Background: DNA sequences called CRMs determine the precise patterns of gene expression.
Results: We identify and characterize the function of six novel hes1 CRMs.
Conclusion: HES1 expression is controlled by multiple distal CRMs in addition to the known promoter.
Significance: A powerful combination of computational and experimental methodologies enhances our knowledge of hes1 transcriptional control.
Keywords: Chromatin Immunoprecipitation (ChiP), DNA-Protein Interaction, Notch, Transcription Enhancers, Transcription Factors, Transcription Regulation, Transcription/Developmental Factors, Transfection, Comparative Genomics
Abstract
The expression profile of a gene is controlled by DNA sequences called cis-regulatory modules (CRMs). CRMs can function over large genomic distances and can be located many kilobases away from their target promoters. hes1 is a key developmental gene that is overexpressed in certain cancers and is a primary target of NOTCH signaling. Despite this, analysis of hes1 transcriptional control has been limited solely to its promoter. Here, we identify seven conserved DNA sequence blocks, representing the hes1 promoter and six novel CRMs, within 57 kb upstream of the mouse hes1 gene. We identify 12 binding sites for the RBP-Jκ NOTCH effector and a single M-CAT motif within these regions. We validate RBP-Jκ and TEAD family occupancy in cells in culture and test the response of each of these CRMs to active NOTCH. We show that two regions, CRM5 and CRM7, function as enhancers, and four can repress transcription. A pair of RBP-Jκ motifs arranged in a tail-tail configuration in CRM5 and the M-CAT motif in CRM7 are necessary for enhancer function. Furthermore, these enhancers are occupied by transcriptional co-activators and loop onto the hes1 promoter within the endogenous hes1 locus. This work demonstrates the power of combining computational genomics and experimental methodologies to identify novel CRMs and characterize their function.
Introduction
The Hairy and Enhancer of Split-1 (HES1) transcriptional repressor is a member of the HES family of basic helix loop helix transcription factors, which contains seven members in the mouse (HES1–7). HES1 plays an important role in the control of cellular proliferation and differentiation during development. This is due, at least in part, to its ability to block the activity of positively acting basic helix loop helix factors involved in lineage determination, such as MASH1 during the process of neurogenesis (1). However, HES1 can also directly interact with co-repressor proteins, such as GROUCHO/TLE/GRG and histone deacetylases, to actively repress target gene expression (1–5). Indeed, HES1 is expressed in most undifferentiated cell types in the developing mouse embryo, and hes1−/− mutant mice show premature differentiation, progenitor cell depletion, and a consequent lethality (reviewed in Ref. 6).
HES1 is an essential effector of activated NOTCH. The NOTCH pathway has many functions during development and in the adult and has been implicated in the control of cell fate decisions and the maintenance of progenitor cell identity. The molecular mechanism of NOTCH signaling via the canonical pathway is well understood. Upon NOTCH receptor activation by binding to the DELTA/JAGGED ligand expressed on adjacent cells, the NOTCH intracellular domain (NICD)2 is cleaved and translocates to the nucleus where it interacts with the RBP-Jκ transcription factor. This leads to dissociation of co-repressor proteins from RBP-Jκ, and recruitment of transcriptional co-activators such as MASTERMIND, p300, and PCAF and the subsequent activation of RBP-Jκ target genes, including hes1. Consistent with this, the hes1 proximal promoter contains two functional RPB-JK-binding sites (BSs), which have been shown to play a role in mediating the response to NOTCH (6–9).
NOTCH activation induces an oscillating expression of HES1 with a periodicity of ∼1.5–3 h in the presomitic mesoderm in the mouse embryo and in a variety of cell types such as myoblasts, fibroblasts, and neural progenitor cells (10–12). These oscillations are regulated by a negative feedback loop in which HES1 represses its own expression by binding to three copies of its recognition motif, the N-box, in its proximal promoter. Oscillatory expression of HES1 plays a role in the control of somite segmentation and the maintenance of cell identity in a number of progenitor cell populations and has been implicated in embryonic stem (ES) cell fate choice. For example, sustained HES1 expression in neural progenitors inhibits both their proliferation and differentiation (12, 13).
HES1 expression can also be regulated by the Sonic hedgehog, c-Jun N-terminal kinase, transforming growth factor-α, extracellular signal-regulated kinase, and tumor necrosis factor-α pathways in a variety of different cell types (14–19). This suggests that multiple signaling inputs control HES1 expression in a context-dependent manner, and it raises the possibility that cross-talk between these pathways generates the correct spatio-temporal pattern of HES1 expression.
In addition to its role during development, HES1 has been implicated in the progression of cancer. Indeed, it has been proposed that a common function of HES1 is to protect cancer cells against differentiation-inducing signals, thus promoting proliferation (20). As such, HES1 overexpression has been detected in breast cancers, lung cancers, ovarian cancers, meningiomas, rhabdomyosarcomas, and medulloblastomas (21–27). Therefore, the identification of the DNA elements and proteins that control HES1 expression levels will enhance our understanding of the molecular pathways operating during development and in disease.
The precise expression pattern of a gene is determined by the interplay between DNA elements called CRMs, which include both enhancers and silencers, their target promoters, the structure of the surrounding chromatin, and the spatial organization of the locus in the nucleus. A CRM is typically several hundred base pairs long and contains BSs for multiple transcription factors. The transcriptional regulatory function of a CRM is therefore determined by the concentration of active interacting transcription factors expressed in a cell at a given time, whereas a single gene can be regulated by multiple CRMs that interact to determine the overall rate of transcription.
In this study, we use a combination of computational and experimental techniques to identify and characterize the function of novel mouse hes1 CRMs in C2C12 myoblasts. We discover seven phylogenetically conserved sequence blocks, including the proximal promoter, within 57 kb upstream of the hes1 transcription start site (TSS). We identify recognition motifs for the NOTCH effector RBP-Jκ in each of these CRMs and an M-CAT motif, a response element for the TEAD family of transcription factors, in the most distal region CRM7, and we verify RBP-Jκ and TEAD2 binding in cells in culture. We show that the newly identified conserved regions play a role in hes1 transcriptional control, and we provide evidence that the RPB-JK-bound CRMs are targets of the NOTCH signaling pathway. Two of the hes1 CRMs, CRM5 and CRM7, function as enhancers, and we demonstrate that the RPB-JK and M-CAT motifs are necessary for this activity. Consistent with an enhancer function, these regions are occupied by transcriptional co-activators in C2C12 cells and are in close genomic proximity to the hes1 promoter in the nucleus.
EXPERIMENTAL PROCEDURES
Plasmid Constructs
pGL3-Venus-NLS, pSV40-Venus-NLS, and pCMV-mCherry-NLS vectors have been described previously (57). Amino acids 1744–2531 of NOTCH1 were PCR-amplified as an XbaI-BamHI fragment from mouse C2C12 cDNA and cloned into pcDNA3.1(−) to generate the NICD expression vector. Fluorescent reporters containing hes1 CRMs cloned upstream of either the SV40 or the endogenous hes1 promoter were generated as follows: the endogenous hes1 promoter sequence (−952 to +122 nucleotides relative to the TSS) was PCR-amplified from mouse genomic DNA and inserted into pGL3-Venus-NLS as a BglII-HindIII fragment to generate pHes1pro-Ven. Individual CRMs were then PCR-amplified as BglII-BamHI fragments (with the exception of CRM6, which was amplified as a BglII-BclI fragment) from mouse genomic DNA and cloned into the BglII site upstream of the hes1 promoter in pHes1pro-Ven or the SV40 promoter in pSV40-Venus-NLS. Mutation of the RBP-JK and M-CAT motifs in the pCRM5-Hes1pro-Ven and pCRM7-Hes1pro-Ven plasmids was carried out using the QuikChange site-directed mutagenesis kit (Stratagene) as described by the manufacturer. The fidelity of all constructs was verified by sequencing. Primer sequences are provided in the supplemental material.
Cells
C2C12 cells were grown in Dulbecco's modified Eagle's medium (DMEM) containing Glutamax without pyruvate and supplemented with 10% fetal bovine serum (FBS) at 37 °C and 5% CO2.
Flow Cytometry
1 × 105 cells per well were seeded in 6-well plates. The next day cells were transfected with 1 μg of reporter plasmid and 250 ng of pCMV-mCherry-NLS (to control for transfection efficiency) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Cells were incubated for 48 h, washed twice with PBS, trypsinized, and then harvested in 1 ml of growth medium. Cells were then washed twice with ice-cold PBS and resuspended in 800 μl of Cell Fix (BD Biosciences). Fluorescent reporter activity was measured in 10,000 transfected cells using the Partec CyFlow® Space-3-Laser 7-Color flow cytometry system with Partec FlowMax software (Partec GmbH). Cells were gated using forward scatter and side scatter to exclude cell debris and clumps. To quantify reporter activity, background fluorescence of untransfected cells was subtracted, and the results were normalized against mCherry. Each experiment was performed in triplicate.
Chromatin Immunoprecipitation (ChIP)
ChIP was performed as described previously (28). Cross-linked chromatin isolated from proliferating C2C12 cells was immunoprecipitated using antibodies against RBP-JK, p300, TEAD2, YAP1 (all Santa Cruz Biotechnology), NOTCH1, TEAD4 (Abcam), TEAD1 (BD Biosciences), and anti-mouse or anti-rabbit GFP antibodies as controls (BD Biosciences and Santa Cruz Biotechnology). We used PCR primers spanning the potential RBP-JK or M-CAT motifs within each CRM to amplify the immunoprecipitated DNA. Primers specific to a hes1 upstream genomic fragment where no RBP-JK- or TEAD-binding sites were detected in silico were used as a control. ChIP data were analyzed by semi-quantitative PCR ensuring that amplification was in the linear range. PCR products were separated on 1.5% agarose gels and quantified using the GeneTools software package (Syngene). Each ChIP experiment was performed at least in triplicate. ChIP primer sequences are provided in the supplemental material.
Chromosome Conformation Capture (3C)
3C experiments to determine the spatial organization of the hes1 locus were carried out using the procedure described previously (29). Approximately 1 × 107 C2C12 cells were trypsinized, resuspended in 12.5 ml of growth medium, and cross-linked using 2% formaldehyde for 10 min at room temperature. 0.125 m glycine was then added to stop the reaction. Nuclei were isolated by resuspending the cells in ice-cold lysis buffer and incubating for 90 min at 4 °C with stirring. The nuclei were pelleted and resuspended in 0.5 ml of 1.2× restriction enzyme buffer containing 0.3% SDS and incubated for 1 h at 37 °C with shaking. Triton X-100 was added to a final concentration of 2%, and the sample was incubated for a further 1 h at 37 °C with shaking. 400 units of BglII restriction enzyme was added, and the cross-linked DNA was digested overnight at 37 °C. 1.6% final concentration of SDS was added, and the sample was incubated for 20 min at 65 °C to inactivate the restriction enzyme. The digested DNA was then diluted to favor intramolecular ligations by the addition of 6.125 ml of 1.15× ligation buffer. 1% final concentration Triton X-100 was added, and the samples were incubated for 1 h at 37 °C. The DNA was then ligated for 4 h at 16 °C using 100 units of T4 ligase. 300 μg of proteinase K was added, and the samples were incubated overnight at 65 °C to reverse the cross-links. Samples were then treated with 300 μg of RNase A for 30 min at 37 °C, and the DNA was purified by phenol extraction and ethanol precipitation.
A bacterial artificial chromosome (BAC) clone (RP23 55P17) covering the hes1 locus was used to generate a control template. 3C ligation products were analyzed using TaqMan quantitative PCR (qPCR). 3C qPCR primers and probe were designed using Primer Express (Applied Biosystems). The sequences of these are shown in the supplemental material. Real time PCR using 400 ng of each 3C product was carried out using an ABI Prism 7000 sequence detection system operated by ABI PRISM 7000 software. All reactions were performed in triplicate. To generate the relative cross-linking frequency C2C12, 3C qPCR values were normalized to a GAPDH internal control and then expressed relative to the BAC. Results are presented as a mean value of three independent experiments.
Bioinformatics
Mouse hes1 CRMs were identified by comparing 140 kb of genomic sequence surrounding the mouse hes1 gene with orthologous sequences from human, rat, Xenopus, stickleback, Tetraodon, and fugu using the Regulatory Module Graphical User Interface (57). The Regulatory Module Graphical User Interface is a modified version of the Needleman-Wunsch alignment algorithm. To identify the RBP-JK and TEAD BSs, we searched position-specific scoring matrices (PSSMs) from the TRANSFAC data base and calculated the likelihood of factor binding using BiFa, a custom discovery tool (30).
RESULTS
Bioinformatic Identification of Putative hes1 cis-Regulatory Modules
Analysis of the DNA sequence elements controlling mouse hes1 expression has been restricted to the proximal promoter. We predicted that this small genomic fragment would not include the complete repertoire of hes1 transcriptional control regions as distal enhancer and silencer elements that operate over large genomic distances are widespread in the mammalian genome. Given that conservation of sequence has been used to identify functional regulatory regions for a growing number of genes, we applied comparative genomics to identify additional putative hes1 CRMs for biochemical and functional testing.
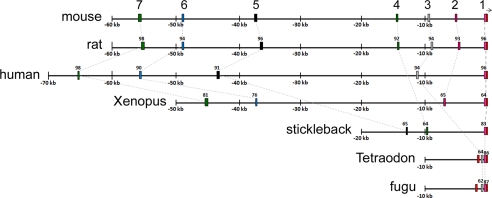
To do this, we used a bioinformatics algorithm3 to compare 140 kb of mouse genomic sequence surrounding the hes1 gene (Ensembl release 47) with orthologous sequences from human, rat, Xenopus, stickleback, Tetraodon, and fugu, a group that includes mammals as well as phylogenetically distant vertebrates. The algorithm uses a 100-bp sliding window to compare mouse sequences to other species in a stepwise manner and compute the percentage similarity. This analysis identified seven conserved DNA sequence blocks ranging from 190 to 495 bp lying within 57 kb upstream of the mouse hes1 TSS (Fig. 1). The complete sequence of the mouse CRMs are provided in the supplemental material. This included the previously identified hes1 core promoter and upstream sequence, annotated as CRM1 (8, 32), and an additional six regions of conservation (CRM2–7) that have not been characterized previously. CRM1 (265 bp) is highly conserved between all the analyzed species. CRM2 (300 bp), CRM6 (305 bp), and CRM7 (413 bp), located ∼4.5, 48, and 56 kb upstream of the hes1 TSS, are conserved down to amphibians, whereas CRM3 (290 bp), CRM4 (190 bp), and CRM5 (495 bp), located ∼9, 14.5, and 37 kb upstream of the hes1 TSS, are conserved between mammals and teleost fishes. We did not identify any conserved sequence blocks within the hes1 intronic sequence, although we only found a single block of ∼880 bp, conserved between mouse and amphibians within ∼50 kb of analyzed downstream hes1 sequence (data not shown). This region was not included for further analysis due to the lack of high probability RBP-JK BSs (see below).
FIGURE 1.
Identification of conserved sequence blocks in the hes1 upstream sequence. Schematic representation of the mouse hes1 upstream sequence and the orthologous regions from the different species used in the comparative analysis. Conserved sequences, identified using a Needleman-Wunsch optimal alignment algorithm, are shown as different colored boxes, and red boxes indicate exon sequence. Numbers above each box indicate percentage sequence similarity to mouse.
Hes1 Conserved Sequence Blocks Function as Transcriptional Regulatory Elements
HES1 plays an important role in the control of proliferation and lineage commitment in mouse C2C12 cells, a well characterized model of myogenic differentiation. HES1 is expressed in an oscillatory manner in proliferating C2C12 cells with a periodicity similar to that observed in the presomitic mesoderm (21), whereas up-regulation of HES1 in C2C12 myoblasts in response to active NOTCH can block myogenic differentiation (33). C2C12 cells therefore represent an ideal model system to study the mechanisms of hes1 transcriptional control.
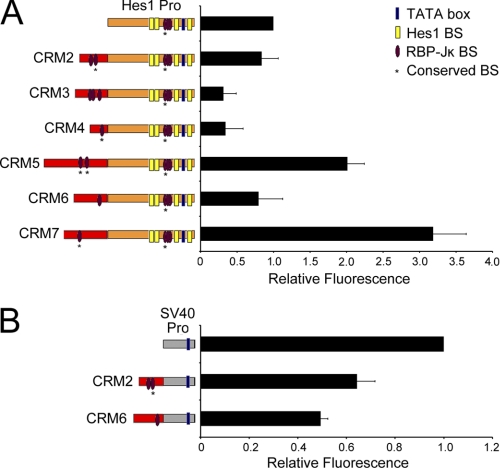
To characterize the transcriptional properties of the hes1 conserved sequence blocks, we generated a series of hes1 CRM promoter Venus reporter constructs (Fig. 2). Individual CRM-Pro constructs were then transiently transfected into C2C12 cells along with an mCherry expression vector to control for transfection efficiency. Venus fluorescence in transfected cells was then compared with that of the hes1 promoter alone using flow cytometry. The results in Fig. 2A show that CRM5 and CRM7 function as enhancers, whereas CRM3 and CRM4 function as repressors of the hes1 promoter in C2C12 cells. CRM5 enhances hes1 promoter activity ∼2-fold and CRM7 enhances 3.2-fold, whereas CRM3 and CRM4 both repress the hes1 promoter ∼3-fold. Although CRM2 and CRM6 had no effect on the hes1 promoter in this assay, it is possible that they may still play a role in the control of hes1 expression in a different cellular or developmental context. Consistent with this, we show that CRM2 and CRM6 can function to repress the activity of the strong heterologous SV40 promoter suggesting that these elements have the ability to regulate gene expression (Fig. 2B). A Hes1CRM2-SV40 reporter is ∼40% less active than the SV40 promoter alone, whereas a Hes1CRM6 reporter has roughly 50% the activity of the SV40 promoter. These assays therefore delineate either transcriptional enhancer or silencer functions for all of the hes1 conserved sequence blocks.
FIGURE 2.
hes1 conserved sequence blocks function as CRMs. A, regulation of the hes1 promoter by the newly identified CRMs. B, hes1 CRM2 and -6 repress the activity of the heterologous SV40 promoter. hes1 CRM-Pro reporter constructs (left) were transiently transfected into proliferating C2C12 cells along with an mCherry expression vector to control for transfection efficiency. Fluorescence activity was measured by flow cytometry. Venus activity was measured in 10,000 cells and normalized against mCherry activity. The data are displayed relative to the activity of the promoter construct alone (set as 1). The results are indicated as a mean value ± S.D. of three independent experiments.
Hes1 CRMs Are Bound by the RBP-JK and TEAD Transcription Factors
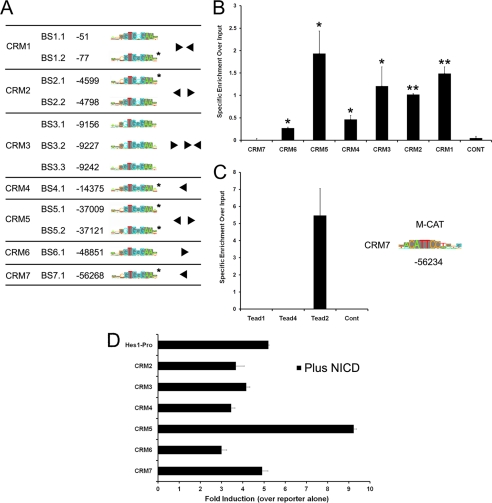
CRM function is mediated by the number and arrangement of different types of transcription factor- binding sites it contains. Given the important role played by NOTCH signaling in the control of hes1 expression, we analyzed the newly identified hes1 CRMs for RBP-JK BSs (Fig. 3A). RBP-JK is a major mediator of the NOTCH signaling pathway. To do this, we searched PSSMs from the TRANSFAC data base and calculated the likelihood of factor binding using BiFa, a custom discovery tool (30). This analysis identified the two known functional RBP-JK-binding sites in hes1 CRM1 validating our approach (7–9). Strikingly, the results also revealed an additional 10 high probability RBP-JK-binding sites within the newly identified mouse CRMs. The organization of these sites within each CRM as well as phylogenetic conservation and sequence similarity to the consensus high affinity PSSM is indicated in Fig. 3A. Our analysis did not predict any RBP-JK BSs with a high binding probability score in the intervening mouse genomic sequences. This suggests that these regions represent additional NOTCH-responsive transcriptional regulatory elements involved in the control of hes1 expression.
FIGURE 3.
hes1 CRMs are bound by the RBP-JK and TEAD2 transcription factors. A, identification of RBP-JK motifs in the hes1 CRMs using the BiFa discovery tool. The position of each RBP-JK motif relative to the hes1 TSS is shown. Sequences matching the consensus high affinity PSSM are marked in gray and the architecture of the RPB-JK BSs within each CRM is indicated. Conserved BSs are marked by an asterisk. B, RBP-JK binds to hes1 CRMs in proliferating C2C12 cells. ChIP assays were performed using either an antibody against RBP-JK or an isotype-specific anti-GFP control (n = 4). The precipitated DNA fragments were PCR-amplified using primers spanning the predicted RBP-JK BSs within each CRM. The control represents an intervening region in the hes1 upstream sequence in which no RBP-JK BSs were detected in silico. Analysis using an unpaired Student's t test shows that RBP-JK occupancy at CRM1–6 is statistically significant compared with a nonbinding control region. * indicates p < 0.05; ** indicates p < 0.01. C, position of the conserved M-CAT motif in hes1 CRM7 and sequence matches to the consensus PSSM are shown. ChIP assays were performed in proliferating C2C12 cells using anti-TEAD1, anti-TEAD2, anti-TEAD4, or anti-GFP (isotype control) antibodies. Precipitated DNA fragments were PCR-amplified using primers spanning the predicted M-CAT motif in CRM7. To calculate specific enrichment over input, the signal for each PCR was quantified and divided by the input, and the background intensity, measured using an IgG isotope control, was subtracted. D, modulation of hes1 CRM-pro reporter activity by active NOTCH. Hes1CRM-reporter constructs were co-transfected into C2C12 cells along with a 1:1 ratio of reporter to NICD overexpression vector or an empty control vector to make the total amount of DNA transfected in each case equal. The results are presented as fold induction relative to the activity of each reporter in the absence of NICD and represent a mean ± S.D. of three independent experiments.
We additionally identified a phylogenetically conserved M-CAT motif as the best hit in CRM7 that was not present in any of the other CRMs (Fig. 3C). This motif was included for further analysis as it is bound by the TEAD family of transcription factors, TEAD1–4, which are known to play important roles in the regulation of muscle-specific gene expression (34–36). In addition, TEAD transcriptional activity can be regulated by HIPPO signaling (37–39), thus implicating an additional NOTCH-independent pathway in the control of hes1 expression.
We carried out ChIP assays using chromatin from proliferating C2C12 cells to test whether RBP-JK and TEAD1, TEAD2 and TEAD4, but not TEAD3, as it is not expressed in C2C12 cells (40, 41), can bind to the endogenous hes1 CRMs as predicted. Cross-linked chromatin was immunoprecipitated using antibodies against RBP-JK, TEAD1, TEAD2, TEAD4, or a nonspecific isotype control, and we used specific PCR primers spanning the potential transcription factor-binding site motifs within each CRM to amplify the immunoprecipitated DNA.
Consistent with published data, the results in Fig. 3B show that RBP-JK binds CRM1. Additionally, we detected specific RBP-JK binding to all newly identified CRMs, except CRM7, compared with the IgG control. A low level of background signal was obtained using primers to amplify a randomly selected control region of the hes1 upstream sequence where no RBP-JK-binding sites were detected in silico. These results therefore verify RBP-JK binding to six of the seven regions that were predicted computationally. Moreover, RBP-JK occupancy is not uniform across these regions and appears higher at the CRMs containing multiple predicted RBP-JK BSs (Fig. 3B). For the TEAD analysis, the results indicate that TEAD2, but not TEAD1 and TEAD4, specifically binds to CRM7 in proliferating C2C12 cells (Fig. 3C). Furthermore, TEAD2 binding was not observed at a control region that does not contain an M-CAT motif. We therefore distinguish which specific member of the TEAD transcription factor family is interacting with the identified regulatory motif in cells in culture, a finding that could not be predicted using sequence analysis alone. Taken together, our bioinformatics and experimental approach has identified additional functional hes1 CRMs containing binding sites for the RBP-JK and TEAD2 transcription factors.
Hes1 CRM2–6 Are Targets for the NOTCH Signaling Pathway
As RBP-JK occupies six of the seven hes1 CRMs, we tested whether the activity of the hes1 CRM-pro reporters can be modulated by canonical NOTCH signaling. To do this, we assayed fold induction for each hes1 CRM-pro construct in response to NICD overexpression and compared this to the promoter alone. We used a 1:1 ratio of reporter to overexpression vector as titration experiments showed that this was in the range of the linear response for the promoter alone (data not shown). We first tested whether NICD overexpression can modulate the enhancer activity of CRM5 and CRM7 and showed that NICD induces hes1 promoter reporter activity ∼5-fold and the CRM5-pro construct 9.2-fold (Fig. 3D). This increased response of the CRM5-pro reporter to NICD overexpression indicates that the enhancer function of this region can be increased by active NOTCH. Furthermore, in agreement with our ChIP data showing that RBP-JK does not bind CRM7, the enhancer activity of the CRM7-pro reporter is not induced relative to the hes1 promoter in response to NICD (Fig. 3D). In addition, this analysis also revealed that the hes1 CRM-pro reporters containing the CRMs that have the ability to act as repressors, i.e. CRM2–4 and -6, display a reduced response to NICD overexpression compared with the promoter alone (Fig. 3D). This is consistent with the observation that a NOTCH-dependent repressive effect has to be derepressed before these elements can positively respond to active NOTCH. Together, these data suggest that, in addition to the hes1 promoter, hes1 CRM2–6 are also able to integrate signals from the NOTCH signaling pathway.
Contribution of RPB-JK and M-CAT Motifs to CRM5 and CRM7 Enhancer Function
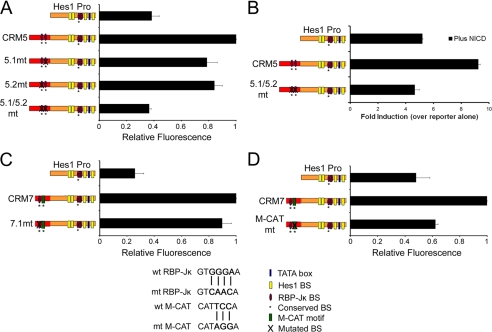
We next tested whether the RBP-JK and M-CAT motifs in CRM5 and CRM7 play a role in mediating enhancer function. To do this, we mutated these motifs within the Hes1CRM5-Pro and Hes1CRM7-Pro reporter constructs, using mutations that have previously been shown to disrupt factor binding (42, 43). The wild-type and mutated constructs along and the hes1 promoter alone were then transiently transfected into C2C12 cells, and fluorescence activity was compared using flow cytometry.
We first analyzed the role of RBP-JK in CRM5 function. CRM5 has the highest levels of RBP-JK binding as determined using ChIP (Fig. 3B) and contains two predicted RBP-JK motifs as follows: BS5.1 at −37,009 represents an exact match to the consensus high affinity GTGGGAA site, and BS5.2 at position −37,121 contains a mutation at position 1 of the consensus (Fig. 3A). These sites are organized in a tail-tail configuration separated by 112 bp of genomic sequence. The results in Fig. 4A reveal that both RBP-Jκ BS5.1 and BS5.2 are necessary for CRM5 enhancer function. Mutation of these sites individually reduced CRM5 enhancer activity by ∼20% when compared with the wild-type Hes1CRM5-Pro construct. However, mutation of both RBP-JK BSs resulted in the complete loss of CRM5 enhancer function suggesting cooperativity between these motifs. Furthermore, the increased response of the Hes1CRM5-Pro construct to NICD overexpression is also abrogated by mutation of both the RBP-JK BSs in CRM5 (Fig. 4B). Together, these data demonstrate that the enhancer activity of CRM5 is regulated by NOTCH signaling and that the RBP-JK BSs in this region mediate the effect.
FIGURE 4.
RBP-JK and M-CAT motifs are necessary for CRM5 and CRM7 enhancer function. A, transcriptional effect of mutating the RBP-JK BSs in the Hes1CRM5-Pro reporter. B, mutation of the RBP-JK BSs in Hes1CRM5-Pro results in a reduced response to NICD overexpression. C, RBP-JK BSs in Hes1CRM7 do not contribute to enhancer activity. D, contribution of the M-CAT motif to Hes1CRM7-Pro reporter activity. The RBP-JK and M-CAT motifs were mutated (verified in Refs. 42, 43) within the Hes1CRM5-Pro and Hes1CRM7-Pro reporters. Constructs containing either the wild-type or mutated RBP-JK and M-CAT motifs were co-transfected into C2C12 cells with an mCherry expression vector. A 1:1 ratio of reporter to NICD expression vector was used in B. Fluorescence activity was measured by flow cytometry. Venus activity was measured and normalized against mCherry activity. A, C, and D, data are displayed relative to the activity of the construct with highest fluorescence intensity (set as 1). B, results are presented as fold induction relative to the activity of each reporter in the absence of NICD. The results are indicated as the mean ± S.D. of at least two independent experiments.
We next assessed the function of the RBP-JK and TEAD2 BSs in CRM7. Consistent with the finding in Fig. 3B that RBP-JK was not detected at CRM7 using ChIP, mutation of the RBP-JK BS within CRM7 had no effect on CRM7 enhancer function (Fig. 4C). However, Fig. 4D shows that mutation of the M-CAT motif strongly reduces CRM7 enhancer activity by ∼75%. Taken together with the ChIP assays, the data suggest that TEAD2 is an important determinant of CRM7 enhancer activity and that CRM7 is able to integrate signals from the HIPPO pathway. These assays therefore demonstrate functionality for several of the in silico predicted transcription factor-binding sites in the hes1 enhancers.
Co-factor Occupancy on CRM5 and CRM7 Is Characteristic of Enhancers
Transcription factors are known to function by recruiting multiprotein co-activator and co-repressor complexes. We therefore performed a series of ChIP experiments on the endogenous hes1 locus to test for occupancy of CRM-specific co-activators as predicted by protein-protein interaction data.
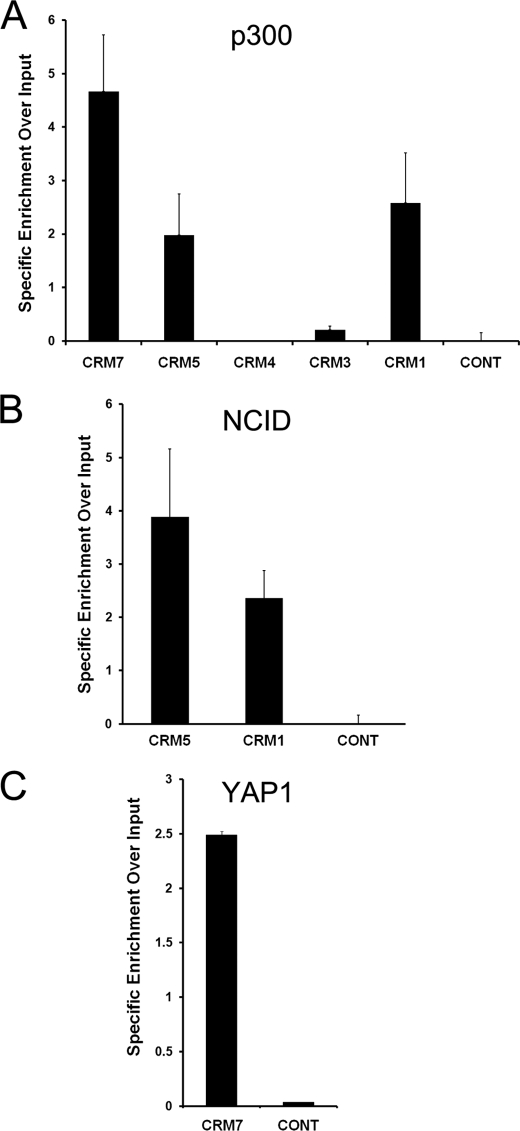
The p300 histone acetyltransferase can be found in complex with both the RBP-Jκ and TEAD2 transcription factors (44, 45). Importantly, p300 occupancy has also been shown to mark transcriptional enhancers (46). We therefore tested for p300 binding at hes1 CRM3–5 and -7, as these regions regulated the activity of the hes1 promoter in our reporter assays (Fig. 2A). In addition, we used p300 occupancy at CRM1 as a positive control. The results in Fig. 5A demonstrate that p300 is enriched on CRM1 as well as both enhancer CRMs, CRM5 and CRM7, compared with a nonspecific control. Furthermore, CRM3 and -4, which functioned to repress the hes1 promoter in our reporter assays, showed markedly reduced levels of p300. These data are consistent with the finding that p300 occupancy can predict enhancer activity in a tissue-specific manner (47) and correlate with the transcriptional activity of these CRMs as measured using reporter assays (Fig. 2).
FIGURE 5.
Enhancer associated co-activators occupy hes1 CRM5 and CRM7 in C2C12 cells. ChIP assays were performed using antibodies against p300 (A), NICD (B), and YAP or a nonspecific control (anti-GFP) (C). Input consists of DNA extracted from sonicated nuclei. Precipitated DNA was PCR-amplified using primers spanning the indicated regions. Results are expressed as specific enrichment over input. To do this, the signal for each PCR quantified, divided by the input and the background value of an IgG isotope control, was then subtracted.
RBP-JK has been shown to interact with the NICD upon NOTCH activation (8, 33, 48). As CRM5 contains functional RBP-Jκ BSs, we carried out a ChIP assay using an anti-NICD antibody to assess NICD occupancy at this region. This revealed that NICD specifically binds to CRM5 as well as CRM1, a known NOTCH-responsive element that we used as a positive control, in proliferating C2C12 cells (Fig. 5B). NICD was not detected above background at a control region in the hes1 upstream sequence containing no RBP-JK motifs. Taken together, these results demonstrate that a RBP-JK-NCID-p300 transcription factor-cofactor complex occupies the CRM5 enhancer in C2C12 cells.
TEAD proteins contain a DNA binding domain but lack an activation domain (49). TEAD2 has been shown to form a complex with the YAP co-activator, a major effector of the HIPPO signaling pathway, to regulate gene expression (50, 51). We therefore tested for YAP occupancy at CRM7 in proliferating C2C12 cells by ChIP. The results show specific enrichment of PCR signal with primers for CRM7 using an anti-YAP antibody (Fig. 5C). In addition, YAP binding was not detected at a control region in the hes1 upstream sequence. This suggests that a TEAD2-YAP-p300 complex is recruited to the functional M-CAT motif within CRM7 to regulate hes1 expression in proliferating C2C12 cells.
The identification of non-DNA-binding transcriptional co-activators at CRM5 and CRM7 provides additional evidence that these regions function as enhancers within the context of the endogenous hes1 locus in cells in culture, and it also suggests that their activity might be regulated in response to specific signaling pathways.
Hes1 Enhancer CRMs Loop onto Endogenous Promoter
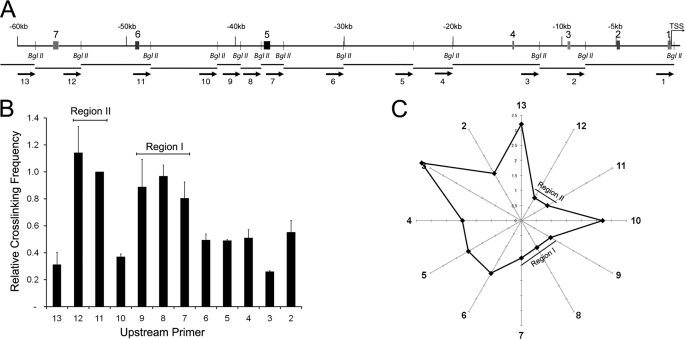
Several different models have been proposed to explain how CRMs can regulate transcription over large genomic distances. The looping model suggests that CRM-promoter communication is mediated by direct interactions between proteins bound to distal regulatory elements and the promoter with a consequent looping out of the intervening DNA (52–54). We therefore predicted that if CRM5 and CRM7 function as enhancers, they would be in close spatial proximity to the hes1 promoter in the nucleus. To test this, we performed 3C experiments to map the genomic architecture of the hes1 locus. We measured genomic interactions between a BglII fragment containing the hes1 promoter and ∼60 kb of upstream sequence encompassing all the newly identified CRMs. To calculate the relative cross-linking frequency, we used TaqMan qPCR to compare the amount of 3C PCR product obtained in HES1 expressing C2C12 cells to a BAC clone covering the hes1 gene and upstream sequence as a control.
The results in Fig. 6 are consistent with the proposed enhancer function of CRM5 and CRM7 as identified using reporter gene assays. Two large genomic regions in the hes1 upstream sequence appear in close proximity to the hes1 promoter as compared with the intervening and flanking sequences suggesting that these distal regions are looping onto the promoter. Region I spans restriction fragment 7 that contains CRM5 as well as fragments 8 and 9, and region II contains fragments 11 and 12 that cover CRM7. A schematic depicting the genomic architecture of the hes1 upstream locus relative to the promoter is shown in Fig. 6C. Furthermore, additional 3C experiments in synchronized C2C12 cells showed that this organization does not change during the different phases of HES1 oscillatory expression (data not shown). Our 3C-based mapping of the spatial organization of the hes1 locus therefore identifies regulatory interactions between genomic regions containing the newly identified functional hes1 CRMs and the endogenous promoter. Our analysis also raises the possibility of combining comparative genomics with high throughput 3C-based assays to identify functional regions of the genome on a large scale.
FIGURE 6.
Long range hes1 CRM-promoter interactions in C2C12 cells. A, schematic representation of the hes1 upstream sequence. Shaded boxes indicate the position of the hes1 CRMs. Small vertical bars represent the positions of the BglII restriction sites. Below, the restriction fragments generated by BglII digestion are annotated. Arrows indicate the location of the 3C qPCR primers. B, quantification of genomic interactions between the hes1 promoter region and the indicated upstream genomic fragments. The y axis shows the relative cross-linking frequency. The x axis indicates the primers used in combination with primer 1. Results are expressed relative to a BAC control containing equimolar amounts of all possible ligation products and are indicated as the mean relative cross-linking frequency ± S.D. of three independent experiments. C, graphical representation of the genomic distances between the hes1 promoter, positioned at the center of the radar, and the indicated BglII fragments. Distance was calculated as 1/cross-linking frequency.
DISCUSSION
In this study, we used a combination of computational and experimental methodologies to further define the mechanisms of hes1 transcriptional control. In addition to the known hes1 promoter region, we identified six previously uncharacterized conserved DNA elements within 57 kb of the hes1 TSS in mouse. We demonstrated a transcriptional regulatory function for four of these regions at the endogenous hes1 promoter in C2C12 myoblasts. Two of the conserved sequence blocks, CRM5 and CRM7, functioned as enhancers and displayed characteristics of functional enhancer regions within the context of the endogenous hes1 locus. These regions were marked by binding of the p300 co-activator, which has been shown to occupy active enhancers in the genome (46), and are in close genomic proximity to the hes1 promoter within the nucleus. Furthermore, the enhancer activity CRM5 could be increased in response to active NOTCH signaling. These data are consistent with the finding that CRMs can be located many kilobases away from the TSS of their target genes and can communicate with target promoters over large genomic distances through direct regulatory interactions. This work therefore extends the repertoire of known hes1 transcriptional control elements to include distal regulatory regions in addition to the previously characterized hes1 proximal promoter.
The number and affinity of transcription factor-binding sites in the control regions of target genes play a role in generating the correct transcriptional response to signaling pathways and interpreting transcription factor gradients. NOTCH signaling is used in a variety of different developmental contexts in the control of binary cell fate decisions, and the levels of the NOTCH receptor and its ligands have been shown to oscillate in a temporal manner in a number of progenitor cell populations (12, 55, 56). We identified a total of 12 RBP-Jκ BSs in the hes1 upstream region that are distributed solely within the seven conserved sequence blocks and are absent from the intervening DNA. Six of the predicted BSs matched the core consensus motif that has been used to define a high affinity RBP-Jκ BS. We then demonstrated that RBP-Jκ protein occupies CRMs1–6 in C2C12 cells in culture and that although the BS within CRM7 represents a high affinity motif, the inability to detect RBP-Jκ at this region correlated with the observation that mutation of this BS did not affect functionality. We suggest that the presence of multiple copies of RBP-Jκ BSs in the hes1 upstream CRMs would enable hes1 to elicit a fast response to variations in NOTCH signal strength at different stages of development and would allow for the fine control of hes1 expression levels.
RBP-Jκ can bind to its response element as a monomer. However, RBP-Jκ BSs commonly occur in clusters in the transcriptional control regions of it targets (57), for example, the Drosophila single-minded (sim) gene contains 10 binding sites for Suppressor of Hairless (Su(H)), the Drosophila RBP-Jκ homologue (58). Our data suggest that RBP-Jκ occupancy is higher at the hes1 CRMs that contain multiple predicted RBP-Jκ BSs as compared with the regions that contain only one (Fig. 3). Moreover, the number and arrangement of RBP-Jκ BSs play a role in the control of RBP-Jκ target gene selectivity and the response to NOTCH. It has been proposed that an inverted repeat of two motifs separated by a short 15–19-nucleotide spacer, the Su(H)-paired site (SPS) configuration, is the preferred recognition sequence (59) and that RBP-JK-mediated transactivation is affected by changes in spacing, orientation, and mutation of the individual BSs within this sequence (60). However, it appears that the SPS motif functions as a core promoter element to increase responsiveness to distal CRMs and cannot act as an enhancer over large genomic distances (9). Our data are consistent with this. We find that the mouse hes1 promoter contains two RBP-Jκ motifs arranged in the SPS configuration, although the RBP-Jκ motifs in the newly identified hes1 CRMs do not contain the SPS architecture. CRM4, CRM6, and CRM7 contain a single RBP-Jκ motif, and CRM3 contains a cluster of three RBP-Jκ BSs with a tail-head and tail-tail organization. The predicted pairs of RBP-Jκ BSs in hes1 CRM2 and CRM5 are organized in a tail-tail conformation separated by an extended spacer of 199 and 112 nucleotides, respectively. Experiments using synthetic reporters have suggested that NOTCH activation of tail-tail RBP-Jκ motifs is dependent on the surrounding DNA sequence (9). Our reporter assays demonstrate that the paired RBP-Jκ BS architecture within CRM5 is functional and is necessary for the enhancer activity of the region. The results also suggest cooperativity between the individual RPB-JK BSs in CRM5 as the transcriptional response of a double mutant construct containing both BSs mutated was greater than the sum of the effects of the constructs containing single BS mutations. In agreement with this, RBP-Jκ proteins in complex with the NICD and Mastermind co-activators have been shown to cooperatively form dimers on DNA templates containing the high affinity SPS sequence as well as several nonconsensus SPS variants (31, 61). These data therefore reveal a novel functional architecture of paired RBP-Jκ motifs within a distal regulatory region in the mouse genome.
TEAD proteins contain a DNA binding domain but lack a transactivation domain (24) and, as such, have been shown to interact with the YAP co-activator, a component of the HIPPO signaling pathway, to activate target gene expression (31). HIPPO signaling has been indirectly implicated in the control of hes1 as YAP1 activation in the mouse intestine results in expansion of undifferentiated progenitor cells and a rapid increase in HES1 expression (38). This effect is partially dependent on NOTCH. In this study we demonstrate that both TEAD2 and YAP occupy hes1 CRM7 in C2C12 cells and that the enhancer activity of this region is dependent on the Tead2 BS. We therefore provide a direct molecular link between HIPPO signaling and HES1 expression.
This work provides an increased understanding of the molecular mechanisms operating to control the expression of hes1, a key developmental gene that is also overexpressed in certain cancers. It also illustrates, using a single gene, the power of combining computational DNA sequence analysis with experimental methodologies such as ChIP, 3C, and reporter assays to identify and functionally characterize genomic regulatory regions.
Supplementary Material
Acknowledgments
We thank Dr. Andrew Bassett for critical reading of the manuscript and Dr. Sascha Ott for bioinformatic support
This work was supported by Wellcome Trust Grants WT 066790/E/02/Z and 066745/Z/01/Z, Human Frontier Science Program Grant RGP0029/2007-C (to G. K.), and a Warwick University Senior Research Fellowship (to K. W. V.).

This article contains supplemental material.
K. W. Vance, D. J. Woodcock, S. Ott, J. E. Reid, D. M. Jeziorska, T. Bretschneider, M. Komorowski, and G. Koentges, submitted for publication.
- NICD
- Notch intracellular domain
- CRM
- cis-regulatory module
- TSS
- transcription start site
- PSSM
- position specific scoring matrix
- qPCR
- quantitative PCR
- BS
- binding site
- 3C
- chromosome conformation capture
- BAC
- bacterial artificial chromosome
- SPS
- Su(H)-paired site.
REFERENCES
- 1. Sasai Y., Kageyama R., Tagawa Y., Shigemoto R., Nakanishi S. (1992) Two mammalian helix-loop-helix factors structurally related to Drosophila hairy and Enhancer of split. Genes Dev. 6, 2620–2634 [DOI] [PubMed] [Google Scholar]
- 2. Paroush Z., Finley R. L., Jr., Kidd T., Wainwright S. M., Ingham P. W., Brent R., Ish-Horowicz D. (1994) Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79, 805–815 [DOI] [PubMed] [Google Scholar]
- 3. Grbavec D., Stifani S. (1996) Molecular interaction between TLE1 and the carboxyl-terminal domain of HES-1 containing the WRPW motif. Biochem. Biophys. Res. Commun. 223, 701–705 [DOI] [PubMed] [Google Scholar]
- 4. Fisher A. L., Ohsako S., Caudy M. (1996) The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16, 2670–2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Takata T., Ishikawa F. (2003) Human Sir2-related protein SIRT1 associates with the bHLH repressors HES1 and HEY2 and is involved in HES1- and HEY2-mediated transcriptional repression. Biochem. Biophys. Res. Commun. 301, 250–257 [DOI] [PubMed] [Google Scholar]
- 6. Kageyama R., Ohtsuka T., Tomita K. (2000) The bHLH gene Hes1 regulates differentiation of multiple cell types. Mol. Cells 10, 1–7 [DOI] [PubMed] [Google Scholar]
- 7. Issack P. S., Ziff E. B. (1998) Genetic elements regulating HES-1 induction in Wnt-1-transformed PC12 cells. Cell Growth Differ. 9, 827–836 [PubMed] [Google Scholar]
- 8. Jarriault S., Brou C., Logeat F., Schroeter E. H., Kopan R., Israel A. (1995) Signaling downstream of activated mammalian Notch. Nature 377, 355–358 [DOI] [PubMed] [Google Scholar]
- 9. Ong C. T., Cheng H. T., Chang L. W., Ohtsuka T., Kageyama R., Stormo G. D., Kopan R. (2006) Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281, 5106–5119 [DOI] [PubMed] [Google Scholar]
- 10. Hirata H., Yoshiura S., Ohtsuka T., Bessho Y., Harada T., Yoshikawa K., Kageyama R. (2002) Oscillatory expression of the bHLH factor Hes1 regulated by a negative feedback loop. Science 298, 840–843 [DOI] [PubMed] [Google Scholar]
- 11. Jouve C., Palmeirim I., Henrique D., Beckers J., Gossler A., Ish-Horowicz D., Pourquié O. (2000) Notch signaling is required for cyclic expression of the hairy-like gene HES1 in the presomitic mesoderm. Development 127, 1421–1429 [DOI] [PubMed] [Google Scholar]
- 12. Shimojo H., Ohtsuka T., Kageyama R. (2008) Oscillations in notch signaling regulate maintenance of neural progenitors. Neuron 58, 52–64 [DOI] [PubMed] [Google Scholar]
- 13. Baek J. H., Hatakeyama J., Sakamoto S., Ohtsuka T., Kageyama R. (2006) Persistent and high levels of Hes1 expression regulate boundary formation in the developing central nervous system. Development 133, 2467–2476 [DOI] [PubMed] [Google Scholar]
- 14. Wall D. S., Mears A. J., McNeill B., Mazerolle C., Thurig S., Wang Y., Kageyama R., Wallace V. A. (2009) Progenitor cell proliferation in the retina is dependent on Notch-independent Sonic hedgehog/Hes1 activity. J. Cell Biol. 184, 101–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ingram W. J., McCue K. I., Tran T. H., Hallahan A. R., Wainwright B. J. (2008) Sonic Hedgehog regulates Hes1 through a novel mechanism that is independent of canonical Notch pathway signaling. Oncogene 27, 1489–1500 [DOI] [PubMed] [Google Scholar]
- 16. Curry C. L., Reed L. L., Nickoloff B. J., Miele L., Foreman K. E. (2006) Notch-independent regulation of Hes-1 expression by c-Jun N-terminal kinase signaling in human endothelial cells. Lab. Invest. 86, 842–852 [DOI] [PubMed] [Google Scholar]
- 17. Sanalkumar R., Indulekha C. L., Divya T. S., Divya M. S., Anto R. J., Vinod B., Vidyanand S., Jagatha B., Venugopal S., James J. (2010) ATF2 maintains a subset of neural progenitors through CBF1/Notch-independent Hes-1 expression and synergistically activates the expression of Hes-1 in Notch-dependent neural progenitors. J. Neurochem. 113, 807–818 [DOI] [PubMed] [Google Scholar]
- 18. Nakayama K., Satoh T., Igari A., Kageyama R., Nishida E. (2008) FGF induces oscillations of Hes1 expression and Ras/ERK activation. Curr. Biol. 18, R332–R334 [DOI] [PubMed] [Google Scholar]
- 19. Aguilera C., Hoya-Arias R., Haegeman G., Espinosa L., Bigas A. (2004) Recruitment of IκBα to the hes1 promoter is associated with transcriptional repression. Proc. Natl. Acad. Sci. U.S.A. 101, 16537–16542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sang L., Roberts J. M., Coller H. A. (2010) Hijacking HES1. How tumors co-opt the anti-differentiation strategies of quiescent cells. Trends Mol. Med. 16, 17–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farnie G., Clarke R. B., Spence K., Pinnock N., Brennan K., Anderson N. G., Bundred N. J. (2007) Novel cell culture technique for primary ductal carcinoma in situ. role of Notch and epidermal growth factor receptor signaling pathways. J. Natl. Cancer Inst. 99, 616–627 [DOI] [PubMed] [Google Scholar]
- 22. Konishi J., Kawaguchi K. S., Vo H., Haruki N., Gonzalez A., Carbone D. P., Dang T. P. (2007) γ-Secretase inhibitor prevents Notch3 activation and reduces proliferation in human lung cancers. Cancer Res. 67, 8051–8057 [DOI] [PubMed] [Google Scholar]
- 23. Fan X., Matsui W., Khaki L., Stearns D., Chun J., Li Y. M., Eberhart C. G. (2006) Notch pathway inhibition depletes stem-like cells and blocks engraftment in embryonal brain tumors. Cancer Res. 66, 7445–7452 [DOI] [PubMed] [Google Scholar]
- 24. Hopfer O., Zwahlen D., Fey M. F., Aebi S. (2005) The Notch pathway in ovarian carcinomas and adenomas. Br. J. Cancer 93, 709–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cuevas I. C., Slocum A. L., Jun P., Costello J. F., Bollen A. W., Riggins G. J., McDermott M. W., Lal A. (2005) Meningioma transcript profiles reveal deregulated Notch signaling pathway. Cancer Res. 65, 5070–5075 [DOI] [PubMed] [Google Scholar]
- 26. Hallahan A. R., Pritchard J. I., Hansen S., Benson M., Stoeck J., Hatton B. A., Russell T. L., Ellenbogen R. G., Bernstein I. D., Beachy P. A., Olson J. M. (2004) The SmoA1 mouse model reveals that notch signaling is critical for the growth and survival of sonic hedgehog-induced medulloblastomas. Cancer Res. 64, 7794–7800 [DOI] [PubMed] [Google Scholar]
- 27. Sang L., Coller H. A., Roberts J. M. (2008) Control of the reversibility of cellular quiescence by the transcriptional repressor HES1. Science 321, 1095–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vance K. W., Carreira S., Brosch G., Goding C. R. (2005) Tbx2 is overexpressed and plays an important role in maintaining proliferation and suppression of senescence in melanomas. Cancer Res. 65, 2260–2268 [DOI] [PubMed] [Google Scholar]
- 29. Hagège H., Klous P., Braem C., Splinter E., Dekker J., Cathala G., de Laat W., Forné T. (2007) Quantitative analysis of chromosome conformation capture assays (3C-qPCR). Nat. Protoc. 2, 1722–1733 [DOI] [PubMed] [Google Scholar]
- 30. Granier C., Gurchenkov V., Perea-Gomez A., Camus A., Ott S., Papanayotou C., Iranzo J., Moreau A., Reid J., Koentges G., Sabéran-Djoneidi D., Collignon J. (2011) Nodal cis-regulatory elements reveal epiblast and primitive endoderm heterogeneity in the peri-implantation mouse embryo. Dev. Biol. 349, 350–362 [DOI] [PubMed] [Google Scholar]
- 31. Nam Y., Sliz P., Pear W. S., Aster J. C., Blacklow S. C. (2007) Cooperative assembly of higher order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. U.S.A. 104, 2103–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Takebayashi K., Sasai Y., Sakai Y., Watanabe T., Nakanishi S., Kageyama R. (1994) Structure, chromosomal locus, and promoter analysis of the gene encoding the mouse helix-loop-helix factor HES-1. Negative autoregulation through the multiple N box elements. J. Biol. Chem. 269, 5150–5156 [PubMed] [Google Scholar]
- 33. Kuroda K., Tani S., Tamura K., Minoguchi S., Kurooka H., Honjo T. (1999) Delta-induced Notch signaling mediated by RBP-J inhibits MyoD expression and myogenesis. J. Biol. Chem. 274, 7238–7244 [DOI] [PubMed] [Google Scholar]
- 34. Azakie A., Lamont L., Fineman J. R., He Y. (2005) Divergent transcriptional enhancer factor-1 regulates the cardiac troponin T promoter. Am. J. Physiol. Cell Physiol. 289, C1522–C1534 [DOI] [PubMed] [Google Scholar]
- 35. Karns L. R., Kariya K., Simpson P. C. (1995) M-CAT, CArG, and Sp1 elements are required for α1-adrenergic induction of the skeletal α-actin promoter during cardiac myocyte hypertrophy. Transcriptional enhancer factor-1 and protein kinase C as conserved transducers of the fetal program in cardiac growth. J. Biol. Chem. 270, 410–417 [DOI] [PubMed] [Google Scholar]
- 36. Swartz E. A., Johnson A. D., Owens G. K. (1998) Two MCAT elements of the SM α-actin promoter function differentially in SM vs. non-SM cells. Am. J. Physiol. 275, C608–C618 [DOI] [PubMed] [Google Scholar]
- 37. Zhao B., Lei Q. Y., Guan K. L. (2008) The Hippo-YAP pathway. New connections between regulation of organ size and cancer. Curr. Opin. Cell Biol. 20, 638–646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Camargo F. D., Gokhale S., Johnnidis J. B., Fu D., Bell G. W., Jaenisch R., Brummelkamp T. R. (2007) YAP1 increases organ size and expands undifferentiated progenitor cells. Curr. Biol. 17, 2054–2060 [DOI] [PubMed] [Google Scholar]
- 39. Dong J., Feldmann G., Huang J., Wu S., Zhang N., Comerford S. A., Gayyed M. F., Anders R. A., Maitra A., Pan D. (2007) Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell 130, 1120–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tomczak K. K., Marinescu V. D., Ramoni M. F., Sanoudou D., Montanaro F., Han M., Kunkel L. M., Kohane I. S., Beggs A. H. (2004) Expression profiling and identification of novel genes involved in myogenic differentiation. FASEB J. 18, 403–405 [DOI] [PubMed] [Google Scholar]
- 41. Shimizu N., Smith G., Izumo S. (1993) Both a ubiquitous factor mTEF-1 and a distinct muscle-specific factor bind to the M-CAT motif of the myosin heavy chain beta gene. Nucleic Acids Res. 21, 4103–4110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hucl T., Brody J. R., Gallmeier E., Iacobuzio-Donahue C. A., Farrance I. K., Kern S. E. (2007) High cancer-specific expression of mesothelin (MSLN) is attributable to an upstream enhancer containing a transcription enhancer factor-dependent MCAT motif. Cancer Res. 67, 9055–9065 [DOI] [PubMed] [Google Scholar]
- 43. Krebs L. T., Iwai N., Nonaka S., Welsh I. C., Lan Y., Jiang R., Saijoh Y., O'Brien T. P., Hamada H., Gridley T. (2003) Notch signaling regulates left-right asymmetry determination by inducing Nodal expression. Genes Dev. 17, 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Oswald F., Täuber B., Dobner T., Bourteele S., Kostezka U., Adler G., Liptay S., Schmid R. M. (2001) p300 acts as a transcriptional coactivator for mammalian Notch-1. Mol. Cell. Biol. 21, 7761–7774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Strano S., Monti O., Pediconi N., Baccarini A., Fontemaggi G., Lapi E., Mantovani F., Damalas A., Citro G., Sacchi A., Del Sal G., Levrero M., Blandino G. (2005) The transcriptional coactivator Yes-associated protein drives p73 gene-target specificity in response to DNA damage. Mol. Cell 18, 447–459 [DOI] [PubMed] [Google Scholar]
- 46. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 47. Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., Afzal V., Ren B., Rubin E. M., Pennacchio L. A. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jarriault S., Le Bail O., Hirsinger E., Pourquié O., Logeat F., Strong C. F., Brou C., Seidah N. G., Isra l A. (1998) Delta-1 activation of notch-1 signaling results in HES-1 transactivation. Mol. Cell. Biol. 18, 7423–7431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hwang J. J., Chambon P., Davidson I. (1993) Characterization of the transcription activation function and the DNA binding domain of transcriptional enhancer factor-1. EMBO J. 12, 2337–2348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vassilev A., Kaneko K. J., Shu H., Zhao Y., DePamphilis M. L. (2001) TEAD/TEF transcription factors utilize the activation domain of YAP65, a Src/Yes-associated protein localized in the cytoplasm. Genes Dev. 15, 1229–1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yagi R., Chen L. F., Shigesada K., Murakami Y., Ito Y. (1999) A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551–2562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Capturing chromosome conformation. Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 53. Tolhuis B., Palstra R. J., Splinter E., Grosveld F., de Laat W. (2002) Looping and interaction between hypersensitive sites in the active β-globin locus. Mol. Cell 10, 1453–1465 [DOI] [PubMed] [Google Scholar]
- 54. Tiwari V. K., McGarvey K. M., Licchesi J. D., Ohm J. E., Herman J. G., Schübeler D., Baylin S. B. (2008) PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 6, 2911–2927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kobayashi T., Mizuno H., Imayoshi I., Furusawa C., Shirahige K., Kageyama R. (2009) The cyclic gene Hes1 contributes to diverse differentiation responses of embryonic stem cells. Genes Dev. 23, 1870–1875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lai E. C. (2004) Notch signaling. Control of cell communication and cell fate. Development 131, 965–973 [DOI] [PubMed] [Google Scholar]
- 57. Rebeiz M., Reeves N. L., Posakony J. W. (2002) SCORE. A computational approach to the identification of cis-regulatory modules and target genes in whole-genome sequence data. Site clustering over random expectation. Proc. Natl. Acad. Sci. U.S.A. 99, 9888–9893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Morel V., Schweisguth F. (2000) Repression by suppressor of hairless and activation by Notch are required to define a single row of single-minded expressing cells in the Drosophila embryo. Genes Dev. 14, 377–388 [PMC free article] [PubMed] [Google Scholar]
- 59. Bailey A. M., Posakony J. W. (1995) Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9, 2609–2622 [DOI] [PubMed] [Google Scholar]
- 60. Cave J. W., Loh F., Surpris J. W., Xia L., Caudy M. A. (2005) A DNA transcription code for cell-specific gene activation by notch signaling. Curr. Biol. 15, 94–104 [DOI] [PubMed] [Google Scholar]
- 61. Arnett K. L., Hass M., McArthur D. G., Ilagan M. X., Aster J. C., Kopan R., Blacklow S. C. (2010) Structural and mechanistic insights into cooperative assembly of dimeric Notch transcription complexes. Nat. Struct. Mol. Biol. 17, 1312–1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.