Background: Mediator is a conserved eukaryotic transcriptional coregulatory complex.
Results: Disruption of two putative Mediator subunits leads to hyperaccumulation of phenylpropanoids, metabolites important to both the fitness of plants and their utility to humans.
Conclusion: Mediator is essential in plants for the maintenance of phenylpropanoid homeostasis.
Significance: Functional dissection of Mediator is critical to understand eukaryotic transcription and to rationally engineer plant metabolic pathways.
Keywords: Gene Regulation, Genetics, Plant Biochemistry, Secondary Metabolism, Transcription Coactivators
Abstract
The plant phenylpropanoid pathway produces an array of metabolites that impact human health and the utility of feed and fiber crops. We previously characterized several Arabidopsis thaliana mutants with dominant mutations in REDUCED EPIDERMAL FLUORESCENCE 4 (REF4) that cause dwarfing and decreased accumulation of phenylpropanoids. In contrast, ref4 null plants are of normal stature and have no apparent defect in phenylpropanoid biosynthesis. Here we show that disruption of both REF4 and its paralog, REF4-RELATED 1 (RFR1), results in enhanced expression of multiple phenylpropanoid biosynthetic genes, as well as increased accumulation of numerous downstream products. We also show that the dominant ref4-3 mutant protein interferes with the ability of the PAP1/MYB75 transcription factor to induce the expression of PAL1 and drive anthocyanin accumulation. Consistent with our experimental results, both REF4 and RFR1 have been shown to physically associate with the conserved transcriptional coregulatory complex, Mediator, which transduces information from cis-acting DNA elements to RNA polymerase II at the core promoter. Taken together, our data provide critical genetic support for a functional role of REF4 and RFR1 in the Mediator complex, and for Mediator in the maintenance of phenylpropanoid homeostasis. Finally, we show that wild-type RFR1 substantially mitigates the phenotype of the dominant ref4-3 mutant, suggesting that REF4 and RFR1 may compete with one another for common binding partners or for occupancy in Mediator. Determining the functions of diverse Mediator subunits is essential to understand eukaryotic gene regulation, and to facilitate rational manipulation of plant metabolic pathways to better suit human needs.
Introduction
The phenylpropanoids are a diverse group of specialized metabolites in plants that are synthesized from phenylalanine and play a variety of roles in growth, reproduction, and interactions with the biotic and abiotic environment. Hydroxycinnamyl alcohols (or monolignols) serve as monomers for the synthesis of the complex secondary wall polymer lignin (1–3), the synthesis of which represents a globally important commitment of fixed carbon, estimated to be on the order of 10 trillion kilograms annually (4). Soluble phenylpropanoids are similarly important in plant ecology, where they act variously to absorb damaging ultraviolet light, defend against pathogens, repel herbivores, or attract pollinators (5). In addition to their essential roles in ensuring plant fitness, phenylpropanoid metabolites also exert a substantial influence on the utility of plants to suit human needs. The inhibitory effect of lignin on polysaccharide extraction from bulk biomass, for instance, is one of the major limitations in the cost effectiveness of cellulosic biofuel production (6–8). Furthermore, products of the phenylpropanoid pathway found in the diet, such as anthocyanins, isoflavonols, lignans, and hydroxycinnamic acids, have been shown to act as antioxidants or modulators of mammalian hormone signaling pathways, and their consumption is correlated with reduced incidence of cancer, heart disease, and other chronic health problems (9–12). An understanding of phenylpropanoid metabolism is therefore essential both to provide greater insight into the basic biology of plants, and to inform bioengineering strategies aimed at the genetic improvement of bioenergy and food crops.
The biochemical pathways responsible for the synthesis of a number of different phenylpropanoids have been elucidated, such as those leading to the production of monolignols, anthocyanins, flavonols, and various hydroxycinnamate esters (1, 5, 13, 14). More recently, the complex hierarchical network of transcriptional activators and repressors regulating the temporal and spatial control of different phenylpropanoids has begun to be unraveled, particularly in the model angiosperm Arabidopsis thaliana (5, 15, 16). The synthesis of anthocyanins, for instance, is controlled by a set of closely related MYB transcription factors (PAP1/MYB75, PAP2/MYB90, MYB113, and MYB114) that stimulate the transcription of both general phenylpropanoid and anthocyanin-specific genes (17). Constitutive expression of PAP1/MYB75 alone is sufficient to induce the expression of the entire anthocyanin biosynthetic pathway, and plants containing an activation-tagged allele of this transcription factor accumulate clearly visible amounts of anthocyanin pigments throughout their tissues (18). Despite substantial recent progress, however, a great deal of the regulation of phenylpropanoid biosynthesis remains poorly understood, especially with respect to the homeostatic mechanisms by which the levels of different metabolites are coordinated and maintained within a narrowly defined target range.
The genetic regulation of many eukaryotic genes requires a large co-regulatory complex known as Mediator, which physically bridges the interaction between gene-specific transcription factors and the basal transcription machinery at the core promoter (19, 20). Found in all eukaryotes, Mediator consists of some 20–30 different protein subunits that associate with one another in four domains, referred to as the head, middle, tail, and kinase domains (21). The composition of Mediator varies between species, with some subunits conserved throughout the eukaryotes and others found only in particular phylogenetic lineages (22). Studies of Mediator function in plants have been undertaken only relatively recently but have already implicated this complex in numerous biological processes, including flowering time control (23, 24), pathogen resistance (25, 26), cold tolerance (27), micro-RNA production (28), and the regulation of cell division and patterning (29). A catalog of putative Arabidopsis Mediator subunits obtained using a co-immunoprecipitation/proteomics approach has recently been reported (23), but the functions of individual subunits and their roles in the regulation of general or gene-specific transcription remain largely unknown.
Among the likely Arabidopsis Mediator subunits identified by Bäckström et al. (23) were two related proteins designated MED33a (At3g23590) and MED33b (At2g48110). Although At3g23590/MED33a and At2g48110/MED33b were originally suggested to be plant-specific Mediator components, more recent phylogenetic evidence has suggested that both are, in fact, orthologous to yeast Med5 and mammalian MED24 (22). We independently reported an allelic series of three Arabidopsis mutants, ref4-1, ref4-2, and ref4-3, which contain semidominant mutations in At2g48110 and display a range of mutant phenotypes indicative of a global reduction in phenylpropanoid biosynthesis (30, 31). Specifically, dominant ref4 mutants exhibit a reduced epidermal fluorescence phenotype due to a deficiency in the synthesis of sinapoylmalate, a UV-absorbent and fluorescent specialized metabolite accumulated in the epidermis of Arabidopsis leaves, as well as dwarfing, decreased lignin deposition, and a pale seed coat. We also identified the same REF46 paralog as Bäckström et al. (23) (At3g23590/MED33a), which we designated RFR1 (31). At the time of the previous report, however, we were unaware of the association of either protein with Mediator and were unable to confidently assign a function to REF4 or RFR1 due to their lack of sufficient sequence similarity to previously characterized proteins. Furthermore, due to the semidominant nature of the previously reported ref4 mutations, we were unable to formally exclude the possibility that the phenotypes they generated were neomorphic. Here, we present evidence that REF4 and RFR1 are required for phenylpropanoid homeostasis in wild-type plants, acting directly or indirectly to repress the transcription of phenylpropanoid biosynthetic genes.
EXPERIMENTAL PROCEDURES
Plant Material and Growth Conditions
Plants were grown in Redi-earth Plug and Seedling Mix (Sun Gro Horticulture, Bellevue, WA) supplemented with Scotts Osmocote Plus controlled release fertilizer (Hummert International, Earth City, MO) at 22 °C, with a 16-h light, 8-h dark photoperiod.
All photographic images presented are unaltered with respect to brightness, color, and contrast. In some cases, portions of single images have been cropped or rearranged for neatness and clarity of presentation.
The ref4-3 mutant and the suppressor screen yielding the R387H ref4-3 suppressor have been described previously (30, 31). Plants containing T-DNA insertions in REF4 (SALK_037472) and RFR1 (SALK_011621) were obtained from the Arabidopsis Biological Resource Center (The Ohio State University, Columbus, OH). Primers CC1446 (left primer), CC1554 (right primer), and CC1662 (border primer) were used to genotype for the presence of the T-DNA insertion in REF4, and primers CC1834 (left primer), CC1776 (right primer), and CC1662 (border primer) were used to genotype for the presence of the T-DNA insertion in RFR1. ref4 rfr1 plants were obtained by crossing ref4 and rfr1 plants and screening F2 progeny for plants homozygous for both T-DNA insertions. The pap1-D mutant has been described previously (18), and the ref4-3 pap1-D double mutant was obtained by crossing.
Cloning and Plasmid Construction
To clone AtREF4, AtRFR1, and SmREF4, we used the Gateway cloning system (Invitrogen). The Gateway donor vector used in all cases was pCC1155, a modified version of pDONR221 in which a 927-bp BspHI fragment containing the kanamycin resistance marker of pDONR221 was replaced by the 1008-bp BspHI fragment containing the ampicillin resistance marker of pGEM-5Zf(+). The Gateway destination vector used in all cases was pCC0996, a derivative of pBI101.2 containing the Arabidopsis CINNAMATE 4-HYDROXYLASE (C4H) promoter, a Gateway cassette, and a Basta/glufosinate resistance marker. pCC0996 was constructed as follows. First, plasmid pBI101.2 was cut with EcoRI, incubated with Klenow DNA polymerase to fill in overhangs, and religated to eliminate the single EcoRI site. The single HindIII site of pBI101.2 was similarly eliminated. The GUS open reading frame was excised from the resulting vector by digesting with SalI and XbaI, and a SalI-XbaI fragment containing the polylinker from pGEM-7Zf(+) was inserted in its place to yield pCC0870. A 2.9-kb segment of genomic DNA upstream of AtC4H was PCR amplified with primers CC1791 and CC1792, which add a SalI and an EcoRI site, respectively, to the resulting amplicon. This amplicon was then digested with SalI and EcoRI and cloned into the same sites of pBluescript KS− to yield pCC0969. A 2.5-kb SacII fragment containing attR1-CmR-ccdB-attR2 (the Gateway cassette) along with an associated OCS transcriptional terminator downstream was excised from plasmid pZKY1 and cloned into the SacII site of pCC0969 to yield pCC1054. pCC1054 was then digested with SacI and KpnI to release a DNA fragment containing the entire C4H promoter-Gateway cassette-OCS terminator, which was then cloned into the same sites of pCC0870 to yield pCC1061. To replace the pBI101.2-derived kanamycin resistance marker of pCC1061 with Basta/glufosinate resistance marker (Bar), the following steps were taken. The NOS terminator of pBI101.2 was amplified with CC1871 and CC1872, which adds a XbaI site to the resulting amplicon, then digested with BamHI and XbaI and inserted into the same sites of pBluescript KS− to yield pCC0999. The NOS promoter and the glufosinate resistance marker were excised from pGPTV-Bar with EcoRI and BamHI and inserted into the same sites of pCC0999 to yield pCC1037. pCC1037 was then digested with NheI and XbaI to release the NOS promoter-Bar-NOS terminator fragment, which was then cloned into the same sites in pCC1061 to yield pCC0996, a Gateway plant expression vector conferring resistance to Basta/glufosinate in which the inserted gene is driven by the Arabidopsis C4H promoter.
AtREF4, AtRFR1, and SmREF4 were amplified from cDNA using the following primers: AtREF4, CC2206 and CC2207; AtRFR1, CC2204 and CC2205; SmREF4, CC2946 and CC2947. The resulting amplicons were cloned into pCC1155 using BP clonase and subsequently cloned into pCC0996 using LR clonase according to the manufacturer's instructions.
Site-directed Mutagenesis
The SmREF4 point mutants described in the text were generated using the QuikChange Multi Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions.
Generation of Transgenic Plants
Transgenic plants described in the text were generated using the floral dip method, as previously described (32). Agrobacterium tumefaciens strain C58 pGV3850 was utilized in all cases. To select for herbicide-resistant transformants, soil-grown T1 seedlings were sprayed with a 1:500 dilution of Finale Concentrate (glufosinate ammonium; Farnam Companies, Phoenix, AZ) in water approximately 7 days after germination. In some cases, a second application of herbicide several days later was utilized to eliminate seedlings that had germinated late or were poorly covered by the initial herbicide treatment.
HPLC Analysis of Soluble Metabolites
To extract and measure sinapate esters, flavonols, and hydroxycinnamyl alcohol coupling products, plant tissues (leaves, inflorescence stems, or roots as indicated in the text) were harvested, crushed, and extracted in 50% methanol (v/v) for 1 h at 65 °C at a tissue concentration of 100 mg/ml. Insoluble material was sedimented by spinning for 5 min at 16,000 × g in a microcentrifuge. 10 μl of the supernatant was then loaded onto a Shim-pack XR-ODS column (Shimadzu, Kyoto, Japan; column dimensions 3.0 × 75 mm, bead size 2.2 μm) and eluted at a flow rate of 0.7 ml/min at an increasing concentration of acetonitrile from 10 to 35% over 8.6 min in 0.1% aqueous formic acid. Sinapic acid was used as a standard for quantification of sinapate esters. A similar protocol was used to extract and measure anthocyanins, with minor modifications. Extractions were performed in 50% methanol (v/v), 4% glacial acetic acid (v/v), and elution from the HPLC column was performed with an increasing concentration of acetonitrile from 10 to 20% over 10.6 min in 10% formic acid.
Liquid Chromatography/Mass Spectrometry
To analyze plant metabolites by LC/MS, plant extracts were prepared as described above for analysis of sinapate esters, flavonols, and hydroxycinnamyl alcohol coupling products. 10 μl of extract was run on the same Shim-pack XR-ODS column as above, but with a flow rate of 0.5 ml/min at an increasing concentration of acetonitrile from 10 to 25% over 14 min and from 25 to 70% over 8 min in 0.1% formic acid. Metabolites were detected and analyzed with an Agilent 1100 series LC/MSD TOF mass spectrometer in negative electrospray ionization (ESI) mode, nebulizer pressure 55 p.s.i., capillary voltage 3200 V, fragmentor voltage 125 V, drying gas flow rate 11 liter/min, temperature 350 °C.
Gene Expression Analysis
Quantitative real-time PCR experiments were performed as previously described (33), using the following primers: PAL1 (At2g37040), CC3280 and CC3281; PAL2 (At3g53260), CC3282 and CC3283; 4CL1 (At1g51680), CC3256 and CC3257; C4H (At2g30490), CC3258 and CC3259; and At1g13320 (reference gene), CC2558 and CC2559. Global expression analysis of phenylpropanoid biosynthetic and regulatory genes was performed using the Affymetrix ATH1 genome array according to the manufacturer's instructions, with RNA from 3-week-old soil-grown rosettes harvested ∼8 h after subjective dawn.
Phylogenetic Analysis
The phylogenetic tree of plant REF4 homologs was constructed using MRBAYES version 3.1.1 (34), with amino acid alignments generated with MEGA version 4 (35). Sequences were retrieved using the BLAST algorithm with the online databases TAIR (arabidopsis.org), JGI (genome.jgi-psf.org), Phytozome (phytozome.net), and GenBankTM and are listed in supplemental Table S1.
RESULTS
Neither rfr1- nor ref4 rfr1-deficient Plants Morphologically Resemble Plants Carrying the ref4-3 Mutation
In contrast to the dominant ref4-3 mutant, which exhibits a severe dwarf phenotype, light-colored seeds, and dark, spatulate leaves, we previously reported that the morphological phenotype of plants containing a T-DNA insertion in REF4 was indistinguishable from that of wild-type plants (31). This observation suggests that the phenotype of the ref4-3 mutant is most likely not the result of inactivation of the REF4 gene. On the other hand, the existence of the paralogous RFR1 gene prevented us from ruling out the possibility that REF4 and RFR1 are redundant in wild-type plants and that the ref4-3 mutant protein acts as a dominant-negative inhibitor of RFR1. To test whether disruption of both REF4 and RFR1 phenocopies ref4-3, we generated plants containing T-DNA disruptions in both genes. Although ref4 rfr1-deficient plants under some growth conditions appear slightly less robust than wild-type plants, neither they nor plants null for rfr1 alone morphologically resemble ref4-3 mutant plants (Fig. 1). Both rfr1-deficient and ref4 rfr1-deficient plants develop normal rosette leaves (Fig. 1A), attain wild-type height at maturity (Fig. 1B), and yield seeds that are dark brown in color (Fig. 1C). On the basis of these observations, we conclude that ref4-3 does not interfere with the wild-type function of RFR1 by acting as a dominant-negative version of the protein.
FIGURE 1.
Arabidopsis mutants containing an inactivating insertion in RFR1 or in both REF4 and RFR1 do not phenotypically resemble the previously reported ref4-3 mutant. A, comparison of 3-week-old rosettes from a wild-type plant and plants containing a single inactivating insertion in the REF4 gene (ref4), RFR1 gene (rfr1), or both (ref4 rfr1), together with plants homozygous for the previously described ref4-3 point mutation. B, mature plants and C, seeds of plants with the genotypes described in A.
Disruption of REF4 and RFR1 Results in Phenylpropanoid Hyperaccumulation
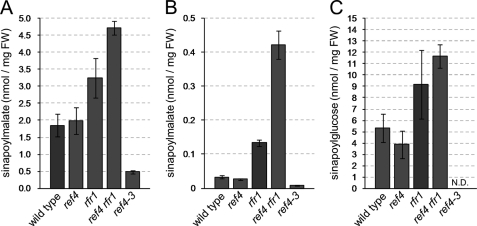
To test whether the lack of a phenylpropanoid phenotype in ref4-deficient plants is due to redundancy of REF4 and RFR1, we used high-performance liquid chromatography (HPLC) to measure the concentration of soluble phenylpropanoids in tissues of rfr1 and ref4 rfr1 mutants. These analyses clearly showed that leaves of both rfr1- and ref4 rfr1-deficient plants contain higher levels of sinapate esters (hydroxycinnamate esters derived from sinapic acid including sinapoylmalate and its biosynthetic precursor sinapoylglucose) than wild-type plants, in contrast to the low sinapate ester levels seen in leaves of ref4-3 plants (Fig. 2). The mean sinapoylmalate concentration in leaves of rfr1-deficient plants was ∼75% higher than in wild-type plants, whereas leaves of ref4 rfr1-deficient plants contained ∼2.5 times more sinapoylmalate than wild-type plants. There was no difference between sinapoylmalate levels in leaves of ref4-deficient plants and wild-type plants, as reported previously (31). The increase in sinapoylmalate levels in rfr1- and ref4 rfr1-deficient plants was also observed in inflorescence stem tissue, where the mutant plants accumulated ∼4- and 12-fold more sinapoylmalate, respectively, than did wild-type plants (Fig. 2B). Sinapoylglucose, the second most abundant sinapate ester in Arabidopsis seeds, followed a less pronounced, but similar trend as sinapoylmalate, with seeds from ref4 rfr1 mutants showing the highest level of sinapoylglucose, and seeds of rfr1 mutant plants intermediate between those of wild-type and ref4 rfr1 mutants (Fig. 2C). The concentration of flavonols (in leaves) and sinapoylcholine (in seeds) of rfr1- and ref4 rfr1-deficient plants was similar to that observed in wild-type plants.
FIGURE 2.
rfr1 and ref4 rfr1 mutant plants hyperaccumulate phenylpropanoids. A, quantification of sinapoylmalate in leaves of wild-type, ref4 null, rfr1 null, ref4 rfr1 null, and ref4-3 mutant plants. B, quantification of sinapoylmalate in inflorescence stems and C, sinapoylglucose in seeds of the plants described in A. Sinapate esters were quantified by HPLC by comparing their absorbance at 330 nm to the absorbance of a sinapic acid standard of known concentration. Values shown for leaves and inflorescence stems are the mean of six biological replicates, and those shown for seeds are the mean of three technical replicates on each of three biological replicates. Error bars represent S.D. in all cases.
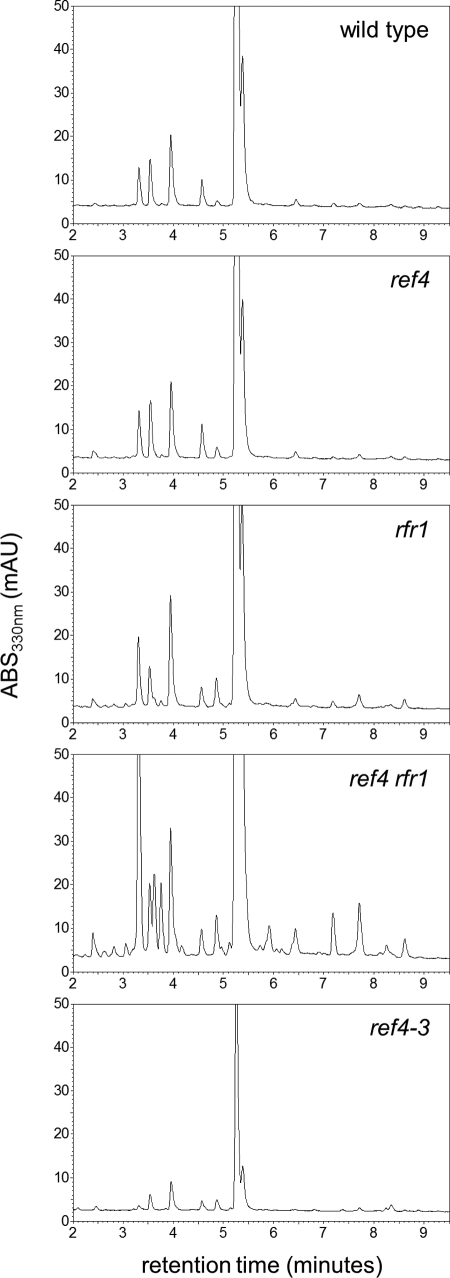
In the course of HPLC analyses of sinapate ester and flavonol levels, we noticed that in addition to these normally abundant soluble metabolites, rosette leaves of rfr1-deficient plants also accumulated a substantial number of other UV-absorbent metabolites that are normally either of very low abundance or below the limit of detection in leaves of wild-type or ref4-deficient plants (Fig. 3). The levels of these metabolites were further elevated in leaves of ref4 rfr1-deficient plants, in some cases more than 50-fold as estimated by HPLC, and similar results were observed in inflorescence stems and roots (supplemental Fig. S1). Using liquid chromatography/mass spectrometry (LC/MS) we obtained masses for at least 15 different compounds that accumulated in ref4 rfr1-deficient plants to at least 2-fold higher levels than in wild-type plants (Table 1). In addition to well-characterized metabolites such as sinapoylmalate (m/z 339), sinapoylglucose (m/z 385), and 1,2-disinapoylglucose (m/z 691), these compounds include phenylpropanoid dimers that arise from the radical coupling of hydroxycinnamyl alcohols with hydroxycinnamate esters (m/z 487, 505, 533, and 551) and glycosylated monolignol dimers (m/z 519 and 521), as well as several currently unidentified compounds.
FIGURE 3.
HPLC analysis reveals increased levels of many UV absorbing compounds in ref4 rfr1 plants relative to wild-type plants. HPLC profiles of methanol-soluble, UV absorbing metabolites from leaves of wild-type, ref4 null, rfr1 null, ref4 rfr1 null, and ref4-3 mutant plants.
TABLE 1.
Compounds that hyperaccumulate in ref4 rfr1-deficient plants
| Mass (M-H) | Putative empirical formula | Putative identity |
|---|---|---|
| 309 | C14H14O8 | Feruloylmalate |
| 339 | C15H16O9 | Sinapoylmalate |
| 355 | C16H20O9 | Feruloylglucose |
| 385 | C17H22O10 | Sinapoylglucose |
| 487 | C24H24O11 | Feruloylmalate-coniferyl alcohol coupling product |
| (dehydroconiferyl feruloylmalate) | ||
| 505 | C24H26O12 | Feruloylmalate-coniferyl alcohol coupling product |
| (guaiacylglycerol feruloylmalate) | ||
| 519 | C26H32O11 | Glycoside of coniferyl alcohol dimer (pinoresinol or dehydrodiconiferyl alcohol) |
| 521 | C26H34O11 | Modified glycoside of coniferyl alcohol dimer |
| (lariciresinol or dihydrodehydrodiconiferyl alcohol) | ||
| 533 | C26H30O12 | Feruloylglucose-coniferyl alcohol coupling product |
| (dehydroconiferyl feruloylglucose) | ||
| 535 | C25H28O13 | Unknown |
| 551 | C26H32O13 | Feruloylglucose-coniferyl alcohol coupling product |
| (guaiacylglycerol feruloylmalate) | ||
| 565 | C27H36O14 | Unknown |
| 583 | C27H34O13 | Unknown |
| 591 | C28H32O14 | 1,2-Disinapoylglucose |
| 681 | C33H34O18 | Possible lignan diglycoside |
RFR1 Complements the Phenylpropanoid Hyperaccumulation Phenotype of ref4 rfr1-deficient Plants
To rule out the possibility that hyperaccumulation of phenylpropanoids in ref4 rfr1-deficient plants is a result of a mutation in another gene in the original ref4 or rfr1 mutant lines, we tested the ability of wild-type RFR1 to complement their phenylpropanoid hyperaccumulation phenotype. We cloned the RFR1 open reading frame and placed it under the control of the promoter of the C4H gene, which is highly expressed in cells carrying out phenylpropanoid biosynthesis (36). Transformation of ref4 rfr1-deficient plants with the resulting C4H:RFR1 transgene was sufficient to substantially reduce phenylpropanoid hyperaccumulation (supplemental Fig. S2), demonstrating that the phenotype of ref4 rfr1-deficient plants is not due to disruption of another gene in either the ref4- or rfr1-deficient T-DNA lines from which they were generated. In contrast to our results with RFR1, transformation of ref4 rfr1-deficient plants with an analogous C4H:REF4 transgene had no discernible effect on phenylpropanoid levels (supplemental Fig. S2). Together with the fact that disruption of RFR1 alone is sufficient to generate a metabolic phenotype, whereas disruption of REF4 alone has no apparent effect, these data suggest that RFR1 may play a more substantial role than REF4 in the repression of phenylpropanoid biosynthesis in wild-type plants. We should note that attempts to define and utilize the endogenous promoters of REF4 and RFR1 for complementation of the ref4 rfr1 phenotype have been unsuccessful to date, suggesting the existence of currently unidentified cis-acting sequences that are essential for the expression of REF4 and RFR1 in wild-type plants.
Disruption of REF4 and RFR1 Leads to Increased Transcript Levels of Early Phenylpropanoid Biosynthetic Genes
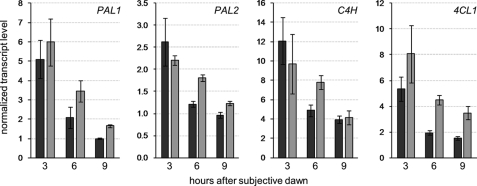
Given that REF4 and RFR1 have been shown to be components of a conserved transcriptional co-regulator (23), we tested whether changes in phenylpropanoid accumulation in ref4 rfr1-deficient plants might be caused by changes in phenylpropanoid biosynthetic gene expression. Using quantitative real-time PCR (RT-PCR), we measured the transcript abundance of the early phenylpropanoid biosynthetic genes PAL1, PAL2, C4H, and 4CL1 in wild-type and ref4 rfr1-deficient plants. Because these genes are all known to be regulated by the circadian clock (37, 38), we measured transcript levels at three different time points, from 3 to 9 h after the start of the light period. No significant differences between wild-type and ref4 rfr1-deficient plants were observed at the earliest time point, but transcript levels of all four genes were significantly lower in wild-type plants than in ref4 rfr1-deficient plants at most later time points (Fig. 4). In no case did we observe higher levels of transcript for any phenylpropanoid biosynthetic gene in wild-type plants than in ref4 rfr1-deficient plants. Microarray analysis of ref4 rfr1-deficient and ref4-3 mutant plants further supported these observations, and revealed that genetic manipulation of REF4 and RFR1 led to substantial changes in transcript levels of multiple phenylpropanoid biosynthetic genes, but not of transcriptional regulators of the pathway (supplemental Fig. S3). Together, these data show that REF4 and RFR1 are involved in maintaining normal expression of phenylpropanoid biosynthetic genes in wild-type plants, and suggest that they are required for the activity, but not the expression, of one or more transcriptional regulators of phenylpropanoid metabolism.
FIGURE 4.
Early phenylpropanoid genes show increased expression in ref4 rfr1-deficient plants. Steady-state transcript levels of PAL1, PAL2, C4H, and 4CL1 genes relative to the reference gene At1g13220 at the indicated times after subjective dawn. Dark gray bars show transcript measurements from wild-type plants, and light gray bars show measurements from ref4 rfr1-deficient plants. Error bars represent the S.D. of three biological replicates.
The Dominant ref4-3 Mutation Severely Impairs the Ability of MYB75 to Drive Anthocyanin Production and PAL1 Expression
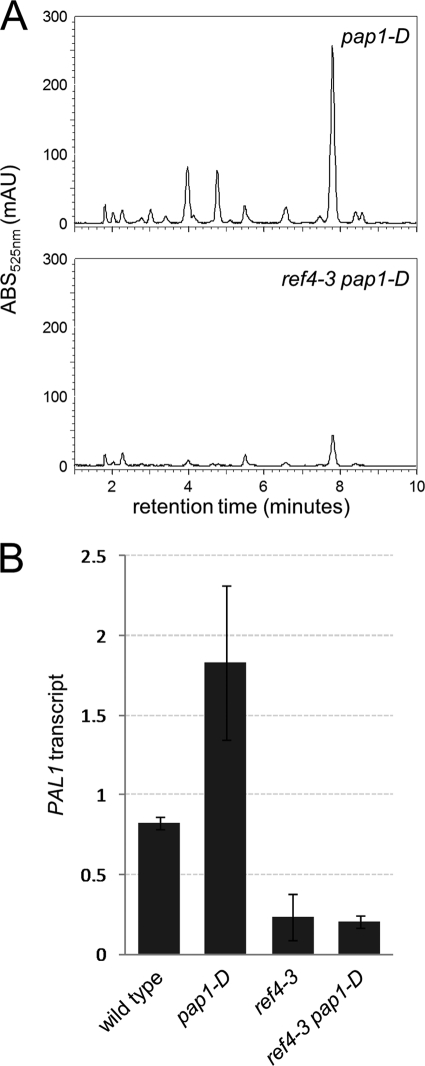
Having shown that deletion of REF4 and RFR1 up-regulates the expression level of phenylpropanoid biosynthetic genes, we wanted to determine whether the protein encoded by the ref4-3 allele acts to down-regulate them. The Arabidopsis mutant pap1-D is an activation tagged line that hyperaccumulates anthocyanins (18). Characterization of this mutant showed that its phenotype could be attributed to constitutive expression of the R2-R3 MYB transcription factor MYB75, which stimulates transcription of multiple genes in the anthocyanin biosynthetic pathway including those encoding phenylalanine-ammonia lyase (PAL), a major point of phenylpropanoid pathway flux control (39, 40). Using this mutant, we tested whether expression of the dominant ref4-3 mutant protein can influence the ability of the MYB75 transcription factor to induce the PAL1 gene and drive the accumulation of anthocyanins. In contrast to pap1-D plants, which accumulated visible levels of purple pigment in leaf veins and inflorescence stems, ref4-3 pap1-D mutant plants were uniformly green (supplemental Fig. S4). This observation was supported by HPLC analysis of anthocyanin content in rosette leaves (Fig. 5A) showing that ref4-3 pap1-D mutants contained substantially less anthocyanins than pap1-D mutants. Under the same growth conditions, rosette leaves of wild-type and ref4-3 plants contained levels of anthocyanins only barely detectable above background. Quantitative RT-PCR analysis of PAL1 showed a clear decrease in the steady-state level of this transcript in both ref4-3 and ref4-3 pap1-D plants, in contrast to the substantial increase seen in the pap1-D mutant relative to wild type (Fig. 5B), suggesting that the mutant protein attenuates anthocyanin accumulation at least partially through this mechanism.
FIGURE 5.
The ref4-3 mutant interferes with the ability of MYB75 to drive anthocyanin production and PAL1 expression. A, HPLC chromatograms of anthocyanins in 3-week-old rosette leaves of pap1-D and ref4-3 pap1-D plants grown in parallel. B, quantification of PAL1 transcript levels from 3-week-old rosette leaves of wild-type, pap1-D, ref4-3, and ref4-3 pap1-D plants, relative to the reference gene At1g13320.
Phylogenetic and Genetic Analysis of REF4/RFR1 Orthologs Supports a Functional Role for Gly-383 and Arg-387 in Repression of Phenylpropanoid Biosynthesis
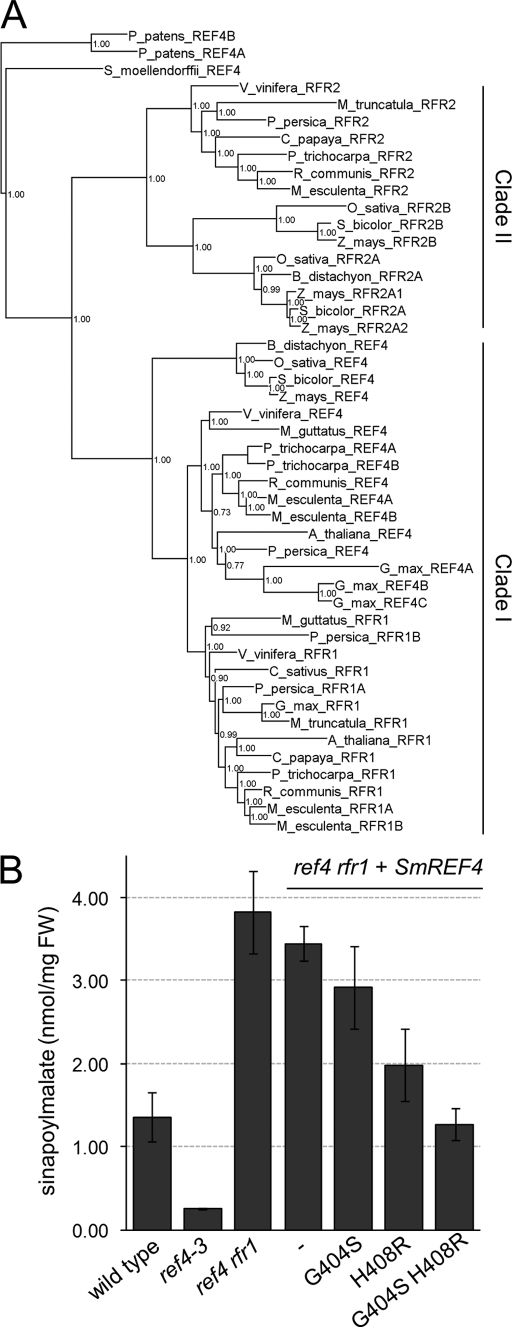
To learn more about their evolution and possible conservation, we used Bayesian analysis to generate a phylogenetic tree of REF4 and RFR1 orthologs from plants with sequenced genomes, including the non-vascular plant Physcomitrella patens, which was used as an outgroup (Fig. 6A). A close inspection of this tree yielded several significant insights. First, all angiosperm REF4 homologs cluster into two large clades (designated Clade I and Clade II in Fig. 6A). Arabidopsis REF4 and Arabidopsis RFR1 are both members of Clade I, and the majority of other dicots included in our analysis also possessed both a REF4 ortholog and an RFR1 ortholog. In contrast, all monocots we examined possessed a single Clade I family member, and these sequences formed an outgroup to the dicot Clade I sequences, suggesting that the genes encoding dicot REF4 and RFR1 are derived from a gene duplication event occurring after the divergence of monocots and dicots. In addition to these Clade I sequences, the majority of species included in our analysis possessed one or more Clade II family members (which we have designated RFR2 sequences). In the case of Clade II, there appears to have been a monocot-specific gene duplication event resulting in two RFR2 sequences in monocots and only a single RFR2 in dicots. The majority of angiosperms possess at least one RFR2 sequence, with the notable exception of Arabidopsis, suggesting a relatively recent loss of RFR2 in the Arabidopsis lineage.
FIGURE 6.
Phylogeny and rescue with modified Selaginella REF4. A, Bayesian phylogenetic tree of plant REF4/Med5p homologs. B, sinapoylmalate concentration in ref4 rfr1-deficient plants transformed with the indicated SmREF4 expression constructs.
We previously reported that a P919L mutation completely rescues the growth phenotype and sinapoylmalate deficiency engendered by the dominant ref4-3 allele (31). Pro-919 lies in a WW protein interaction domain that is conserved among REF4/RFR1/RFR2 homologs from other species, suggesting that this motif may be important for the ability of REF4 family members to associate with an interaction partner essential for their function. We have subsequently discovered that another suppressor isolated from the same genetic screen is also intragenic, containing an R387H substitution, only four residues downstream of the original G383S mutation in ref4-3 (supplemental Fig. S5). The R387H mutation largely alleviates the dwarf phenotype of ref4-3 without completely restoring its ability to accumulate wild-type levels of sinapoylmalate, indicating that unlike the P919L mutation, the R387H mutation does not completely eliminate the ability of the ref4-3 protein to repress phenylpropanoid biosynthesis.
As we reported previously, an alignment of REF4/RFR1/RFR2 sequences showed that the Gly-383 residue of Arabidopsis REF4 (the site mutated in the ref4-3 protein) is universally conserved throughout the plant kingdom (31). In contrast, the Arg-387 residue of Arabidopsis REF4 is conserved throughout all dicot REF4 and RFR1 sequences, but is replaced with a histidine residue in all dicot RFR2 sequences and in all other REF4 homologs from other plants, including the lycophyte Selaginella moellendorffii and the moss P. patens. This observation suggests that the R387H intragenic suppressor of ref4-3 is, in fact, a reversion to the ancestral sequence of all REF4 homologs.
To test the functional significance of the Gly-383 and Arg-387 residues of Arabidopsis REF4 (AtREF4), as well as the possible conservation of REF4 function among vascular plants, we cloned the coding sequence of Selaginella REF4 (SmREF4) and used site-directed mutagenesis to modify the SmREF4 residues corresponding to AtREF4 Gly-383 and Arg-387 (SmREF4 Gly-404 and His-408, respectively). We then tested the ability of these SmREF4 variants to repress the phenylpropanoid hyperaccumulation of ref4 rfr1-deficient plants when expressed under control of the Arabidopsis C4H promoter. Transformation of ref4 rfr1-deficient plants with wild-type SmREF4 had no effect, but introduction of either the H408R or G404S mutation into the SmREF4 coding sequence led to a significant reduction in sinapoylmalate levels (Fig. 6B). Combining both mutations in the same construct led to restoration of wild-type levels of sinapoylmalate. These data support a role for Gly-383 and Arg-387 of Arabidopsis REF4 in the repression of phenylpropanoids, and show that with minor modification SmREF4 can functionally substitute for AtREF4, suggesting that despite 400 million years of intervening evolution, the SmREF4 protein can still interact with other Arabidopsis Mediator subunits to form a functional complex in vivo and that SmREF4 may be a regulator of phenylpropanoid metabolism in Selaginella. The conservation of REF4/RFR1 function over such great evolutionary distance in turn increases the likelihood that these gene products represent viable targets for genetic engineering of economically important plant species.
The Phenotypic Impact of the ref4-3 Mutation Is Exacerbated in Absence of Wild-type RFR1
REF4 and RFR1 appear to be partially redundant with respect to their influence on phenylpropanoid biosynthesis, and disruption of both genes is required for the full increase in phenylpropanoids observed in ref4 rfr1-deficient plants. Given that the dominant ref4-3 mutation was identified in an otherwise wild-type genetic background, we hypothesized that the presence of wild-type RFR1 could exert a significant influence on its mutant phenotype. To test this hypothesis, we began by crossing ref4-3 plants to ref4 rfr1-deficient plants. Interestingly, the F1 progeny of this cross (genotype ref4-3/ref4 RFR1/rfr1) exhibited a dwarf phenotype that was significantly more severe than that of the ref4-3 mutant. F2 progeny from these plants (Fig. 7) that did not inherit the ref4-3 allele (i.e. were homozygous null for ref4) appeared identical to either wild-type or ref4 rfr1 plants, as expected. F2 progeny that did inherit the ref4-3 allele fell into three phenotypic categories, depending on their genotype at the RFR1 locus. Those with the genotype RFR1/RFR1 were predictably identical to the original ref4-3 mutant; those with the genotype RFR1/rfr1 were more severely dwarfed than ref4-3 plants, similar to their F1 parents; and those with the genotype rfr1/rfr1 were most severely affected, developing rosettes only a few millimeters in diameter that failed to grow to maturity or to flower. These data show that the presence of wild-type alleles of RFR1 in the ref4-3 mutant mitigates its mutant phenotype, and that elimination of RFR1 in this genetic background leads to extreme dwarfing and developmental arrest.
FIGURE 7.
The presence of wild-type RFR1 alleles substantially mitigates the mutant phenotype resulting from the ref4-3 mutation. Representative plants with the phenotypes observed in the F2 progeny of a cross between ref4 rfr1 null and ref4-3 mutant plants. The genotype of each plant is indicated, and a close-up view of the most severely dwarfed plant on the far right is provided in the inset.
DISCUSSION
It now appears that the enzymes of the core phenylpropanoid biosynthetic pathway have all been identified, along with many of the transcription factors controlling their expression, but the mechanisms by which the levels of different metabolites are coordinated remain far from completely understood. We have presented evidence here that two subunits of Mediator, REF4/MED33b and RFR1/MED33a, are essential for the maintenance of phenylpropanoid homeostasis in wild-type Arabidopsis, and their disruption leads to phenylpropanoid hyperaccumulation and increased expression of multiple phenylpropanoid biosynthetic genes.
Phenylpropanoid hyperaccumulation in ref4 rfr1-deficient plants is not restricted to one organ and was observed in leaves, inflorescence stems, seeds, and roots. The same organs all showed decreased phenylpropanoids in ref4-3 mutant plants, consistent with publicly available data showing that REF4 and RFR1 are ubiquitously expressed. The dominant ref4-3 mutant is affected in the synthesis of all phenylpropanoids, suggesting that it acts on the general phenylpropanoid pathway. On the other hand, phenylpropanoids such as sinapoylcholine and flavonols did not show increased levels in ref4 rfr1-deficient plants. One possible explanation for this observation is that the product of another biochemical pathway becomes limiting for the synthesis of these metabolites in ref4 rfr1-deficient plants, such as malonyl-CoA in the case of flavonols or choline in the case of sinapoylcholine. Alternatively, it could be that REF4 and RFR1 are sufficient but not necessary to prevent hyperaccumulation of some phenylpropanoids, and that particular branches of the pathway can be repressed by a REF4/RFR1-independent mechanism. In contrast with the highly reproducible phenotype of ref4 rfr1-deficient plants with respect to soluble phenylpropanoids, measurements of lignin content have yielded intriguing but as yet inconsistent results. Some experiments showed a 20% increase in lignin content in ref4 rfr1 mutants, whereas others showed no difference with wild-type plants, similar to our findings with sinapoylcholine and flavonols described above. Given the key role of Mediator in integrating intrinsic and extrinsic cues to modulate gene expression, we suspect that these inconsistencies may be due to as yet unidentified differences in the growth conditions between different experiments, but we are at present unable to confidently conclude whether disruption of REF4 and RFR1 has a bona fide effect on lignin biosynthesis.
The most significant fold-changes in metabolite levels we observed in ref4 rfr1-deficient plants were on a diverse set of compounds that appear to arise from radical coupling reactions of monolignols either with one another or with hydroxycinnamate esters. For instance, we observed a set of compounds with masses (m/z 487 and 505) and fragmentation patterns consistent with β-5 and β-O-4 coupling products of feruloylmalate with coniferyl alcohol, respectively, that have been described previously (41, 42). Extracted ion chromatograms at both m/z 505 and 487 show multiple peaks, likely due to the different stereochemistries with which hydroxycinnamate ester-hydroxycinnamyl alcohol coupling can occur. A similar set of compounds showed masses m/z 533 and 551 consistent with the analogous coupled glucose esters of the malate esters described above, which to our knowledge have not been reported previously. Lignans are compounds that are formed from β-β coupling of two molecules of coniferyl alcohol, and glucoside derivatives of lignans such as pinoresinol (m/z 519) and lariciresinol (m/z 521) are synthesized in Arabidopsis roots (43). We observed compounds with these masses in rosette leaves of ref4 rfr1-deficient plants, again at levels much greater than in wild-type plants. It should be noted, however, that we cannot currently rule out the possibility that these compounds result from β-5 dimerization of coniferyl alcohol, and that the m/z 519 and 521 compounds are dehydrodiconiferyl alcohol glucoside and dihydrodehydrodiconiferyl alcohol glucoside, respectively.
All of the available data suggest that the mechanism by which REF4 and RFR1 influence phenylpropanoid accumulation is transcriptional. First, Bäckström et al. (23) have convincingly shown that both REF4 and RFR1 are physically associated with the conserved transcriptional co-regulator complex Mediator, as both gene products were immunoprecipitated in independent experiments using antibodies raised against MED6 and MED7, two different subunits of the complex. Consistent with a role for REF4 and RFR1 in the control of phenylpropanoid biosynthetic gene expression, we have found that ref4 rfr1-deficient plants show increased steady-state transcript levels of the early phenylpropanoid biosynthetic genes PAL1, PAL2, C4H, and 4CL1 (Fig. 4). These changes in transcript levels are limited to time points in the circadian clock where wild-type plants are actively down-regulating steady-state levels of phenylpropanoid mRNAs, suggesting that REF4 and RFR1 are directly or indirectly required for this process. Furthermore, plants containing the dominant ref4-3 mutation show decreased steady-state levels of PAL1 mRNA, and are relatively unresponsive to constitutive expression of MYB75 (Fig. 5), suggesting that the ref4-3 mutant protein may interfere with its ability to drive transcription of phenylpropanoid genes. Interestingly, microarray analysis revealed that MYB75 was substantially up-regulated in ref4-3 mutant plants, perhaps indicating the activation of an apparently futile mechanism to compensate for the loss of MYB75 function.
Although the conclusion in both our previous report (31) and that of Bäckström et al. (23) was that REF4/MED33b and RFR1/MED33a are plant-specific gene products, more recent phylogenetic evidence has suggested that they are in fact homologs of Saccharomyces cerevisiae Med5p and human MED24/TRAP100 (22), both of which had been previously thought to be lineage-specific. Interestingly, yeast Med5p is also involved in the regulation of metabolic processes and has been shown by interaction studies to localize to a region of Mediator connecting the middle and tail domains of the complex (44). Based on the mass of a subcomplex of yeast Mediator that can be assembled in vitro, as well as the available cryo-EM structure of the RNA pol II holoenzyme, it appears that the yeast Mediator complex contains only a single Med5p polypeptide (44–46). Assuming that the structure of Mediator is broadly conserved, this would suggest that REF4 and RFR1 cannot coexist together in the same Mediator complex.
Based on the experimental results described above, we propose the following model. REF4 and RFR1 play a role in Mediator similar to the yeast Med5p protein and act to suppress the expression of phenylpropanoid biosynthetic genes in response to environmental or developmental cues. They appear to act directly, for instance, functioning as transcriptional co-repressors at the phenylpropanoid genes themselves, but we cannot rule out that REF4/RFR1 could interact with an uncharacterized transcriptional activator or repressor further upstream in the network of transcription factors. In the absence of REF4 and RFR1, phenylpropanoid biosynthetic gene expression is inappropriately derepressed, leading to hyperaccumulation of various pathway products, the synthesis of which can be supported by the rest of the cellular metabolic apparatus. Conversely, repression is inappropriately maintained in the ref4-3 mutant, perhaps as a result of G383S substitution stabilizing the interaction between REF4 and one of its binding partners. Consistent with this idea, constitutive repression can be mitigated by mutation of the nearby Arg-387 to histidine, which might be expected to be involved in the same interactions as Gly-383. The divergence of the residues equivalent to Arg-387 in Selaginella and Clade 2 MED33/Med5p proteins in other species suggests that REF4-, RFR1-, and RFR2-containing Mediator complexes may regulate different sets of genes, potentially ones outside of phenylpropanoid metabolism. Alternatively, the binding partners of these other REF4 family members may contain compensatory amino acid changes that allow them to maintain their interaction in this sequence context.
The constitutive repression executed by ref4-3 is also apparently mitigated by the presence of wild-type RFR1, consistent with a model in which REF4 and RFR1 are partially redundant and compete with one another for occupancy in Mediator or for binding partners. According to this model, Mediator complexes containing the ref4-3 mutant protein constitutively repress phenylpropanoid biosynthesis, whereas those containing RFR1 can be regulated normally. Elimination of RFR1 thus increases the proportion of constitutively repressive complexes and exacerbates the phenotypic effects stemming from the ref4-3 mutation. Determining the binding partners of REF4 and its paralogs, the details of the mechanism by which they influence phenylpropanoid biosynthesis, and their possible involvement in other Mediator-dependent phenomena in plants all remain important questions for future study.
Supplementary Material
This work was supported by grants from the Division of Energy Biosciences, United States Department of Energy Grant DE-FG02-07ER15905, the Global Climate and Energy Program (GCEP), a fellowship from the Life Sciences Research Foundation (to N. D. B.), and a summer research fellowship from the Howard Hughes Medical Institute (to M. R. B.).

This article contains supplemental Tables S1 and S2 and Figs. S1–S5.
- REF4
- reduced epidermal fluorescence 4
- C4H
- cinnamate 4-hydroxylase
- PAL
- phenylalanine-ammonia lyase
- RFR1
- REF4-related 1.
REFERENCES
- 1. Bonawitz N. D., Chapple C. (2010) The genetics of lignin biosynthesis. Connecting genotype to phenotype. Annu. Rev. Genet. 44, 337–363 [DOI] [PubMed] [Google Scholar]
- 2. Ralph J. (2004) Lignins: Natural polymers from oxidative coupling of 4-hydroxyphenylpropanoids. Phytochem. Rev. 3, 29–60 [Google Scholar]
- 3. Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010) Lignin biosynthesis and structure. Plant Physiol. 153, 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Field C. B., Behrenfeld M. J., Randerson J. T., Falkowski P. (1998) Primary production of the biosphere. Integrating terrestrial and oceanic components. Science 281, 237–240 [DOI] [PubMed] [Google Scholar]
- 5. Umezawa T. (2010) The cinnamate/monolignol pathway. Phytochem. Rev. 9, 1–17 [Google Scholar]
- 6. Boudet A. M., Kajita S., Grima-Pettenati J., Goffner D. (2003) Lignins and lignocellulosics. A better control of synthesis for new and improved uses. Trends Plant Sci. 8, 576–581 [DOI] [PubMed] [Google Scholar]
- 7. Vanholme R., Morreel K., Ralph J., Boerjan W. (2008) Lignin engineering. Curr. Opin. Plant Biol. 11, 278–285 [DOI] [PubMed] [Google Scholar]
- 8. Weng J. K., Li X., Bonawitz N. D., Chapple C. (2008) Emerging strategies of lignin engineering and degradation for cellulosic biofuel production. Curr. Opin. Biotechnol. 19, 166–172 [DOI] [PubMed] [Google Scholar]
- 9. Adlercreutz H. (2002) Phyto-oestrogens and cancer. Lancet Oncol. 3, 364–373 [DOI] [PubMed] [Google Scholar]
- 10. Butelli E., Titta L., Giorgio M., Mock H. P., Matros A., Peterek S., Schijlen E. G., Hall R. D., Bovy A. G., Luo J., Martin C. (2008) Enrichment of tomato fruit with health-promoting anthocyanins by expression of select transcription factors. Nat. Biotechnol. 26, 1301–1308 [DOI] [PubMed] [Google Scholar]
- 11. Kitts D. D., Yuan Y. V., Wijewickreme A. N., Thompson L. U. (1999) Antioxidant activity of the flaxseed lignan secoisolariciresinol diglycoside and its mammalian lignan metabolites enterodiol and enterolactone. Mol. Cell. Biochem. 202, 91–100 [DOI] [PubMed] [Google Scholar]
- 12. Niggeweg R., Michael A. J., Martin C. (2004) Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22, 746–754 [DOI] [PubMed] [Google Scholar]
- 13. Humphreys J. M., Chapple C. (2002) Rewriting the lignin roadmap. Curr. Opin. Plant Biol. 5, 224–229 [DOI] [PubMed] [Google Scholar]
- 14. Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 126, 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rogers L. A., Campbell M. M. (2004) The genetic control of lignin deposition during plant growth and development. New Phytol. 164, 17–30 [DOI] [PubMed] [Google Scholar]
- 16. Zhong R., Ye Z. H. (2007) Regulation of cell wall biosynthesis. Curr. Opin. Plant Biol. 10, 564–572 [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez A., Zhao M., Leavitt J. M., Lloyd A. M. (2008) Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827 [DOI] [PubMed] [Google Scholar]
- 18. Borevitz J. O., Xia Y., Blount J., Dixon R. A., Lamb C. (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Flanagan P. M., Kelleher R. J., 3rd, Sayre M. H., Tschochner H., Kornberg R. D. (1991) A mediator required for activation of RNA polymerase II transcription in vitro. Nature 350, 436–438 [DOI] [PubMed] [Google Scholar]
- 20. Kim Y. J., Björklund S., Li Y., Sayre M. H., Kornberg R. D. (1994) A multiprotein mediator of transcriptional activation and its interaction with the C-terminal repeat domain of RNA polymerase II. Cell 77, 599–608 [DOI] [PubMed] [Google Scholar]
- 21. Bourbon H. M., Aguilera A., Ansari A. Z., Asturias F. J., Berk A. J., Bjorklund S., Blackwell T. K., Borggrefe T., Carey M., Carlson M., Conaway J. W., Conaway R. C., Emmons S. W., Fondell J. D., Freedman L. P., Fukasawa T., Gustafsson C. M., Han M., He X., Herman P. K., Hinnebusch A. G., Holmberg S., Holstege F. C., Jaehning J. A., Kim Y. J., Kuras L., Leutz A., Lis J. T., Meisterernest M., Naar A. M., Nasmyth K., Parvin J. D., Ptashne M., Reinberg D., Ronne H., Sadowski I., Sakurai H., Sipiczki M., Sternberg P. W., Stillman D. J., Strich R., Struhl K., Svejstrup J. Q., Tuck S., Winston F., Roeder R. G., Kornberg R. D. (2004) A unified nomenclature for protein subunits of mediator complexes linking transcriptional regulators to RNA polymerase II. Mol. Cell 14, 553–557 [DOI] [PubMed] [Google Scholar]
- 22. Bourbon H. M. (2008) Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res. 36, 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bäckström S., Elfving N., Nilsson R., Wingsle G., Björklund S. (2007) Purification of a plant mediator from Arabidopsis thaliana identifies PFT1 as the Med25 subunit. Mol. Cell 26, 717–729 [DOI] [PubMed] [Google Scholar]
- 24. Cerdán P. D., Chory J. (2003) Regulation of flowering time by light quality. Nature 423, 881–885 [DOI] [PubMed] [Google Scholar]
- 25. Dhawan R., Luo H., Foerster A. M., Abuqamar S., Du H. N., Briggs S. D., Mittelsten Scheid O., Mengiste T. (2009) Histone monoubiquitination1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kidd B. N., Edgar C. I., Kumar K. K., Aitken E. A., Schenk P. M., Manners J. M., Kazan K. (2009) The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Boyce J. M., Knight H., Deyholos M., Openshaw M. R., Galbraith D. W., Warren G., Knight M. R. (2003) The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 34, 395–406 [DOI] [PubMed] [Google Scholar]
- 28. Kim Y. J., Zheng B., Yu Y., Won S. Y., Mo B., Chen X. (2011) The role of Mediator in small and long noncoding RNA production in Arabidopsis thaliana. EMBO J. 30, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Autran D., Jonak C., Belcram K., Beemster G. T., Kronenberger J., Grandjean O., Inzé D., Traas J. (2002) Cell numbers and leaf development in Arabidopsis. A functional analysis of the STRUWWELPETER gene. EMBO J. 21, 6036–6049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ruegger M., Chapple C. (2001) Mutations that reduce sinapoylmalate accumulation in Arabidopsis thaliana define loci with diverse roles in phenylpropanoid metabolism. Genetics 159, 1741–1749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Stout J., Romero-Severson E., Ruegger M. O., Chapple C. (2008) Semidominant mutations in reduced epidermal fluorescence 4 reduce phenylpropanoid content in Arabidopsis. Genetics 178, 2237–2251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Clough S. J., Bent A. F. (1998) Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 33. Li X., Bonawitz N. D., Weng J. K., Chapple C. (2010) The growth reduction associated with repressed lignin biosynthesis in Arabidopsis thaliana is independent of flavonoids. Plant Cell 22, 1620–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Huelsenbeck J. P., Ronquist F. (2001) MRBAYES. Bayesian inference of phylogenetic trees. Bioinformatics 17, 754–755 [DOI] [PubMed] [Google Scholar]
- 35. Tamura K., Dudley J., Nei M., Kumar S. (2007) MEGA4, Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 36. Bell-Lelong D. A., Cusumano J. C., Meyer K., Chapple C. (1997) Cinnamate-4-hydroxylase expression in Arabidopsis. Regulation in response to development and the environment. Plant Physiol. 113, 729–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Harmer S. L., Hogenesch J. B., Straume M., Chang H. S., Han B., Zhu T., Wang X., Kreps J. A., Kay S. A. (2000) Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290, 2110–2113 [DOI] [PubMed] [Google Scholar]
- 38. Rogers L. A., Dubos C., Cullis I. F., Surman C., Poole M., Willment J., Mansfield S. D., Campbell M. M. (2005) Light, the circadian clock, and sugar perception in the control of lignin biosynthesis. J. Exp. Bot. 56, 1651–1663 [DOI] [PubMed] [Google Scholar]
- 39. Hahlbrock K., Knobloch K. H., Kreuzaler F., Potts J. R., Wellmann E. (1976) Coordinated induction and subsequent activity changes of two groups of metabolically interrelated enzymes. Light-induced synthesis of flavonoid glycosides in cell suspension cultures of Petroselinum hortense. Eur. J. Biochem. 61, 199–206 [DOI] [PubMed] [Google Scholar]
- 40. Ro D. K., Douglas C. J. (2004) Reconstitution of the entry point of plant phenylpropanoid metabolism in yeast (Saccharomyces cerevisiae). Implications for control of metabolic flux into the phenylpropanoid pathway. J. Biol. Chem. 279, 2600–2607 [DOI] [PubMed] [Google Scholar]
- 41. Mir Derikvand M., Sierra J. B., Ruel K., Pollet B., Do C. T., Thévenin J., Buffard D., Jouanin L., Lapierre C. (2008) Redirection of the phenylpropanoid pathway to feruloyl malate in Arabidopsis mutants deficient for cinnamoyl-CoA reductase 1. Planta 227, 943–956 [DOI] [PubMed] [Google Scholar]
- 42. Rohde A., Morreel K., Ralph J., Goeminne G., Hostyn V., De Rycke R., Kushnir S., Van Doorsselaere J., Joseleau J. P., Vuylsteke M., Van Driessche G., Van Beeumen J., Messens E., Boerjan W. (2004) Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell 16, 2749–2771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nakatsubo T., Mizutani M., Suzuki S., Hattori T., Umezawa T. (2008) Characterization of Arabidopsis thaliana pinoresinol reductase, a new type of enzyme involved in lignan biosynthesis. J. Biol. Chem. 283, 15550–15557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Béve J., Hu G. Z., Myers L. C., Balciunas D., Werngren O., Hultenby K., Wibom R., Ronne H., Gustafsson C. M. (2005) The structural and functional role of Med5 in the yeast Mediator tail module. J. Biol. Chem. 280, 41366–41372 [DOI] [PubMed] [Google Scholar]
- 45. Davis J. A., Takagi Y., Kornberg R. D., Asturias F. A. (2002) Structure of the yeast RNA polymerase II holoenzyme. Mediator conformation and polymerase interaction. Mol. Cell 10, 409–415 [DOI] [PubMed] [Google Scholar]
- 46. Kornberg R. D. (2007) The molecular basis of eukaryotic transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.