans of the diabetic cat are derived from a neuropeptide-like protein also present in normal islet cells. Proc. Natl. Acad. Sci. U.S.A. 84, 3881–3885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Westermark P., Wilander E., Johnson K. H. (1987) Islet amyloid polypeptide. Lancet 2, 623. [DOI] [PubMed] [Google Scholar]
- 6. Ahrén B., Oosterwijk C., Lips C. J., Höppener J. W. (1998) Transgenic overexpression of human islet amyloid polypeptide inhibits insulin secretion and glucose elimination after gastric glucose gavage in mice. Diabetologia 41, 1374–1380 [DOI] [PubMed] [Google Scholar]
- 7. Gebre-Medhin S., Mulder H., Pekny M., Westermark G., Törnell J., Westermark P., Sundler F., Ahrén B., Betsholtz C. (1998) Increased insulin secretion and glucose tolerance in mice lacking islet amyloid polypeptide (amylin). Biochem. Biophys. Res. Commun. 250, 271–277 [DOI] [PubMed] [Google Scholar]
- 8. Kahn S. E., D'Alessio D. A., Schwartz M. W., Fujimoto W. Y., Ensinck J. W., Taborsky G. J., Jr., Porte D., Jr. (1990) Evidence of co-secretion of islet amyloid polypeptide and insulin by β-cells. Diabetes 39, 634–638 [DOI] [PubMed] [Google Scholar]
- 9. Young A. (2005) Inhibition of food intake. Adv. Pharmacol. 52, 79–98 [DOI] [PubMed] [Google Scholar]
- 10. Butler A. E., Jang J., Gurlo T., Carty M. D., Soeller W. C., Butler P. C. (2004) Diabetes due to a progressive defect in β-cell mass in rats transgenic for human islet amyloid polypeptide (HIP Rat): a new model for type 2 diabetes. Diabetes 53, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 11. Cooper G. J., Willis A. C., Clark A., Turner R. C., Sim R. B., Reid K. B. (1987) Purification and characterization of a peptide from amyloid-rich pancreases of type 2 diabetic patients. Proc. Natl. Acad. Sci. U.S.A. 84, 8628–8632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Janson J., Soeller W. C., Roche P. C., Nelson R. T., Torchia A. J., Kreutter D. K., Butler P. C. (1996) Spontaneous diabetes mellitus in transgenic mice expressing human islet amyloid polypeptide. Proc. Natl. Acad. Sci. U.S.A. 93, 7283–7288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kahn S.E., Andrikopoulos S., Verchere C.B. (1999) Islet amyloid: a long-recognized but underappreciated pathological feature of type 2 diabetes. Diabetes 48, 241–253 [DOI] [PubMed] [Google Scholar]
- 14. Higham C. E., Jaikaran E. T., Fraser P. E., Gross M., Clark A. (2000) Preparation of synthetic human islet amyloid polypeptide (IAPP) in a stable conformation to enable study of conversion to amyloid-like fibrils. FEBS Lett. 470, 55–60 [DOI] [PubMed] [Google Scholar]
- 15. Radovan D., Smirnovas V., Winter R. (2008) Effect of pressure on islet amyloid polypeptide aggregation: revealing the polymorphic nature of the fibrillation process. Biochemistry 47, 6352–6360 [DOI] [PubMed] [Google Scholar]
- 16. Sumner Makin O., Serpell L. C. (2004) Structural characterization of islet amyloid polypeptide fibrils. J. Mol. Biol. 335, 1279–1288 [DOI] [PubMed] [Google Scholar]
- 17. Jayasinghe S. A., Langen R. (2004) Identifying structural features of fibrillar islet amyloid polypeptide using site-directed spin labeling. J. Biol. Chem. 279, 48420–48425 [DOI] [PubMed] [Google Scholar]
- 18. Goldsbury C. S., Cooper G. J., Goldie K. N., Müller S. A., Saafi E. L., Gruijters W. T., Misur M. P., Engel A., Aebi U., Kistler J. (1997) Polymorphic fibrillar assembly of human amylin. J. Struct. Biol. 119, 17–27 [DOI] [PubMed] [Google Scholar]
- 19. Luca S., Yau W. M., Leapman R., Tycko R. (2007) Peptide conformation and supramolecular organization in amylin fibrils: constraints from solid-state NMR. Biochemistry 46, 13505–13522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wiltzius J. J., Sievers S. A., Sawaya M. R., Cascio D., Popov D., Riekel C., Eisenberg D. (2008) Atomic structure of the cross-β-spine of islet amyloid polypeptide (amylin). Protein Sci. 17, 1467–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kajava A. V., Aebi U., Steven A. C. (2005) The parallel superpleated β-structure as a model for amyloid fibrils of human amylin. J. Mol. Biol. 348, 247–252 [DOI] [PubMed] [Google Scholar]
- 22. Jao C. C., Hegde B. G., Chen J., Haworth I. S., Langen R. (2008) Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proc. Natl. Acad. Sci. U.S.A. 105, 19666–19671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chiang Y. W., Borbat P. P., Freed J. H. (2005) The determination of pair distance distributions by pulsed ESR using Tikhonov regularization. J. Magn. Reson. 172, 279–295 [DOI] [PubMed] [Google Scholar]
- 24. Jeschke G., Chechik V., Ionita P., Godt A., Zimmermann H., Banham J., Timmel C. R., Hilger D., Jung H. (2006) DeerAnalysis2006: a comprehensive software package for analyzing pulsed ELDOR data Appl. Magn. Reson. 30, 473–498 [Google Scholar]
- 25. Mchaourab H. S., Lietzow M. A., Hideg K., Hubbell W. L. (1996) Motion of spin-labeled side chains in T4 lysozyme: correlation with protein structure and dynamics. Biochemistry 35, 7692–7704 [DOI] [PubMed] [Google Scholar]
- 26. Hatmal M. M., Li Y., Hegde B. G., Hegde P. B., Jao C. C., Langen R., Haworth I. S. (2012) Computer modeling of nitroxide spin labels on proteins. Biopolymers 97, 35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 29. Antzutkin O. N., Leapman R. D., Balbach J. J., Tycko R. (2002) Supramolecular structural constraints on Alzheimer β-amyloid fibrils from electron microscopy and solid-state nuclear magnetic resonance. Biochemistry 41, 15436–15450 [DOI] [PubMed] [Google Scholar]
- 30. Lührs T., Ritter C., Adrian M., Riek-Loher D., Bohrmann B., Döbeli H., Schubert D., Riek R. (2005) Three-dimensional structure of Alzheimer amyloid-β(1–42) fibrils. Proc. Natl. Acad. Sci. U.S.A. 102, 17342–17347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Petkova A. T., Ishii Y., Balbach J. J., Antzutkin O. N., Leapman R. D., Delaglio F., Tycko R. (2002) A structural model for Alzheimer β-amyloid fibrils based on experimental constraints from solid-state NMR. Proc. Natl. Acad. Sci. U.S.A. 99, 16742–16747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petkova A. T., Yau W. M., Tycko R. (2006) Experimental constraints on quaternary structure in Alzheimer β-amyloid fibrils. Biochemistry 45, 498–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Török M., Milton S., Kayed R., Wu P., McIntire T., Glabe C. G., Langen R. (2002) Structural and dynamic features of Alzheimer Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J. Biol. Chem. 277, 40810–40815 [DOI] [PubMed] [Google Scholar]
- 34. Jao C. C., Hegde B. G., Gallop J. L., Hegde P. B., McMahon H. T., Haworth I. S., Langen R. (2010) Roles of amphipathic helices and the bin/amphiphysin/rvs (BAR) domain of endophilin in membrane curvature generation. J. Biol. Chem. 285, 20164–20170 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.