Background: Intracellular pathogens avoid lysosomal targeting but recruit lysosome-associated membrane protein 1 (LAMP1), the mechanism of which remains unclear.
Results: SipC binds with Syntaxin6 and acquires LAMP1 on the phagosome by fusing with Golgi-derived vesicles.
Conclusion: This is a novel mechanism by which Salmonella acquires LAMP1 from Golgi.
Significance: This is the first demonstration showing how an intracellular pathogen like Salmonella recruits LAMP1 without fusing with lysosomes.
Keywords: Golgi, Host-Pathogen Interactions, Intracellular Trafficking, Phagocytosis, SNARE, LAMP1, Macrophage, Salmonella, SipC, Syntaxin6
Abstract
Several intracellular pathogens have developed diverse strategies to avoid targeting to lysosomes. However, they universally recruit lysosome-associated membrane protein 1 (LAMP1); the mechanism of LAMP1 recruitment remains unclear. Here, we report that a Salmonella effector protein, SipC, specifically binds with host Syntaxin6 through its C terminus and thereby recruits Syntaxin6 and other accessory molecules like VAMP2, Rab6, and Rab8 on Salmonella-containing phagosomes (SCP) and acquires LAMP1 by fusing with LAMP1-containing Golgi-derived vesicles. In contrast, sipC knock-out:SCP (sipC−:SCP) or sipCM398K:SCP fails to obtain significant amounts of Syntaxin6 and is unable to acquire LAMP1. Moreover, phagosomes containing respective knock-out Salmonella like sipA−, sipB−, sipD−, sopB−, or sopE− recruit LAMP1, demonstrating the specificity of SipC in this process. In addition, depletion of Syntaxin6 by shRNA in macrophages significantly inhibits LAMP1 recruitment on SCP. Additionally, survival of sipC−:Salmonella in mice is found to be significantly inhibited in comparison with WT:Salmonella. Our results reveal a novel mechanism showing how Salmonella acquires LAMP1 through a SipC-Syntaxin6-mediated interaction probably to stabilize their niche in macrophages and also suggest that similar modalities might be used by other intracellular pathogens to recruit LAMP1.
Introduction
Several intracellular pathogens like Mycobacterium, Salmonella, Legionella, and Toxoplasma use their effectors to modulate the endocytic pathway of host cells and avoid transport to the lysosomes (1–4). However, pathogen-containing phagosomes recruit LAMP1; the mechanism of which remains to be elucidated. Lysosome-associated membrane proteins form a continuous carbohydrate lining on the inner leaflet of the lysosomal membrane (5) and are required for phagosome maturation (6) suggesting that recruitment of lysosome-associated membrane protein is possibly required to maintain the structural integrity of the phagosome. Newly synthesized LAMP1 is trafficked from the trans-Golgi network (TGN)3 to lysosomes via endosomes (7), and therefore, lysosomes are enriched with LAMP1. It is tempting to speculate that pathogen-containing phagosomes might recruit LAMP1 by fusing with LAMP1-containing vesicles originating from the TGN during its trafficking through the secretory pathway. However, the interaction of phagosomes with the secretory pathway is not well characterized (8). Previous studies have shown (9) that disruption of Golgi by brefeldin A or overexpression of ARF1:T31N inhibits Salmonella replication in host cells indicating that transport from the Golgi is required. Additionally, pathogens like Legionella, Salmonella, Chlamydia, and Brucella translocate near the Golgi (10–12) in host cells, but the physiological significance of this localization is still unclear. Here, we have shown that SCP recruits host Syntaxin6 through its effector protein, SipC, and acquires LAMP1 by fusing with LAMP1-containing Golgi derived vesicles.
EXPERIMENTAL PROCEDURES
Antibodies
Antibodies against SopE, SopB, and SipC were kindly provided by Dr. E. E. Galyov from the Institute for Animal Health, Berkshire, UK. Antibodies against mammalian EEA-1 and Rab5 were received as kind gifts from Dr. Marino Zerial (Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany) and Dr. A. Wandinger-Ness (University of New Mexico, Albuquerque, NM), respectively. Antibodies against GM130, Vti1a, Vti1b, LAMP1, and Rab8 were purchased from BD Biosciences. Anti-His antibody was purchased from Amersham Biosciences. Anti-Rab6 and anti-GAPDH were obtained from Santa Cruz Biotechnology. Antibodies against Rab7, transferrin receptor, and VAMP2 were purchased from Cell Signaling, Zymed Laboratories Inc., and Abcam, respectively. Salmonella antiserum H, which predominantly detects flagellin, was purchased from BD Biosciences.
Cells
The Salmonella typhimurium (SL1344 strain) was generously provided by Dr. David W. Holden, Imperial College of Sciences, London, UK. The Salmonella mutant strains, invA−, ssaR−, sipA−, sipB−, sipD−, sopB−, and sopE−, were kind gifts from Dr. J. Brumell (Hospital for Sick Children, Toronto, Canada), Dr. Wolf-Dietrich Hardt (Institute of Microbiology, ETH, Z÷rich, Switzerland), Dr. David W. Holden (Imperial College of Sciences, London), and Dr. Dipshikha Chakravortty (Department of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, India). Dr. Samuel I. Miller (University of Washington, Seattle) kindly provided sipCM398K:Salmonella and sipCR315Z:Salmonella strains. Bacteria were routinely grown overnight in Luria broth containing appropriate antibiotics at 37 °C, and late log phase cells were harvested by centrifugation for experimental purposes.
J774E, a murine macrophage cell line, was kindly provided by Dr. Philip Stahl of Washington University (St. Louis). RAW 264.7, a murine macrophage cell line, was obtained from American Type Culture Collection (ATCC). These cells were maintained in RPMI 1640 medium and supplemented with 10% fetal calf serum and gentamycin (50 μg/ml) at 37 °C in 5% CO2, 95% air atmosphere.
Plasmids
SipC plasmid was received as a kind gift from Dr. Bobby J. Cherayil of Massachusetts General Hospital, Charlestown, MA. SipC(1–120) and SipC(200–409) constructs were kindly provided by Dr. J. E. Casanova (University of Virginia, Charlottesville). Salmonella expression vectors, pFPV25.1 and pIZ1590, for constitutive expression of GFP and RFP were kindly provided by Dr. Raphael Valdivia (Duke Center for microbial pathogenesis, Durham, NC) and Dr. Francisco Ramos-Morales (Universidad de Sevilla, Spain), respectively. pBAD24, arabinose-inducible expression vector, was kindly provided by Dr. A. Surolia of National Institute of Immunology. LAMP1-GFP construct was obtained from Dr. Alberto Luini (Consorzio Mario, Negrusid, Italy).
Preparation of SCP
SCP were purified from J774E macrophages using a method described previously (13). Briefly, J774E macrophages (1 × 108) were incubated with Salmonella (2 × 109) for 5 min at 37 °C to restrict them in the early compartment. Cells were washed (300 × g for 6 min) three times to remove uninternalized bacteria. One aliquot of the cell suspension was processed for the preparation of early SCP. The rest of the cell suspension was further incubated for the indicated periods of time at 37 °C to prepare SCP at the respective time points. Subsequently, cells were washed and homogenized in homogenization buffer, and post-nuclear supernatants were prepared by low speed (400 × g for 10 min) centrifugation. Finally, phagosomes were purified from post-nuclear supernatants through a 12% sucrose cushion.
Detection of Levels of Various Markers on Phagosomes
To detect the levels of various proteins on respective SCP during its maturation in J774E macrophages, phagosomes containing WT or mutant Salmonella were purified at the indicated time points of their maturation. Subsequently, phagosomal proteins (40 μg) were resolved on a SDS-PAGE, and Western blot analyses were carried out using specific antibodies as indicated.
Protein Purification
Full-length Syntaxin6(1–255) and its truncations (1–76 and 176–230) were cloned into pGEX4T2 and transformed into Escherichia coli (BL21 strain). Cells were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for 3 h at 37 °C, and the respective GST fusion proteins were affinity-purified as per the manufacturer's instructions (Amersham Biosciences) under native conditions using glutathione-Sepharose 4B. Briefly, bacterial cells were sonicated and lysed in phosphate-buffered saline, pH 7.2 (PBS), containing 1% Triton X-100 in the presence of DTT (1 mg/ml) at 4 °C. Lysates were incubated with glutathione-Sepharose beads for 1 h at 4 °C. Following extensive washes with PBS, recombinant proteins were eluted from the beads in 50 mm Tris-HCl containing 30 mm glutathione, pH 9.0, and dialyzed against PBS.
To purify SipC as a His6-tagged fusion protein, full-length SipC was subcloned into pET28a and transformed into E. coli (BL21 strain). Cells were induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 4 h. Cells were harvested, and the protein was purified under denaturing conditions using nickel-nitrilotriacetic acid-agarose as per the manufacturer's instructions (Qiagen). Briefly, the inclusion bodies were solubilized in 20 mm sodium phosphate, pH 7.8, 500 mm NaCl, and 6 m guanidine HCl. Subsequently, lysates were incubated with nickel-nitrilotriacetic acid-agarose in 20 mm sodium phosphate, pH 7.8, 500 mm NaCl, and 8 m urea. Following extensive washes with 20 mm sodium phosphate, pH 6.0, containing 500 mm NaCl and 8 m urea, recombinant His6-SipC was eluted from the beads in 20 mm sodium phosphate, pH 4.0, 500 mm NaCl, and 8 m urea. The eluted protein was diluted 1:10 in 20 mm Tris-Cl, pH 8.0, 150 mm NaCl, and 2 m urea and step-dialyzed against this buffer with reducing amounts of urea from 2 to 0 m to renature the protein.
Similarly, cells containing His6SipC(1–120) or His6SipC(200–409) plasmid were induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 37 °C for 4 h, and the proteins were purified as described previously (14).
Identification of Syntaxin6-binding Protein from Salmonella
To identify the Salmonella effector molecules interacting with Syntaxin6, bacteria were grown overnight at 37 °C in Luria broth. Cells were removed by centrifugation, and Salmonella secretory proteins present in the spent medium were concentrated and biotinylated as described previously (15). To detect the Syntaxin6-binding protein, 10 μg of GST-Syntaxin6 was immobilized on glutathione-Sepharose beads and incubated in the presence of biotinylated secretory proteins (5 mg) of Salmonella in PBS, pH 7.2, for 2 h at 24 °C. Beads were washed three times with PBS containing 1% Triton X-100, followed by three washes with PBS to remove unbound proteins. The proteins were separated on a 12% SDS-PAGE and were transferred onto nitrocellulose membrane. Finally, Western blot analysis was carried out with avidin-HRP (Vector Laboratories) to detect the biotinylated Salmonella proteins bound with Syntaxin6. Identical experiments were carried out using nonbiotinylated secretory proteins of Salmonella, and the proteins were identified by Western blot analysis using the indicated antibodies against different Salmonella effector proteins. An equimolar amount of immobilized GST was used as a control.
Immunoprecipitation of Syntaxin6 from Macrophage Lysate by SipC
To detect the binding of SipC with Syntaxin6 from the macrophage cell lysate, SipC was immobilized by incubating anti-SipC-coated protein G beads with Salmonella secretory proteins (2 mg). Macrophages (1 × 107) were lysed by incubating the cells in 1 ml of PBS containing 0.1% Triton X-100 for 45 min at 4 °C, and cellular debris were separated by centrifugation (12,000 × g using an Eppendorf rotor FA-45-24-11) at 4 °C for 15 min. Subsequently, 4 mg of this cell lysate was incubated with immobilized SipC for 8 h at 4 °C in the same buffer. Beads were washed as mentioned earlier, and proteins were separated on a 12% SDS-PAGE. Finally, Western blot analysis was carried out using anti-Syntaxin6 or anti-Syntaxin8 (Synaptic Systems) antibody.
Binding of SipC with Syntaxin6
To determine the region of binding of Syntaxin6 with SipC, 2.5 μg of GST-Syntaxin6 or an equimolar amount of GST-Syntaxin6(1–76) or GST-Syntaxin6(176–230) was immobilized on glutathione-Sepharose beads. These beads were incubated with 1 μg of His6-SipC or equimolar concentration of His6-SipC(1–120) or His6-SipC(200–409) for 2 h at 4 °C in PBS. Beads were washed as described previously, and the binding of SipC was detected by Western blot analysis using the indicated antibody.
Uptake of Salmonella by Macrophages
To determine the phagocytosis of sipC−:Salmonella and WT:Salmonella by macrophages, bacteria were metabolically labeled with [35S]methionine for 4 h (A600 of 0.8–0.9). Subsequently, J774E cells were incubated with respective [35S]methionine-labeled Salmonella (10 multiplicities of infection) at 37 °C for indicated periods of time. Finally, cells were washed extensively with PBS containing 2% BSA, and the radioactivity associated with the cells was measured to determine the uptake of bacteria.
Immunofluorescence
To compare the intracellular route of WT:Salmonella and sipC−:Salmonella in J774E macrophages, cells were infected with respective bacteria overexpressing either GFP or RFP as described previously (13). Briefly, 0.5 × 106 J774E macrophages were seeded on a sterile glass coverslip placed in a 6-well plate 12 h before infection. Cells were then infected with GFP:WT or GFP:sipC− Salmonella at a multiplicity of infection of 10 for 5 min at 37 °C in FCS-free RPMI 1640 medium. The cells were then washed three times with PBS to remove uninternalized bacteria and incubated for the indicated times. Subsequently, infected cells were washed, fixed, and permeabilized as described previously (13). The permeabilized cells were probed with specific antibodies against GM130 or LAMP1 and incubated with Alexa Fluor 546-labeled goat anti-mouse secondary antibody (1:1000). Slides were mounted in prolong gold antifade reagent (Molecular Probes) and viewed in an LSM 510 meta confocal microscope using an oil immersion objective.
The shortest distance of bacteria from Golgi was calculated taking the different Z-sections into consideration. The bacteria that were within 1 μm of Golgi were considered as near Golgi. Colocalization between bacteria and LAMP1 was considered when bacteria were surrounded by a ring-like staining indicating that the membrane of the SCP is positive for LAMP1.
To determine the targeting of WT or sipC−:Salmonella to the lysosomes, J774E cells were incubated with 10 μg/ml DQ-BSA Red (Molecular Probes) for 1 h at 37 °C in RPMI 1640 medium containing 10% FCS. The cells were washed and incubated with fluorescent latex beads (Polysciences), GFP:WT, or GFP:sipC−:Salmonella for 5 min at 37 °C, followed by chase for 2 h at 37 °C in FCS free RPMI 1640 medium. Cells were washed with PBS and visualized in a confocal microscope.
Recruitment of LAMP1-GFP by SCP from Golgi
RAW 264.7 macrophages were grown to 80% confluency. Prior to transfection, cells were washed with FCS-free RPMI 1640 medium, and 1 × 107 cells were resuspended in 400 μl of the same medium. Cells were transferred to a 4-mm electroporation cuvette, and 20 μg of purified LAMP1-GFP construct was added. After gentle mixing of the contents, the DNA was electroporated into the cells at 300 V, 975 microfarads using a GenePulser (Bio-Rad). Transfected cells were plated on glass coverslips placed in a 6-well plate and incubated in a 5% CO2, 95% air atmosphere for 6 h at 37 °C in FCS-containing medium. Cells were washed and further incubated for an additional 14 h at 37 °C under similar conditions before checking the expression of LAMP1-GFP by confocal microscopy. To synchronize the transport of LAMP1-GFP in Golgi, LAMP1-GFP-overexpressing cells were incubated at 15 °C for 20 min followed by a chase at 37 °C for 20 min based on the fact that t½ of newly synthesized LAMP1 was shown to be 13 min (16). Cells were fixed and stained with GM130 antibody to visualize the presence of LAMP1-GFP in Golgi.
To monitor the acquisition of LAMP1 by WT or sipC−:SCP from Golgi-derived vesicles, RAW 264.7 macrophages overexpressing LAMP1-GFP were infected with RFP:WT or RFP:sipC− Salmonella for 5 min at 37 °C, 18 h post-transfection. After infection, uninternalized bacteria were removed by washing with FCS-free RPMI 1640 medium, and the infected cells were further incubated at 37 °C for 120 min to allow the transport of bacteria toward the appropriate late compartments. Subsequently, LAMP1 was synchronized in the Golgi by an appropriate temperature block as indicated. Finally, cells were shifted at 37 °C to allow the budding of LAMP1-GFP-containing vesicles from Golgi. At the indicated times after vesicle budding, the cells were fixed, mounted, and analyzed by confocal microscopy. Percentage colocalization of SCP with LAMP1-GFP was calculated.
shRNA-mediated Silencing of Syntaxin6 and LAMP1 Recruitment
To understand the importance of Syntaxin6 in LAMP1 recruitment, Syntaxin6 shRNA (5′-CUGAAUUGAGCAUAAGAAA-3′) was designed and cloned into pSIREN-RetroQ-ZsGreen RNAi vector (Clontech). RAW 264.7 macrophages were electroporated with 20 μg of shRNA as described above. Depletion of Syntaxin6 in RAW 264.7 cells was checked by Western blot analysis and immunofluorescence using specific antibody 48 h after transfection. Luciferase shRNA-transfected cells were used as control. Subsequently, shRNA-transfected cells were infected with RFP:WT Salmonella for 5 min at 37 °C. After infection, uninternalized bacteria were removed by washing with FCS-free RPMI 1640 medium, and the infected cells were further incubated at 37 °C for 120 min to allow the transport of bacteria toward the appropriate late compartments. Subsequently, cells were fixed and stained with LAMP1 antibody followed by Alexa Fluor 633-labeled rabbit anti-mouse secondary antibody (1:1000). Cells were viewed in an LSM 510 meta confocal microscope using an oil immersion objective. Percentage colocalization of SCP with LAMP1 was calculated in the respective cells.
Intracellular Survival of Salmonella in Mice
Salmonella (1000 bacteria in 100 μl of PBS) were injected intraperitoneally into 8–10-week-old BALB/c mice. Animals were sacrificed either on days 2 or 4 post-infection, and spleen was dissected out from each infected mouse. Subsequently, the spleen was homogenized, and the cells were suspended in RPMI 1640 medium. The cells were lysed in PBS containing 0.2% Triton X-100, and an aliquot of the cell lysate was plated on Salmonella shigella agar plates to determine the number of viable bacteria present in spleen as colony-forming units.
RESULTS
Salmonella Effector Protein SipC Specifically Binds with Host Syntaxin6
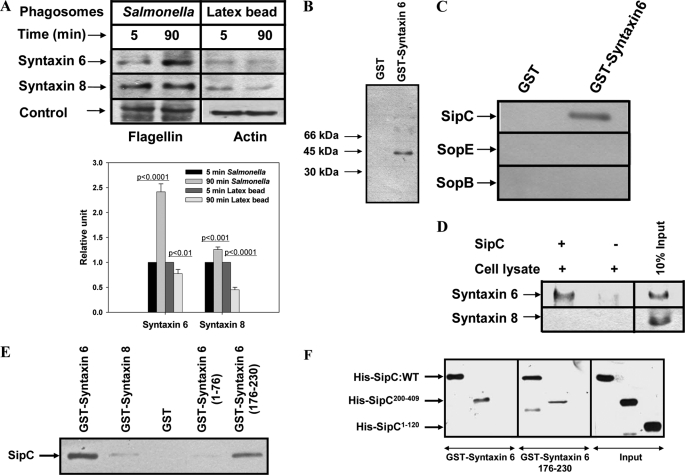
To characterize the maturation of SCP in macrophages, we analyzed the presence of different syntaxins on purified SCP at various stages of their internalization. The results presented in Fig. 1A demonstrated that SCP after 90 min of maturation specifically recruit higher amounts of Syntaxin6 than early (5 min) phagosomes. Biochemical characterization revealed that SCP are devoid of contamination with other intracellular components (supplemental Fig. S1). To identify the bacterial protein involved in the recruitment of Syntaxin6, GST-Syntaxin6 was immobilized and incubated with biotinylated Salmonella secretory proteins. Western blot analysis using avidin-HRP detected a 40-kDa Salmonella protein (Fig. 1B) that was subsequently identified as SipC by Western blot analysis using specific antibody (Fig. 1C) and confirmed by mass spectrometry (supplemental Fig. S2). Immunoprecipitation studies using immobilized SipC specifically recognized Syntaxin6 from the macrophage cell lysate (Fig. 1D). In addition, our results showed that Syntaxin6 (2.5 μg) binds with SipC in a concentration-dependent way, and optimal binding was observed with 1 μg of SipC (supplemental Fig. S3). Therefore, we used equimolar amounts of respective proteins in subsequent binding experiments. Interestingly, our results showed that SipC specifically binds with truncated Syntaxin6(176–230), the SNARE motif of Syntaxin6, but not with Syntaxin6(1–76) (Fig. 1E). We further mapped the region of SipC important for this interaction and found that both Syntaxin6 and Syntaxin6(176–230) interact specifically with the C-terminal end of SipC, SipC(200–409), and not the N-terminal end SipC(1–120) (Fig. 1F). Taken together, our results demonstrated that the C-terminal end of SipC specifically interacts with Syntaxin6 through its SNARE motif.
FIGURE 1.
SCP bind host Syntaxin6 through a SipC-mediated interaction. A, Western blot analyses were carried out to compare the levels of various syntaxins on live Salmonella or latex beads containing phagosomes at early and late stages of their maturation in macrophages. Flagellin and actin were used as controls for different phagosomal preparations as indicated. Lower panel of the figure indicates relative levels of syntaxin normalized to the respective loading control. Results are represented as mean ± S.D. of three independent preparations, and the level of the respective marker present on early phagosomes has been arbitrarily chosen as 1 unit. Levels of significance calculated by t test are indicated by p values. B, to identify Syntaxin6-binding protein from Salmonella, GST-Syntaxin6 was immobilized and incubated with biotinylated secretory proteins of Salmonella. Biotinylated protein was detected by Western blot using avidin-HRP. C, identification of the Salmonella protein by Western blot using antibodies against various Salmonella effector proteins. D, immunoprecipitation of Syntaxin6 from macrophage lysate by immobilized SipC. Syntaxin8 was used as a control. E, to detect the direct binding of Syntaxin6 with SipC, GST-Syntaxin6, GST-Syntaxin8, GST-Syntaxin6(1–76), or GST-Syntaxin6(176–230) was immobilized and incubated with His6-SipC. Binding of SipC with Syntaxin6 was detected by Western blot analysis using anti-SipC antibody. Immobilized GST was used as a control. F, to determine the binding region of SipC, GST-Syntaxin6 or GST-Syntaxin6(176–230) was immobilized and incubated with His6-SipC, His6-SipC(200–409), or His6-SipC(1–120). Binding of SipC with Syntaxin6 was detected by anti-His antibody. Input shows the amount of respective SipC used in the experiment. All results are representative of three independent experiments.
Internalization of sipC−:Salmonella into Macrophages
To determine the role of SipC in the trafficking of Salmonella in macrophages, sipC knock-out Salmonella (sipC−) was generated (supplemental Fig. S4, A–C), and its trafficking was compared with WT:Salmonella. Previous studies have shown that SipC is required for bacterial invasion into epithelial cells (17). Therefore, we determined the ability of sipC−:Salmonella to internalize into macrophages. The results presented in Fig. 2 showed that [35S]methionine-labeled sipC−:Salmonella is taken up by macrophages; however, the efficiency of uptake of mutant bacteria by macrophages is relatively lower in comparison with WT:Salmonella. In addition, our results showed that WT:bacteria induce actin rearrangements and membrane ruffling in close vicinity of SCP in macrophages, whereas actin localization was more diffuse and distant from sipC−:SCP (supplemental Fig. S4D) as observed in HeLa cells (17).
FIGURE 2.
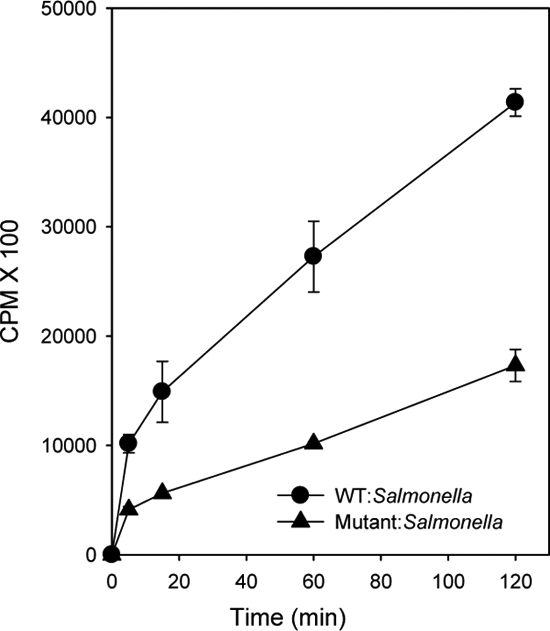
Uptake of WT:Salmonella or sipC−:Salmonella into macrophages. Uptake of [35S]methionine-labeled WT:Salmonella or sipC−:Salmonella by J774E macrophages is shown. Results are represented as mean ± S.D. of three independent experiments.
Intracellular Trafficking of WT and sipC−:Salmonella in Macrophages
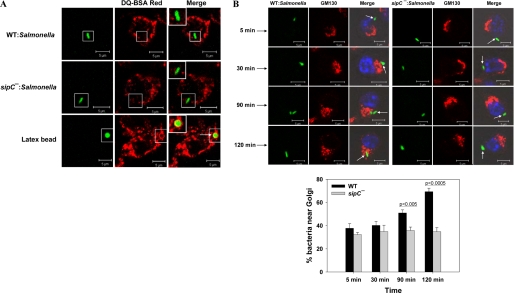
J774E cells were infected with respective GFP-Salmonella or fluorescent latex beads for 5 min and chased for 120 min at 37 °C, and their localization with DQ-BSA labeled lysosomes was determined. Our results showed that both WT and sipC−:Salmonella do not colocalize with DQ-BSA Red-labeled lysosomes (Fig. 3A). In contrast, latex bead-containing phagosomes were found to be colocalized with DQ-BSA. These results indicated that WT and sipC−:Salmonella are not targeted to lysosomes. This finding was further supported by the fact that both WT:SCP and sipC−:SCP retain Rab5 and show reduced levels of Rab7 at a later time point (90 min) in their maturation (supplemental Fig. S5). However, latex bead-containing late phagosomes showed reduced levels of Rab5 and enhanced content of Rab7 in comparison with early phagosomes.
FIGURE 3.
Comparison of intracellular trafficking of WT and sipC−:Salmonella in macrophages. A, colocalization of DQ-BSA Red-labeled lysosomes with GFP expressing WT:Salmonella, sipC−:Salmonella, or fluorescent latex beads 2 h after internalization into macrophages. Arrows indicate colocalization. B, confocal micrographs showing the localization of GFP-Salmonella (green) near the Golgi (red) nucleus is labeled with Hoechst (blue). Images represent a single x/y plane of a Z-section. Lower panel represents the quantification of bacteria in the vicinity of Golgi for each time point after analyzing at least 100 Salmonella in infected cells. Data from three independent experiments were analyzed by t test, and levels of significance are indicated by p values. Inset shows the enlarged region.
Previous studies have shown that Salmonella-containing vacuoles at later stages of their maturation in epithelial cells are in close vicinity of Golgi membranes (9). However, not much is known about the targeting of Salmonella toward Golgi in macrophages. Therefore, J774E cells were infected with GFP-Salmonella and chased for the indicated periods of time followed by Golgi labeling. Our results indicated that both WT:Salmonella and sipC−:Salmonella enter into an early endocytic compartment (Rab5 and transferrin receptor-positive) by 5 min of internalization into macrophages (supplemental Fig. S5). However, by 120 min post-internalization (Fig. 3B), about 70% of the WT:Salmonella were found in the vicinity of the Golgi compartment (less than 1 μm from Golgi). In contrast, less than 35% of sipC−:Salmonella was found near the Golgi under similar conditions suggesting the possible role of SipC in targeting of the Salmonella toward the Golgi in macrophages.
SipC-mediated Recruitment of LAMP1 by SCP in Macrophages
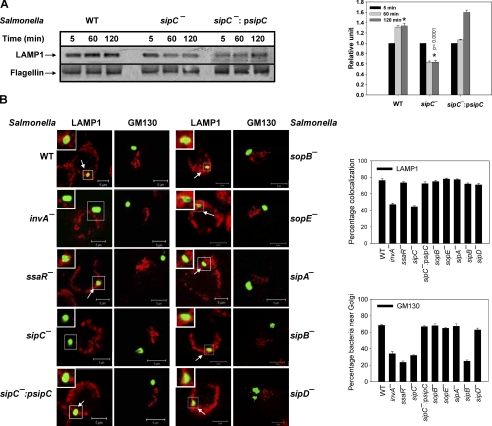
To compare the levels of LAMP1 between WT:SCP and sipC−:SCP, Western blot analysis of purified phagosomes at different time points of their maturation in macrophages was carried out. We found that sipC−:SCP progressively loses LAMP1, whereas WT:SCP acquire significantly higher amounts of this marker as they mature (Fig. 4A). Interestingly, phagosomes containing sipC complemented in sipC−:Salmonella (sipC−:psipC) restored the kinetics of LAMP1 recruitment like WT:bacteria.
FIGURE 4.
Recruitment of LAMP1 on SCP. A, Western blot showing levels of LAMP1 on indicated purified phagosomes (80 μg of protein per lane). Flagellin was used as a loading control. Right panel of the figure indicates quantification of the data by densitometry. Results are represented as mean ± S.D. of three observations, normalized to the loading control. The amount of LAMP1 present on respective 5-min phagosome is arbitrarily chosen as 1 unit. Data from three independent experiments were analyzed by t test, and the significance is indicated by p values between the respective 120 min phagosomes (*). B, confocal image showing the localization of respective Salmonella (green) near Golgi (labeled with GM130-red) or their colocalization with LAMP1 (red), after 120 min of internalization into macrophages. Salmonella was visualized by probing with Salmonella H antiserum followed by Alexa 488-labeled anti-rabbit secondary antibody. Image represents a single x/y plane of a Z-section, and arrows indicate colocalization. Inset shows the enlarged region. The graphs represent the percentage of SCP positive for LAMP1 or in vicinity of Golgi from at least 50 observations.
To unequivocally prove the role of SipC in the recruitment of LAMP1 on SCP, macrophages were infected with various mutant Salmonella, and the presence of LAMP1 on the respective phagosomes was determined after 120-min post-internalization. Our results showed (Fig. 4B) that only ∼45% invA−:SCP (defective in translocation in all SPI-1 TTSS effectors) are able to recruit LAMP1 in comparison with ∼80% of WT:SCP, whereas no significant change (∼75%) in the LAMP1 localization was detected on ssaR−:SCP (defective in SPI-2 TTSS effector translocation). These results suggest that SPI-1 TTSS effectors are involved in the LAMP1 recruitment on SCP. To explore this possibility, we used various SPI-1 TTSS effector deletion mutant bacteria and determined LAMP1 recruitment on their phagosomes. We found that like WT:SCP, about 75% of phagosomes containing sipA−, sipB−, sipD−, sopB−, or sopE− bacteria were positive for LAMP1. In contrast, only 44% of sipC−:SCP were positive for LAMP1. Complementation of sipC in sipC−:Salmonella (sipC−:psipC) significantly enhanced the recruitment of LAMP1 (∼72%) on phagosomes demonstrating the specific role of SipC in LAMP1 recruitment.
We also determined the localization of various mutant Salmonella near GM130-labeled Golgi after 120 min post-internalization (Fig. 4B), and we found that both invA−:Salmonella (∼35%) and ssaR−:Salmonella (∼25%) are defective in targeting toward Golgi as compared with WT:Salmonella (∼70%). Among the various mutant bacteria, sipB− and sipC− were not targeted toward Golgi (∼25 and ∼30% respectively), whereas the majority (65%) of sipA−, sipD−, sopB−, and sopE− bacteria moved near Golgi-like WT:Salmonella. In addition, complementation of sipC in sipC−:Salmonella (sipC−:psipC) restored the targeting of this bacteria near Golgi (∼70%). These results demonstrated that possibly SipB and SipC are necessary for the movement of Salmonella toward Golgi along with SPI-2 TTSS effectors.
Role of SipC Mutant Salmonella in Recruitment of LAMP1 on SCP
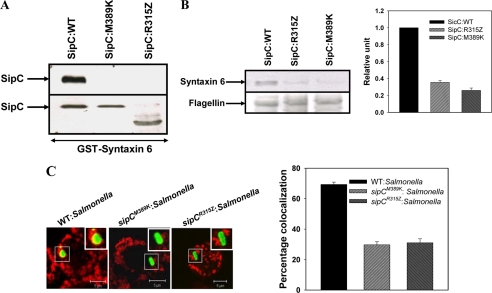
To confirm the importance of the C-terminal region of SipC in the Syntaxin6-mediated recruitment of LAMP1, we used two Salmonella mutants, sipCR315Z and sipCM389K. These mutants were initially reported to disrupt the interaction of SipC with cytokeratin-8 (18). Subsequently, secretory proteins from sipCR315Z and sipCM389K:Salmonella were used to determine the binding of mutated SipC with Syntaxin6, and our results showed that binding of sipCR315Z and sipCM389K proteins with Syntaxin6 is disrupted (Fig. 5A). Interestingly, we also observed that phagosomes containing sipCM389K and sipCR315Z Salmonella do not recruit significant amounts of Syntaxin6 after 90 min of internalization in comparison with WT:SCP (Fig. 5B). Consequently, we determined the recruitment of LAMP1 by these SipC mutants that are deficient in the Syntaxin6 binding after 120 min of internalization into macrophages. Interestingly, we found that only about 30% of both sipCM389K:SCP and sipCR315Z:SCP are colocalized with LAMP1 in comparison with 70% by WT:SCP (Fig. 5C).
FIGURE 5.
Role of C terminus of SipC in the recruitment of Syntaxin6 and LAMP1. A, immobilized GST-Syntaxin6 was incubated with secretory proteins from indicated Salmonella, and binding of respective SipC with Syntaxin6 was detected by anti-SipC antibody. Lower panel shows 5% of the total input of secretory proteins. B, respective phagosomes were purified after 90 min of internalization into macrophages, and Western blot shows the levels of Syntaxin6 on indicated SCP. Flagellin was used as control. Right panel of the figure indicates relative levels of Syntaxin6 normalized to the WT:SCP arbitrarily chosen as 1 unit. Results are represented as mean ± S.D. of three independent preparations. C, confocal micrographs showing colocalization between LAMP1 (red) and respective Salmonella (green) after 120 min of internalization in macrophages. The graph represents the percentage of SCP positive for LAMP1 at least from 50 observations. The image represents a single x/y plane of a Z-section where arrows indicate colocalization and inset shows the enlarged region. All results are representative of three independent experiments.
Role of Syntaxin6 in Recruitment of LAMP1 on SCP in Macrophages
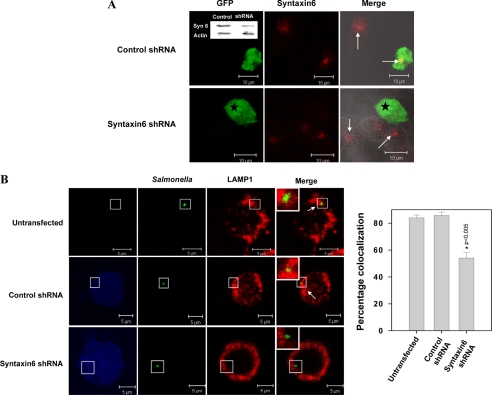
To establish the role of Syntaxin6 in LAMP1 recruitment on SCP, we used specific shRNA to silence endogenous Syntaxin6 in RAW macrophages. Silencing of Syntaxin6 was confirmed by Western blotting and immunostaining using the Syntaxin6-specific antibody (Fig. 6A). Subsequently, we analyzed the recruitment of LAMP1 by SCP in Syntaxin6-depleted cells and observed (Fig. 6B) that silencing of Syntaxin6 in RAW cells led to a significant reduction in LAMP1 recruitment (∼50%) on WT:SCP at later stages of maturation (120 min) in comparison with control shRNA-transfected cells (85%).
FIGURE 6.
Recruitment of LAMP1 on SCP in Syntaxin6-depleted macrophages. A, confocal micrographs showing depletion of Syntaxin6 in Syntaxin6 shRNA-transfected cells. Luciferase shRNA-transfected cells were used as control. GFP expression indicates transfected cells (star) and Syntaxin6 appears in red (indicated by arrows). Inset, Western blot showing levels of Syntaxin6 in indicated cells. B, RAW 264.7 macrophages were transfected with respective shRNA and infected with WT:Salmonella as described under “Experimental Procedures.” The confocal image represents a single x/y plane of a Z-section where transfected cells are represented in blue; green shows the Salmonella and LAMP1 appears in red. Arrow indicates colocalization. Inset shows the enlarged region. The graph represents the percentage of SCP positive for LAMP1 from at least 50 observations. The level of significance was calculated by a t test, and the p value is as indicated.
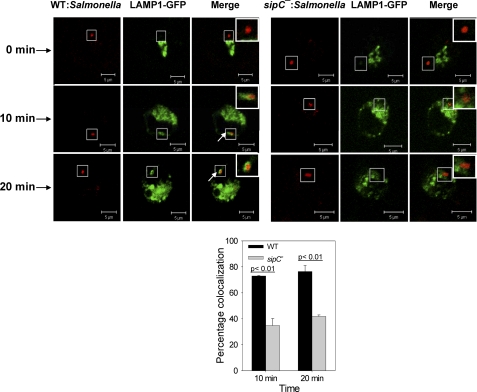
Recruitment of LAMP1 by SCP from Golgi
To unequivocally prove that the WT:SCP recruit LAMP1 from Golgi, LAMP1-GFP was overexpressed in RAW macrophages and synchronized in the Golgi (supplemental Fig. S6). To include Salmonella infection in this experimental setup, RAW cells were infected with RFP:WT or RFP:sipC−:Salmonella after 18 h of transfection with LAMP1-GFP and incubated at 37 °C for 2 h to allow transport of bacteria toward the late compartment. Thereafter, LAMP1 was synchronized in the Golgi by appropriate temperature block. Confocal images showed that neither WT:SCP nor sipC−:SCP possesses the LAMP1-GFP at this stage (Fig. 7, upper panel). Finally, cells were incubated at 37 °C for various periods of time to allow the budding of LAMP1-containing vesicles from the Golgi, and the presence of LAMP1 on respective SCP was determined. The results presented in Fig. 7 demonstrated that WT:SCP acquire LAMP1 by fusing with Golgi-derived vesicles, whereas a significantly smaller number of sipC−:SCP recruit LAMP1. Quantification of the confocal data suggested that about 80% of WT:SCP are positive for LAMP1, whereas less than 40% of sipC−:SCP are colocalized with LAMP1 after 20 min of LAMP1-containing vesicle budding from the Golgi. Fusion of SCP with Golgi-derived vesicles in macrophages was further confirmed by overexpression and synchronization of vesicular stomatitis virus glycoprotein (VSVG). Our results also showed that WT:SCP recruit exocytic transport vesicles containing VSVG, whereas sipC−:SCP fail to acquire VSVG (supplemental Fig. S7).
FIGURE 7.
Recruitment of LAMP1 on SCP by fusion with Golgi-derived LAMP1-containing vesicles. The image represents a single x/y plane of a Z-section, where green shows the LAMP1 and Salmonella appears in red. A ring-like staining around Salmonella indicates LAMP1-positive phagosomes as indicated by arrows. The graph below represents the percentage of SCP positive for LAMP1 where at least 100 phagosomes were analyzed for each time point. Data from three independent experiments was analyzed by t test, and levels of significance are indicated by p values. Inset shows the enlarged region.
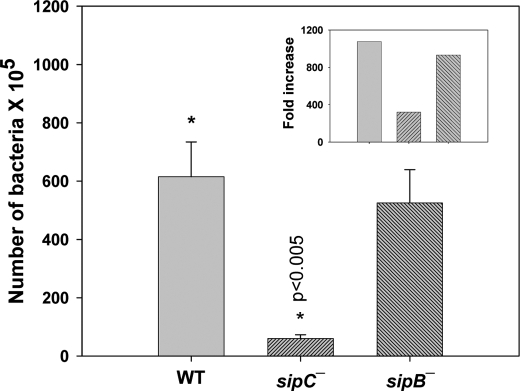
Comparison of Survival of WT and sipC−:Salmonella in Mice
To determine whether SipC-mediated recruitment of LAMP1 is important for the survival of Salmonella in vivo, mice were intraperitoneally infected with an equal number (1 × 103) of Salmonella (WT, sipB−, or sipC−) on day 0, and the bacterial load in the spleen was checked on day 4 (Fig. 8). We observed that about 6 × 107 bacteria were present in the spleen 4 days after infection with WT:Salmonella, whereas only 6 × 106 bacteria were found when mice were infected with sipC−:Salmonella. Interestingly, sipB−:Salmonella that efficiently recruits LAMP1 survived in mice almost like WT:Salmonella. To determine that the differences in the survival of various bacteria are not due to their rate of internalization in the target cells, we measured the splenic load of different bacteria on days 2 and 4 and compared it with the rate of intracellular multiplication. Our results showed about 1000-fold increase in the load of both WT and sipB−:Salmonella in mice between days 2 and 4, whereas only about 300-fold was observed with sipC−:Salmonella (Fig. 8, inset) demonstrating that survival of sipC−:Salmonella in mice is compromised.
FIGURE 8.
Survival of WT or sipC−:Salmonella in mice. Mice were infected with different strains of Salmonella, and bacterial load in infected spleen on day 4 (n = 5) was calculated by measuring colony-forming unit. Data were analyzed by t test, and levels of significance are indicated by p values (*). Inset shows the fold increase in the number of bacteria from days 2 to 4 post-infection.
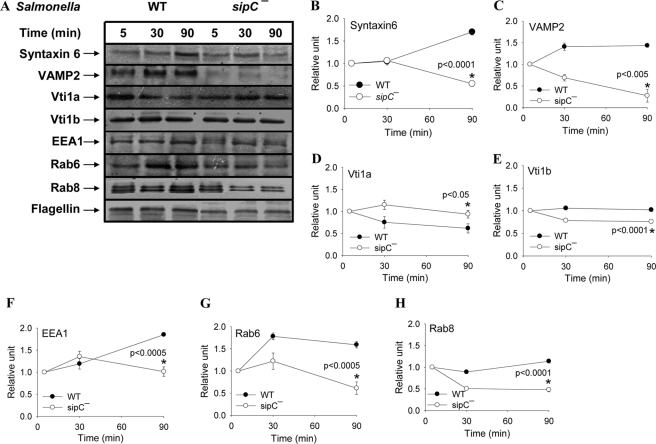
Mechanism of Recruitment of LAMP1 by SCP from Golgi
To understand the mechanism of SipC-mediated recruitment of LAMP1 from the Golgi by SCP, we analyzed the recruitment of Syntaxin6 and its interacting partners on these phagosomes. The results presented in Fig. 9 showed that WT:SCP progressively acquired higher amounts of Syntaxin6 and VAMP2 during its maturation inside macrophages, and the maximum amount of these proteins on the phagosomes are found at 90 min post-internalization. Although Vti1b levels remained unchanged during maturation of SCP, Vti1a levels peaked at the early time point of internalization and gradually declined. In contrast, sipC−:SCP showed reduced levels of Syntaxin6, VAMP2, and Vti1b and enhanced content of Vti1a particularly at later time points in their maturation. Interestingly, higher levels of EEA1 were also observed on WT:SCP than sipC−:SCP. We also compared the recruitment of two Golgi-associated Rabs like Rab6 and Rab8 on respective SCP and found (Fig. 9) that WT:SCP recruit significantly higher levels of both Rab6 and Rab8 in comparison with sipC−:SCP at a later time point of their maturation in macrophages. Moreover, complementation of sipC into sipC−:Salmonella (sipC−:psipC Salmonella) recruited similar levels of Syntaxin6, VAMP2, and Rab6 on their phagosomes as observed with WT:SCP (supplemental Fig. S8).
FIGURE 9.
Recruitment of different SNAREs and Rabs on SCP. A, Western blot showing the levels of various markers on respective SCP at indicated time points of their internalization in macrophages. Flagellin was used as a loading control. B–H, quantification of the data by densitometry. Results are represented as mean ± S.D. of three independent observations, normalized to the loading control. Level of each marker present on respective 5-min phagosomes is arbitrarily chosen as 1 unit. The level of significance was calculated by a t test, and the indicated p values are between the respective 90-min phagosomes (*).
DISCUSSION
S. typhimurium causes gastroenteritis in humans and systemic typhoid-like disease in mice (1). We and others have demonstrated that Salmonella initially enters into a Rab5-positive early compartment and subsequently dissociates from the endocytic pathway to avoid fusion with lysosomes (2, 13, 19). Similarly, Mycobacterium, Legionella, and Toxoplasma, etc., have also evolved diverse strategies to avoid fusion with lysosomes (20–24). However, they universally recruit LAMP1, the mechanism of which is not known.
Intracellular trafficking is regulated by Rabs and syntaxins (25–27). Previous studies have shown that intracellular pathogens either mimic or target these proteins to establish their niche in the host cells (20, 21, 24). Similarly, maturation of SCP also depends on the sequential removal and recruitment of different Rabs and syntaxins (2, 3, 15, 19, 28). In this study, we have determined the recruitment of different syntaxins on SCP at various stages of their maturation in macrophages and have shown that SCP specifically recruit higher amounts of Syntaxin6 after 90 min of internalization than at 5 min of internalization. Syntaxin6 regulates the post-Golgi membrane fusion events (29); thus, our results suggest that SCP might interact with Golgi or secretory vesicles originating from Golgi. However, the interaction of phagosomes with secretory pathway is not well characterized (8).
Salmonella use their effector molecules to modulate the trafficking pathway of host cells (20, 21, 30). Initial studies have shown that SPI-1-encoded genes are required for bacterial entry, although SPI-2 is necessary for intracellular survival. However, the boundaries between SPI-1 and SPI-2 have gradually diminished as several SPI-1 TTSS effectors are now found to contribute toward the intracellular survival of the bacteria (31–35). Thus, efforts were made to identify the effector molecule involved in the recruitment of Syntaxin6 on SCP in macrophages. Three lines of evidence have demonstrated that SipC, a SPI-1 effector of Salmonella, specifically interacts with host Syntaxin6. (i) Immobilized GST-Syntaxin6 specifically binds with a 40-kDa secretory protein from Salmonella, which is subsequently identified as SipC. (ii) Immunoprecipitation studies using immobilized SipC specifically recognize Syntaxin6 from the macrophage lysate. (iii) GST-Syntaxin6 specifically binds with purified His6-SipC. Topological analysis of SipC reveals that first 120 amino acids of the N terminus and the last 209 amino acids of the C terminus extend into the host cytoplasm (14). Therefore, we determined the binding of Syntaxin6 with the N- or C-terminal fragment of SipC. Our results have shown that Syntaxin6 specifically binds with the C terminus of SipC through its SNARE motif.
Previous studies have shown that SipC is required for bacterial invasion into epithelial cells (17). However, our results show that sipC−:Salmonella is also phagocytosed and enters into macrophages. This observation is supported by the fact that noninvasive Salmonella are also phagocytosed by macrophages (36). The higher efficiency of internalization of WT:Salmonella in comparison with mutant Salmonella could be attributed to a SipC-mediated bacterium-driven process besides phagocytosis. In addition, we have shown that WT:bacteria immediately after their internalization in macrophages induce actin rearrangements and membrane ruffling in close vicinity to the bacteria, whereas actin localization is more diffuse and distant from sipC−:SCP. These results are consistent with previous findings in HeLa cells that SipC modulates actin dynamics (37) and induces membrane ruffling (17). Therefore, the function of SipC in terms of actin rearrangements is well conserved in macrophages and epithelial cells.
Movement of intracellular pathogens toward the Golgi appears to be a generalized phenomenon (10–12), and inhibition of this process is detrimental to the pathogens. The movement of Salmonella toward the Golgi in epithelial cells has previously been shown to be dependent on the SPI-2 TTSS effectors like SseG, SseF, and SifA (38). However, our results using ssaR−:Salmonella (defective in SPI-2 TTSS effectors translocation) and invA−:Salmonella (defective in SPI-1 TTSS effectors translocation) have shown that both SPI-1 and SPI-2 TTSS effectors are required for targeting of Salmonella near the Golgi in macrophages. Interestingly, our results have shown that most of the WT:Salmonella translocate near the Golgi compartment after 120 min post-internalization in macrophages, whereas a significantly lower number of sipC−:Salmonella are targeted near the Golgi under similar conditions suggesting a possible role of SipC in this process. Among the different SPI-1 effectors, SipB, SipC, and SipD are collectively required to form the TTSS translocation pore, and the absence of any of these three proteins in Salmonella inhibits the translocation of other SPI-1 effectors (39). Therefore, targeting of sipD−:Salmonella near the Golgi and the inhibition of targeting of sipB− and sipC−:Salmonella toward the Golgi as observed in this investigation clearly indicates that among various SPI-1 TTSS effectors, only SipB and SipC are needed for Golgi targeting. These results are further confirmed by the fact that other SPI-1 TTSS effectors that deleted mutant Salmonella like sipA−:Salmonella, sopB−:Salmonella, and sopE−:Salmonella are targeted near the Golgi. Finally, complementation of sipC in sipC−:Salmonella restores the targeting of Salmonella near the Golgi confirming the role of SipC in this process. Inhibition of sipB−:Salmonella targeting toward the Golgi is possibly due to disruption of SipB-SipC interaction (40).
Subsequently, we have found that both WT:Salmonella and sipC−:Salmonella follow a similar endocytic route and are not targeted to the lysosomes. However, WT:SCP recruit LAMP1. Our results have shown that ssaR−:Salmonella recruit LAMP1, whereas invA−:Salmonella are unable to acquire significant amounts of LAMP1 demonstrating that only SPI-1 effectors are involved in the recruitment of LAMP1 on SCP in macrophages. Interestingly, we have found that sipC−:SCP progressively lose LAMP1 during their maturation in macrophages in comparison with WT:SCP. However, the presence of relatively small amounts of LAMP1 on sipC−:SCP is due to their fusion with early endosomes (15). These results suggest the possible role of SipC in the recruitment of LAMP1 on SCP. As SipC is part of the TTSS translocon complex along with SipB and SipD, sipC−:Salmonella might inhibit the translocation of other SPI-1 TTSS effectors, which may play a role in LAMP1 recruitment on SCP. However, recruitment of LAMP1 on sipB−:SCP and sipD−:SCP rules out the role of other SPI-1 effectors except for SipC in this process. To unambiguously determine the role of the C-terminal end of SipC in the recruitment of LAMP1, we have determined the recruitment of LAMP1 by sipCM398K:Salmonella and sipCR315Z:Salmonella in macrophages. Our results have shown that both sipCM398K:Salmonella and sipCR315Z:Salmonella are unable to bind significant amounts of Syntaxin6 in comparison with WT:SCP and thereby fail to recruit LAMP1 on their phagosomes. These results clearly demonstrate the specific role of SipC in the LAMP1 recruitment. This is supported by the fact that phagosomes containing sipA−, sopB−, or sopE−:Salmonella also recruit LAMP1. Recent studies have shown (41) that in epithelial cells LAMP1 recruitment on SCV is dependent on SopB and SNX3 interaction. SNX3 regulates trafficking in both the endocytic and secretory pathways (42–44), and depletion of SNX3 by siRNA only partially inhibits the recruitment of LAMP1 on SCV (41) suggesting that other host proteins could be involved in this process. Our results have shown that SipC is specifically required for LAMP1 recruitment, and SopB has no role in this process in macrophages. Depletion of Syntaxin6 in macrophages inhibits LAMP1 recruitment on WT:SCP, and Salmonella mutants deficient in the Syntaxin6 binding (sipCM398K:Salmonella) neither recruit Syntaxin6 nor acquire LAMP1 on SCP, collectively demonstrating that SipC-mediated recruitment of Syntaxin6 is required to acquire LAMP1 on SCP.
To unambiguously prove that WT:SCP recruit LAMP1 from the secretory pathway through a SipC-mediated process, we have overexpressed and synchronized LAMP1-GFP in the Golgi of RAW cells and finally determined the presence of LAMP1 on WT or sipC−:SCP after the budding of LAMP1-containing vesicles from the Golgi. Our results show that WT:SCP acquire LAMP1 by fusing with the Golgi-derived vesicles, whereas a significantly lower number of sipC−:SCP recruit LAMP1. The higher number of LAMP1-positive WT:SCP could be attributed to the high affinity fusion of LAMP1-containing vesicles budded from the TGN through the SipC-Syntaxin6 interaction. This is supported by the fact that depletion of endogenous Syntaxin6 led to a significant reduction in LAMP1 recruitment by WT:SCP. We have also used VSVG, a commonly used marker to follow protein transport from the Golgi to the plasma membrane. Consistent with previous observations in epithelial cells (36), our results have shown that WT:SCP recruit exocytic transport vesicles containing VSVG. In contrast, sipC−:SCP fail to acquire VSVG in macrophages. These results clearly demonstrate that SCP recruit LAMP1 by fusing with LAMP1-containing Golgi-derived vesicles. Subsequently, we have found that survival of sipC−:Salmonella in mice is significantly compromised in comparison with WT:Salmonella. In contrast, another SPI-1 effector translocation-defective mutant, sipB−:Salmonella, that efficiently recruits LAMP1 and successfully survives like WT:Salmonella in mice demonstrated the specificity of SipC in this process. Taken together, these results indicate that SipC-mediated recruitment of LAMP1 on SCP provides an appropriate niche for bacterial multiplication within phagosomes.
To address the mechanism of fusion of Golgi-derived vesicles with SCP, we have analyzed the recruitment of Syntaxin6 and its interacting partners on these phagosomes. Previous studies have shown that Syntaxin6, a Qc-SNARE, regulates multiple post-Golgi membrane trafficking events in the secretory pathway by interacting with appropriate R-SNAREs and Q-SNAREs (45). Among the different Syntaxin6 (45)-associated R-SNAREs, VAMP2 has been shown to regulate post-Golgi fusion events in the exocytic pathway (46). Similarly, Syntaxin6 also interacts with Qb-SNAREs like Vti1a and Vti1b (47, 48) in macrophages. It has been shown that Vti1a along with VAMP4 participates in the endosome fusion (47), whereas Vti1b is shown to regulate trafficking between the Golgi and plasma membrane (48). Therefore, we have compared the recruitment of VAMP2, Vti1a, and Vti1b between WT:SCP and sipC−:SCP. We have found that WT:SCP progressively acquire higher amounts of Syntaxin6 and VAMP2 during its maturation inside the macrophages, and the maximum amount of these proteins on these phagosomes are found at 90 min post-internalization. Although Vti1b levels remain constant, Vti1a levels peak early and gradually decline as the SCP mature. In contrast, sipC−:SCP show reduced levels of Syntaxin6, VAMP2, Vti1b, and enhanced content of Vti1a particularly at a later time point of their maturation. These findings demonstrate that SCP at later stages of their maturation recruit Syntaxin6 and VAMP2 and retain Vti1b to promote the fusion with LAMP1-containing Golgi-derived vesicles. This is supported by the fact that Syntaxin6 is required for the fusion of immature secretory vesicles originating from the TGN (49), and it also binds VAMP2 (50). Syntaxin6 also interacts with EEA1 and regulates transport of post-Golgi vesicles from the TGN to early endosomes in the secretory pathway (51). Therefore, higher levels of EEA1 present on WT:SCP than sipC−:SCP are possibly due to the presence of enhanced levels of Syntaxin6 on SCP. This result suggests that EEA1 possibly forms a pre-fusion complex to regulate the trafficking of LAMP1 from the Golgi to SCP. Specificity of membrane fusion is also regulated by Rab GTPases (25); therefore, we have compared the recruitment of two Golgi-associated Rabs like Rab6 and Rab8 by respective SCP. Previous studies have shown that Rab6 regulates traffic to and from the Golgi (52), whereas inactivation of Rab8 is found to disrupt the post-Golgi trafficking of lysosomal enzymes and VSVG proteins (53, 54). In concordance with these findings, our results show that WT:SCP recruit significantly higher levels of both Rab6 and Rab8 at a later time point of their maturation in macrophages. On the contrary, sipC−:SCP show relatively lower amounts of these Rabs. Recruitment of Rab6 on WT:SCP is possibly due to a higher amount of Syntaxin6 as it has been shown that Ypt6p (yeast homologue of Rab6) interacts with the Tlg1p (yeast homologue of Syntaxin6) to facilitate the post-Golgi membrane fusion (55). However, the mechanism of Rab8 recruitment by SCP needs to be addressed. In addition, sipC−:psipC SCP restores the recruitment of some of these proteins to WT:SCP levels confirming the role of SipC in the recruitment of these proteins.
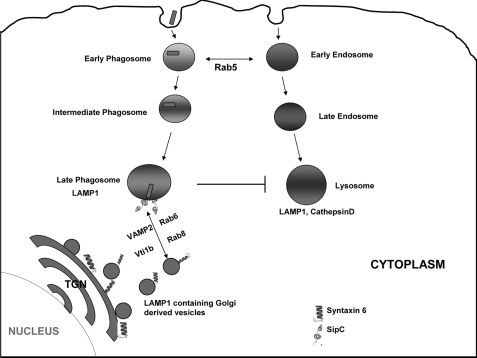
In correlation with our studies, recent reports suggest that SipC is linked to docking and fusion of exocytic vesicles (56), and a Chlamydia protein is required to obtain eukaryotic lipids from TGN by recruitment of Syntaxin6 (57). It is also suggested that some effector molecules of various bacteria mimic the structure of SNARE proteins and bind with cognate SNARE partners in host cells (58, 59). The C terminus of SipC has a large number of hydrophobic residues interspersed by 2–3 polar residues. Such sequence pattern is characteristic of coiled coil motifs that also resemble SNARE motifs. In silico structure prediction studies of SipC also suggest that its C-terminal stretch adopts a coiled coil helix (data not shown). Hence, interaction of SipC with the SNARE motif of Syntaxin6 is likely to involve interactions between coiled coil regions of both proteins through formation of helix bundles. Such interactions involving helix bundles are known to be stabilized by hydrophobic packing. Therefore, mutation of hydrophobic methionine at position 389 in the C-terminal region to positively charged lysine is likely to destabilize helix bundle interaction between coiled coil regions of SipC and Syntaxin6, and therefore sipCM398K:SCP are unable to recruit Syntaxin6. Hence, it is possible that the coiled coil region of SipC recruits Syntaxin6 on the phagosomal membrane by mimicking a cognate SNARE. The current model of vesicle fusion suggests that specific interaction of R-SNARE along with cognate Qa-, Qb-, and Qc-SNAREs mediates docking and fusion between donor and acceptor compartments (60). Thus, we speculate from our results that Syntaxin6 (Qc-SNARE)-positive LAMP1-containing Golgi-derived vesicles bind with SipC as a cognate SNARE partner present on SCP and recruits associated SNARE partners like Vti1b (Qb-SNARE) and VAMP2 (R-SNARE) to promote fusion between these vesicles to acquire LAMP1 on the phagosome (Fig. 10). Interestingly, a variety of effectors from different intracellular pathogens exhibit coiled coil motifs as observed in SipC and might mimic different host SNAREs. Therefore, it will be important to look for structural or functional homologues of SipC in other intracellular pathogens that might be involved in recruiting cognate SNAREs to acquire important molecules from the secretory pathway of the host cells for their survival.
FIGURE 10.
Model of SipC-mediated recruitment of LAMP1 on SCP. In macrophages, S. typhimurium first enter into Rab5-positive early phagosomes and then divert from the endocytic pathway and move toward the Golgi. SipC present on the SCP interacts with host Syntaxin6 to facilitate fusion with LAMP1-containing vesicles originating from Golgi.
This is the first demonstration of how an intracellular pathogen like Salmonella recruits LAMP1 from Golgi-derived vesicles through its effector protein without fusing with lysosomes. This study also suggests that similar strategies might be used by other intracellular pathogens to acquire LAMP1 on their phagosomes to maintain the structural integrity of the phagosomal membrane so that the phagosome provides an appropriate niche for their replication.
Supplementary Material
Acknowledgments
We are grateful to Professor Sandip K. Basu (National Institute of Immunology, New Delhi, India) for critically reviewing the manuscript.
This work was supported in part by a grant from National Institute of Immunology, Government of India, New Delhi, India.

This article contains supplemental Figs. S1–S8 and additional references.
- TGN
- trans-Golgi network
- SCP
- Salmonella-containing phagosome
- VSVG
- vesicular stomatitis virus glycoprotein
- RFP
- red fluorescent protein
- TTSS
- type III secretion system.
REFERENCES
- 1. Haraga A., Ohlson M. B., Miller S. I. (2008) Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6, 53–66 [DOI] [PubMed] [Google Scholar]
- 2. Steele-Mortimer O. (2008) The Salmonella-containing vacuole. Moving with the times. Curr. Opin. Microbiol. 11, 38–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. McGhie E. J., Brawn L. C., Hume P. J., Humphreys D., Koronakis V. (2009) Salmonella takes control. Effector-driven manipulation of the host. Curr. Opin. Microbiol. 12, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Russell D. G. (2001) Mycobacterium tuberculosis. Here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2, 569–577 [DOI] [PubMed] [Google Scholar]
- 5. Eskelinen E. L., Tanaka Y., Saftig P. (2003) At the acidic edge. Emerging functions for lysosomal membrane proteins. Trends Cell Biol. 13, 137–145 [DOI] [PubMed] [Google Scholar]
- 6. Huynh K. K., Eskelinen E. L., Scott C. C., Malevanets A., Saftig P., Grinstein S. (2007) LAMP proteins are required for fusion of lysosomes with phagosomes. EMBO J. 26, 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cook N. R., Row P. E., Davidson H. W. (2004) Lysosome-associated membrane protein 1 (Lamp1) traffics directly from the TGN to early endosomes. Traffic 5, 685–699 [DOI] [PubMed] [Google Scholar]
- 8. Salcedo S. P., Holden D. W. (2005) Bacterial interactions with the eukaryotic secretory pathway. Curr. Opin. Microbiol. 8, 92–98 [DOI] [PubMed] [Google Scholar]
- 9. Salcedo S. P., Holden D. W. (2003) SseG, a virulence protein that targets Salmonella to the Golgi network. EMBO J. 22, 5003–5014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kagan J. C., Roy C. R. (2002) Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat. Cell Biol. 4, 945–954 [DOI] [PubMed] [Google Scholar]
- 11. Hackstadt T., Scidmore M. A., Rockey D. D. (1995) Lipid metabolism in Chlamydia trachomatis-infected cells. Directed trafficking of Golgi-derived sphingolipids to the chlamydial inclusion. Proc. Natl. Acad. Sci. U.S.A. 92, 4877–4881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Celli J., de Chastellier C., Franchini D. M., Pizarro-Cerda J., Moreno E., Gorvel J. P. (2003) Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198, 545–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Parashuraman S., Madan R., Mukhopadhyay A. (2010) NSF-independent fusion of Salmonella-containing late phagosomes with early endosomes. FEBS Lett. 584, 1251–1256 [DOI] [PubMed] [Google Scholar]
- 14. Ly K. T., Casanova J. E. (2007) Mechanisms of Salmonella entry into host cells. Cell. Microbiol. 9, 2103–2111 [DOI] [PubMed] [Google Scholar]
- 15. Mukherjee K., Siddiqi S. A., Hashim S., Raje M., Basu S. K., Mukhopadhyay A. (2000) Live Salmonella recruits N-ethylmaleimide-sensitive fusion protein on phagosomal membrane and promotes fusion with early endosome. J. Cell Biol. 148, 741–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Akasaki K., Michihara A., Fujiwara Y., Mibuka K., Tsuji H. (1996) Biosynthetic transport of a major lysosome-associated membrane glycoprotein 2, lamp-2. A significant fraction of newly synthesized lamp-2 is delivered to lysosomes by way of early endosomes. J. Biochem. 120, 1088–1094 [DOI] [PubMed] [Google Scholar]
- 17. Chang J., Chen J., Zhou D. (2005) Delineation and characterization of the actin nucleation and effector translocation activities of Salmonella SipC. Mol. Microbiol. 55, 1379–1389 [DOI] [PubMed] [Google Scholar]
- 18. Scherer C. A., Cooper E., Miller S. I. (2000) The Salmonella type III secretion translocon protein SspC is inserted into the epithelial cell plasma membrane upon infection. Mol. Microbiol. 37, 1133–1145 [DOI] [PubMed] [Google Scholar]
- 19. Hashim S., Mukherjee K., Raje M., Basu S. K., Mukhopadhyay A. (2000) Live Salmonella modulate expression of Rab proteins to persist in a specialized compartment and escape transport to lysosomes. J. Biol. Chem. 275, 16281–16288 [DOI] [PubMed] [Google Scholar]
- 20. Weber S. S., Ragaz C., Hilbi H. (2009) Pathogen trafficking pathways and host phosphoinositide metabolism. Mol. Microbiol. 71, 1341–1352 [DOI] [PubMed] [Google Scholar]
- 21. Brumell J. H., Scidmore M. A. (2007) Manipulation of rab GTPase function by intracellular bacterial pathogens. Microbiol. Mol. Biol. Rev. 71, 636–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dowse T., Soldati D. (2004) Host cell invasion by the apicomplexans. The significance of microneme protein proteolysis. Curr. Opin. Microbiol. 7, 388–396 [DOI] [PubMed] [Google Scholar]
- 23. Rohde K., Yates R. M., Purdy G. E., Russell D. G. (2007) Mycobacterium tuberculosis and the environment within the phagosome. Immunol. Rev. 219, 37–54 [DOI] [PubMed] [Google Scholar]
- 24. Hubber A., Roy C. R. (2010) Modulation of host cell function by Legionella pneumophila type IV effectors. Annu. Rev. Cell Dev. Biol. 26, 261–283 [DOI] [PubMed] [Google Scholar]
- 25. Stenmark H. (2009) Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 10, 513–525 [DOI] [PubMed] [Google Scholar]
- 26. Wickner W., Schekman R. (2008) Membrane fusion. Nat. Struct. Mol. Biol. 15, 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zerial M., McBride H. (2001) Rab proteins as membrane organizers. Nat. Rev. Mol. Cell Biol. 2, 107–117 [DOI] [PubMed] [Google Scholar]
- 28. Smith A. C., Cirulis J. T., Casanova J. E., Scidmore M. A., Brumell J. H. (2005) Interaction of the Salmonella-containing vacuole with the endocytic recycling system. J. Biol. Chem. 280, 24634–24641 [DOI] [PubMed] [Google Scholar]
- 29. Bock J. B., Klumperman J., Davanger S., Scheller R. H. (1997) Syntaxin6 functions in trans-Golgi network vesicle trafficking. Mol. Biol. Cell 8, 1261–1271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Drecktrah D., Knodler L. A., Howe D., Steele-Mortimer O. (2007) Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8, 212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bakowski M. A., Braun V., Lam G. Y., Yeung T., Heo W. D., Meyer T., Finlay B. B., Grinstein S., Brumell J. H. (2010) The phosphoinositide phosphatase SopB manipulates membrane surface charge and trafficking of the Salmonella-containing vacuole. Cell Host Microbe 7, 453–462 [DOI] [PubMed] [Google Scholar]
- 32. Brawn L. C., Hayward R. D., Koronakis V. (2007) Salmonella SPI1 effector SipA persists after entry and cooperates with a SPI2 effector to regulate phagosome maturation and intracellular replication. Cell Host Microbe 1, 63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Giacomodonato M. N., Uzzau S., Bacciu D., Caccuri R., Sarnacki S. H., Rubino S., Cerquetti M. C. (2007) SipA, SopA, SopB, SopD, and SopE2 effector proteins of Salmonella enterica serovar Typhimurium are synthesized at late stages of infection in mice. Microbiology 153, 1221–1228 [DOI] [PubMed] [Google Scholar]
- 34. Dukes J. D., Lee H., Hagen R., Reaves B. J., Layton A. N., Galyov E. E., Whitley P. (2006) The secreted Salmonella dublin phosphoinositide phosphatase, SopB, localizes to PtdIns(3)P-containing endosomes and perturbs normal endosome to lysosome trafficking. Biochem. J. 395, 239–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Humphreys D., Hume P. J., Koronakis V. (2009) The Salmonella effector SptP dephosphorylates host AAA+ ATPase VCP to promote development of its intracellular replicative niche. Cell Host Microbe 5, 225–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kuhle V., Abrahams G. L., Hensel M. (2006) Intracellular Salmonella enterica redirect exocytic transport processes in a Salmonella pathogenicity island 2-dependent manner. Traffic 7, 716–730 [DOI] [PubMed] [Google Scholar]
- 37. Zhou D., Mooseker M. S., Galán J. E. (1999) Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283, 2092–2095 [DOI] [PubMed] [Google Scholar]
- 38. Ramsden A. E., Holden D. W., Mota L. J. (2007) Membrane dynamics and spatial distribution of Salmonella-containing vacuoles. Trends Microbiol. 15, 516–524 [DOI] [PubMed] [Google Scholar]
- 39. Cornelis G. R. (2006) The type III secretion injectisome. Nat. Rev. Microbiol. 4, 811–825 [DOI] [PubMed] [Google Scholar]
- 40. Hayward R. D., Koronakis V. (1999) Direct nucleation and bundling of actin by the SipC protein of invasive Salmonella. EMBO J. 18, 4926–4934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Braun V., Wong A., Landekic M., Hong W. J., Grinstein S., Brumell J. H. (2010) Sorting nexin 3 (SNX3) is a component of a tubular endosomal network induced by Salmonella and involved in maturation of the Salmonella-containing vacuole. Cell. Microbiol. 12, 1352–1367 [DOI] [PubMed] [Google Scholar]
- 42. Carlton J., Bujny M., Rutherford A., Cullen P. (2005) Sorting nexins. Unifying trends and new perspectives. Traffic 6, 75–82 [DOI] [PubMed] [Google Scholar]
- 43. Voos W., Stevens T. H. (1998) Retrieval of resident late-Golgi membrane proteins from the prevacuolar compartment of Saccharomyces cerevisiae is dependent on the function of Grd19p. J. Cell Biol. 140, 577–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xu Y., Hortsman H., Seet L., Wong S. H., Hong W. (2001) SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat. Cell Biol. 3, 658–666 [DOI] [PubMed] [Google Scholar]
- 45. Wendler F., Tooze S. (2001) Syntaxin6. The promiscuous behavior of a SNARE protein. Traffic 2, 606–611 [DOI] [PubMed] [Google Scholar]
- 46. Lin R. C., Scheller R. H. (2000) Mechanisms of synaptic vesicle exocytosis. Annu. Rev. Cell Dev. Biol. 16, 19–49 [DOI] [PubMed] [Google Scholar]
- 47. Kreykenbohm V., Wenzel D., Antonin W., Atlachkine V., von Mollard G. F. (2002) The SNAREs vti1a and vti1b have distinct localization and SNARE complex partners. Eur. J. Cell Biol. 81, 273–280 [DOI] [PubMed] [Google Scholar]
- 48. Murray R. Z., Wylie F. G., Khromykh T., Hume D. A., Stow J. L. (2005) Syntaxin6 and Vti1b form a novel SNARE complex, which is up-regulated in activated macrophages to facilitate exocytosis of tumor necrosis factor-α. J. Biol. Chem. 280, 10478–10483 [DOI] [PubMed] [Google Scholar]
- 49. Wendler F., Page L., Urbé S., Tooze S. A. (2001) Homotypic fusion of immature secretory granules during maturation requires syntaxin 6. Mol. Biol. Cell 12, 1699–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bethani I., Lang T., Geumann U., Sieber J. J., Jahn R., Rizzoli S. O. (2007) The specificity of SNARE pairing in biological membranes is mediated by both proofreading and spatial segregation. EMBO J. 26, 3981–3992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Simonsen A., Gaullier J. M., D'Arrigo A., Stenmark H. (1999) The Rab5 effector EEA1 interacts directly with syntaxin-6. J. Biol. Chem. 274, 28857–28860 [DOI] [PubMed] [Google Scholar]
- 52. Mallard F., Tang B. L., Galli T., Tenza D., Saint-Pol A., Yue X., Antony C., Hong W., Goud B., Johannes L. (2002) Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J. Cell Biol. 156, 653–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. del Toro D., Alberch J., Lázaro-Diéguez F., Martín-Ibáñez R., Xifró X., Egea G., Canals J. M. (2009) Mutant huntingtin impairs post-Golgi trafficking to lysosomes by delocalizing optineurin/Rab8 complex from the Golgi apparatus. Mol. Biol. Cell 20, 1478–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Huber L. A., Pimplikar S., Parton R. G., Virta H., Zerial M., Simons K. (1993) Rab8, a small GTPase involved in vesicular traffic between the TGN and the basolateral plasma membrane. J. Cell Biol. 123, 35–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Liewen H., Meinhold-Heerlein I., Oliveira V., Schwarzenbacher R., Luo G., Wadle A., Jung M., Pfreundschuh M., Stenner-Liewen F. (2005) Characterization of the human GARP (Golgi-associated retrograde protein) complex. Exp. Cell Res. 306, 24–34 [DOI] [PubMed] [Google Scholar]
- 56. Nichols C. D., Casanova J. E. (2010) Salmonella-directed recruitment of new membrane to invasion foci via the host exocyst complex. Curr. Biol. 20, 1316–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Moore E. R., Mead D. J., Dooley C. A., Sager J., Hackstadt T. (2011) The trans-Golgi SNARE syntaxin 6 is recruited to the chlamydial inclusion membrane. Microbiology 157, 830–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Delevoye C., Nilges M., Dehoux P., Paumet F., Perrinet S., Dautry-Varsat A., Subtil A. (2008) SNARE protein mimicry by an intracellular bacterium. PLoS Pathog. 4, e1000022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Delahay R. M., Frankel G. (2002) Coiled-coil proteins associated with type III secretion systems. A versatile domain revisited. Mol. Microbiol. 45, 905–916 [DOI] [PubMed] [Google Scholar]
- 60. Jahn R., Scheller R. H. (2006) SNAREs. Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.