Background: Nfe2 restricts Gcm1 expression, placental vascularization, and embryonic growth.
Results: Nfe2 induces hypoacetylation of JunD, thus limiting JunD binding to the Gcm1 promoter (at −1441).
Conclusion: In trophoblast cells Nfe2 negatively controls Gcm1 expression and syncytiotrophoblast formation by repressing JunD-binding activity.
Significance: This identifies a novel, acetylation dependent interaction of bZip transcription factors regulating placental and embryonic development.
Keywords: AP1 Transcription Factor, Development, Embryo, Placenta, Trophoblast, Gcm1, JunD, Nfe2, Acetylation, Syncytiotrophoblast Differentiation
Abstract
We recently demonstrated that the bZip transcription factor nuclear factor erythroid-derived 2 (Nfe2) represses protein acetylation and expression of the transcription factor glial cell missing 1 (Gcm1) in trophoblast cells, preventing excess syncytiotrophoblast formation and permitting normal placental vascularization and embryonic growth. However, the Gcm1 promoter lacks a Nfe2-binding site and hence the mechanisms linking Nfe2 and Gcm1 expression remained unknown. Here we show that Nfe2 represses JunD DNA-binding activity to the Gcm1 promoter during syncytiotrophoblast differentiation. Interventional studies using knockdown and knockin approaches show that enhanced JunD DNA-binding activity is required for increased expression of Gcm1 and syncytiotrophoblast formation as well as impaired placental vascularization and reduced growth of Nfe2−/− embryos. Induction of Gcm1 expression requires binding of JunD to the −1441 site within the Gcm1 promoter, which is distinct from the −1314 site previously shown to induce Gcm1 expression by other bZip transcription factors. Nfe2 modulates JunD binding to the Gcm1 promoter via acetylation, as reducing JunD acetylation using the histone acetyltransferase inhibitor curcumin reverses the increased JunD DNA-binding activity observed in the absence of Nfe2. This identifies a novel mechanism through which bZip transcription factors interact. Within the placenta this interaction regulates Gcm1 expression, syncytiotrophoblast formation, placental vascularization, and embryonic growth.
Introduction
We recently identified a novel function of the bZip transcription factor Nfe2 in trophoblast cells (1). In trophoblast cells Nfe2 prevents excess syncytiotrophoblast formation during the later pregnancy stages in the mouse (after day 14.5 p.c.,2 corresponding to the 3rd trimester), thus ensuring sufficient vascularization of the placenta and normal embryonic growth (1). This new function of Nfe2 may provide novel insight into mechanisms underlying placental dysfunction and intrauterine growth restriction. These are frequent but incompletely understood pregnancy complications associated not only with increased peripartal morbidity and mortality of the newborn, but also with diseases such as diabetes mellitus or cardiovascular diseases later on in adult life (2, 3). Hence, deciphering the mechanism through which Nfe2 modulates trophoblast differentiation and placental development and function may reveal new diagnostic or therapeutic targets for placental dysfunction and intrauterine growth restriction.
Nfe2 is the larger (45 kDa) subunit of the heterodimeric transcription factor NFE2. Until recently expression of Nfe2 was thought to be restricted to hematopoietic cells (cells of the megakaryocyte, erythrocyte, or mast cell lineage) (4). We identified a function of Nfe2 in nonhematopoietic cells (1). In trophoblast cells Nfe2 represses Gcm1 (also referred to as GCMa) activity (1). Gcm1 belongs to a unique family of transcription factors hallmarked by a highly conserved zinc-coordinated DNA-binding domain (5). Gcm1 is required for branching and syncytiotrophoblast fusion during placental development, and both loss and gain of Gcm1 function disturb syncytiotrophoblast formation and placental development (1, 6). Loss of Nfe2 function increases acetylation of histone H4 within the Gcm1 promoter and of Gcm1 itself, enhancing Gcm1 expression in murine placenta and trophoblast cells (1). Although these studies identified a novel function of Nfe2 as a repressor during syncytiotrophoblast differentiation, the positive gene regulators repressed by Nfe2 during normal trophoblast and placental differentiation remain unknown. Based on in silico analyses of the Gcm1 promoter the 6000-bp 5′ of the ATG lack potential Nfe2-binding sites, indicating that Nfe2 must regulate Gcm1 expression through an indirect mechanism. Interaction of Nfe2 with CBP or other bZip transcription factors has been reported and may be mechanistically linked to the developmental function of Nfe2 (1, 7, 8).
Indeed, bZip transcription factors have established functions for Gcm1 expression and placental development. Thus, the bZip transcription factors CREB and OASIS stimulate Gcm1 expression in vitro and enhance expression of syncytiotrophoblast markers in vivo (9). The relevant cAMP response element (CRE)-binding site was located at −1337 within the mouse Gcm1 promoter. In vivo, expression of CREB was replaced by expression of OASIS around late midgestation (E13.5), at which time the hemochorial placenta is fully established. This demonstrates a dynamic regulation of bZip transcription factors during placental development.
Other bZip transcription factors with known function during placentation belong to the AP-1 proteins. Knockout studies in mice revealed embryonic lethal placental defects in JunB and Fra1-deficient embryos (10, 11), whereas placental development was normal in mice lacking other AP-1 proteins. Placental JunB or Fra1 deficiency impairs vascularization of the labyrinthine layer, causing early embryonic lethality between days 8.5 and 10.5 p.c. (10, 11).
Whether Nfe2 regulates syncytiotrophoblast formation and vascularization of the placental labyrinthine layer through an interaction with TORC1, OASIS, JunB, Fra1, or yet to be identified transcription factors remained hitherto unknown. Within the current study we explored the interaction partners of Nfe2 during placental development, identifying a so far unknown interaction of Nfe2 with JunD. During placental development Nfe2 regulates through an acetylation dependent mechanism JunD-binding activity to the −1441 site of the Gcm1 promoter, regulating syncytiotrophoblast formation, placental vascularization, and embryonic growth.
EXPERIMENTAL PROCEDURES
Materials
Heparin sodium salt from porcine intestinal mucosa, human acidic fibroblast growth factor 4 (aFGF4), trichostatin A (TSA), Polybrene, and puromycin dihydrochloride from Streptomyces alboniger were obtained from Sigma. All cell culture media, FBS, glutamine, penicillin/streptomycin mixture, neuromycin, nonessential amino acids, and sodium pyruvate were from PAA, Cölbe, Germany. P24 tag antigen ELISA and the CHIP assay kit were from BioCat, Heidelberg, Germany. ECL reagent was from Amersham Biosciences. Murine Nfe2 and JunD shRNA constructs were from Open Biosystems, Huntsville, AL. GoTaq DNA polymerase, AP-1, and CREB consensus oligonucleotides for EMSA were obtained from Promega. Rabbit polyclonal IgG antibodies against Nfe2, c-Jun, JunD, JunB, FosB, and Fra1 were purchased from Santa Cruz Biotechnology. Rabbit polyclonal IgG antibody against acetylated lysine was purchased from Cell Signaling Technology, Boston, MA. CellMask Deep Red stain was bought from Invitrogen. QuikChange XL Site-directed Mutagenesis kit was purchased from Stratagene. All primers were synthesized by Thermo Scientific, Langenselbold, Germany.
Plasmids
Murine JunD expression (pCMV-JunD-SPORT6) and JunD knockdown (pLKO1-JunD) constructs were purchased from Open Biosystems. The JunD expression plasmid pLV-JunD was generated by replacing the EGFP fragment (EcoRI/XhoI) of the plasmid pLV-EGFP (kindly provided by Masahito Ikawa, Osaka, Japan) (12) with the EcoRI/XhoI fragment of pCMV-JunD-SPORT6. To demonstrate specificity of the knockdown experiment six nucleotides within the JunD shRNA hybridization sequence (5′-ACT CGA GTT GAG CAG CCC AAG ATC CGG CTT TTT G-3′) were replaced in pLV-JunD (5′-ACT CGA GTT GAG CAA CCG AAA ATC CGC CTG TTC G-3′) by site-directed mutagenesis. This resulted in silent mutations yielding a JunD resistant to the shRNA used (pLV-JunD-shRes).
Reporter constructs consisting of various fragments of the murine Gcm1 promoter (starting at −4041, −2892, −1637, and −396 relative to ATG) linked to the luciferase coding sequence (Promega, Mannheim, Germany) have been previously described (9). Gcm1 promoter constructs containing mutated AP-1-binding sites were generated by site-directed mutagenesis at positions −1572/−1571 (AC to TG), −1434/−1433 (TC to AG), and −1310/−1309 (AC to TG). The Gcm1 −1637 PGL2 luciferase reporter construct was used as the template. The following primers were used: gcm-1 −1572, AC to TG (5′-GGT TGT CTG TGA TGC TTG TGT GTA CAG TTT TAT TTC TCA-3′ and 5′-TGA GAA ATA AAA CTG TAC ACA CAA GCA TCA CAG ACA ACC-3′); gcm-1 −1434, TC to AG (5′-CCC AGC GTG GCC TCT GAT AGA TTG TAA GAA TTC AGT CAA-3′ and 5′-TTG ACT GAA TTC TTA CAA TCT ATC AGA GGC CAC GCT GGG-3′); gcm-1 −1310, AC to TG (5′-GCC TGG TCT ATA GGA GGT CTG TCC CAT CTC AAA AAA CTA-3′ and 5′-TAG TTT TTT GAG ATG GGA CAG ACC TCC TAT AGA CCA GGC-3′). All mutations were generated using the QuikChange® II Site-directed Mutagenesis kit (Stratagene) and identity of all mutations was confirmed by sequencing.
Mice
Nfe2-deficient mice were kindly provided by R. Shivdasani (Harvard Medical School, Cambridge, MA). All animals were housed and the experiments were performed at the Interdisciplinary Biomedical Research institution (IBF) of the University of Heidelberg, Germany. For in vivo intervention studies we used TSA (15 μg/mouse (13)) or valproic acid (sodium salt, 155 mm, 25 ml/kg of body weight (14)) as HDAC (histone deacetylase) inhibitors and epigallocatechin-3-gallate (50 mg/kg in saline (15)) or curcumin (0.5 mg/kg (16)) as HAT inhibitors. Animal experiments were conducted following standards and procedures approved by the local Animal Care and Use Committee (Regierungspräsidium Karlsruhe, Germany).
Cell Culture
HEK293T and HEK293 cells were kindly provided by Dr. Peter Seeburg (Max-Planck-Institute for Medical Research, Heidelberg, Germany). They were used to generate lentiviral particles and perform the luciferase assay. These cells were maintained in DMEM supplemented with 4.5 g/ml of glucose, 10% FBS, 2 mm l-glutamine, 50 μg/ml of penicillin/streptomycin, and 4 μg/ml of neomycin at 37 °C in a humidified incubator with 5% CO2. For transfection experiments medium without antibiotics was used.
Mouse trophoblast stem (TS) cells were obtained from J. Rossant (Hospital for Sick Children, Toronto, Ontario, Canada) and were induced to differentiate in plain TS cell medium (RPMI 1640 supplemented with 25 mm HEPES, 2.0 g/liter of NaHCO3, 20% FBS, 1 mm sodium pyruvate, 100 μm β-mercaptoethanol, 2 mm l-glutamine, and 50 μg/ml of penicillin/streptomycin) or maintained as stem cells in complete TS cell medium (30% plain TS cell medium plus 70% fibroblast-conditioned TS cell medium, 25 ng/ml of FGF4, and 1 μg/ml of heparin) as described previously (1, 17–19). In some experiments TS cells were treated with 100 nm TSA or 2.5 mm sodium butyrate for HDAC inhibition (20) or 30 nm curcumin or 5 μm epigallocatechin-3-gallate for HAT inhibition (21–23).
In Vitro Syncytium Formation
Syncytiotrophoblast formation was monitored in nearly confluent TS cells differentiated for 6 days, washed with PBS, incubated with 2.5 μm CellMask Deep Red for 5 min at 37 °C, and then fixed for 3 min in 3.75% formaldehyde at RT. Cells were counterstained with 0.5 μg/ml of DAPI in PBS for 10 min at RT in the dark. For quantification of syncytiotrophoblasts at least 30 random images per slide were captured (University of Texas Health Science Center San Antonio ImageTool software) and a blinded investigator determined the frequency of syncytial cells in these random images (1).
Production of Lentiviral Particles
Highly purified plasmid DNA prepared by alkaline lysis preparation was used for lentiviral particle production. Vesicular stomatitis virus-G pseudotyped lentiviral particles were generated as described (1, 24). Briefly, HEK 293T cells were transfected with pLKO1-JunD, pLV-JunD, or pLV-JunD-shRes plasmids together with the packaging plasmid (pCMV-dR8.91) and vesicular stomatitis virus-G-expressing plasmid using the standard calcium phosphate method. Lentiviral particles were harvested 2 and 3 days after transfection and concentrated by ultracentrifugation (50,000 × g, 2 h, 2 times). After resuspension in HBSS buffer the viral particle concentration was determined by measuring the p24 gag antigen by ELISA according to the manufacturer's instruction (BioCat, Heidelberg, Germany). Viral particles were stored at −80 °C.
Luciferase Assay
The HEK293 cells were transfected in 60-mm dishes with 1 μg of luciferase reporter construct and 0.1 μg of CMV driven expression vectors using a standard calcium phosphate method. At 48 h post-transfection, cells were harvested for luciferase assays using the luciferase assay system according to the manufacturer's instruction.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear proteins were isolated and binding activity of CREB and AP-1 was determined as described previously (1, 25). Briefly, for isolation of nuclear proteins cells were washed in PBS and resuspended in 200 μl of ice-cold buffer A (10 mm HEPES-KOH, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT, 0.2 mm PMSF). For tissue extracts the tissue samples were homogenized in 400 μl of ice-cold buffer B (10 mm HEPES-KOH, pH 7.9, 1.5 mm MgCl2, 10 mm KCl, 0.5 mm DTT 0.2 mm PMSF, 1 mm EDTA, 0.6% Nonidet P-40), vortexed for 10 s, and incubated on ice for 10 min. Samples were centrifuged at 12,000 × g for 2 min and the supernatant was discarded. The pellet was either resuspended in 100 μl of buffer C (cell lysates: 20 mm HEPES-KOH, pH 7.9, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5 mm DTT, 0.2 mm PMSF) or 200 μl of buffer D (tissue lysates: 20 mm HEPES-KOH, pH 7.9, 25% glycerol, 420 mm NaCl, 1.5 mm MgCl2, 0.2 mm EDTA, 0.5 mm DTT, 0.2 mm PMSF, 0.2 mm benzamidine, 5 μg/ml of leupeptin) and incubated on ice for 20 min. After centrifugation (12,000 × g, 5 min, 4 °C) the supernatant was transferred to a new tube, protein concentration was measured by colorimetric assay (BSA), and samples were quick-frozen at −80 °C. Binding of transcription factors to 1 ng of radiolabeled oligonucleotides (AP-1 consensus: 5′-CGC TTG ATG ACT CAG CCG GAA-3′, JunD-binding site at −1441 derived from Gcm1: 5′-G CGT GGC CTC TGA TTC ATT GTA AG-3′, JunD mutated (TC to AG), derived from Gcm1 −1441: 5′-G CGT GGC CTC TGA TAG ATT GTA AG-3′, CREB consensus: 5′-AGA GAT TGC CTG ACG TCA GAG AGC TAG-3′) was performed for 20 min at RT in 10 mm HEPES, pH 7.5, 0.5 mm EDTA, 100 mm KCl, 2 mm DTT, 2% glycerol, 4% Ficoll, 0.25% Nonidet P-40, 1 mg/ml of BSA, and 0.1 mg/ml of poly(dI/dC). Protein-DNA complexes were separated on 5% native polyacrylamide gels containing 3.25% glycerol and 0.5× Tris/boric acid/EDTA buffer. Identity of AP-1 subunits was determined by supershift experiments. To this end 4 μg of antibody against AP-1 subunits (Fra1, JunB, c-Jun, JunD, and FosB) were preincubated with 10 μg of protein extract for 20 min at RT before adding radiolabeled nucleotides.
Transduction of Preimplantation Embryos
Female Nfe2−/− or wild type (WT) mice were superovulated by intraperitoneal injection of pregnant mare's serum gonadotropin (5 units) followed by human chorionic gonadotropin (5 units) 48 h later and then mated with Nfe2−/− or WT males, respectively. Two-cell stage embryos were collected from the females at day 1.5 p.c. and incubated for 2 days to obtain blastocysts. The zona pellucida was removed with acidic Tyrode solution. Zona pellucida-free embryos were incubated individually in 50 μl of KSOM medium containing JunD with either shRNA or JunD expressing lentiviral particles (1 × 103 ng of p24/ml) for 4 h (1, 12). Transduced blastocysts were transferred into pseudo-pregnant females. The transfer day was counted as day 3.5 p.c. and analyses were performed at day 14.5 or 16.5 p.c. Embryos generated from blastocysts transduced with lentiviral particles generated from nonspecific shRNA or empty vector served as controls.
Morphological Analyses of Placenta
Placental tissues were obtained from Nfe2−/− or WT females after lentiviral transduction at day 14.5 p.c. Half of the placental tissue was fixed in 4% buffered formalin, whereas the other half was further divided and stored either in RNAlater or quick frozen in liquid nitrogen for protein isolation. The fixed tissue was embedded in paraffin, sectioned, and stained with hematoxylin and eosin. For vascular space quantification randomly selected microscopic fields (magnification ×10) of the labyrinthine region from 3 nonconsecutive placental sections (each section ≥10 images) were acquired at 1280 × 960 resolution using a digital camera (Nikon digital sight DS-U1) connected to a light microscope (Nikon Eclipse TE2000). We implemented an image analysis routine using NIS Elements AR2.30 imaging software. Briefly, after acquisition, the images underwent an automated analysis procedure and were given a color threshold to cover the area corresponding to the lumen of blood spaces. The coverage percentage was calculated as the ratio between the number of pixels covered by the area defined by the threshold and the overall number of pixels in the image (1). A blinded investigator performed analyses.
Transmission Electron Microscopic Analyses of Placenta
Placenta tissues obtained at day 16.5 p.c. were fixed with 2.5% glutaraldehyde, 2.5% polyvinylidone 25, 0.1 m sodium cacodylate, pH 7.4. After washing with 0.1 m sodium cacodylate buffer, pH 7.4, samples were post-fixed with 2% osmium tetroxide, 1.5% potassium ferrocyanide in 0.1 m sodium cacodylate buffer, pH 7.4, for 1 h, washed, contrasted en bloc with uranyl acetate, dehydrated with an ascending series of ethanol, and embedded in glycid ether 100-based resin. Ultrathin sections were cut with a Reichert ULTRACUT S ultramicrotome (Leica Microsystems, Wetzlar, Germany), contrasted with uranyl acetate and lead citrate, and viewed with an EM 10 CR electron microscope (Carl Zeiss NTS, Oberkochen, Germany).
Gene Knockdown and Knockin in Mouse Trophoblast Cells
Generation of stably transfected Nfe2 knockdown mouse trophoblast cells (Nfe2kd) has been previously described (1). Transient knockdown or knockin of JunD in control, Nfe2kd, or Nfe2kd JunDkd TS cells was performed by lentiviral transduction. Briefly, pLKO1-JunD, pLV-JunD-shRes, or pLV-JunD viral particles (5 × 105 TU/ml) were generated as described above and used to infect Nfe2kd (yielding Nfe2kdJunDkd cells), Nfe2kd JunDkd (yielding Nfe2kdJunDkdRes cells), or control (yielding Nfe2cJunD+ cells) TS cells, respectively, in the presence of 8 μg/ml of Polybrene in TS cell medium. The efficiency of the JunD knockdown or knockin was determined after 3 days of differentiation by RT-PCR and EMSA. In all experiments cells transduced with viral particles generated from nonspecific shRNA or empty pLV-CAG1.1 plasmid served as controls.
Quantitative Real Time PCR (qRT-PCR)
RNA was isolated from mouse placental tissue or TS cells. Placental tissues were stored initially in RNAlater and placed into TRIzol for RNA isolation, whereas TRIzol was added directly to TS cells. Following incubating with TRIzol (5 min at RT), 0.2 ml of chloroform/1 ml of TRIzol was added, mixed, and centrifuged (12,000 × g, 15 min, 4 °C). The aqueous phase containing the RNA was transferred to a fresh tube and RNA was precipitated by adding 0.5 ml of 2-propanol/1 ml of TRIzol reagent initially used. Samples were incubated at RT (10 min) and centrifuged (12,000 × g, 10 min, 4 °C). Supernatant was removed and RNA was washed by adding 1 ml of 75% ethanol/1 ml of TRIzol. Samples were vortexed, centrifuged (7,500 × g, 5 min, 4 °C), the RNA pellet was air dried (5 min), dissolved in 20 μl of diethyl pyrocarbonate/water, and incubated for 10 min at 55 °C. The RNA concentration was measured in a photometer, and RNA purity and integrity were verified on a 1.8% agarose gel. cDNA was synthesized according to the manufacturer's protocol (SuperScript First-Strand Synthesis System). All primers were custom synthesized by Thermo Fisher Scientific and quantitative RT-PCR was performed as described previously (1, 26).
The following primer sequences were used for amplification: actin, 5′-CTA GAC TTC GAG CAG GAG ATG G-3′ and 5′-GCT AGG AGC CAG AGC AGT AAT C-3′; Cebpa, 5′-CGC TGG TGA TCA AAC AAG AG-3′ and 5′-GTC ACT GGT CAA CTC CAG CA-3′; Gcm1, 5′-GAC TGC TCC ACA GAG GAA GG-3′ and 5′-GGA GAG CCA TAG GTG AGC AG-3′; Synb, 5′-TGA CCT TGA AGT GGG TAG GG-3′ and 5′-TGA CCT TGA AGT GGG TAG GG-3′; Syna, 5′-GGT TCG TCC TTG GGT TTT CT-3′ and 5′-GGA GTG GGG AGT CAA TGG T-3′; and Cstq, 5′-TTC CTG TGA CTG GTG CCA TA-3′ and 5′-CCA GAC CTC CAT TGT GCA AA-3′).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed on placental tissue extracts using the Epiquik ChIP assay kit for tissue following the manufacturer's instructions. Briefly, tissue samples were minced into small fragments using a scalpel and exposed to 1% formaldehyde in TS cell medium for cross-linking (20 min at RT on a rocking platform). Tissue extracts were washed in 125 mm glycine in TS cell medium, centrifuged, and the pellet was disaggregated using a Dounce homogenizer in 1 ml of homogenization buffer/200 mg of tissue. Following centrifugation the pellet was resuspended in lysis buffer CP3 containing protease inhibitors and incubated on ice for 10 min. DNA was sheared by sonication (5 intervals, each 7 s with 5-s breaks). Following centrifugation (8,000 × g, 10 min, 4 °C) the supernatant was diluted with buffer CP4, an aliquot was removed as “input” control, and from the remaining solution proteins were precipitated using rabbit polyclonal anti-JunD IgG antibody. Following incubation (60 min at RT) supernatants were removed and the precipitates were washed 6 times (buffer CP1). DNA was released using proteinase K in buffer CP5 (15 min at 65 °C) and then captured, washed, and finally eluted using spin columns. Purified DNA was stored at −20 °C until final analyses. Recovered DNA was analyzed by PCR using the following primers for the various mouse Gcm1 promoter regions: −3150 to −2950 (5′-GAA GAA ACC ACC TAA AGC TA-3′ and 5′-GGG AAG ACT CAC CCA ACT CA-3′), −2930 to −2720 (5′-AGG ATG CTG TAT AAG CCC TTC-3′ and 5′-CCT CAG GCA TGC TAT GGG TGA-3′), −2140 to −1930 (5′-TGG AAC ACT AGT TGA AAT AG-3′ and 5′-GCA TTC ATA ACC CAG GCA TC-3′), −1910 to −1720 (5′-CAG AAT GTA ATA TCT AGG GA-3′ and 5′-GGA GTA GCT GAC CTT ACC TG-3′), −1630 to −1460 (5′-ACG GAG GCG ACA AAG AC-3′ and 5′-CTA AAT TAC GCT TGA TGC GC-3′), −1450 to −1300 (5′-CAG CGT GGC CTC TGA TTC AT-3′ and 5′-GAG ATG GGA GTG ACC TCC TA-3′), and −1100 to −900 (5′-AAA AAC CCT TCG ACT CAG AG-3′ and 5′-TTA TCA TTG TGG CTA ATC CA-3′). Cycling conditions were 35 cycles with an annealing temperature of 58 °C.
Statistical Analysis
All in vitro experiments were performed at least three times in duplicates. All data were expressed as mean ± S.E. Statistical analyses were performed using Student's t test or analysis of variance as appropriate. Post hoc comparisons of analysis of variance were corrected using the method of Tukey. Excel and StatistiXL software were used for all statistical analyses. Statistical significance was accepted at p < 0.05.
RESULTS
Nfe2 Modulates AP-1 Binding in Murine Placenta
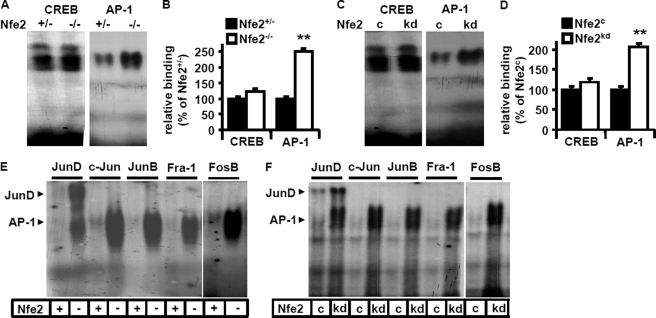
We have recently shown that Nfe2 suppresses Gcm1 expression and syncytiotrophoblast formation in trophoblast cells. Considering the negative gene regulatory function of Nfe2 in regard to Gcm1 and the absence of a Nfe2-binding site within the first 6000 bp of the Gcm1 promoter (both in humans and mice) we hypothesized that bZip transcription factor Nfe2 regulates Gcm1 indirectly, potentially by interacting with other bZip transcription factors. To this end we analyzed binding activity to two bZip transcription factor-binding sites, which have been previously shown to regulate placental development and/or Gcm1 expression (CREB and AP-1) (9–11, 27). We observed a marked induction of AP-1 DNA-binding activity (251 versus 100% in Nfe2+/− embryos, p < 0.001), but not of CREB DNA-binding activity (121 versus 100% in Nfe2+/− embryos, p = 0.8, Fig. 1, A and B), in placental tissue nuclear extracts obtained from day 14.5 p.c. Nfe2−/− murine placenta. Next, we performed EMSA using cellular nuclear extracts obtained from control (Nfe2c) and Nfe2 knockdown (Nfe2kd) trophoblast cells differentiated for 3 days. Again, AP-1 DNA-binding activity (206 versus 100% in Nfe2c, p < 0.01), but not CREB DNA-binding activity (117 versus 100% in Nfe2c, p = 0.12, Fig. 1, C and D), was markedly enhanced in Nfe2kd samples. These results suggest that in the absence of Nfe2 expression of Gcm1 is induced through an AP-1-dependent mechanism.
FIGURE 1.
Nfe2 inhibits JunD-binding activity in murine placenta. A–D, enhanced AP-1-binding activity in the absence of Nfe2. Representative EMSA images of protein lysates obtained from Nfe2+/− and Nfe2−/− placenta (A) and cell lysates obtained from 3-day differentiated control (Nfe2c) or Nfe2-deficient (Nfe2kd) TS cells (C) using radiolabeled oligonucleotides for the detection of AP-1 or CREB. Bar graph summarizing results of placental (B, n = 10) and trophoblast cell (D, 5 independent repeat experiments) analyses. E and F, binding of the JunD subunit is predominately increased in the absence of Nfe2. Analyses of AP-1 subunits (JunD, c-Jun, JunB, Fra1, or FosB) by supershift using protein lysates obtained from Nfe2+/− (+) or Nfe2−/− (−) placenta (E) or from 3-day differentiated Nfe2 control (c) or Nfe2-deficient (kd) TS cells (F). Mean value ± S.E., **, p < 0.01 (t test).
Nfe2 Modulates JunD Binding in Murine Placenta and Trophoblast Cells
Given the strong induction of AP-1-binding activity in the absence of Nfe2 and the established role of some AP-1 family members (JunB and Fra1) during placental development (10, 11, 27) we next performed supershift analyses. These revealed enhanced binding activity particularly of the JunD subunit in the absence of Nfe2 both ex vivo (Nfe2−/−, Fig. 1E) and in vitro (Nfe2kd, Fig. 1F). No relevant contribution of c-Jun, JunB, Fra1, or FosB to the enhanced AP-1-binding activity could be detected. Thus, in the absence of Nfe2, binding activity of JunD is markedly increased. This indicates that Nfe2 may reduce Gcm1 expression and trophoblast syncytium formation by repressing JunD, an AP-1 protein not previously known to be relevant for placental development.
JunD Is Required for Nfe2-dependent Syncytiotrophoblast Formation in Vitro
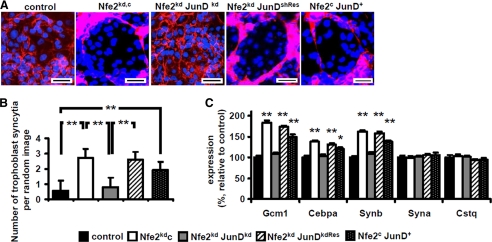
To explore whether enhanced syncytiotrophoblast formation in the absence of Nfe2 depends on JunD we reduced expression of JunD in trophoblast cells in vitro. Consistent with our previous observation, syncytiotrophoblast formation and expression of Gcm1, Cebpa, and Synb were increased in Nfe2-deficient trophoblast cells (Fig. 2, A–C) (1). Following inhibition of JunD using shRNA (supplemental Fig. S1) the frequency of syncytial trophoblast cells in 6-day differentiated TS cells was normalized (2.7 syncytial trophoblast cells/random image in Nfe2kd versus 0.8 in Nfe2kdJunDkd, p < 0.01, Fig. 2, A and B). Likewise, expression of Gcm1 (109 versus 183%, p < 0.001), Cebpa (103 versus 138%, p < 0.001), and Synb (108 versus 162%, p < 0.001, Fig. 2C) were normalized in Nfe2 and JunD double knockdown trophoblast cells as compared with Nfe2kd cells after 3 days of differentiation. In agreement with our previous observation, other differentiation markers, e.g. Syna or Cstq, were not dependent on Nfe2 or JunD levels in trophoblast cells (Fig. 2C)(1).
FIGURE 2.
The interaction of JunD and Nfe2 regulates syncytiotrophoblast formation in vitro. A–C, reduction of JunD expression in Nfe2-deficient trophoblast cells (Nfe2kdJunDkd, gray bars) reduces syncytiotrophoblast formation and expression of syncytiotrophoblast markers, which are characteristically increased in Nfe2-deficient (Nfe2kdc, white bars) trophoblast cells, to levels observed in control trophoblast cells (control, black bars). Overexpression of a shRNA-resistant JunD in Nfe2kdJunDkd trophoblast cells reverses the phenotype to that observed in Nfe2kdc cells (Nfe2kd JunDkdRes cells; increased syncytiotrophoblast formation and increased expression of Gcm1, Cebpa, and Synb, striped bars) and isolated gain of JunD function (Nfe2c JunD+) is sufficient to increase syncytiotrophoblast formation and expression of Gcm1, Cebpa, and Synb (dotted bars). Representative images of 6-day differentiated trophoblast cells stained with CellMask (red) and counterstained with DAPI (blue) (A, scale bar: 20 μm) and the bar graph summarizing results (B). C, bar graph summarizing results of quantitative RT-PCR analyses of the syncytiotrophoblast markers Gcm1, Cepba, Synb, Syna, and Cstq. Mean value ± S.E., **, p < 0.01 (analysis of variance).
Additional experiments were conducted to confirm the role of JunD in regulating syncytiotrophoblast formation. First, transient overexpression of a shRNA-resistant JunD mutant in Nfe2kdJunDkd trophoblast cells (Nfe2kd JunDkdRes) reversed the phenotype to that observed in single Nfe2kd trophoblast cells. Thus, syncytiotrophoblast formation increased to 2.6 per random image (Fig. 2, A and C) and the expression levels of Gcm1 (173%), Cebpa (131%), and Synb (158%) increased to levels not different from those in single Nfe2kd trophoblast cells (Fig. 2C). Second, isolated overexpression of JunD in trophoblast cells (Nfe2c JunD+) increased syncytiotrophoblast formation (to 1.93 syncytial trophoblast cells per random image versus 0.56 in Nfe2c, p < 0.01, Fig. 2, A and B) and induced expression of Gcm1 (149%, p < 0.001), Cebpa (120%, p < 0.001), and Synb (138%, p < 0.001, Fig. 2C). These experiments establish that gain of JunD function is sufficient for increased syncytiotrophoblast formation, confirming the mechanistic relevance of increased JunD activity for the enhanced syncytiotrophoblast formation in the absence of Nfe2. Furthermore, these in vitro studies demonstrate that syncytiotrophoblast formation depends on a cell autonomous cross-talk between Nfe2 and JunD.
Nfe2 and JunD Coordinately Control Placental Vascularization and Syncytiotrophoblast Formation in Vivo
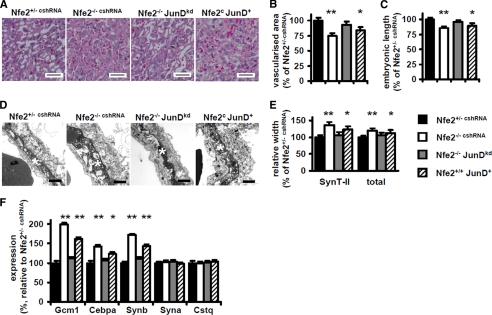
We have recently shown that Nfe2 restricts syncytiotrophoblast formation, thus regulating vascularization of the labyrinthine layer of the placenta and embryonic growth (1). Given the before mentioned results we next explored whether cross-coupling of Nfe2 with JunD in trophoblast cells represses Gcm1 in vivo. We reduced JunD expression in cells of the trophectodermal lineage by transducing Nfe2−/− blastocysts with JunD shRNA expressing lentiviral particles. This resulted in a marked reduction of JunD expression and of JunD DNA-binding activity within the placenta of Nfe2−/−JunDkd embryos as compared with Nfe2−/− embryos (see supplemental Fig. S2). Reduction of JunD expression normalizes placental vascularization within the labyrinthine layer of day 14.5 p.c. placental tissues (93% in Nfe2−/− JunDkd versus 75% in Nfe2−/− embryos, p < 0.001, Fig. 3, A and B). Restoration of placental vascularization was associated with a normalization of growth in Nfe2−/− JunDkd embryos (weight 97% and length 95% of that of control and Fig. 3C). Furthermore, electron microscopic analyses revealed a normalization of the total width of the feto-maternal placental barrier (108% in Nfe2−/− JunDkd versus 122% in Nfe2−/− embryos, p < 0.001, Fig. 3, D and E) and syncytiotrophoblast layer II (110 versus 138%, p < 0.001, Fig. 3, D and E). Using quantitative RT-PCR we observed a normalization of Gcm1 (112 versus 192%, p < 0.001), Cepba (107 versus 142%, p < 0.001), and Synb (110 versus 172%, p < 0.001, Fig. 3F) expression in Nfe2−/− placenta following JunD knockdown in cells of the trophectodermal lineage. In agreement with our previous observations we did not observe any morphological change in syncytiotrophoblast layers I and III, in endothelial cells, or in the expression of Syna or Cstq (Fig. 3F and data not shown) (1).
FIGURE 3.
The interaction of Nfe2 and JunD regulates placental vascularization and syncytiotrophoblast formation in vivo. A and B, impaired placental vascularization, which is observed in the absence of embryonic Nfe2 (Nfe2−/−,cshRNA, white bars), is normalized following suppression of JunD expression in cells of the trophectodermal cell lineage (compare Nfe2−/− JunDkd, gray bars, with control Nfe2+/− cshRNA, black bars). Isolated overexpression of JunD is sufficient to impair placental vascularization (Nfe2c JunD+, striped bars, compare with Nfe2+/− cshRNA, black bars). Representative hematoxylin and eosin-stained sections of placental tissue (A) and bar graph summarizing results of vascularization analyses (B, n = 6 per group). C, embryonic length of Nfe2−/− embryos is normalized following reduction of JunD expression in cells of the trophectodermal cell lineage, whereas isolated gain of JunD function (Nfe2c JunD+, striped bars) is sufficient to impair embryonic length. D and E, the characteristic enhanced width of syncytiotrophoblast layer II (SynT-II, marked with a white star) in the absence of Nfe2 (Nfe2−/− cshRNA), which is associated with an increased width of the total embryonic-maternal barrier (total), is reversed following reduction of JunD expression in Nfe2−/− embryos (Nfe2−/− JunDkd, gray bars). Isolated gain of JunD function is sufficient to increase the total width of the embryonic-maternal barrier and the width of syncytiotrophoblast layer II (Nfe2c JunD+, striped bars). For control Nfe2−/− embryos were infected with lentiviral particles expressing control shRNA (Nfe2−/− cshRNA). EM images of the maternal embryonic barrier (D) and bar graph summarizing results of EM analyses (E, 6 embryos per group, for each embryo at least 50 images were analyzed) are shown. F, reduction of JunD normalizes expression of syncytiotrophoblast markers in Nfe2−/− placenta (gray bars), whereas isolated overexpression of JunD is sufficient to increase expression of these markers (striped bars). Bar graph summarizing results of qRT-PCR analyses of Gcm1, Cebpa, Synb, Syna, and Cstq expression in placenta tissue samples (n = 6 embryos each) is shown. Mean value ± S.E., size bars are 50 μm (A) and 1 μm (D); *, p < 0.05, **, p < 0.01 (compared with Nfe2+/− cshRNA, analysis of variance).
We next explored whether gain of JunD function is sufficient for the changes observed in Nfe2−/− placenta. To this end we transduced wild type blastocysts with JunD expressing lentiviral particles (Nfe2+/+ JunD+). This resulted in a marked induction of JunD expression and JunD DNA-binding activity within the placenta (see supplemental Fig. S2). An isolated gain of JunD function had in essence the same consequences as the gain of JunD function observed in Nfe2+/+ placenta. Thus, placental vascularization within the labyrinthine layer of day 14.5 p.c. placental tissues was reduced (84% in Nfe2+/+ JunD+ versus 100% in Nfe2c embryos, p = 0.03, Fig. 3, A and B), embryonic growth was impaired (weight 94% and length 89% of that of controls, data not shown and Fig. 3C), the total width of the feto-maternal placental barrier (113% in Nfe2+/+ JunD+ versus 100% in Nfe2c embryos, p = 0.04, Fig. 3, D and E) and syncytiotrophoblast layer II (123% versus 100%, p = 0.03, Fig. 3, D and E) was increased. In addition, expression of Gcm1 (162%, p < 0.001), Cepba (124%, p < 0.03), and Synb (143%, p < 0.001, Fig. 3F) were increased in Nfe2+/+ JunD+ compared with Nfe2+/+ placenta. Hence, the enhanced syncytiotrophoblast formation, resulting in impaired vascularization of the labyrinthine layer and impaired growth of Nfe2-deficient embryos, depends on cross-coupling of Nfe2 with JunD.
Nfe2 Regulates JunD Binding to Gcm1 Promoter Particularly at −1441
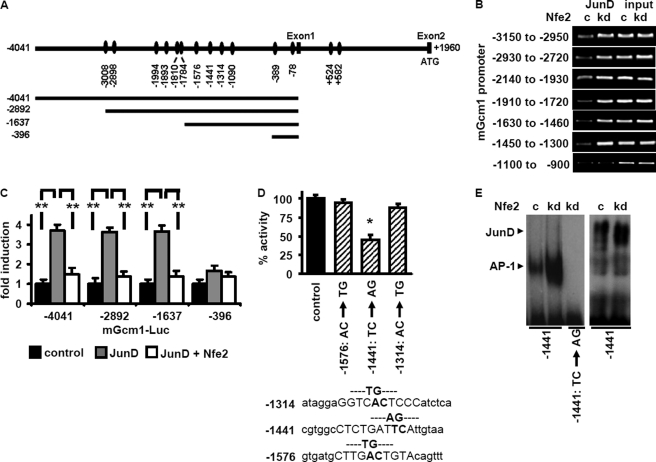
To define the promoter region required for JunD-dependent Gcm1 expression we conducted further in vitro experiments. Using computational analyses we identified several potential AP-1/JunD-binding sites with the Gcm1 promoter (Fig. 4A). ChIP analyses revealed increased JunD binding in 3 days differentiated Nfe2kd trophoblast cells to all in silico identified binding regions except the most proximal one (−1100 to −900, Fig. 4B).
FIGURE 4.
Nfe2 regulates JunD binding to the −1441 site within the mGcm1 promoter. A, graphical illustration of potential JunD bindings sites within the mGcm1 promoter, identified by in silico analyses (top), and of the promoter deletion constructs used (bottom). B, binding of JunD to the mGcm1 promoter is increased in the absence of Nfe2. Potential JunD-binding sites identified in silico were analyzed by ChIP assay. In the absence of Nfe2 enhanced binding of JunD can be detected in all regions except −1100 to −900. Representative images of semiquantitative RT-PCR using Nfe2 expressing control (c) or Nfe2-deficient (kd) trophoblast cells; input DNA serves as positive control. C, the −1637 to −396 Gcm1 promoter region contains a functional relevant JunD-binding site, which is negatively controlled by Nfe2. Analyses of promoter activity using mGcm1 promoter deletion constructs fused to a luciferase reporter gene and transfected into HEK293 cells. Cells were co-transfected with empty vector (control), JunD alone (JunD), or JunD and Nfe2 (JunD+ Nfe2) expression constructs. D, the functional JunD-binding site within the mGcm1 promoter is at position −1441. The three potential AP-1/JunD-binding sites within the −1637 to −1430 promoter fragment were inactivated by targeted mutagenesis. Only inactivation of the −1441 JunD-binding site results in impaired promoter activity. HEK293 cells were transfected with the Gcm1 −1637 promoter/luciferase reporter constructs (nonmutated control; −1576 AC/TG mutant; −1441 TC/AG mutant; and −1314 AC/TG mutant) and a JunD expression construct. E, JunD-binding activity determined using a probe corresponding to the mGcm1 promoter region −1426 to −1448 with the JunD-binding site at −1441 intact or mutated (TC → AG). Binding to the intact but not to the mutated Gcm1 probe is enhanced in Nfe2kd trophoblast cells (kd) compared with control (c) cells (left image). Supershift analyses demonstrating binding of JunD to the Gcm1-derived oligonucleotide probe (right image) are shown. Mean value ± S.E., **, p < 0.01 (C and D, analysis of variance).
To analyze the functional relevance of these promoter regions in the context of JunD binding and Gcm1 expression we next performed luciferase reporter assays in HEK293 cells. HEK293 cells were transfected with reporter constructs consisting of Gcm1 promoter fragments of various lengths cloned to the luciferase reporter gene (9). These cells were co-transfected either with empty control vector (control), a JunD expression construct (JunD), or both JunD and Nfe2 expression constructs (JunD + Nfe2). The −4041, −2982, and −1637 reporter constructs, but not the −396 reporter construct, contained inducible JunD-binding site(s). Hence, the functional relevant JunD-binding site must be within the −1637 to −396 promoter fragment. Of note, co-transfection with Nfe2 suppressed JunD-binding activity, directly demonstrating the repressive effect of Nfe2 in regard to JunD driven Gcm1 promoter activation.
Considering that the −1637 to −396 region contained inducible JunD-binding site(s) and that, based on the above ChIP analyses, JunD binds to the −1630 to −1460 and the −1450 to −1300 promoter regions, but not to the −1100 to −900 promoter fragment (see above, Fig. 4B), we hypothesized that the functional relevant JunD-binding site(s) must reside within the −1637 to −1300 region of the Gcm1 promoter. This region contains three potential JunD-binding sites (at −1576, −1441 and −1314). To determine their functional relevance for Gcm1 expression these sites were inactivated by targeted point mutations. JunD-mediated induction of the reporter construct containing the mutated JunD-binding site at −1441 was reduced to 45% (p = 0.02, Fig. 4D), whereas induction of the reporter construct containing the mutated JunD-binding site at −1576 or −1314 was not significantly reduced (94 and 88% compared with the control construct, p = 0.48 and 0.26, respectively, Fig. 4D).
To establish whether JunD binds to the endogenous Gcm1 promoter sequence we next performed EMSA using an oligonucleotide derived from the Gcm1 sequence (−1426 to −1448). Binding of nuclear extracts to this oligonucleotide was enhanced in the absence of Nfe2 (Nfe2kd, Fig. 4E). Using a mutated oligonucleotide (CT → AG) no binding activity could be detected (Fig. 4E). Supershift analyses confirmed binding of JunD to the Gcm1-derived oligonucleotide (Fig. 4E). Taken together, JunD binds to the Gcm1 promoter at −1441, which is required for JunD-mediated Gcm1 promoter activation.
Regulation of JunD-binding Activity by Nfe2 Is Acetylation Dependent
We have previously shown that the regulation of Gcm1 by Nfe2 is acetylation dependent (1). Given the current finding that JunD is crucially involved in the regulation of Gcm1 we next evaluated the role of acetylation for the Nfe2-dependent regulation of JunD.
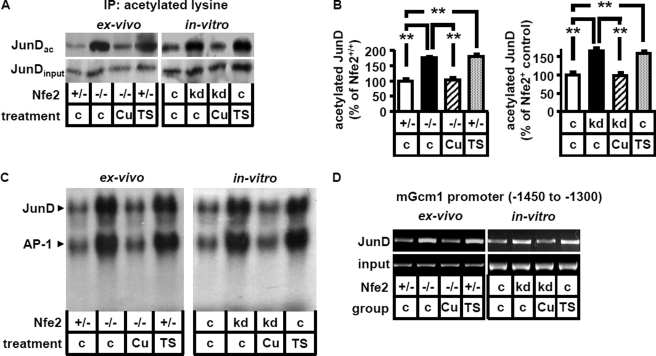
Using immunoprecipitation we detected a marked enhancement of JunD acetylation in Nfe2-deficient placenta (ex vivo: Nfe2−/−, c, Fig. 5, A and B) or trophoblast cells (in vitro: Nfe2kd, c, Fig. 5, A and B). Inhibition of acetylation using the HAT inhibitors curcumin or epigallocatechin-3-gallate reduced acetylation of JunD (103 versus 175% in Nfe2−/− placenta, p < 0.001, 98 versus 165% in Nfe2kd trophoblast cells, p < 0.001, Fig. 5, A and B, and data not shown) and JunD-binding activity in Nfe2-deficient placenta (Nfe2−/−) and trophoblast (Nfe2kd) cells (Fig. 5C). Conversely, enhancement of acetylation by inhibition of HDAC activity using trichostatin A (TSA, TS), valproic acid (in vivo, data not shown), or sodium butyrate (in vitro, data not shown) increased acetylation of JunD (182 versus 100% in Nfe2+/− placenta, p < 0.001, 158 versus 100% in Nfe2c trophoblast cells, p < 0.001, Fig. 5, b and C) and JunD-binding activity in Nfe2 expressing placenta (Nfe2+/+) and trophoblast cells (Nfe2+) (Fig. 5C). Modulation of JunD acetylation was associated with corresponding changes of JunD binding to the Gcm1 promoter. Thus, inhibition of acetylation using curcumin reduced JunD binding to the −1450 to −1300 Gcm1 promoter region in Nfe2-deficient placenta (Nfe2−/−, ex vivo) and in Nfe2kd trophoblast cells (in vitro), whereas enhancing acetylation in Nfe2 expressing trophoblast cells using TSA was sufficient to enhance JunD binding to the −1450 to −1300 Gcm1 promoter region both in Nfe2 expressing placenta (Nfe2+/−, ex vivo) and trophoblast cells (Nfe2c, in vitro, Fig. 5D). These data provide experimental evidence that Nfe2 regulates JunD DNA-binding activity to the Gcm1 promoter via acetylation.
FIGURE 5.
Modulation of JunD-binding activity by Nfe2 is acetylation dependent. A and B, acetylation of JunD is enhanced in the absence of Nfe2 (ex vivo, −/−; in vitro, kd). Enhanced acetylation is reversed following intervention with the HAT inhibitor curcumin (Cu), both in placental samples (ex vivo) and in trophoblast cells (in vitro). Treatment with the HDAC inhibitor TSA (TS) is sufficient to increase JunD acetylation both ex vivo and in vitro. Immunoprecipitation using an antibody against acetylated lysine and immunoblotting for JunD (JunDac, A, top). JunD immunoblot without prior immunoprecipitation serves as input control (JunDinput, A, bottom). B, bar graph summarizing results. C and D, enhanced JunD DNA-binding activity (supershift analyses, C) and JunD binding to the mGcm1 promoter (analyses of the −1450 to −1300 mGcm1 promoter region by ChIP assay, D) in the absence of Nfe2 is acetylation dependent. Enhanced JunD DNA-binding activity and binding to the mGcm1 promoter in the absence of Nfe2 (ex vivo, −/−; in vitro, kd) is reversed following treatment with the HAT inhibitor curcumin in placental samples (C and D, ex vivo) and trophoblast cells (C and D, in vitro). Conversely, increasing acetylation using the HDAC inhibitor TSA (TS) is sufficient to increase JunD DNA-binding activity and JunD binding to the mGcm1 promoter both ex vivo (C and D) and in vitro (C and D). Representative images of JunD supershifts (C) and ChIP assay using an anti-JunD antibody for DNA precipitation (D) are shown.
DISCUSSION
Previously we identified a novel function of the bZip transcription factor Nfe2 during trophoblast differentiation. We demonstrated that Nfe2 represses Gcm1 expression, a key regulator of syncytiotrophoblast differentiation and placental vascularization, in trophoblast cells through an unknown modus operandi. In the present study we establish that the AP-1 protein JunD is a positive gene regulator during syncytiotrophoblast differentiation. Nfe2 prevents excess syncytiotrophoblast formation, which would impair placental vascularization and embryonic growth (1) by limiting JunD DNA-binding activity. The Nfe2-dependent repression of JunD DNA-binding activity is mediated via acetylation.
The acetylation dependent cross-coupling of Nfe2 with JunD during trophoblast differentiation provides a novel mechanism controlling Gcm1 expression, syncytiotrophoblast formation, placental vascularization, and embryonic growth. A function of other members of the AP-1 protein family, namely JunB and Fra1, for placental development has been previously described (10, 11). The function of JunD must be different from JunB or Fra1. Thus, loss of JunB or Fra1 function results in embryonically lethal defects, whereas in mice with a gain of JunB or Fra1 function no embryonic phenotype has been reported (28). Contrary, loss of JunD function has no impact on embryonic development (29), whereas gain of function impairs placental vascularization and embryonic growth (this study).
In contrast to the nonlethal consequences of increased JunD-binding activity in Nfe2−/− embryos after day 14.5 p.c. the absence of JunB or Fra1 results in a lethal placental defect associated with defective yolk sac vascularization during establishment of the hemochorial placenta (day 8.5 p.c. in JunB−/− and 10.5 p.c. in Fra1−/− embryos) (10, 11). Thus, whereas loss of function of JunB and Fra1 results in early fetal loss, bZip transcription factors Nfe2 and JunD coordinately regulate placental development and embryonic growth at later developmental stages. Such diverse functions of distinct Jun family proteins likely reflect their ability to form various heterodimers with other bZiP family proteins resulting in either positive or negative regulation of target genes (28, 30). The newly identified interaction of JunD and Nfe2 expands our knowledge about the role of bZip transcription factors during placental development and demonstrates that this expanding network conveys crucial roles for placentation at various developmental stages.
Transcriptional activation of AP-1 proteins is regulated by MAPKs, in particular by the c-Jun NH2-terminal kinases (JNKs) and the extracellular signal-regulated kinases (ERKs), which phosphorylate AP-1 proteins (31, 32). The molecular mechanism of transcriptional activation of AP-1 proteins by MAPKs remained unknown until recently. Aguilera and colleagues (27) demonstrated that transcriptional activation of the AP-1 protein c-Jun is linked to increased histone acetylation within AP-1-dependent promoters. Nonphosphorylated c-Jun recruits the Mbd3/nucleosome remodeling and histone deacetylation (NuRD) repressor complex through an interaction with its transactivation domain, resulting in gene repression of AP-1 target genes. This repression is relieved by JNK-mediated c-Jun NH2-terminal phosphorylation. In the current study we demonstrate that DNA-binding and transcriptional activity of JunD is likewise modulated by acetylation, indicating that the mechanism identified by Aguilera and colleagues (27) is not specific for c-Jun. We previously demonstrated that acetylation of histone H4 within the Gcm1 promoter is modulated by Nfe2 in trophoblast cells and placenta (1), which is entirely consistent with the observation made by Aguilera et al. (27). In addition, we demonstrate now that the AP-1 protein JunD itself is modulated by acetylation. Whether other AP-1 proteins are directly modulated by acetylation and the relative importance of either histone or AP-1 protein acetylation for transcriptional activation needs to be explored in future studies.
Whether Nfe2-dependent acetylation of JunD depends on MAPK activity remains currently unknown. Unlike c-Jun, JunD lacks a functional JNK docking site within the NH2-terminal region (28). However, following heterodimerization with c-Jun, the serine residues of JunD can be phosphorylated (33), indicating that MAPK-mediated JunD phosphorylation may be involved in the regulation of acetylation. However, whereas regulation of Nfe2 by JNK has been demonstrated (34), the reverse, e.g. regulation of MAPK activity by Nfe2, has to the best of our knowledge not been reported. Hence the mechanism underlying Nfe2-dependent regulation of acetylation remains presently unknown. Based on previous and the current findings we propose a MAPK independent mechanism through which Nfe2 may inhibit acetylation. Interaction of Nfe2 with CBP in hematopoietic and trophoblast cells is established (1, 7, 35). CBP/p300 are coactivators generally influencing gene expression by functioning as scaffolding proteins facilitating protein-protein interactions and/or through their intrinsic acetyltransferase activity toward histone proteins and other nearby nuclear factors (36). Considering that Nfe2 acts as a repressor during trophoblast differentiation we speculate that Nfe2 may inhibit the intrinsic acetylation activity of CBP in trophoblast cells, thus inducing hypoacetylation of JunD and histones within the Gcm1 promoter (1) (and current results). The Nfe2-dependent repression of acetylation may result from a competition of bZip transcription factors for a limited CBP pool (37). Indeed, a competitive interaction between CBP and bZip proteins, including Nfe2 and c-Jun, has been previously reported (38). In addition, we observed a markedly increased interaction between CBP and Gcm1 in the absence of Nfe2 (1), which is in agreement with a competitive interaction of CBP with various transcription factors during trophoblast differentiation. Such competitive interaction with CBP may be the mechanism underlying the functional antagonism between bZip transcription factors like Nfe2 and JunD during trophoblast differentiation and may constitute an alternative, MAPK independent pathway through which AP-1 proteins regulate acetylation and gene expression. The existence of such a competitive mechanism, as proposed, needs to be explored in future studies.
Given the current and previous findings Nfe2 acts as an epigenetic and post-translational repressor during placental development, providing a mechanism limiting syncytiotrophoblast formation. It appears that all mechanisms through which Nfe2 modulates placental vascularization converge at the regulatory level of Gcm1. Thus, the current finding underscores a crucial function of Gcm1 in regulating placental labyrinthine formation (6). This novel, Nfe2-dependent pathway controlling Gcm1 activity is important to ensure sufficient placental vascularization and embryonic growth during later pregnancy stages (1) (and current findings).
Future studies are required to determine whether the same pathway is of functional relevance in the human placenta. We observed expression of Nfe2 in human placenta, specifically in human syncytiotrophoblast cells3 and expression of Nfe2 within human syncytiotrophoblast is depicted on the human protein atlas webpage. Interestingly, JunD, but not other AP-1 proteins, is specifically expressed in human syncytiotrophoblast (39). Hence it appears possible, but remains to be shown, that cross-coupling of Nfe2 and JunD via acetylation constitutes a novel mechanism regulating placental development in humans as well. Providing that the relevance of this pathway can be confirmed in humans this may identify new diagnostic and therapeutic targets for placental diseases associated with altered expression of Gcm1, such as pre-eclampsia.
Supplementary Material
Acknowledgments
Nfe2-deficient mice were kindly provided by R. Shivdasani (Harvard Medical School, Cambridge, MA). We thank S. Schmidt and F. Zimmermann for technical support. We are grateful to Hilmar Bading, Department of Neurobiology, Interdisciplinary Center for Neurosciences, University of Heidelberg, for providing the opportunity to carry out the electron microscopy work in his laboratory.
This work was supported by Deutsche Forschungsgemeinschaft Grants IS 67/4-1 and IS 67/4-2 (to B. I.) and a grant of the German Israeli Foundation (to B. I. and P. P. N.).

This article contains supplemental Figs. S1 and S2.
M. Kashif and B. Isermann, unpublished observation.
- p.c.
- post coitus
- Nfe2
- nuclear factor erythroid-derived 2
- CREB
- cAMP-response element-binding protein
- TSA
- trichostatin A
- HDAC
- histone deacetylase
- TS
- trophoblast stem
- HAT
- histone acetyltransferase
- kd
- knockdown.
REFERENCES
- 1. Kashif M., Hellwig A., Kolleker A., Shahzad K., Wang H., Lang S., Wolter J., Thati M., Vinnikov I., Bierhaus A., Nawroth P. P., Isermann B. (2011) p45NF-E2 represses Gcm1 in trophoblast cells to regulate syncytium formation, placental vascularization, and embryonic growth. Development 138, 2235–2247 [DOI] [PubMed] [Google Scholar]
- 2. McIntire D. D., Bloom S. L., Casey B. M., Leveno K. J. (1999) Birth weight in relationship to morbidity and mortality among newborn infants. N. Engl. J. Med. 340, 1234–1238 [DOI] [PubMed] [Google Scholar]
- 3. Gluckman P. D., Hanson M. A., Cooper C., Thornburg K. L. (2008) Effect of in utero and early-life conditions on adult health and disease. N. Engl. J. Med. 359, 61–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen Z., Hu M., Shivdasani R. A. (2007) Expression analysis of primary mouse megakaryocyte differentiation and its application in identifying stage-specific molecular markers and a novel transcriptional target of NF-E2. Blood 109, 1451–1459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hashemolhosseini S., Schmidt K., Kilian K., Rodriguez E., Wegner M. (2004) Conservation and variation of structure and function in a newly identified GCM homolog from chicken. J. Mol. Biol. 336, 441–451 [DOI] [PubMed] [Google Scholar]
- 6. Anson-Cartwright L., Dawson K., Holmyard D., Fisher S. J., Lazzarini R. A., Cross J. C. (2000) The glial cells missing-1 protein is essential for branching morphogenesis in the chorioallantoic placenta. Nat. Genet. 25, 311–314 [DOI] [PubMed] [Google Scholar]
- 7. Hung H. L., Kim A. Y., Hong W., Rakowski C., Blobel G. A. (2001) Stimulation of NF-E2 DNA binding by CREB-binding protein (CBP)-mediated acetylation. J. Biol. Chem. 276, 10715–10721 [DOI] [PubMed] [Google Scholar]
- 8. Johnson K. D., Grass J. A., Boyer M. E., Kiekhaefer C. M., Blobel G. A., Weiss M. J., Bresnick E. H. (2002) Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl. Acad. Sci. U.S.A. 99, 11760–11765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schubert S. W., Abendroth A., Kilian K., Vogler T., Mayr B., Knerr I., Hashemolhosseini S. (2008) bZIP-type transcription factors CREB and OASIS bind and stimulate the promoter of the mammalian transcription factor GCMa/Gcm1 in trophoblast cells. Nucleic Acids Res. 36, 3834–3846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schorpp-Kistner M., Wang Z. Q., Angel P., Wagner E. F. (1999) JunB is essential for mammalian placentation. EMBO J. 18, 934–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Schreiber M., Wang Z. Q., Jochum W., Fetka I., Elliott C., Wagner E. F. (2000) Placental vascularization requires the AP-1 component fra1. Development 127, 4937–4948 [DOI] [PubMed] [Google Scholar]
- 12. Okada Y., Ueshin Y., Isotani A., Saito-Fujita T., Nakashima H., Kimura K., Mizoguchi A., Oh-Hora M., Mori Y., Ogata M., Oshima R. G., Okabe M., Ikawa M. (2007) Complementation of placental defects and embryonic lethality by trophoblast-specific lentiviral gene transfer. Nat. Biotechnol. 25, 233–237 [DOI] [PubMed] [Google Scholar]
- 13. Nervi C., Borello U., Fazi F., Buffa V., Pelicci P. G., Cossu G. (2001) Inhibition of histone deacetylase activity by trichostatin A modulates gene expression during mouse embryogenesis without apparent toxicity. Cancer Res. 61, 1247–1249 [PubMed] [Google Scholar]
- 14. Göttlicher M., Minucci S., Zhu P., Krämer O. H., Schimpf A., Giavara S., Sleeman J. P., Lo Coco F., Nervi C., Pelicci P. G., Heinzel T. (2001) Valproic acid defines a novel class of HDAC inhibitors inducing differentiation of transformed cells. EMBO J. 20, 6969–6978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Giunta B., Obregon D., Hou H., Zeng J., Sun N., Nikolic V., Ehrhart J., Shytle D., Fernandez F., Tan J. (2006) EGCG mitigates neurotoxicity mediated by HIV-1 proteins gp120 and Tat in the presence of IFN-γ. Role of JAK/STAT1 signaling and implications for HIV-associated dementia. Brain Res. 1123, 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pan Y., Chen C., Shen Y., Zhu C. H., Wang G., Wang X. C., Chen H. Q., Zhu M. S. (2008) Curcumin alleviates dystrophic muscle pathology in mdx mice. Mol. Cells 25, 531–537 [PubMed] [Google Scholar]
- 17. Tanaka S., Kunath T., Hadjantonakis A. K., Nagy A., Rossant J. (1998) Promotion of trophoblast stem cell proliferation by FGF4. Science 282, 2072–2075 [DOI] [PubMed] [Google Scholar]
- 18. Isermann B., Sood R., Pawlinski R., Zogg M., Kalloway S., Degen J. L., Mackman N., Weiler H. (2003) The thrombomodulin-protein C system is essential for the maintenance of pregnancy. Nat. Med. 9, 331–337 [DOI] [PubMed] [Google Scholar]
- 19. Himeno E., Tanaka S., Kunath T. (2008) Curr. Protoc. Stem Cell. Biol., Chapter 1, Unit 1E 4 [DOI] [PubMed] [Google Scholar]
- 20. Maltepe E., Krampitz G. W., Okazaki K. M., Red-Horse K., Mak W., Simon M. C., Fisher S. J. (2005) Hypoxia-inducible factor-dependent histone deacetylase activity determines stem cell fate in the placenta. Development 132, 3393–3403 [DOI] [PubMed] [Google Scholar]
- 21. Balasubramanyam K., Varier R. A., Altaf M., Swaminathan V., Siddappa N. B., Ranga U., Kundu T. K. (2004) Curcumin, a novel p300/CREB-binding protein-specific inhibitor of acetyltransferase, represses the acetylation of histone/nonhistone proteins and histone acetyltransferase-dependent chromatin transcription. J. Biol. Chem. 279, 51163–51171 [DOI] [PubMed] [Google Scholar]
- 22. Tachibana H., Fujimura Y., Yamada K. (2004) Tea polyphenol epigallocatechin-3-gallate associates with plasma membrane lipid rafts. Lipid rafts mediate anti-allergic action of the catechin. Biofactors 21, 383–385 [DOI] [PubMed] [Google Scholar]
- 23. Tachibana H., Koga K., Fujimura Y., Yamada K. (2004) A receptor for green tea polyphenol EGCG. Nat. Struct. Mol. Biol. 11, 380–381 [DOI] [PubMed] [Google Scholar]
- 24. Follenzi A., Bakovic S., Gual P., Stella M. C., Longati P., Comoglio P. M. (2000) Cross-talk between the proto-oncogenes Met and Ron. Oncogene 19, 3041–3049 [DOI] [PubMed] [Google Scholar]
- 25. Bierhaus A., Schiekofer S., Schwaninger M., Andrassy M., Humpert P. M., Chen J., Hong M., Luther T., Henle T., Klöting I., Morcos M., Hofmann M., Tritschler H., Weigle B., Kasper M., Smith M., Perry G., Schmidt A. M., Stern D. M., Häring H. U., Schleicher E., Nawroth P. P. (2001) Diabetes-associated sustained activation of the transcription factor nuclear factor-κB. Diabetes 50, 2792–2808 [DOI] [PubMed] [Google Scholar]
- 26. Rudofsky G., Jr., Reismann P., Grafe I. A., Konrade I., Djuric Z., Tafel J., Buchbinder S., Zorn M., Humpert P. M., Hamann A., Morcos M., Nawroth P. P., Bierhaus A. (2007) Improved vascular function upon pioglitazone treatment in type 2 diabetes is not associated with changes in mononuclear NF-κB binding activity. Horm. Metab. Res. 39, 665–671 [DOI] [PubMed] [Google Scholar]
- 27. Aguilera C., Nakagawa K., Sancho R., Chakraborty A., Hendrich B., Behrens A. (2011) c-Jun N-terminal phosphorylation antagonises recruitment of the Mbd3/NuRD repressor complex. Nature 469, 231–235 [DOI] [PubMed] [Google Scholar]
- 28. Reddy S. P., Mossman B. T. (2002) Role and regulation of activator protein-1 in toxicant-induced responses of the lung. Am. J. Physiol. Lung Cell Mol. Physiol. 283, L1161–1178 [DOI] [PubMed] [Google Scholar]
- 29. Thépot D., Weitzman J. B., Barra J., Segretain D., Stinnakre M. G., Babinet C., Yaniv M. (2000) Targeted disruption of the murine JunD gene results in multiple defects in male reproductive function. Development 127, 143–153 [DOI] [PubMed] [Google Scholar]
- 30. Francastel C., Augery-Bourget Y., Prenant M., Walters M., Martin D. I., Robert-Lézénès J. (1997) c-Jun inhibits NF-E2 transcriptional activity in association with p18/maf in Friend erythroleukemia cells. Oncogene 14, 873–877 [DOI] [PubMed] [Google Scholar]
- 31. Dhanasekaran N., Premkumar Reddy E. (1998) Signaling by dual specificity kinases. Oncogene 17, 1447–1455 [DOI] [PubMed] [Google Scholar]
- 32. Davis R. J. (2000) Signal transduction by the JNK group of MAP kinases. Cell 103, 239–252 [DOI] [PubMed] [Google Scholar]
- 33. Kallunki T., Deng T., Hibi M., Karin M. (1996) c-Jun can recruit JNK to phosphorylate dimerization partners via specific docking interactions. Cell 87, 929–939 [DOI] [PubMed] [Google Scholar]
- 34. Lee T. L., Shyu Y. C., Hsu P. H., Chang C. W., Wen S. C., Hsiao W. Y., Tsai M. D., Shen C. K. (2010) JNK-mediated turnover and stabilization of the transcription factor p45/NF-E2 during differentiation of murine erythroleukemia cells. Proc. Natl. Acad. Sci. U.S.A. 107, 52–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Johnson K. D., Norton J. E., Bresnick E. H. (2002) Requirements for utilization of CREB-binding protein by hypersensitive site two of the β-globin locus control region. Nucleic Acids Res. 30, 1522–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nandiwada S. L., Li W., Zhang R., Mueller D. L. (2006) p300/cyclic AMP-responsive element-binding protein mediates transcriptional coactivation by the CD28 T cell costimulatory receptor. J. Immunol. 177, 401–413 [DOI] [PubMed] [Google Scholar]
- 37. Kamei Y., Xu L., Heinzel T., Torchia J., Kurokawa R., Gloss B., Lin S. C., Heyman R. A., Rose D. W., Glass C. K., Rosenfeld M. G. (1996) A CBP integrator complex mediates transcriptional activation and AP-1 inhibition by nuclear receptors. Cell 85, 403–414 [DOI] [PubMed] [Google Scholar]
- 38. Cheng X., Reginato M. J., Andrews N. C., Lazar M. A. (1997) The transcriptional integrator CREB-binding protein mediates positive cross-talk between nuclear hormone receptors and the hematopoietic bZip protein p45/NF-E2. Mol. Cell. Biol. 17, 1407–1416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bamberger A. M., Bamberger C. M., Aupers S., Milde-Langosch K., Löning T., Makrigiannakis A. (2004) Expression pattern of the activating protein-1 family of transcription factors in the human placenta. Mol. Hum. Reprod. 10, 223–228 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.