Background: Protein O-glycosylation is an evolutionarily conserved modification that is initiated by a family of enzymes.
Results: RNA interference to the genes encoding each enzyme of the family identified 4 genes that are essential for viability.
Conclusion: Protein O-glycosylation is required for eukaryotic development and viability.
Significance: Certain members of this enzyme family serve unique and essential functions in specific tissues during eukaryotic development.
Keywords: Drosophila, Glycosylation, Glycosyltransferases, Post-translational Modification, RNA Interference (RNAi), <I>pgant</I>, Mucin-type O-Glycosylation
Abstract
Mucin-type O-glycosylation represents a major form of post-translational modification that is conserved across most eukaryotic species. This type of glycosylation is initiated by a family of enzymes (GalNAc-Ts in mammals and PGANTs in Drosophila) whose members are expressed in distinct spatial and temporal patterns during development. Previous work from our group demonstrated that one member of this family is essential for viability and another member modulates extracellular matrix composition and integrin-mediated cell adhesion during development. To investigate whether other members of this family are essential, we employed RNA interference (RNAi) to each gene in vivo. Using this approach, we identified 4 additional pgant genes that are required for viability. Ubiquitous RNAi to pgant4, pgant5, pgant7, or the putative glycosyltransferase CG30463 resulted in lethality. Tissue-specific RNAi was also used to define the specific organ systems and tissues in which each essential family member is required. Interestingly, each essential pgant had a unique complement of tissues in which it was required. Additionally, certain tissues (mesoderm, digestive system, and tracheal system) required more than one pgant, suggesting unique functions for specific enzymes in these tissues. Expanding upon our RNAi results, we found that conventional mutations in pgant5 resulted in lethality and specific defects in specialized cells of the digestive tract, resulting in loss of proper digestive system acidification. In summary, our results highlight essential roles for O-glycosylation and specific members of the pgant family in many aspects of development and organogenesis.
Introduction
Post-translational modifications provide added levels of complexity and diversity to the encoded genome by influencing many aspects of protein structure, function, and stability. In the post-genomic era, understanding how these protein modifications operate is essential for a comprehensive understanding of all factors involved in various developmental events. Glycosylation, or the addition of sugars to proteins, is an abundant and diverse set of co- and post-translational modifications that imparts a multitude of effects in vivo (reviewed in Refs. 1–8). Various types of glycosylation have been shown to influence protein processing, stability, and transport, as well as modulate protein interactions with other molecules, thereby affecting diverse cellular processes, including signaling events, proteolytic cascades, and the unfolded protein response (UPR).
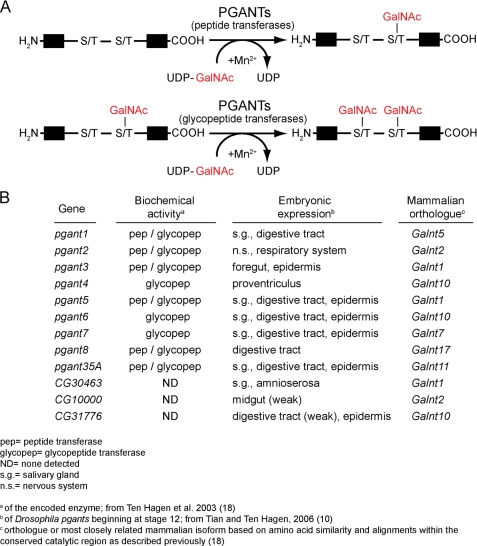
Mucin-type O-linked glycosylation is an evolutionarily conserved form of glycosylation found across diverse species and in most developing organ systems (6, 9–13). This type of glycosylation is initiated by UDP-GalNAc: polypeptide N-acetylgalactosaminyltransferase family of enzymes (PGANTs in Drosophila or GalNAc-Ts in mammals) (Fig. 1) whose members transfer a GalNAc sugar onto serine or threonine residues in protein substrates. The genes encoding members of this enzyme family have unique developmental expression patterns (10, 11) (Fig. 1B), suggesting specific functional roles in certain developing tissues. In vitro biochemical assays have revealed unique peptide preferences for certain family members that are conserved across species (14, 15). Additionally, members of this family form 2 subgroups: 1) those that perform the initial addition of GalNAc to unmodified substrates (peptide transferases) and 2) those that will only add GalNAc to previously glycosylated substrates (glycopeptide transferases) (Fig. 1, A and B) (see Refs. 16–18 and reviewed in Refs. 6 and 12). In summary, the unique expression patterns and substrate specificities of the many members of this family suggest unappreciated biological complexity associated with this type of glycosylation.
FIGURE 1.
Protein O-linked glycosylation. A, the enzymatic addition of N-acetylgalactosamine (GalNAc) to serine or threonine of protein substrates is catalyzed by the PGANT family of enzymes. The PGANT family is divided into 2 subgroups: peptide transferases, which catalyze the addition of GalNAc to unmodified substrates; and glycopeptide transferases, which only add GalNAc to previously glycosylated substrates. B, summarized is the pgant gene family from Drosophila, their embryonic expression patterns, the biochemical activity of the encoded enzymes, and the most closely related mammalian isoforms.
The importance of mucin-type O-linked glycosylation in human health has been demonstrated by the discovery that defects in O-glycosylation are responsible for Tn syndrome (19) and familial tumoral calcinosis, a disease characterized by hyperphosphatemia and the development of subdermal calcified tumors (20, 21). Additionally, genome wide association studies and linkage studies have identified the genes controlling O-glycosylation among those associated with alterations in HDL cholesterol and triglyceride levels (22–24), congenital heart disease (25), and increased risk of colon cancer (26). However, the specific roles of O-glycosylation in these instances remains unknown.
Studies investigating the role of mucin-type O-glycosylation in Drosophila melanogaster provided the first demonstration that one member of this gene family (pgant35A) is essential for viability (27, 28). Indeed, pgant35A was shown to affect the establishment of apical-basal polarity and diffusion barrier formation in the developing respiratory system (29). Another member, pgant3, was found to affect integrin-mediated cell adhesion during Drosophila development by influencing the secretion of an extracellular matrix protein (30–31). In addition, studies in mice demonstrated roles for O-glycosylation and the gene Galnt1 in lymphocyte homing and blood coagulation (32). Thus, the use of genetically tractable model organisms can provide crucial insight into the diversity of cellular and biological functions of this complex post-translational modification.
In vivo RNA interference (RNAi) has been used successfully in Drosophila to decipher the biological role of many genes by specifically targeting the gene of interest and faithfully reproducing phenotypes seen with conventional gene mutations (31, 33–35). Here we demonstrate that 4 additional pgant family members are essential for viability in Drosophila by employing RNAi in vivo. Additionally, we show that individual family members are required in specific organs and tissues by employing tissue-specific RNAi. Finally, we expand upon our RNAi results by examining the effect of conventional mutations in pgant5 on digestive system function. Our results suggest that the PGANT family of enzymes serve diverse functions throughout Drosophila development and provide further insight into the roles of O-glycosylation in mammalian development and disease.
EXPERIMENTAL PROCEDURES
Fly Strains Used
The Gal4 driver stocks used in this study are listed in Table 1 (36). The wild type stocks used were either Oregon-R or w1118 (VDRC 60000). Stocks from the Vienna Drosophila RNAi Center (VDRC)2 (37) that were used to perform RNAi in vivo are listed in Table 2. Additionally, VDRC 60008 (w1118; P{UAS-dicer2w+}) was used to recombine with UAS-pgant35AIR#7 to generate the line UAS-pgant35AIR#7,dicer2D (to enhance levels of pgant35A knock-down). VDRC 60009 (w1118; P{UAS-dicer2w+}) was used to recombine with VDRC 26163 to generate the line VDRC 26163,dicer2 (to enhance knock-down of pgant2). Bloomington stocks 8283 (w1118; CyO, P{w+mc = FRT (w+)Tub-PBac/T}2/wgsp-1), 8674 (w1118; Df(2L)BSC109/CyO), and 1522 (w*; P{UAS-GFP.S65T}T10) were used. The transposon insertion in pgant5 (PBacpgant5c03193/CyO) was from the Drosophila Exelixis Collection (38). The w; Dr/TM3, Sb1, twi-2XGFP stock was the kind gift of Dr. D. Andrew. The following stocks were the kind gifts of Dr. J. Kennison: w; TM6C, cu, Sb, e, ca/Su(Tpl)s1, red, e stock; w; Sco/SM6a stock, and cn1 bw1 sp1 stock.
TABLE 1.
Gal4 driver lines and expression patterns
| Gal4 driver line | Expression pattern | Source | Reference |
|---|---|---|---|
| Tub | Ubiquitous | Bloomington No. 5138 | Flybasea |
| MS1096 | Dorsal wing disc | Bloomington No. 25706 | Flybase |
| 6-4 | Larval hemocytes | Bloomington No. 6396 | Flybase |
| c381 | Amnioserosa, PNS | Bloomington No. 3734 | Flybase |
| 69B | Embryonic epidermis, imaginal discs | Bloomington No. 1774 | Flybase |
| 48Y | Embryonic endoderm | Bloomington No. 4935 | Flybase |
| 24B | Embryonic mesoderm | Bloomington No. 1767 | Flybase |
| c179 | Embryonic mesoderm, larval muscles, wing imaginal discs | Bloomington No. 6450 | Flybase |
| btl | Tracheal cells/respiratory system | D. Andrew | Shiga et al. (36) |
| c135 | Proventriculus, midgut, salivary gland, fat body, reproductive tract | Bloomington No. 6978 | Flybase |
| elav | Nervous system | Bloomington No. 8765 | Flybase |
a Flybase.org.
TABLE 2.
Viability of flies expressing dsRNA to each pgant under the control of the Tub-Gal4 driver
Crosses are described under “Experimental Procedures.”
| Gene knocked down | CG number | IR lines used | Viabilitya | nb |
|---|---|---|---|---|
| pgant2 | CG3254 | VDRC 26163, dicer2 | 99% | 257 |
| pgant3 | CG4445 | UAS-pgant3IR2#2 | 97% | 298 |
| pgant4 | CG31956 | UAS-pgant4IR#8 | 0% | 174 |
| pgant5 | CG31651 | VDRC 2629 | 0% | 226 |
| pgant6 | CG2103 | VDRC 106203 | 41% | 207 |
| pgant7 | CG6394 | VDRC 105160 | 0% | 528 |
| pgant35A | CG7480 | UAS-pgant35AIR#7,dicer2D | 0% | 224 |
| CG30463 | CG30463 | UAS-CG30463IR#5 | 0% | 202 |
| CG10000 | CG10000 | UAS-FlyKIR#2 | 92% | 232 |
| CG31776 | CG31776 | VDRC 7283 | 92% | 296 |
a Relative to siblings not expressing dsRNA.
b n, total number of flies scored.
Construction of Gal4-inducible pgantIR Vectors and Transgenic Lines
The pWIZ plasmid (39) was used to generate Gal4-inducible constructs expressing double-stranded RNA (dsRNA) to each pgant, which were then used to create transgenic flies. To generate each Gal4-inducible pgant construct (UAS-pgantIR), sense and antisense primers (supplemental Table S1) were used to amplify an ∼500 bp fragment from each pgant. The PCR product was then cloned stepwise into the AvrII and NheI sites on either side of the white intron in the vector pWIZ (39) to generate a vector containing two inversely oriented fragments or inverted repeats (IR) from the gene of interest flanking the white intron. Transformants were produced by Genetic Services Inc. (Cambridge, MA) using methodology based on the procedure described previously (40, 41).
Fly Crosses
Drosophila crosses were performed as described previously (30, 31). Briefly, crosses to generate expression of dsRNA were performed using flies from a UAS-pgantIR transgenic line and the Gal4-driver stocks described herein (Table 1). Crosses to generate RNAi to each pgant gene ubiquitously were performed by crossing UAS-pgantIR transgenic lines to a tubulin-Gal4-driver line (P{tubP-GAL4}LL7/TM3, Sb1, twi-2XGFP); viability of flies expressing dsRNA to each pgant was determined by quantitating the number of progeny with and without Sb1. Tissue-specific knockdown of each pgant was performed by crossing each UAS-pgantIR transgenic line to the tissue-specific Gal4 driver lines listed in Table 1 and quantitating the percentage of surviving progeny that express dsRNA to the pgant of interest. Crosses to the wing-specific driver (MS1096-Gal4) were performed using homozygous w1118, P{w+mW.hs = GawB}BxMS1096 females crossed to UAS-pgantIR transgenic males; wing blistering was quantitated in progeny expressing dsRNA to each pgant. All Drosophila crosses were kept on MM media (KD Medical, Inc.) at 25 °C unless specified otherwise.
Quantitative RT-PCR
Quantitative RT-PCR (qPCR) to determine expression levels of all pgant family members when RNAi was induced was performed using the PCR primers listed in supplemental Table S2. cDNA prepared from first instar larvae from Tub-Gal4>pgantIR or the controls Tub-Gal4>VDRC60000 (w1118) or Oregon-R was used in qPCR. Briefly, RNA was isolated using the FastRNA Pro Green kit (Q-BIOgene). cDNA synthesis was performed using the iScript cDNA Synthesis Kit (Bio-Rad). PCR primers were designed using Beacon Designer software (Bio-Rad). qPCR was performed on a CFX96 real time PCR thermocycler (Bio-Rad) using the SYBR Green PCR Master Mix (Bio-Rad). RNA levels were normalized to 18S rRNA.
Western Blotting
Protein extracts were prepared from midguts dissected from third instar wandering larvae. Tissues were dissected in PBS and immediately transferred to lysis buffer (PBS, 1% Triton X-100, 5 mm EDTA) containing protease inhibitors (Thermo Scientific). Tissues were then homogenized on ice. Protein extracts from an equivalent number of wild type, c135>pgant5IR, pgant5c03193/Df (2L) BSC109, or pgant5c03193(excision)/Df(2L)BSC109 midguts were electrophoresed under reducing conditions in 4–12% SDS-PAGE gradient gels and then transferred to nitrocellulose membranes. The membranes were blocked with Odyssey Blocking Buffer (LI-COR) and incubated with IRDye 680LT-conjugated peanut agglutinin (PNA) (1:5000) and rabbit α-tubulin antibody (Cell Signaling, 1:1000). IRDye 800CW-conjugated goat anti-rabbit secondary antibody was used (1:10,000). Membranes were scanned using a LI-COR Odyssey Infrared Imaging System.
Tissue Fixation, Staining, and Immunofluorescence
Midguts from third instar wandering larvae were dissected in PBS and immediately transferred to 4% paraformaldehyde in PBS. Midguts were fixed for 20 min at 4 °C. Following fixation, samples were washed three times in PBST (PBS, 0.1% Triton X-100) and transferred to blocking buffer (2% goat serum/PBS, 0.1% Triton X-100) for 1 h on a rocker at room temperature. To visualize actin staining in copper cells, samples were incubated with either TRITC-conjugated phalloidin (Sigma, 1:100) or Alexa 488-conjugated phalloidin (Invitrogen, 1:100) for 3 h at room temperature. Samples were incubated with fluorescein- or rhodamine-labeled PNA (Vector Laboratories, 1:100) for 3 h at room temperature to visualize core 1 O-glycans (Galβ1–3GalNAcα1-S/T). Samples were imaged using a Zeiss Axiovert microscope.
RESULTS
Additional pgant Family Members Are Essential for Viability
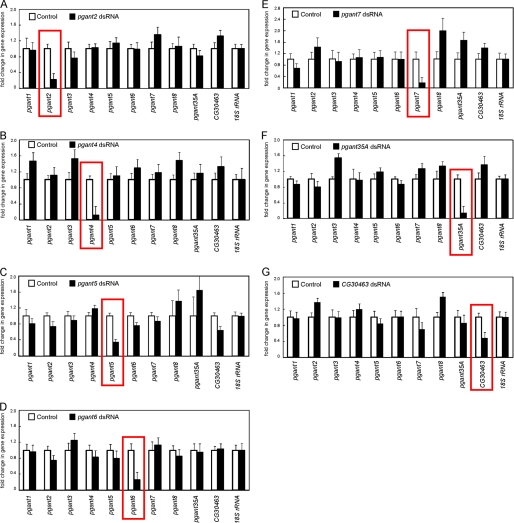
The Drosophila family of pgants consists of 9 genes encoding enzymes that have been biochemically confirmed to function as glycosyltransferases, along with 3 putative family members based on sequence conservation (10, 18) (Fig. 1B). Previous studies have identified unique developmental expression patterns for each family member (10) (summarized in Fig. 1B). To comprehensively investigate the in vivo role of each remaining member of this multigene family, we performed RNAi in vivo by crossing transgenic lines containing Gal4-inducible IR for each gene to a line expressing the Gal4 transcriptional activator under the control of the ubiquitously expressed tubulin promoter (Tub-Gal4). Progeny from these crosses will express dsRNA to the gene of interest in all cells and tissues throughout development, thus inducing RNAi ubiquitously. RNA from progeny expressing dsRNA was collected and qPCR was performed to verify that the expression of the targeted gene was specifically decreased. As shown in Fig. 2, induction of dsRNA for pgant2, pgant4, pgant5, pgant6, pgant7, pgant35A, or the putative pgant, CG30463, resulted in a specific and significant decrease in expression of the targeted gene. Specific knockdown of pgant3 expression via RNAi was also seen and has been reported previously (31). Expression levels of CG10000 and CG31776 (2 putative members of the pgant family) were below levels of detection in all wild type tissues and stages examined, thus we were unable to quantitate specific knockdown of these putative glycosyltransferase genes. Finally, we were unable to see a specific decrease in gene expression for pgant1 or pgant8 with multiple different RNAi lines, so these genes were not pursued further in this study.
FIGURE 2.
Quantitative PCR reveals specific decreases in gene expression of each pgant in progeny expressing dsRNA. Progeny expressing dsRNA to (A) pgant2, (B) pgant4, (C) pgant5, (D) pgant6, (E) pgant7, (F) pgant35A, and (G) CG30463 are shown. Total RNA was isolated and qPCR was performed for all pgant family members to verify specific knockdown of gene expression (red boxes). RNA levels were normalized to 18 S rRNA. Standard deviations are shown.
Progeny from each cross was then scored to determine the effects on viability (Table 2). Interestingly, RNAi to pgant4, pgant5, pgant7, or CG30463 using the Tub-Gal4 driver resulted in complete loss of viability, indicating an essential role for these genes in Drosophila (Table 2). As expected, RNAi to pgant35A (an essential gene in Drosophila (27–29)) also resulted in lethality. Additionally, whereas RNAi to pgant6 did not cause complete lethality, it did result in reduced viability. No effect on viability was seen after RNAi to pgant2, pgant3, CG10000, or CG31776.
pgant Genes Are Required in Specific Tissues during Development
To investigate the specific role of the newly identified essential pgants, we performed tissue-specific RNAi to each gene using the Gal4 driver lines listed in Table 1. Lines expressing Gal4 specifically in the endoderm, mesoderm, ectoderm, respiratory system, nervous system, digestive system, salivary gland, amnioserosa, and hemocytes were used. Interestingly, tissue-specific requirements were seen for each essential pgant (Table 3). RNAi to pgant4 or pgant5 in the digestive system resulted in almost complete lethality, indicating an essential role for each in the developing gut. pgant5 was also required in the embryonic mesoderm and the respiratory system. RNAi to pgant35A in the respiratory system resulted in a dramatic reduction in viability, supporting previous studies indicating a role for this transferase in tracheal system development (29). Interestingly, pgant35A was also found to be required in the mesoderm. Tissue-specific RNAi to pgant7 revealed a role for it in respiratory system development. And finally, knockdown of the putative family member, CG30463, in the mesoderm, the respiratory system, or the amnioserosa resulted in lethality. RNAi to CG30463 in the digestive system also resulted in a reduction in viability. These in vivo results further suggest that this conceptual gene may encode a functional glycosyltransferase that is performing unique roles in specific developing tissues. Taken together, our results indicate that multiple members of the pgant family are essential for viability and are performing functions in specific developing tissues. Additionally, certain tissues (the respiratory system, digestive system, and mesoderm) require more than one pgant.
TABLE 3.
Viability (relative to siblings not expressing dsRNA) of flies expressing dsRNA to essential pgants in specific tissues
| Gal4 driver line (expression pattern) | IR lines used |
||||
|---|---|---|---|---|---|
| pgant4IR | pgant5IR | pgant35AIR | pgant7IR | CG30463IR | |
| 6-4 (hemocytes) | 100% (n = 271)a | 100% (n = 331) | 98% (n = 269) | 100% (n = 485) | 95% (n = 393) |
| c381 (amnioserosa, PNS) | 82% (n = 317) | 100% (n = 291) | 67% (n = 369) | 100% (n = 675) | 11% (n = 211) |
| 69B (epiderm/ectoderm) | 100% (n = 227) | 51% (n = 292) | 45% (n = 635) | 97% (n = 834) | 36% (n = 336) |
| 48Y (endoderm) | 100% (n = 327) | 100% (n = 280) | 69% (n = 499) | 76% (n = 576) | 62% (n = 281) |
| 24B (mesoderm) | 100% (n = 271) | 81% (n = 336) | 0% (n = 275) | 69% (n = 429) | 0% (n = 190) |
| c179 (mesoderm) | 100% (n = 308) | 0% (n = 280) | 19% (n = 298) | 100% (n = 825) | 0% (n = 203) |
| btl (respiratory system) | 100% (n = 309) | 17% (n = 327) | 12% (n = 254) | 12% (n = 444) | 0% (n = 159) |
| c135 (digestive system, reproductive tract) | 2% (n = 402) | 0% (n = 177) | 25% (n = 208) | 100% (n = 721) | 23% (n = 178) |
| elav (nervous system) | 100% (n = 295) | 100% (n = 289) | 86% (n = 290) | 85% (n = 636) | 97% (n = 297) |
a n, total number of flies scored.
pgant5 Is Required for Proper Gut Acidification
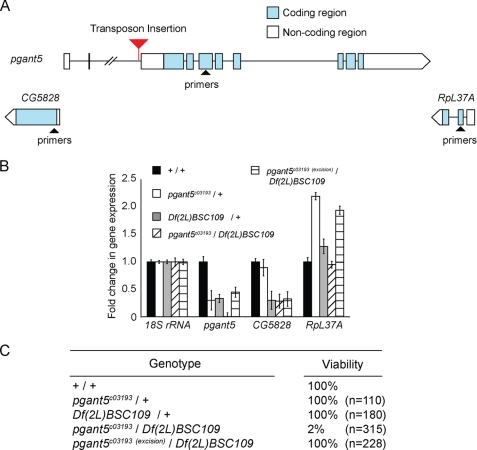
To follow up on the RNAi results presented above, we examined conventional mutations in pgant5. For these studies we utilized a piggyBac transposon insertion line from the Exelixis Drosophila stock collection that contains a transposon in the second intron of the pgant5 gene (PBacpgant5c03193, hereafter designated as pgant5c03193) (Fig. 3A). pgant5c03193 flies were crossed to a deficiency line that uncovers the pgant5 gene (Df(2L)BSC109) to assess the effect of the transposon on viability and pgant5 gene expression. Indeed, pgant5c03193/Df(2L)BSC109 progeny were not viable when compared with either the heterozygous transposon insertion (pgant5c03193/+) or heterozygous deficiency (Df(2L)BSC109/+) (Fig. 3C). qPCR confirmed that the transposon in pgant5 abrogated pgant5 expression, but did not decrease the expression of the flanking genes (CG5828 and RpL37A) (Fig. 3B). To confirm that the transposon insertion lethality is specifically due to its effect on pgant5 gene expression, we precisely excised the transposon from the pgant5 gene using the piggyBac transposase line (Bloomington number 8283), as described previously (38). Excision of the transposon (pgant5c03193(excision)) resulted in restoration of pgant5 expression and viability (Fig. 3, B and C). These results confirm that pgant5 is required for viability and provide an independent validation of our in vivo RNAi results.
FIGURE 3.
Transposon insertion mutation in pgant5 causes lethality and confirms pgant5 is an essential gene. A, the position of the transposon insertion in intron 2 of pgant5 is shown. Exons are represented as boxes and introns are represented as lines. Blue boxes are coding regions and white boxes are noncoding regions of pgant5. Flanking genes are also shown. Regions used to generate PCR primers are represented as triangles. B, real-time PCR analysis of pgant5 transcript levels using the primer pairs shown in A reveals a significant decrease in pgant5 gene expression in pgant5c03193/Df(2L)BSC109 relative to pgant5c03193/+ heterozygotes, Df(2L)BSC109/+ heterozygotes, pgant5c03193(excision)/Df(2L)BSC109, or wild type. RNA was normalized to 18S rRNA. C, loss of pgant5 results in lethality.
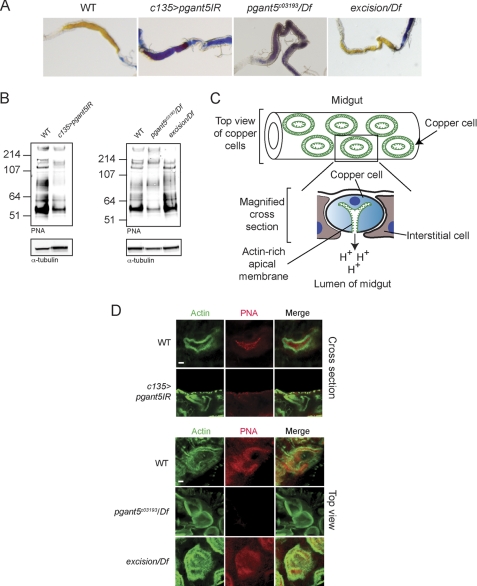
Building upon our tissue-specific RNAi results that indicated a role for pgant5 in the digestive system, we next examined the larval digestive tracts of pgant5 mutants. Interestingly, both conventional pgant5 mutations (pgant5c03193/Df(2L)BSC109) as well as RNAi to pgant5 (c135>pgant5IR) resulted in midguts with an unusually basic pH, suggesting a role for pgant5 in the proper digestive system acidification (Fig. 4A). Western blots of midguts probed with the lectin PNA, which detects core 1 O-glycan structures (Galβ1–3GalNAcα1-S/T), revealed the loss or reduction in intensity of many bands in pgant5 mutants (Fig. 4B), indicating that pgant5 is responsible for the glycosylation of numerous proteins within the larval midgut.
FIGURE 4.
Loss of pgant5 results in loss of gut acidification and disruption of O-glycosylation in copper cells of the digestive tract. A, larval gut acidification as detected with bromophenol blue shows acidified midguts (yellow) in wild type and pgant5 transposon excision (excision/Df), but basic midguts (blue) in pgant5 mutants (pgant5c03193/Df) or larvae expressing dsRNA to pgant5 in the digestive system (c135>pgant5IR). B, Western blots of proteins from wild type, c135>pgant5IR, pgant5c03193/Df, and excision/Df third instar larval midguts probed with the lectin PNA to detect O-glycosylated proteins or with α-tubulin as a loading control. C, diagram of midgut and copper cell morphology. Shown is a diagram of a section of the midgut where copper cells stain for actin (green) in concentric circles. Below is a magnified cross-section of a copper cell, highlighting the unique invaginated actin-rich apical surface (green). D, copper cells from wild type, c135>pgant5IR, pgant5c03193/Df, and excision/Df third instar larval midguts were stained with PNA (red) and phalloidin/actin (green). In wild type copper cells, phalloidin staining labels actin-based microvilli unique to the apical region of copper cells. PNA-reactive O-glycoproteins are found along the apical and luminal regions of wild type copper cells (shown in upper panel cross-sections). All apical and luminal PNA staining is lost from c135>pgant5IR (upper panel) and pgant5c03193/Df copper cells (lower panel). Additionally, apical actin staining is irregular in size and shape in pgant5c03193/Df copper cells. PNA staining and gut acidification are restored upon excision of the transposon (excision/Df). Scale bars = 4 μm.
We next examined the effects of pgant5 on copper cell O-glycosylation and morphology. Copper cells are specialized cells of the Drosophila midgut that regulate gut acidification (42, 43). These cells have a unique morphology, with an invaginated apical surface rich in actin-based microvilli (Fig. 4C) (42, 43). Copper cells from wild type and pgant5 mutants were stained with fluorescently labeled PNA to detect O-glycoproteins, and with phalloidin to detect the actin-rich apical surface (Fig. 4D). Interestingly, wild type copper cells displayed abundant O-glycoproteins throughout the apical and luminal regions of copper cells (Fig. 4D). However, loss of pgant5 (c135>pgant5IR or pgant5c03193/Df(2L)BSC109) was accompanied by a dramatic loss of O-glycosylation normally found on copper cells (Fig. 4D). Although phalloidin staining of the actin-rich apical surface was normal in c135>pgant5IR copper cells (Fig. 4D), it was irregular in shape and size in pgant5c03193/Df(2L)BSC109 copper cells (Fig. 4D), indicating a more severe apical/cytoskeletal phenotype for the conventional pgant5 mutants. Copper cell O-glycosylation, gut acidification, and actin staining were restored upon excision of the transposon from pgant5 (pgant5c03193(excision)/Df(2L)BSC109) (Fig. 4, A, B, and D). These results indicate that PGANT5 is responsible for glycosylating apical and luminal proteins within copper cells and that loss of PGANT5 results in loss of proper gut acidification. These results highlight a previously unknown role for pgant5 in specialized cells of the digestive tract responsible for gut acidification.
pgant3 Is Uniquely Responsible for Wing Adhesion
We further employed this RNAi system to address unique roles for individual pgant family members in a specific developmental process. Previous work from our laboratory demonstrated that loss of pgant3 results in defects in the secretion of an extracellular matrix protein, with effects on integrin-mediated cell adhesion, resulting in wing blistering. To investigate whether other pgant family members are also involved in integrin-mediated cell adhesion in the developing wing, we performed RNAi to each in the developing wing using the wing-specific Gal4 driver, MS1096. Although RNAi to pgant3 resulted in wing blistering, no blistering was seen upon induction of RNAi to the remaining family members (Table 4). These results suggest a potentially unique role for pgant3 in specific cell adhesion processes occurring in the developing wing and are supported by previous results indicating that the expression of another family member cannot compensate for the loss of pgant3 in vivo (31).
TABLE 4.
Wing blistering in flies expressing dsRNA to pgant genes in the developing wing
| Gene knocked down | Blistereda | nb |
|---|---|---|
| % | ||
| pgant2 | 0 | 253 |
| pgant3 | 18 | 200 |
| pgant4 | 0 | 114 |
| pgant5 | 0 | 120 |
| pgant6 | 0 | 178 |
| pgant7 | 0 | 160 |
| pgant35A | 0 | 172 |
| CG30463 | 0 | 178 |
| CG10000 | 0 | 235 |
| CG31776 | 0 | 210 |
a Represents the (number of flies displaying blistered wings/total number of flies of denoted genotype) × 100.
b n, total number of flies of denoted genotype scored.
DISCUSSION
Here, we identify 4 additional members of this multigene glycosyltransferase family that are essential for viability by taking advantage of the highly efficient and specific in vivo RNAi system in Drosophila. This work serves to highlight the biological importance of O-linked glycosylation and of specific family members responsible for initiating this conserved post-translational modification. Additionally, these studies indicate that at least 5 family members in the fly are serving unique and nonredundant functions during development. Thus, whereas these members all catalyze the transfer of GalNAc to serine or threonine, they are doing so on specific substrates and/or at specific positions within these substrates. Previous enzymatic studies demonstrated that many mammalian GalNAc-Ts and Drosophila PGANTs have unique substrate preferences and sites of GalNAc addition in vitro, suggesting that certain substrates can only be glycosylated by certain enzymes (17, 18, 44, 45) (reviewed in Refs. 6, 12, and 15). Our in vivo results support this hypothesis and suggest a highly complex system for proper O-glycosylation of proteins.
The in vivo RNAi system also offers the unique advantage of allowing the knockdown of genes in specific tissues to determine exactly where a particular gene is required. We used Gal4 driver lines that induced RNAi in most major tissue types and developing organ systems to determine where each essential pgant was required. In agreement with previous studies demonstrating that conventional pgant35A mutants affect respiratory system development, we found that RNAi to pgant35A in the tracheal system resulted in significantly reduced viability. Interestingly, each essential gene was found to be required in a specific subset of tissues and organ systems, suggesting unique functional roles for each. Developing organs known for the abundant production of O-glycoproteins (digestive and respiratory systems) (11) were most significantly affected by the knockdown of pgant family members. Knockdown of pgant4, pgant5, pgant35A, or the putative transferase CG30463 in the digestive system resulted in significant losses of viability. In addition to pgant35A, knockdown of pgant5, pgant7, or CG30463 in the respiratory system also affected viability. CG30463 was also found to be required in the amnioserosa, an embryonic tissue involved in developmentally regulated cell migration events. Finally, 3 of the 5 essential pgants were required in the mesoderm. The results presented here will form the basis for future studies investigating the mechanistic role of these essential pgants in diverse tissues and developmental stages with the hope of gaining insight into the role of their mammalian orthologues.
To begin to address the specific role of one of the novel essential genes identified in this screen, we further characterized the phenotypes associated with the loss of pgant5. Interestingly, loss of pgant5 resulted in the disruption of the proper structure and function of specialized cells of the digestive tract responsible for gut acidification. Our studies revealed that the loss of pgant5 caused loss of O-glycoproteins along the apical and luminal surfaces of copper cells of the midgut, along with an irregular apical actin-based microvillar structure in pgant5c03193/Df(2L)BSC109. Restoration of pgant5 expression resulted in restoration of O-glycosylation, actin-based microvilli, and proper gut acidification. Western blots revealed that the loss of pgant5 affected the glycosylation of many proteins, making it difficult to discern what the primary target(s) might be. It remains possible that PGANT5 could be glycosylating subunits of the ion transporters responsible for gut acidification. Alternatively, PGANT5 could be glycosylating other proteins involved in the localization of ion transporters or the establishment of proper apical polarity within the copper cells. In support of this, previous work on PGANT35A in the respiratory tract demonstrated that it also glycosylates apical and luminal proteins and affects apical-basal polarity when mutated (29). As O-glycoproteins are abundant along the apical regions of many developing tissues (11), it is possible that they are performing functions related to maintenance of the unique characteristics of apical surfaces.
Previous work on the mammalian orthologue of pgant5 (Galnt1) did not examine the digestive system but did find lymphocyte homing defects (due to decreased presence of selectin ligands on lymphocytes and endothelial cells) and bleeding disorders (due to decreased plasma levels of blood coagulation factors) in Galnt1−/− mice (32). However, the specific substrates involved and the mechanism(s) by which these phenotypes occur remain unknown (32). Nonetheless, the results presented here highlight a previously unknown role for pgant5 in digestive tract function and provide a basis for examining the role of Galnt1 in mammalian digestive system function. There is precedent for mucins and mucin-type O-glycosylation having functional roles in the digestive system as the loss of a major O-glycosylated mucin (Muc2) or the disruption of certain O-glycan structures results in digestive tract abnormalities, including increased rates of infection, decreased barrier function, and increased susceptibility to colitis and colon cancer (26, 46–48). Future studies on PGANT5 in the Drosophila gut will identify direct in vivo targets of this enzyme and define how O-glycosylation regulates their function.
As mentioned above, the putative glycosyltransferase CG30463 was found to be essential for viability and required in multiple developing tissues. Based on sequence conservation, CG30463 is assumed to be a member of the pgant family. However, in vitro biochemical assays have failed to detect glycosyltransferase activity for the purified recombinant CG30463 protein (data not shown), thus its biological relevance has been unclear. The data shown here, indicating that knockdown of CG30463 in specific tissues results in lethality, demonstrates a crucial in vivo role for this gene. It is possible that the CG30463 protein has very specific substrate preferences and thus its in vitro enzymatic activity may not have been detectable with the panel of substrate peptides used. Future experiments will use this RNAi system to investigate in vivo changes in O-glycosylation upon CG30463 knockdown.
Finally, we used this in vivo RNAi system to address the role of each family member in a specific developmental process, integrin-mediated cell adhesion. Here we demonstrate that pgant3 has a unique role in integrin-mediated wing blade adhesion, as RNAi to other family members had no effect on wing blistering. Our results, demonstrating a unique function for pgant3 in the wing, are also supported by genetic data indicating that expression of another family member in the wing could not compensate for loss of pgant3 (31). These studies indicate that PGANT3 may be uniquely responsible for glycosylating the extracellular matrix protein Tiggrin in the wing disc, further supporting the notion of unique substrate preferences in vivo. The role for other pgants in specific cell adhesion events in other tissues will be investigated using this methodology.
In summary, we have identified 4 additional members of this glycosyltransferase family that are essential for viability in Drosophila, highlighting the importance of this protein modification and unique requirements for individual family members. Future work will define the specific functions and substrates of each member in Drosophila to gain insight into the roles of O-glycosylation in development and disease.
Supplementary Material
Acknowledgments
We thank our colleagues for many helpful discussions. We thank Drs. J. Kennison and D. Andrew for the kind gift of fly stocks. We also thank the Bloomington Stock Center, the Developmental Studies Hybridoma Bank, and the Vienna Drosophila RNAi Center for providing fly stocks, antibodies, and other reagents.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program of the NIDCR.

This article contains supplemental Tables S1 and S2.
- VDRC
- Vienna Drosophila RNAi Center
- GalNAc-T or PGANT or pgant
- UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase
- qPCR
- quantitative PCR
- dsRNA
- double-stranded RNA
- IR
- inverted repeat
- PNA
- peanut agglutinin
- TRITC
- tetramethylrhodamine isothiocyanate.
REFERENCES
- 1. Hart G. W., Copeland R. J. (2010) Glycomics hits the big time. Cell 143, 672–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Liu L., Xu Y. X., Hirschberg C. B. (2010) The role of nucleotide sugar transporters in development of eukaryotes. Semin. Cell Dev. Biol. 21, 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Love D. C., Krause M. W., Hanover J. A. (2010) O-GlcNAc cycling. Emerging roles in development and epigenetics. Semin. Cell Dev. Biol. 21, 646–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nakamura N., Lyalin D., Panin V. M. (2010) Protein O-mannosylation in animal development and physiology. From human disorders to Drosophila phenotypes. Semin. Cell Dev. Biol. 21, 622–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Schachter H. (2010) Mgat1-dependent N-glycans are essential for the normal development of both vertebrate and invertebrate metazoans. Semin. Cell Dev. Biol. 21, 609–615 [DOI] [PubMed] [Google Scholar]
- 6. Tabak L. A. (2010) The role of mucin-type O-glycans in eukaryotic development. Semin. Cell Dev. Biol. 21, 616–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takeuchi H., Haltiwanger R. S. (2010) Role of glycosylation of Notch in development. Semin. Cell Dev. Biol. 21, 638–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kramer K. L. (2010) Specific sides to multifaceted glycosaminoglycans are observed in embryonic development. Semin. Cell Dev. Biol. 21, 631–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kingsley P. D., Hagen K. G., Maltby K. M., Zara J., Tabak L. A. (2000) Diverse spatial expression patterns of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family member mRNAs during mouse development. Glycobiology 10, 1317–1323 [DOI] [PubMed] [Google Scholar]
- 10. Tian E., Ten Hagen K. G. (2006) Expression of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family is spatially and temporally regulated during Drosophila development. Glycobiology 16, 83–95 [DOI] [PubMed] [Google Scholar]
- 11. Tian E., Ten Hagen K. G. (2007) O-Linked glycan expression during Drosophila development. Glycobiology 17, 820–827 [DOI] [PubMed] [Google Scholar]
- 12. Tian E., Ten Hagen K. G. (2009) Recent insights into the biological roles of mucin-type O-glycosylation. Glycoconj. J. 26, 325–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Young W. W., Jr., Holcomb D. R., Ten Hagen K. G., Tabak L. A. (2003) Expression of UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase isoforms in murine tissues determined by real-time PCR. A new view of a large family. Glycobiology 13, 549–557 [DOI] [PubMed] [Google Scholar]
- 14. Gerken T. A., Ten Hagen K. G., Jamison O. (2008) Conservation of peptide acceptor preferences between Drosophila and mammalian polypeptide-GalNAc-transferase ortholog pairs. Glycobiology 18, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ten Hagen K. G., Fritz T. A., Tabak L. A. (2003) All in the family. The UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferases. Glycobiology 13, 1R–16R [DOI] [PubMed] [Google Scholar]
- 16. Perrine C. L., Ganguli A., Wu P., Bertozzi C. R., Fritz T. A., Raman J., Tabak L. A., Gerken T. A. (2009) Glycopeptide-preferring polypeptide GalNAc-transferase 10 (ppGalNAc T10), involved in mucin-type O-glycosylation, has a unique GalNAc-O-Ser/Thr-binding site in its catalytic domain not found in ppGalNAc T1 or T2. J. Biol. Chem. 284, 20387–20397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ten Hagen K. G., Tetaert D., Hagen F. K., Richet C., Beres T. M., Gagnon J., Balys M. M., VanWuyckhuyse B., Bedi G. S., Degand P., Tabak L. A. (1999) Characterization of a UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase that displays glycopeptide N-acetylgalactosaminyltransferase activity. J. Biol. Chem. 274, 27867–27874 [DOI] [PubMed] [Google Scholar]
- 18. Ten Hagen K. G., Tran D. T., Gerken T. A., Stein D. S., Zhang Z. (2003) Functional characterization and expression analysis of members of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family from Drosophila melanogaster. J. Biol. Chem. 278, 35039–35048 [DOI] [PubMed] [Google Scholar]
- 19. Ju T., Cummings R. D. (2005) Protein glycosylation. Chaperone mutation in Tn syndrome. Nature 437, 1252. [DOI] [PubMed] [Google Scholar]
- 20. Ichikawa S., Guigonis V., Imel E. A., Courouble M., Heissat S., Henley J. D., Sorenson A. H., Petit B., Lienhardt A., Econs M. J. (2007) Novel GALNT3 mutations causing hyperostosis-hyperphosphatemia syndrome result in low intact fibroblast growth factor 23 concentrations. J. Clin. Endocrinol. Metab. 92, 1943–1947 [DOI] [PubMed] [Google Scholar]
- 21. Topaz O., Shurman D. L., Bergman R., Indelman M., Ratajczak P., Mizrachi M., Khamaysi Z., Behar D., Petronius D., Friedman V., Zelikovic I., Raimer S., Metzker A., Richard G., Sprecher E. (2004) Mutations in GALNT3, encoding a protein involved in O-linked glycosylation, cause familial tumoral calcinosis. Nat. Genet. 36, 579–581 [DOI] [PubMed] [Google Scholar]
- 22. Kathiresan S., Manning A. K., Demissie S., D'Agostino R. B., Surti A., Guiducci C., Gianniny L., Burtt N. P., Melander O., Orho-Melander M., Arnett D. K., Peloso G. M., Ordovas J. M., Cupples L. A. (2007) A genome-wide association study for blood lipid phenotypes in the Framingham Heart Study. BMC Med. Genet. 8, Suppl. 1, S17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Teslovich T. M., Musunuru K., Smith A. V., Edmondson A. C., Stylianou I. M., Koseki M., Pirruccello J. P., Ripatti S., Chasman D. I., Willer C. J., Johansen C. T., Fouchier S. W., Isaacs A., Peloso G. M., Barbalic M., Ricketts S. L., Bis J. C., Aulchenko Y. S., Thorleifsson G., Feitosa M. F., Chambers J., Orho-Melander M., Melander O., Johnson T., Li X., Guo X., Li M., Shin Cho Y., Jin Go M., Jin Kim Y., Lee J. Y., Park T., Kim K., Sim X., Twee-Hee Ong R., Croteau-Chonka D. C., Lange L. A., Smith J. D., Song K., Hua Zhao J., Yuan X., Luan J., Lamina C., Ziegler A., Zhang W., Zee R. Y., Wright A. F., Witteman J. C., Wilson J. F., Willemsen G., Wichmann H. E., Whitfield J. B., Waterworth D. M., Wareham N. J., Waeber G., Vollenweider P., Voight B. F., Vitart V., Uitterlinden A. G., Uda M., Tuomilehto J., Thompson J. R., Tanaka T., Surakka I., Stringham H. M., Spector T. D., Soranzo N., Smit J. H., Sinisalo J., Silander K., Sijbrands E. J., Scuteri A., Scott J., Schlessinger D., Sanna S., Salomaa V., Saharinen J., Sabatti C., Ruokonen A., Rudan I., Rose L. M., Roberts R., Rieder M., Psaty B. M., Pramstaller P. P., Pichler I., Perola M., Penninx B. W., Pedersen N. L., Pattaro C., Parker A. N., Pare G., Oostra B. A., O'Donnell C. J., Nieminen M. S., Nickerson D. A., Montgomery G. W., Meitinger T., McPherson R., McCarthy M. I., McArdle W., Masson D., Martin N. G., Marroni F., Mangino M., Magnusson P. K., Lucas G., Luben R., Loos R. J., Lokki M. L., Lettre G., Langenberg C., Launer L. J., Lakatta E. G., Laaksonen R., Kyvik K. O., Kronenberg F., Konig I. R., Khaw K. T., Kaprio J., Kaplan L. M., Johansson A., Jarvelin M. R., Janssens A. C., Ingelsson E., Igl W., Kees Hovingh G., Hottenga J. J., Hofman A., Hicks A. A., Hengstenberg C., Heid I. M., Hayward C., Havulinna A. S., Hastie N. D., Harris T. B., Haritunians T., Hall A. S., Gyllensten U., Guiducci C., Groop L. C., Gonzalez E., Gieger C., Freimer N. B., Ferrucci L., Erdmann J., Elliott P., Ejebe K. G., Doring A., Dominiczak A. F., Demissie S., Deloukas P., de Geus E. J., de Faire U., Crawford G., Collins F. S., Chen Y. D., Caulfield M. J., Campbell H., Burtt N. P., Bonnycastle L. L., Boomsma D. I., Boekholdt S. M., Bergman R. N., Barroso I., Bandinelli S., Ballantyne C. M., Assimes T. L., Quertermous T., Altshuler D., Seielstad M., Wong T. Y., Tai E. S., Feranil A. B., Kuzawa C. W., Adair L. S., Taylor H. A., Jr., Borecki I. B., Gabriel S. B., Wilson J. G., Holm H., Thorsteinsdottir U., Gudnason V., Krauss R. M., Mohlke K. L., Ordovas J. M., Munroe P. B., Kooner J. S., Tall A. R., Hegele R. A., Kastelein J. J., Schadt E. E., Rotter J. I., Boerwinkle E., Strachan D. P., Mooser V., Stefansson K., Reilly M. P., Samani N. J., Schunkert H., Cupples L. A., Sandhu M. S., Ridker P. M., Rader D. J., van Duijn C. M., Peltonen L., Abecasis G. R., Boehnke M., Kathiresan S. (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466, 707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., Strait J., Duren W. L., Maschio A., Busonero F., Mulas A., Albai G., Swift A. J., Morken M. A., Narisu N., Bennett D., Parish S., Shen H., Galan P., Meneton P., Hercberg S., Zelenika D., Chen W. M., Li Y., Scott L. J., Scheet P. A., Sundvall J., Watanabe R. M., Nagaraja R., Ebrahim S., Lawlor D. A., Ben-Shlomo Y., Davey-Smith G., Shuldiner A. R., Collins R., Bergman R. N., Uda M., Tuomilehto J., Cao A., Collins F. S., Lakatta E., Lathrop G. M., Boehnke M., Schlessinger D., Mohlke K. L., Abecasis G. R. (2008) Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40, 161–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fakhro K. A., Choi M., Ware S. M., Belmont J. W., Towbin J. A., Lifton R. P., Khokha M. K., Brueckner M. (2011) Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proc. Natl. Acad. Sci. U.S.A. 108, 2915–2920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guda K., Moinova H., He J., Jamison O., Ravi L., Natale L., Lutterbaugh J., Lawrence E., Lewis S., Willson J. K., Lowe J. B., Wiesner G. L., Parmigiani G., Barnholtz-Sloan J., Dawson D. W., Velculescu V. E., Kinzler K. W., Papadopoulos N., Vogelstein B., Willis J., Gerken T. A., Markowitz S. D. (2009) Inactivating germ-line and somatic mutations in polypeptide N-acetylgalactosaminyltransferase 12 in human colon cancers. Proc. Natl. Acad. Sci. U.S.A. 106, 12921–12925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Schwientek T., Bennett E. P., Flores C., Thacker J., Hollmann M., Reis C. A., Behrens J., Mandel U., Keck B., Schäfer M. A., Haselmann K., Zubarev R., Roepstorff P., Burchell J. M., Taylor-Papadimitriou J., Hollingsworth M. A., Clausen H. (2002) Functional conservation of subfamilies of putative UDP-N-acetylgalactosamine:polypeptide N-acetylgalactosaminyltransferases in Drosophila, Caenorhabditis elegans, and mammals. One subfamily composed of l(2)35Aa is essential in Drosophila. J. Biol. Chem. 277, 22623–22638 [DOI] [PubMed] [Google Scholar]
- 28. Ten Hagen K. G., Tran D. T. (2002) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is essential for viability in Drosophila melanogaster. J. Biol. Chem. 277, 22616–22622 [DOI] [PubMed] [Google Scholar]
- 29. Tian E., Ten Hagen K. G. (2007) A UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase is required for epithelial tube formation. J. Biol. Chem. 282, 606–614 [DOI] [PubMed] [Google Scholar]
- 30. Zhang L., Tran D. T., Ten Hagen K. G. (2010) An O-glycosyltransferase promotes cell adhesion during development by influencing secretion of an extracellular matrix integrin ligand. J. Biol. Chem. 285, 19491–19501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang L., Zhang Y., Hagen K. G. (2008) A mucin-type O-glycosyltransferase modulates cell adhesion during Drosophila development. J. Biol. Chem. 283, 34076–34086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tenno M., Ohtsubo K., Hagen F. K., Ditto D., Zarbock A., Schaerli P., von Andrian U. H., Ley K., Le D., Tabak L. A., Marth J. D. (2007) Initiation of protein O-glycosylation by the polypeptide GalNAcT-1 in vascular biology and humoral immunity. Mol. Cell. Biol. 27, 8783–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pastor-Pareja J. C., Xu T. (2011) Shaping cells and organs in Drosophila by opposing roles of fat body-secreted collagen IV and perlecan. Dev. Cell 21, 245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neumüller R. A., Richter C., Fischer A., Novatchkova M., Neumüller K. G., Knoblich J. A. (2011) Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell 8, 580–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zacharogianni M., Kondylis V., Tang Y., Farhan H., Xanthakis D., Fuchs F., Boutros M., Rabouille C. (2011) EMBO J. 30, 3684–3700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shiga Y., Tanaka-Matakatsu M., Hayashi S. (1996) A nuclear GFP/β-galactosidase fusion protein as a marker for morphogenesis in living Drosophila. Dev. Growth Differ. 38, 99–106 [Google Scholar]
- 37. Dietzl G., Chen D., Schnorrer F., Su K. C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S., Couto A., Marra V., Keleman K., Dickson B. J. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature 448, 151–156 [DOI] [PubMed] [Google Scholar]
- 38. Thibault S. T., Singer M. A., Miyazaki W. Y., Milash B., Dompe N. A., Singh C. M., Buchholz R., Demsky M., Fawcett R., Francis-Lang H. L., Ryner L., Cheung L. M., Chong A., Erickson C., Fisher W. W., Greer K., Hartouni S. R., Howie E., Jakkula L., Joo D., Killpack K., Laufer A., Mazzotta J., Smith R. D., Stevens L. M., Stuber C., Tan L. R., Ventura R., Woo A., Zakrajsek I., Zhao L., Chen F., Swimmer C., Kopczynski C., Duyk G., Winberg M. L., Margolis J. (2004) A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat. Genet. 36, 283–287 [DOI] [PubMed] [Google Scholar]
- 39. Lee Y. S., Carthew R. W. (2003) Making a better RNAi vector for Drosophila. Use of intron spacers. Methods 30, 322–329 [DOI] [PubMed] [Google Scholar]
- 40. Rubin G. M., Spradling A. C. (1982) Genetic transformation of Drosophila with transposable element vectors. Science 218, 348–353 [DOI] [PubMed] [Google Scholar]
- 41. Spradling A. C., Rubin G. M. (1982) Transposition of cloned P elements into Drosophila germ line chromosomes. Science 218, 341–347 [DOI] [PubMed] [Google Scholar]
- 42. Dubreuil R. R. (2004) Copper cells and stomach acid secretion in the Drosophila midgut. Int. J. Biochem. Cell Biol. 36, 745–752 [DOI] [PubMed] [Google Scholar]
- 43. Shanbhag S., Tripathi S. (2009) Epithelial ultrastructure and cellular mechanisms of acid and base transport in the Drosophila midgut. J. Exp. Biol. 212, 1731–1744 [DOI] [PubMed] [Google Scholar]
- 44. Ten Hagen K. G., Bedi G. S., Tetaert D., Kingsley P. D., Hagen F. K., Balys M. M., Beres T. M., Degand P., Tabak L. A. (2001) Cloning and characterization of a ninth member of the UDP-GalNAc:polypeptide N-acetylgalactosaminyltransferase family, ppGaNTase-T9. J. Biol. Chem. 276, 17395–17404 [DOI] [PubMed] [Google Scholar]
- 45. Wandall H. H., Hassan H., Mirgorodskaya E., Kristensen A. K., Roepstorff P., Bennett E. P., Nielsen P. A., Hollingsworth M. A., Burchell J., Taylor-Papadimitriou J., Clausen H. (1997) Substrate specificities of three members of the human UDP-N-acetyl-α-d-galactosamine:polypeptide N-acetylgalactosaminyltransferase family, GalNAc-T1, -T2, and -T3. J. Biol. Chem. 272, 23503–23514 [DOI] [PubMed] [Google Scholar]
- 46. Van der Sluis M., De Koning B. A., De Bruijn A. C., Velcich A., Meijerink J. P., Van Goudoever J. B., Büller H. A., Dekker J., Van Seuningen I., Renes I. B., Einerhand A. W. (2006) Muc2-deficient mice spontaneously develop colitis, indicating that MUC2 is critical for colonic protection. Gastroenterology 131, 117–129 [DOI] [PubMed] [Google Scholar]
- 47. Velcich A., Yang W., Heyer J., Fragale A., Nicholas C., Viani S., Kucherlapati R., Lipkin M., Yang K., Augenlicht L. (2002) Colorectal cancer in mice genetically deficient in the mucin Muc2. Science 295, 1726–1729 [DOI] [PubMed] [Google Scholar]
- 48. An G., Wei B., Xia B., McDaniel J. M., Ju T., Cummings R. D., Braun J., Xia L. (2007) Increased susceptibility to colitis and colorectal tumors in mice lacking core 3-derived O-glycans. J. Exp. Med. 204, 1417–1429 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.