Background: Hsp110, an Hsp70 homolog, is highly efficient in preventing protein aggregation but lacks the folding activity seen in Hsp70s.
Results: In contrast to Hsp70s, Hsp110s exhibit distinct peptide substrate binding properties.
Conclusion: The peptide substrate binding properties determine the chaperone activity differences between Hsp70s and Hsp110s.
Significance: Our studies shed light on the molecular mechanism of the chaperone activities of Hsp70s and Hsp110s.
Keywords: Chaperone Chaperonin, Heat-shock Protein, Molecular Chaperone, Protein Aggregation, Protein Folding, Hsp110, Hsp70, Peptide Substrate Binding
Abstract
The molecular chaperone 70-kDa heat-shock proteins (Hsp70s) play essential roles in maintaining protein homeostasis. Hsp110, an Hsp70 homolog, is highly efficient in preventing protein aggregation but lacks the hallmark folding activity seen in Hsp70s. To understand the mechanistic differences between these two chaperones, we first characterized the distinct peptide substrate binding properties of Hsp110s. In contrast to Hsp70s, Hsp110s prefer aromatic residues in their substrates, and the substrate binding and release exhibit remarkably fast kinetics. Sequence and structure comparison revealed significant differences in the two peptide-binding loops: the length and properties are switched. When we swapped these two loops in an Hsp70, the peptide binding properties of this mutant Hsp70 were converted to Hsp110-like, and more impressively, it functionally behaved like an Hsp110. Thus, the peptide substrate binding properties implemented in the peptide-binding loops may determine the chaperone activity differences between Hsp70s and Hsp110s.
Introduction
Molecular chaperone 70-kDa heat-shock proteins (Hsp70s)3 play a crucial role in maintaining cellular protein homeostasis under both normal and stress conditions by assisting protein folding, assembly, translocation into organelles, and degradation (1–3). In this role, Hsp70s are inextricably linked to many diseases, such as cancers and neurodegenerative diseases; thus, they are potential targets for treating these diseases (4). Consistent with their essential roles, Hsp70s are ubiquitous and abundant. They are present in the cytoplasm of prokaryotic organisms and all the cellular compartments of eukaryotic organisms.
The domain organization of Hsp70s is highly conserved (1, 2, 5, 6). All Hsp70s have two functional domains, the nucleotide-binding domain (NBD) and the substrate-binding domain (SBD), corresponding to two intrinsic activities. The N-terminal NBD binds ATP or ADP and also possesses an ATPase activity. A number of crystal structures of the isolated NBD have revealed the structural basis of nucleotide binding (7–10). The C-terminal SBD is the site of peptide substrate binding and is further subdivided into α and β subdomains. Previous biochemical and structural studies demonstrated that Hsp70s prefer binding to stretches of hydrophobic segments, which normally fold inside native proteins (11–14). A short and highly conserved linker segment, the interdomain linker, connects the NBD and SBD. Although the binding functions of the NBD and SBD are separable, Hsp70 chaperone activity strictly depends on the tight coupling of these two domains upon ATP binding, which leads to modulation of the two intrinsic activities. ATP binding dramatically reduces the affinity for peptide substrates by accelerating both the binding and especially the release rates (15). At the same time, peptide substrate binding stimulates the ATP hydrolysis rate (16). Thus, this ATP-induced tight coupling assures that the energy from ATP hydrolysis is efficiently used to regulate peptide substrate binding and release, thereby conferring efficient chaperone activity. In summary, the two intrinsic activities of the NBD and SBD as well as their allosteric coupling are at the core of the Hsp70 chaperone activity. Moreover, the chaperone activities of Hsp70s are facilitated by two classes of co-chaperones: Hsp40s and nucleotide exchange factors (NEFs) (1, 3, 17, 18).
Two classes of large homologs of Hsp70 have been discovered thus far: Hsp110 and Grp170 (19, 20). Present only in the cytosol of eukaryotes, Hsp110s have the same domain organization as classic Hsp70s but are larger in size due to an insertion in SBDβ and a longer C-terminal extension, both of which are dispensable for function (21, 22). Although crystal structures of the isolated domains from Hsp70s have been known for more than a decade, we are just beginning to understand the structures of Hsp110s. In 2007, we reported the first x-ray crystal structure of a full-length Hsp110 from yeast, Sse1 (22), which revealed the extensive contacts between the domains. The subdomain structures in this Sse1 structure are highly similar to those of Hsp70s from the isolated domain structures, confirming that Hsp110s and Hsp70s are indeed relatives.
Hsp110s are essential in eukaryotes as demonstrated by genetic studies in both yeast and Caenorhabditis elegans (21, 23) (WormBase); however, their exact cellular roles are poorly defined. A number of studies suggested that Hsp110s participate in many processes associated with cytosolic Hsp70s, including de novo protein folding, refolding under stress, translocation into the endoplasmic reticulum, degradation, and prion formation (24–31). Consistent with these observations, Hsp110s have recently been shown to form complexes with cytosolic Hsp70s for which they function as the major NEF (25, 32–35). Moreover, the corresponding mechanism of this nucleotide exchange activity was recently revealed by a number of biochemical and crystallographic studies (36–39). However, the function of Hsp110s extends beyond serving as NEFs for Hsp70s. They also show chaperone activity of their own. Unlike Hsp70s, Hsp110s lack the hallmark activity to assist in protein folding; however, they exhibit high activity in preventing aggregation of denatured proteins for which they are even more efficient than Hsp70s (40, 41). For this reason, Hsp110s are called “holdases” in comparison with “foldases” for Hsp70s. More recently, this NEF-independent chaperone activity has been shown to directly contribute to the role of Hsp110s in prion formation and propagation (31), endoplasmic reticulum-associated degradation (42), and tumor antigen presentation (43, 44). Therefore, Hsp110s serve dual roles as chaperones of their own and as co-chaperones for Hsp70s.
Because of the lack of biochemical and structural information, the chaperone activity of Hsp110s remains poorly understood. It is not clear what determines the chaperone activity differences between Hsp70s and Hsp110s. To approach this question, we began by analyzing the peptide substrate binding properties of Hsp110s. Although a previous study on the yeast Hsp110 Sse1 suggested that Hsp110s most likely have a different preference from Hsp70s for peptide substrates (45), the peptide substrate binding specificity and kinetics of Hsp110s are still largely unknown. In the present study, we demonstrated that Hsp110s have a strong preference for aromatic side chains in their substrates in contrast to Hsp70s, which prefer aliphatic residues. Surprisingly, Hsp110s bind peptide substrates with exceptionally fast kinetics. Comparing the sequences and structures of Hsp70s and Hsp110s, we observed significant differences in the two peptide substrate-binding loops. Our mutagenesis assays suggested that the peptide substrate-binding loops govern the unique peptide binding qualities in both Hsp70s and Hsp110s, which contribute to their distinct chaperone activity to serve as holdases or foldases.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The full-length DnaK proteins were overexpressed from a dnak expression plasmid pBB46 (ampR) in dnak deletion strain BB205 (camR kanR) (46). To facilitate protein purification, a six-histidine tag was added at the C terminus of DnaK; the tag had no observable influence on the in vivo function of DnaK. After induction at 30 °C for 5 h, the cells were harvested by centrifugation, resuspended in ice-cold 2× PBS (20 mm Na2HPO4, 1.76 mm KH2PO4, 274 mm NaCl, and 5.4 mm KCl), and sonicated for 3 min in an ice-water bath. After centrifugation at 12,000 × g for 1 h, the supernatant was loaded onto a HisTrap column (GE Healthcare) and eluted with a linear gradient of imidazole. The eluted DnaK protein was further purified on a HiTrap Q column and concentrated to >20 mg/ml in a buffer containing 10 mm Hepes-KOH, pH 7.5 and 50 mm KCl before flash freezing in liquid nitrogen.
The full-length Sse1 protein was purified as described before (22). The expression and purification of the full-length Sse2 and human Hsp110 (hHsp110) proteins were similar to that of Sse1 with modifications. Briefly, the open reading frames (ORFs) of Sse2 and hHsp110 were PCR-amplified from yeast genomic DNA and plasmid pOTB7_hHSP110 (ordered from ATCC), respectively, and cloned into pSMT3 vector for expression as Smt3 fusion proteins. The Smt3 fusion proteins were first purified on a HisTrap column. For Sse2, the Smt3 tag was removed by treating with Ulp1 protease overnight, and the resulting Sse2 was further purified using HiTrap Q and Superdex 200 16/60 columns (GE Healthcare). For hHsp110, it was difficult to remove the Smt3 tag; thus, we further purified the Smt3-hHsp110 fusion protein.
The full-length Ssa1 protein was expressed and purified from yeast Pichia pastoris strain GS115 as described previously with modification (47). The P. pastoris strain expressing Ssa1 was a generous gift from Dr. Johannes Buchner. Briefly, after breaking open yeast with Avestin Emulsiflex (Molecular Biology Core Facility at Virginia Commonwealth University), the cell lysate was cleared by centrifugation at 12,000 × g. The supernatant was first subjected to ammonium sulfate precipitation. The pellets from 40–50% ammonium sulfate precipitation were dialyzed overnight and loaded on a HiTrap Q column. After further purification on butyl-Sepharose and Superdex 200 16/60 columns, the Ssa1 protein was concentrated and flash frozen in liquid nitrogen. The isolated SBD domains of DnaK (amino acids 389–607), Sse1 (amino acids 389–659 and 396–659), Sse2 (amino acids 389–659), and hHsp110 (amino acids 391–719) were expressed and purified in a similar way as the full-length Sse1 and Sse2 proteins as described above.
Fluorescence Anisotropy Peptide Substrate Binding Assay
All peptides were labeled at the N terminus with fluorescein and ordered from NEOBioscience with greater than 95% purity. To determine peptide binding affinity, serial dilutions of each protein were incubated with a 20 nm concentration of the corresponding peptides in buffer A (25 mm Hepes-KOH, pH 7.5, 100 mm KCl, 10 mm Mg(OAc)2, and 10% glycerol). After binding reached equilibrium, fluorescence anisotropy measurements were made on a Beacon 2000 instrument (Invitrogen), and data were fitted to a one-site binding equation using PRISM (GraphPad) to deduce dissociation constants (Kd). Each sample was read at least three times to get an average reading. All the binding curves were repeated at least three times with more than two different protein preparations.
Peptide Substrate Binding Kinetics Measurements
The binding kinetics of each chaperone protein were measured by mixing fluorescein-labeled peptide (20 nm) with various concentrations of chaperone protein. Peptides alone were used for initial readings. After adding the chaperone proteins, measurements were made every 10 s. For the binding in the presence of ATP, each chaperone protein was first incubated with ATP (2 mm final concentration) for 2 min before adding the peptide.
Native Gel Analysis on Stability of Chaperone-Peptide Complexes
Chaperone proteins (1.5 mg/ml) were mixed with fluorescein-labeled peptides at a 1:1 molar ratio in buffer A and incubated at room temperature for 1 h. A 3-μl sample was loaded onto an 8–25% gradient native gel (PhastGel from GE Healthcare) and electrophoresed. Fluorescein-labeled peptides were visualized with a Typhoon phosphorimaging system (GE Healthcare), and proteins were stained with Coomassie Blue.
Mutagenesis and Growth Test
Peptide-binding loop mutations in both DnaK and Sse1 were induced by the QuikChange Lightning Mutagenesis kit (Stratagene). The growth phenotype of the DnaK mutations were tested in dnak deletion strain BB205 as described previously (22).
Luciferase Refolding Assay
Firefly luciferase (Promega) was diluted with buffer B (25 mm Hepes-KOH, pH 7.5, 100 mm KOAc, 10 mm Mg(OAc)2, 2 mm DTT, and 3 mm ATP) to a final concentration of 100 nm in the presence of either WT DnaK or DnaK_Loop12/34 mutant protein (3 μm). After incubating at 42 °C for 20 min, refolding reactions were carried out by diluting the heated luciferase into a reaction mixture containing 3 μm DnaK, 0.67 μm DnaJ, and 0.33 μm GrpE in buffer B. At various time points, luciferase activity was measured in a luminometer (Berthold LB9507) by mixing 2 μl of refolding reaction with 50 μl of luciferase substrate (Promega). The activity of unheated luciferase in buffer B was set as 100%.
In Vitro Luciferase Aggregation Assay or Holdase Assay
To analyze the holdase activity, firefly luciferase (Promega) was used as a model substrate. The aggregation of luciferase in the presence of different chaperones was monitored by UV absorbance at 320 nm over incubation time at 41 °C. Measurements were taken on a UV-visible spectrophotometer with a water-jacketed cuvette holder. The final concentration of luciferase was kept at 200 nm for all the reactions. The chaperones were added at a 1:4 or 4:1 ratio to luciferase. We tested a set of molecular ratios of chaperones over luciferase, and these ratios were optimal under our experimental condition. Luciferase or chaperone proteins alone were used as controls.
RESULTS
Hsp70s and Hsp110s Prefer Different Hydrophobic Sequences in Peptide Substrates
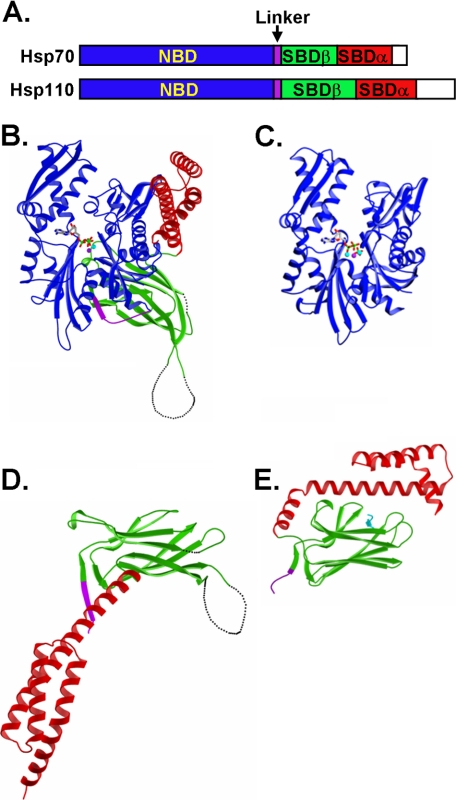
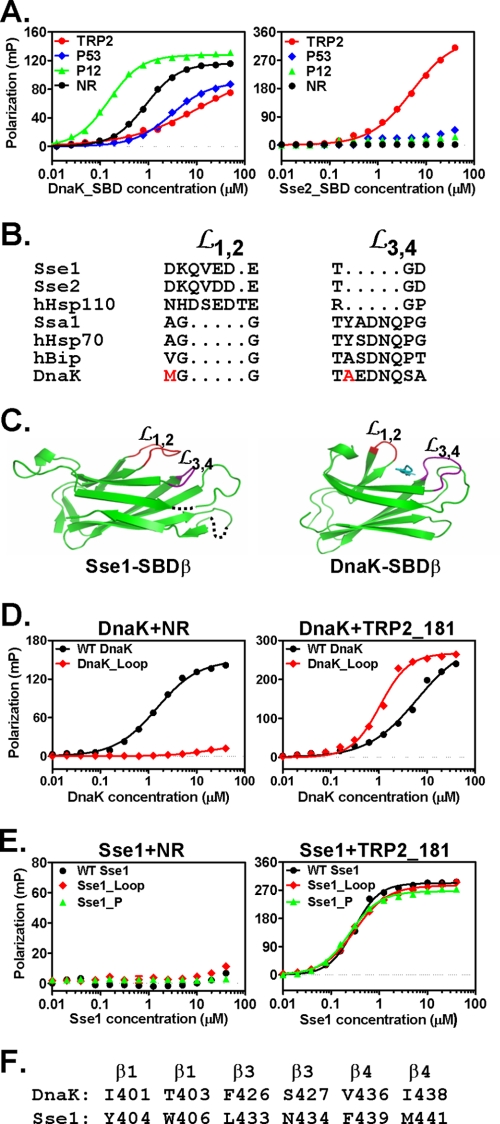
As distant homologs of Hsp70s, Hsp110s demonstrate different chaperone activities. Specifically, Hsp110s are characterized by high activity in preventing denatured proteins from aggregation but lack normal folding activities observed in Hsp70s. Like Hsp70s, Hsp110s contain two functional domains, the NBD and SBD (Fig. 1A). Although Hsp110s share high sequence and structure similarity to Hsp70s at the NBD (Fig. 1, B and C), the SBD is more divergent. As described previously (22), structural comparison revealed a number of significant differences in the SBD (Fig. 1, D and E) especially within the peptide-binding loops (as described in more details below). Hence, to understand the mechanistic differences between these two classes of related chaperones, we first explored the peptide substrate binding properties.
FIGURE 1.
Structural comparison of Hsp70s and Hsp110s. A, schematics of Hsp70 and Hsp110 sequences. NBD is in blue. SBD is divided into two subdomains: SBDα and SBDβ are in red and green, respectively. The interdomain linker is shown in purple, whereas the extreme C-terminal end is shown in white. B and C, ribbon diagrams of the Sse1-ATP structure (Protein Data Bank code 2QXL) and bHsc70 isolated NBD structure (Protein Data Bank code 1HPM), respectively, with the NBD aligned. The coloring is the same as in A. ATP in Sse1 and ADP in bHsc70 are shown as bonds. The associated ions, Mg2+ and K+, are shown as balls. D and E, ribbon diagrams of SBD structures of Sse1 (Protein Data Bank code 2QXL) and DnaK (Protein Data Bank code 1DKX), respectively, with the SBDβ aligned. The coloring is the same as in A with the peptide substrate NR in DnaK shown in cyan.
For initial studies, we synthesized four different peptides containing hydrophobic sequences, NR, p12, p53, and tyrosinase-related protein 2 (TRP2) (Table 1). NR is a well characterized peptide substrate for DnaK, a major Hsp70 in Escherichia coli that has been studied extensively (13, 14). The sequences of p12, p53, and TRP2 peptides were derived from the chicken aspartate aminotransferase (48), the tumor suppressor p53 (49), and TRP2, a tumor antigen (50), respectively. The crystal structure of DnaK SBD in complex with NR peptide revealed the molecular basis of NR binding and established the central roles of the three key Leu residues in NR (Table 1) (13). Like NR, the sequence of p12 also has three consecutive Leu residues and has been shown to bind DnaK with higher affinity than NR due to its longer sequence. Previous studies suggested that peptides p53 and TRP2 potentially bind to mitochondrial Hsp70 and Grp170, another larger Hsp70 homolog, respectively (49, 51).
TABLE 1.
Peptide substrate binding affinities of full-length Hsp70s and Hsp110s
ND, affinity too low to be determined; NB, no binding detected; —, not tested. Bold indicates residues changed in TRP2.
| Peptide (sequence) |
Kd of peptide substrates |
||||
|---|---|---|---|---|---|
| DnaK | Ssa1 | Sse1 | Sse2 | hHsp110 | |
| μm | |||||
| TRP2 (SVYDFFVWL) | ND | ND | 1.27 ± 0.03 | 3.15 ± 0.11 | 5.64 ± 0.32 |
| p53 (LDGEYFTLQIRGRER) | ND | ND | ND | ND | ND |
| NR (NRLLLTG) | 1.43 ± 0.02 | 0.76 ± 0.03 | NB | NB | NB |
| p12 (LQSRLLLSAPRR) | 0.26 ± 0.02 | 0.60 ± 0.03 | NB | NB | NB |
| TRP2_F5L/F6L (SVYDLLVWL) | 1.76 ± 0.04 | 1.12 ± 0.06 | 3.62 ± 0.12 | 8.45 ± 0.30 | ND |
| TRP2_W8L (SVYDFFVLL) | 6.54 ± 0.23 | ND | 14.92 ± 0.43 | 12.62 ± 0.86 | ND |
| TRP2_181 (VYDFFVWLHYY) | 6.27 ± 0.94 | — | 0.29 ± 0.01 | 1.78 ± 0.06 | 1.50 ± 0.05 |
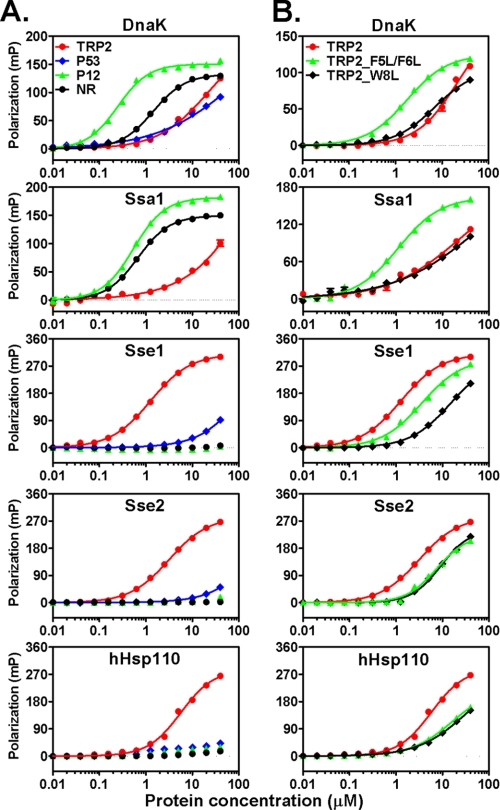
There are two closely related Hsp110s in yeast, Sse1 and Sse2, with Sse1 being the predominant form. To study the peptide substrate binding properties of Hsp110s, we purified both yeast Hsp110s as well as an hHsp110. For comparison, we also purified two classic Hsp70s, DnaK from E. coli and Ssa1 from yeast, as representative Hsp70s from prokaryotes and eukaryotes, respectively. We used a fluorescence polarization assay to detect peptide substrate binding. All peptides were labeled at the N terminus with the fluorescent dye fluorescein. Corresponding to peptide binding, the fluorescence polarization increased dramatically (Fig. 2A). We determined the dissociation constants (Kd) of these peptides for purified Hsp70s and Hsp110s (Fig. 2A and Table 1). Consistent with previous reports (46, 52), DnaK and Ssa1 bind NR and p12 with high affinity. However, both DnaK and Ssa1 have much lower affinity for the hydrophobic TRP2 and p53 peptides. In contrast, all three Hsp110s bound only the TRP2 peptide with high affinity and showed little binding to either the NR or p12 peptide. For p53, all Hsp110s showed low but significant binding. Thus, compared with Hsp70s, Hsp110s have different binding selectivity for peptide substrates. Although hHsp110 shares low sequence identity with Sse1 and Sse2, all three Hsp110s showed similar preference for the same peptides, suggesting a functional conservation among Hsp110s.
FIGURE 2.
Hsp70s and Hsp110s have different peptide substrate preference. A, chaperone binding to four different peptides analyzed by fluorescence anisotropy assay. The peptides are TRP2 (red circle), p53 (blue diamond), p12 (green triangle), and NR (black circle). From top to bottom, the plots are DnaK, Ssa1, Sse1, Sse2, and hHsp110, respectively. B, chaperone binding to the mutated TRP2 peptides TRP2_FF (green triangle) and TRP2_W (black diamond) with TRP2 peptide (red filled circle) as a control. The sequence of the plots is the same as in A. mP, millipolarization level.
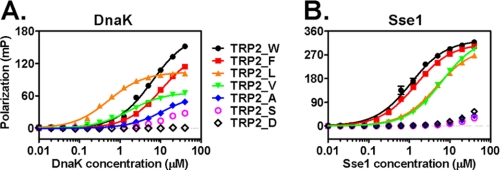
One common feature of the Hsp110 binding peptides TRP2 and p53 is the presence of aromatic residues (Table 1). TRP2 is especially rich in aromatic residues with four aromatic residues of a total of nine residues. We proceeded to test the roles of these aromatic residues in binding to Hsp110s using two mutant versions of TRP2, TRP2_F5L/F6L (with the two Phe residues changed to Leu) and TRP2_W8L (with the Trp changed to Leu) (Fig. 2B and Table 1). To our satisfaction, the affinity of Hsp110s for both mutant peptides was significantly reduced, supporting an essential role of these aromatic residues in the molecular recognition by Hsp110s. Conversely, the affinity of TRP2_F5L/F6L to both Hsp70s was significantly increased (∼8–10-fold). Moreover, replacing the three central Leu residues in the NR peptide with FVW significantly increased its binding to Hsp110s, whereas its affinity for DnaK was reduced (supplemental Fig. S1). To further test the roles of these three aromatic residues, we replaced them with a series of residues that vary in hydrophobicity and aromaticity (Table 2) and tested their binding to DnaK and Sse1. To facilitate comparison, we put these residues in the middle of the TRP2 peptide. Consistent with the above observations, Sse1 prefers Trp and Phe, whereas DnaK prefers Leu and Val (Fig. 3 and Table 2). In contrast, there was no significant binding to peptides with Ala, Ser, or Asp substitutions for either DnaK or Sse1. All these results are consistent with the crystal structure of the DnaK SBD in complex with the NR peptide that suggests that DnaK prefers aliphatic side chains, whereas bulky aromatic side chains are not favored. Thus, for the first time, our data suggest that aromatic residues within a peptide shift the binding from the aliphatic residue-preferring Hsp70s to the aromatic residue-preferring Hsp110s.
TABLE 2.
Binding affinities of TRP2 mutant peptides
ND, affinity too low to be determined; NB, no binding detected. Bold indicates residues that are changed.
| Peptide (sequence) |
Kd of peptide substrates |
|
|---|---|---|
| DnaK | Sse1 | |
| μm | ||
| TRP2_W (VYDFFVWLHY) | 6.25 ± 0.26 | 1.04 ± 0.05 |
| TRP2_F (VYDFFVFLHY) | 10.76 ± 0.29 | 1.45 ± 0.05 |
| TRP2_L (VYDLLVLLHY) | 0.53 ± 0.02 | 4.68 ± 0.07 |
| TRP2_V (VYDVVVVLHY) | 1.76 ± 0.09 | 6.44 ± 0.31 |
| TRP2_A (VYDAAVALHY) | ND | ND |
| TRP2_S (VYDSSVSLHY) | ND | ND |
| TRP2_D (VYDDDVDLHY) | NB | ND |
FIGURE 3.
Binding of DnaK and Sse1 to mutant TRP2 peptides. A, DnaK. B, Sse1. The mutant TRP2 peptides are labeled between A and B: TRP2_W (black filled circle), TRP2_F (red filled square), TRP2_L (orange filled triangle), TRP2_V (green filled triangle), TRP2_A (blue filled diamond), TRP2_S (blue open circle), and TRP2_D (black open diamond). mP, millipolarization level.
Characterization of Peptide Substrate Binding Properties of Hsp110s
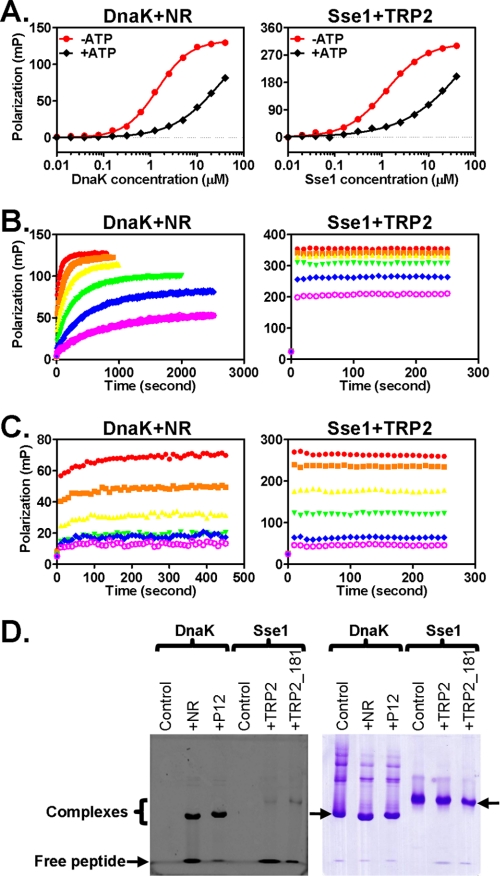
The identification of the TRP2 peptide as a high affinity substrate for Hsp110s allowed us to study the peptide substrate binding properties of Hsp110s. We first tested the allosteric control of peptide binding by ATP. For classic Hsp70s, peptide substrate binding affinity is controlled by ATP, which is an essential part of the allosteric coupling between the two functional domains, the NBD and SBD. ATP binding to Hsp70s results in an ∼2 order of magnitude decrease in peptide substrate binding affinity (15, 53, 54). Consistent with previous studies, we observed a significant reduction of the affinity of DnaK for NR in the presence of ATP (Fig. 4A). Incubation of Sse1 with ATP also resulted in a significantly reduced binding affinity with a comparable change in Kd obtained for the ATP-induced binding of DnaK to the NR peptide (Fig. 4A). For Sse2 and hHsp110, we also observed a similar ATP sensitivity in TRP2 binding (supplemental Fig. S2). Thus, peptide substrate binding to Hsp110 is also controlled by ATP, indicating that like Hsp70s there is allosteric coupling in Hsp110s between their two functional domains.
FIGURE 4.
Kinetics of Hsp110 peptide substrate binding and stability of Hsp110-peptide complexes. A, ATP reduces the peptide substrate binding affinity in Sse1. Left, DnaK protein binding to the NR peptide. Right, Sse1 binding to the TRP2 peptide. B and C, peptide binding kinetics in the absence and presence of ATP, respectively. Left, DnaK binding to the NR peptide. Right, Sse1 binding to the TRP2 peptide. The concentrations of DnaK and Sse1 used were 40 (red), 20 (orange), 10 (yellow), 5 (green), 2.5 (blue), and 1.25 μm (magenta). D, the stability of the Hsp70-peptide and Hsp110-peptide complexes were analyzed using native PAGE. The fluorescein-labeled peptides were visualized using a phosphorimaging system (left panel), and the chaperone proteins on the same gel were stained with Coomassie Blue (right panel). The positions of the free peptides and the chaperone-peptide complexes, respectively, are indicated in the left panel. The arrowheads in the right panel point to the monomeric forms of DnaK and Sse1, respectively. The oligomeric forms of DnaK ran above the monomeric form. mP, millipolarization level.
It is well established that in the absence of ATP Hsp70s stably bind peptide substrates with slow binding (kon) and release (koff) rates (15). ATP binding dramatically speeds up both rates with the koff being accelerated to a greater extent. Consistent with previous studies, in the absence of ATP, the binding of NR peptide to DnaK was relatively slow with a kon value of 663.6 ± 8.0 s−1 m−1 and a koff value of 0.001232 ± 0.000021 s−1 as determined in our studies by fitting association kinetics with six different concentrations of DnaK protein (Fig. 4B); in contrast, when DnaK was preincubated with ATP, both rates were dramatically accelerated (Fig. 4C). Because there is no prior report on the peptide binding kinetics of Hsp110s, we proceeded to analyze the kinetics of the binding of Hsp110s to TRP2. Surprisingly, all three Hsp110s bound TRP2 with extraordinarily fast kinetics regardless of the presence or absence of ATP (shown for Sse1 in Fig. 4, B and C). These binding rates were even faster than DnaK in the presence of ATP and well beyond the detection limit of our instrument. Because DnaK and Sse1 have comparable Kd values for the NR and TRP2 peptides, respectively, the release rate for Sse1 (koff) should also be much higher than that of DnaK based on the equation Kd = koff/kon. Moreover, the decrease of TRP2 binding to Sse1 in the presence of ATP observed in Fig. 4C is consistent with the ATP-sensitive nature of TRP2 binding to Sse1 as shown above in Fig. 3A. Thus, compared with the Hsp70 peptide substrate binding, Hsp110s exhibit much faster binding and release rates.
The fast kinetics of Hsp110 peptide substrate binding suggest that Hsp110s may bind peptide substrates transiently even in the absence of ATP. This is markedly different from Hsp70s, which bind peptide substrates stably in the absence of ATP. To test this hypothesis, we analyzed the stability of the Hsp110-peptide complex using native gel electrophoresis. It has been shown that, like many Hsp70s, DnaK self-associates into low ordered oligomers, and binding of peptide substrates dissociates the oligomers into monomers (46, 55). Consistent with these observations, DnaK by itself ran as a ladder on native gel, and adding either NR or p12 peptides shifted some of the oligomeric species into monomers (Fig. 4D, right panel). When DnaK and fluorescein-labeled NR were mixed in a 1:1 ratio, ∼55% of NR co-migrated with DnaK monomer on the native gel (Fig. 4D), indicating a stable complex formed between DnaK and NR. Although Sse1 has an affinity for TRP2 similar to that of DnaK for NR in the above fluorescence polarization assays, the amount of complex was much lower (only ∼3.5% of total TRP2 peptide used), and the majority of the TRP2 peptide was free peptide, suggesting that the Sse1-TRP2 complex is much less stable than the DnaK-NR complex. To further expand on this result, we used a longer TRP2 peptide with increased affinity for Sse1. We called this new peptide TRP2_181 because it starts at residue 181 in TRP2 protein. Peptide TRP2_181 has severalfold higher affinity than TRP2 peptide for all Hsp110s used in our assay (Table 1). For Sse1, TRP2_181 has an affinity similar to that of DnaK for the p12 peptide (Table 1). Consistent with the higher affinities, the amount of chaperone-peptide complexes increased for both DnaK-p12 and Sse1-TRP2_181. Now, almost all of the p12 is bound to DnaK (∼90% of total peptide used). In contrast, only about 25% TRP2_181 formed a complex with Sse1, which is much lower than the DnaK-p12 complex and even lower than the DnaK-NR complex. Hence, consistent with the kinetics data, Hsp110s bind peptide only transiently.
Peptide-binding Loops Govern Peptide Binding Properties in Hsp70s
Our experiments above suggested that Hsp110s have peptide substrate binding properties distinct from Hsp70s: a strong preference for aromatic side chains and exceptionally fast kinetics. These differences between Hsp70s and Hsp110s must be rooted in their primary sequences and three-dimensional structures. For Hsp70s, it is well established that the SBD is the site for peptide substrate binding (13, 14, 56). However, the peptide-binding site on Hsp110s is still controversial. Some studies suggest that, like Hsp70s, the SBD is the site for peptide substrate binding because the isolated SBD is sufficient for the holdase activity (41, 57). Other studies suggest that the peptide-binding site resides on a rift between the NBD and SBDβ (37, 58). The identification of TRP2 peptide as a high affinity peptide substrate for Hsp110s gave us an advantage to test directly whether the SBD is the peptide substrate-binding site. To determine the peptide-binding site on Hsp110s, we purified isolated SBD fragments from all three Hsp110s used in our studies. As a control, the SBD from DnaK was purified, and its peptide substrate binding was determined. As shown in Fig. 5A, the DnaK SBD bound peptide substrates with essentially the same affinity as the full-length DnaK protein. Like DnaK, the SBDs from Sse2 and hHsp110 bound the TRP2 peptide with an affinity similar to that of the full-length protein (Fig. 5A for Sse2 SBD and supplemental Fig. S3A for hHsp110 SBD), suggesting that in Sse2 and hHsp110 the peptide-binding site is on the SBD as in Hsp70s. Surprisingly, the SBD of Sse1 showed little binding to any peptide tested, including TRP2 and TRP2_181 peptide (supplemental Fig. S3B). Thus, it is possible that the SBD of Sse1 is essential but not sufficient to bind peptide substrates independently as suggested previously (37). The NBD may be required to stabilize the conformation of the SBD. Taken together, most likely, Hsp110s possess peptide-binding sites analogous to those of DnaK.
FIGURE 5.
Peptide-binding loops determine peptide binding properties in Hsp70. A, peptide substrate binding to the isolated SBDs. The peptide binding of DnaK SBD (left) and Sse2 SBD (right) were measured with fluorescence anisotropy assays. The peptides are TRP2 (red circle), p53 (blue diamond), p12 (green triangle), and NR (black circle). B, sequence alignment of Loop1,2 (L1,2) and Loop3,4 (L3,4) regions from a number of Hsp110s and Hsp70s. The hydrophobic residues that form direct contacts with the bound NR peptide in the DnaK crystal structure are highlighted in red. C, ribbon diagrams of the SBDβ structures from Sse1 (left) and DnaK (right), respectively, with the β strands aligned. Loop1,2 (L1,2) and Loop3,4 (L3,4) are highlighted in red and purple, respectively. The NR peptide in DnaK is in cyan. D, the loop-swapping mutation in DnaK (DnaK_Loop) changed the peptide substrate binding properties. The binding of WT DnaK (black circle) and DnaK_Loop mutant protein (red diamond) to NR (left) and TRP2_181 (right) peptides was determined after binding reached equilibrium. E, swapping the peptide-binding loops or mutating the peptide binding cleft has little effect on the peptide binding of Sse1. The binding of the NR peptide (left) and the TRP2_181 peptide (right) to Sse1_Loop (red diamond) and Sse1_P (green triangle) mutants was determined and compared with that of the WT Sse1 (black circle). F, sequence alignment of peptide binding cleft residues that form hydrophobic interactions with the NR peptide in the structure of isolated DnaK SBD. mP, millipolarization level.
Sequence alignments of the SBDs from a number of Hsp70s and Hsp110s showed that Hsp110s bear significant differences from Hsp70s in their peptide substrate-binding loops (Fig. 5B) (22). As shown in the structure of the SBD of DnaK in complex with the NR peptide, NR peptide is bound between two peptide-binding loops, Loop1,2 and Loop3,4 (Fig. 5C) (13). One hydrophobic residue (highlighted in red) from each loop forms a direct contact with the bound peptide (Fig. 5B). However, in Hsp110s, the length and characteristics of these two loops are switched. The difference between the two loops is apparent when examining our previously published Sse1-ATP structure (Fig. 5C). Differences in the peptide-binding loops could be the structural basis for the different peptide substrate binding properties between Hsp70s and Hsp110s. To test this hypothesis, we swapped the peptide-binding loops in both DnaK and Sse1 to generate DnaK_Loop and Sse1_Loop mutants, respectively, and then analyzed their peptide substrate binding properties. To construct the DnaK_Loop mutant, we switched the sequences of Loop1,2 and Loop3,4 in DnaK. Thus, the mutated Loop1,2 has the sequence of the WT Loop3,4 (TAEDNQS), and the mutated Loop3,4 has the WT Loop1,2 sequence (MGG). For the Sse1_Loop mutant, the cloning to switch its own loops turned out to be difficult. Thus, we used the sequences from DnaK to replace the corresponding loops with MGG replacing DKQVEDE for Loop1,2 and TAEDNQSA replacing TGD for Loop3,4.
For the DnaK_Loop mutant, the binding to the NR peptide was almost completely abolished; in contrast, the binding affinity to the TRP2_181 peptide was increased by severalfold (Fig. 5D and Table 3). Therefore, consistent with our hypothesis, this loop-swapping DnaK mutant behaves like Hsp110s in terms of peptide substrate binding. Furthermore, when either Loop1,2 or Loop3,4 was swapped individually, the resulting mutants, DnaK_Loop12 and DnaK_Loop34, both showed reduced affinity for the NR peptide and increased affinity for the TRP2_181 peptide (Table 3 and supplemental Fig. S4) with the change being more pronounced for the DnaK_Loop34 mutant. Thus, the peptide substrate binding specificity change in the DnaK_Loop mutant was mainly due to the change of Loop3,4, although both loops contributed.
TABLE 3.
Binding affinities of DnaK and Sse1 mutants
NB, no binding detected.
| Protein |
Kd of peptide substrates |
|
|---|---|---|
| NR | TRP2_181 | |
| μm | ||
| WT DnaK | 1.49 ± 0.05 | 6.27 ± 0.94 |
| DnaK_Loop | NB | 1.06 ± 0.04 |
| DnaK_Loop12 | 6.20 ± 0.25 | 4.42 ± 0.16 |
| DnaK_Loop34 | NB | 1.47 ± 0.07 |
| WT Sse1 | NB | 0.30 ± 0.007 |
| Sse1_Loop | NB | 0.31 ± 0.009 |
| Sse1_P | NB | 0.25 ± 0.006 |
In contrast to these results, we detected little change in the peptide substrate binding for the Sse1_Loop mutant (Fig. 5E and Table 3). Moreover, we obtained identical results for hHsp110 when we switched the peptide-binding loops as in the DnaK_Loop mutant (data not shown). It is possible that in Hsp110s the loops alone are not sufficient to control the peptide binding specificity. Thus, we proceeded to test the peptide binding cleft. As shown in the structure of the DnaK SBD (13), a number of residues in the peptide binding cleft form hydrophobic interactions with the bound NR peptide (Fig. 5, C and F). We changed all the corresponding residues in Sse1 to those of DnaK to construct the Sse1_P mutant and performed a peptide binding assay. Surprisingly, little change in peptide substrate binding was observed for this mutant (Fig. 5E). One possible explanation is that these residues are conserved enough between Hsp70s and Hsp110s to account for specificity.
Peptide Binding Properties Determine Holdase and Foldase Activity in Hsp70
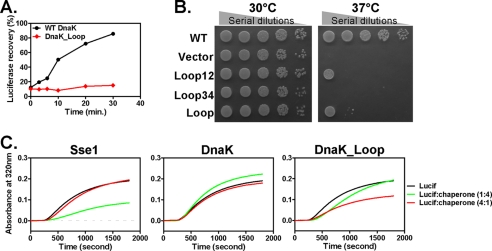
Our result that a mere swap of the peptide-binding loops changed the peptide substrate binding properties of DnaK to mimic those of Hsp110s raised the question of whether the biological activity of DnaK is changed accordingly. It is well established that unlike Hsp110s classic Hsp70s, such as DnaK, display folding activity (19, 20). In contrast, Hsp110s display a higher activity than Hsp70s in preventing protein aggregation. This is why Hsp70s are called foldases, whereas Hsp110s are called holdases. To test the folding activity of DnaK, we used a standard luciferase refolding assay. Firefly luciferase was used as a folding substrate and denatured by incubation at the elevated temperature of 42 °C. As shown in Fig. 6A, in the presence of WT DnaK, heat-denatured luciferase regained activity in a time-dependent manner. In contrast, we observed little recovery of luciferase activity for the DnaK_Loop mutant protein, suggesting that this mutant compromised the folding activity of DnaK almost completely. Consistent with the loss of folding activity, this mutant protein lost the in vivo chaperone activity in supporting growth at an elevated temperature of 37 °C (Fig. 6B). This result further confirmed that the folding activity of DnaK is essential for its in vivo chaperone activity. Moreover, swapping either peptide-binding loop compromised growth, supporting the importance of the peptide binding activity in DnaK function (Fig. 6B).
FIGURE 6.
Influence of loop swapping on chaperone activity of DnaK. A, the DnaK_Loop mutant lost folding activity. After heat denaturation, refolding of denatured firefly luciferase by WT DnaK (black circle) and DnaK_Loop mutant (red diamond) was monitored over time. B, mutating peptide-binding loops in DnaK resulted in loss of in vivo function. Serial dilutions of fresh E. coli cultures were spotted on LB agar plates with empty vector and WT DnaK as negative and positive controls, respectively. 30 °C growth was used as culture control. C, the DnaK_Loop mutant gained holdase activity. Holdase activity was assayed by analyzing the aggregation of firefly luciferase (Lucif) in the presence of various chaperones, Sse1 (left), WT DnaK (middle), and DnaK_Loop (right), at a 1:4 (green) and 4:1 (red) ratio to luciferase. The final concentration of luciferase was kept at 200 nm for all the reactions. The aggregation of luciferase by itself (black) was used as a no chaperone control.
To test whether the DnaK_Loop mutant displays increased activity in preventing protein aggregation, we performed a holdase assay using firefly luciferase as a model substrate (40, 41). Luciferase was heated to 41 °C, which causes it to unfold and aggregate. This aggregation results in increased apparent absorbance at wavelength 320 nm due to light scattering of protein aggregates. Neither DnaK nor Sse1 protein by itself showed any obvious aggregation in our assay (supplemental Fig. S5). As shown previously (41), when Sse1 was mixed with luciferase before heating, the aggregation was significantly reduced (Fig. 6C, left panel). However, when WT DnaK was incubated with luciferase, as expected, we did not observe significant holdase activity under our experimental condition (Fig. 6C, middle panel). Impressively, the DnaK_Loop mutant protein consistently showed much higher holdase activity than the WT DnaK in preventing the aggregation of luciferase (Fig. 6C, right panel), although it was less effective than Sse1 and showed optimal activity at a different ratio. The holdase activity correlates well with the peptide binding affinity toward the TRP2_181 peptide shown above. Therefore, the different peptide binding properties implemented in the peptide-binding loops between Hsp70 and Hsp110 are directly correlated with their biological activities.
DISCUSSION
Our study elucidates novel differences in substrate binding between two major classes of molecular chaperones, the Hsp70s and Hsp110s. We first showed that Hsp110s prefer aromatic residues in their substrates in contrast to Hsp70s, which prefer aliphatic residues. Second, Hsp110s show extremely fast kinetics for peptide substrate binding, even faster than Hsp70s in the ATP state. The sequence homology between hHsp110 and the two yeast Hsp110s Sse1 and Sse2 is low (38 and 37%, respectively), consistent with some functional differences between them (58, 59). However, their peptide binding properties are conserved, suggesting conservation in their function. More importantly, our study for the first time has provided mechanistic insights into the functional differences between Hsp70 and Hsp110. It has been well documented that Hsp110s serve as holdases, whereas the Hsp70s serve as foldases; however, little is known about the molecular basis of this difference. We were able to convert the peptide binding properties of DnaK to be Hsp110-like by swapping the peptide-binding loops. This conversion resulted in a loss of the foldase activity of DnaK and a gain in holdase activity, suggesting that the peptide substrate binding properties are a major determinant of the functional differences between the foldase and holdase activities. How do the peptide substrate preference and binding kinetics contribute to this functional difference? It is possible that aromatic residues are more hydrophobic and thus tend to aggregate more than aliphatic residues. Efficient binding to aromatic residues by Hsp110s will lead to efficient holdase activity. The exceptionally fast kinetics in Hsp110s are also consistent with the high holdase activity. Although the fast kinetics result in transient binding, we hypothesize that the fast peptide binding and release cycles give Hsp110s the competitive advantage in binding polypeptide substrates and preventing their aggregation. It is interesting to point out that the overall content of aromatic residues in eukaryotic proteins is about 8%, whereas the overall content of aliphatic residues is close to 30% (61). For efficient folding, it is conceivable that Hsp70s need to work on the majority of the hydrophobic residues, such as aliphatic residues. Hsp70s use the energy from ATP hydrolysis to power their folding activity (1). During the ATP hydrolysis, Hsp70s convert their fast kinetics and low affinity to slow kinetics and high affinity for peptide substrate binding. Although the affinity change is conserved in Hsp110s, the change in kinetics is missing, especially the slow kinetics and stable binding following ATP hydrolysis. Thus, it is tempting to postulate that during this transition Hsp70s generate some force for protein folding while binding to unfolded polypeptides.
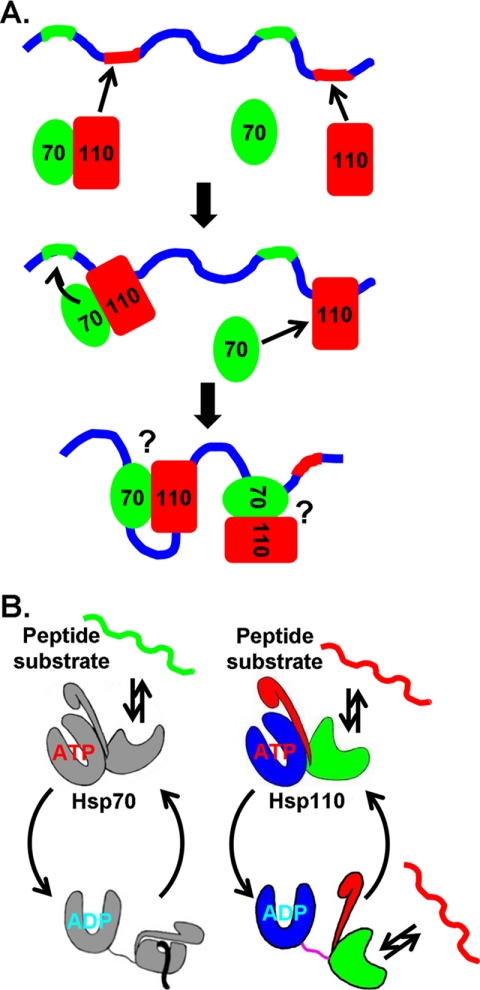
Hsp110 has recently been established as the major NEF for Hsp70 in the eukaryotic cytosol and has been shown to dramatically enhance the folding activity of the eukaryotic Hsp70 chaperone machinery (32, 40). Although other NEFs have been found for eukaryotic Hsp70s, such as Fes1 and HspBP1, they have failed to facilitate Hsp70-mediated protein folding (32). The major difference between Hsp110s and all the other classes of nucleotide exchange factors is that Hsp110s are also chaperones on their own. This chaperone activity of Hsp110s is likely to contribute to the efficient folding activity of the Hsp70-Hsp110 chaperone machinery (32, 33). The fast peptide binding kinetics in Hsp110s suggest that when both Hsp110s and Hsp70s encounter an unfolded polypeptide chain the Hsp110s will most likely first bind to segments rich in aromatic residues (Fig. 7A). This binding may facilitate the binding of Hsp70s to nearby aliphatic residue-rich segments by bringing Hsp70s in close vicinity through the Hsp70-Hsp110 interaction. Although both Hsp70s and Hsp110s could bind the same unfolded polypeptide chain at different segments, they may cooperate in protein folding. This proposed cooperative mechanism needs to be further investigated. At the same time, Hsp110s facilitate the nucleotide exchange in Hsp70s to speed up the chaperone activity of Hsp70. As pointed out above, the content of aromatic residues is far less than that of aliphatic residues in eukaryotic proteins; this correlates with the lower cellular level of Hsp110s than Hsp70s.
FIGURE 7.
Model of Hsp70-Hsp110 chaperone machinery. A, model of the cooperative actions of Hsp70 and Hsp110 in protein folding. Hsp70s and Hsp110s prefer different segments in polypeptide substrates. Based on this study, we speculate that the segments preferred by Hsp70s (highlighted in green) are rich in aliphatic residues, whereas the segments preferred by Hsp110s (highlighted in red) are rich in aromatic residues. B, comparison of the chaperone cycles of Hsp70 and Hsp110. Peptide substrates are bound to and released from Hsp70 in the ATP-bound state due to fast kinetics; whereas, in the ADP-bound state, Hsp70 holds the bound peptide stably. For Hsp110, peptide substrates are bound and released constantly regardless of the nucleotide-bound state.
What is the molecular basis for the different peptide substrate binding properties between Hsp70s and Hsp110s? The crystal structure of the DnaK SBD in complex with NR peptide offers a structural basis for its preference for aliphatic residues in substrates (13). Large aromatic side chains are unfavorable in this peptide binding pocket due to steric clash, which explains why DnaK and Ssa1 do not show high affinity for the TRP2 peptide even though the TRP2 peptide is highly hydrophobic. On the other hand, Hsp110s appear to have the opposite peptide preference, indicating that the peptide binding pocket is different from that of Hsp70s. Indeed, we observed significant differences in the two peptide-binding loops: their length and properties are switched. Interestingly, when we switched these two loops, we changed the peptide binding preference of DnaK into becoming Hsp110-like, suggesting that the peptide-binding loops govern peptide binding preference. However, when we swapped the peptide-binding loops in Sse1 and hHsp110, we observed little change in peptide preference. This result suggests that a mere swap of the loops is not sufficient (as discussed below).
How do the peptide-binding loops determine the peptide binding properties? In the DnaK SBD structure, one residue from each loop forms hydrophobic contacts with the peptide substrate (Fig. 5, B and C). Interestingly, these two residues are not conserved in classic Hsp70s. A previous mutational analysis suggested that they contribute to the substrate specificity (62). Their contribution is minor at best when one considers that DnaK and Ssa1 have almost identical peptide substrate preference despite the two residues being completely different. Therefore, these residues could only partially, if at all, account for the dramatic effects observed in the loop-swapping DnaK mutant in our study. Furthermore, SBDα covers up the peptide binding pocket through interactions with the tips of both peptide-binding loops (Fig. 1E). If we swap these two loops in Hsp70s, the contacts between SBDα and the tips of these two loops will be lost, although the peptide binding pocket still remains hydrophobic. Thus, SBDα may not be able to cover up the peptide binding pocket appropriately. Moreover, this new Loop1,2 is larger than normal; hence, it may further prevent SBDα from covering the peptide binding pocket. This change would leave the peptide binding pocket more open. Without the lid and with a much shorter Loop3,4, the peptide binding pocket may be more flexible to accommodate the larger aromatic residues, a prediction that is consistent with the fast kinetics in Hsp110s and the DnaK loop mutant. This hypothesis is further supported by a number of observations that disrupting the contacts between SBDα and the peptide-binding loops resulted in lower peptide binding affinity and faster kinetics when classic peptides for Hsp70s were used (63–66). In the cases of Sse1 and hHsp110, loop swapping showed little change in peptide binding specificity. One explanation might be that when we just swapped the peptide-binding loops most likely we were not able to restore the interaction between the SBDα and peptide-binding loops for SBDα to cover the peptide binding pocket appropriately. Taken together, SBDα in Hsp110s may never cover the peptide binding pocket as in Hsp70s (Fig. 7B). Furthermore, this open peptide binding pocket in Hsp110s may also explain the difference of Hsp70s and Hsp110s in presenting tumor antigens: Hsp110s can bind and present large tumor antigens, whereas Hsp70s mainly presents small peptides (67). Consistent with our hypothesis, two recent studies suggest that when Hsp70s bind to native substrates the SBDα does not close onto SBDβ like it does in the DnaK SBD structure (60, 68). Alternatively, peptide substrates may bind Hsp110s at a different site in the SBD because mutating either the peptide-binding loops or the peptide binding cleft has little influence in peptide substrate binding. A crystal structure of an Hsp110 in complex with peptide substrates will ultimately reveal the molecular basis for their special peptide substrate binding properties.
It is also interesting to point out our finding that ATP binding reduced peptide binding affinity in Hsp110s just like in Hsp70s, suggesting that Hsp110s also undergo similar allosteric coupling. This observation is consistent with our previous limited proteolysis analysis and another study using tryptophan fluorescence (22, 59). However, an earlier study showed that the binding of Sse1 to another high affinity peptide is not ATP-sensitive (45). This discrepancy could be due to the fact that different peptide substrates were used. Another possible difference is that our Sse1 protein was highly purified. The Sse1 protein by itself did not exhibit ATPase activity (supplemental Fig. S6), which is consistent with previous crystallographic analyses (22, 37). In contrast, the Sse1 protein used in the other study has an ATPase activity close to Ssa1, which raises the possibility of an Ssa1 contamination in the protein preparation used. Furthermore, in our previous mutational analysis based on the Sse1 structure, we isolated a number of Sse1 mutations on the interdomain interfaces of the ATP state (22). We purified one mutant protein with a strong in vivo defect, and we found that the ATP-induced decrease in peptide binding affinity was abolished in this mutant protein (data not shown), suggesting the importance of this allosteric coupling in Hsp110s. Future studies will need to characterize the mechanism and role of this allosteric coupling.
Supplementary Material
Acknowledgments
We thank Drs. Elizabeth Craig, Wayne Hendrickson, Diomedes Logothetis, Gea-Ny Tseng, Jason Rife, and Catherine Fox for critically reading the manuscript and providing insightful suggestions. We are grateful to Dr. Johannes Buchner for providing the Ssa1-expressing Pichia strain. We thank Dr. Vanessa Ledesma for technical support. We thank Dr. Andrew Larner for the UV-visible spectrophotometer.
This work was supported by startup funds from the Virginia Commonwealth University School of Medicine (to Q. L.), a New Scholar Award in Aging from the Ellison Medical Foundation (to Q. L.) and a grant-in-aid award from the American Heart Association (to Q. L.).

This article contains supplemental Figs. S1–S6.
- Hsp
- heat-shock protein
- NBD
- nucleotide-binding domain
- SBD
- substrate-binding domain
- NEF
- nucleotide exchange factor
- hHsp110
- human Hsp110
- TRP2
- tyrosinase-related protein 2.
REFERENCES
- 1. Mayer M. P., Bukau B. (2005) Hsp70 chaperones: cellular functions and molecular mechanism. Cell. Mol. Life Sci. 62, 670–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bukau B., Weissman J., Horwich A. (2006) Molecular chaperones and protein quality control. Cell 125, 443–451 [DOI] [PubMed] [Google Scholar]
- 3. Hartl F. U., Hayer-Hartl M. (2009) Converging concepts of protein folding in vitro and in vivo. Nat. Struct. Mol. Biol. 16, 574–581 [DOI] [PubMed] [Google Scholar]
- 4. Evans C. G., Chang L., Gestwicki J. E. (2010) Heat shock protein 70 (hsp70) as an emerging drug target. J. Med. Chem. 53, 4585–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mayer M. P. (2010) Gymnastics of molecular chaperones. Mol. Cell 39, 321–331 [DOI] [PubMed] [Google Scholar]
- 6. Bukau B., Horwich A. L. (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92, 351–366 [DOI] [PubMed] [Google Scholar]
- 7. Flaherty K. M., DeLuca-Flaherty C., McKay D. B. (1990) Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature 346, 623–628 [DOI] [PubMed] [Google Scholar]
- 8. Sriram M., Osipiuk J., Freeman B., Morimoto R., Joachimiak A. (1997) Human Hsp70 molecular chaperone binds two calcium ions within the ATPase domain. Structure 5, 403–414 [DOI] [PubMed] [Google Scholar]
- 9. Wisniewska M., Karlberg T., Lehtiö L., Johansson I., Kotenyova T., Moche M., Schüler H. (2010) Crystal structures of the ATPase domains of four human Hsp70 isoforms: HSPA1L/Hsp70-hom, HSPA2/Hsp70-2, HSPA6/Hsp70B′, and HSPA5/BiP/GRP78. PLoS One 5, e8625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jiang J., Maes E. G., Taylor A. B., Wang L., Hinck A. P., Lafer E. M., Sousa R. (2007) Structural basis of J cochaperone binding and regulation of Hsp70. Mol. Cell 28, 422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Rüdiger S., Germeroth L., Schneider-Mergener J., Bukau B. (1997) Substrate specificity of the DnaK chaperone determined by screening cellulose-bound peptide libraries. EMBO J. 16, 1501–1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Blond-Elguindi S., Cwirla S. E., Dower W. J., Lipshutz R. J., Sprang S. R., Sambrook J. F., Gething M. J. (1993) Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell 75, 717–728 [DOI] [PubMed] [Google Scholar]
- 13. Zhu X., Zhao X., Burkholder W. F., Gragerov A., Ogata C. M., Gottesman M. E., Hendrickson W. A. (1996) Structural analysis of substrate binding by the molecular chaperone DnaK. Science 272, 1606–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gragerov A., Zeng L., Zhao X., Burkholder W., Gottesman M. E. (1994) Specificity of DnaK-peptide binding. J. Mol. Biol. 235, 848–854 [DOI] [PubMed] [Google Scholar]
- 15. Schmid D., Baici A., Gehring H., Christen P. (1994) Kinetics of molecular chaperone action. Science 263, 971–973 [DOI] [PubMed] [Google Scholar]
- 16. Flynn G. C., Chappell T. G., Rothman J. E. (1989) Peptide binding and release by proteins implicated as catalysts of protein assembly. Science 245, 385–390 [DOI] [PubMed] [Google Scholar]
- 17. Hartl F. U., Hayer-Hartl M. (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295, 1852–1858 [DOI] [PubMed] [Google Scholar]
- 18. Kampinga H. H., Craig E. A. (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11, 579–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Easton D. P., Kaneko Y., Subjeck J. R. (2000) The hsp110 and Grp1 70 stress proteins: newly recognized relatives of the Hsp70s. Cell Stress Chaperones 5, 276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Shaner L., Morano K. A. (2007) All in the family: atypical Hsp70 chaperones are conserved modulators of Hsp70 activity. Cell Stress Chaperones 12, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shaner L., Trott A., Goeckeler J. L., Brodsky J. L., Morano K. A. (2004) The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 279, 21992–22001 [DOI] [PubMed] [Google Scholar]
- 22. Liu Q., Hendrickson W. A. (2007) Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell 131, 106–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mukai H., Kuno T., Tanaka H., Hirata D., Miyakawa T., Tanaka C. (1993) Isolation and characterization of SSE1 and SSE2, new members of the yeast HSP70 multigene family. Gene 132, 57–66 [DOI] [PubMed] [Google Scholar]
- 24. Yam A. Y., Albanèse V., Lin H. T., Frydman J. (2005) Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280, 41252–41261 [DOI] [PubMed] [Google Scholar]
- 25. Shaner L., Wegele H., Buchner J., Morano K. A. (2005) The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280, 41262–41269 [DOI] [PubMed] [Google Scholar]
- 26. Albanèse V., Yam A. Y., Baughman J., Parnot C., Frydman J. (2006) Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell 124, 75–88 [DOI] [PubMed] [Google Scholar]
- 27. Liu X. D., Morano K. A., Thiele D. J. (1999) The yeast Hsp110 family member, Sse1, is an Hsp90 cochaperone. J. Biol. Chem. 274, 26654–26660 [DOI] [PubMed] [Google Scholar]
- 28. Mandal A. K., Gibney P. A., Nillegoda N. B., Theodoraki M. A., Caplan A. J., Morano K. A. (2010) Hsp110 chaperones control client fate determination in the hsp70-Hsp90 chaperone system. Mol. Biol. Cell 21, 1439–1448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fan Q., Park K. W., Du Z., Morano K. A., Li L. (2007) The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics 177, 1583–1593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kryndushkin D., Wickner R. B. (2007) Nucleotide exchange factors for Hsp70s are required for [URE3] prion propagation in Saccharomyces cerevisiae. Mol. Biol. Cell 18, 2149–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sadlish H., Rampelt H., Shorter J., Wegrzyn R. D., Andréasson C., Lindquist S., Bukau B. (2008) Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS One 3, e1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Dragovic Z., Broadley S. A., Shomura Y., Bracher A., Hartl F. U. (2006) Molecular chaperones of the Hsp110 family act as nucleotide exchange factors of Hsp70s. EMBO J. 25, 2519–2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Raviol H., Sadlish H., Rodriguez F., Mayer M. P., Bukau B. (2006) Chaperone network in the yeast cytosol: Hsp110 is revealed as an Hsp70 nucleotide exchange factor. EMBO J. 25, 2510–2518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang X. Y., Chen X., Oh H. J., Repasky E., Kazim L., Subjeck J. (2000) Characterization of native interaction of hsp110 with hsp25 and hsc70. FEBS Lett. 465, 98–102 [DOI] [PubMed] [Google Scholar]
- 35. Hatayama T., Yasuda K. (1998) Association of HSP105 with HSC70 in high molecular mass complexes in mouse FM3A cells. Biochem. Biophys. Res. Commun. 248, 395–401 [DOI] [PubMed] [Google Scholar]
- 36. Schuermann J. P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L. E., Jin S., Taylor A. B., Demeler B., Morano K. A., Hart P. J., Valpuesta J. M., Lafer E. M., Sousa R. (2008) Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell 31, 232–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Polier S., Dragovic Z., Hartl F. U., Bracher A. (2008) Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell 133, 1068–1079 [DOI] [PubMed] [Google Scholar]
- 38. Andréasson C., Fiaux J., Rampelt H., Druffel-Augustin S., Bukau B. (2008) Insights into the structural dynamics of the Hsp110-Hsp70 interaction reveal the mechanism for nucleotide exchange activity. Proc. Natl. Acad. Sci. U.S.A. 105, 16519–16524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Andréasson C., Fiaux J., Rampelt H., Mayer M. P., Bukau B. (2008) Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283, 8877–8884 [DOI] [PubMed] [Google Scholar]
- 40. Oh H. J., Chen X., Subjeck J. R. (1997) Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J. Biol. Chem. 272, 31636–31640 [DOI] [PubMed] [Google Scholar]
- 41. Goeckeler J. L., Stephens A., Lee P., Caplan A. J., Brodsky J. L. (2002) Overexpression of yeast Hsp110 homolog Sse1p suppresses ydj1-151 thermosensitivity and restores Hsp90-dependent activity. Mol. Biol. Cell 13, 2760–2770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hrizo S. L., Gusarova V., Habiel D. M., Goeckeler J. L., Fisher E. A., Brodsky J. L. (2007) The Hsp110 molecular chaperone stabilizes apolipoprotein B from endoplasmic reticulum-associated degradation (ERAD). J. Biol. Chem. 282, 32665–32675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Manjili M. H., Henderson R., Wang X. Y., Chen X., Li Y., Repasky E., Kazim L., Subjeck J. R. (2002) Development of a recombinant HSP110-HER-2/neu vaccine using the chaperoning properties of HSP110. Cancer Res. 62, 1737–1742 [PubMed] [Google Scholar]
- 44. Wang X. Y., Kazim L., Repasky E. A., Subjeck J. R. (2001) Characterization of heat shock protein 110 and glucose-regulated protein 170 as cancer vaccines and the effect of fever-range hyperthermia on vaccine activity. J. Immunol. 166, 490–497 [DOI] [PubMed] [Google Scholar]
- 45. Goeckeler J. L., Petruso A. P., Aguirre J., Clement C. C., Chiosis G., Brodsky J. L. (2008) The yeast Hsp110, Sse1p, exhibits high-affinity peptide binding. FEBS Lett. 582, 2393–2396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Burkholder W. F., Zhao X., Zhu X., Hendrickson W. A., Gragerov A., Gottesman M. E. (1996) Mutations in the C-terminal fragment of DnaK affecting peptide binding. Proc. Natl. Acad. Sci. U.S.A. 93, 10632–10637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wegele H., Haslbeck M., Buchner J. (2003) Recombinant expression and purification of Ssa1p (Hsp70) from Saccharomyces cerevisiae using Pichia pastoris. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 786, 109–115 [DOI] [PubMed] [Google Scholar]
- 48. Schmid D., Jaussi R., Christen P. (1992) Precursor of mitochondrial aspartate aminotransferase synthesized in Escherichia coli is complexed with heat-shock protein DnaK. Eur. J. Biochem. 208, 699–704 [DOI] [PubMed] [Google Scholar]
- 49. Kaul S. C., Aida S., Yaguchi T., Kaur K., Wadhwa R. (2005) Activation of wild type p53 function by its mortalin-binding, cytoplasmically localizing carboxyl terminus peptides. J. Biol. Chem. 280, 39373–39379 [DOI] [PubMed] [Google Scholar]
- 50. Parkhurst M. R., Fitzgerald E. B., Southwood S., Sette A., Rosenberg S. A., Kawakami Y. (1998) Identification of a shared HLA-A*0201-restricted T-cell epitope from the melanoma antigen tyrosinase-related protein 2 (TRP2). Cancer Res. 58, 4895–4901 [PubMed] [Google Scholar]
- 51. Wang X. Y., Sun X., Chen X., Facciponte J., Repasky E. A., Kane J., Subjeck J. R. (2010) Superior antitumor response induced by large stress protein chaperoned protein antigen compared with peptide antigen. J. Immunol. 184, 6309–6319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pfund C., Huang P., Lopez-Hoyo N., Craig E. A. (2001) Divergent functional properties of the ribosome-associated molecular chaperone Ssb compared with other Hsp70s. Mol. Biol. Cell 12, 3773–3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Montgomery D. L., Morimoto R. I., Gierasch L. M. (1999) Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J. Mol. Biol. 286, 915–932 [DOI] [PubMed] [Google Scholar]
- 54. Liu Q., Krzewska J., Liberek K., Craig E. A. (2001) Mitochondrial Hsp70 Ssc1: role in protein folding. J. Biol. Chem. 276, 6112–6118 [DOI] [PubMed] [Google Scholar]
- 55. Shi L., Kataoka M., Fink A. L. (1996) Conformational characterization of DnaK and its complexes by small-angle x-ray scattering. Biochemistry 35, 3297–3308 [DOI] [PubMed] [Google Scholar]
- 56. Wang T. F., Chang J. H., Wang C. (1993) Identification of the peptide binding domain of hsc70. 18-Kilodalton fragment located immediately after ATPase domain is sufficient for high affinity binding. J. Biol. Chem. 268, 26049–26051 [PubMed] [Google Scholar]
- 57. Oh H. J., Easton D., Murawski M., Kaneko Y., Subjeck J. R. (1999) The chaperoning activity of hsp110. Identification of functional domains by use of targeted deletions. J. Biol. Chem. 274, 15712–15718 [DOI] [PubMed] [Google Scholar]
- 58. Polier S., Hartl F. U., Bracher A. (2010) Interaction of the Hsp110 molecular chaperones from S. cerevisiae with substrate protein. J. Mol. Biol. 401, 696–707 [DOI] [PubMed] [Google Scholar]
- 59. Raviol H., Bukau B., Mayer M. P. (2006) Human and yeast Hsp110 chaperones exhibit functional differences. FEBS Lett. 580, 168–174 [DOI] [PubMed] [Google Scholar]
- 60. Marcinowski M., Höller M., Feige M. J., Baerend D., Lamb D. C., Buchner J. (2011) Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat. Struct. Mol. Biol. 18, 150–158 [DOI] [PubMed] [Google Scholar]
- 61. Bogatyreva N. S., Finkelstein A. V., Galzitskaya O. V. (2006) Trend of amino acid composition of proteins of different taxa. J. Bioinform. Comput. Biol. 4, 597–608 [DOI] [PubMed] [Google Scholar]
- 62. Rüdiger S., Mayer M. P., Schneider-Mergener J., Bukau B. (2000) Modulation of substrate specificity of the DnaK chaperone by alteration of a hydrophobic arch. J. Mol. Biol. 304, 245–251 [DOI] [PubMed] [Google Scholar]
- 63. Mayer M. P., Schröder H., Rudiger S., Paal K., Laufen T., Bukau B. (2000) Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat. Struct. Biol. 7, 586–593 [DOI] [PubMed] [Google Scholar]
- 64. Fernández-Sáiz V., Moro F., Arizmendi J. M., Acebrón S. P., Muga A. (2006) Ionic contacts at DnaK substrate binding domain involved in the allosteric regulation of lid dynamics. J. Biol. Chem. 281, 7479–7488 [DOI] [PubMed] [Google Scholar]
- 65. Moro F., Fernández-Sáiz V., Muga A. (2004) The lid subdomain of DnaK is required for the stabilization of the substrate-binding site. J. Biol. Chem. 279, 19600–19606 [DOI] [PubMed] [Google Scholar]
- 66. Buczynski G., Slepenkov S. V., Sehorn M. G., Witt S. N. (2001) Characterization of a lidless form of the molecular chaperone DnaK: deletion of the lid increases peptide on- and off-rate constants. J. Biol. Chem. 276, 27231–27236 [DOI] [PubMed] [Google Scholar]
- 67. Wang X. Y., Li Y., Yang G., Subjeck J. R. (2005) Current ideas about applications of heat shock proteins in vaccine design and immunotherapy. Int. J. Hyperthermia 21, 717–722 [DOI] [PubMed] [Google Scholar]
- 68. Schlecht R., Erbse A. H., Bukau B., Mayer M. P. (2011) Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat. Struct. Mol. Biol. 18, 345–351 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.