Background: SepCysS catalyzes the conversion of tRNA-bound O-phosphoserine to Cys.
Results: Three Cys residues are essential for SepCysS activity, and two of them form disulfide and persulfide intermediates.
Conclusion: SepCysS transfers sulfur through generating disulfide and persulfide enzyme adducts.
Significance: Elucidation of the sulfur transfer mechanism of SepCysS is crucial for understanding the tRNA-dependent Cys biosynthesis and sulfur metabolism in methanogens.
Keywords: Archaea, Disulfide, Enzyme Mechanisms, Sulfur, Transfer RNA (tRNA), Sep-tRNA:Cys-tRNA Synthase, Cysteine, Methanogen, Persulfide
Abstract
Sep-tRNA:Cys-tRNA synthase (SepCysS) catalyzes the sulfhydrylation of tRNA-bound O-phosphoserine (Sep) to form cysteinyl-tRNACys (Cys-tRNACys) in methanogens that lack the canonical cysteinyl-tRNA synthetase (CysRS). A crystal structure of the Archaeoglobus fulgidus SepCysS apoenzyme provides information on the binding of the pyridoxal phosphate cofactor as well as on amino acid residues that may be involved in substrate binding. However, the mechanism of sulfur transfer to form cysteine was not known. Using an in vivo Escherichia coli complementation assay, we showed that all three highly conserved Cys residues in SepCysS (Cys64, Cys67, and Cys272 in the Methanocaldococcus jannaschii enzyme) are essential for the sulfhydrylation reaction in vivo. Biochemical and mass spectrometric analysis demonstrated that Cys64 and Cys67 form a disulfide linkage and carry a sulfane sulfur in a portion of the enzyme. These results suggest that a persulfide group (containing a sulfane sulfur) is the proximal sulfur donor for cysteine biosynthesis. The presence of Cys272 increased the amount of sulfane sulfur in SepCysS by 3-fold, suggesting that this Cys residue facilitates the generation of the persulfide group. Based upon these findings, we propose for SepCysS a sulfur relay mechanism that recruits both disulfide and persulfide intermediates.
Introduction
Aminoacyl-tRNAs are normally synthesized by direct attachment of specific amino acids to their cognate tRNAs. However, at least four aminoacyl-tRNAs (Asn-tRNAAsn, Gln-tRNAGln, Cys-tRNACys, and Sec-tRNASec) can be generated through tRNA-dependent modification reactions (1). For the biosynthesis of Cys-tRNACys in methanogenic archaea, tRNACys is initially aminoacylated with Sep3 by SepRS; then Sep-tRNACys is converted to Cys-tRNACys by SepCysS (2). The SepRS/SepCysS pathway is the primary pathway for Cys biosynthesis in methanococci (3), which do not possess O-acetylserine sulfhydrylase, the bacterial cysteine biosynthesis enzyme (4). Furthermore, the canonical CysRS is either absent (5) or nonessential in these organisms (6). SepRS and SepCysS are conserved in most methanogenic archaea (except the Methanobrevibacter and Methanosphaera species) and the Archaeoglobus species, which are sulfur-reducing archaea and have many enzymes of the methanogenesis pathway (7). Phylogenetic analysis suggested that the SepRS/SepCysS pathway is perhaps ancient and was already present at the time of the last universal common ancestor (7).
SepCysS is a pyridoxal 5′-phosphate (PLP)-dependent enzyme converting the tRNA-bound Sep moiety to cysteine using an undefined sulfur donor (2). Sulfide, cysteine, and thiophosphate were sufficient sulfur donors in in vitro assays. However, all of them support only a very slow formation of cysteine (8). Therefore, the natural sulfur donor and the sulfur delivery mechanism of SepCysS remain unclear. A persulfide group is proposed to be a sulfur donor for cysteine biosynthesis, mainly based upon the fact that SepCysS is most closely related to cysteine desulfurase in terms of primary sequence (7) and overall structure (9). Cysteine desulfurase catalyzes sulfur transfer by mobilizing the thiol group of free cysteine, yielding a persulfide enzyme adduct and alanine (10). The persulfidic sulfur (R-S-SH), which is covalently linked to a conserved Cys residue of cysteine desulfurase, is then donated to other sulfur carrier proteins for the biosynthesis of Fe-S clusters, thionucleotides, or sulfur-containing vitamins (11). Analogous to cysteine desulfurase, SepCysS may mobilize sulfur from a sulfur source, form a persulfide group on a Cys residue, and then donate the sulfur to Sep-tRNACys to generate Cys-tRNACys.
Three Cys residues of SepCysS (Cys39, Cys42, and Cys247 of the Archaeoglobus fulgidus enzyme) have been proposed to be candidates for persulfidation according to two observations (9). First, the three Cys residues are completely conserved in SepCysS, indicating that they are possibly essential for activity. Second, the crystal structure of A. fulgidus SepCysS showed that, similar to cysteine desulfurase, the enzyme forms a dimer, and the active cleft is located at the dimer interface. Cys247 is located near the active site in the crystal structure. Cys39 and Cys42, although located within a disordered region in the crystal structure, could be modeled proximal to the active site (9). However, whether the three Cys residues are involved in sulfur delivery awaits further investigation.
Here, we present that three Cys residues (Cys64, Cys67, and Cys272 of Methanocaldococcus jannaschii SepCysS corresponding to Cys39, Cys42, and Cys247 of the A. fulgidus enzyme) are essential for the sulfhydrylation reaction in vivo. More specifically, Cys64 and Cys67 are responsible for forming a disulfide linkage and carrying a sulfane sulfur. Therefore, we propose that through disulfide formation and disulfide-persulfide rearrangement, sulfur from a persulfide sulfur donor is transferred to SepCysS and in turn delivered to the tRNA-bound Sep for Cys-tRNACys formation.
EXPERIMENTAL PROCEDURES
Materials and Reagents
Oligonucleotide synthesis and DNA sequencing were performed by the Keck Foundation Biotechnology Resource Laboratory at Yale University. The maltose-binding protein-tagged IscS clone was a gift from Eugene Mueller (University of Louisville). N-(iodoacetamino-ethyl)-1-naphthylamine-5-sulfonic acid (1,5-I-AEDANS), 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), PLP, and crystalline sodium sulfide (Na2S·9H2O) were purchased from Sigma. Dithiothreitol (DTT), isopropyl β-d-1-thiogalactopyranoside, and imidazole were purchased from American Bioanalytical.
Cloning, Expression, and Purification of Recombinant His6-SepCysS
The SepCysS gene from M. jannaschii (MJ1678) and A. fulgidus (AF0028) were cloned into pET15b vector as described (12). The mutations of MJ1678, including six single mutants (K234A, C64A, C67A, C113A, C209A, and C272A), three double mutants (C64A/C67A, C64A/C272A, and C67A/C272A), and one triple mutant (C64A/C67A/C272A), were constructed using the QuikChange mutagenesis kit (Stratagene, La Jolla, CA). The same set of mutants was also constructed for A. fulgidus SepCysS. The DNA oligonucleotides used for mutagenesis were designed using a web-based program primerX or a program provided by Stratagene. The manufacturer's recommended protocol was followed except that electrocompetent DH10β cells were used for transformation. All mutations were confirmed by plasmid DNA sequencing.
The resulting plasmids were individually transformed into the Escherichia coli BL21-CodonPlus (DE3) strain (Stratagene) for expression of recombinant proteins. The transformed cells were grown in 1 liter of Luria-Bertani (LB) medium supplemented with 100 μg/ml ampicillin at 37 °C with shaking until they reached an absorbance at 600 nm of 0.6∼0.8. Isopropyl β-d-1-thiogalactopyranoside was added to a final concentration of 1 mm to induce the production of recombinant proteins. After an additional 5-h incubation at 30 °C or overnight incubation at 15 °C, the cells were harvested by centrifugation at 5,000 × g for 20 min at 4 °C. For aerobic protein purification, cells were disrupted by sonication, and His-tagged SepCysS was purified with nickel-nitrilotriacetic acid resin (Qiagen) as described (2). For anaerobic purification of M. jannaschii SepCysS, the harvested E. coli cells were transferred into the anaerobic chamber (atmosphere of 95% N2 and 5% H2) and resuspended in 20 ml of buffer A (20 mm sodium phosphate, 0.5 m NaCl, 20 mm imidazole, pH 7.4). The cells were disrupted by the addition of 2 ml of BugBuster (Novagen). RNA and DNA were digested with 10 μl of Benzonase (Sigma) by incubation at room temperature for 30 min. E. coli host proteins were then denatured by heating at 75 °C for 30 min and separated from soluble proteins by centrifugation at 100,000 × g for 30 min at 4 °C. For further purification of recombinant SepCysS, the supernatant was applied to 1 ml of TALON Metal Affinity Resins (Clontech) equilibrated with buffer A. Proteins bound to the column were eluted with 5 ml of buffer B (20 mm sodium phosphate, 0.5 m NaCl, 200 mm imidazole, pH 7.4). The elution fractions of the desired proteins were analyzed by SDS-PAGE, and the purified proteins were then dialyzed against buffer C (50 mm HEPES-NaOH, 20 mm MgCl2, pH 8.0), concentrated to a volume of 1 ml with a 30-kDa cutoff centrifugal filter (Millipore), and stored at −20 °C until use. Protein concentrations were determined with the bicinchoninic acid assay (13).
Complementation of E. coli ΔselA Mutant
The M. jannaschii wild-type (MJ1678) and mutant SepCysS genes (K234A, C64A, C67A, C272A, C209A, and C113A) were transformed into the E. coli ΔselA strain JS1 together with the M. jannaschii phosphoseryl-tRNASec kinase gene (MJ1538). The complementation experiment was performed as described (14). Briefly, the transformed strains were plated on glucose-minimum medium agar plates and grown for 2 days at 30 °C. The plates were then overlaid with agar containing 1 mg/ml benzyl viologen, 0.25 m sodium formate, and 25 mm KH2PO4, adjusted to pH 7.0. The appearance of blue/purple colonies is an indication of the production of active formate dehydrogenase H (15) and therefore a complementation with active SepCysS.
Spectroscopic Analysis of PLP
Absorption spectra of purified recombinant M. jannaschii SepCysS (20 μm) were measured with the spectrophotometer system (Shimadzu, UV-2401PC) aerobically in cuvettes with a 1.0-cm light path at room temperature. The absorption spectra of the wild-type enzyme were compared with the mutant SepCysS (K234A) with and without potassium borohydride (20 mm) treatment.
Sulfur Transfer Assay with Radioactive Sulfur
M. jannaschii SepCysS was purified anaerobically as described above. The following procedure was carried out under anaerobic conditions. Purified maltose binding protein-tagged IscS (2.5–10 μm) was incubated with SepCysS (20 μm) in the presence of 35[S]Cys (150 μm) at 37 °C in reaction buffer containing 50 mm HEPES, pH 7.3, 150 mm KCl, 10 mm MgCl2, and 10 μm PLP for 10 min, 30 min, 1 h, 2 h, and 3 h (supplemental Fig. S1). The reaction was stopped by the addition of nonreducing SDS loading dye. The protein mixture was then separated by SDS-PAGE, and the radioactivity retained on the gel was followed by autoradiography.
Determination of Disulfide Content of SepCysS by Alkaline Sulfide/Cyanolysis Assay
The alkaline sulfide/cyanolysis assay (16) was used to determine disulfide linkages in SepCysS. The A. fulgidus wild-type SepCysS and five mutants including C247A, C39A/C42A, C39A/C247A, C42A/C247A, and C39A/C42A/C247A were purified in the presence of 10 mm DTT to remove intrinsic persulfidic sulfur. The protein preparations were then dialyzed extensively against an anoxic buffer (50 mm HEPES-NaOH, 20 mm MgCl2, pH 8.0, degassed with N2 for 1 h) to remove DTT and thus allow generation of disulfide linkages with trace amount of oxygen or other oxidants in the buffer. Circular dichroism spectra showed that no structural changes are introduced to these mutant proteins (data not shown). The wild-type and mutant SepCysS proteins (5 μg/μl) were then treated with sulfide (2 mm) at pH 9.0 anaerobically at room temperature for 3 h to allow disulfide linkages to react with sulfide and in turn form persulfide groups. Persulfide groups in the proteins were then reacted with cyanide (10 mm) at room temperature to produce thiocyanate (16). The amount of thiocyanate was measured by absorbance at 470 nm after reaction with the ferric reagent (17).
Determination of Thiol Content of SepCysS by DTNB Titration
The anaerobically purified M. jannaschii wild-type SepCysS and three mutants including C64A, C67A, and C272A were subjected to DTNB titrations under denaturing condition to determine the presence of disulfide bond in the resting state of the enzyme. The enzyme sample (0.5–2 nmol) in 200 μl of denaturing buffer containing 0.1 m sodium phosphate, pH 7.3, 1 mm disodium EDTA, and 6.4 m guanidinium chloride was mixed with 200 μl of 10 mm DTNB. The mixture (400 μl) was incubated for 30 min at room temperature, and the production of 2-nitro-5-thiobenzoate anion (TNB2−) was quantified by absorbance at 412 nm using an extinction coefficient of 13,700 m−1 cm−1 (18). The mixture without addition of protein was used as a blank control. All reactions were performed in triplicate.
Determination of Persulfide Content of SepCysS by Fluorescent Labeling
For detection of persulfide groups, the anaerobically purified M. jannaschii SepCysS (∼2 nmol) in 100 μl of buffer C (50 mm HEPES-NaOH, 20 mm MgCl2, pH 8.0) was incubated anaerobically with 20 nmol of the fluorescent dye 1,5-I-AEDANS at 37 °C for 30 min to derivatize thiol groups in the protein (19). The reaction mixture was then applied to a Microcon YM-10 Centrifugal Filter unit (Millipore) and centrifuged at 14,000 × g for 15 min. The protein in the retentate was washed four times with 350 μl of buffer C to remove unreacted fluorescent dye. The protein sample was then reduced to a volume of 50 μl by centrifugal filtration, supplemented with 50 μl of 10 mm DTT, and incubated at 37 °C for 20 min. In this step, persulfide groups, if reduced by DTT, would release the fluorescent dye into the solution. Finally, the reduced protein sample was applied to a Microcon YM-10 Centrifugal Filter and centrifuged at 14,000 × g to dryness. The free fluorescent dye collected in the filtrate was quantified using the QuantaMaster Fluorescence Spectrofluorometer (Photon Technology International) with λex 337 nm and λem 498 nm. The standard curve was generated with 0.01–0.5 nmol of 1,5-I-AEDANS in 100 μl of buffer C with 5 mm DTT. As a negative control, the M. jannaschii SepCysS protein (∼2 nmol) in 100 μl of buffer C was incubated with 50 nmol of DTT for 30 min at 37 °C to reduce persulfide groups before reaction with the fluorescent dye.
Identification of Disulfide Linkage and Sulfane Sulfur in SepCysS by Mass Spectrometry
The buffer of the anaerobically purified recombinant M. jannaschii SepCysS (MJ1678) was exchanged with 0.1 m ammonium bicarbonate-formic acid buffer, pH 7.2, by passage through a 0.5-ml Zeba Desalt Spin Column (Thermo Scientific). The protein (200 μg) was digested with 2 μg of trypsin (Promega) in a volume of 100 μl at 37 °C overnight, and then the samples were dried in vacuum. For the control experiment, the protein was incubated with 10 mm DTT at 60 °C for 1 h to remove persulfide group before trypsin digestion.
The peptide samples obtained from trypsin digestion were resuspended in 30 μl of deionized H2O containing 3% acetonitrile and 0.1% formic acid, and 8 μl of sample was analyzed on an Agilent 1100 Capillary LC (Palo Alto, CA) interfaced directly to a linear ion trap (LTQ) mass spectrometer (Thermo Electron, San Jose, CA). Mobile phases A and B were H2O-0.1% formic acid and acetonitrile-0.1% formic acid, respectively. Peptides eluted from the C18 column were injected into the mass spectrometer during a 60-min linear gradient from 5 to 60% mobile phase B at a flow rate of 4 μl/min. The instrument was set to acquire MS/MS spectra on the nine most abundant precursor ions from each MS scan with a repeat count of 3 and repeat duration of 5 s. Dynamic exclusion was enabled for 60 s. Generated raw MS/MS were converted into the mzXML format and then into the PKL format using ReAdW followed by mzMXL2Other (20). The peak lists were then searched using the Mascot 2.2 software (Matrix Science, Boston, MA) against the sequence of MJ1678 with the following parameters: full tryptic enzymatic cleavage with two possible missed cleavages, peptide tolerance of 1000 parts per million, fragment ion tolerance of 0.6 Da, and variable modifications with oxidation of methionyl residues (+16 Da), disulfide linkage of Cys residues (−2 Da), and persulfide modification of Cys residues (+32 Da).
RESULTS
Three Conserved Cys Residues Are Essential for SepCysS Activity
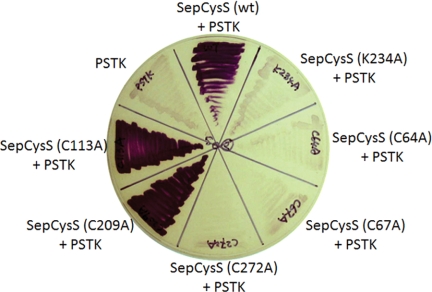
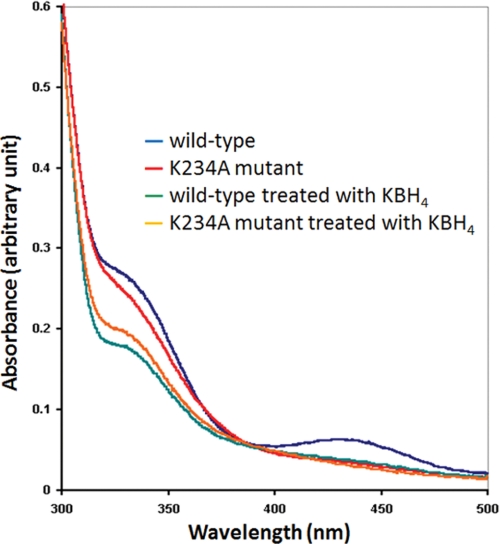
SepCysS can utilize Sep-tRNASec as a substrate and generate Cys-tRNASec in E. coli (14). In the E. coli ΔselA strain JS1, active formate dehydrogenase H cannot be produced due to the lack of selenocysteine incorporation (14). SepCysS in combination with phosphoseryl-tRNASec kinase complements the ΔselA strain and converts Ser-tRNASec → Cys-tRNASec to produce the cysteine homolog of formate dehydrogenase H, which has reduced activity compared with the native selenocysteine enzyme (14). Therefore, the in vivo activity of SepCysS can be tested by measuring formate dehydrogenase H activity, which reduces benzyl viologen with formate as the electron donor and results in blue/purple colonies (14). Using this method, the activities of six M. jannaschii SepCysS mutant enzymes were examined. The K234A mutant, which contains an Ala substitution of Lys234 (corresponding to Lys209 in A. fulgidus SepCysS), was inactive in the benzyl viologen assay (Fig. 1). This result is in agreement with the absorption spectra characterization (Fig. 2) and the crystal structure data (9) showing that Lys234 forms a Schiff base with PLP and therefore is functionally essential.
FIGURE 1.
Complementation of the E. coli ΔselA mutant with phosphoseryl-tRNASec kinase (PSTK) and SepCysS. The in vivo activities of M. jannaschii wild-type and mutant (K234A, C64A, C67A, C272A, C209A, and C113A) SepCysS converting Sep-tRNASec to Cys-tRNASec were tested by their ability to restore formate dehydrogenase H activity in the E. coli ΔselA mutant. The complementation was observed by formation of purple colonies resulted from reduction of benzyl viologen with formate. The complementation required co-expression of phosphoseryl-tRNASec kinase, which converts Ser-tRNASec to Sep-tRNASec.
FIGURE 2.
Spectra of the M. jannaschii wild-type and K234A mutant SepCysS. The absorption between 400–430 nm was used as an indication of internal Schiff base formed between PLP and a Lys residue of the enzyme (30).
To identify the Cys residues involved in sulfur delivery, five cysteines in M. jannaschii SepCysS were individually altered to Ala. The three Cys→Ala mutants of the conserved Cys residues (Cys64, Cys67, and Cys272 corresponding to Cys39, Cys42, and Cys272 in A. fulgidus SepCysS) lost their ability to complement the ΔselA strain (Fig. 1), indicating that these three cysteines are necessary for in vivo SepCysS activity. Interestingly, they are probably located near the SepCysS active site based upon the crystal structure and have been proposed as candidates for persulfidation (9). On the other hand, the other two Cys mutants (C113A and C209A) exhibited activity similar to that of wild-type SepCysS (Fig. 1), indicating that these two Cys residues are dispensable.
SepCysS Can Receive Sulfur from IscS
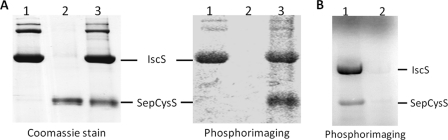
The conversion of Sep-tRNASec → Cys-tRNASec by SepCysS requires a sulfur donor in E. coli; therefore, we tested whether SepCysS can receive sulfur from another sulfur-carrying protein. The cysteine desulfurase IscS, which generates a persulfide enzyme adduct at the expense of free cysteine, is a central physiological sulfur donor for the biosynthesis of sulfur-containing cofactors and tRNAs in bacteria (10, 21, 22). To investigate whether E. coli IscS can donate sulfur to SepCysS, anaerobically purified recombinant M. jannaschii SepCysS was incubated with [35S]Cys in the absence or presence of the MBP-tagged IscS. The protein(s) was then subjected to SDS-PAGE analysis under nonreducing conditions, and the sulfur transfer from [35S]Cys to protein(s) was monitored by autoradiography. As shown in lane 1 of Fig. 3A, no significant labeling of SepCysS was observed when incubated with [35S]Cys by itself, indicating that sulfur was not able to transfer from free Cys directly to SepCysS. However, radioactivity was observed in the band corresponding to IscS when incubated with [35S]Cys (lane 2 of Fig. 3A), indicating that 35S was able to transfer from Cys directly to IscS. When SepCysS was incubated with IscS and [35S]Cys, radioactivity was detected in two bands that corresponded to IscS and SepCysS, respectively (lane 3 of Fig. 3A). Addition of the reducing reagent β-mercaptoethanol (1%, v/v) to the protein mixture removed the radioactive labeling (Fig. 3B); this suggests that 35S was transferred in the sulfane sulfur form. Therefore, these results indicate that SepCysS is able to accept IscS-associated sulfane sulfur.
FIGURE 3.
Sulfur transfer from the cysteine desulfurase IscS to SepCysS. A, E. coli IscS (8 μm; lane 1), M. jannaschii SepCysS (20 μm; lane 2), and the two proteins together (lane 3) were incubated with 150 μm [35S]Cys at 37 °C for 3 h, and then the incubation mixtures were analyzed by SDS-PAGE under nonreducing condition. The left and right panels show the Coomassie-stained gel and the PhosphorImager scan, respectively. The positions of maltose-binding protein-tagged IscS (∼90 kDa) and His-tagged SepCysS (∼45 kDa) are labeled. B, E. coli IscS (4 μm) and M. jannaschii SepCysS (20 μm) were incubated with 150 μm [35S]Cys at 37 °C for 30 min, and then the incubation mixtures were analyzed by SDS-PAGE under nonreducing condition (lane 1) and reducing condition with 1% (v/v) β-mercaptoethanol (lane 2).
SepCysS Carries Sulfane Sulfur
Because sulfur transfer from IscS to SepCysS occurred in vitro, we tested whether SepCysS expressed in E. coli carries sulfane sulfur. Recombinant M. jannaschii SepCysS was purified anaerobically in the absence of reducing reagents; then the free sulfhydryl groups of the purified protein were labeled with I-AEDANS, a fluorescent derivative of iodoacetamide (19). After removal of unreacted fluorescent dye by centrifugal filtration, the protein was treated with 100-fold excess of DTT. If persulfide (R-S-SH) or another sulfane sulfur containing group (e.g. polysulfide, RSSnSH) is present in the protein, the fluorophore would be released from the protein into solution upon reduction with DTT. In three independent treatments of 2 nmol of wild-type protein, 30 ± 10 pmol of the fluorophore were released; this suggests that ∼1.5% of the purified protein contained sulfane sulfur. For either of the C64A or the C67A mutant, no detectable amount of fluorophore (<5 pmol) was released after reduction with DTT, suggesting that Cys64 and Cys67 are essential for the formation of the sulfane sulfur-containing group. For the C272A mutant, only 10 ± 5 pmol of the fluorophore (∼0.5%) was released from 2 nmol of protein, which accounted for about one third of the wild-type level. The lower amount of sulfane sulfur in the C272A mutant suggests that Cys272 stimulates the formation or stabilizes the sulfane sulfur-containing group.
SepCysS Forms Disulfide Linkage
Intramolecular disulfide bonds are often formed between the sulfane sulfur-carrying Cys and a second catalytic Cys as a result of sulfur donation by sulfur transfer enzymes, such as ThiI (23, 24) and MnmA (25, 26). Two methods were used to determine whether SepCysS forms disulfide bonds.
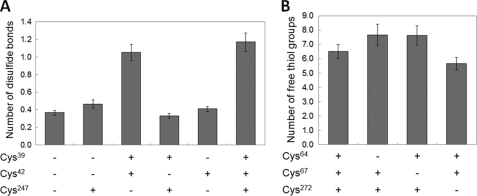
To test whether a disulfide bond can be formed by SepCysS in vitro, the A. fulgidus wild-type and mutant proteins were incubated with Na2S under alkaline conditions (16). In this process, disulfide bonds of the protein would be cleaved by sulfide to produce persulfide groups, which can then be quantified by the cyanolysis assay (16). As a control experiment, the protein was subjected to cyanolysis without alkaline sulfide treatment, and no significant amount of persulfide was detected. This control suggests that intrinsic persulfide was removed during the purification with DTT and would not interfere with the following assay. After removal of DTT by extensive dialysis with a nonreducing anoxic buffer, we tested whether disulfide linkages can be generated with trace amount of oxygen or other oxidants in the buffer. As a result, 1.2 ± 0.1 disulfide linkages were observed in the wild-type SepCysS after dialysis (Fig. 3). The background level was determined with the triple mutant, which has all three conserved cysteines (Cys39, Cys42, and Cys247) mutated to Ala. When either Cys39 or Cys42 (corresponding to Cys64 or Cys67 of M. jannaschii SepCysS, respectively) was substituted with Ala, the absorbance decreased to the background level (Fig. 4A), indicating that both cysteines are involved in disulfide bond formation. On the other hand, when Cys247 (corresponding to Cys272 of M. jannaschii SepCysS) was substituted with Ala, the absorbance was similar to that of the wild-type enzyme (Fig. 4A), suggesting that Cys247 is not involved in forming intramolecular disulfide.
FIGURE 4.
Determination of disulfide linkages in SepCysS. A, disulfide content in A. fulgidus SepCysS was determined with the alkaline sulfide/cyanolysis assay. B, free thiol content in M. jannaschii SepCysS was determined with DTNB titration. + indicates that the Cys residue is intact; − indicates that the Cys residue is altered to Ala. Error bars represent S.D. from three independent assays.
To test whether SepCysS contains disulfide bonds in the resting state, the M. jannaschii wild-type and mutant proteins were purified anaerobically in the absence of reducing reagents and then reacted anaerobically with DTNB. Under denaturing conditions, 6.5 ± 0.5 free thiol groups were observed in the wild-type SepCysS (Fig. 4B), which contains 9 Cys residues according to the protein sequence. When Cys64 and Cys67 were individually substituted with Ala, the number of free thiol groups increased to 7.7 ± 0.7 and 7.6 ± 0.7 (Fig. 4B), respectively, suggesting that these two cysteines were inaccessible to DTNB in the wild-type protein and potentially formed a disulfide bond. The C272A mutation decreased the number of free thiol groups to 5.7 ± 0.4 in the protein (Fig. 4B), suggesting that Cys272 is not involved in forming disulfide. These observations were consistent with those obtained from the alkaline sulfide/cyanolysis assay, which suggests that Cys64 and Cys67 naturally form a disulfide bond.
Identification of Disulfide Linkage and Sulfane Sulfur in SepCysS by Mass Spectrometry
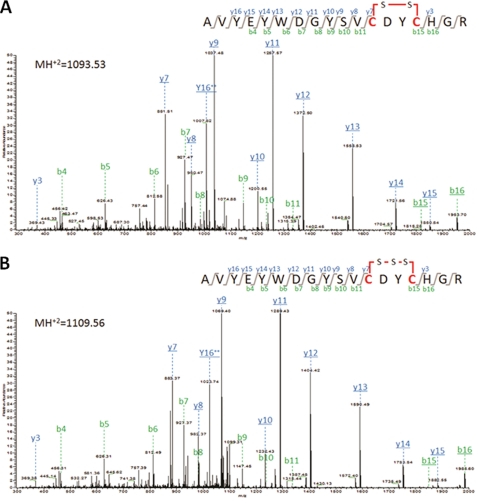
To confirm the presence of disulfide linkage and sulfane sulfur in SepCysS, the protein was purified anaerobically in the absence of reducing reagents and digested with trypsin, and the resulting peptides were then analyzed by LC-MS/MS (Fig. 5). For the peptide 53AVYEYWDGYSVCDYCHGR70, the unmodified precursor ion (2187.4 Da) was not detected. However, the precursor ions can be observed with either −2 Da (mass of −2H) or +30 Da (mass of −2H + 1S) shift. When the −2 Da shift was considered as the loss of dihydrogen caused by the formation of an intrapeptide disulfide between Cys64 and Cys67, analysis of the MS/MS spectra of the −2 Da shifted precursor ion confirmed the expected amino acid sequence (Fig. 5A). The fragmentation of the +30 Da precursor ion identified +32 Da shifts of the disulfide bond-containing fragment ions, suggesting that a sulfane sulfur was added to the disulfide bond (Fig. 5B). This fragmentation pattern presumably resulted from an oxidation (loss of dihydrogen) between a persulfide group on either Cys64 or Cys67 and a free thiol group from the other Cys to form a trisulfide linkage, which occurred either in vivo or during the protein purification and the mass spectrometry processes. No persulfide modification or disulfide linkage with another Cys-containing peptide was observed for Cys272.
FIGURE 5.
Identification of disulfide and sulfane sulfur modification of the M. jannaschii SepCysS by LC-MS/MS. Fragment b- and y-ions derived from the tryptic peptide 53AVYEYWDGYSVCDYCHGR70 (theoretical MH2+ = 1094.20) are labeled in green and blue, respectively. A, MS/MS fragmentation spectra of the −2 Da shifted precursor ion (MH2+ = 1093.53). Detected fragment b- and y-ions containing Cys64 and Cys67 with −2 Da shift are underlined. B, MS/MS fragmentation spectra of the +30 Da shifted precursor ion (MH2+ = 1109.56). Detected fragment b- and y-ions containing Cys64 and Cys67 with +30 Da shift are underlined.
DISCUSSION
The data presented here demonstrate that all three conserved Cys residues of SepCysS (Cys64, Cys67, and Cys272 in M. jannaschii SepCysS) are required for the conversion of the tRNA-bound Sep moiety to Cys. Furthermore, the formation of disulfide and trisulfide (presumably derived from an oxidation of a persulfide and a thiol group or from a reaction between two persulfide group to expel a bisulfide) linkages between Cys64 and Cys67 suggests that SepCysS employs both disulfide and persulfide intermediates for sulfur transfer.
The requirement of multiple cysteines for sulfur transfer by SepCysS resembles the sulfur relay enzymes for tRNA thiolation, e.g. ThiI for 4-thiouridine biosynthesis (23, 24) and MnmA for 2-thiouridine biosynthesis (25, 26). For both ThiI and MnmA, two cysteines are required for catalysis: one cysteine receives a sulfane sulfur from cysteine desulfurase and forms a persulfide enzyme adduct, and the other cysteine attacks the bridging sulfur of the persulfide to liberate the terminal sulfur and consequently forms a disulfide with the first cysteine. Based upon our findings that a disulfide is present in the resting state of SepCysS and both Cys64 and Cys67 are required for the formation of a persulfide, it is conceivable that a sulfur donor (equivalent of S2−) may attack one sulfur of the disulfide linkage to generate a persulfide on either Cys64 or Cys67, leaving the other Cys as a free thiol (Fig. 6). Then, a thiolate derived from deprotonation of the free thiol can attack the bridging sulfur of the persulfide to liberate the terminal sulfur for cysteine biosynthesis and consequently regenerate the disulfide for the next catalytic cycle (Fig. 6). This reaction scheme can also explain the in vitro activity with sulfide, which may directly attack the disulfide of SepCysS or the double bond of the dehydroalanyl moiety (8).
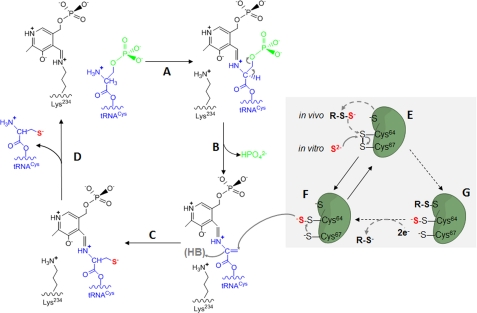
FIGURE 6.
Proposed mechanism of sulfur delivery by SepCysS. A–D, proposed PLP-dependent Sep → Cys conversion is based upon the mechanism of Sep → Sec conversion catalyzed by SepSecS (31). The electron movement through PLP has been omitted. E and F, proposed sulfur relay mechanism is based upon the disulfide and persulfide intermediates of SepCysS presented in this study. A, amino group of Sep-tRNACys attacks the Schiff base (internal aldimine) between Lys234 and PLP to form an external aldimine. B, electron delocalization leads to β-elimination of phosphate and formation of dehydroalanyl-tRNACys. C, double bond of dehydroalanyl-tRNACys is attacked by a persulfide sulfur donor to form the Cys moiety. D, Lys234 forms the Schiff base with PLP, leading to the release of Cys-tRNACys and regeneration of the active site of SepCysS. E, sulfur donor (S2− equivalent) attacks the disulfide between Cys64 and Cys67, and the sulfur (in red) is consequently transferred to either Cys64 or Cys67 (Cys64 shown in the scheme here) to generate a persulfide enzyme adduct of SepCysS. F, thiolate formed by the other Cys (Cys67 shown in the scheme here) attacks the bridging sulfur of the persulfide to liberate the terminal sulfur (in red) as a formal equivalent of S2− for Cys moiety biosynthesis. The intramolecular disulfide is consequently regenerated. G, a proposal of in vivo sulfur transfer from a persulfide sulfur donor (R-S-S−) to SepCysS is presented here with dashed arrows. The sulfur transfer results in an intermolecular disulfide between the sulfur donor and SepCysS and a persulfide on SepCysS. An exogenous reductant is required to resolve the intermolecular disulfide and release the sulfur donor (R-S−).
Although a S2− equivalent is likely a sulfur donor for the persulfidation of SepCysS (described above), the low in vitro sulfhydrylation activity of SepCysS with sodium sulfide suggested that bisulfide is unlikely to be the physiological sulfur donor (2, 8). Because SepCysS was able to receive sulfur from the IscS persulfide, an unidentified protein-carrying persulfide possibly provides the sulfur for the persulfidation of SepCysS. The nature of the protein sulfur carrier is still under investigation. In analogy to the sulfur transfer from IscS to IscU, which forms disulfide-bridged IscS-IscU complex during iron-sulfur cluster assembly (27), if the persulfide of a sulfur carrier protein nucleophilically attacks the disulfide of SepCysS, then an intermolecular disulfide between SepCysS and the sulfur carrier protein would be expected as a result of persulfide group transfer. In this scenario, an exogenous reductant would be required to dissociate the sulfur carrier protein from SepCysS and regenerate free thiol groups.
Although a disulfide or sulfane sulfur was not observed on Cys272 in our experiments, the reduced level of persulfide group in the C272A mutant suggested that Cys272 may facilitate the formation of the persulfide enzyme adduct. Moreover, replacement of Cys272 with Ser also abolished the activity of SepCysS (32), suggesting that a covalent modification on Cys272 is likely required for in vivo sulfur transfer. Scenarios such as that Cys272 serves as an initial nucleophilic group that attacks on a persulfide sulfur donor or as a general acid-base that deprotonates Cys64 or Cys67 can account for the observations, but further investigations are required to clarify its function.
This study provides a framework for understanding the sulfur transfer for the formation of Cys-tRNACys by SepCysS. SepCysS is a first example that recruits both persulfide and disulfide intermediates in methanogens. Given the obligately anaerobic lifestyle of methanogens, the mechanism of maintaining the relatively oxidized persulfide and disulfide species in the reducing cytoplasm is unclear. Because methanogens lack glutathione (28, 29), other thiol-containing compounds (e.g. coenzyme M, coenzyme B, and the heterodisulfide of coenzyme M and coenzyme B) may function as the redox-buffering molecules. Furthermore, the enzyme that delivers sulfur to SepCysS remains an open question for future research. Because cysteine desulfurase is not encoded in M. jannaschii and many other methanogens (3) and cysteine should not be the ultimate sulfur donor for Cys-tRNACys biosynthesis, a new type of enzyme that can initiate the generation of persulfide from a noncysteine sulfur source would be responsible for the persulfidation of SepCysS. Identification of this enzyme and its intermediates would more thoroughly elucidate the Cys-tRNACys biosynthetic system and may shed light on a general mechanism of sulfur metabolism in methanogenic archaea.
Supplementary Material
Acknowledgments
We thank Drs. William B. Whitman, Jiqiang Ling, Patrick O'Donoghue, and Michael J. Hohn for helpful discussions of this research and critical reviewing of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grant P41-RR-018502 (to R. O.). This work was also supported by Department of Energy Office of Basic Energy Sciences Grant DE-FG02-98ER20311 and NIGMS/National Institutes of Health Grant GM22854 (to D. S.) and National Science Foundation Grant MCB-071770 (to D. R. D.).

This article contains supplemental Fig. S1.
- Sep
- O-phosphoserine
- 1,5-I-AEDANS
- N-(iodoacetamino-ethyl)-1-naphthylamine-5-sulfonic acid
- DTNB
- 5,5′-dithiobis(2-nitrobenzoic acid
- MS/MS
- tandem MS
- PLP
- pyridoxal 5′-phosphate
- RS
- tRNA synthetase
- SepCysS
- Sep-tRNA:Cys-tRNA synthase.
REFERENCES
- 1. Sheppard K., Yuan J., Hohn M. J., Jester B., Devine K. M., Söll D. (2008) From one amino acid to another: tRNA-dependent amino acid biosynthesis. Nucleic Acids Res. 36, 1813–1825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sauerwald A., Zhu W., Major T. A., Roy H., Palioura S., Jahn D., Whitman W. B., Yates J. R., 3rd, Ibba M., Söll D. (2005) RNA-dependent cysteine biosynthesis in archaea. Science 307, 1969–1972 [DOI] [PubMed] [Google Scholar]
- 3. Liu Y., Sieprawska-Lupa M., Whitman W. B., White R. H. (2010) Cysteine is not the sulfur source for iron-sulfur cluster and methionine biosynthesis in the methanogenic archaeon Methanococcus maripaludis. J. Biol. Chem. 285, 31923–31929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ambrogelly A., Kamtekar S., Sauerwald A., Ruan B., Tumbula-Hansen D., Kennedy D., Ahel I., Söll D. (2004) Cys-tRNACys formation and cysteine biosynthesis in methanogenic archaea: two faces of the same problem? Cell Mol. Life Sci. 61, 2437–2445 [DOI] [PubMed] [Google Scholar]
- 5. Li T., Graham D. E., Stathopoulos C., Haney P. J., Kim H. S., Vothknecht U., Kitabatake M., Hong K. W., Eggertsson G., Curnow A. W., Lin W., Celic I., Whitman W., Söll D. (1999) Cysteinyl-tRNA formation: the last puzzle of aminoacyl-tRNA synthesis. FEBS Lett. 462, 302–306 [DOI] [PubMed] [Google Scholar]
- 6. Stathopoulos C., Kim W., Li T., Anderson I., Deutsch B., Palioura S., Whitman W., Söll D. (2001) Cysteinyl-tRNA synthetase is not essential for viability of the archaeon Methanococcus maripaludis. Proc. Natl. Acad. Sci. U.S.A. 98, 14292–14297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. O'Donoghue P., Sethi A., Woese C. R., Luthey-Schulten Z. A. (2005) The evolutionary history of Cys-tRNACys formation. Proc. Natl. Acad. Sci. U.S.A. 102, 19003–19008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hauenstein S. I., Perona J. J. (2008) Redundant synthesis of cysteinyl-tRNACys in Methanosarcina mazei. J. Biol. Chem. 283, 22007–22017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fukunaga R., Yokoyama S. (2007) Structural insights into the second step of RNA-dependent cysteine biosynthesis in archaea: crystal structure of Sep-tRNA:Cys-tRNA synthase from Archaeoglobus fulgidus. J. Mol. Biol. 370, 128–141 [DOI] [PubMed] [Google Scholar]
- 10. Kessler D. (2006) Enzymatic activation of sulfur for incorporation into biomolecules in prokaryotes. FEMS Microbiol. Rev. 30, 825–840 [DOI] [PubMed] [Google Scholar]
- 11. Mueller E. G. (2006) Trafficking in persulfides: delivering sulfur in biosynthetic pathways. Nat. Chem. Biol. 2, 185–194 [DOI] [PubMed] [Google Scholar]
- 12. Yuan J., Palioura S., Salazar J. C., Su D., O'Donoghue P., Hohn M. J., Cardoso A. M., Whitman W. B., Söll D. (2006) RNA-dependent conversion of phosphoserine forms selenocysteine in eukaryotes and archaea. Proc. Natl. Acad. Sci. U.S.A. 103, 18923–18927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Smith P. K., Krohn R. I., Hermanson G. T., Mallia A. K., Gartner F. H., Provenzano M. D., Fujimoto E. K., Goeke N. M., Olson B. J., Klenk D. C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem. 150, 76–85 [DOI] [PubMed] [Google Scholar]
- 14. Yuan J., Hohn M. J., Sherrer R. L., Palioura S., Su D., Söll D. (2010) A tRNA-dependent cysteine biosynthesis enzyme recognizes the selenocysteine-specific tRNA in Escherichia coli. FEBS Lett. 584, 2857–2861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lacourciere G. M., Levine R. L., Stadtman T. C. (2002) Direct detection of potential selenium delivery proteins by using an Escherichia coli strain unable to incorporate selenium from selenite into proteins. Proc. Natl. Acad. Sci. U.S.A. 99, 9150–9153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cavallini D., Federici G., Barboni E. (1970) Interaction of proteins with sulfide. Eur. J. Biochem. 14, 169–174 [DOI] [PubMed] [Google Scholar]
- 17. Sörbo B. (1957) Enzymic transfer of sulfur from mercaptopyruvate to sulfate or sulfinates. Biochim. Biophys. Acta 24, 324–329 [DOI] [PubMed] [Google Scholar]
- 18. Riddles P. W., Blakeley R. L., Zerner B. (1983) Reassessment of Ellman's reagent. Methods Enzymol. 91, 49–60 [DOI] [PubMed] [Google Scholar]
- 19. Hudson E. N., Weber G. (1973) Synthesis and characterization of two fluorescent sulfhydryl reagents. Biochemistry 12, 4154–4161 [DOI] [PubMed] [Google Scholar]
- 20. Pedrioli P. G., Eng J. K., Hubley R., Vogelzang M., Deutsch E. W., Raught B., Pratt B., Nilsson E., Angeletti R. H., Apweiler R., Cheung K., Costello C. E., Hermjakob H., Huang S., Julian R. K., Kapp E., McComb M. E., Oliver S. G., Omenn G., Paton N. W., Simpson R., Smith R., Taylor C. F., Zhu W., Aebersold R. (2004) A common open representation of mass spectrometry data and its application to proteomics research. Nat. Biotechnol. 22, 1459–1466 [DOI] [PubMed] [Google Scholar]
- 21. Hidese R., Mihara H., Esaki N. (2011) Bacterial cysteine desulfurases: versatile key players in biosynthetic pathways of sulfur-containing biofactors. Appl. Microbiol. Biotechnol. 91, 47–61 [DOI] [PubMed] [Google Scholar]
- 22. Mihara H., Esaki N. (2002) Bacterial cysteine desulfurases: their function and mechanisms. Appl. Microbiol. Biotechnol. 60, 12–23 [DOI] [PubMed] [Google Scholar]
- 23. Wright C. M., Christman G. D., Snellinger A. M., Johnston M. V., Mueller E. G. (2006) Direct evidence for enzyme persulfide and disulfide intermediates during 4-thiouridine biosynthesis. Chem. Commun. 29, 3104–3106 [DOI] [PubMed] [Google Scholar]
- 24. Mueller E. G., Palenchar P. M., Buck C. J. (2001) The role of the cysteine residues of ThiI in the generation of 4-thiouridine in tRNA. J. Biol. Chem. 276, 33588–33595 [DOI] [PubMed] [Google Scholar]
- 25. Kambampati R., Lauhon C. T. (2003) MnmA and IscS are required for in vitro 2-thiouridine biosynthesis in Escherichia coli. Biochemistry 42, 1109–1117 [DOI] [PubMed] [Google Scholar]
- 26. Numata T., Ikeuchi Y., Fukai S., Suzuki T., Nureki O. (2006) Snapshots of tRNA sulfuration via an adenylated intermediate. Nature 442, 419–424 [DOI] [PubMed] [Google Scholar]
- 27. Kato S., Mihara H., Kurihara T., Takahashi Y., Tokumoto U., Yoshimura T., Esaki N. (2002) Cys-328 of IscS and Cys-63 of IscU are the sites of disulfide bridge formation in a covalently bound IscS/IscU complex: implications for the mechanism of iron-sulfur cluster assembly. Proc. Natl. Acad. Sci. U.S.A. 99, 5948–5952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Newton G. L., Javor B. (1985) γ-Glutamylcysteine and thiosulfate are the major low-molecular-weight thiols in halobacteria. J. Bacteriol. 161, 438–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McFarlan S. C., Terrell C. A., Hogenkamp H. P. (1992) The purification, characterization, and primary structure of a small redox protein from Methanobacterium thermoautotrophicum, an archaebacterium. J. Biol. Chem. 267, 10561–10569 [PubMed] [Google Scholar]
- 30. Johnson R. J., Metzler D. E. (1970) Analyzing spectra of vitamin B6 derivatives. Methods Enzymol. 18, 433–471 [Google Scholar]
- 31. Palioura S., Sherrer R. L., Steitz T. A., Söll D., Simonovic M. (2009) The human SepSecS-tRNASec complex reveals the mechanism of selenocysteine formation. Science 325, 321–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Helgadóttir S., Sinapah S., Söll D., Ling J. (2012) Mutational analysis of Sep-tRNA: Cys-tRNA synthase reveals critical residues for tRNA-dependent cysteine formation. FEBS Lett. 586, 60–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.