Background: The SID family is a highly conserved group of transmembrane channel-like proteins.
Results: SIDT1 facilitates rapid contact-dependent intercellular small RNA transfer and mediates chemoresistance driven by microRNA-21 in human adenocarcinoma cells.
Conclusion: By mediating small RNA transfer, SIDT1 contributes to cancer chemoresistance mechanisms.
Significance: A better understanding of non-cell-autonomous RNA-based intercellular communication may yield novel anticancer therapeutics.
Keywords: Cancer; Chemoresistance; MicroRNA; RNA interference (RNAi); siRNA; SIDT1, Systemic, miR-21, Pancreatic, Cancer, Adenocarcinoma, Gemcitabine
Abstract
Locally initiated RNA interference (RNAi) has the potential for spatial propagation, inducing posttranscriptional gene silencing in distant cells. In Caenorhabditis elegans, systemic RNAi requires a phylogenetically conserved transmembrane channel, SID-1. Here, we show that a human SID-1 orthologue, SIDT1, facilitates rapid, contact-dependent, bidirectional small RNA transfer between human cells, resulting in target-specific non-cell-autonomous RNAi. Intercellular small RNA transfer can be both homotypic and heterotypic. We show SIDT1-mediated intercellular transfer of microRNA-21 to be a driver of resistance to the nucleoside analog gemcitabine in human adenocarcinoma cells. Documentation of a SIDT1-dependent small RNA transfer mechanism and the associated phenotypic effects on chemoresistance in human cancer cells raises the possibility that conserved systemic RNAi pathways contribute to the acquisition of drug resistance. Mediators of non-cell-autonomous RNAi may be tractable targets for novel therapies aimed at improving the efficacy of current cytotoxic agents.
Introduction
RNA interference (RNAi) is initiated locally by double-stranded RNA (dsRNA) but has the capacity to propagate systemically (sysRNAi),2 inducing non-cell-autonomous posttranscriptional gene silencing in distant cells. Although best described as an antiviral mechanism in plants (1–3), sysRNAi also occurs in animals (4). In Caenorhabditis elegans, sysRNAi is dependent on a member of the systemic RNA interference-defective (SID) family of channels, SID-1 (5). SID-1 was initially identified following a screen of C. elegans mutants lacking the wild-type sysRNAi phenotype (5, 6). However, a range of organisms, including mice and humans, exhibit striking SID gene conservation (7–9).
SID channels have relative specificity for small RNA molecules (10, 11). Although organism-wide sysRNAi phenomena are not apparent in mammals, both SID-1 and its human orthologue SIDT1 (SID-1 transmembrane family member 1) have been shown to facilitate small interfering RNA (siRNA) uptake in human systems (12–14). The increased uptake of extracellular siRNA into human cells that SIDT1 mediates can result in highly specific posttranscriptional gene silencing (12, 13). We previously demonstrated that SIDT1 functions as a transmembrane channel for siRNA and localizes to the plasma membrane in human cells (12). This observation led us to hypothesize that SIDT1 might also play a role in the complex contact-dependent intercellular communication that is not only essential for normal histogenesis but, when dysregulated, also drives malignant progression and therapeutic resistance.
Small RNAs have a capacity to convey highly specific sequence-encoded signaling information (15). The microRNA (miRNA) system plays critical roles in the genesis, progression, and cytotoxic drug resistance of a range of human malignancies (16). Both the functional complexity of the “miRNome” and the diversity of miRNA targets suggest that regulation of gene function by miRNAs can be extremely subtle and adaptable (17). Within the tumor microenvironment, contact-dependent intercellular communication that is critical to the development of chemoresistance (18–20) is directly influenced by perturbation of the miRNome (21). This form of intercellular communication may represent an opportunity for novel targeted therapies.
MicroRNA-21 (miR-21), a relatively well characterized “oncogenic” miRNA, is widely overexpressed in human cancer and promotes therapeutic resistance in a number of human cancers (22–25). Pancreatic ductal adenocarcinoma is almost universally resistant to the nucleoside analog gemcitabine, the agent that remains the mainstay of non-surgical therapy for this cancer. In vitro, in vivo, and clinical resistance to gemcitabine is closely associated with levels of miR-21, which targets key apoptotic regulators (e.g. p53, PTEN (phosphatase and tensin homolog), Akt, and repressors of Ras signaling) (26, 27). An improved understanding of the mechanisms through which miRNAs drive chemoresistance will guide the development of urgently needed novel approaches that may increase the efficacy of existing cytotoxic agents, such as gemcitabine. Here we show that, by mediating rapid contact-dependent intercellular transfer of small RNAs, SIDT1 facilitates non-cell-autonomous sequence-specific gene silencing in human cells and can influence the development of chemoresistance in human adenocarcinoma cell populations.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
Human HEK293 and BxPC3 cells were purchased from the American Type Culture Collection (ATCC, Teddington, UK) and maintained as described previously (28). Gemcitabine (Lilly) and 18-α-glycerretinic acid (Sigma) were dissolved in phosphate-buffered saline (PBS). RNase blend (Cambio) pretreatment was performed using 5 units at 37 °C for 30 min. Trypsinization using 0.25% trypsin with EDTA (Invitrogen) was stopped by the addition of complete medium containing 10% fetal bovine serum (FBS). Human pancreatic stellate cells were obtained from surgical resection specimens under the ethically approved Edinburgh Pancreatic Biorepository scheme, using the outgrowth technique, detailed elsewhere (29).
Stable Far Red Membrane Linker
Cell surface labeling was performed in accordance with the manufacturer's instructions (Sigma). In brief, 106 cells were washed in PBS, and the pellet was resuspended in 1 ml of Diluent C in a 15-ml tube. Fluorescent CellVue Claret lipophilic probe was diluted at a final concentration of 3 μm in 1 ml of Diluent C in a 15-ml tube, and the solution was rapidly added to cells and incubated for 5 min at room temperature with occasional agitation. 2 ml of FBS was added, and after 1 min of incubation at room temperature, cells were washed three times in complete culture medium.
shRNA, Oligonucleotides, Plasmids, and Transfection and Electroporation
pCMV6-AC, pCMV6-AC-tGFP, pCMV6-AC-tGFP-SIDT1 (NM_017699), and pCMV6-Connexin-43/GJA1 (NM_000165) plasmids originated from Origene. Turbo green fluorescent protein (tGFP) was excised by NotI/PmeI digestion, fill-in, and ligation to derive pCMV-AC-SIDT1. Virus-incompetent pTRIPZ-based shRNA vectors (Open Biosystems) were used for microRNA expression. A miR-21 dual luciferase reporter construct was engineered using oligonucleotides designed to include SgfI and PmeI sites (see Table 1 for oligonucleotide sequences). A miR-21-resistant single base mismatch insert served as a control. Oligonucleotides were directionally cloned into the corresponding sites of the psiCHECK2 vector (Promega), in accordance with the manufacturer's protocol. Renilla luciferase substrate luminescence was normalized to that of firefly luciferase substrate to allow quantification of Renilla luciferase-miR-21 target sequence mRNA degradation by miR-21. The Dual-Glo luciferase assay system (Promega) was read using a VICTOR3-1420 multilabel reader (PerkinElmer Life Sciences). Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Stable cell lines were derived using G418 (0.3 mg/ml) or puromycin (5 μg/ml; both from Sigma) selection, as appropriate. All constructs were verified by sequencing. SIDT1-specific siRNA and mismatch control and Cy3-siRNA (supplemental material) were obtained from Dharmacon, Sigma, and Eurogentec. Lucifer yellow introduction was performed by electroporation in accordance with the manufacturer's cell type-specific protocols using the NucleofectorTM system (Lonza). Gap junction intercellular communication was quantified by flow cytometric quantification of Lucifer yellow transfer as described previously (30). Recipient cells were labeled with far red membrane linker as described above.
TABLE 1.
Oligonucleotide sequences
| Oligonucleotide | Sequence |
|---|---|
| miR-21 | 5′-UAGCUUAUCAGACUGAUGUUGA-3′ |
| miR-21 reporter oligonucleotides | 5′-CGCAGTAGAGCTCTAGTTCAACATCAGTCTGATAAGCTAGTTT-3′ |
| 5′-AAACTAGCTTATCAGACTGATGTTGAACTAGAGCTCTACTGCGAT-3′. | |
| miR-21-resistant single-base mismatch control oligonucleotides | 5′-GCAGTAGAGCTCTAGTTCAACATCAGAAGATAAGCTAGTTT-3′ |
| 5′-AAACTAGCTTATCTTCTGATGTTGAACTAGAGCTCTACTGCGAT-3′. | |
| SIDT1-specific siRNA 1 target sequence | 5′-CUCUCAGGAUGAACGUAUU-3′ |
| SIDT1-specific siRNA 2 target sequence | 5′-CUACUGGGAUAGAUGUUUU-3′ |
| SIDT1-specific siRNA 3 target sequence | 5′-GAGCAAUUAUGGGACAAUAUU-3′ |
| Cy3/control siRNA | 5′-UAGCGACUAAACACAUCAAUU-3′ |
Direct Coculture and Flow Cytometric Analysis of Intercellular siRNA Transfer
Cocultured labeled cell subpopulations were encouraged to conjugate by centrifugation at 500 rpm for 1 min and cocultured at 37 °C or 4 °C. Following coculture, cells were washed and resuspended in 5 mm EDTA/PBS and kept on ice. Multiparametric flow cytometry data were obtained from 10,000 single cell events with stringent doublet exclusion gating FACSAriaTM II using FACSDiva software (BD Biosciences). Flow cytometry data were analyzed using FlowJo V8 (TreeStar). Viable single cells were identified based on forward scatter and side scatter characteristics (width, height, and area).
Indirect Transwell Coculture and Conditioned Medium
Upper or lower compartments of each Transwell chamber (0.4-μm pore diameter, 12-well format) were populated with cells at the specified ratios, in accordance with the manufacturer's instructions (Corning). Cells were incubated for the specified times at 37 °C, collected using 5 mm EDTA/PBS, and analyzed for Cy3-siRNA acquisition by flow cytometry, as described above.
Immunoblotting and Antibodies
Cell extracts and gels were prepared and used as previously described (28). Goat anti-SIDT1 polyclonal antibody was obtained from Cambridge BioScience, rabbit anti-SIDT1 polyclonal and anti-α-smooth muscle actin monoclonal antibodies were obtained from Sigma, and mouse anti-β-actin monoclonal antibody was obtained from AbCam. Signal intensities were quantified and normalized to that of β-actin. Blots were performed in triplicate. Mean densitometric values ± S.D. are shown.
Proliferation, Cytotoxicity, Colony Formation, and Apoptosis Assays
Cellular proliferation and cytotoxic effects of gemcitabine were quantified using the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay (Trevigen, Gaithersburg, MD), as described previously (31). MTT correlates closely with [3H]thymidine incorporation in pancreatic cancer cell lines (32). Gemcitabine-induced cytotoxicity was determined after 24 h of exposure. Plates were read at a wavelength of 570 nm, corrected to 560 nm, and normalized to controls. Readings were obtained from four biological replicates, with 10 determinations for each condition tested. The concentration of gemcitabine required to inhibit cellular proliferation by 50% (IC50) was calculated using Microsoft Excel software with semilog curve fitting regression analysis. Caspase 3 activity was quantified using the ApoTarget colorimetric assay in accordance with the manufacturer's protocol. Relative absorbance at 405 nm was quantified using a VICTOR3-1420 multilabel reader (PerkinElmer Life Sciences). Apoptotic cells were quantified by fluorescent terminal deoxynucleotidyltransferase-mediated nick end labeling, as described previously (33). Colony formation in the presence of 1 μm gemcitabine was quantified as described elsewhere (34) following Giemsa staining. tGFP was quantified using a microplate reader (excitation, 485 nm; emission, 530 nm), as described elsewhere (27). tGFP was normalized to MTT A560 with background subtraction.
RESULTS
Analysis of Stable HEK293-derived Transfectant Cell Lines
HEK293 cells were transfected with pCMV-based plasmids encoding SIDT1 alone or in combination with tGFP and the aminoglycoside 3′-phosphotransferase neomycin resistance selection marker. Levels of SIDT1 expression were quantified by Western blotting in the following stable transfectants, which were derived using G418 selection: HEKSIDT1, which overexpresses SIDT1; HEKSIDT1/tGFP, which overexpresses SIDT1 and tGFP; and HEKSIDT1-tGFP, which overexpresses a fusion protein comprising SIDT1 and C terminus tGFP. Stable tGFP (HEKtGFP) and empty vector transfectants (HEKVector) served as controls. The electrophoretic migration of SIDT1 and SIDT1-tGFP fusion protein was consistent with predicted respective molecular masses of 94 and 120 kDa (Fig. 1a). The transfectant cell lines demonstrated no significant differences in respective rates of cellular proliferation or fraction of apoptotic cells (TUNEL) under standard culture conditions (supplemental Fig. S1).
FIGURE 1.
SIDT1 facilitates rapid contact-dependent intercellular Cy3-siRNA transfer between donor and acceptor subpopulations. a, characterization of SIDT1 protein expression in HEK293-derived stable transfectant cell lines. Shown is representative Western blot analysis of SIDT1 expression in control HEK293 cells (HEKVector), tGFP control (HEKtGFP), SIDT1 (HEKSIDT1), SIDT1 and tGFP (SIDT1/tGFP, clones 1 and 2), and SIDT1-tGFP fusion (HEKSIDT1-tGFP) transfectants (clones 1–3) and β-actin loading control. Electrophoretic mobility of SIDT-1 with C-terminal fusion tGFP protein (SIDT1-tGFP) corresponded to a predicted molecular mass of 120 kDa versus 94 kDa for SIDT1. b, schematic representation of siRNA transfer coculture experimental design. Donor subpopulations comprised stable HEKVector or HEKSIDT1 transfectants. Acceptor subpopulations comprised HEKtGFP or HEKSIDT1/tGFP transfectants. The donor subpopulation was tagged with far red membrane linker, and Cy3-siRNA was introduced by electroporation. Donor and acceptor subpopulations were directly cocultured for 90 min at 37 °C, and single cell suspensions were generated and analyzed by flow cytometry. Quantification of tGFP+/Cy3-siRNA+/FarRed− cells allowed donor to acceptor Cy3-siRNA transfer to be measured. c, I, in the absence of SIDT1 overexpression, Cy3-siRNA transfer was negligible. II–IV, SIDT1 overexpression resulted in a marked increase in Cy3-siRNA transfer. SIDT1 overexpression in either donor or acceptor subpopulation was sufficient for this increase in Cy3-siRNA transfer to occur, indicating that SIDT1 facilitates bidirectional siRNA transfer. V, preincubation with AGA had no effect on Cy3-siRNA transfer efficiency, indicating the effects of SIDT1 overexpression on Cy3-siRNA transfer to be independent of GJIC. Indirect coculture and conditioned medium exposure (90 min in each case) did not result in significant acceptor Cy3-siRNA acquisition (supplemental Fig. S3b). VI, Cy3-siRNA transfer was abolished by preincubation with anti-SIDT1 antibody. VII, time course of Cy3-siRNA transfer from HEKSIDT1 donor to HEKSIDT1/tGFP acceptor cells. Shown is a histogram representation of tGFP+/Cy3-siRNA+/FarRed− acceptor cells at specified time points. VIII, data presented are typical of quadruplicate biological repeat experiments. In each sample, 10,000 single cell events were recorded.
SIDT1 Facilitates Rapid Contact-dependent siRNA Transfer between Human Cells
A direct coculture assay was used to investigate the role of SIDT1 in contact-dependent siRNA transfer. “Donor” (HEKSIDT1 or HEKVector) and “acceptor” (HEKSIDT1/tGFP or HEKtGFP) cell subpopulations were subjected to direct coculture, allowing cell-cell contact as schematized (Fig. 1b). Cy3-labeled 21-mer siRNA was introduced into donor cells by electroporation alone to eliminate potentially confounding effects of persisting transfection reagent. To mitigate against donor epifluorescence signal decay, as might occur due to Cy3-siRNA degradation, and to control for trogocytosis or cell fusion events, donor cells were co-labeled with a far red fluorescent plasma membrane linker. This linker is highly persistent (t½ = 12 days), is biochemically inert, does not affect cell viability or membrane function, and has an emission spectrum that is readily distinguishable from that of Cy3 (35–37). Far red label transfer to unlabeled cells was not observed during any direct or indirect coculture experiments. Potential artifact arising from contact-independent medium-borne siRNA transfer was minimized by postelectroporation washing and RNase treatment, which degrades extracellular RNA to nucleoside monophosphates (supplemental Fig. S2a) (38, 39).
SIDT1-overexpressing (HEKSIDT1 and HEKSIDT1/tGFP) and control (HEKVector and HEKtGFP) donor and acceptor cells were cocultured at 1:1 ratios. Following coculture for 90 min, cell conjugates were disrupted to form single cell suspensions by EDTA treatment and agitation. Additional RNase treatment ensured removal of cell surface-associated Cy3-siRNA and free RNA that may have been released from lysed cells. Intercellular Cy3-siRNA transfer to acceptor cells was quantified by flow cytometry using stringent doublet exclusion. We quantified Cy3-siRNA-positive, tGFP-positive, far red-negative acceptor cells, defining a new subset of acceptor cells that had acquired Cy3-siRNA from donor cells (Fig. 1c, I–VI). Transfer of Cy3-siRNA between HEKSIDT1 donor and HEKSIDT1/tGFP acceptor cells was insensitive to RNase treatment and occurred rapidly (Fig. 1c, VII). In contrast, transfer of Cy3-siRNA between HEKVector donor and HEKtGFP acceptor cells was negligible (Fig. 1c, I). Direct coculture using HEKSIDT1 donor and HEKtGFP acceptor cells (Fig. 1c, II), as well as HEKVector donor and HEKSIDT1/tGFP acceptor cells (Fig. 1c, III), resulted in no significant difference in siRNA transfer (i.e. SIDT1 increased Cy3-siRNA acquisition regardless of whether it was overexpressed by donor or acceptor cells, indicating that facilitation of intercellular siRNA transfer by SIDT1 overexpression is bidirectional). Transfer of Cy3-siRNA from HEKSIDT1 to HEKSIDT1/tGFP was abolished by preincubation with polyclonal anti-SIDT1 (10 μg/ml; Fig. 1c, VI).
Although free Cy3-siRNA was eliminated from the culture medium, we were cognizant that nascent or RNase-resistant exosome-borne Cy3-siRNA arising from donor cells could also potentially contribute to the acceptor Cy3-siRNA signal, through contact-independent acquisition. To control for contact-independent Cy3-siRNA transfer, we performed indirect coculture of identical donor and acceptor cell groups, separated by permeable (0.4-μm diameter pore) Transwell insert membranes. In addition, acceptor cells were exposed to cell-free (0.4-μm filtered) donor conditioned medium. After either 90 min of indirect coculture or 90 min of exposure to donor conditioned medium, Cy3 epifluorescence was quantified by flow cytometry, as described above. No transfer of Cy3-siRNA to either HEKSIDT1/tGFP or HEKtGFP acceptor cells was detected either following indirect coculture with donor cells or following exposure to donor conditioned medium (supplemental Fig. S2b). Contact-independent uptake of extracellular Cy3-siRNA therefore did not account for the Cy3 signal acquired by the acceptor cell subpopulation.
SIDT1-mediated Cy3-siRNA Transfer Is Gap Junction-independent
The contribution of GJIC to Cy3-siRNA acquisition was predicted to be small in HEK293 cells, given their low levels of connexin junction formation and GJIC (40–42). However, gap junction-mediated intercellular transfer of small RNAs has been reported in some cell types (43, 44). We therefore took steps to distinguish the contribution of SIDT1 to the acquisition of Cy3-siRNA by acceptor cells from that of GJIC. We reasoned that enhanced intercellular Cy3-siRNA transfer could result from either native gap junction-mediated siRNA transfer or facilitation of GJIC by SIDT1 overexpression.
GJIC was quantified in the direct coculture system by flow cytometric measurement of Lucifer yellow transfer following electroporation-mediated Lucifer yellow loading, a quantitative approach that correlates with the scratch-loading Lucifer yellow transfer assay (30, 45). Intercellular Lucifer yellow transfer was minimal in all combinations of HEKSIDT1, HEKSIDT1-tGFP, HEKSIDT1/tGFP, HEKtGFP, and HEKVector following a 90-min coculture. GJIC was inhibited without cytotoxicity in all transfectant HEK- and BxPC3-derived cell lines following pretreatment with the specific small molecule gap junction inhibitor 18-α-glycyrrhetinic acid (AGA (25 μm); supplemental Fig. S3) (46, 47). Coculture and flow cytometric analysis of Cy3-siRNA transfer was repeated following pretreatment with AGA prior to a 90-min coculture at 37 °C, as described above. Cy3-siRNA transfer was not significantly affected by AGA. Together, these observations indicate that SIDT1 overexpression does not facilitate GJIC; nor is the resulting increase in Cy3-siRNA transfer dependent on functional GJIC.
Induction of Non-autonomous miRNA Activity through Physical Contact with Subpopulation of miR-21-overexpressing BxPC3 Cells
miR-21 is a critical driver of resistance to the nucleoside analog gemcitabine in human pancreatic adenocarcinoma, an exemplar of a chemoresistant human malignancy (26, 27). The human pancreatic ductal adenocarcinoma cell line BxPC3 was selected as a model system because it exhibits relatively low levels of miR-21 activity under standard culture conditions (26).
We stably transfected a subpopulation of BxPC3 cells with a non-viral pTRIPZ-derived miR-21 expression construct. These cells (BxPC3miR21) generate miR-21 in a doxycycline-inducible manner. Irrelevant miRNA-generating transfectants (BxPC3miRN/S, derived from RHS4346) served as controls. A miR-21 reporter cell line (BxPC3CkmiR21) was derived from BxPC3 by stable transfection of a psiCHECK-2-based miR-21 reporter construct. This dual luciferase reporter system allows Renilla luciferase activity, which decreases in the presence of miR-21, to be normalized to firefly luciferase, controlling for variations in reporter construct abundance. A single nucleotide mismatch reporter cell line (BxPC3CkmiR21 mm) was employed to confirm the specificity of the reporter system for miR-21 (supplemental Fig. S4).
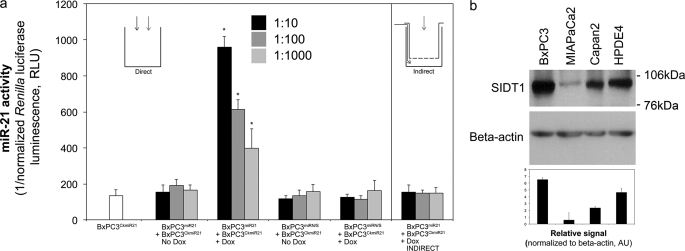
BxPC3miR21 and BxPC3CkmiR21 cells were subjected to direct coculture at high total cell density (80–90% cell-cell contact) at BxPC3miR21/BxPC3CkmiR21 ratios ranging from 1:10 to 1:1000, in the presence or absence of 1 μg/ml doxycycline. Doxycycline-induced BxPC3miR21 activation (confirmed by RFP epifluorescence) led to a decrease in normalized Renilla luciferase activity of directly cocultured BxPC3CkmiR21 reporter cells, reflecting increased miR-21 activity within the BxPC3CkmiR21 reporter cell subpopulation. Direct coculture with BxPC3miRN/S had no effect on normalized Renilla activity of BxPC3CkmiR21 cells, confirming specificity for the miR-21 sequence. Significant decreases in normalized Renilla luciferase activity were observed even when BxPC3miR21 cells were present as a minority of 0.1% (Fig. 2a). This non-autonomous increase in miR-21 activity within the BxPC3CkmiR21 subpopulation was not observed when BxPC3CkmiR21 cells were subjected to indirect coculture with BxPC3miR21 cells (0.4-μm pore diameter; Transwell; Fig. 2a).
FIGURE 2.
A minority subpopulation increases non-autonomous miR-21 activity in adenocarcinoma cells. a, BxPC3miR21 (doxycycline-inducible miR-21) or irrelevant miRNA (BxPC3miRN/S) controls were directly cocultured with miR-21 reporter cells (BxPC3CkmiR21) at the indicated ratios (±1 μg/ml doxycycline). Renilla luciferase luminescence (levels decreased by miR-21) was normalized to firefly luciferase luminescence to allow quantitative comparison (relative luminescence units (RLU)). Doxycycline-induced BxPC3miR21 activation increased miR-21 activity in BxPC3CkmiR21 reporter cells. Coculture of BxPC3CkmiR21 with BxPC3miRN/S had no effect on normalized Renilla activity in BxPC3CkmiR21. Indirect coculture was insufficient to induce non-autonomous miR-21 activity. The first white column indicates BxPC3CkmiR21 in standard monoculture. *, p < 0.05 versus BxPC3miR21 + BxPC3CkmiR21, no doxycycline by multifactorial analysis of variance, n = 4. b, representative Western blot analysis of SIDT1 expression in pancreatic adenocarcinoma (BxPC3, MIAPaCa2, and Capan2) and immortalized normal ductal epithelial cells (HPDE4), demonstrating differential expression of SIDT1. Densitometric quantification (means ± S.D. (error bars)) of SIDT1 signal, normalized to that of β-actin. Shown are mean values ± S.D. (n = 3). AU, arbitrary absorbance units.
Non-autonomous miR-21 Activity Is SIDT1-dependent
Our previous observations led us to hypothesize that the SIDT1 channel could facilitate miR-21 transfer between contacting cells. SIDT1 channel protein expression varies considerably between pancreatic adenocarcinoma and immortalized normal ductal epithelial cell (HPDE4) lines. Among adenocarcinoma cells, BxPC3 expresses relatively high levels of SIDT1 (Fig. 2b). Increased miR-21 activity in BxPC3CkmiR21 induced by the doxycycline-activated minority BxPC3miR21 subpopulation was abrogated by pretreatment with siRNAs directed against different regions of the SIDT1 sequence, including the SIDT1 3′-untranslated region (3′-UTR), but not control siRNA (Fig. 3). The specificity of this siRNA-induced effect was further confirmed by a “rescue step” in which a SIDT1 expression construct lacking the SIDT1 3′-UTR target sequence or empty vector control was co-transfected with SIDT1–3′-UTR siRNA 1. SIDT1 overexpression “rescued” the abrogation of miR-21 induction in the BxPC3CkmiR21 reporter subpopulation that was observed with SIDT1–3′-UTR siRNA 1 treatment (Fig. 3).
FIGURE 3.
SIDT1 is required for intercellular miR-21 transfer. a, knockdown of SIDT1 expression by siRNA was quantified by Western blotting. Three siRNAs targeting different regions of the SIDT1 sequence were compared. Mean ± S.D. (error bars) densitometric values are presented (n = 3). Levels of SIDT1 were most effectively decreased by siRNA 1, which targets the 3′-UTR, but could be rescued by co-transfection of the SIDT1 expression vector (which lacks the siRNA 1 3′-UTR target) but not by control vector. b, BxPC3CkmiR21 reporter activation by the minority (1%) BxPC3miR21 subpopulation was abrogated by transfection of siRNA 1 but not control siRNA prior to coculture. SIDT1 rescue restored BxPC3CkmiR21 miR-21 induction despite siRNA 1. Triplicate means ± S.D. are shown. SIDT1 signal was normalized to that of β-actin. *, p < 0.05 versus N/S siRNA; **, p < 0.05 versus 3′-UTR siRNA 1 + vector, by multifactorial analysis of variance (n = 5). RFU, relative fluorescence units.
Minority Subpopulation of miR-21-overexpressing Adenocarcinoma Cells Increases Global Cellular Chemoresistance to Gemcitabine
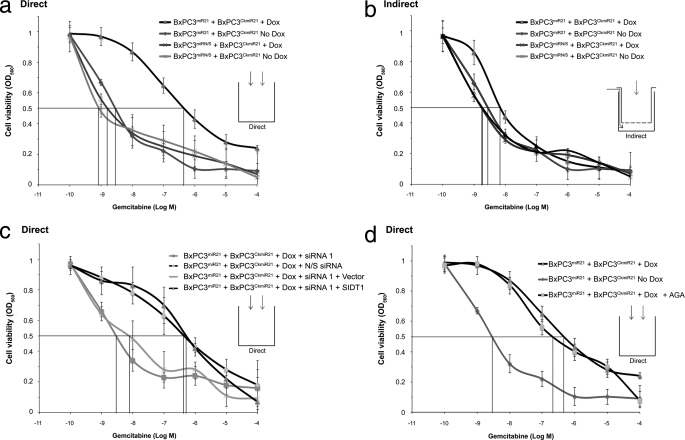
Given the ability of a minority subpopulation of BxPC3miR21 cells to increase miR-21 activity in physically contacting BxPC3CkmiR21 cells, we examined the effect of a 1% subpopulation of BxPC3miR21 on the gemcitabine IC50 for the total mixed cell population. Cells were exposed to clinically relevant concentrations of gemcitabine (48–52), and the IC50 was derived for the whole cell population. The gemcitabine IC50 was increased from 6 × 10−8 to 3 × 10−6 m by direct coculture of doxycycline-activated BxPC3miR21 with BxPC3CkmiR21 cells (ratio 1:100). Importantly, firefly luciferase activity normalized to total cell number remained constant, indicating preservation of the BxPC3miR21/BxPC3CkmiR21 ratio over the duration of the experiment. The gemcitabine IC50 approximated control levels when doxycycline was withheld and was unaffected by direct coculture of BxPC3miRN/S with BxPC3CkmiR21, confirming specificity of the effect to the miR-21 sequence (Fig. 4a). Transfer of mir-21 and IC50 were unaffected by 25 μm AGA (Fig. 4d and supplemental Fig. S3).
FIGURE 4.
SIDT1 deficiency abrogates chemoresistance induced by a minority miR-21-expressing subpopulation of pancreatic adenocarcinoma cells. Chemoresistance was quantified by MTT assay, and IC50 values were derived. a, the IC50 of gemcitabine was significantly increased by direct coculture of doxycycline-activated BxPC3miR21 with BxPC3CkmiR21 cells (ratio 1:100). b, the alteration in IC50 was minimal when the minority subpopulation was indirectly cocultured (Transwell). The gemcitabine IC50 was unaffected by coculture of BxPC3miRN/S with BxPC3CkmiR21. Means ± S.D. (error bars) are presented (n = 3). c, the increase in IC50 induced by miR-21 was attenuated when cells were treated with SIDT1–3′-UTR siRNA 1 but not by control mismatch siRNA. SIDT1 rescue abrogated the effect of SIDT1–3′-UTR siRNA 1. d, gap junction inhibition using 25 μm AGA did not significantly affect the increase in IC50 induced by miR-21.
SIDT1 Deficiency Attenuates Chemoresistance Induced by Minority miR-21-overexpressing Subpopulation of Adenocarcinoma Cells
The increase in global cellular chemoresistance to gemcitabine that miR-21 induced was cell contact-dependent, the IC50 being unaffected when BxPC3miR21 and BxPC3CkmiR21 cells were subjected to indirect coculture (BxPC3miR21/BxPC3CkmiR21 = 1:100; Fig. 4b). The miR-21-induced increase in IC50 was significantly attenuated when cells were treated with SIDT1–3′-UTR siRNA 1 but not control siRNA. A SIDT1 re-expression rescue step, as described above, confirmed that the decrease in gemcitabine IC50 induced by SIDT1–3′-UTR siRNA 1 could be abolished by restoring levels of SIDT1 (Fig. 4c). Corresponding impairment of colony forming capacity and increased caspase 3 activities were observed when BXPC3miR21 and BxPC3CkmiR21 were cocultured in the presence of gemcitabine following treatment with SIDT1–3′-UTR siRNA 1. Reintroduction of SIDT1 abolished these effects on colony formation and caspase activities (Fig. 5).
FIGURE 5.
SIDT1 knockdown abrogates miR-21-induced chemoresistance. a, caspase 3 activities were quantified 24 h following exposure to 1 μm gemcitabine by a colorimetric caspase 3 assay. Relative absorbances at 405 nm with background subtraction are presented. Mean values ± S.D. (error bars) from four biological replicates are shown. b, representative images of colony formation capacity for each condition.
SIDT1 Contributes to Cell Adhesion-mediated Drug Resistance (CAM-DR)
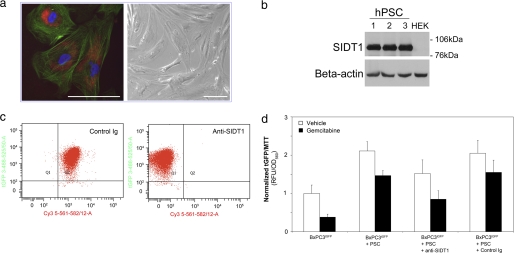
Given the capacity for small RNA transfer observed between adenocarcinoma cells, we examined whether a similar process might contribute to the complex tumor-stromal cell interactions that can enhance chemoresistance in a range of human cancers (53). Human pancreatic stellate cells (hPSC) were isolated from surgical resection specimens, and their morphology and immunophenotype (54) were confirmed (Fig. 6a). SIDT1 was relatively highly expressed in all three hPSC lines tested (Fig. 6b). We repeated the flow cytometric Cy3-siRNA transfer assay, as before, and observed rapid Cy3-siRNA transfer between adenocarcinoma and hPSC cells that was sensitive to anti-SIDT1 antibody treatment (Fig. 6c). In order to quantify CAM-DR, we subjected BxPC3tGFP cells to direct coculture with hPSC in the presence of 1 μm gemcitabine and measured tGFP fluorescence, which correlates with adenocarcinoma cell number (27), normalizing GFP fluorescence to MTT-based total cell quantification. BxPC3tGFP proliferation in the presence of clinically relevant gemcitabine levels was markedly increased by hPSC coculture. Furthermore, this effect was abrogated by treatment with 10 μg/ml anti-SIDT1 antibody but not control-matched immunoglobulin. Although this aspect of SIDT1 biology requires further investigation, this result demonstrates that heterotypic intercellular small RNA transfer can occur and that SIDT1 contributes to this process.
FIGURE 6.
Pancreatic stellate cell-induced CAM-DR is attenuated by anti-SIDT1 antibody. a, hPSC, expanded from pancreatic resection specimens, were immunocytochemically characterized. Shown is a representative image of PSC immunophenotype. Green, smooth muscle actin; red, vitamin A granule autofluorescence; blue, DAPI. Magnification is ×40. A phase-contrast image of PSC is shown (magnification ×20). b, patient-derived hPSC express SIDT1 protein. Shown is Western analysis of SIDT1 expression in patient-derived PSC cells (1–3). HEK293 cells serve as a negative control. c, small RNA transfer between adenocarcinoma cells and hPSC is disrupted by anti-SIDT1 antibody. Flow cytometric analysis of Cy3-siRNA transfer between BxPC3 cells and hPSC. BxPC3 cells loaded with Cy3-siRNA (electroporation) and labeled with far red fluorescent linker were directly cocultured with hPSC (green fluorescence) for 90 min. The subpopulation of Cy3-positive, far red-negative cells (indicating Cy3-siRNA acquisition) was significantly reduced by anti-SIDT1 antibody. d, stellate cell-induced CAM-DR is inhibited by anti-SIDT1 treatment. 5 × 103 BxPCtGFP cells were directly cocultured with confluent hPSC for 72 h in the presence of 1 μm gemcitabine with anti-SIDT1 antibody or immunoglobulin control. tGFP fluorescence, attributable to the BxPC3 adenocarcinoma subpopulation, was measured (excitation, 485 nm; emission, 530 nm) and normalized to MTT colorimetric readout of cell viability (A560, with hPSC background subtraction). Direct coculture with hPSC increased adenocarcinoma cellular proliferation in the presence of gemcitabine, relative to standard monoculture. Anti-SIDT1 significantly decreased hPSC-induced CAM-DR. Shown are mean values ± S.D. (error bars) (n = 4).
In summary, SIDT1 facilitates rapid bidirectional, contact-dependent, RNase-insensitive transfer of Cy3-siRNA that is independent of GJIC. Contact-independent siRNA transfer was insignificant in comparison with SIDT1-mediated contact-dependent Cy3-siRNA acquisition. SIDT1-dependent Cy3-siRNA intercellular transfer is not restricted to adenocarcinoma cells and can occur between stromal cells, influencing CAM-DR.
DISCUSSION
Although organism-wide sysRNAi is not apparent in mammals, significant phylogenetic molecular conservation suggests that sysRNAi pathways may be relevant to human physiology and pathophysiology (7). The C. elegans orthologue of SIDT1, SID1, has recently been shown to be a dsRNA-gated channel capable of selective bidirectional intercellular dsRNA transfer (11). Our findings demonstrate first that SIDT1 facilitates contact-dependent small RNA transfer and non-cell-autonomous posttranscriptional regulation; second, they support the assertion that small RNA-based signaling represents a further level of adaptive capacity and complexity within the tumor microenvironment; and third, they indicate that disruption or exploitation of sysRNAi pathways may have therapeutic utility, particularly as a means of impairing the acquisition of resistance to cytotoxic agents.
Contact-dependent intercellular communication not only maintains normal tissue organization but can also drive neoplasia. However, to date, studies of SID family proteins have generally focused on these proteins as conduits for the contact-independent uptake of free small RNAs from the extracellular milieu (12, 13). In addition to characterizing the role of SIDT1 in the context of contact-dependent small RNA intercellular transfer, this study provides new evidence that contact-dependent non-cell-autonomous RNAi can shape therapeutic resistance in pancreatic cancer and that SIDT1 can act as a mediator of this form of RNA-based intercellular communication.
The miRNome is a highly complex and adaptable system, with each miRNA exerting pleiotropic effects. Recent studies illustrate miRNA biogenesis to be exquisitely sensitive to cell context, miRNA levels increasing in a contact-dependent manner (55). miR-21 was the focus of this study because it promotes chemoresistance to gemcitabine in human adenocarcinoma cells (22–25). The ability of a minority subpopulation of miR-21-overexpressing cells to influence global chemoresistance through a contact-mediated, SIDT1-dependent mechanism raises the intriguing possibility that subgroups of cells within a heterogeneous tumor population can influence resistance within the wider tumor microenvironment through contact-dependent non-cell-autonomous RNAi. Subpopulations of drug-tolerant tumor cells employ dynamic survival strategies that result in therapeutic resistance (34). Parallels can be drawn with microbial resistance, in which a small number of tolerant organisms can influence the “fitness” of the populations as a whole. Similarly, cells exposed to cytotoxic drug may, through small RNA-based communication, influence survival pathways within contacting cells over significant distances.
The rapid nature of SIDT1-mediated small RNA transfer is particularly striking and significantly precedes RNA transfer via contact-independent mechanisms, such as exosomal shuttling and free RNA transfer (38). Tumor cells that are capable of rapid adaptation are more likely to gain selective advantage in the presence of a toxic perturbation. Rapid small RNA transfer and resulting posttranscriptional gene regulation would allow more timely adaptive changes than those resulting from “classic” genetic mutation. This study supports the premise that small RNAs have the capacity to act as signaling intermediaries. The absence of disseminating fluorescent protein expression to acceptor cells in longer term coculture studies confirms a degree of RNA type and size specificity (i.e. SIDT1 does not facilitate functional mRNA transfer). These observations are in keeping with the relative specificity for dsRNA exhibited by SID1 (11).
Our results demonstrate SIDT1-mediated siRNA transfer to be independent of GJIC. GJIC can increase the susceptibility of cancer cells to cytotoxic agents through connexin-mediated “bystander effects” in pancreatic cancer (56). BxPC3 cells in which GJIC is artificially increased exhibit bystander cytotoxicity when exposed to gemcitabine (56–58). Interestingly, connexin expression and GJIC are frequently decreased in cancer, re-expression of connexins commonly suppressing tumorigenicity (59). In contrast to the relatively non-selective nature of connexin-mediated communication, SIDT1 overexpression does not increase Lucifer yellow transfer. SIDT1 may therefore represent a means by which tumor cells can adapt to maintain small RNA-based intercellular communication, without experiencing greater bystander cytotoxicity that increased GJIC would incur.
Recent data from other groups indicate that non-cell-autonomous small RNA effects may be of general relevance to human cancer. Katakowski et al. (60) reported microvesicle-independent microRNA transfer between U87 human glioma cells. Although this study did not directly examine chemoresistance, the authors demonstrated non-autonomous microRNA effects in contacting cells. Zhao et al. (61) have demonstrated that SNB19 glioma cells can undergo intercellular transfer of PTEN-silencing siRNA in coculture. Interestingly, this effect was only observed in direct (contact-dependent) but not indirect (contact-independent) coculture. The PTEN tumor suppressor mRNA is a target of miR-21 and a number of other microRNAs. PTEN deficiency is clinically associated with chemoresistance in a range of human cancers. These data also suggest that non-autonomous gene silencing can result in molecular events that promote clinical chemoresistance.
We have demonstrated that SIDT1-dependent small RNA transfer can also operate between adenocarcinoma and stromal cells, in this case pancreatic stellate cells. The important chemoprotective effects of direct tumor-stromal cell contact are increasingly recognized (27). CAM-DR that develops in the tumor microenvironment may represent a future therapeutic opportunity. Follicular dendritic cells can protect B-cell lymphoma cells from drug-induced apoptosis through contact-mediated microRNA-dependent mechanisms (21, 62). miR-181a was found to be increased by direct contact between dendritic and lymphoma cells but not when cells were cultured under indirect coculture conditions, suggesting that free RNA, RNA bound to proteins (e.g. Argonaute or lipoproteins), or exosomal RNA transfer is less likely to mediate the effects on chemoresistance in this setting. This is in keeping with our observations. Although the possibility that the non-autonomous increase in miR-181a levels could result from intercellular RNA transfer was not explored, interestingly, SIDT1 is also relatively overexpressed in dendritic cells.
The clinical implications of SIDT1 expression levels are likely to be complex and will be influenced by the prevailing miRNome within the tumor. Preliminary studies of primary breast cancer mRNA and microRNA expression arrays are consistent with the hypothesis that SIDT1 may have a “permissive” role in some of the phenotypic effects of miR-21 overexpression. High levels of miR-21 expression result in poorer survival for patients with SIDT1-overexpressing tumors relative to those with low SIDT1 expression (supplemental Fig. S5). Further clinical studies are ongoing to address this question.
In conclusion, SIDT1 mediates contact-dependent siRNA and miRNA transfer and non-cell-autonomous RNAi, which can enhance pancreatic adenocarcinoma chemoresistance to gemcitabine that is driven by miR-21. SIDT1-dependent small RNA transfer may also contribute to CAM-DR. Although sysRNAi in humans appears not to be the organism-wide phenomenon observed in C. elegans, comparable processes may support adaptation to perturbations and selective pressures within the tumor microenvironment, such as those induced by cytotoxic therapy. Therapeutic exploitation of sysRNAi pathways may have utility in human cancer and warrants further experimental evaluation.
Supplementary Material
Acknowledgments
We thank Drs. Nick Gilbert and Bernard Ramsahoye, Prof. John Iredale, and members of the Frame and Brunton laboratories for helpful advice and reagents. We thank Dr. Jacqueline Dickson for the pTRIPZ-IRES-GFP construct, Elizabeth Freyer (Flow Cytometry Facility, Medical Research Council Human Genetics Unit, Edinburgh) for technical assistance with flow cytometry, and Dr. Andy Sims for bioinformatics advice.
This work was supported by Chief Scientist Office Project Grant CZB/4/693 and by the Melville Trust for the Care and Cure of Cancer.

This article contains supplemental Figs. S1–S5.
- sysRNAi
- systemic RNAi
- GJIC
- gap junction intercellular communication
- SID
- systemic RNA interference-defective
- tGFP
- turbo green fluorescent protein
- AGA
- α-glycyrrhetinic acid
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- CAM-DR
- cell adhesion-mediated drug resistance
- PSC
- pancreatic stellate cell(s)
- hPSC
- human PSC
- miRNA
- microRNA
- miR-21 and miR-181a
- microRNA 21 and 181a, respectively.
REFERENCES
- 1. Chitwood D. H., Timmermans M. C. (2010) Small RNAs are on the move. Nature 467, 415–419 [DOI] [PubMed] [Google Scholar]
- 2. Herr A. J., Baulcombe D. C. (2004) RNA silencing pathways in plants. Cold Spring Harb. Symp. Quant. Biol. 69, 363–370 [DOI] [PubMed] [Google Scholar]
- 3. Baulcombe D. (2004) RNA silencing in plants. Nature 431, 356–363 [DOI] [PubMed] [Google Scholar]
- 4. Jose A. M., Hunter C. P. (2007) Transport of sequence-specific RNA interference information between cells. Annu. Rev. Genet. 41, 305–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Winston W. M., Molodowitch C., Hunter C. P. (2002) Systemic RNAi in C. elegans requires the putative transmembrane protein SID-1. Science 295, 2456–2459 [DOI] [PubMed] [Google Scholar]
- 6. Feinberg E. H., Hunter C. P. (2003) Transport of dsRNA into cells by the transmembrane protein SID-1. Science 301, 1545–1547 [DOI] [PubMed] [Google Scholar]
- 7. Li H., Coghlan A., Ruan J., Coin L. J., Hériché J. K., Osmotherly L., Li R., Liu T., Zhang Z., Bolund L., Wong G. K., Zheng W., Dehal P., Wang J., Durbin R. (2006) TreeFam. A curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Tomoyasu Y., Miller S. C., Tomita S., Schoppmeier M., Grossmann D., Bucher G. (2008) Exploring systemic RNA interference in insects. A genome-wide survey for RNAi genes in Tribolium. Genome Biol. 9, R10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Su A. I., Wiltshire T., Batalov S., Lapp H., Ching K. A., Block D., Zhang J., Soden R., Hayakawa M., Kreiman G., Cooke M. P., Walker J. R., Hogenesch J. B. (2004) A gene atlas of the mouse and human protein-encoding transcriptomes. Proc. Natl. Acad. Sci. U.S.A. 101, 6062–6067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hunter C. P., Winston W. M., Molodowitch C., Feinberg E. H., Shih J., Sutherlin M., Wright A. J., Fitzgerald M. C. (2006) Systemic RNAi in Caenorhabditis elegans. Cold Spring Harb. Symp. Quant. Biol. 71, 95–100 [DOI] [PubMed] [Google Scholar]
- 11. Shih J. D., Hunter C. P. (2011) SID-1 is a dsRNA-selective dsRNA-gated channel. RNA 17, 1057–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Duxbury M. S., Ashley S. W., Whang E. E. (2005) RNA interference. A mammalian SID-1 homologue enhances siRNA uptake and gene silencing efficacy in human cells. Biochem. Biophys. Res. Commun. 331, 459–463 [DOI] [PubMed] [Google Scholar]
- 13. Wolfrum C., Shi S., Jayaprakash K. N., Jayaraman M., Wang G., Pandey R. K., Rajeev K. G., Nakayama T., Charrise K., Ndungo E. M., Zimmermann T., Koteliansky V., Manoharan M., Stoffel M. (2007) Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 25, 1149–1157 [DOI] [PubMed] [Google Scholar]
- 14. Tsang S. Y., Moore J. C., Huizen R. V., Chan C. W., Li R. A. (2007) Ectopic expression of systemic RNA interference-defective protein in embryonic stem cells. Biochem. Biophys. Res. Commun. 357, 480–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dinger M. E., Mercer T. R., Mattick J. S. (2008) RNAs as extracellular signaling molecules. J. Mol. Endocrinol. 40, 151–159 [DOI] [PubMed] [Google Scholar]
- 16. Macfarlane L. A., Murphy P. R. (2010) MicroRNA. Biogenesis, Function, and Role in Cancer. Curr. Genomics 11, 537–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inui M., Martello G., Piccolo S. (2010) MicroRNA control of signal transduction. Nat. Rev. Mol. Cell Biol. 11, 252–263 [DOI] [PubMed] [Google Scholar]
- 18. Patek P. Q., Lin Y., Case P. G. (1989) Cell lines cultured at high density are resistant to lysis by tumor necrosis factor and natural cytotoxic cells. Proc. Soc. Exp. Biol. Med. 190, 234–239 [DOI] [PubMed] [Google Scholar]
- 19. Kobayashi H., Man S., Graham C. H., Kapitain S. J., Teicher B. A., Kerbel R. S. (1993) Acquired multicellular-mediated resistance to alkylating agents in cancer. Proc. Natl. Acad. Sci. U.S.A. 90, 3294–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Green S. K., Frankel A., Kerbel R. S. (1999) Adhesion-dependent multicellular drug resistance. Anticancer Drug Des. 14, 153–168 [PubMed] [Google Scholar]
- 21. Lwin T., Lin J., Choi Y. S., Zhang X., Moscinski L. C., Wright K. L., Sotomayor E. M., Dalton W. S., Tao J. (2010) Follicular dendritic cell-dependent drug resistance of non-Hodgkin lymphoma involves cell adhesion-mediated Bim down-regulation through induction of microRNA-181a. Blood 116, 5228–5236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Krichevsky A. M., Gabriely G. (2009) miR-21. A small multifaceted RNA. J. Cell Mol. Med. 13, 39–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bai H., Xu R., Cao Z., Wei D., Wang C. (2011) Involvement of miR-21 in resistance to daunorubicin by regulating PTEN expression in the leukaemia K562 cell line. FEBS Lett. 585, 402–408 [DOI] [PubMed] [Google Scholar]
- 24. Misawa A., Katayama R., Koike S., Tomida A., Watanabe T., Fujita N. (2010) AP-1-dependent miR-21 expression contributes to chemoresistance in cancer stem cell-like SP cells. Oncol. Res. 19, 23–33 [DOI] [PubMed] [Google Scholar]
- 25. Pan X., Wang Z. X., Wang R. (2011) MicroRNA-21. A novel therapeutic target in human cancer. Cancer Biol. Ther. 10, 1224–1232 [DOI] [PubMed] [Google Scholar]
- 26. Giovannetti E., Funel N., Peters G. J., Del Chiaro M., Erozenci L. A., Vasile E., Leon L. G., Pollina L. E., Groen A., Falcone A., Danesi R., Campani D., Verheul H. M., Boggi U. (2010) MicroRNA-21 in pancreatic cancer. Correlation with clinical outcome and pharmacologic aspects underlying its role in the modulation of gemcitabine activity. Cancer Res. 70, 4528–4538 [DOI] [PubMed] [Google Scholar]
- 27. Fujita H., Ohuchida K., Mizumoto K., Egami T., Miyoshi K., Moriyama T., Cui L., Yu J., Zhao M., Manabe T., Tanaka M. (2009) Tumor-stromal interactions with direct cell contacts enhance proliferation of human pancreatic carcinoma cells. Cancer Sci. 100, 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Duxbury M. S., Ito H., Ashley S. W., Whang E. E. (2004) CEACAM6 cross-linking induces caveolin-1-dependent, Src-mediated focal adhesion kinase phosphorylation in BxPC3 pancreatic adenocarcinoma cells. J. Biol. Chem. 279, 23176–23182 [DOI] [PubMed] [Google Scholar]
- 29. Bachem M. G., Schünemann M., Ramadani M., Siech M., Beger H., Buck A., Zhou S., Schmid-Kotsas A., Adler G. (2005) Pancreatic carcinoma cells induce fibrosis by stimulating proliferation and matrix synthesis of stellate cells. Gastroenterology 128, 907–921 [DOI] [PubMed] [Google Scholar]
- 30. Kiang D. T., Kollander R., Lin H. H., LaVilla S., Atkinson M. M. (1994) Measurement of gap junction conduction by fluorescence activated cell sorting. In Vitro Cell. Dev. Biol. Anim. 30A, 796–802 [DOI] [PubMed] [Google Scholar]
- 31. Duxbury M. S., Whang E. E. (2007) RRM2 induces NF-κB-dependent MMP-9 activation and enhances cellular invasiveness. Biochem. Biophys. Res. Commun. 354, 190–196 [DOI] [PubMed] [Google Scholar]
- 32. Raitano A. B., Scuderi P., Korc M. (1990) Binding and biological effects of tumor necrosis factor and γ interferon in human pancreatic carcinoma cells. Pancreas 5, 267–277 [DOI] [PubMed] [Google Scholar]
- 33. Duxbury M. S., Ito H., Benoit E., Waseem T., Ashley S. W., Whang E. E. (2004) A novel role for carcinoembryonic antigen-related cell adhesion molecule 6 as a determinant of gemcitabine chemoresistance in pancreatic adenocarcinoma cells. Cancer Res. 64, 3987–3993 [DOI] [PubMed] [Google Scholar]
- 34. Sharma S. V., Lee D. Y., Li B., Quinlan M. P., Takahashi F., Maheswaran S., McDermott U., Azizian N., Zou L., Fischbach M. A., Wong K. K., Brandstetter K., Wittner B., Ramaswamy S., Classon M., Settleman J. (2010) A chromatin-mediated reversible drug-tolerant state in cancer cell subpopulations. Cell 141, 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bantly A. D., Gray B. D., Breslin E., Weinstein E. G., Muirhead K. A., Ohlsson-Wilhelm B. M., Moore J. S. (2007) CellVue Claret, a new far-red dye, facilitates polychromatic assessment of immune cell proliferation. Immunol. Invest. 36, 581–605 [DOI] [PubMed] [Google Scholar]
- 36. Horan P. K., Slezak S. E. (1989) Stable cell membrane labeling. Nature 340, 167–168 [DOI] [PubMed] [Google Scholar]
- 37. Wallace P. K., Palmer L. D., Perry-Lalley D., Bolton E. S., Alexander R. B., Horan P. K., Yang J. C., Muirhead K. A. (1993) Mechanisms of adoptive immunotherapy. Improved methods for in vivo tracking of tumor-infiltrating lymphocytes and lymphokine-activated killer cells. Cancer Res. 53, 2358–2367 [PubMed] [Google Scholar]
- 38. Rechavi O., Erlich Y., Amram H., Flomenblit L., Karginov F. V., Goldstein I., Hannon G. J., Kloog Y. (2009) Cell contact-dependent acquisition of cellular and viral nonautonomously encoded small RNAs. Genes Dev. 23, 1971–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Rocha J. J., Korolchuk V. I., Robinson I. M., O'Kane C. J. (2011) A phagocytic route for uptake of double-stranded RNA in RNAi. PLoS One 6, e19087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Toyofuku T., Yabuki M., Otsu K., Kuzuya T., Hori M., Tada M. (1998) Intercellular calcium signaling via gap junction in connexin-43-transfected cells. J. Biol. Chem. 273, 1519–1528 [DOI] [PubMed] [Google Scholar]
- 41. John S. A., Kondo R., Wang S. Y., Goldhaber J. I., Weiss J. N. (1999) Connexin-43 hemichannels opened by metabolic inhibition. J. Biol. Chem. 274, 236–240 [DOI] [PubMed] [Google Scholar]
- 42. McSpadden L. C., Kirkton R. D., Bursac N. (2009) Electrotonic loading of anisotropic cardiac monolayers by unexcitable cells depends on connexin type and expression level. Am. J. Physiol. Cell Physiol. 297, C339–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wolvetang E. J., Pera M. F., Zuckerman K. S. (2007) Gap junction-mediated transport of shRNA between human embryonic stem cells. Biochem. Biophys. Res. Commun. 363, 610–615 [DOI] [PubMed] [Google Scholar]
- 44. Valiunas V., Polosina Y. Y., Miller H., Potapova I. A., Valiuniene L., Doronin S., Mathias R. T., Robinson R. B., Rosen M. R., Cohen I. S., Brink P. R. (2005) Connexin-specific cell-to-cell transfer of short interfering RNA by gap junctions. J. Physiol. 568, 459–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. el-Fouly M. H., Trosko J. E., Chang C. C. (1987) Scrape-loading and dye transfer. A rapid and simple technique to study gap junctional intercellular communication. Exp. Cell Res. 168, 422–430 [DOI] [PubMed] [Google Scholar]
- 46. Goldberg G. S., Moreno A. P., Bechberger J. F., Hearn S. S., Shivers R. R., MacPhee D. J., Zhang Y. C., Naus C. C. (1996) Evidence that disruption of connexon particle arrangements in gap junction plaques is associated with inhibition of gap junctional communication by a glycyrrhetinic acid derivative. Exp. Cell Res. 222, 48–53 [DOI] [PubMed] [Google Scholar]
- 47. Lahlou H., Fanjul M., Pradayrol L., Susini C., Pyronnet S. (2005) Restoration of functional gap junctions through internal ribosome entry site-dependent synthesis of endogenous connexins in density-inhibited cancer cells. Mol. Cell. Biol. 25, 4034–4045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Abbruzzese J. L., Grunewald R., Weeks E. A., Gravel D., Adams T., Nowak B., Mineishi S., Tarassoff P., Satterlee W., Raber M. N. (1991) A phase I clinical, plasma, and cellular pharmacology study of gemcitabine. J. Clin. Oncol. 9, 491–498 [DOI] [PubMed] [Google Scholar]
- 49. Bhargava P., Marshall J. L., Fried K., Williams M., Lefebvre P., Dahut W., Hanfelt J., Gehan E., Figuera M., Hawkins M. J., Rizvi N. A. (2001) Phase I and pharmacokinetic study of two sequences of gemcitabine and docetaxel administered weekly to patients with advanced cancer. Cancer Chemother. Pharmacol. 48, 95–103 [DOI] [PubMed] [Google Scholar]
- 50. van Riel J. M., Peters G. J., Mammatas L. H., Honeywell R. J., Laan A. C., Ruyter R., van den Berg F. G., Giaccone G., van Groeningen C. J. (2009) A phase I and pharmacokinetic study of gemcitabine given by 24-h hepatic arterial infusion. Eur. J. Cancer 45, 2519–2527 [DOI] [PubMed] [Google Scholar]
- 51. Grunewald R., Abbruzzese J. L., Tarassoff P., Plunkett W. (1991) Saturation of 2′,2′-difluorodeoxycytidine 5′-triphosphate accumulation by mononuclear cells during a phase I trial of gemcitabine. Cancer Chemother. Pharmacol. 27, 258–262 [DOI] [PubMed] [Google Scholar]
- 52. Veltkamp S. A., Beijnen J. H., Schellens J. H. (2008) Prolonged versus standard gemcitabine infusion. Translation of molecular pharmacology to new treatment strategy. Oncologist 13, 261–276 [DOI] [PubMed] [Google Scholar]
- 53. Dalton W. S. (1999) The tumor microenvironment as a determinant of drug response and resistance. Drug Resist. Updat. 2, 285–288 [DOI] [PubMed] [Google Scholar]
- 54. Bachem M. G., Schneider E., Gross H., Weidenbach H., Schmid R. M., Menke A., Siech M., Beger H., Grünert A., Adler G. (1998) Identification, culture, and characterization of pancreatic stellate cells in rats and humans. Gastroenterology 115, 421–432 [DOI] [PubMed] [Google Scholar]
- 55. Hwang H. W., Wentzel E. A., Mendell J. T. (2009) Cell-cell contact globally activates microRNA biogenesis. Proc. Natl. Acad. Sci. U.S.A. 106, 7016–7021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Garcia-Rodríguez L., Pérez-Torras S., Carrió M., Cascante A., García-Ribas I., Mazo A., Fillat C. (2011) Connexin-26 is a key factor mediating gemcitabine bystander effect. Mol. Cancer Ther. 10, 505–517 [DOI] [PubMed] [Google Scholar]
- 57. Cottin S., Ghani K., de Campos-Lima P. O., Caruso M. (2010) Gemcitabine intercellular diffusion mediated by gap junctions. New implications for cancer therapy. Mol. Cancer 9, 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ammerpohl O., Trauzold A., Schniewind B., Griep U., Pilarsky C., Grutzmann R., Saeger H. D., Janssen O., Sipos B., Kloppel G., Kalthoff H. (2007) Complementary effects of HDAC inhibitor 4-PB on gap junction communication and cellular export mechanisms support restoration of chemosensitivity of PDAC cells. Br. J. Cancer 96, 73–81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Zhang Z. Q., Zhang W., Wang N. Q., Bani-Yaghoub M., Lin Z. X., Naus C. C. (1998) Suppression of tumorigenicity of human lung carcinoma cells after transfection with connexin43. Carcinogenesis 19, 1889–1894 [DOI] [PubMed] [Google Scholar]
- 60. Katakowski M., Buller B., Wang X., Rogers T., Chopp M. (2010) Functional microRNA is transferred between glioma cells. Cancer Res. 70, 8259–8263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhao T. Y., Zou S. P., Alimova Y. V., Wang G., Hauser K. F., Ghandour M. S., Knapp P. E. (2006) Short interfering RNA-induced gene silencing is transmitted between cells from the mammalian central nervous system. J. Neurochem. 98, 1541–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lin J., Lwin T., Zhao J. J., Tam W., Choi Y. S., Moscinski L. C., Dalton W. S., Sotomayor E. M., Wright K. L., Tao J. (2011) Follicular dendritic cell-induced microRNA-mediated up-regulation of PRDM1 and down-regulation of BCL-6 in non-Hodgkin's B-cell lymphomas. Leukemia 25, 145–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.