Background: The mechanistic role of MTA1 in tumor aggressiveness is yet to be deciphered.
Results: RNF144A is a direct target of transcriptional repression by MTA1 and inhibits migration and invasion.
Conclusion: Transcriptional repression of RNF144A by MTA1 confers a migratory and invasive phenotype of cancer cells.
Significance: This study provides novel mechanistic insights into regulation of tumor progression by MTA1.
Keywords: Invasion, Migration, Transcription Regulation, Tumor Cell Biology, Tumor Metastases, MTA1, RING Finger Protein
Abstract
Metastasis-associated protein 1 (MTA1), a component of the nucleosome-remodeling and histone deacetylase complex, is widely up-regulated in human cancers and significantly correlated with tumor invasion and metastasis, but the mechanisms involved remain largely unknown. Here, we report that MTA1 transcriptionally represses the expression of RING finger protein 144A (RNF144A), an uncharacterized gene whose protein product possesses potential E3 ubiquitin ligase activity, by recruiting the histone deacetylase 2 (HDAC2) and CCAAT/enhancer-binding protein α (c/EBPα) co-repressor complex onto human RNF144A promoter. Furthermore, an inverse correlation between the expression levels of MTA1 and RNF144A was demonstrated in publicly available breast cancer microarray datasets and the MCF10 breast cancer progression model system. To address functional aspects of MTA1 regulation of RNF144A, we demonstrate that RNF144A is a novel suppressor of cancer migration and invasion, two requisite steps of metastasis in vivo, and knockdown of endogenous RNF144A by small interfering RNAs accelerates the migration and invasion of MTA1-overexpressing cells. These results suggest that RNF144A is partially responsible for MTA1-mediated migration and invasion and that MTA1 overexpression in highly metastatic cancer cells drives cell migration and invasion by, at least in part, interfering with the suppressive function of RNF144A through transcriptional repression of RNF144A expression. Together, these findings provide novel mechanistic insights into regulation of tumor progression and metastasis by MTA1 and highlight a previously unrecognized role of RNF144A in MTA1-driven cancer cell migration and invasion.
Introduction
Although advances have been made in tumor diagnosis and therapies, over 90% of cancer-associated mortality is attributable to tumor metastasis (1, 2). One of the critical steps during tumor metastasis is tumor cell migration and invasion, which are responsible for the entry of tumor cells into lymphatic and blood vessels as well as the extravasation of tumor cells into the secondary organs (2, 3). The identification of molecular pathways that contribute to tumor cell motility and invasion is therefore essential for understanding the mechanism of metastasis and for providing new therapeutic targets to treat metastatic cancer (4).
Accumulating evidence over the past two decades has identified the metastasis-associated protein 1 (MTA1)4 as one of the most important players in the multistep invasion-metastasis cascade (5–7). Originally, MTA1 was isolated by a differential cDNA library screening using a rat mammary adenocarcinoma metastatic model as an up-regulated gene in highly metastatic cells (7). Subsequent studies demonstrate that MTA1 is up-regulated in a variety of human cancers, and its expression is closely correlated with tumor invasion and metastasis as well as poor patient survival (5, 8, 9). In this context, it has been shown that overexpression of MTA1 promotes tumor motility and invasiveness (10–12), whereas silencing of MTA1 expression significantly suppresses tumor migration and invasion in various cancer models (13–15). Yet, the molecular details of how MTA1 drives tumor migration and invasion, important steps of metastatic cascades, are still largely unknown.
Recently, in an attempt to uncover novel gene targets and functions of MTA1, we carried out high-throughput microarray-based expression profiling analyses of mouse embryonic fibroblasts (MEFs) under the condition of depletion or overexpression of MTA1 (16) and discovered that MTA1 modulates the expression of RING finger protein 144A (RNF144A) (also known as KIAA0161, RNF144, or ubiquitin-conjugating enzyme 7-interacting protein 4) in a p53-independent manner (16, 17). RNF144A is an uncharacterized gene whose encoding protein contains a RING finger motif that is known to be involved in protein-protein interaction as well as protein ubiquitination and degradation (18). As many RING finger proteins, such as breast cancer type 1 susceptibility protein (BRCA1; also known as RING finger protein 53), are implicated in cancer development and progression (19), in the present study we attempted to detail the regulatory mechanism of RNF144A by MTA1 and to investigate the functional implication of the MTA1-RNF144A pathway in cancer progression.
Here, we provide convincing evidence that MTA1 transcriptionally represses RNF144A expression by recruiting the histone deacetylase 2 (HDAC2) and CCAAT/enhancer-binding protein α (c/EBPα) co-repressor complex onto human RNF144A promoter. Moreover, we demonstrate that RNF144A is a novel suppressor of cancer migration and invasion. Consequently, MTA1 overexpression in highly metastatic tumor cells enhances the ability of cancer cells to migrate and invade by, at least in part, interfering with RNF144A function through transcriptional repression of RNF144A. Thus, the dysregulated MTA1-RNF144A pathway is intimately linked to the migratory and invasive phenotype of cancer cells and may represent a promising target for cancer therapy.
EXPERIMENTAL PROCEDURES
Cell Lines and Cell Culture
Human MCF-7 breast adenocarcinoma cells and human HeLa epithelial cervical carcinoma cells were obtained from American Type Culture Collection (Manassas, VA). Wild-type (MTA1+/+) and MTA1-knock-out (MTA1−/−) MEFs were generated in our laboratory from embryos at day 9 of development using a standard protocol (20) and were maintained in Dulbecco's Modified Eagle's Medium/Nutrient Ham's Mixture F-12 (DMEM/F-12) (Mediatech, Manassas, VA) supplemented with 10% fetal bovine serum (Atlanta Biologicals, Lawrenceville, GA) and 1× antibiotic-antimycotic solution. The immortalized normal breast epithelial MCF10A, pre-malignant MCF10AT, comedo ductal carcinoma in situ MCF10DCIS, metastatic MCF10CA1D cell lines have been described previously (21–25) and were cultured in DMEM/F-12 medium supplemented with 5% horse serum, 10 ng/ml epidermal growth factor, 500 ng/ml hydrocortisone, 100 ng/ml cholera toxins, and 10 μg/ml insulin. Stable clones of MCF-7 cells expressing pcDNA3.1 empty vector (MCF-7/pcDNA) and T7-MTA1 (MCF-7/T7-MTA1) were established in our laboratory (26) and maintained in DMEM/F-12 medium containing 200 μg/ml Geneticin (G418). All of the cell lines used were incubated in a humidified 5% CO2 chamber at 37 °C, and all cell culture reagents were purchased from Invitrogen (Carlsbad, CA) unless specifically stated otherwise.
Antibodies, Western Blotting Analysis, and Immunoprecipitation
Rabbit polyclonal anti-MTA1 (BL1805) and anti-T7 antibodies were purchased from Bethyl Laboratories (Montgomery, TX). Rabbit polyclonal anti-HDAC2 (H-54) antibody was purchased from Santa Cruz Biotechnology (Santa Cruz Biotechnology). Rabbit monoclonal anti-c/EBPα [EP709Y] and rabbit polyclonal anti-RNF144A antibodies were obtained from Abcam (Cambridge, MA). Mouse monoclonal anti-β-actin (AC40) and anti-vinculin (hVIN-1) were obtained from Sigma-Aldrich. Horseradish peroxidase (HRP)-conjugated secondary antibodies and enhanced chemiluminescence (ECL) reagents were purchased from Amersham Biosciences (Piscataway, NJ). All of the primary antibody dilutions were used according to manufacturer's instructions, and all reagents were obtained from Sigma-Aldrich unless otherwise stated.
Protein extracts were prepared by lysing the cells with RIPA (radio-immunoprecipitation assay) buffer containing 50 mm Tris-HCl, pH 7.4, 1% Nonidet P-40, 0.25% sodium deoxycholate, 150 mm NaCl, 1 mm EDTA, 1×protease inhibitor mixture (Roche Applied Science, Indianapolis, IN), and 1× phosphatase inhibitor mixture I and II (Sigma-Aldrich), and protein concentrations were determined using Bio-Rad DC Protein Assay reagents (Bio-Rad). Cell extracts were then resolved by SDS-PAGE, transferred to nitrocellulose membranes, and incubated with the indicated antibodies. Corresponding antibody specific signal detections were performed using the ECL reagents and protein band densities were quantified using NIH image processing and analysis software ImageJ following manufacturer's instructions. For quantification of Western blot signal, a set area around the band of interest was selected, the average pixel intensity was measured, and the results were normalized to the signal for loading controls.
For immunoprecipitation (IP) analysis of the interaction of MTA1 with c/EBPα, nuclear extracts were prepared from MCF-7 cells using high salt extraction method as described previously (27) with some modifications. Briefly, cells were washed twice with ice-cold PBS containing 1 mm phenylmethylsulfonyl fluoride (PMSF) and then scraped into PBS. Cells were harvested by centrifugation at 2,000 rpm for 10 min. Cell pellet was re-suspended in one packed cell pellet volume of buffer A (20 mm HEPES, pH7.9, 1.5 mm MgCl2, 10 mm KCl, 1 mm DTT, 300 mm sucrose, 1×protease inhibitor mixture, and 1× phosphatase inhibitor mixture I and II) and incubated on ice for 5 min. Equal volume of buffer B (buffer A supplemented with Nonidet P-40 at a final concentration of 0.25%) was added dropwise to the cell suspension and incubated on ice for 5–10 min. Nuclei were pelleted by centrifugation at 2,200 rpm at 4 °C for 10 min, and then incubated with high salt extraction buffer C (20 mm HEPES, pH7.9, 1.5 mm MgCl2, 420 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, 25% glycerol, 1× protease inhibitor mixture, and 1× phosphatase inhibitor mixture I and II) on ice for 1 h with intermittent tapping for efficient lysis. Nuclear lysates were harvested by centrifugation at 14,000 rpm for 20 min at 4 °C and diluted with buffer D (buffer C without KCl) to dilute KCl concentration to final working concentration of 150 mm. The diluted fraction was incubated with 1 μg of primary antibody and corresponding control IgG overnight at 4 °C on a rocker platform, followed by incubation with total 30 μl of Trueblot IP beads (eBioscience, San Diego, CA) for 2 h at 4 °C. The immunoprecipitates were collected by centrifugation in a microcentrifuge at 4,000 rpm for 5 min, and then the supernatant was discarded, whereupon the pellet was washed with buffer E (20 mm HEPES, pH7.9, 1.5 mm MgCl2, 150 mm KCl, 0.2 mm EDTA, 0.5 mm DTT, 25% glycerol, 1×protease inhibitor mixture, and 1× phosphatase inhibitor mixture I and II) for three to five times and then dissolved in a sample buffer for SDS-PAGE. To reduce interference by the ∼55 kDa heavy and ∼23 kDa light chains of the immunoprecipitating antibody, HRP-conjugated TrueBlot secondary antibodies (eBioscience) were used in the IP/immunoblotting applications.
Quantitative Real-time PCR (qRT-PCR)
Total RNA was isolated from cultured cells using TRIzol reagent (Invitrogen) according to the manufacturer's protocol, and 2 μg of extracted RNA was converted to cDNA using the SuperScriptTM III First-Strand Synthesis System for RT-PCR (Invitrogen). The resultant cDNA was subjected to qRT-PCR by using the iQTM SYBR® Green Supermix (Bio-Rad) on a CFX96™ Real-Time PCR Detection System (Bio-Rad). The values for specific genes were normalized to β-actin as a housekeeping control gene. Mean values are displayed as ± standard deviations. All of qRT-PCR primers were synthesized by Sigma-Aldrich, and the sequences of the primers are available in the supplemental Table S1.
siRNA and Transfections
Specific siRNAs targeting human MTA1 and RNF144A were purchased from Dharmacon (Lafayette, CO), and the specificity of these MTA1 siRNAs has been verified previously (28). The transfection of siRNA was performed twice at 24-h intervals with OligofectamineTM reagent (Invitrogen) according to the manufacturer's protocol. Cells were subjected to further analyses after 48 h of second round of transfection.
Cloning of Human RNF144A Promoter Construct, Luciferase, and β-Galactosidase Assays
Human RNF144A promoter construct (−2130 to −1613) was amplified from human genomic DNA using primers CTAGCCCGGGCTCGAGTCA CTCAGTATTTATTGAGCACTT (forward) and CCGGAATGCCAAGCTTTCTCTGAGCT GGTGTAATACTTAG (reverse), and the resulting fragment was cloned into the pGL3 luciferase reporter vector (Promega) as XhoI and HindIII insert using In-Fusion Dry-Down PCR Cloning kit (Clontech, Mountain View, CA). The sequence of amplified fragment was confirmed by DNA sequencing.
Plasmid DNA was purified using Qiagen Plasmid Midi Kit (Qiagen, Valencia, CA), and transfections were carried out using FuGENE HD Transfection Reagent (Roche Applied Science, Indianapolis, IN) according to manufacturer's instructions. Briefly, cells were seeded in six-well plates 24 h prior to transfection, and transfected with the indicated expression vectors. The mammalian reporter pCMVβ vector expressing extremely high levels of β-galactosidase from the human cytomegalovirus immediate early promoter (Clontech, Mountain View, CA) was used as an internal control for transfection efficiency. The pGL3 basic vector (Promega) was used as a negative control. Cells were harvested after 48 h of transfection, and luciferase as well as β-galactosidase activities were determined using the Luciferase Assay System (Promega) on a luminometer according to the manufacturer's instructions. Luciferase activity was normalized for β-galactosidase activity in cell lysate and expressed as average of three independent experiments. Mean values are displayed as ± S.D., and statistical analysis was performed using a Student's t test to calculate the significance of the difference between different experimental conditions.
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared using a Nonidet P-40 lysis method (27) as described above and 2 μg of nuclear extracts was used for each reaction. EMSA for RNF144A DNA binding was performed using the annealed and [γ-32P]ATP end-labeled oligonucleotides or PCR product in a 20 μl reaction mixture for 15 min at 20 °C. Samples were run on a nondenaturing 5% polyacrylamide gel and imaged by autoradiography. Specific competitions were performed by adding a 100-fold excess of cold probe to the incubation mixture, and supershift EMSAs were performed by adding 100 ng of either c/EBPα or MTA1 antibodies.
Chromatin Immunoprecipitation (ChIP) Assay
The ChIP assays were performed as described previously (26). Briefly, ∼106 cells were treated with 1% formaldehyde (final concentration) for 10 min at 37 °C to cross-link histones to DNA and then washed twice with phosphate-buffered saline containing protease inhibitors mixture. Cells were lysed by sonication on ice and centrifuged at 14,000 rpm for 15 min at 4 °C. The supernatant was immunoprecipitated with specific antibodies (1 μg of primary antibody per 1 mg of protein extracts) and corresponding IgG controls. After the immunoprecipitates were washed twice, the DNA was eluted off the beads, and purified DNA (phenol-chloroform extraction) was subjected to PCR analysis. PCR primers for ChIP assays are provided in supplemental Table S2.
Wound-healing Assay and Invasion Assay
For wound-healing assay, MCF-7/pcDNA and MCF-7/T7-MTA1 stable clone cells (26) were seeded in 6-well plates for 24 h and then transfected with control siRNAs or specific siRNAs targeting human RNF144A as described above. After 24 h of transfection, each plate was received multiple “wounds” with a 200 μl pipette tip. 0 h measurements were acquired to calculate the width of the wound. After additional 24 h, each plate was examined by phase contrast microscopy for the amount of wound closure by measuring the physical separation remaining between the original wound widths using the Olympus DP2-BSW digital camera software (Olympus, Center Valley, PA). Ten separate measurements were made per plate, and each experiment was performed in triplicates.
For invasion assay, after 24 h of siRNA transfection, cells were trypsinized, washed with PBS, and resuspended in DMEM medium containing 0.1% bovine serum albumin. Then, cells were loaded onto the upper well of an matrigel coated Boyden chamber (BD Biosciences) at a concentration of 1 × 105 cells per well. The lower side of the separating filter was filled with conditioned medium prepared from NIH3T3 cells which serves as a source of chemoattractant. Cells were stained with DAPI, and imaged using Olympus IX71 inverted microscope with DP2-BSW application software (Olympus Imaging America Inc., Center Valley, PA). Cell numbers for invasion were then determined by counting the number of cells present in 15 microscope fields at 20× magnification per insert.
Data Mining
Two publicly available microarray datasets containing primary breast tumor samples GSE22093 (29) and GSE3494 (30) were used for analyzing the correlation between MTA1 and RNF144A expression levels. They can be accessed from Gene Expression Omnibus (GEO). The probes used to detect the transcript levels of MTA1 were 202247_s_at and 211783_s_at and that used to detect RNF144A levels was 204040_at. The intensity values for each of the probes were summarized and normalized with GeneSpring GX 11.0 using RMA algorithm. The log transformed values for each probe were then averaged and the mean was used to analyze the correlation between MTA1 and RNF144A in the chosen dataset. Based on the mean expression levels of MTA1, patients were divided into three groups. The patients with mean value of MTA1 higher than mean ± S.D. were defined as MTA1 high expression group, which lower than mean ± S.D. were defined as MTA1 low expression group, others patients were defined as medium expression group. Student's t test was used to compare the RNF144A transcript levels in patients expressing higher and lower MTA1 levels. A p value of less than 0.05 was considered as significant in all the analyses performed.
RESULTS
Bioinformatic Analysis of the Correlation of the Transcript Levels between MTA1 and RNF144A in Multiple Published Datasets
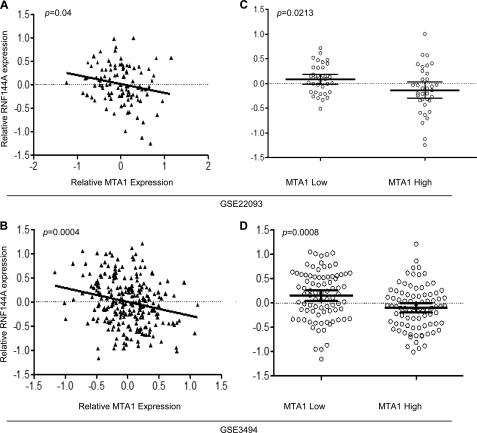
We have shown previously that MTA1 has an obligatory role in regulating RNF144A expression in a p53-independent manner using microarray-based expression profiling analysis (16). To further validate the significance of these findings, we next analyzed correlation of the transcription levels of MTA1 and RNF144A in two publicly available microarray datasets containing primary breast tumor samples (GSE22093 and GSE3494), and found a significant negative correlation between transcript levels of MTA1 and RNF144A (Fig. 1, A and B). Student's t test analysis showed a significant difference between the means of RNF144A transcript levels in samples expressing lower and higher levels of MTA1 mRNA levels (Fig. 1, C and D). The mean RNF144A expression in samples containing low MTA1 level was found to be higher than mean RNF144A mRNA level in samples with high MTA1 levels (Fig. 1, C and D). These results demonstrate an inverse correlation of the transcript levels between MTA1 and RNF144A in multiple microarray datasets.
FIGURE 1.
A and B, correlation between transcript levels of MTA1 and RNF144A in breast cancer dataset sGSE22093 (n = 103, Pearson's r = −0.20, p = 0.04) (A) and GSE3494 (n = 251, Pearson's r = −0.22, p = 0.0004) (B). C and D, correlation between transcript levels of RNF144A levels in patients with high and low MTA1 levels in breast cancer datasets GSE22093 (p = 0.0213) (C) and GSE3494 (p = 0.0008) (D).
Expression Patterns of Endogenous MTA1 and RNF144A in a Well Established Breast Cancer Progression Model System
To demonstrate the physiological significance of MTA1 regulation of RNF144A, we next analyzed the expression levels of endogenous MTA1 and RNF144A proteins in a well-established breast cancer progression model system, including the non-malignant MCF10A, weakly tumorigenic MCF10AT, highly proliferative and invasive MCF10DCIS, and undifferentiated metastatic MCF10CA1D cell lines (21–25). We found that the protein levels of MTA1 were up-regulated, while the levels of RNF144A protein were gradually down-regulated from non-malignant (lane 1) to highly metastatic (lane 4) breast cancer cell lines (Fig. 2, A and B). Following these observations, we next investigated whether the change of MTA1 and RNF144A in protein levels is resultant of increased corresponding mRNA levels. qRT-PCR analysis demonstrate that the expression of RNF144A and MTA1 mRNA levels followed a reverse correlation in the MCF10 progression panel cell lines corresponding to their metastatic potential (Fig. 2C). Together, these results suggest a reverse correlation of MTA1 and RNF144A in human breast cancer progression model system, highlighting a potential role of MTA1-RNF144A pathway in tumorigenesis and tumor progression.
FIGURE 2.
Expression of MTA1 and RNF144A in the MCF10 breast cancer progression model system. A and B, Western blot analysis of the expression of MTA1 and RNF144A in the MCF10 breast cancer progression model with the indicated antibodies (A) and quantitative results of Western blots (B) using the ImageJ software as mentioned above. C, qRT-PCR analysis of the expression of MTA1 and RNF144A mRNA levels in the MCF10 breast cancer progression model.
MTA1 Regulates RNF144A Expression at mRNA Levels
We next evaluated the status of RNF144A expression under the condition of MTA1 up-regulation or knockdown to test whether MTA1 could regulate RNF144A expression. In agreement with the above observations, we found that forced expression of MTA1 in MCF-7 stable clone cells (26) led to a significant decrease in the RNF144A protein expression (Fig. 3, A and B). As human HeLa cells express high levels of endogenous MTA1 protein according to the results from our laboratory (31) and others (32), we next knocked down endogenous MTA1 in HeLa cells using a specific MTA1 siRNA (28) to verify that the observation of an inverse correlation of MTA1 and RNF144A is general but not unique phenomenon in MCF-7 cells only. As shown in Fig. 3, C and D, knockdown of endogenous of MTA1 resulted in an increase in the protein levels of RNF144A in MTA1 siRNA-transfected cells as compared with control siRNA-treated cells with a given degree of MTA1 knockdown. These results suggest that MTA1 negatively regulates RNF144A expression.
FIGURE 3.
MTA1 negatively regulates RNF144A expression at mRNA level. A and B, Western blot analysis of the protein extracts from MCF-7 cells stably expressing pcDNA empty vector (MCF-7/pcDNA) and T7-MTA1 (MCF-7/T7-MTA1) with the indicated antibodies (A) and quantitative results of Western blots (B) using the ImageJ software. C, HeLa cells were transfected with control siRNAs or specific siRNAs targeting human MTA1. After 48 h of the second-round transfection, protein extracts were prepared and subjected to Western blot analysis with the indicated antibodies. Vinculin was shown as a loading control. D, quantitative results of Western blots shown in C using the ImageJ software. E and F, qRT-PCR analysis of the expression of RNF144A and MTA1 mRNA levels in the MCF-7/pcDNA and MCF-7/T7-MTA1 stable clone cells (E) and Hela cells transfected with control siRNAs or specific siRNAs targeting human MTA1 (F). G, MCF-7 cells were transfected with control siRNAs or specific siRNAs targeting human MTA1. After 36 h of the second-round transfection, cells were treated with or without 250 ng/ml of Actinomycin-D (Act-D) for another 12 h, and then subjected to qRT-PCR analysis of the expression of RNF144A and MTA1 mRNA levels as described above.
To figure out the mechanism of MTA1 regulation of RNF144A, we next examined the possibility that MTA1 affects the mRNA levels of RNF144A. In support of this notion, we found that MTA1 overexpression in MCF-7 cells decreases (Fig. 3E), whereas MTA1 silencing by siRNA in HeLa cells increases (Fig. 3F), the mRNA levels of RNF144A. Moreover, we further demonstrate that Actinomycin-D (Act-D), a putative transcriptional inhibitor (33, 34), effectively blocked MTA1-mediated repression of RNF144A mRNA levels (Fig. 3G), suggesting that MTA1 regulates RNF144A expression, at least in part, at transcriptional level.
MTA1 Is Recruited onto Human RNF144A Promoter and Inhibits Its Promoter Activity
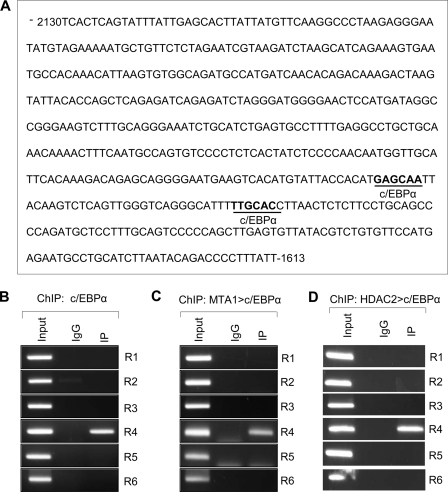
To gain a deeper insight into MTA1 regulation of RNF144A transcription, we next examined the recruitment of MTA1 onto human RNF144A promoter using a ChIP-based promoter-walk assay (35). We found that MTA1 was recruited onto regions R2 (−906 to −661), R3 (−1366 to −1124), R4 (−1788 to −1613), R5 (−2130 to −1953), and R6 (−2914 to −2668) of human RNF144A promoter in MCF-7 cells (Fig. 4, A and B). Given the fact that MTA1 is a dual-functional transcriptional co-regulator, which can not directly bind to DNA, and that MTA1 interacts with histone deacetylase 2 (HDAC2) and both MTA1 and HDAC2 are components of the NuRD co-repressor complex (26, 36), we next tested whether HDAC2 and MTA1/HDAC2 complex were recruited to human RNF144A promoter. Indeed, ChIP assay demonstrated that HDAC2 alone (Fig. 4C) and the MTA1/HDAC2 complex (Fig. 4D) were recruited to the regions R4 (−1788 to −1613) and R5 (−2130 to −1953) of human RNF144A promoter.
FIGURE 4.
MTA1 is recruited onto human RNF144A promoter and inhibits RNF144A promoter activity. A, line diagram showing the regions of human RNF144A promoter analyzed. Nomenclature uses first and last nucleotide number, with the translational start being +1. R, region. B and C, ChIP analysis for the recruitment of MTA1 (A) or HDAC2 (B) onto human RNF144A promoter in MCF-7 cells. D, double ChIP analysis for the recruitment of the MTA1/HDAC2 complex onto human RNF144A promoter. The first ChIP with an anti-MTA1 antibody was followed by the second ChIP with an anti-HDAC2 antibody. E and F, MTA1+/+ and MTA1−/− MEFs (E) or MCF-7/pcDNA and MCF-7/T7-MTA1 stable clone cells (F) were transfected with human RNF144A luciferase reporter plasmid. After 36–48 h of transfection, RNF144A luciferase activity was determined as described under “Experimental Procedures.”
In light of these findings, we next cloned these two regions (R4 and R5; −2130 to −1613) of human RNF144A promoter into a pGL3 basic luciferase reporter vector and examined whether MTA1 influences RNF144A promoter activity. As shown in Fig. 4E, we found a significantly increased RNF144A promoter activity in the MTA1-knock-out (MTA1−/−) MEFs relative to its wild-type (MTA1+/+) controls. In contrast, the RNF144A promoter activity was reduced in the MTA1-overexpressing MCF-7 stable clone cells (MCF-7/T7-MTA1) (26) as compared with its empty vector-expressing controls (MCF-7/pcDNA) (Fig. 4F). These results collectively suggest that MTA1 is recruited onto human RNF144A promoter and inhibits its promoter activity, and further strengthen the notion that MTA1 regulates RNF144A at least in part at transcriptional level.
Recruitment of the MTA1/HDAC2/c/EBPα Co-repressor Complex onto Human RNF144A Promoter
Because MTA1 is a transcriptional coregulator that interacts with transcription factors to either activate or repress the transcription of specific genes (36), we next performed a bioinformatic analysis using the AliBaba 2.1 software program to examine the nature of the putative transcription factor binding sites in the MTA1/HDAC2 complex-RNF144A promoter interacting regions (−2130 to −1613). Interestingly, we found 2 putative consensus binding sequences for the CCAAT/enhancer binding protein α (c/EBPα) transcription factor in these regions (Fig. 5A).
FIGURE 5.
Recruitment of the MTA1/HDAC2/c/EBPα complex onto human RNF144A promoter. A, sequence analysis of the MTA1/HDAC2 complex-RNF144A promoter interacting regions (−2130 to −1613) (R4 and R5) using the AliBaba 2.1 program revealed the presence of two putative consensus binding sequences for transcription factor c/EBPα. B, ChIP analysis for the recruitment of c/EBPα onto human RNF144A promoter. C, double ChIP analysis for the recruitment of the MTA1/c/EBPα complex onto human RNF144A promoter. The first ChIP with an anti-MTA1 antibody was followed by the second ChIP with an anti-c/EBPα antibody. D, double ChIP analysis for the recruitment of the HDAC2/c/EBPα complex onto human RNF144A promoter. The first ChIP was carried out using an anti-HDAC2 antibody, followed by the second ChIP with an anti-c/EBPα antibody.
We next tested the hypothesis that MTA1 is a transcriptional co-repressor of RNF144A gene through the recruitment of c/EBPα transcription factor onto RNF144A promoter. Indeed, ChIP assay revealed that c/EBPα was recruited to only region R4 (−1788 to −1613) of RNF144 promoter (Fig. 5B). Moreover, the results of a double ChIP assay, in which the initial ChIP analysis was performed with an anti-MTA1 antibody followed by the second ChIP analysis using an anti-c/EBPα antibody, demonstrate that the MTA1/c/EBPα complex was co-recruited onto region R4 (−1788 to −1633) of human RNF144A promoter (Fig. 5C). Given the fact that HDAC2 interacts with c/EBPα (37), we next showed that the HDAC2/c/EBPα complex was also recruited to the same region (Fig. 5D).
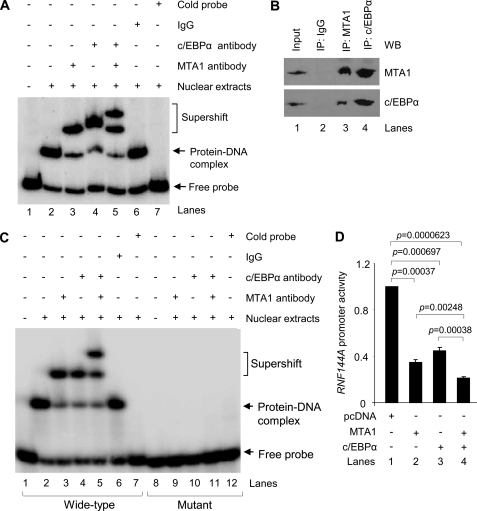
These findings were further confirmed by electrophoretic mobility shift assay (EMSA) analysis using nuclear extracts from MCF-7 cells. As shown in Fig. 6A, we found that the noted protein/RNF144A DNA complex could be super-shifted by incubation of the nuclear extracts with a specific antibody against MTA1 (lane 3) or c/EBPα (lane 4) but not control IgG (lane 6) at the expense of basal protein/RNF144A DNA complexes (lane 2), suggesting that both MTA1 and c/EBPα proteins may interact with RNF144A promoter. This notion was firmly supported by the finding that co-incubation of anti-MTA1 and anti- c/EBPα antibodies resulted in the formation of further higher molecular weight protein/DNA complexes (Fig. 6A, lane 5). In agreement with these observations, we further demonstrated that endogenous MTA1 interacts with endogenous c/EBPα protein in MCF-7 cells revealed by sequential immunoprecipiation (IP)/Western blot analysis with the indicated antibodies (Fig. 6B).
FIGURE 6.
A, EMSA analysis of the binding of c/EBPα and MTA1 to human RNF144A promoter using nuclear extract from the MCF-7 cells. B, nuclear extracts from MCF-7 cells were subjected to immunoprecipitation (IP) analysis with an anti-MTA1 (lane 3), anti-c/EBPα (lane 4) antibody or control IgG (lane 2), followed by Western blotting with the indicated antibodies. WB, Western blotting. C, EMSA analysis of the binding of MTA1 and c/EBPα to the oligos with wild-type or mutant c/EBPα binding motif on human RNF144A promoter. D, MCF-7 cells were transfected with a pGL3-RNF144A luciferase reporter plasmid along with the indicated expression vectors, and RNF144A luciferase activity was determined as described above.
In light of these findings, we next mutated c/EBPα consensus binding motifs in human RNF144A promoter, and demonstrated that there was a drastic reduction in the binding of the MTA1/c/EBPα complex to human RNF144A promoter DNA (Fig. 6C), indicating that the MTA1/c/EBPα complex binds to the RNF144A promoter through c/EBPα consensus binding motifs on human RNF144A promoter. Because MTA1/c/EBPα complex is recruited onto the RNF144A promoter, we next investigated the possibility of potential functional cooperation between MTA1 and c/EBPα in the regulation of the RNF144A gene transcription. As shown in Fig. 6D, we found that co-expression of MTA1 and c/EBPα (lane 4) resulted in a stronger cooperative inhibition of RNF144A promoter activity as compared with MTA1 (lane 2) or c/EBPα (lane 3) expression alone. Collectively, these results suggest that MTA1 represses RNF144A gene transcription by recruiting the MTA1/HDAC2/c/EBPα co-repressor complex onto its promoter.
Role of RNF144A in MTA1-mediated Tumor Cell Migration and Invasion
Given the fact that MTA1 has been identified as one of the critical players in cancer cell migration and invasion (10, 38, 39), we next determined the functional role of the MTA1-RNF144A pathway in the migratory and invasive phenotype of cancer cells. As the biological function of RNF144A is not known so far, we first determined whether RNF144A has any role in cell migration and invasion. To this end, MCF-7 cells stably expressing empty vector (MCF-7/pcDNA) or T7-MTA1 (MCF-7/T7-MTA1) (26) were transfected with specific siRNAs targeting human RNF144A or control siRNA. As shown in supplemental Fig. S1, RNF144A expression was substantially reduced in RNF144A siRNA-transfected cells as compared with control siRNA-treated cells. Wound-healing migration assay shows that the RNF144A siRNA-transfected cells exhibited an increase in migratory activity as compared with control siRNA-transfected cells (Fig. 7A, upper panel; Fig. 7B, compare column 2 with column 1). Boyden chamber invasion assays similarly indicates an increase in cell invasiveness in the RNF144A-depeted cells as compared with control cells (Fig. 7C, upper panel; Fig. 7D, compare column 2 with column 1). These results suggest that RNF144A, by itself, is a novel suppressor of migration and invasion (Fig. 7E).
FIGURE 7.
RNF144A is a novel inhibitor of migration and invasion and the effect of MTA1 on tumor migration and invasion is dependent on RNF144A expression. A and B, effect of depletion of endogenous RNF144A by specific RNF144A siRNAs on cell migration capacity. MCF-7/pcDNA and MCF-7/T7-MTA1 stable clone cells were transfected with control siRNAs or RNF144A siRNAs and then subjected to wounding-healing assay as described under “Experimental Procedures.” The representative images of migrated cells after 24 h and the quantification of the average difference in wound closure between 0 and 24 h are shown in A and B, respectively. C and D, effect of depletion of endogenous RNF144A by specific RNF144A siRNAs on cell invasive capacity. MCF-7/pcDNA and MCF-7/T7-MTA1 stable clone cells were transfected with control siRNAs or RNF144A siRNAs and subjected to Boyden chamber invasion assay. The representative images of cells that invaded the Matrigel Invasion chambers (scale bars, 0.5 mm) and the quantification of the total number of invaded cells are shown in C and D, respectively. Error bars indicate S.D. E, an abstract model for the role of the MTA1-RNF144A pathway in cell migration and invasion.
In support of the promoting role of MTA1 in tumor migration and invasion (10, 38, 39), wound-healing and Boyden chamber invasion assays revealed that forced expression of MTA1 in MCF-7 cells promotes these cells' migratory (Fig. 7A, left panel; Fig. 7B, compare column 3 with column 1) and invasive (Fig. 7C, left panel; Fig. 7D, compare column 3 with column 1) potential. In addition, we noticed that the degree of the effect of MTA1 overexpression on cell migration and invasion is larger than that of RNF144A knockdown (Fig. 7, B and D, compare column 3 with 2), raising the possibility that MTA1 may regulate multiple targets other than RNF144A which also contribute to the noticed migratory and invasive phenotype in MCF-7-overexpressing cells.
To determine whether the effect of MTA1 on cell migration and invasion are dependent on RNF144A expression, we next demonstrated that siRNA-mediated RNF144A silencing renders cells a further more migratory (Fig. 7A, bottom panel; Fig. 7B, compare column 4 with column 3) and invasive (Fig. 7C, bottom panel; Fig. 7D, compare column 4 with column 3) phenotype in MTA1-overexpressing cells, suggesting that RNF144A is functional and partially responsible for MTA1-induced migration and invasion in these MTA1-overexpressing cells. This is in-line with the notion that the net amounts of RNF144A are repressed by both RNF144A siRNAs and MTA1 overexpression. Given the evidence that induced expression of MTA1 in MCF-7 cells results in a significant decrease in the levels of RNF144A protein, mRNA and promoter activity (Figs. 3, A, B, E, and 4F) and that RNF144A is a potential inhibitor of migration and invasion (above), we conclude that MTA1 overexpression in highly metastatic cancer cells facilitates the acquisition of a highly invasive and metastatic phenotype of cancer cells by, at least in part, compromising the suppressive function of RNF144A in tumor migration and invasion through transcriptional repression of RNF144A expression (Fig. 7E).
DISCUSSION
A critical step in the development of a metastatic tumor is characterized by a gain in the tumor cells' migratory and invasive capabilities (40). In the present study, we investigated the molecular mechanism of transcriptional regulation of RNF144A expression by MTA1 and its attributed functions in cancer migration and invasion, hallmarks of tumor metastasis in vivo. First, we made a systematic attempt to understand the correlation between the expression levels of MTA1 and RNF144A in breast cancer microarray data sets and a breast cancer model system (Figs. 1 and 2). Next, we further demonstrated a reverse correlation between MTA1 and RNF144A at the level of mRNA and protein expression in both human breast cancer MCF-7 cells and human cervical carcinoma HeLa cells under the condition of knockdown or overexpression of MTA1 (Fig. 3). These results firmly established a negative correlation between the expression levels of MTA1 and RNF144A. On the mechanistic level, substantial evidence showed that MTA1 transcriptionally represses RNF144A expression by recruiting MTA1/HDAC2/c/EBPα co-repressor complex onto human RNF144A promoter (Fig. 8). Although MTA1 has been shown previously to activate and repress gene transcription in the appropriate cellular and genetic context (9, 16, 36), in this study we provide the molecular details of how MTA1 drives tumor progression through the delicate transcriptional control of its target genes.
FIGURE 8.
Summary of the findings presented in this study. MTA1 promotes tumor progression by, at least in part, interfering with the suppressive function of RNF144A in tumor migration and invasion through transcriptional repression of RNF144A expression via recruiting the MTA1/HDAC2/c/EBPα co-repressor complex onto human RNF144A promoter.
Another novel finding presented here is that we demonstrate that transcriptional repression of RNF144A by MTA1 is functionally linked to the malignant phenotype of cancer cells. In this context, we discovered that siRNA-mediated knockdown of human RNF144A, which had not previously been implicated in cancer, increased the migration and invasiveness of MCF-7 cells, suggesting that RNF144A is a novel suppressor of tumor migration and invasion (Fig. 7). This novel finding supports the observations that the expression levels of RNA144A are negatively correlated with those of MTA1 in various tumor models (Figs. 1 and 2). Although the mechanism by which RNF144A suppresses the migratory and invasive ability of MCF-7 cells is currently unknown, the mechanistic role of other members of the RNF gene family in cancer invasion and metastasis has been documented. For example, RING finger protein 5 (RNF5) has been shown to inhibit cell motility by targeting cytoskeletal protein paxillin ubiquitination and altered localization (41). In contrast, RNF13 and RNF55 (also known as proto-oncogene c-Cbl) promote pancreatic cancer and glioma invasion by enhancing the activity of extracellular matrix metallopeptidase-9 (MMP-9) and MMP-2, respectively (42, 43), which are required for degrading structural extracellular matrix proteins to promote invasion and metastasis (44). Similarly, RNF45 (also known as tumor autocrine motility factor receptor or gp78) promote sarcoma metastasis by targeting the metastasis suppressor KAI1 for degradation (45). Originally, RNF45 was isolated as a membrane glycoprotein from murine melanoma cells and was implicated in cell migration (46, 47). Subsequent studies identified RNF45 as the tumor autocrine motility factor receptor mediating tumor invasion and metastasis (47, 48). In addition, expression of RNF188 (also known as HAKAI or CBLL1) also increases epithelial cell invasion (49). Given that many RING finger proteins have intrinsic E3 ligase activities (18), it is possible that RNF144A exerts its effect on cell migration and invasion through ubiquitylating, possibly causing degradation of key effector proteins of the migration/invasion machinery.
In support of its promoting role in metastasis (10–12), we demonstrated that MTA1 expression enhances cell migration and invasion ability in MCF-7 cells (Fig. 7). More interestingly, we demonstrated that siRNA-mediated knockdown of endogenous RNF144A accelerates the motility and invasion of MTA1-overexpressing cells, suggesting that the effect of MTA1 on cell migratory and invasive functions is dependent on RNF144A expression. Thus, MTA1 facilitates the acquisition of an invasive and metastatic phenotype of cancer cells by, at least in part, compromising the suppressive function of RNF144A in tumor migration and invasion through transcriptional repression of RNF144A expression, and the dysregulated MTA1-RNF144A pathway augments the capacity of cancer cells for metastatic dissemination.
In summary, we demonstrate that RNF144A is a direct target of transcriptional repression by MTA1 and is a novel potential inhibitor of cell migration and invasion. MTA1 overexpression in highly metastatic tumor cells inhibits RNF144A expression and function, facilitating the acquisition of a highly invasive and metastatic phenotype of cancer cells. In addition, as RNF144A protein contains a putative RING finger domain and MTA1 is an ubiquitinated protein (50), it will be interesting to determine whether MTA1 would be targeted by RNF144A as a potential E3 ubiquitin-protein ligase for the ubiquitin-dependent degradation. Thus, MTA1 and RNF144A may form a double vicious circle, contributing to tumor aggressiveness and progression. Collectively, the results presented here provide novel mechanistic insights into regulation of tumor progression by MTA1 and highlight a previously unrecognized role of RNF144A in MTA1-driven cancer cell migration and invasiveness. These findings open the possibility that development of specific modifiers of the MTA1-RNF144A pathway may lead to novel therapeutic approaches for targeting the metastatic process in the future.
Supplementary Material
Acknowledgments
We thank all the members of the Kumar laboratory for technical assistance and fruitful discussion and are grateful to Dr. Lei Wang (Department of Pathology and Immunology, Baylor College of Medicine, Houston, TX) for microarray data mining.
This work was supported, in whole or in part, by National Institutes of Health Grant CA98823 (to R. K.).

This article contains supplemental Fig. S1 and Tables S1 and S2.
- MTA1
- metastasis-associated protein 1
- c/EBPα
- CCAAT/enhancer-binding protein α
- EMSA
- electrophoretic mobility shift assay
- HDAC2
- histone deacetylase 2
- MEFs
- mouse embryonic fibroblasts
- qRT-PCR
- quantitative real-time PCR
- RNF144A
- RING finger protein 144A
- siRNA
- small interfering RNA.
REFERENCES
- 1. Valastyan S., Weinberg R. A. (2011) Tumor metastasis: molecular insights and evolving paradigms. Cell 147, 275–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gupta G. P., Massagué J. (2006) Cancer metastasis: building a framework. Cell 127, 679–695 [DOI] [PubMed] [Google Scholar]
- 3. Steeg P. S. (2006) Tumor metastasis: mechanistic insights and clinical challenges. Nat. Med. 12, 895–904 [DOI] [PubMed] [Google Scholar]
- 4. Wang W., Eddy R., Condeelis J. (2007) The cofilin pathway in breast cancer invasion and metastasis. Nat. Rev. Cancer 7, 429–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Toh Y., Nicolson G. L. (2009) The role of the MTA family and their encoded proteins in human cancers: molecular functions and clinical implications. Clin. Exp. Metastasis 26, 215–227 [DOI] [PubMed] [Google Scholar]
- 6. Kumar R., Wang R. A., Mazumdar A., Talukder A. H., Mandal M., Yang Z., Bagheri-Yarmand R., Sahin A., Hortobagyi G., Adam L., Barnes C. J., Vadlamudi R. K. (2002) A naturally occurring MTA1 variant sequesters oestrogen receptor-α in the cytoplasm. Nature 418, 654–657 [DOI] [PubMed] [Google Scholar]
- 7. Toh Y., Pencil S. D., Nicolson G. L. (1994) A novel candidate metastasis-associated gene, mta1, differentially expressed in highly metastatic mammary adenocarcinoma cell lines. cDNA cloning, expression, and protein analyses. J. Biol. Chem. 269, 22958–22963 [PubMed] [Google Scholar]
- 8. Kumar R., Wang R. A., Bagheri-Yarmand R. (2003) Emerging roles of MTA family members in human cancers. Semin Oncol. 30, 30–37 [DOI] [PubMed] [Google Scholar]
- 9. Manavathi B., Singh K., Kumar R. (2007) MTA family of coregulators in nuclear receptor biology and pathology. Nucl. Recept. Signal 5, e010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mahoney M. G., Simpson A., Jost M., Noé M., Kari C., Pepe D., Choi Y. W., Uitto J., Rodeck U. (2002) Metastasis-associated protein (MTA)1 enhances migration, invasion, and anchorage-independent survival of immortalized human keratinocytes. Oncogene 21, 2161–2170 [DOI] [PubMed] [Google Scholar]
- 11. Nicolson G. L., Nawa A., Toh Y., Taniguchi S., Nishimori K., Moustafa A. (2003) Tumor metastasis-associated human MTA1 gene and its MTA1 protein product: role in epithelial cancer cell invasion, proliferation, and nuclear regulation. Clin. Exp. Metastasis 20, 19–24 [DOI] [PubMed] [Google Scholar]
- 12. Hofer M. D., Menke A., Genze F., Gierschik P., Giehl K. (2004) Expression of MTA1 promotes motility and invasiveness of PANC-1 pancreatic carcinoma cells. Br. J. Cancer 90, 455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Qian H., Yu J., Li Y., Wang H., Song C., Zhang X., Liang X., Fu M., Lin C. (2007) RNA interference of metastasis-associated gene 1 inhibits metastasis of B16F10 melanoma cells in a C57BL/6 mouse model. Biol. Cell. 99, 573–581 [DOI] [PubMed] [Google Scholar]
- 14. Kai L., Wang J., Ivanovic M., Chung Y. T., Laskin W. B., Schulze-Hoepfner F., Mirochnik Y., Satcher R. L., Jr., Levenson A. S. (2011) Targeting prostate cancer angiogenesis through metastasis-associated protein 1 (MTA1). Prostate 71, 268–280 [DOI] [PubMed] [Google Scholar]
- 15. Jiang Q., Zhang H., Zhang P. (2011) ShRNA-mediated gene silencing of MTA1 influenced on protein expression of ER α, MMP-9, CyclinD1 and invasiveness, proliferation in breast cancer cell lines MDA-MB-231 and MCF-7 in vitro. J. Exp. Clin. Cancer Res. 30, 60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ghanta K. S., Li D. Q., Eswaran J., Kumar R. (2011) Gene profiling of MTA1 identifies novel gene targets and functions. PLoS One 6, e17135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li D. Q., Pakala S. B., Reddy S. D., Ohshiro K., Peng S. H., Lian Y., Fu S. W., Kumar R. (2010) Revelation of p53-independent function of MTA1 in DNA damage response via modulation of the p21 WAF1-proliferating cell nuclear antigen pathway. J. Biol. Chem. 285, 10044–10052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Joazeiro C. A., Weissman A. M. (2000) RING finger proteins: mediators of ubiquitin ligase activity. Cell 102, 549–552 [DOI] [PubMed] [Google Scholar]
- 19. Lipkowitz S., Weissman A. M. (2011) RINGs of good and evil: RING finger ubiquitin ligases at the crossroads of tumour suppression and oncogenesis. Nat. Rev. Cancer 11, 629–643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Manavathi B., Peng S., Rayala S. K., Talukder A. H., Wang M. H., Wang R. A., Balasenthil S., Agarwal N., Frishman L. J., Kumar R. (2007) Repression of Six3 by a corepressor regulates rhodopsin expression. Proc. Natl. Acad. Sci. U.S.A. 104, 13128–13133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Santner S. J., Dawson P. J., Tait L., Soule H. D., Eliason J., Mohamed A. N., Wolman S. R., Heppner G. H., Miller F. R. (2001) Malignant MCF10CA1 cell lines derived from premalignant human breast epithelial MCF10AT cells. Breast Cancer Res. Treat. 65, 101–110 [DOI] [PubMed] [Google Scholar]
- 22. Dawson P. J., Wolman S. R., Tait L., Heppner G. H., Miller F. R. (1996) MCF10AT: a model for the evolution of cancer from proliferative breast disease. Am. J. Pathol. 148, 313–319 [PMC free article] [PubMed] [Google Scholar]
- 23. Miller F. R., Santner S. J., Tait L., Dawson P. J. (2000) MCF10DCIS.com xenograft model of human comedo ductal carcinoma in situ. J. Natl. Cancer Inst. 92, 1185–1186 [DOI] [PubMed] [Google Scholar]
- 24. Hurst D. R., Xie Y., Edmonds M. D., Welch D. R. (2009) Multiple forms of BRMS1 are differentially expressed in the MCF10 isogenic breast cancer progression model. Clin. Exp. Metastasis 26, 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soule H. D., Maloney T. M., Wolman S. R., Peterson W. D., Jr., Brenz R., McGrath C. M., Russo J., Pauley R. J., Jones R. F., Brooks S. C. (1990) Isolation and characterization of a spontaneously immortalized human breast epithelial cell line, MCF-10. Cancer Res. 50, 6075–6086 [PubMed] [Google Scholar]
- 26. Mazumdar A., Wang R. A., Mishra S. K., Adam L., Bagheri-Yarmand R., Mandal M., Vadlamudi R. K., Kumar R. (2001) Transcriptional repression of oestrogen receptor by metastasis-associated protein 1 corepressor. Nat. Cell Biol. 3, 30–37 [DOI] [PubMed] [Google Scholar]
- 27. Dignam J. D., Lebovitz R. M., Roeder R. G. (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 11, 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Li D. Q., Ohshiro K., Khan M. N., Kumar R. (2010) Requirement of MTA1 in ATR-mediated DNA damage checkpoint function. J. Biol. Chem. 285, 19802–19812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Iwamoto T., Bianchini G., Booser D., Qi Y., Coutant C., Shiang C. Y., Santarpia L., Matsuoka J., Hortobagyi G. N., Symmans W. F., Holmes F. A., O'Shaughnessy J., Hellerstedt B., Pippen J., Andre F., Simon R., Pusztai L. (2011) Gene pathways associated with prognosis and chemotherapy sensitivity in molecular subtypes of breast cancer. J. Natl. Cancer Inst. 103, 264–272 [DOI] [PubMed] [Google Scholar]
- 30. Miller L. D., Smeds J., George J., Vega V. B., Vergara L., Ploner A., Pawitan Y., Hall P., Klaar S., Liu E. T., Bergh J. (2005) An expression signature for p53 status in human breast cancer predicts mutation status, transcriptional effects, and patient survival. Proc. Natl. Acad. Sci. U.S.A. 102, 13550–13555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reddy S. D., Pakala S. B., Ohshiro K., Rayala S. K., Kumar R. (2009) MicroRNA-661, a c/EBPα target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Res. 69, 5639–5642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khaleque M. A., Bharti A., Gong J., Gray P. J., Sachdev V., Ciocca D. R., Stati A., Fanelli M., Calderwood S. K. (2008) Heat shock factor 1 represses estrogen-dependent transcription through association with MTA1. Oncogene 27, 1886–1893 [DOI] [PubMed] [Google Scholar]
- 33. Sobell H. M. (1985) Actinomycin and DNA transcription. Proc. Natl. Acad. Sci. U.S.A. 82, 5328–5331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Flamée P. A. (1985) The action of actinomycin D on the transcription of T7 coliphage DNA by Escherichia coli RNA polymerase. Biochem. J. 230, 557–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li D. Q., Pakala S. B., Reddy S. D., Ohshiro K., Zhang J. X., Wang L., Zhang Y., Moreno de Alborán I., Pillai M. R., Eswaran J., Kumar R. (2011) Bidirectional autoregulatory mechanism of metastasis-associated protein 1-alternative reading frame pathway in oncogenesis. Proc. Natl. Acad. Sci. U.S.A. 108, 8791–8796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Manavathi B., Kumar R. (2007) Metastasis tumor antigens, an emerging family of multifaceted master coregulators. J. Biol. Chem. 282, 1529–1533 [DOI] [PubMed] [Google Scholar]
- 37. Duan H., Heckman C. A., Boxer L. M. (2005) Histone deacetylase inhibitors down-regulate bcl-2 expression and induce apoptosis in t(14;18) lymphomas. Mol. Cell Biol. 25, 1608–1619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Qian H., Lu N., Xue L., Liang X., Zhang X., Fu M., Xie Y., Zhan Q., Liu Z., Lin C. (2005) Reduced MTA1 expression by RNAi inhibits in vitro invasion and migration of esophageal squamous cell carcinoma cell line. Clin. Exp. Metastasis 22, 653–662 [DOI] [PubMed] [Google Scholar]
- 39. Rao Y., Wang H., Fan L., Chen G. (2011) Silencing MTA1 by RNAi reverses adhesion, migration and invasiveness of cervical cancer cells (SiHa) via altered expression of p53, and E-cadherin/β-catenin complex. J. Huazhong. Univ. Sci. Technolog. Med. Sci. 31, 1–9 [DOI] [PubMed] [Google Scholar]
- 40. Wicki A., Lehembre F., Wick N., Hantusch B., Kerjaschki D., Christofori G. (2006) Tumor invasion in the absence of epithelial-mesenchymal transition: podoplanin-mediated remodeling of the actin cytoskeleton. Cancer Cell 9, 261–272 [DOI] [PubMed] [Google Scholar]
- 41. Didier C., Broday L., Bhoumik A., Israeli S., Takahashi S., Nakayama K., Thomas S. M., Turner C. E., Henderson S., Sabe H., Ronai Z. (2003) RNF5, a RING finger protein that regulates cell motility by targeting paxillin ubiquitination and altered localization. Mol. Cell Biol. 23, 5331–5345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhang Q., Meng Y., Zhang L., Chen J., Zhu D. (2009) RNF13: a novel RING-type ubiquitin ligase overexpressed in pancreatic cancer. Cell Res. 19, 348–357 [DOI] [PubMed] [Google Scholar]
- 43. Lee H., Tsygankov A. Y. (2010) c-Cbl regulates glioma invasion through matrix metalloproteinase 2. J. Cell Biochem. 111, 1169–1178 [DOI] [PubMed] [Google Scholar]
- 44. Egeblad M., Werb Z. (2002) New functions for the matrix metalloproteinases in cancer progression. Nat. Rev. Cancer 2, 161–174 [DOI] [PubMed] [Google Scholar]
- 45. Tsai Y. C., Mendoza A., Mariano J. M., Zhou M., Kostova Z., Chen B., Veenstra T., Hewitt S. M., Helman L. J., Khanna C., Weissman A. M. (2007) The ubiquitin ligase gp78 promotes sarcoma metastasis by targeting KAI1 for degradation. Nat. Med. 13, 1504–1509 [DOI] [PubMed] [Google Scholar]
- 46. Nabi I. R., Raz A. (1987) Cell shape modulation alters glycosylation of a metastatic melanoma cell surface antigen. Int. J. Cancer 40, 396–402 [DOI] [PubMed] [Google Scholar]
- 47. Fang S., Ferrone M., Yang C., Jensen J. P., Tiwari S., Weissman A. M. (2001) The tumor autocrine motility factor receptor, gp78, is a ubiquitin protein ligase implicated in degradation from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 98, 14422–14427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Nabi I. R., Watanabe H., Raz A. (1992) Autocrine motility factor and its receptor: role in cell locomotion and metastasis. Cancer Metastasis Rev. 11, 5–20 [DOI] [PubMed] [Google Scholar]
- 49. Rodríguez-Rigueiro T., Valladares-Ayerbes M., Haz-Conde M., Aparicio L. A., Figueroa A. (2011) Hakai reduces cell-substratum adhesion and increases epithelial cell invasion. BMC Cancer 11, 474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Li D. Q., Ohshiro K., Reddy S. D., Pakala S. B., Lee M. H., Zhang Y., Rayala S. K., Kumar R. (2009) E3 ubiquitin ligase COP1 regulates the stability and functions of MTA1. Proc. Natl. Acad. Sci. U.S.A. 106, 17493–17498 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.