Background: Many GPCRs including mGluR5 are expressed on cell surface and intracellular membranes although their intracellular functions are unknown.
Results: Activation of intracellular mGluR5 in neurons up-regulates many genes associated with synaptic plasticity like Arc.
Conclusion: Intracellular mGluR5 is critical for synaptic plasticity.
Significance: Learning how intracellular mGluR5 signals is crucial for understanding neurodevelopmental disorders with disrupted synaptic plasticity like fragile X and autism.
Keywords: Calcium; G protein-coupled Receptors (GPCR); Glutamate; Glutamate Receptors Metabotropic; Inositol 1,4,5-Trisphosphate; Arc; IEG; SRF
Abstract
The G-protein coupled receptor, metabotropic glutamate receptor 5 (mGluR5), is expressed on both cell surface and intracellular membranes in striatal neurons. Using pharmacological tools to differentiate membrane responses, we previously demonstrated that cell surface mGluR5 triggers rapid, transient cytoplasmic Ca2+ rises, resulting in c-Jun N-terminal kinase, Ca2+/calmodulin-dependent protein kinase, and cyclic adenosine 3′,5′-monophosphate-responsive element-binding protein (CREB) phosphorylation, whereas stimulation of intracellular mGluR5 induces long, sustained Ca2+ responses leading to the phosphorylation of extracellular signal-regulated kinase (ERK1/2) and Elk-1 (Jong, Y. J., Kumar, V., and O'Malley, K. L. (2009) J. Biol. Chem. 284, 35827–35838). Using pharmacological, genetic, and bioinformatics approaches, the current findings show that both receptor populations up-regulate many immediate early genes involved in growth and differentiation. Activation of intracellular mGluR5 also up-regulates genes involved in synaptic plasticity including activity-regulated cytoskeletal-associated protein (Arc/Arg3.1). Mechanistically, intracellular mGluR5-mediated Arc induction is dependent upon extracellular and intracellular Ca2+ and ERK1/2 as well as calmodulin-dependent kinases as known chelators, inhibitors, and a dominant negative Ca2+/calmodulin-dependent protein kinase II construct block Arc increases. Moreover, intracellular mGluR5-induced Arc expression requires the serum response transcription factor (SRF) as wild type but not SRF-deficient neurons show this response. Finally, increased Arc levels due to high K+ depolarization is significantly reduced in response to a permeable but not an impermeable mGluR5 antagonist. Taken together, these data highlight the importance of intracellular mGluR5 in the cascade of events associated with sustained synaptic transmission.
Introduction
Glutamate, the major excitatory amino acid neurotransmitter present in the central nervous system, exerts its effects by binding and activating receptors that are classified as ionotropic (ligand-gated ion channels) or metabotropic glutamate receptors (mGluRs2; G-protein coupled receptors GPCRs). One such GPCR, mGluR5, is not only a key player in many aspects of neuronal development, synaptic plasticity, and learning and memory but is also implicated in various neurological disorders such as epilepsy, fragile X syndrome, neuropathic pain, and Parkinson disease (2–5).
Observations from this laboratory have shown that a large percentage of mGluR5 is expressed on intracellular membranes; ligand stimulation of endogenous mGluR5 on isolated, striatal nuclei leads to rapid, sustained Ca2+ responses that can be blocked by receptor-specific antagonists (1, 6, 7). Studies using tagged molecules and impermeable agonists and antagonists indicate that the ligand binding domain is within the nuclear lumen such that glutamate or other permeable agonists such as quisqualate (Quis) must cross both the plasma and nuclear membranes to activate receptors (1, 6–8). Mechanistically, this is accomplished via the excitatory amino acid transporters or the cystine-glutamate exchanger (1, 6, 7). Using optical, pharmacological, and genetic techniques, we have also demonstrated that nuclear mGluR5 couples to Gq/11 to activate nuclear phosphatidylinositol-phospholipase C, hydrolysis of phosphoinositol 4,5-bisphosphate, and generation of nuclear inositol 1,4,5-trisphosphate (IP3) (7). Thus, nuclear mGluR5s play a dynamic role in mobilizing Ca2+ in a specific, localized fashion.
What are the functional consequences of activating endogenous mGluR5 expressed on striatal cell membranes versus those expressed intracellularly? Using the permeable and impermeable mGluR5 ligands, our recent data show that activation of cell surface receptors via the impermeable agonist (S)-3,5-dihydroxyphenylglycine (DHPG) induces rapid, transient Ca2+ responses, whereas activation of intracellular mGluR5 with the permeable agonist Quis in the presence of impermeable antagonists leads to sustained Ca2+ responses. Membrane-specialized mGluR5-mediated Ca2+ responses lead to distinct cellular responses as well. For instance, stimulation of cell surface mGluR5 results in phosphorylation of critical signaling entities such as c-Jun N-terminal kinase, CaMK, and CREB, whereas intracellular mGluR5 activation leads to a cascade of molecular events starting with the phosphorylation of ERK1/2 and Elk-1 followed by the enhanced expression of synaptic plasticity genes like c-fos, egr-1, Fras, and FosB. Thus stimulation of intracellular mGluR5 initiates a cascade of events underlying processes with hallmarks of synaptic plasticity (1).
In neurons, one IEG discovered in a screen for genes rapidly induced by synaptic stimulation and linked to long term synaptic adaptations is activity-regulated cytoskeletal-associated protein, Arc/Arg3.1 (hereafter termed Arc) (9, 10). Arc, a protein enriched at the post synaptic density, is involved in multiple forms of neuronal plasticity: long term potentiation (LTP), long term depression (LTD), and homeostatic plasticity (10). One important function attributed to Arc is AMPA receptor endocytosis leading to reduced AMPA currents and LTD (9, 10). Arc is also involved in local actin polymerization and LTP consolidation (10) as well as regulation of Notch1 signaling in response to neural activity (11). High levels of Arc are also found in the nucleus where it appears to associate with so-called promyelocytic leukemia bodies (12). These subnuclear structures are primarily composed of proteins involved in gene regulation and DNA repair (13). Inasmuch as studies have shown that mGluR5 activation up-regulates Arc in the hippocampus and that this up-regulation may be critical in synaptic plasticity and in disorders such as fragile X syndrome (14, 15), it is important to determine whether this is true for other regions of the brain and whether intracellular mGluR5 plays a role in Arc induction. Here we used an unbiased bioinformatics approach to determine what genes, if any, were rapidly changed in response to cell surface and/or intracellular mGluR5 activation. These studies reveal that in striatal neurons, intracellular mGluR5, not cell surface receptors, up-regulates various IEGs including Arc, that are important in sustained synaptic transmission. Taken together these studies expand our current understanding of the roles of intracellular GPCRs and their involvement in essential cellular functions.
EXPERIMENTAL PROCEDURES
Materials
Quis, (S)-3, 5-dihydroxyphenylglycine (DHPG), 2-methyl-6-(phenylethynyl)-pyridine (MPEP), dizocilpine/(5S,10R)-methyl-10, 11-dihydro-5H-dibenzo [a,d]cyclohepten-5,10-imine maleate (MK801), 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX), (±)-4-(4-Aminophenyl)-1,2-dihydro-1-methyl-2-propylcarb amoyl-6,7-methylenedioxyphthalazine (SYM2206), 7-(hydroxyimino)cyclopropa[b]chromen-1a-carboxylate ethyl ester (CPCCOEt), d-(−)-2-amino-5-phosphonopentanoic acid (d-APV), 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one hydrochloride (LY294002), and (1S,6br,9aS,11R,11bR)11-(acetyloxy)-1,6b,7,8,9a,10,11,11b-octahydro-1-(methoxymethyl)-9a,11b-dimethyl-3H-furo[4,3,2-de]indeno[4,5,-h]-2-h]-2-benzopyran-3,6,9-trione (wortmannin) were purchased from Tocris Cookson Inc. (Ellisville, MO). 2-Amino-2-(4-carboxyphenyl)-3- (9H-thioxanthen-9-yl) propanoic acid (LY367366) and 2-amino-2-(3-cis and trans-carboxycyclobutyl)-3-(9H-thioxanthen-9-yl) propionic acid (LY393053) were obtained from Lilly. 1,4-Diamino-2,3-dicyano-1,4-bis-(2-′aminophenylthio)butadiene (U0126), 1-[N,O-bis-(5-isoquinolinesulfonyl)-N-methyl-l-tyrosyl]-4-phenylpiperazine (KN62), 2-[N-(2-hydroxyethyl)]-N-(4-methoxybenzenesulfonyl)]amino-N-(4-chlorocinnamyl)-N-methylbenzylamine) (KN93), 7H-Benz[de]benzimidazo[2,1-a]isoquinoline-7-one-3-carboxylic acid acetate (STO609), 2-[1-(3-dimethylaminopropyl)-1H-indol-3-yl]-3-(1H-indol-3-yl)-maleimide (GF109203X), cyclosporin A, and cycloheximide were purchased from EMD Biosciences, Inc. (La Jolla, CA). Rapamycin was from L. C. Laboratories (Woburn, MA). Unless otherwise indicated all other chemicals were from Sigma.
Cell Culture
Dissociated striatal and hippocampal neuronal cultures were prepared using neonatal 1–2-day-old rat or mouse pups as described (6, 16). Cells were cultured for 14–18 days before use. All antagonists were added 30 min before and during the time treated with agonists or additional inhibitors. Reagents were added at the concentrations indicated in the figure legends.
Animals, Plasmids, and Transfection
Srff/f (wild type) and Srff/+; NesCre (heterozygote) and Srff/f; NesCre (knockout) animals were obtained from Dr. N. Ramanan (Washington University School of Medicine, St. Louis, MO) (17). IP3 receptor (IP3R) buffer constructs RFP-IP3R-NLS and RFP-NES-IP3R were from Dr. M. Nathanson (Yale University School of Medicine, New Haven, CT) (18), the “kinase dead” dominant-negative (dn) CaMK constructs dnCaMKI, dnCaMKIV, and dnCaMKK were obtained from Dr. T. R. Soderling (Oregon Health and Science University, Portland, OR), and the dnCaMKII was obtained from Dr. U. Bayer (University of Colorado, Denver) (19). Cultures (50,000 cells/dish) were transfected with vectors using Lipofectamine 2000 (Invitrogen). Treatment and subsequent analysis was done 24 h post-transfection.
Immunocytochemistry and Western Blotting
Striatal or hippocampal neurons were fixed, blocked, and incubated with antibody as described (1, 6–8, 20). Primary antibodies included polyclonal anti-C-terminal mGluR5 (1:250; Millipore), mouse monoclonal C-7 anti-Arc/Arg3.1 (1:100; Santa Cruz Biotechnology), rabbit polyclonal anti-SRF (1:1000, Santa Cruz), anti-NeuN (1:100, Millipore), monoclonal anti-lamin B2 (1:100; Zymed Laboratories Inc.), polyclonal anti-EAAT3 (1:1000; Chemicon International, Temecula, CA), polyclonal anti-quisqualate antibody (1:250; Dr. J. Koerner, University of Minnesota), and polyclonal anti-pERK1/2 (1:200; Cell Signaling Technology, Beverly, MA). Secondary antibodies included goat anti-mouse Alexa 488 IgG2A and goat anti-mouse Alexa 647 IgG1 (both at 1:500, Molecular Probes, Eugene, OR), goat anti-rabbit or anti-mouse Cy3 (both at 1:300; Jackson ImmunoResearch, West Grove, PA), and goat anti-rabbit or mouse Alexa 488 (1:300, Molecular Probes). Western blotting was performed using whole cell extracts from 14 days in vitro hippocampal cultures. Protein concentrations were determined using the Bradford assay (Bio-Rad). Proteins were separated by SDS-PAGE, blotted, and probed with polyclonal anti-pERK1/2 (1:2000) and monoclonal anti-ERK (1:1000, Cell Signaling Technology). A horseradish peroxidase conjugated with goat anti-rabbit immunoglobulin G (IgG; 1:2000, Cell Signaling Technology) or anti-mouse IgG (1:2000, Sigma) was used in conjunction with enhanced chemiluminescence (Amersham Biosciences) to detect the signal followed by densitometric analysis (Storm 860 Imager, GE Healthcare, together with associated software).
Gene Expression Profiling
DIV14 striatal neurons were treated with either DHPG or Quis at 37 °C for 1 h in triplicate. Because these agonists would also activate AMPA receptors and mGluR1, they were always bath-applied in the presence of 25 μm SYM2206, an AMPA receptor antagonist, and 20 μm CPCCOEt, an mGluR1 antagonist. Total cellular RNA was extracted from untreated and treated neurons (3 × 106 neurons per sample) using the RNeasy Mini kit (Qiagen). Ten μg of RNA per sample was submitted to the Multiplexed Gene Analysis Core Facility, Washington University School of Medicine for labeling, hybridization, scanning, and software services. The GeneChip Rat Genome 230v2.0 Array (Affymetrix) was utilized. The raw fluorescence data were analyzed using the MAS 5 algorithm within Affymetrix Expression Console software, and all arrays were scaled to a mean signal intensity of 1500. Data mining was performed using Spotfire DecisionSite for Functional Genomics Version 8.2.1 (Somerville, MA) and Partek Genomics Suite 6.08.0414 (St. Louis, MO). Principal Component Analysis was performed to assess the quality of the data. To determine which probe sets were changed between the two conditions, DHPG versus control or Quis versus control, a fold change of at least 2.0 and a present call in all 3 chips were required before making an assignment. In addition, a two-tailed t test with p < 0.05 was applied. Supplemental Tables S1 and S2 show the genes that were up-regulated by Quis and DHPG, respectively. Annotations were retrieved from Affymetrix GeneChip; Entrez Gene (NCBI) and AmiGO were used to search for Gene Ontology terms for the genes identified.
Quantitative Reverse Transcriptase Polymerase Chain Reaction
Two-step quantitative reverse transcriptase PCR was performed using the ABI Prism 7000 Sequence Detection System (Applied Biosystems, Foster City, CA) as described previously (1). Total RNA was isolated from striatal neurons using TRIzol reagent (Invitrogen) according to the manufacturer's instructions. Gene-specific primers for RT-PCR were designed using Primer3 Version 0.4.0 software (21) according to the Applied Biosystems guidelines (supplemental Table S3). The expression levels of the target mRNA were normalized to the expression of gapdh mRNA. The results, calculated as fold change compared with the untreated control samples, are expressed as the mean ± S.E. Student's t test was performed, and p < 0.05 was considered statistically significant.
Cellular Imaging Using High Content Imager and Analysis
To quantitate Arc protein up-regulation with different treatments, dissociated striatal neurons were treated with mGluR5 agonists or antagonists, 15 mm KCl, or antagonists for different signaling pathways and then fixed and stained with anti-Arc and anti-NeuN antibody as described above. Images from a total of 36–64 sites were acquired per treatment using ImageXpressMICRO Automated Acquisition and Analysis System (Molecular Devices Corp, Sunnyvale, CA). These images were processed using MetaExpress image analysis and informatics package (Molecular Devices) that utilizes Adaptive Background CorrectionTM (ABC). The application used was “multi-wavelength cell scoring.” The criteria chosen for Arc-positive neurons were a nuclear diameter range of 5–15 μm and NeuN and Arc staining with average pixel intensity at least 60 and 240 gray levels above background, respectively. The results were calculated as Arc-positive neurons/total neurons. Results from at least three independent experiments were expressed as the mean ± S.E., and Student's t test was performed; p < 0.05 was considered statistically significant. Data for this study were collected using the Chemical Genetics Screening Core Facility at Washington University School of Medicine.
Fluorescent Measurements of Intracellular Ca2+
DIV12-14 striatal neurons were loaded with Ca2+ fluorophore, imaged, and quantitated as described (1, 6, 7).
[3H]Quis Uptake Assay
Assays were performed in striatal neurons and acutely isolated nuclei as described (6). Different buffers that were used are the same as described previously (6).
RESULTS
Cell Surface and Intracellular mGluR5 Exhibit Differential Gene Expression Profiles
To determine the functional consequences of activating intracellular mGluR5, we took advantage of agonists known to activate Group 1 mGluRs: DHPG and Quis. Using radiolabeled uptake studies as well as biochemical, cellular, and pharmacological techniques, we have previously shown that DHPG cannot be transported into the cell and, therefore, only activates cell surface receptors, whereas Quis activates both cell surface and intracellular receptor pools (1, 6, 7). To confirm and extend these findings, immunocytochemical and radiolabeled uptake studies were used to show that Quis is taken up into striatal neurons via sodium and chloride-dependent processes (supplemental Fig. S1, A and B). Furthermore, we demonstrate the presence of the neuronal-specific, sodium-dependent transporter, EAAT3, on purified striatal nuclei, where it colocalizes with mGluR5 and lamin B2 (supplemental Fig. S2). Finally, we demonstrate that the half-maximal Quis concentration to stimulate a rapid transient Ca2+ response (cell surface; Ref. 1) is 0.45 μm, whereas the half-maximal concentration to induce both a rapid transient and sustained plateau Ca2+ response (intracellular; Ref. 1) is 5.9 μm (supplemental Fig. S1C). Presumably, the increased EC50 value associated with intracellular mGluR5 reflects properties of the uptake mechanisms involved in Quis transport into the cell (1, 6).
Because our earlier study demonstrated agonist up-regulation of several IEGs within an hour (1), striatal neurons were treated with vehicle, DHPG (100 μm), or Quis (20 μm) for 1 h. As stated under “Experimental Procedures,” these agonists were always bath-applied in the presence of 25 μm SYM2206, an AMPA receptor antagonist, and 20 μm CPCCOEt, an mGluR1 antagonist. Previously we have shown using wild type and mGluR5-deficient murine striatal cultures that Quis application in the presence of SYM2206, CPCCOEt, or APV led to a rise in intracellular Ca2+ in wild type but not null cultures (1). These data rule out the possibility that intracellular AMPA, N-methyl-d-aspartate (NMDA) or mGluR1 receptors mediate the Quis effects. Total RNA was isolated and then hybridized with the Affymetrix Rat 230V2 gene chip containing the entire transcribed rat genome. Results showed that at this time point 29 different genes were significantly up-regulated in Quis-treated versus DHPG-treated cells. A 1-h treatment with DHPG led to the up-regulation of 20 genes of which 16 were in common with Quis. Not surprisingly, many of these are IEGs or other types of signaling modulators (supplemental Tables S1 and S2). Inasmuch as our focus was on determining genes specific for intracellular versus cell surface mGluR5 responses and because the four DHPG-only induced transcripts exhibited only modest responses, these four genes were not further validated at this time.
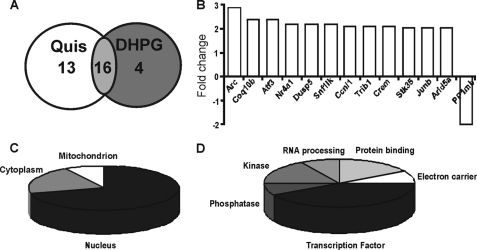
Thirteen genes were up-regulated as a consequence of intracellular mGluR5 activation via Quis (Fig. 1A, supplemental Tables S1 and S2). Of these, one probe (AI578037; supplemental Table S1) still lacks a Unigene annotation, and BLAST searches failed to turn up any homologs. The remaining 12 genes that were up-regulated by Quis treatment but not DHPG are shown in Fig. 1B. Only one gene was consistently down-regulated; protein phosphatase 1b showed a 2-fold decrease in response to Quis versus DHPG. Genes that were differentially expressed upon Quis treatment were further analyzed using Gene Ontology to categorize the likely cellular location of each gene (Fig. 1C) as well as its molecular function (Fig. 1D). Like the genes up-regulated by both agonists, almost 40% of the Quis-up-regulated transcripts were themselves transcription factors (Fig. 1D).
FIGURE 1.
Microarray analysis reveals both common and distinct transcriptional changes induced by Quis and DHPG. Striatal neurons were treated with Quis or DHPG for 1 h, and total RNA was isolated, further processed, and used for Affymetrix Rat 230V2 gene chip hybridization. Data were analyzed as described under “Experimental Procedures.” A, shown is the number of genes with increased expression in both treatments. B, shown are genes that were specifically up-regulated or down-regulated by Quis treatment but not DHPG. Bars represents -fold change as compared with the control (n = 3). C and D, pie charts show the cellular localization and molecular function distribution, respectively, of 12 known genes specifically up-regulated by Quis treatment but not by DHPG.
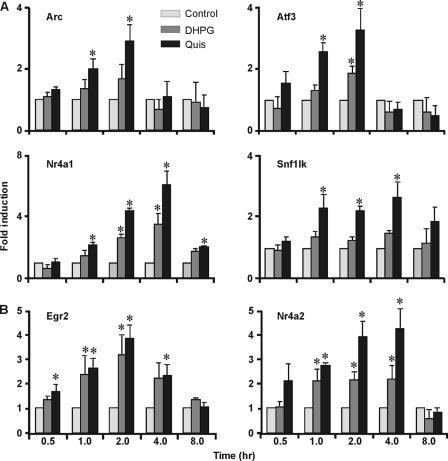
To verify induction by independent methodologies and to extend the time course, differentially regulated genes were examined by quantitative real time PCR using gene-specific primers. Overall, microarray results were consistent with data derived from post hoc testing of individual genes using striatal RNA prepared at various time points (Fig. 2 and supplemental Fig. S3). As predicted from the genechip results, the IEG Arc, the transcription factors Atf3 (activating transcription factor 3) and nuclear receptor subfamily 4 group A member 1 (Nr4a1/Nurr77), and the kinase SNF1-like kinase (SIK1) were all significantly increased at 1 h in response to Quis but not DHPG (Fig. 2A). DHPG did up-regulate some of these genes, albeit at later time points (Fig. 2A). Genes that were up-regulated by both agonists as assessed by microarray analysis, early growth response 2 (Egr2), and nuclear receptor subfamily 4 group A member 2 (Nr4a2), were also increased by both drugs as determined by quantitative real time PCR (Fig. 2B). Overall, microarray results were consistent with data derived from independent striatal time courses at least at the time point used for the original experiment. Given that many of these genes are known to be involved in synaptic plasticity (Arc, Atf3, DUSP5, SIK1, cyclin L1, Trb-1, Crem, STK35, Arid5a, JunB) and because mGluR5 plays an important role in synaptic changes (22), these data provide new evidence that genes involved in synaptic plasticity can be regulated by intracellular mGluR5 in this model of sustained synaptic transmission.
FIGURE 2.
Quis-specific up-regulated genes validated by quantitative RT-PCR. Striatal neurons were treated with Quis (black) or DHPG (gray) for indicated time points, total RNA was isolated, and the expression levels of genes by Quis only (A) or both DHPG and Quis (B) were measured by quantitative reverse transcriptase-PCR normalized to the expression levels of the reference gene, Gapdh. Bars represent fold change compared with control (mean ± S.E.) from at least three independent experiments performed in triplicate. *, p < 0.05 versus control.
Intracellular but Not Cell Surface mGluR5 Increases Expression of IEG Effector, Arc
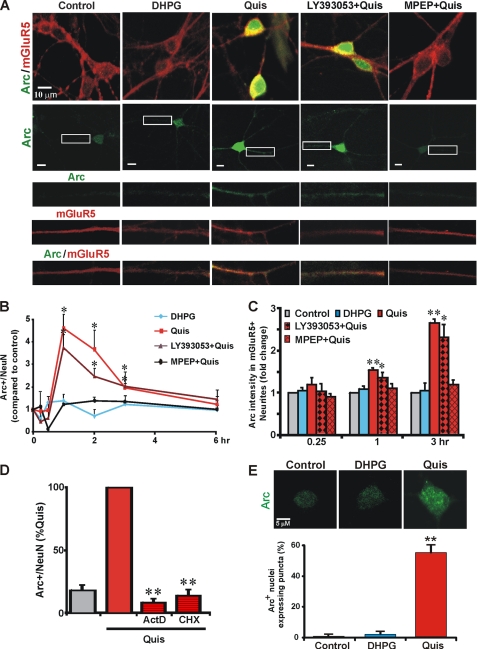
Consistent with the finding that Quis differentially activates Arc mRNA, Arc protein was also dramatically up-regulated in neuronal nuclei, cell bodies, and dendrites after Quis treatment from times ranging from 1 to 3 h (Fig. 3). Up-regulation was due to intracellular receptors because 1) DHPG did not elicit significant Arc induction at these time points, and 2) blocking cell surface mGluR5 with the impermeable antagonist LY393053 did not prevent Arc protein increase in the presence of Quis, although the permeable antagonist MPEP blocked this effect (Fig. 3, A and B). Arc protein increases were most evident in the cell body but readily apparent in dendrites as well after Quis treatment (Fig. 3, A and C). Pretreating striatal neurons with the transcription suppressor, actinomycin D, and the translation inhibitor cycloheximide reduced Quis-mediated Arc induction by 86–91%, indicating intracellular mGluR5-mediated Arc induction is both transcription- and translation-dependent (Fig. 3D).
FIGURE 3.
Activation of intracellular, but not cell surface mGluR5, up-regulates immediate early gene Arc. A, DIV17–18 striatal neurons were treated with the indicated drugs for 3 h before staining for Arc (green) and mGluR5 (red). Quis but not DHPG increased Arc expression in mGluR5-positive cell bodies and neurites. MPEP (10 μm) but not LY393053 (20 μm) abolished Quis-induced Arc levels. B, quantitation of Arc immunofluorescence in neurons (expressed as mean ± S.E., compared with control) in the absence or presence of various ligands treated for indicated times; more than 2500 neurons per treatment condition from 3–6 independent experiments were assessed using High Content Imaging; *, p < 0.05 versus basal level. C, quantitation of Arc staining intensity in mGluR5-positive neurites was measured up to 50 μm from the cell body. Bars represent the mean ± S.E. from at least three independent experiments with more than 140 neurites per treatment condition analyzed. *, p < 0.05; **, p < 0.001 versus basal levels. D, Quis-mediated Arc induction is dependent on transcription and translation. Quantitation of Arc immunofluorescence in neurons (expressed as % of Quis treatment) in the absence or presence of the indicated antagonists (ActD, actinomycin D, transcription inhibitor, 40 μm; CHX, cycloheximide, translation inhibitor, 80 μm) treated for 1 h; ∼700 neurons per treatment condition from n = 4 independent experiments. **, p < 0.001 versus Quis. E, Quis but not DHPG induced Arc+ nuclear puncta. Bars represent mean ± S.E. from three independent experiments: >200 neurons were analyzed per treatment. **, p < 0.05.
Increased Nuclear Arc Is Associated with Nuclear Puncta
Up-regulation of Arc has been associated with nuclear puncta in hippocampal neuronal nuclei (12). Similarly, puncta were also observed in Quis-treated mGluR5-positive striatal nuclei but not in control or DHPG-treated cultures (Fig. 3E). Such puncta are thought to be sites of transcriptional regulation, thus these findings suggest that Quis-mediated Arc induction is in turn associated with downstream signaling cascades.
Cytoplasmic and Nuclear Ca2+ Is Required for Intracellular mGluR5-mediated Arc Induction
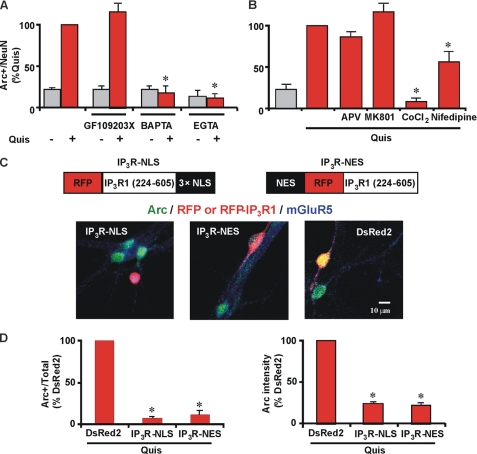
We made use of pharmacological, genetic, and molecular tools combined with optical imaging to identify the pathway involved in mGluR5-mediated Arc induction. Specifically, the protein kinase C inhibitor GF109203X did not abolish intracellular mGluR5-mediated Arc induction, whereas the intracellular Ca2+ chelator BAPTA-1AM reduced Arc to basal levels (Fig. 4A; 17.5% ± 9.2; n = 3). The extracellular Ca2+ chelator, EGTA, also blocked Quis-mediated Arc increases by ∼87% (Fig. 4A), suggesting that extracellular Ca2+ is necessary for intracellular mGluR5-mediated Arc induction in striatal neurons. To test whether NMDA receptors or voltage-gated Ca2+ channels (VGCC) were involved, we pretreated neurons with specific inhibitors of both. The general VGCC blocker cobalt led to a 90% reduction in Quis-induced Arc levels, whereas the specific l-type VGCC antagonist nifedipine only partially inhibited (∼45% reduction) this response (Fig. 4B). In contrast, neither APV nor MK801 (NMDA antagonists) had any effect on Quis-increased Arc levels (Fig. 4B).
FIGURE 4.
Quis-mediated Arc induction is dependent on Ca2+. A, striatal neurons were treated with Quis (red) in the absence or presence of indicated antagonists (GF109203X, protein kinase C inhibitor, 1 μm; BAPTA-1AM, intracellular Ca2+ chelator, 30 μm; EGTA, extracellular Ca2+ chelator, 5 mm) for 1 h, fixed, and stained for Arc and neuronal nuclei (NeuN). Bars represent quantitation of Arc immunofluorescence in neurons (expressed as % of Quis treatment) from n = 3 independent experiments; *, p < 0.001 versus Quis. A total of ∼1300 neurons per treatment condition was assessed. Gray bars indicate untreated neurons or neurons treated with indicated antagonists in absence of Quis. B, quantitation of Arc immunofluorescence in neurons (expressed as % of Quis treatment) from n = 3 independent experiments; *, p < 0.001 versus Quis. A total of ∼1100 neurons per treatment condition were assessed. Gray bars indicate untreated control. APV, NMDA antagonist, 100 μm; MK801, NMDA antagonist, 5 μm; CoCl2, nonspecific Ca2+ channel blocker, 1 mm; Nifedipine, specific L-type Ca2+ channel blocker, 10 μm. C, shown is a schematic representation of IP3R buffer constructs (18). A plasmid encoding DsRed2 alone, RFP-IP3R-NLS, or NES-RFP-IP3R was transiently transfected into striatal neurons (red) 24 h post-transfection, and neurons were treated with Quis for 1 h, fixed, and stained with Arc (green) and mGluR5 (blue). D, shown is quantitation of Arc immunofluorescence in terms of the number of Arc-positive neurons (expressed as % of DsRed2 alone transfected neurons, left graph) and Arc intensity in transfected neurons (expressed as % of DsRed2 alone transfected neurons, right graph) from n = 3 independent experiments. A total of >85 neurons/plasmid was assessed. *, p < 0.05 compared with DsRed2 alone.
Because Ca2+ is required for Quis-mediated Arc induction, we used IP3R “buffer” constructs to determine whether cytoplasmic or nuclear Ca2+ or both play a role. As described (18), these constructs target IP3 binding domains fused with RFP to either the nucleus (IP3R-NLS; IP3R with nuclear localization signal) or the cytoplasm (IP3R-NES; IP3R with nuclear export signal). Because the IP3 binding site has sufficient affinity to compete with the native receptor, these constructs act like a buffer to remove IP3 from the compartment in which they are expressed (18). As seen in Fig. 4, C and D, Quis does not increase Arc levels in neurons transfected with either of the IP3 buffer constructs (IP3R-NLS or IP3R-NES), whereas it does so in neurons transfected with red fluorescent protein DsRed2 alone. The reduction occurs both in the number of neurons showing Arc-positive staining as well as in the average intensity of Arc expression in the neuronal cell body (Fig. 4, C and D). Taken together, these data indicate that both cytoplasmic as well as nuclear Ca2+ is required for intracellular mGluR5-mediated Arc induction.
Intracellular mGluR5-mediated Arc Induction Is CaMKII-, ERK1/2-, and Serum Response Factor (SRF)-dependent
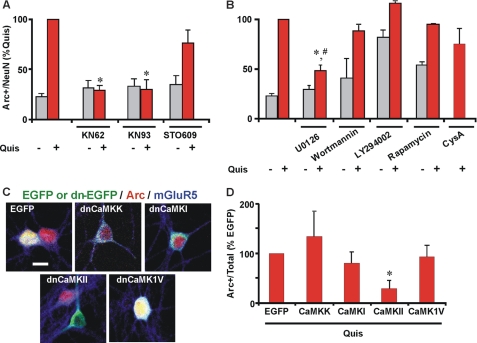
We have previously shown that intracellular mGluR5 selectively activates the CaMKII, ERK1/2, and Elk-1 pathway and induction of IEGs such as c-Fos and Egr1 (1). Here we tested if the same cascade is also involved in intracellular mGluR5-mediated Arc induction. Using various pharmacological inhibitors, we found that the Quis-mediated Arc protein increase could be blocked by the general CaMK inhibitor, KN62 (∼70% reduction) and the specific CaMKII inhibitor, KN93 (65–70% reduction) but not by the CaMKK inhibitor, STO609 (Fig. 5A). We also used GFP-tagged dominant negative CaM kinase constructs as an independent approach in determining which CaM kinase family members were involved. Only dnCaMKII blocked intracellular mGluR5-mediated Arc increases, whereas dnCaMKK, dnCaMKI, and dnCaMKIV did not (Fig. 5, C and D).
FIGURE 5.
CaMKII and ERK1/2 are involved in intracellular mGluR5-mediated Arc up-regulation. A and B, striatal neurons were treated with Quis (red) in the absence or presence of indicated antagonists (KN62, general CaMK antagonist, 10 μm; KN93, specific CaMKII antagonist, 10 μm; STO609, CaMKK inhibitor, 1.5 μm; U0126, MEK/ERK1/2 antagonist, 1 μm; LY294002, PI3K antagonist, 50 μm; wortmannin, PI3K antagonist, 500 nm; rapamycin, mTOR antagonist, 5 nm; CysA, cyclosporin A, calcineurin antagonist, 4 μm) for 1 h, fixed, and stained for Arc and neuronal nuclei (NeuN). Bars represent quantitation of Arc immunofluorescence in neurons (expressed as % of Quis treatment) from n = 3–5 independent experiments; *, p < 0.001 versus Quis; #, p < 0.001 versus control. More than 3000 neurons per treatment condition were assessed. Gray bars indicate untreated neurons or neurons treated with indicated antagonists in absence of Quis. C, plasmid encoding EGFP alone or EGFP-labeled dominant negative CaMK constructs as indicated were transiently transfected in striatal neurons (green), and 24 h post-transfection neurons were treated with Quis for 1 h, fixed, and stained with Arc (red) and mGluR5 (blue). D, shown is quantitation of Arc immunofluorescence in terms of number of Arc-positive neurons (expressed as % of EGFP alone transfected neurons) from n = 5 independent experiments; a total of >250 neurons/plasmid was assessed. *, p < 0.05 compared with EGFP alone.
To test what other proteins might be involved in intracellular mGluR5-mediated Arc induction, we used inhibitors of ERK1/2, phosphoinositide-3-kinase (PI3K) and mammalian target of rapamycin (mTOR). Interestingly, only the MEK inhibitor U0126 partially inhibited Arc induction (∼ 55% reduction) whereas PI3K inhibitors LY294002 and Wortmannin did not (Fig. 5B). In addition, the mTOR antagonist rapamycin did not affect Quis-mediated Arc induction (Fig. 5B). Surprisingly, LY294002 and rapamycin seemed to increase Arc levels on their own (Fig. 5B). Collectively, the most parsimonious model for these data is that activation of intracellular but not cell surface mGluR5 triggers a CaMKII/MEK/ERK1/2 cascade leading to Arc mRNA and protein induction.
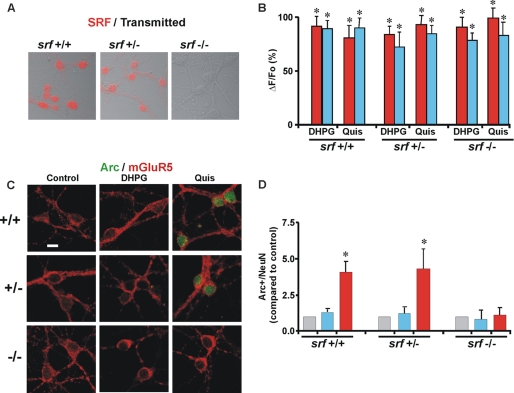
Two transcription factors, myocyte enhancer factor 2 (MEF2) and SRF, up-regulate Arc transcription in neurons (17, 23–26). MEF2 is activated via dephosphorylation by the calcium/calmodulin-regulated phosphatase calcineurin, which in turn is activated by Ca2+ influx (23). Because Quis leads to sustained intracellular Ca2+ levels in striatal neurons, we tested whether MEF2 is involved by pretreating neurons with an antagonist of calcineurin, cyclosporin A (25). However, cyclosporin A did not inhibit Quis-mediated Arc induction (Fig. 5B). To test if SRF is involved, we prepared striatal cultures from Srff/f (wild type) and Srff/+; NesCre (heterozygote) and Srff/f; NesCre (knockout) animals. As expected, SRF immunoreactivity was only seen in cultures prepared from Srf wild type or heterozygous mice but not in Srf deficiency mice (Fig. 6A). Treatment of these cultures with either DHPG or Quis led to a rise in Ca2+ in both the cytoplasm as well as the nucleus in all genotypes, indicating that upstream Ca2+ signaling by cell surface or intracellular mGluR5 is unaffected by the loss of SRF (Fig. 6B). DHPG did not increase Arc in any of the genotypes (Fig. 6, C and D), whereas Quis increased Arc protein levels only in the neurons prepared from Srf wild type or heterozygous mice, not in Srf deficient neurons (Fig. 6, C and D). Taken together, these findings are consistent with the pathway we previously established in which intracellular mGluR5 is coupled to a Ca2+/CaMKII/ERK/Elk1-SRF signaling cascade (1).
FIGURE 6.
Intracellular mGluR5-orchestrated Arc protein increase depends on serum response factor. A, mouse striatal neurons prepared from Srff/f (wild type) and Srff/+; NesCre (heterozygote) and Srff/f; NesCre (knockout) animals were stained for SRF immunoreactivity. For simplicity, in the figure cultures are labeled Srf+/+, Srf+/−, and Srf−/−. B, shown are compiled data from the maximal initial Ca2+ response (ΔF/Fo, %) from n > 26 Srf+/+, n > 58 Srf+/−, and n > 53 Srf−/− neurons, where red bars represent nuclear and blue bar represent cytoplasmic responses. *, p < 0.0001 for all responses compared with basal Ca2+. C, striatal neurons prepared from Srf+/+, Srf+/−, and Srf−/− were treated with the indicated drugs for 1 h before staining for Arc (green) and mGluR5 (red). Quis increased Arc expression in mGluR5-positive cell bodies in Srf+/+ and Srf+/− but not in Srf−/−. D, shown is quantitation of Arc immunofluorescence in neurons (expressed as the mean ± S.E., compared with control) in the absence (gray) or presence of the indicated ligands (blue for DHPG and red for Quis) treated for 1 h; n = 3. *, p < 0.05 versus control.
Intracellular mGluR5 Plays a Critical Role in Glutamate and Neuronal Activity-induced Arc
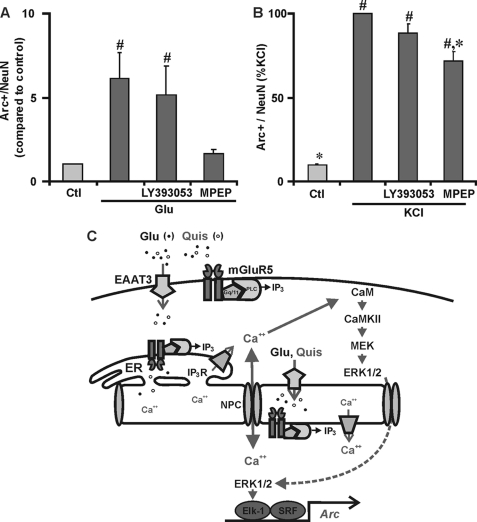
Previously we have shown that the endogenous ligand, glutamate, induces the same Ca2+ response as does Quis in Grm5+/+ but not in null cultures (1). Consistent with those results, like Quis, glutamate induced Arc protein levels about 6-fold even in the presence of the non-transported mGluR5 antagonist, LY393053. Only the permeable antagonist MPEP blocked glutamate-induced Arc in striatal neurons (Fig. 7A).
FIGURE 7.
Intracellular mGluR5 plays a critical role in glutamate and neuronal activity-induced Arc. A, striatal neurons were pretreated with NMDA antagonist MK801 (5 μm), the AMPA/kainate receptor antagonist, 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX; 10 μm), and mGluR1 antagonist CPCCOEt (20 μm) followed by treatment with glutamate (100 μm, 1 h). Glutamate increased Arc expression that was blocked by MPEP but not LY393053. Bars indicate Arc-positive/total neurons compared with control. #, p < 0.05 versus control; n = 4, more than 2500 neurons assessed per treatment. Ctl, control. B, bath application of 15 mm KCl-induced Arc in ∼30% striatal neurons. This KCl-regulated induction was decreased ∼28% by MPEP, whereas LY395053 + KCl was not significantly different from KCl alone. Bars indicate Arc positive/total neurons expressed as the percentage of KCl treatment. More than 2000 neurons per treatment from four independent experiments were assessed for Arc immunofluorescence using the High Content Imager. #, p < 0.0001 compared with untreated control. *, p < 0.005 compared with KCl treated alone. C, the proposed model for intracellular mGluR5-mediated transcriptional activation of Arc/Arg3.1 is shown. NPC, nuclear pore complex.
Depolarization with KCl in the presence or absence of mGluR5 cell surface and/or intracellular antagonists was used to determine whether intracellular receptors play a critical role in neuronal activity-induced Arc. After 1 h of KCl stimulation, Arc expression dramatically increased, supporting a role for activity-dependent induction of this IEG (Fig. 7B). This response was significantly inhibited by MPEP (∼28% reduction, n = 4, p < 0.005 versus KCl treated neurons) but not by LY395053 (Fig. 7B). The nonspecific Ca2+ channel blocker, cobalt chloride (1 mm), significantly decreased KCl-induced Arc expression levels, whereas the NMDA receptor antagonist MK801 (5 μm) or the Na+ channel blocker tetrodotoxin (2 μm) did not (not shown). Taken together, these data show that activation of intracellular mGluR5 contributes to important components of sustained synaptic transmission.
DISCUSSION
Mounting evidence shows that GPCRs signal not only from the cell surface but also from intracellular locations such as endosomes/clathrin-coated pits, the endoplasmic reticulum, Golgi, nuclear membranes, and even the nucleoplasm itself (27). Despite the growing number of GPCRs found in each location, few studies have addressed intracellular GPCR function. Using pharmacological, genetic, and unbiased bioinformatic approaches, the present findings show that activation of endogenous intracellular mGluR5 leads to the up-regulation of many IEGs and signaling molecules associated with synaptic plasticity. One such plasticity effector, Arc, is increased 4-fold within the nucleus, where it forms puncta-like structures and almost 3-fold in mGluR5+ neurites (Fig. 3). Mechanistically, Arc is induced via an intracellular mGluR5-mediated pathway involving Ca2+, CaMKII, ERK1/2, and SRF (Fig. 4–6). Consistent with the notion that Arc is up-regulated in striatal neurons via intracellular but not cell surface mGluR5, depolarization-induced Arc was significantly reduced in response to a permeable but not an impermeable mGluR5 antagonist. Given that mGluR5 plays an active role in modulating synaptic transmission and as such is implicated in a large number of neurological and developmental disorders, these data underscore the importance of intracellular mGluR5 in the cascade of events underlying long term processes associated with sustained synaptic transmission (Fig. 7C).
Intracellular mGluR5 Up-regulates Markers of Sustained Synaptic Transmission
Because we were interested in the most upstream events after receptor activation and because our earlier study using a candidate gene approach demonstrated that transcription factors such as Elk-1 and CREB were activated within 15–30 min of agonist application (1), we chose a 1-h time frame. Given that brief window of time, it is not surprising that only a small number of genes were reliably changed, and almost half (48%) were IEGs themselves (Fig. 1; supplemental Tables S1 and S2). Sixteen genes were up-regulated by both agonists, presumably due to cell surface mGluR5 activation as well as 13 genes, which were altered only in response to Quis, hence representing intracellular mGluR5 responses. IEGs up-regulated by both treatment paradigms included transcription factors such as: Csrnp1, a highly conserved cysteine-serine-rich nuclear protein required for neural progenitor proliferation and survival in zebra fish (28); Egr2, a zinc finger transcription factor required for general development and for the development of certain brain structures in particular (29); Klf4, one of four so-called reprogramming factors that can differentiate fibroblasts into functional neural stem cells (30); Nr4A2 (NurrI) and Nr4A3 (Nor1), members of the nuclear hormone receptor superfamily involved in differentiation, proliferation, and various metabolic processes (31); v-maf musculoaponeurotic fibrosarcoma oncogene homolog F (Maff), involved in neuronal differentiation and regulation of NGF-mediated neurite outgrowth (32); TIS11, an important regulator of cellular differentiation and inflammation (33).
Many of the transcripts up-regulated by Quis alone are also stimuli-responsive transcription factors involved in neuronal survival and growth (Atf3, Nr4a1, Trib1, CREM, JunB, and Arid5a) as well as effector proteins such as Arc, which as described is involved in gene regulation and synaptic plasticity (22). Interestingly, many of these genes are regulated by ERK1/2. For example, Dusp5, an inactivator of MAPK pathways, is itself induced in response to growth factors dependent on ERK1/2 activation, and the Dusp5 protein is phosphorylated at three sites by ERK1/2 in vitro and in vivo (34). These observations are consistent with our previous studies showing that intracellular but not cell surface mGluR5 triggers MAPK signaling pathways in striatal neurons (1). These in turn activate the IEG Elk-1, which can partner with SRF to activate promoters carrying serum response elements or function independently to up-regulate transcription machinery as a whole (35). In support of this model, Arc, Nr4A1, Dusp5, and JunB are all regulated by SRF (36, 37). Other genes such as Snf1lk (SIK1) repress CREB-dependent transcriptional activity both in the nucleus and in the cytoplasm (38). Indeed, Crem acts as a natural antagonist of cAMP-response element (CRE)-mediated gene transcription (39). Thus, one initial outcome of intracellular mGluR5 activation is to damp down CREB signaling and promote SRF-mediated IEGs.
What are the consequences of intracellular mGluR5 induction of IEGs? As described, Arc is critical for various forms of LTP and LTD (10). Crem is also involved in the regulation of long term plasticity underlying learning and memory (39). Finally, Nr4a1 (Nurr77), JunB (supplemental Table S1), c-Fos, and Egr1 (1) have all been linked to synaptic remodeling and, in some cases, learning and memory (25, 40). Taken together the present findings point to a critical role for intracellular mGluR5 in shaping synaptic processes.
Intracellular mGluR5 Induces Arc in Striatal Neurons
Several groups have shown that DHPG activates ERK1/2 and Arc. Many of those responses are derived from in vivo experiments (41), from hippocampal slice preparations (14, 15), or in different neuronal cultures such as cortex (42). In the latter case, DHPG up-regulated Arc via a CREB- and NMDA-dependent process versus VGCCs (42). Using striatal slices, Pisani et al. (43) showed that DHPG had no effect on medium spiny neurons unless it was coupled with NMDA activation. Finally, it is well documented in peripheral pain pathways that DHPG activates presynaptic Group1 receptors to increase glutamate release (44–46). Glutamate of course would activate post-synaptic receptors and depending upon the transporters present has the potential to activate intracellular receptors as well. Based on in vivo microdialysis experiments in the striatum (47), the nucleus tractus solitaries (48), and forebrain (47, 49), a similar facilitator role at excitatory synapses in the CNS might also occur. Indeed, this laboratory has previously reported evidence for presynaptic mGluR5 (50) as have others (51). Finally it should be noted that most DHPG “users” do not add additional antagonists to control for non-mGluR5 effects; thus, conceivably ERK1/2 activation in previous studies might be generated from mGluR1 activation. Indeed, without these inhibitors, DHPG-induced ERK1/2 phosphorylation is seen in our cultures as well (supplemental Fig. S4). Consistent with the notion that striatal, hippocampal, and cortical neurons will exhibit cell type-specific responses, we also see DHPG activation of ERK1/2 in dissociated hippocampal cultures (supplemental Fig. S4). Interestingly, p-ERK1/2 is seen in the cytoplasm, not the nucleus in hippocampal cultures, whereas Quis treatment leads to an ∼4-fold induction in the nucleus of dissociated striatal cultures. Collectively, these data point to distinct neuron-specific responses to DHPG that presumably trigger unique downstream sequelae.
Intracellular mGluR5-mediated Arc Induction Requires High Levels of Ca2+
Either in the hippocampus, cortex, or in the striatum, group 1 mGluR-mediated Arc induction requires Ca2+, although the channels by which Ca2+ enters the cell are different. In cortical and hippocampal neurons, NMDA receptors are required for group I mGluR-dependent Arc transcription (14, 20, 42), whereas in striatal neurons mGluR5-mediated Arc induction is unaffected by NMDA antagonists (Fig. 4B). In contrast, striatal Arc induction was completely abolished by the general Ca2+ channel blocker, cobalt, or partially blocked in the presence of the L-type Ca2+ channel blocker, nifedipine (Fig. 4B).
How would intracellular mGluR5 trigger VGCC opening? VGCCs are complex proteins involved in numerous processes and as such are modulated by multiple pathways. Functional coupling between group I mGluRs and L-type VGCCs has been described in cortical and hippocampal neurons as well as cerebellar granule cells (45). Based on electrophysiological and immunohistochemical experiments, these interactions are coupled to the refilling of endoplasmic reticulum Ca2+ stores (52). Recent advances in defining proteins associated with this process are consistent with that hypothesis. For example, depletion of endoplasmic reticulum Ca2+ stores trigger endoplasmic reticulum sensors (STIM1 and -2), which send signals to the plasma membrane to activate two types of store operated Ca2+ channels: Ca2+ release-activated Ca2+ (CRAC) channels, which are composed of Orai proteins, and STIMs (53, 54) as well as SOC channels, which incorporate Transient Receptor Potential Channels (55–57). In turn formation of STIM/Orai channels inhibits VGCCs and even leads to their internalization (53, 54). Depletion of extracellular Ca2+ via EGTA chelation or blockade of VGCCs via cobalt chloride might affect store refilling by 1) simply blocking Ca2+ influx and SERCA uptake, 2) altering the ratio of STIM to Orai leading to non-optimal Orai activation and decreased store refilling (58), or 3) preventing TRPC1 Ca2+-mediated recruitment to the plasma membrane where it complexes with Orai and STIM to form SOC channels (56). The latter are inhibited when Ca2+ entry is blocked or external Ca2+ is removed (56). Which of these possibilities contributes to intracellular mGluR5-mediated Arc induction awaits further studies.
Inasmuch as DHPG elicits a rapid, transient Ca2+ peak in dissociated striatal neurons versus a long sustained response following Quis (1), it would appear that more prolonged Ca2+ responses are required for Arc induction in striatal neurons. This interpretation is consistent with the current results using IP3 buffers to block Quis-mediated nuclear or cytoplasmic Ca2+ levels. In either case, Arc induction was also blocked (Fig. 4, C and D), indicating Ca2+ in either compartment is essential for Arc up-regulation.
CaMKII and ERK1/2 Are Important Mediators of Intracellular mGluR5-induced Arc
CaMK and MAPK signaling pathways appear to underlie Arc induction in other systems. For example, in neuroblastoma cells Arc increases were completely blocked by the general CaMK inhibitor, KN62 (59), and in rat dorsal striatum, stimuli such as spatial exploration induced Arc only in GABAergic/αCaMKII-positive neurons (60). In vivo activation of Arc in dentate granule cells is mediated in part by MAP kinase activation (61), suggesting that other signaling pathways also play a role. Moreover, Arc mRNA transport into dendrites requires MAPK signaling as local application of U0126 blocked newly synthesized Arc transcripts from being transported into activated synapses (62). Our data confirm and extend these findings showing that both CaMKII and ERK1/2 phosphorylation are critical for mGluR5-mediated Arc induction in striatal neurons (Fig. 5),
SRF Is a Critical Component of Arc Induction by Intracellular mGluR5
In hippocampal, cortical, and cerebellar Purkinje cells, Arc is regulated in an SRF-dependent fashion (17, 24, 26) via one or both of its serum response elements (63). For example in Purkinje cells, SRF binding to the most distal serum response element in the Arc promoter is essential for Arc induction and LTD in this system (26). Our studies are consistent with these published reports as intracellular mGluR5-mediated Arc induction is seen only in striatal cultures prepared from Srf wild type not in Srf-deficient mice (Fig. 6). The transcription factor MEF2 is also involved in Arc induction. For instance, activity-driven Ca2+ influx into hippocampal neurons induces the activation of the Ca2+/calmodulin-regulated phosphatase calcineurin, which dephosphorylates and activates MEF2; activated MEF2 increases the transcription of Arc (23). Similarly, sustained depolarization of striatal neurons led to calcineurin activation, MEF2 up-regulation, and Arc induction (25). MEF2 may not play its predicted role in the case of mGluR5-induced Arc levels however, as inclusion of the calcineurin inhibitor cyclosporin A failed to block Quis-induced Arc (Fig. 5B).
Intracellular mGluR5 Is a Key Component in Activity-dependent Arc Changes
Decreased Arc levels in response to MPEP but not LY395053 treatment after depolarization using KCl suggests an important role for intracellular mGluR5 in activity-dependent pathways underlying synaptic plasticity (Fig. 7B). Although NMDA receptors are thought to play a key role in the regulation of Arc mRNA localization and translation, MK801 did not block the induction of Arc by KCl in our paradigm (data not shown). Although further studies are required to support this idea, the fact that Arc is induced even in the presence of the impermeable antagonist suggests that a large component of mGluR5 signaling is being overlooked in models activated only by DHPG.
What Is the Role of Intracellular mGluR5-mediated Arc Induction in Striatal Neurons?
Numerous studies have shown that Arc is critical for long term memory and synaptic plasticity in the hippocampus (10). For example, increased Arc expression in slices and cultures decreases AMPA receptor-mediated synaptic currents presumably via its ability to regulate AMPA receptor trafficking on the synaptic membrane (64–66). In hippocampal neurons, DHPG activation of mGluR5 increases Arc, leading to AMPA receptor endocytosis, resulting in LTD (14, 15). It remains to be tested whether the dramatic induction of Arc in this study may similarly precede AMPA receptor internalization.
Arc is also thought to play a role in transcriptional regulation (12). This is based in part on increased Arc protein levels in the nucleus, where in the dentate gyrus and now in the striatum (Fig. 3) it interacts with a spectrin variant, βSpIVSigma5, in so-called promyelocytic leukemia bodies (12). Thus Arc appears to play an as yet poorly defined role in nuclear structures known to be highly involved in transcriptional regulation. It remains to be determined what the downstream effects of Arc induction are in striatal neurons.
In summary, these studies suggest a major role for intracellular mGluR5 receptors in the transcriptional up-regulation of genes associated with sustained synaptic transmission. Understanding long term consequences of intracellular mGluR5 activation may provide better insight into the many disorders where synaptic plasticity is disrupted.
Supplementary Material
Acknowledgments
We thank Steve Harmon, Taiwo Aderibigbe, Dennis Oakley, Bill Nolan, and Beth Gohara for technical assistance. We also thank Dr. M. Nathanson for inositol 1,4,5-trisphosphate buffer constructs RFP-IP3R-NLS and RFP-NES-IP3R, Dr. T. R. Soderling and Dr. U. Bayer for the dominant-negative CaMK constructs, and Dr. J. Koerner for anti-quisqualate antibody.
This work was supported, in whole or in part, by National Institutes of Health Grants MH57817 and MH69646 (to K. L. O.) and NS057105 (a Neuroscience Blueprint Core grant to Washington University). This work was also supported by a FRAXA Research Grant (to K. L. O. and V. K.), the Simons Foundation, and by the Bakewell Family Foundation, Bioinformatics Core and Chemical Genetics Screening Core facilities at Washington University School of Medicine.

This article contains supplemental Tables S1–S3 and Figs. S1–S4.
- mGluR5
- metabotropic glutamate receptor 5
- CaMK
- Ca2+/calmodulin-dependent protein kinase
- CREB
- cyclic adenosine 3′,5′-monophosphate-responsive element binding
- IEG
- immediate early gene
- Arc/Arg3.1
- activity-regulated cytoskeletal-associated protein
- SRF
- serum response factor
- GPCR
- G-protein coupled receptor
- IP3
- inositol 1,4,5-trisphosphate
- IP3
- IP3 receptor
- DHPG
- (S)-3, 5-dihydroxyphenylglycine
- Quis
- quisqualate
- LTP
- long term potentiation
- LTD
- long term depression
- MEF2
- myocyte enhancer factor 2
- NMDA
- N-methyl-d-aspartate
- VGCC
- voltage-gated Ca2+ channels
- CPCCOEt
- 7-(hydroxyimino)cyclopropa[b]chromen-1a-carboxylate ethyl ester
- d-APV
- d-(−)-2-amino-5-phosphonopentanoic acid
- RFP
- red fluorescent protein
- dn
- dominant-negative
- Atf3
- activating transcription factor 3
- Nr4a1
- nuclear receptor subfamily 4 group A member 1
- SIK1
- SNF1-like kinase
- Egr2
- early growth response 2
- MPEP
- 2-methyl-6-(phenylethynyl)-pyridine
- NeuN
- neuronal nuclei
- EGFP
- enhanced GFP
- NES
- nuclear export signal.
REFERENCES
- 1. Jong Y. J., Kumar V., O'Malley K. L. (2009) Intracellular metabotropic glutamate receptor 5 (mGluR5) activates signaling cascades distinct from cell surface counterparts. J. Biol. Chem. 284, 35827–35838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hermans E., Challiss R. A. (2001) Structural, signaling and regulatory properties of the group I metabotropic glutamate receptors. Prototypic family C G-protein-coupled receptors. Biochem. J. 359, 465–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Catania M. V., D'Antoni S., Bonaccorso C. M., Aronica E., Bear M. F., Nicoletti F. (2007) Group I metabotropic glutamate receptors. A role in neurodevelopmental disorders? Mol. Neurobiol. 35, 298–307 [DOI] [PubMed] [Google Scholar]
- 4. Dölen G., Bear M. F. (2008) Role for metabotropic glutamate receptor 5 (mGluR5) in the pathogenesis of fragile X syndrome. J. Physiol. 586, 1503–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gasparini F., Bilbe G., Gomez-Mancilla B., Spooren W. (2008) mGluR5 antagonists. Discovery, characterization, and drug development. Curr. Opin. Drug Discov. Devel. 11, 655–665 [PubMed] [Google Scholar]
- 6. Jong Y. J., Kumar V., Kingston A. E., Romano C., O'Malley K. L. (2005) Functional metabotropic glutamate receptors on nuclei from brain and primary cultured striatal neurons. Role of transporters in delivering ligand. J. Biol. Chem. 280, 30469–30480 [DOI] [PubMed] [Google Scholar]
- 7. Kumar V., Jong Y. J., O'Malley K. L. (2008) Activated nuclear metabotropic glutamate receptor mGlu5 couples to nuclear Gq/11 proteins to generate inositol 1,4,5-trisphosphate-mediated nuclear Ca2+ release. J. Biol. Chem. 283, 14072–14083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. O'Malley K. L., Jong Y. J., Gonchar Y., Burkhalter A., Romano C. (2003) Activation of metabotropic glutamate receptor mGlu5 on nuclear membranes mediates intranuclear Ca2+ changes in heterologous cell types and neurons. J. Biol. Chem. 278, 28210–28219 [DOI] [PubMed] [Google Scholar]
- 9. Tzingounis A. V., Nicoll R. A. (2006) Arc/Arg3.1. Linking gene expression to synaptic plasticity and memory. Neuron 52, 403–407 [DOI] [PubMed] [Google Scholar]
- 10. Bramham C. R., Worley P. F., Moore M. J., Guzowski J. F. (2008) The immediate early gene arc/arg3.1. Regulation, mechanisms, and function. J. Neurosci. 28, 11760–11767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Alberi L., Liu S., Wang Y., Badie R., Smith-Hicks C., Wu J., Pierfelice T. J., Abazyan B., Mattson M. P., Kuhl D., Pletnikov M., Worley P. F., Gaiano N. (2011) Activity-induced Notch signaling in neurons requires Arc/Arg3.1 and is essential for synaptic plasticity in hippocampal networks. Neuron 69, 437–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bloomer W. A., VanDongen H. M., VanDongen A. M. (2007) Activity-regulated cytoskeleton-associated protein Arc/Arg3.1 binds to spectrin and associates with nuclear promyelocytic leukemia bodies. Brain Res. 1153, 20–33 [DOI] [PubMed] [Google Scholar]
- 13. Bauer D. C., Willadsen K., Buske F. A., Lê Cao K. A., Bailey T. L., Dellaire G., Bodén M. (2011) Sorting the nuclear proteome. Bioinformatics 27, i7–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Park S., Park J. M., Kim S., Kim J. A., Shepherd J. D., Smith-Hicks C. L., Chowdhury S., Kaufmann W., Kuhl D., Ryazanov A. G., Huganir R. L., Linden D. J., Worley P. F. (2008) Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron 59, 70–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Waung M. W., Pfeiffer B. E., Nosyreva E. D., Ronesi J. A., Huber K. M. (2008) Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron 59, 84–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Mennerick S., Que J., Benz A., Zorumski C. F. (1995) Passive and synaptic properties of hippocampal neurons grown in microcultures and in mass cultures. J. Neurophysiol. 73, 320–332 [DOI] [PubMed] [Google Scholar]
- 17. Ramanan N., Shen Y., Sarsfield S., Lemberger T., Schütz G., Linden D. J., Ginty D. D. (2005) SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 8, 759–767 [DOI] [PubMed] [Google Scholar]
- 18. Gomes D. A., Rodrigues M. A., Leite M. F., Gomez M. V., Varnai P., Balla T., Bennett A. M., Nathanson M. H. (2008) c-Met must translocate to the nucleus to initiate calcium signals. J. Biol. Chem. 283, 4344–4351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wayman G. A., Impey S., Marks D., Saneyoshi T., Grant W. F., Derkach V., Soderling T. R. (2006) Activity-dependent dendritic arborization mediated by CaM kinase I activation and enhanced CREB-dependent transcription of Wnt-2. Neuron 50, 897–909 [DOI] [PubMed] [Google Scholar]
- 20. Bloomer W. A., VanDongen H. M., VanDongen A. M. (2008) Arc/Arg3.1 translation is controlled by convergent N-methyl-d-aspartate and Gs-coupled receptor signaling pathways. J. Biol. Chem. 283, 582–592 [DOI] [PubMed] [Google Scholar]
- 21. Rozen S., Skaletsky H. J. (2000) in Bioinformatics Methods and Protocols: Methods in Molecular Biology (Krawetz S. A., Misener S., eds) pp. 365–386, Humana Press Inc., Totowa, NJ [Google Scholar]
- 22. López de Maturana R., Sánchez-Pernaute R. (2010) Regulation of corticostriatal synaptic plasticity by G protein-coupled receptors. CNS Neurol. Disord. Drug Targets 9, 601–615 [DOI] [PubMed] [Google Scholar]
- 23. Flavell S. W., Cowan C. W., Kim T. K., Greer P. L., Lin Y., Paradis S., Griffith E. C., Hu L. S., Chen C., Greenberg M. E. (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 [DOI] [PubMed] [Google Scholar]
- 24. Pintchovski S. A., Peebles C. L., Kim H. J., Verdin E., Finkbeiner S. (2009) The serum response factor and a putative novel transcription factor regulate expression of the immediate-early gene Arc/Arg3.1 in neurons. J. Neurosci. 29, 1525–1537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tian X., Kai L., Hockberger P. E., Wokosin D. L., Surmeier D. J. (2010) MEF-2 regulates activity-dependent spine loss in striatopallidal medium spiny neurons. Mol. Cell. Neurosci. 44, 94–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Smith-Hicks C., Xiao B., Deng R., Ji Y., Zhao X., Shepherd J. D., Posern G., Kuhl D., Huganir R. L., Ginty D. D., Worley P. F., Linden D. J. (2010) SRF binding to SRE 6.9 in the Arc promoter is essential for LTD in cultured Purkinje cells. Nat. Neurosci. 13, 1082–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boivin B., Vaniotis G., Allen B. G., Hébert T. E. (2008) G protein-coupled receptors in and on the cell nucleus. A new signaling paradigm? J. Recept. Signal Transduct. Res. 28, 15–28 [DOI] [PubMed] [Google Scholar]
- 28. Feijóo C. G., Sarrazin A. F., Allende M. L., Glavic A. (2009) Cysteine-serine-rich nuclear protein 1, Axud1/Csrnp1, is essential for cephalic neural progenitor proliferation and survival in zebrafish. Dev. Dyn. 238, 2034–2043 [DOI] [PubMed] [Google Scholar]
- 29. Pérez-Cadahía B., Drobic B., Davie J. R. (2011) Activation and function of immediate-early genes in the nervous system. Biochem. Cell Biol. 89, 61–73 [DOI] [PubMed] [Google Scholar]
- 30. Kim J., Efe J. A., Zhu S., Talantova M., Yuan X., Wang S., Lipton S. A., Zhang K., Ding S. (2011) Direct reprogramming of mouse fibroblasts to neural progenitors. Proc. Natl. Acad. Sci. U.S.A. 108, 7838–7843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhao Y., Bruemmer D. (2010) NR4A orphan nuclear receptors. Transcriptional regulators of gene expression in metabolism and vascular biology. Arterioscler. Thromb. Vasc. Biol. 30, 1535–1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Töröcsik B., Angelastro J. M., Greene L. A. (2002) The basic region and leucine zipper transcription factor MafK is a new nerve growth factor-responsive immediate early gene that regulates neurite outgrowth. J. Neurosci. 22, 8971–8980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zanocco-Marani T. (2010) TIS11/TTP gene family. It's never too late for tumor suppressors. Cell Cycle 9, 4771. [DOI] [PubMed] [Google Scholar]
- 34. Kucharska A., Rushworth L. K., Staples C., Morrice N. A., Keyse S. M. (2009) Regulation of the inducible nuclear dual-specificity phosphatase DUSP5 by ERK MAPK. Cell. Signal. 21, 1794–1805 [DOI] [PubMed] [Google Scholar]
- 35. Galbraith M. D., Espinosa J. M. (2011) Lessons on transcriptional control from the serum response network. Curr. Opin. Genet. Dev. 21, 160–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chai J., Tarnawski A. S. (2002) Serum response factor. Discovery, biochemistry, biological roles, and implications for tissue injury healing. J. Physiol. Pharmacol. 53, 147–157 [PubMed] [Google Scholar]
- 37. Tullai J. W., Schaffer M. E., Mullenbrock S., Kasif S., Cooper G. M. (2004) Identification of transcription factor binding sites upstream of human genes regulated by the phosphatidylinositol 3-kinase and MEK/ERK signaling pathways. J. Biol. Chem. 279, 20167–20177 [DOI] [PubMed] [Google Scholar]
- 38. Katoh Y., Takemori H., Min L., Muraoka M., Doi J., Horike N., Okamoto M. (2004) Salt-inducible kinase-1 represses cAMP response element-binding protein activity both in the nucleus and in the cytoplasm. Eur. J. Biochem. 271, 4307–4319 [DOI] [PubMed] [Google Scholar]
- 39. Borlikova G., Endo S. (2009) Inducible cAMP early repressor (ICER) and brain functions. Mol. Neurobiol. 40, 73–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Abraham W. C., Dragunow M., Tate W. P. (1991) The role of immediate early genes in the stabilization of long term potentiation. Mol. Neurobiol. 5, 297–314 [DOI] [PubMed] [Google Scholar]
- 41. Choe E. S., Wang J. Q. (2002) CREB and Elk-1 phosphorylation by metabotropic glutamate receptors in striatal neurons (review). Int. J. Mol. Med. 9, 3–10 [PubMed] [Google Scholar]
- 42. Wang Y., Zheng F., Zhou X., Sun Z., Wang H. (2009) Converging signal on ERK1/2 activity regulates group I mGluR-mediated Arc transcription. Neurosci. Lett. 460, 36–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Pisani A., Bernardi G., Bonsi P., Centonze D., Giacomini P., Calabresi P. (2000) Cell-type specificity of mGluR activation in striatal neuronal subtypes. Amino Acids 19, 119–129 [DOI] [PubMed] [Google Scholar]
- 44. Lefebvre C., Fisher K., Cahill C. M., Coderre T. J. (2000) Evidence that DHPG-induced nociception depends on glutamate release from primary afferent C-fibers. Neuroreport 11, 1631–1635 [DOI] [PubMed] [Google Scholar]
- 45. Fagni L., Chavis P., Ango F., Bockaert J. (2000) Complex interactions between mGluRs, intracellular Ca2+ stores, and ion channels in neurons. Trends Neurosci. 23, 80–88 [DOI] [PubMed] [Google Scholar]
- 46. Lorrain D. S., Correa L., Anderson J., Varney M. (2002) Activation of spinal group I metabotropic glutamate receptors in rats evokes local glutamate release and spontaneous nociceptive behaviors. Effects of 2-methyl-6-(phenylethynyl)-pyridine pretreatment. Neurosci. Lett. 327, 198–202 [DOI] [PubMed] [Google Scholar]
- 47. Thomas L. S., Jane D. E., Harris J. R., Croucher M. J. (2000) Metabotropic glutamate autoreceptors of the mGlu(5) subtype positively modulate neuronal glutamate release in the rat forebrain in vitro. Neuropharmacology 39, 1554–1566 [DOI] [PubMed] [Google Scholar]
- 48. Jones N. M., Lawrence A. J., Beart P. M. (1998) In vivo microdialysis reveals facilitatory metabotropic glutamate receptors regulating excitatory amino acid release in rat nucleus tractus solitarius. Neurochem. Int. 32, 31–38 [DOI] [PubMed] [Google Scholar]
- 49. Thomas L. S., Jane D. E., Gasparini F., Croucher M. J. (2001) Glutamate release inhibiting properties of the novel mGlu(5) receptor antagonist 2-methyl-6-(phenylethynyl)-pyridine (MPEP). Complementary in vitro and in vivo evidence. Neuropharmacology 41, 523–527 [DOI] [PubMed] [Google Scholar]
- 50. Romano C., van den Pol A. N., O'Malley K. L. (1996) Enhanced early developmental expression of the metabotropic glutamate receptor mGluR5 in rat brain. Protein, mRNA splice variants, and regional distribution. J. Comp. Neurol. 367, 403–412 [DOI] [PubMed] [Google Scholar]
- 51. Alvarez F. J., Villalba R. M., Carr P. A., Grandes P., Somohano P. M. (2000) Differential distribution of metabotropic glutamate receptors 1a, 1b, and 5 in the rat spinal cord. J. Comp. Neurol. 422, 464–487 [DOI] [PubMed] [Google Scholar]
- 52. Chavis P., Fagni L., Lansman J. B., Bockaert J. (1996) Functional coupling between ryanodine receptors and L-type calcium channels in neurons. Nature 382, 719–722 [DOI] [PubMed] [Google Scholar]
- 53. Park C. Y., Shcheglovitov A., Dolmetsch R. (2010) The CRAC channel activator STIM1 binds and inhibits L-type voltage-gated calcium channels. Science 330, 101–105 [DOI] [PubMed] [Google Scholar]
- 54. Wang Y., Deng X., Mancarella S., Hendron E., Eguchi S., Soboloff J., Tang X. D., Gill D. L. (2010) The calcium store sensor, STIM1, reciprocally controls Orai and CaV1.2 channels. Science 330, 105–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Murtazina D. A., Chung D., Ulloa A., Bryan E., Galan H. L., Sanborn B. M. (2011) TRPC1, STIM1, and ORAI influence signal-regulated intracellular and endoplasmic reticulum calcium dynamics in human myometrial cells. Biol. Reprod. 85, 315–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Cheng K. T., Liu X., Ong H. L., Swaim W., Ambudkar I. S. (2011) Local Ca2+ entry via Orai1 regulates plasma membrane recruitment of TRPC1 and controls cytosolic Ca2+ signals required for specific cell functions. PLoS Biol. 9, e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Salido G. M., Jardín I., Rosado J. A. (2011) The TRPC ion channels. Association with Orai1 and STIM1 proteins and participation in capacitative and non-capacitative calcium entry. Adv. Exp. Med. Biol. 704, 413–433 [DOI] [PubMed] [Google Scholar]
- 58. Hoover P. J., Lewis R. S. (2011) Stoichiometric requirements for trapping and gating of Ca2+ release-activated Ca2+ (CRAC) channels by stromal interaction molecule 1 (STIM1). Proc. Natl. Acad. Sci. U.S.A. 108, 13299–13304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Donai H., Sugiura H., Ara D., Yoshimura Y., Yamagata K., Yamauchi T. (2003) Interaction of Arc with CaM kinase II and stimulation of neurite extension by Arc in neuroblastoma cells expressing CaM kinase II. Neurosci. Res. 47, 399–408 [DOI] [PubMed] [Google Scholar]
- 60. Vazdarjanova A., Ramirez-Amaya V., Insel N., Plummer T. K., Rosi S., Chowdhury S., Mikhael D., Worley P. F., Guzowski J. F., Barnes C. A. (2006) Spatial exploration induces ARC, a plasticity-related immediate-early gene, only in calcium/calmodulin-dependent protein kinase II-positive principal excitatory and inhibitory neurons of the rat forebrain. J. Comp. Neurol. 498, 317–329 [DOI] [PubMed] [Google Scholar]
- 61. Chotiner J. K., Nielson J., Farris S., Lewandowski G., Huang F., Banos K., de Leon R., Steward O. (2010) Assessment of the role of MAP kinase in mediating activity-dependent transcriptional activation of the immediate early gene Arc/Arg3.1 in the dentate gyrus in vivo. Learn. Mem. 17, 117–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Huang F., Chotiner J. K., Steward O. (2007) Actin polymerization and ERK phosphorylation are required for Arc/Arg3.1 mRNA targeting to activated synaptic sites on dendrites. J. Neurosci. 27, 9054–9067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stritt C., Knöll B. (2010) Serum response factor regulates hippocampal lamination and dendrite development and is connected with reelin signaling. Mol. Cell. Biol. 30, 1828–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Shepherd J. D., Rumbaugh G., Wu J., Chowdhury S., Plath N., Kuhl D., Huganir R. L., Worley P. F. (2006) Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron 52, 475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Rial Verde E. M., Lee-Osbourne J., Worley P. F., Malinow R., Cline H. T. (2006) Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron 52, 461–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Chowdhury S., Shepherd J. D., Okuno H., Lyford G., Petralia R. S., Plath N., Kuhl D., Huganir R. L., Worley P. F. (2006) Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron 52, 445–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.