Background: WaaL mediates the ligation of O-antigen onto lipid A-core.
Results: This ligation was reconstituted in vitro using synthetic donor substrates and donor mimics bearing structural variations. All of them were accepted as substrates by WaaL.
Conclusion: WaaL exhibits relaxed donor substrate specificity.
Significance: This work, together with other previously published studies, lays important foundations for dissecting the mechanism of WaaL enzymes.
Keywords: Carbohydrate Biosynthesis, Escherichia coli, Lipopolysaccharide (LPS), Membrane Enzymes, Protein Purification, Chemoenzymatic Synthesis, O-Antigen Ligase, In Vitro Assay
Abstract
The WaaL-mediated ligation of O-antigen onto the core region of the lipid A-core block is an important step in the lipopolysaccharide (LPS) biosynthetic pathway. Although the LPS biosynthesis has been largely characterized, only a limited amount of in vitro biochemical evidence has been established for the ligation reaction. Such limitations have primarily resulted from the barriers in purifying WaaL homologues and obtaining chemically defined substrates. Accordingly, we describe herein a chemical biology approach that enabled the reconstitution of this ligation reaction. The O-antigen repeating unit (O-unit) of Escherichia coli O86 was first enzymatically assembled via sequential enzymatic glycosylation of a chemically synthesized GalNAc-pyrophosphate-undecaprenyl precursor. Subsequent expression of WaaL through use of a chaperone co-expression system then enabled the demonstration of the in vitro ligation between the synthesized donor (O-unit-pyrophosphate-undecaprenyl) and the isolated lipid A-core acceptor. The previously reported ATP and divalent metal cation dependence were not observed using this system. Further analyses of other donor substrates revealed that WaaL possesses a highly relaxed specificity toward both the lipid moiety and the glycan moiety of the donor. Lastly, three conserved amino acid residues identified by sequence alignment were found essential for the WaaL activity. Taken together, the present work represents an in vitro systematic investigation of the WaaL function using a chemical biology approach, providing a system that could facilitate the elucidation of the mechanism of WaaL-catalyzed ligation reaction.
Introduction
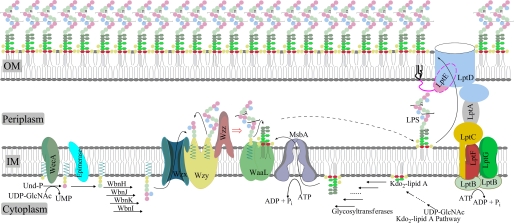
Bacterial cell surfaces are decorated with various types of glycoconjugates that play critical roles in the interactions between bacteria and the environment (1). For example, the outer leaflet of the outer membrane of Gram-negative bacteria is primarily composed of lipopolysaccharide (LPS) that plays critical roles in bacterial cell physiology (2) and bacterial pathogenicity (3–5). A typical LPS molecule consists of three structural components: lipid A (endotoxin), a non-repeating core oligosaccharide, and O-antigen (4). Over the past several decades, considerable efforts have been made to fully understand the LPS biogenesis that involves the LPS biosynthesis at the inner membrane (IM),4 transport across the periplasmic space, and insertion into the outer leaflet of the outer membrane (Fig. 1). The LPS biosynthesis at the IM involves the respective synthesis and export to the periplasmic surface of the IM of lipid A-core and O-antigen-pyrophosphate-undecaprenyl (O-antigen-PP-Und) and the final ligation of O-antigen onto the core region of the lipid A-core block (4, 6–9). Upon the completion of the LPS biosynthesis, a transenvelope complex formed by seven essential Lipopolysaccharide transport (Lpt) system proteins extracts LPS from the IM, transports it across the periplasmic space, and finally inserts it into the outer leaflet of the outer membrane (10–14). Diverse covalent modifications of the lipid A moiety may occur during the LPS transit from the outer surface of the IM to the outer membrane (6).
FIGURE 1.
Models for LPS biogenesis. Three assembly strategies (the Wzy-, ABC transporter- and synthase-dependent pathways) have been identified for the synthesis and export to the periplasmic surface of the IM of O-antigen-PP-Und. The Wzy-dependent pathway of E. coli O86 is presented here as an example. OM, outer membrane.
WaaL is currently the only enzyme presumed to be required for the O-antigen ligation reaction (4). Numerous WaaL homologues from various bacterial species have been identified. Sequence analyses indicate that these WaaL homologues are all integral membrane proteins. Furthermore, although the primary amino acid sequences of these homologues show significant divergence, they exhibit similar membrane topology characteristics highlighted by the presence of multiple membrane-spanning domains and one large periplasmic loop with variable lengths (4). The experiment-based membrane topology maps of three WaaL homologues have been reported (15–17). Several functionally critical amino acid residues located in the large periplasmic loop or in the adjacent small periplasmic loop of these three WaaL homologues were also identified (15–17). These critical residues, probably forming a part of a putative catalytic center that was suggested to participate in the chemical reaction(s) required for the release of O-antigen from the PP-Und lipid carrier (15), have been hypothesized to be involved in the binding of the pyrophosphate group of O-antigen-PP-Und (15, 16).
Although considerable knowledge regarding the WaaL function has been obtained from in vivo-based studies, only a limited amount of in vitro biochemical evidence has been reported (16, 18). This lack of in vitro evidence is directly related to the difficulties in handling integral membrane protein WaaL as well as obtaining donor substrates (PP-Und-linked glycan). In this study, a WaaL homologue from Escherichia coli O86:B7 (the R3-type core) was successfully expressed and purified with high purity. This advancement, coupled with the use of chemoenzymatically synthesized donor substrates, allowed an in vitro WaaL assay to be established. This assay then enabled the elucidation of the specificity of WaaL toward the donor substrate as well as the identification of three functionally critical amino acid residues through the incorporation of mutant WaaL proteins into the assay.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Materials
E. coli O86:B7 ATCC 12701 was purchased from the American Type Culture Collection. The E. coli O86:B7 waaL knock-out strain was previously prepared (7). GroES/EL expression plasmid pGro7 was a gift from J. Liu (University of North Carolina). The E. coli O86 O-antigen antiserum was acquired from the China Institute of Veterinary Control. n-Dodecyl β-d-maltoside was purchased from Anatrace. The site-directed mutagenesis kit was purchased from Agilent Technologies. All kits or enzymes were used following manufacturers' instructions. All other chemical reagents were obtained from Sigma-Aldrich unless otherwise noted.
Chemoenzymatic Synthesis of O-unit-PP-Lipid Substrates
The expression and purification of each of the four glycosyltransferases (WbnH, WbnJ, WbnK, and WbnI), the chemical synthesis of GalNAc/GlcNAc-PP-lipid compounds, and the subsequent stepwise assembly of the O-unit pentasaccharide on these compounds followed the previously published procedures (7, 19–21). The chemical synthesis procedures and product characterization are presented in detail in the supplemental text and supplemental Figs. S1 and S2.
Cloning, Expression, and Purification of WaaL
To clone the waaL gene from E. coli O86:B7, this gene was first sequenced in our laboratory. The primers used to clone the gene were designed based on the sequence (supplemental Table S1). The gene was then amplified by PCR from the genomic DNA of E. coli O86:B7. The PCR product was digested with NcoI and HindIII and subsequently cloned into the NcoI and HindIII sites of the pBAD/Myc-His A vector (Invitrogen), forming a recombinant plasmid, pBAD-waaL10×His. E. coli C43(DE3), which has been broadly employed for the expression of toxic proteins or integral membrane proteins (22), and which was used to express WaaL. Prior to its use in this expression, however, E. coli C43(DE3) was pretransformed with pGro7, a GroES/EL chaperone expression plasmid. Chaperones have been demonstrated to be involved in the protein folding process, and the co-expression of a target protein with chaperone molecules increases the recovery of expressed proteins in the correctly folded form (23). For protein expression, E. coli C43(DE3) bearing both plasmids was incubated in 3 liters of Luria-Bertani medium at 37 °C until A600 reached 0.8. l(+)-Arabinose was added to a final concentration of 0.2%. The protein expression was allowed to continue for 20 h at 20 °C. Cells were harvested and resuspended in lysis buffer (25 mm Tris-HCl, pH 7.5, 1 mm EDTA) and then disrupted by sonication as described previously (24). Cell lysates were centrifuged at 10,000 × g for 10 min at 4 °C to remove the cell debris. The supernatant was ultracentrifuged at 150,000 × g for 1 h at 4 °C. The membrane pellets were solubilized in membrane solubilization buffer (25 mm Tris-HCl, pH 7.5, 300 mm NaCl, 0.2 mm PMSF, 10 mm imidazole, and 2% n-dodecyl β-d-maltoside) overnight at 4 °C. The insolubilized materials were removed by centrifugation (150,000 × g, 1 h, at 4 °C). n-Dodecyl β-d-maltoside-bound WaaL in the supernatant was purified by using HisTrap HP column (GE Healthcare) as described previously (24). Finally, eluted WaaL from the HisTrap HP column was applied to the PD-10 column (GE Healthcare) to desalt and then eluted with storage buffer (50 mm Tris-HCl, pH 7.5, 100 mm NaCl, 0.05% n-dodecyl β-d-maltoside, and 20% glycerol). Purified WaaL was monitored by SDS-PAGE and Western blotting and kept at −20 °C.
WaaL-catalyzed in Vitro Ligation Assay
Lipid A-core was extracted from E. coli O86:B7 waaL knock-out cells using the phenol, chloroform, and petroleum ether method as described previously (25). The WaaL ligation reaction was performed with a 50-μl reaction mixture containing 50 mm Tris, pH 7.5, 2.5 μm lipid A-core, 10 μm O-unit-PP-lipid, and purified WaaL (∼1–2 μg). The reaction was allowed to continue for 12 h at 37 °C and then quenched with the SDS-PAGE loading buffer. Proteinase K was added to digest WaaL. 20 μl of reaction mixture was applied to a 4–12% Bis-Tris gradient gel (Invitrogen). LPS molecules were blotted onto the nitrocellulose membrane and visualized using the E. coli O86 O-antigen antiserum and the HRP-linked anti-rabbit IgG antibody.
To plot the time course of the WaaL ligation reaction, the in vitro assays for different donor substrates were run at the same conditions (i.e. 2.5 μm lipid A-core, 10 μm glycan-PP-lipid, 1 μg of purified WaaL, and 37 °C). At each of eight time points (0 min, 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 12 h), an aliquot of reaction mixture was removed out and immediately quenched with the SDS-PAGE loading buffer. The samples from all time points were applied to one SDS-PAGE gel and visualized by Western blotting. The band intensities were quantified by using ImageQuant TL software (GE Healthcare) and plotted over the reaction time.
Site-directed Mutagenesis of WaaL
Three WaaL mutants, WaaLR209A, WaaLR268A, and WaaLH319A, were created by using the QuikChange site-directed mutagenesis kit (Agilent Technologies). Briefly, oligonucleotide primers (supplemental Table S1) were designed to create the desired amino acid change. The pBAD-waaL10×His plasmid was used as the DNA template. PCR products were digested with DpnI and transformed into XL1-Blue Supercompetent cells. Plasmids were isolated and sequenced to confirm the desired substitution of amino acid residues. The WaaL mutants were expressed, purified, and applied to the in vitro ligation assay.
RESULTS
Assembly of O-unit-PP-Lipid Substrates
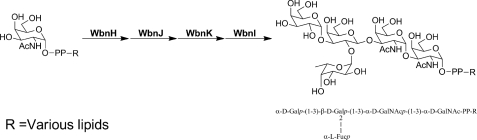
Most of E. coli O-antigen polymers are ligated to Und-P using a GlcNAc or GalNAc reducing-end residue that is attached separately in the initial step(s) of the O-antigen biosynthesis. The WecA protein catalyzes the formation of GlcNAc-PP-Und by transferring GlcNAc-1-P from UDP-GlcNAc onto Und-P. GalNAc-PP-Und, however, is formed via a two-step pathway, i.e. the initial WecA-catalyzed synthesis of GlcNAc-PP-Und followed by the epimerization of the 4-OH of GlcNAc catalyzed by a novel epimerase (26). Our initial attempts to express and purify integral membrane proteins WecA and the epimerase of E. coli to synthesize GlcNAc-PP-Und and GalNAc-PP-Und were not successful. Accordingly, an alternative methodology was employed for obtaining the O-unit-PP-Und molecule. Specifically, we exploited a chemoenzymatic approach in which the chemically synthesized GalNAc-PP-Und precursor was elaborated to the E. coli O86 pentasaccharide repeating unit via successive enzymatic glycosylation using four glycosyltransferases cloned from E. coli O86 following the established protocols (7, 19–21) (Fig. 2).
FIGURE 2.
Strategy for chemoenzymatic synthesis of O-unit-PP-lipid substrates. See Table 1 for structures of lipid moieties.
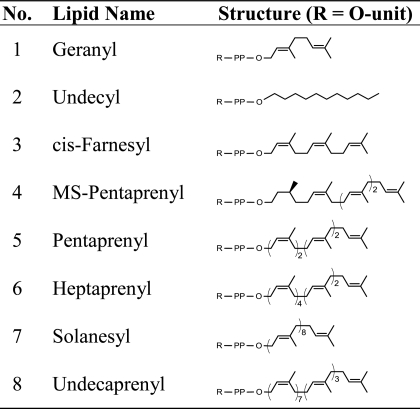
Some evidence has demonstrated that bacterial glycosyltransferases acting on Und-linked acceptors are relaxed in their recognition of lipid moiety (21, 27, 28). Such flexibility allows for the in vitro construction of an oligosaccharide on an unnatural lipid mimic rather than Und. Herein, a total of seven other unique O-unit-PP-lipid molecules were synthesized on a milligram scale, wherein the lipid moieties exhibit variations in the isoprenyl geometry (i.e. cis or trans double bond configuration), the saturation degree, and the chain length (Table 1). The acquisition of these various lipids appended with the same oligosaccharide moiety provided us with a comprehensive library to probe the donor-lipid specificity of WaaL using an in vitro ligation assay. It should be noted that the enzymatic glycosylation reactions are routinely highly efficient with all the intermediates nearly completely converted into products as monitored by ESI-MS analysis of each reaction step.
TABLE 1.
O-unit-PP-lipid compounds used in this study
Expression and Purification of Integral IM Protein WaaL
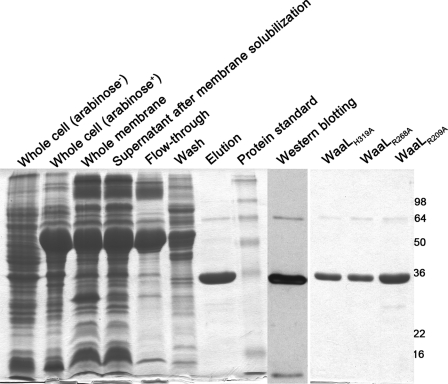
In E. coli, five core oligosaccharide variants have been identified, termed R1–R4 and K12, in which structural differences are primarily located in the outer core region (4). Each of the five core variants has its own specific WaaL ligase. The waaL gene of E. coli O86:B7 was sequenced in our laboratory. The sequence BLAST analysis indicated that the strain encodes R3 WaaL. A recombinant plasmid (pBAD-waaL10×His) was prepared to overexpress this WaaL, and the construct encodes full-length E. coli O86:B7 WaaL with a C terminus 10×His epitope. The pBAD-waaL10×His recombinant plasmid and the pGro7 GroES/EL chaperone expression vector were co-transformed into E. coli C43(DE3). The use of E. coli C43(DE3), in combination with the co-expression of chaperones, provided sufficient active WaaL-His10 fusion protein that was purified to near homogeneity (Fig. 3). Purified WaaL was monitored by SDS-PAGE and Western blotting (Fig. 3). Two main bands were found on SDS-PAGE, and further analysis using Western blotting indicated that both bands contain the His epitope. One band (32 kDa) corresponds to the monomer of WaaL, and the other band (64 kDa) was believed to be the dimer of WaaL. The finding that WaaL forms a dimer agrees with previously observed dimer formation of Pseudomonas aeruginosa PAO1 WaaL (16). Furthermore, although the apparent molecular mass of 32 kDa for the monomer is obviously considerably lower than the predicted value of 47.7 kDa, such aberrations in molecular masses have frequently been encountered in SDS-PAGE analyses of integral membrane proteins (16).
FIGURE 3.
SDS-PAGE and Western blotting analyses of purified WaaL (wild type and mutants).
Development of in Vitro O-Antigen Ligation Assay
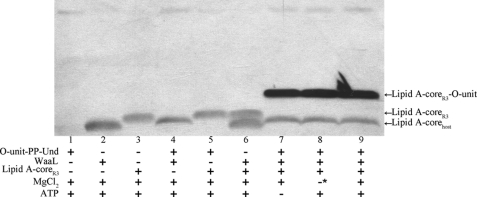
The lipid A-core molecule was extracted from E. coli O86:B7 waaL knock-out cells. Upon the acquisition of the synthesized donor substrate, isolated lipid A-core, and purified WaaL, we then sought to develop an in vitro WaaL assay. A series of reactions was accordingly performed in which all components were present or one or more were absent. Following these reactions, the samples were analyzed using Western blotting that employed the E. coli O86 O-antigen antiserum to visualize LPS molecules. The results are shown in Fig. 4. The band observed in lane 3 of Fig. 4 represents the lipid A-core acceptor (hereafter referred to as lipid A-coreR3). The observation that the O-antigen antiserum can visualize LPS molecules indicates that the antiserum also contains core epitope-specific antibodies. This phenomenon is also observed with an E. coli O86 O-antigen antiserum from a different manufacturer (Statens Serum Institute of Denmark). Interestingly, a band migrating slightly faster than lipid A-coreR3 was observed when only WaaL, ATP, and MgCl2 were present (Fig. 4, lane 2). We reasoned that it is most likely that it corresponds to lipid A-core produced by E. coli C43(DE3) (hereafter referred to as lipid A-corehost). Lipid A-corehost might bind to WaaL and be co-purified with WaaL. The migration discrepancy observed between lipid A-coreR3 and lipid A-host host might be ascribed to the molecular weight difference of these two molecules.
FIGURE 4.
Western blotting analyses of in vitro ligation reaction samples. Development of the in vitro WaaL assay using synthesized O-unit-PP-Und is shown. *, 25 mm EDTA was supplemented to remove any endogenous residual divalent metal cation.
Surprisingly, we also observed that lipid A-corehost migrates slightly slower when O-unit-PP-Und is included in the reaction mixture, as can be seen by comparison of lanes 2 and 4 of Fig. 4. This trend was also observed in all of the reactions where the O-unit-PP-Und donor is present (Fig. 4, lanes 7–9; Fig. 5A, lane 9; Fig. 5B, lanes 1–5). However, the migration of lipid A-corehost remains unaffected when O-unit-PP-heptaprenyl (supplemental Fig. S3) was used instead. The reason behind the phenomenon is still unclear, but it should be emphasized that such subtle migration discrepancies do not interfere with the interpretation of the results.
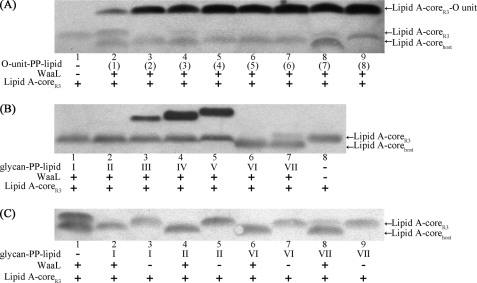
FIGURE 5.
Western blotting analyses of in vitro ligation reaction samples. A, investigation of the WaaL specificity toward the lipid moiety of the donor. The digit in parentheses denotes the number for each O-unit-PP-lipid substrate as shown in Table 1. B, investigation of the WaaL specificity toward the reducing-end sugar of the donor. I, GalNAc-PP-Und. II, GalNAc-α1,3-GalNAc-PP-Und. III, Gal-β1,3-GalNAc-α1,3-GalNAc-PP-Und. IV, Fuc-α1,2-Gal-β1,3-GalNAc-α1,3-GalNAc-PP-Und. V, Gal-α1,3-(Fuc-α1,2)-Gal-β1,3-GalNAc-α1,3-GalNAc-PP-Und. VI, GlcNAc-PP-pentaprenyl. VII, Gal-α1,3-Glc-PP-undecyl. C, in vitro ligation assays showing the consumption of lipid A-coreR3 when the mono- or disaccharide-PP-lipid substrate was used.
As shown in lane 9 of Fig. 4, an additional strong band with a higher molecular weight than that observed in the negative control (Fig. 4, lane 5) was detected. It was thus concluded that this band represents the ligation product, i.e. the lipid A-coreR3-O-unit conjugate. In our initial attempts to establish the in vitro WaaL assay, ATP was supplemented because it was reported that P. aeruginosa PAO1 WaaL requires ATP for its activity (16). However, as can be seen in lane 7 of Fig. 4, although no ATP was supplemented, WaaL still exhibited the enzymatic activity, indicating that E. coli R3 WaaL is ATP-independent, a fact that is consistent with the recently observed ATP independence of Helicobacter pylori WaaL and E. Coli K-12 WaaL (18, 38). So far, it is still unclear why these WaaL homologues exhibit different ATP requirements. Furthermore, we found that E. coli R3 WaaL does not require a divalent metal cation either for its activity (Fig. 4, lane 8). This observation further corroborated the ATP independence of E. coli R3 WaaL as the previously reported ATP hydrolysis by P. aeruginosa PAO1 WaaL requires a divalent metal cation (16). ATP and divalent metal cations were accordingly removed from the reaction mixture in later experiments. Thus, the final reaction mixture for the in vitro WaaL assay contains: 50 mm Tris-HCl, pH 7.5, 10 μm O-unit-PP-Und, 2.5 μm lipid A-coreR3, and 1–2 μg purified WaaL.
It should also be noted that a weak high molecular weight band was observed in lane 1 of Fig. 4 in which only O-unit-PP-lipid, ATP, and MgCl2 were included. We reasoned that this band corresponds to O-unit-PP-Und given that in all reactions in which O-unit-PP-Und was included, this band was detected (Fig. 4, lanes 1, 4, 5, 7, 8, and 9). In principle, O-unit-PP-Und is expected to migrate faster than lipid A-core because of its smaller molecular mass (∼1800 Da versus ∼4000 Da). This observation, however, underscores the notion that O-unit-PP-Und might form a complex molecule instead of a single monomer as we expected.
In summary, through a series of experiments as shown in Fig. 4, we unambiguously confirmed that the in vitro ligation reaction took place only in the presence of the purified WaaL enzyme. Thus, an in vitro WaaL functional assay was successfully established.
WaaL Exhibits Relaxed Specificity to Lipid Moiety of Donor Substrate
Having established the in vitro WaaL ligation assay using synthesized O-unit-PP-Und, the specificity of WaaL toward the donor was further investigated. As illustrated in Fig. 5A, a total of eight O-unit-PP-lipid molecules were assayed using the in vitro ligation system. Surprisingly, we found that WaaL exhibits a highly relaxed specificity toward the lipid moiety of the donor substrate. All of the synthesized mimics were found to be substrates for WaaL, even with the geranyl-PP-linked substrate that contains only two isoprene units in the lipid moiety. It should be noted, however, that only a weak product band was observed in this case (Fig. 5A, lane 2). The changes in isoprenyl geometry do not abolish the WaaL activity, as demonstrated by the observation that undecaprenyl-, solanesyl-, heptaprenyl-, pentaprenyl-, and cis-farnesyl-PP-linked molecules all can serve as substrates for the in vitro ligation. The variations in saturation degree of lipid moieties do not abolish the ligation activity either, as exemplified with the MS-pentaprenyl- and unbranched undecyl alkyl-based substrates. WaaL also exhibits a high degree of tolerance toward variations in the lipid length, although an obviously much weaker product band was observed with the geranyl-PP-linked substrate. The substrates with lipids in which the isoprenyl geometry, the saturation degree, and the lipid length are highly variable were all found to be accepted by WaaL. On the basis of these results, it was concluded that WaaL exhibits a highly relaxed specificity toward the lipid moiety of the donor substrate.
Further Comparison of WaaL Activities on Donors Harboring Different Lipid Moieties
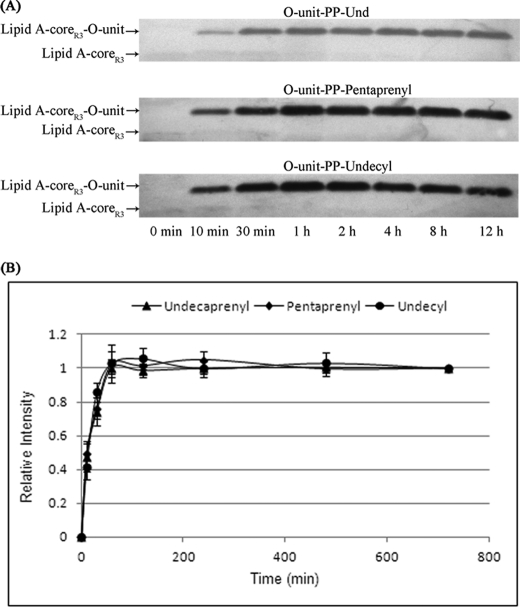
The time course of the WaaL ligation reaction was plotted for three donor substrates (Und-, pentaprenyl- and undecyl-linked glycans) to further probe the difference of WaaL activities on substrates harboring different lipid moieties. The in vitro assays for these substrates were run at the same conditions, and aliquots of reaction mixture from eight time points during 0–12 h (see “Experimental Procedures”) were applied to one SDS-PAGE gel and visualized by Western blotting (Fig. 6A). At the time point of 12 h, the acceptor (i.e. lipid A-coreR3) could be completely converted to product (i.e. lipid-coreR3-O-unit) (Fig. 6A). The band intensity at this time point (12 h) was set to 1, and those from other time points were accordingly normalized and plotted over the reaction time. As illustrated in Fig. 6B, three similar time course curves were obtained for these substrates. The lipid structures of these donor substrates are quite different. The natural lipid, undecaprenyl, has 11 isoprene units (seven cis and three trans double bonds), whereas pentaprenyl has only five isoprene units (two cis and two trans double bonds). The undecyl lipid is the most distinct one in that it is a saturated unbranched C11 alkyl-based lipid (Table 1). Despite the large structural differences between these lipid moieties, WaaL displayed similar time course curves on them, indicating that the enzyme does not exhibit significant activity difference on structurally distinct lipids.
FIGURE 6.
Plotting time courses of WaaL reaction for three donor substrates (i.e. Und-, pentaprenyl-, and undecyl-linked glycans). A, Western blotting analyses of aliquots of reaction mixtures from eight time points during 0–12 h (0 min, 10 min, 30 min, 1 h, 2 h, 4 h, 8 h, and 12 h). B, time course curves for three donor substrates based on the intensities of bands. Error bars indicate S.E.
WaaL Exhibits Relaxed Specificity to Glycan Moiety of Donor Substrate
To further test whether the glycan moiety of the donor plays a critical role in the donor recognition by WaaL, we investigated the impact of variations in the glycan moiety on WaaL activity. Some E. coli O157 strains such as O157:H7 also possess an R3-type core (29) and thus also encode R3 WaaL proteins. The structure of the E. coli O157 O-unit, however, is quite different from that of E. coli O86 with variability in the second residue of the reducing end through the non-reducing end residue (supplemental Fig. S4). Such variability suggests that residues beyond the reducing-end sugar do not play a crucial role in the recognition and binding of donor by WaaL.
To investigate whether the reducing-end residue, GalNAc, is itself critical for the ligation activity, two more molecules, GlcNAc-PP-pentaprenyl and Gal-α1,3-Glc-PP-undecyl, were also synthesized and applied to the in vitro ligation assay (Fig. 5B). The band of acceptor lipid A-coreR3 disappeared when GlcNAc-PP-pentaprenyl was used (Fig. 5B, lane 6), suggesting that lipid A-coreR3 had been consumed. Interestingly, however, the band of desired product lipid A-coreR3-GlcNAc was not observed. The previous observation that the distally ligated glycan moiety can block the accessibility of the LPS core epitope(s) to core-specific antibodies has frequently been reported in Western blotting analyses of LPS (30, 31). Therefore, we reasoned that newly ligated GlcNAc may prevent core-specific antibodies from binding to the core epitope(s), and furthermore, GlcNAc is itself not able to form the O-antigen epitope(s). To confirm this, a set of reactions was performed in which the glycan moiety of the donor ranged from the monosaccharide to the pentasaccharide (Fig. 5B, lanes 1–5). The mono-, di-, tri-, and tetrasaccharide represent truncated O-units of E. coli O86, whereas the pentasaccharide is a complete O-unit. As expected, although the acceptor band disappeared when either monosaccharide-PP-Und or disaccharide-PP-Und were applied, the desired product band was not observed (Fig. 5B, lanes 1 and 2). Interestingly, when trisaccharide-, tetrasaccharide- or pentasaccharide-PP-Und was applied, stair-like bands corresponding to their respective ligation products were clearly observed (Fig. 5B, lanes 3–5), indicating that newly linked sugar residues form the O-antigen epitope(s), which can be visualized by O-antigen-specific antibodies. This observation also suggested that a moiety of as few as three sugar residues is sufficient to form the antiserum-detectable O-antigen epitope(s) of E. coli O86. In the reaction in which Gal-α1,3-Glc-PP-undecyl was used, the acceptor was observed partially consumed as compared with the control experiment (Fig. 5B, lanes 7 and 8).
To more clearly show the consumption of the lipid A-coreR3 acceptor in the reactions where the monosaccharide- or disaccharide-PP-lipid substrate was used, we have performed WaaL assays along with the respective control experiments including WaaL-negative controls and a donor-negative control (Fig. 5C). In the cases where GalNAc-PP-Und, GalNAc-α1,3-GalNAc-PP-Und, or GlcNAc-PP-pentaprenyl was used, the lipid A-coreR3 acceptor was observed completely consumed (Fig. 5C, lanes 2, 4, and 6), whereas lipid A-coreR3 was partially consumed (Fig. 5C, lane 8) when Gal-α1,3-Glc-PP-undecyl was assayed.
Taken together, all of these results suggested that E. coli R3 WaaL is relaxed in its recognition of the glycan moiety of the donor. This conclusion is consistent with the observed flexibility of E. coli K-12 WaaL that can in vivo ligate O-antigen polysaccharides with diverse structures to its cognate lipid A-core acceptor (32–35). Of these polysaccharides, colanic acid is impressive as its reducing-end sugar is glucose rather than GlcNAc or GalNAc (35).
Identification of Three Functionally Critical Amino Acid Residues
To identify conserved amino acid residues, a multiple sequence alignment analysis was performed with 24 WaaL homologues using MAFFT version 6. The alignment analysis revealed three highly conserved amino acid residues: arginine, histidine, and glycine (supplemental Fig. S5). Of these residues, arginine and histidine have been shown functionally critical for the WaaL function in some bacterial species (15–17), whereas glycine was found not essential for the full functionality of E. coli K-12 WaaL (15). In addition, a second arginine residue (Arg-288) of E. coli K-12 WaaL is also critical (15), and most of the arginine residues of other WaaL homologues at the equivalent position are within the GXR motif or its variants (supplemental Fig. S5). For those homologues that lack a GXR motif or its variants, such as E. coli R3 WaaL, an arginine residue is also present at this position (supplemental Fig. S5). To investigate whether these conserved amino acid residues are functionally critical for E. coli R3 WaaL, three WaaL mutants (WaaLR209A, WaaLR268A, and WaaLH319A) were prepared (Fig. 3) and then applied to the well established in vitro WaaL assay using two different donor substrates. As shown in Fig. 7, wild type WaaL could transfer O-unit onto lipid A-coreR3, whereas the mutants did not show any detectable ligation activities, indicating the functional importance of Arg-209, Arg-268, and His-319 for E. coli R3 WaaL. Based on the previously established topology maps of WaaL homologues of E. coli K-12 and Vibrio cholerae O1 (15, 17), as well as the multiple sequence alignment analysis (supplemental Fig. S5), Arg-268 and His-319 are located in the large periplasmic loop of E. coli R3 WaaL, whereas Arg-209 is in the adjacent small periplasmic loop. These three positively charged amino acid residues might form a part of a putative catalytic center, and their roles likely involve the binding of the pyrophosphate group of the donor according to the previously proposed model for E. coli K-12 WaaL (15).
FIGURE 7.
Western blotting analyses of in vitro ligation assays of three WaaL mutants. The digit in parentheses denotes the number for each O-unit-PP-lipid substrate as shown in Table 1.
DISCUSSION
It is widely accepted that Und-P is the predominant lipid carrier involved in the biosynthesis of various bacterial polysaccharides (36). Therefore, a reliable and efficient approach to access PP-Und-linked glycan substrates becomes a key point on the way to dissect the mechanism of the bacterial polysaccharide biosynthesis. Three previous studies used so-called crude (impure) substrates, which were prepared from bacterial cell membrane, to develop the in vitro WaaL assay (16, 18, 38). Nevertheless, PP-Und-linked glycan intermediates are produced by bacteria in a limited amount, making the large scale isolation of pure substrates unrealistic. To address these problems, we employed an efficient chemoenzymatic approach to afford these elusive substrates. Moreover, by taking advantage of the relaxed specificity of the glycosyltransferases, the oligosaccharide could be constructed on a wide spectrum of shorter lipid-PP carriers. These synthesized compounds can be applied to establish an in vitro enzyme assay, to investigate the substrate specificity, or to set up the co-crystallization of the key proteins with substrates. In fact, some of these compounds have been successfully applied to the Wzy study in our laboratory, revealing important aspects of the Wzy-catalyzed polymerization (7), such as the discovery that Wzy possesses a limitedly relaxed specificity toward the lipid moiety of the substrate. Interestingly, in this study, we found that WaaL is much more flexible in its recognition of the lipid moiety than Wzy. It should be noted that the donor-binding site and acceptor-binding site of Wzy are both involved in the recognition and binding of PP-Und-linked substrates. The observed difference in lipid specificity between WaaL and Wzy might indicate that at least one of two sites of Wzy possesses a more stringent requirement for the lipid moiety than the donor-binding site of WaaL.
This study suggested that E. coli R3 WaaL is divalent metal cation-independent. This conclusion might provide additional hints on the possible mechanism by which WaaL binds the donor substrate. Although there is no relationship between WaaL and classic sugar nucleotide-dependent glycosyltransferases, these two types of enzymes essentially catalyze similar reactions in which they both catalyze the glycosidic bond formation. WaaL uses the PP-Und-linked O-antigen polysaccharide as the donor, whereas classic glycosyltransferases act on sugar nucleotides. Two structural folds, GT-A and GT-B, have been identified for sugar nucleotide-dependent glycosyltransferases (37). Most of the GT-A glycosyltransferases possess a DXD motif, by which they can coordinate a divalent metal cation, which in turn facilitates the departure of the nucleoside pyrophosphate leaving group from the donor. In contrast, the GT-B glycosyltransferases were found to be divalent metal cation-independent, and they instead use the helix dipole effect or positively charged side chains to interact with the pyrophosphate of the leaving group (37). The divalent metal cation independence of WaaL prompted us to consider whether it employs a mechanism analogous to that of GT-B glycosyltransferases to bind the pyrophosphate group of the donor. Several functionally critical amino acid residues have been identified for several WaaL homologues, including Arg-186/His-309/His-311 of V. cholerae O1 WaaL (17), Arg-208/His-321 of Salmonella enterica sv. Arizonae IIIA WaaL (17), His-303 of P. aeruginosa PAO1 WaaL (16), and Arg-186/Arg-288/His-337 of E. coli K-12 WaaL (15). In this study, Arg-209, Arg-268, and His-319 were also shown functionally critical for E. coli R3 WaaL. Interestingly, positively charged arginine is frequently used by GT-B glycosyltransferases to bind the pyrophosphate group of sugar nucleotides (37). Due to the high degree of conservation and functional importance of these residues, it has been proposed that these residues may play roles in the binding of the pyrophosphate group of O-antigen-PP-Und (15, 16). The divalent metal cation independence of E. coli R3 WaaL provides additional evidence for such a notion.
Despite these advances, dissecting the WaaL reaction mechanism in detail still awaits the determination of the WaaL structure in both apo forms and donor-bound or acceptor-bound forms. Undecaprenyl-, heptaprenyl-, and solanesyl-PP-linked donor substrates possess long lipid moieties and are thus highly prone to aggregate. Although the assays utilizing these substrates have been developed, it should be mentioned that the organic solvent (dimethyl sulfoxide (DMSO)) and the detergent (Triton X-100) must be supplemented to help dissolve the aggregates in the aqueous medium. In light of these issues, it is not practically efficient to co-crystallize WaaL with such lipid-linked glycans for structural investigations. Pentaprenyl- or other shorter lipid-containing substrates, in contrast, can be easily dissolved in the aqueous solution and potentially serve as promising candidates for such a purpose. The observed flexibility of WaaL in recognition of the lipid moiety of the donor reduces hydrophobicity problems and could potentially facilitate co-crystallization of WaaL with more easily accessible substrates.
In summary, the present work represents the first study on WaaL using a variety of synthesized donor substrates. This work provided some valuable insights into the details of WaaL ligation activity. Future studies focusing on the in vitro reconstitution of whole LPS biosynthesis by the combined use of glycosyltransferases, Wzy, Wzz, and WaaL are currently under investigation in our laboratory.
Supplementary Material
Acknowledgment
We are grateful to C. Whitfield (University of Guelph) for sharing the protocol for the isolation of lipid A-core.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 GM085267 (to P. G. W.).

This article contains supplemental text, Table S1, and Figs. S1–S5.
- IM
- inner membrane
- PP
- pyrophosphate
- Und
- undecaprenyl
- Und-P
- undecaprenyl phosphate
- O-unit
- O-antigen repeating unit
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Whitfield C. (2006) Biosynthesis and assembly of capsular polysaccharides in Escherichia coli. Annu. Rev. Biochem. 75, 39–68 [DOI] [PubMed] [Google Scholar]
- 2. Nikaido H. (2003) Molecular basis of bacterial outer membrane permeability revisited. Microbiol. Mol. Biol. Rev. 67, 593–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Nesper J., Lauriano C. M., Klose K. E., Kapfhammer D., Kraiss A., Reidl J. (2001) Characterization of Vibrio cholerae O1 El TorgalU and galE mutants: influence on lipopolysaccharide structure, colonization, and biofilm formation. Infect Immun. 69, 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Raetz C. R., Whitfield C. (2002) Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71, 635–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung H. S., Raetz C. R. (2011) Dioxygenases in Burkholderia ambifaria and Yersinia pestis that hydroxylate the outer Kdo unit of lipopolysaccharide. Proc. Natl. Acad. Sci. U.S.A. 108, 510–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Raetz C. R., Reynolds C. M., Trent M. S., Bishop R. E. (2007) Lipid A modification systems in Gram-negative bacteria. Annu. Rev. Biochem. 76, 295–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Woodward R., Yi W., Li L., Zhao G., Eguchi H., Sridhar P. R., Guo H., Song J. K., Motari E., Cai L., Kelleher P., Liu X., Han W., Zhang W., Ding Y., Li M., Wang P. G. (2010) In vitro bacterial polysaccharide biosynthesis: defining the functions of Wzy and Wzz. Nat. Chem. Biol. 6, 418–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cuthbertson L., Kos V., Whitfield C. (2010) ABC transporters involved in export of cell surface glycoconjugates. Microbiol. Mol. Biol. Rev. 74, 341–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Islam S. T., Gold A. C., Taylor V. L., Anderson E. M., Ford R. C., Lam J. S. (2011) Dual conserved periplasmic loops possess essential charge characteristics that support a catch-and-release mechanism of O-antigen polymerization by Wzy in Pseudomonas aeruginosa PAO1. J. Biol. Chem. 286, 20600–20605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tran A. X., Dong C., Whitfield C. (2010) Structure and functional analysis of LptC, a conserved membrane protein involved in the lipopolysaccharide export pathway in Escherichia coli. J. Biol. Chem. 285, 33529–33539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silhavy T. J., Kahne D., Walker S. (2010) The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2, a000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Freinkman E., Chng S. S., Kahne D. (2011) The complex that inserts lipopolysaccharide into the bacterial outer membrane forms a two-protein plug-and-barrel. Proc. Natl. Acad. Sci. U.S.A. 108, 2486–2491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chng S. S., Gronenberg L. S., Kahne D. (2010) Proteins required for lipopolysaccharide assembly in Escherichia coli form a transenvelope complex. Biochemistry 49, 4565–4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chimalakonda G., Ruiz N., Chng S. S., Garner R. A., Kahne D., Silhavy T. J. (2011) Lipoprotein LptE is required for the assembly of LptD by the β-barrel assembly machine in the outer membrane of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 108, 2492–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pérez J. M., McGarry M. A., Marolda C. L., Valvano M. A. (2008) Functional analysis of the large periplasmic loop of the Escherichia coli K-12 WaaL O-antigen ligase. Mol. Microbiol. 70, 1424–1440 [DOI] [PubMed] [Google Scholar]
- 16. Abeyrathne P. D., Lam J. S. (2007) WaaL of Pseudomonas aeruginosa utilizes ATP in in vitro ligation of O-antigen onto lipid A-core. Mol. Microbiol. 65, 1345–1359 [DOI] [PubMed] [Google Scholar]
- 17. Schild S., Lamprecht A. K., Reidl J. (2005) Molecular and functional characterization of O-antigen transfer in Vibrio cholerae. J. Biol. Chem. 280, 25936–25947 [DOI] [PubMed] [Google Scholar]
- 18. Hug I., Couturier M. R., Rooker M. M., Taylor D. E., Stein M., Feldman M. F. (2010) Helicobacter pylori lipopolysaccharide is synthesized via a novel pathway with an evolutionary connection to protein N-glycosylation. PLoS Pathog. 6, e1000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Li M., Shen J., Liu X., Shao J., Yi W., Chow C. S., Wang P. G. (2008) Identification of a new α1,2-fucosyltransferase involved in O-antigen biosynthesis of Escherichia coli O86:B7 and formation of H-type 3 blood group antigen. Biochemistry 47, 11590–11597 [DOI] [PubMed] [Google Scholar]
- 20. Yi W., Shao J., Zhu L., Li M., Singh M., Lu Y., Lin S., Li H., Ryu K., Shen J., Guo H., Yao Q., Bush C. A., Wang P. G. (2005) Escherichia coli O86 O-antigen biosynthetic gene cluster and stepwise enzymatic synthesis of human blood group B antigen tetrasaccharide. J. Am. Chem. Soc. 127, 2040–2041 [DOI] [PubMed] [Google Scholar]
- 21. Yi W., Yao Q., Zhang Y., Motari E., Lin S., Wang P. G. (2006) The wbnH gene of Escherichia coli O86:H2 encodes an α-1,3-N-acetylgalactosaminyl transferase involved in the O-repeating unit biosynthesis. Biochem. Biophys. Res. Commun. 344, 631–639 [DOI] [PubMed] [Google Scholar]
- 22. Wagner S., Klepsch M. M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K. J., Slotboom D. J., Persson J. O., de Gier J. W. (2008) Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U.S.A. 105, 14371–14376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Thomas J. G., Ayling A., Baneyx F. (1997) Molecular chaperones, folding catalysts, and the recovery of active recombinant proteins from E. coli. To fold or to refold. Appl. Biochem. Biotechnol. 66, 197–238 [DOI] [PubMed] [Google Scholar]
- 24. Guo H., Lokko K., Zhang Y., Yi W., Wu Z., Wang P. G. (2006) Overexpression and characterization of Wzz of Escherichia coli O86:H2. Protein Expr. Purif. 48, 49–55 [DOI] [PubMed] [Google Scholar]
- 25. Galanos C., Lüderitz O., Westphal O. (1969) A new method for the extraction of R lipopolysaccharides. Eur. J. Biochem. 9, 245–249 [DOI] [PubMed] [Google Scholar]
- 26. Rush J. S., Alaimo C., Robbiani R., Wacker M., Waechter C. J. (2010) A novel epimerase that converts GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia coli O157. J. Biol. Chem. 285, 1671–1680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Brown S., Zhang Y. H., Walker S. (2008) A revised pathway proposed for Staphylococcus aureus wall teichoic acid biosynthesis based on in vitro reconstitution of the intracellular steps. Chem. Biol. 15, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Brockhausen I., Riley J. G., Joynt M., Yang X., Szarek W. A. (2008) Acceptor substrate specificity of UDP-Gal GlcNAc-R β1,3-galactosyltransferase (WbbD) from Escherichia coli O7:K1. Glycoconj. J. 25, 663–673 [DOI] [PubMed] [Google Scholar]
- 29. Amor K., Heinrichs D. E., Frirdich E., Ziebell K., Johnson R. P., Whitfield C. (2000) Distribution of core oligosaccharide types in lipopolysaccharides from Escherichia coli. Infect. Immun. 68, 1116–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Palusiak A., Dzieciatkowska M., Sidorczyk Z. (2008) Application of two different kinds of sera against the Proteus penneri lipopolysaccharide core region in search of epitopes determining cross-reactions with antibodies. Arch. Immunol. Ther. Exp. (Warsz.) 56, 135–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Yokota S., Terashima M., Chiba J., Noguchi H. (1992) Variable cross-reactivity of Pseudomonas aeruginosa lipopolysaccharide code-specific monoclonal antibodies and its possible relationship with serotype. J. Gen. Microbiol. 138, 289–296 [DOI] [PubMed] [Google Scholar]
- 32. al-Hendy A., Toivanen P., Skurnik M. (1991) Expression cloning of Yersinia enterocolitica O:3 rfb gene cluster in Escherichia coli K12. Microb. Pathog. 10, 47–59 [DOI] [PubMed] [Google Scholar]
- 33. Bastin D. A., Romana L. K., Reeves P. R. (1991) Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O-antigen of an E. coli O111 strain. Mol. Microbiol. 5, 2223–2231 [DOI] [PubMed] [Google Scholar]
- 34. Goldberg J. B., Hatano K., Meluleni G. S., Pier G. B. (1992) Cloning and surface expression of Pseudomonas aeruginosa O-antigen in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 89, 10716–10720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meredith T. C., Mamat U., Kaczynski Z., Lindner B., Holst O., Woodard R. W. (2007) Modification of lipopolysaccharide with colanic acid (M-antigen) repeats in Escherichia coli. J. Biol. Chem. 282, 7790–7798 [DOI] [PubMed] [Google Scholar]
- 36. Valvano M. A. (2008) Undecaprenyl phosphate recycling comes out of age. Mol. Microbiol. 67, 232–235 [DOI] [PubMed] [Google Scholar]
- 37. Lairson L. L., Henrissat B., Davies G. J., Withers S. G. (2008) Glycosyltransferases: structures, functions, and mechanisms. Annu. Rev. Biochem. 77, 521–555 [DOI] [PubMed] [Google Scholar]
- 38. Ruan X., Loyola D. E., Marolda C. L., Perez-Donoso J. M., Valvano M. A. (2011) The WaaL O-antigen Lipopolysaccharide ligase has features in common with metal ion-independent inverting glycosyltransferases. Glycobiology, in press [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.