Background: Tah1 and Pih1 are Hsp90 interactors that form a ternary complex with the chaperone.
Results: NMR structure of Tah1 revealed the presence of two tetratricopeptide repeat motifs followed by a C helix and an unstructured region.
Conclusion: Tah1 can bind simultaneously two other proteins using different interaction modes.
Significance: The study provides important insights into protein complex assembly.
Keywords: Chaperone Chaperonin, NMR, Protein Complexes, Protein-Protein Interactions, Structural Biology, Hsp90, Pih1, Tah1, Tetratricopeptide Repeat
Abstract
Tah1 and Pih1 are novel Hsp90 interactors. Tah1 acts as a cofactor of Hsp90 to stabilize Pih1. In yeast, Hsp90, Tah1, and Pih1 were found to form a complex that is required for ribosomal RNA processing through their effect on box C/D small nucleolar ribonucleoprotein assembly. Tah1 is a minimal tetratricopeptide repeat protein of 111 amino acid residues that binds to the C terminus of the Hsp90 molecular chaperone, whereas Pih1 consists of 344 residues of unknown fold. The NMR structure of Tah1 has been solved, and this structure shows the presence of two tetratricopeptide repeat motifs followed by a C helix and an unstructured region. The binding of Tah1 to Hsp90 is mediated by the EEVD C-terminal residues of Hsp90, which bind to a positively charged channel formed by Tah1. Five highly conserved residues, which form a two-carboxylate clamp that tightly interacts with the ultimate Asp-0 residue of the bound peptide, are also present in Tah1. Tah1 was found to bind to the C terminus of Pih1 through the C helix and the unstructured region. The C terminus of Pih1 destabilizes the protein in vitro and in vivo, whereas the binding of Tah1 to Pih1 allows for the formation of a stable complex. Based on our data, a model for an Hsp90-Tah1-Pih1 ternary complex is proposed.
Introduction
Tah1 (TPR7-containing protein associated with Hsp90) is a small protein of 111 amino acids (12.5 kDa). We discovered this previously uncharacterized protein during a proteomic screen for Hsp90 interactors (1). We demonstrated that Tah1 is a novel Hsp90 cofactor that modulates the chaperone activity. Tah1 was found to interact with Hsp90 as well as with another protein that we termed Pih1 (also called Nop17; 344 residues) (1). Hsp90-Tah1 function to stabilize Pih1 and to promote the formation of an Rvb1-Rvb2-Tah1-Pih1 complex, which we named the R2TP complex (2). Rvb1 and Rvb2 are two highly conserved AAA+ helicases involved in many different critical complexes in the cell (3, 4). The R2TP complex is highly conserved from yeast to mammalian cells and has been shown to be required for the proper assembly of box C/D small nucleolar ribonucleoproteins (5, 2), for the assembly of RNA polymerase II (6, 7), and for the stability of the phosphatidylinositol 3-kinase-related kinases through binding to TEL2 (8). Furthermore, both human Pih1 (PIH1D1) and human Tah1 (RPAP3) have been shown to regulate apoptosis (9, 10).
A TPR motif typically consists of 34 amino acids that adopt a helix-turn-helix structure. The motif is defined by a pattern of small and large hydrophobic amino acids, with no positions being completely invariant. The motif was initially discovered in cell cycle regulatory proteins Cdc23 and Nuc2 (11, 12). Most TPR proteins contain between 3 and 16 TPR repeats (13, 14), with adjacent TPR motifs packed in a parallel fashion, resulting in a spiral of α-helices forming a right-handed superhelical arrangement that is typically capped by a C-terminal hydrophilic helix. As a result, a concave and a convex ligand binding interface are formed, making TPR proteins ideal for mediating protein-protein interactions and for acting as scaffolds for the assembly of multiprotein complexes (15, 16). TPR proteins have been implicated in a wide range of cellular activities, such as cell cycle regulation, transcriptional control, protein transport, and protein folding (17, 18).
Several TPR proteins are known to bind the C termini of the Hsp70 and Hsp90 chaperones and to act as cofactors that modulate the function of these chaperones (19, 20). Several groups have reported the crystal structures of TPR domains in complex with peptides that include the C-terminal EEVD residues of Hsp90 and Hsp70 (21–24, 16). These structures provide insights into the molecular basis of TPR-ligand recognition. Typically, the TPR domains in these structures consist of three TPR motifs and a C-terminal cap helix (see below). In this regard, Tah1 is rather unusual because, based on secondary structure analysis programs, it has only two predicted TPR motifs but can still interact with Hsp90 (1, 2). We had demonstrated that Tah1 interaction with Hsp90 is mediated by the C terminus of the chaperone (2). We also showed that the C-terminal fragment of Tah1, consisting of residues 76–111, can bind to Pih1 (2).
In order to understand the basis by which this minimal TPR domain protein attains a folded and stable structure and how it interacts with Pih1 and Hsp90, the NMR structure of Tah1 was solved, and its interaction with Pih1 and Hsp90 was investigated. Our findings show that Tah1 is a stable two-TPR repeat protein with a C-terminal cap helix and an unstructured region. Tah1 forms a positively charged channel in which the MEEVD model peptide binds. Furthermore, the C terminus of Tah1 was found to bind to the C terminus of Pih1. Biochemical and biophysical studies suggest a model of the Hsp90-Tah1-Pih1 ternary complex.
EXPERIMENTAL PROCEDURES
Plasmid Construction
The construction, expression, and purification of full-length Tah1 and Pih1 have been described previously (2). Pih1(1–230), Pih1(1–284), Pih1(231–344), Tah1(1–74), and Tah1(1–93) were amplified from yeast Saccharomyces cerevisiae S288C genome, cloned into p11 expression vector (25), and expressed with an N-terminal His6 tag followed by tobacco etch virus protease cleavage site. To make Pih1-Tah1, Pih1-Tah1(75–111), and Pih1-Tah1(94–111) fusion constructs, the Pih1 coding sequence was amplified using forward and reverse primers carrying an NdeI and a HindIII site, respectively. The Tah1, Tah1(75–111), and Tah1(94–111) were amplified using primers carrying HindIII and BamHI sites. The NdeI-HindIII Pih1 coding sequence was then ligated together with the HindIII-BamHI Tah1 coding sequence constructs and inserted into the NdeI/BamHI sites in p11.
Site-directed mutagenesis was carried out following protocols described in the Stratagene QuikChange site-directed mutagenesis kit instruction manual (Stratagene, La Jolla, CA). Each mutation was confirmed by DNA sequencing.
Protein Purification
All His6-tagged proteins were expressed in E. coli BL21(DE3) gold (pRIL) and purified using Ni2+-NTA resin (Qiagen) according to the manufacturer's protocols. For labeled samples required for NMR measurements, cells were grown in M9 medium supplemented with [15N]NH4Cl and/or [13C]glucose as nitrogen and carbon sources, respectively. The purified proteins were typically dialyzed and stored in buffer A (25 mm Tris-HCl, pH 7.5, 100 mm KCl, 10% glycerol, and 1 mm DTT). Tobacco etch virus (TEV) protease was used to remove the His6 tag. The concentrations of purified proteins were determined using the Bradford assay (26).
Size Exclusion Chromatography
Size exclusion chromatography was performed using a calibrated Superdex 200 or 75 HR 10/30 column (GE Healthcare) attached to an AKTA FPLC system (GE Healthcare). The column was equilibrated with buffer B (25 mm Tris-HCl, pH 7.5, 100 mm KCl, 10% glycerol, and 1 mm DTT). To test the oligomeric state of the purified proteins, at least 500 μg of protein in a total volume of 500 μl were loaded onto Superdex 200. To test the interaction between Tah1/Tah1 fragments with Pih1/Pih1 fragments, 250 μg of each protein were mixed in a total volume of 250 μl and loaded onto Superdex 75. Where indicated, WT Tah1 was preincubated with 450 μm of MEEVD peptide overnight at 4 °C prior to the addition of Pih1. Molecular weight standards used were purchased from Sigma or Bio-Rad: thryoglobulin (669 kDa), apoferritin (443 kDa), α-amylase (200 kDa), alcohol dehydrogenase (150 kDa), bovine serum albumin (66 kDa), ovalbumin (44 kDa), carbonic anhydrase (29 kDa), myoglobin (17 kDa), cytochrome c (12.4 kDa), aprotinin (6.5 kDa), and vitamin B12 (1.4 kDa). All experiments were performed at 4 °C, and absorbance was monitored at 280 nm.
Isothermal Titration Calorimetry
Purified Tah1 and its mutants were dialyzed overnight at 4 °C in buffer C (20 mm Tris-HCl, pH 8.0, 1 mm EDTA, and 5 mm NaCl). Protein concentrations were determined by absorbance at 280 nm using an ND-1000 Spectrophotometer (Thermo Scientific). The MEEVD peptide (663.7 daltons), containing an acetyl moiety at the N terminus and a free C-terminal carboxylate group, was synthesized at the Synthesis of Peptide Service at the Centro de Investigación Príncipe Felipe (Valencia, Spain) using a 433A Applied Biosystems synthesizer and Fmoc (N-(9-fluorenyl)methoxycarbonyl) chemistry. The peptide was weighed on an analytical balance (Mettler) and dissolved in buffer C. Samples were filtered using a 0.45-μm pore syringe filter (Pall Life Sciences) and degassed.
Isothermal Titration Calorimetry (ITC) measurements were performed at 4 °C in buffer C using a VP-ITC microcalorimeter (Microcal) with a cell volume of 1.458 ml. Peptide concentration in the needle was 450 μm, and Tah1 concentration in the cell was 30 μm. The syringe speed was set at 310 rpm, and a 150-s delay time was maintained between each injection. Heat of dilution was determined in a separate experiment by injecting peptide into buffer in the same sequence. The enthalpy value of the first injection was omitted due to experimental errors, and thermodynamic parameters were derived using a non-linear least square curve-fitting algorithm (Microcal Origin) to an n-identical independent binding model with three variables: association constant (Kb), enthalpy (ΔH), and stoichiometry (n). Other thermodynamic parameters and their S.D. values, namely dissociation constant (Kd), Gibbs free energy (ΔG), and entropy (ΔS), were derived from Kb and ΔH.
NMR Experiments
Purified proteins were dialyzed in buffer D (25 mm NaH2PO4/Na2HPO4, pH 8, and 100 mm NaCl) and concentrated to a final concentration between 400 μm and 1 mm, depending on the NMR experiment. 10% D2O was present in the final solution. All two- and three-dimensional NMR experiments were acquired at 293 K using Bruker Avance II spectrometers equipped with a TCI cryoprobe and working at 600.13 or 899.58 MHz for 1H. Tah1-MEEVD backbone assignment was achieved using a set of three-dimensional experiments based on J couplings: HNCO, HNCA, HNCACB, CBCACONH, and HBHACONH. Side chain assignments were based on the analysis of hCCH-TOCSY and HcCH-TOCSY experiments together with 1H,15N-NOESY-HSQC, aliphatic 1H,13C-NOESY-HSQC, and aromatic 1H,13C-NOESY-HSQC. Additionally, 13C direct detection experiments CC-COSY and CACO were used for the assignment of Asx and Glx residue side chains. Upper distance limit constraints were calculated from two-dimensional 1H,1H NOESY, two-dimensional 15N-filtered 1H,1H NOESY, three-dimensional 1H,13C NOESY-HSQC, and 1H,15N NOESY-HSQC. Spectral windows of 12 ppm for 1H, 28 ppm for 15N, and 16 ppm (HNCO), 30 ppm (HNCA), or 62 ppm (CBCA(CO)NH) for 13C were used with their center set at 4.7, 115, 175, 53, and 39 ppm, respectively. The relaxation delay used was 1.1 s, and 8–32 scans were collected. The 13C direct detection experiments were acquired with 60 scans, relaxation delays of 1.4 s, and acquisition times of 72 ms. NMR spectra were acquired and processed using TOPSPIN (version 2.1, Bruker BioSpin, Rheinstetten, Germany) and analyzed using CARA software (ETH, Zurich, Switzerland). Two-dimensional 1H,15N HSQC experiments were used to follow the binding of Tah1 to MEEVD peptide. 15N-labeled Tah1 was concentrated to 0.6 mm, and the peptide was titrated to a final concentration of 0, 0.3, 0.6, 0.89, 1.17, 1.7, and 2.2 mm. Chemical shift changes for the combined 1H and 15N nuclei (ΔδavgHN) and signal broadening were used to map the interactions with peptide.
Structure Calculations
Assignment of the NOESY spectrum of Tah1 was performed using the algorithm ATNOS-CANDID integrated in the UNIO routine (27). Manual modifications were performed after inspection of the results. TALOS software (28) was used to calculate angle restraints derived from the chemical shifts in addition to the constraints already calculated by the UNIO routine. Hydrogen bond constraints were also obtained by solvent exchange and adequately introduced into the structure calculations. HADDOCK (29) was used to model the interaction between Tah1 and MEEVD using the data from NMR titrations, single point mutagenesis, and the solvent accessibility surfaces. Intermolecular NOEs were obtained from the 15N-edited two-dimensional NOESY and introduced into the structure calculations (supplemental Fig. 1). 500 random structures were minimized in the simulated annealing procedure using CYANA 2.1 (27), and the 20 conformers with lowest energy were selected. A summary of the constraints utilized can be found in Table 1. Energy minimization of this family of structures was performed using AMBER 10.0 in a water box of 10 Å (30) and the AMPS-NMR portal within the WeNMR gateway (available on the World Wide Web), which includes a molecular dynamics step for the structure minimization. Evaluation of the structure was done using PSVS (31) and CING (available on the World Wide Web). Validation outcome is summarized in Table 2. The protein secondary structure was assigned using TALOS (28). The full resonance assignment of the complex has been deposited in the BMRB data base with entry number 17312.
TABLE 1.
Summary of the experimental constraints used in the structure determination of Tah1-MEEVD
| Residues | All (116, Tah1 + MEEVD) | Structured (2–92, Tah1) |
|---|---|---|
| NOE-based distance constraints | ||
| Total | 2051 | 1896 |
| Intraresidue (i = j) | 359 | 321 |
| Sequential (|i − j| = 1) | 502 | 446 |
| Medium range (1 < |i − j| < 5) | 680 | 651 |
| Long range (|i − j| ≥ 5) | 505 | 478 |
| Intermolecular | 5 | |
| NOEs/restrained residue | 18.5 | 20.8 |
| Hydrogen bond constraints | 8 | 8 |
| Dihedral angle constraints | ||
| ϕ (degrees) | 78 | 78 |
| ψ (degrees) | 78 | 78 |
| Total no. of constraints | 2215 | 2051 |
| Total no. of constraints/residue | 20.0 | 22.5 |
| Long range constraints/residue | 4.6 | 5.3 |
TABLE 2.
Summary of the quality statistics for the ensemble of 20 structures calculated for Tah1-MEEVD
| Residues | All (116, Tah1 + MEEVD) | Structured (2–92, Tah1) |
|---|---|---|
| Root mean square deviation values | ||
| All backbone atoms | 3.8 Å | 0.56 Å |
| All heavy atoms | 4.1 Å | 0.96 Å |
| Ramachandran plot from Procheck | ||
| Most favored regions | 91.9% | 93.2% |
| Additionally allowed regions | 8.1% | 6.8% |
| Generously allowed regions | 0.0% | 0.0% |
| Disallowed regions | 0.0% | 0.0% |
| Ramachandran plot from Richardson's laboratory | ||
| Most favored regions | 97.6% | 98.2% |
| Allowed regions | 2.3% | 1.8% |
| Disallowed regions | 0.1% | 0.0% |
| Structure quality factors: Overall statistics (mean score ± S.D.) | ||
| Procheck G-factor (ϕ/ψ only) | 0.22 | 0.26 |
| Procheck G-factor (all dihedral angles) | −0.09 | −0.07 |
| Verify3D | 0.19 ± 0.03 | 0.26 ± 0.04 |
| ProsaII (-ve) | 0.43 ± 0.05 | 0.64 ± 0.05 |
| MolProbity clashscore | 1.24 ± 0.96 | 1.14 ± 1.9 |
| Structure quality factors: Overall statistics (Z-score) | ||
| Procheck G-factor (ϕ/ψ only) | 1.18 | 1.34 |
| Procheck G-factor (all dihedral angles) | −0.53 | −0.41 |
| Verify3D | −4.33 | −3.21 |
| ProsaII (-ve) | −0.91 | −0.04 |
| MolProbity clashscore | 1.31 | 1.33 |
Confocal Microscopy and in Vivo Analysis of Pih1 Stability
pAG415GPD-EGFP-Pih1 plasmid was constructed using Pih1 ORF obtained from the Yeast FLEXGene collection (32). Pih1 ORF was first subcloned into Gateway® donor vector pDONR201 and then subcloned into the EGFP fusion expression vector pAG415GPD-EGFP following a published protocol (33). The plasmids p416ADH-GFP and p416ADH-GFP-Pih1(282–344) were constructed by amplifying the relevant Pih1 fragment from pET22-Pih1 (2) and GFP from pRSET-S65T (Clontech) and then ligating the fragments into the yeast expression vector p416ADH (34). pChFP-VHL was a gift from Dr. Judith Frydman (Stanford University).
S288C WT cells and rpt6-25 mutant cells (a gift from Dr. Charles Boone, University of Toronto) transformed with either EGFP-Pih1 or GFP-Pih1(282–344) were grown in appropriate synthetic media to A600 of 0.5 at room temperature, and then the temperature was shifted to 37 °C. At the indicated time points, cultures were pelleted, and a 1.5-μl suspension was spotted onto a glass slide for image analysis. Images were captured using the Quorum WaveFX Spinning Disc Confocal System.
To test the stability of GFP and GFP fusion proteins in vivo, the plasmids p416ADH-GFP and p416ADH-GFP-Pih1(282–344) were transformed into yeast strain W303. The cells were grown to midlog phase, and then cycloheximide was added to the culture at a final concentration of 50 μg/ml. Equal volumes of cell cultures were then withdrawn at different time points and lysed. Proteins were separated on 12% SDS-polyacrylamide gels, followed by immunoblotting using anti-GFP antibody (G1544, Sigma).
RESULTS
NMR Structure of Tah1-MEEVD
Initial attempts to determine the NMR structure of free Tah1 were unsuccessful because the protein was not stable long enough at the concentrations needed for the heteronuclear NMR experiments. The addition of MEEVD, resembling the C-terminal residues of Hsp90/Hsp70, stabilized the protein and allowed the determination of the solution NMR structure of Tah1 in complex with the peptide (see “Experimental Procedures”). Such instability of apo-TPR domains has been observed before for other proteins, such as for protein phosphatase 5 (35). The MEEVD peptide used in our studies was acetylated at the N terminus (mimicking a peptide bond). The structure calculations relied on 2051 meaningful upper distance limit values derived from the 1H,1H NOE intensities and 156 dihedral angle constraints (Table 1). The average total target function for the family of 20 conformers with the lowest energy was 0.93 ± 0.06 Å2 (CYANA calculations), and the root mean square deviation for residues 2–92 calculated with respect to the mean structure was 0.47 ± 0.08 Å for the backbone atoms and 0.86 ± 0.07 Å for the heavy atoms. After energy minimization with AMBER, the final cluster had a target function of 0.64 ± 0.09 Å2, and the root mean square deviation increased to 0.56 ± 0.15 Å for the backbone and 0.96 ± 0.15 Å for the heavy atoms (Table 2).
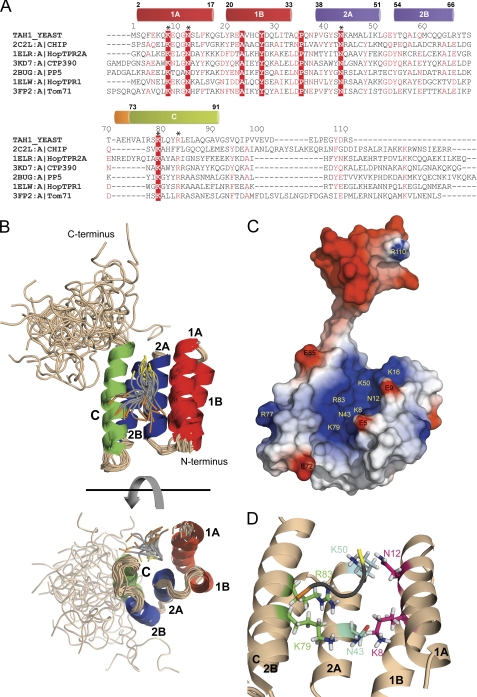
The final family of 20 conformers of Tah1 obtained after energy minimization is shown in Fig. 1. The protein consists of five antiparallel α-helices (Fig. 1, A and B). Residues 2–33 (in red) form the first TPR motif, residues 38–66 (in blue) form the second TPR motif, and residues 73–91 (in green) form a long C helix that interacts mainly with helix 2B. The last 20 C-terminal residues of Tah1 (residues 92–111) are unstructured, with no long range NOEs involving protons in this segment of the protein (Table 1). The mobility of this unstructured region does not follow the general tumbling of the molecule. Some of the conformers of the family of structures present a 310 helix encompassing residues 71–73 (shown in orange in the secondary structure schematic of Fig. 1A). The protein is stabilized by hydrophobic interactions mainly involving the following residues: Gly-11, Leu-14, Ala-23, Tyr-27, Leu-30, Ile-31, Ala-45, Leu-48, Ala-57, Cys-61, Gly-64, Leu-65, Leu-80, and Leu-84.
FIGURE 1.
Tah1 solution structure. A, alignment of yeast Tah1 with TPR domains of mouse CHIP (22), human HOP (21), the engineered protein CTP390 (16), G83R human serine/threonine protein phosphatase 5 (23), and S. cerevisiae Tom71 (24). The structures of all of these domains have been solved with a bound EEVD peptide. The secondary structure sequence of Tah1 is highlighted on top with the helical boundaries indicated. The five highly conserved residues forming the dicarboxylate clamp are indicated by an asterisk. In the alignment, identical residues are shown in white and highlighted in red, whereas highly similar residues are shown in red. The alignment was done using ClustalW2 (43) with default parameters followed by manual inspection and drawn using ESPript (44). B, ribbon representation of the 20 lowest energy conformers of the NMR solution structure of Tah1 bound to MEEVD. The first TPR motif is colored in red, the second in blue, and the C helix in green. Helices are labeled according to the common TPR nomenclature. The last 20 amino acids of Tah1 are unstructured. The MEEVD peptide is in gray with its N terminus in orange and its C terminus in yellow. C, the electrostatic surface potential (red, <−3 kT/e; blue, >3 kT/e) of the lowest energy conformer of Tah1 calculated using DelPhi (45) is shown with the positively charged channel facing the reader. Some residues of interest are highlighted. D, a close-up view of the Tah1 channel of the lowest energy conformer. Side chains of the residues contributing to the positively charged channel are shown in stick representation with nitrogen in blue, oxygen in red, and hydrogen in white. Carbon is colored pink for Lys-8 and Asn-12 in helix 1A, cyan for Asn-43 and Lys-50 in helix 2A, and bright green for Lys-79 and Arg-83 in helix C. All structure figures were generated using PyMOL (Schrodinger LLC, New York).
A positively charged channel ∼11 Å wide and 21 Å long is formed by residues belonging to helices 1A, 2A, and C (Fig. 1, C and D). The MEEVD peptide resides in this channel and stabilizes Tah1. 1H,15N HSQC spectra of Tah1 in the presence and absence of MEEVD are very similar, indicating that the overall fold of the protein is not significantly affected by the presence of the peptide. The interaction between Tah1 and the pentapeptide is characterized by few low intensity NOEs (supplemental Fig. 1), which translates into a disordered ensemble of conformers for the peptide (Fig. 1B). However, the information obtained from these NOE values together with the docking calculations as well as the titration experiments described below provide a clear orientation of the peptide inside the Tah1 binding channel as shown in Fig. 1, B–D, with the peptide lying generally parallel to the helices forming the pocket.
Tah1 Binding to MEEVD
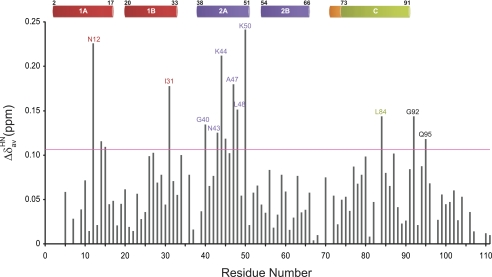
To identify Tah1 residues that interact with MEEVD, 1H,15N HSQC spectra were acquired for labeled Tah1 in the presence of increasing concentrations of MEEVD peptide (Tah1 at 0.6 mm and MEEVD titrated from 0 to 2.2 mm final concentration). According to these measurements and assuming a single binding site, the measured affinity of Tah1 for the peptide is on the order of 1 μm. By mapping the chemical shift changes observed in the titration to the Tah1 structure, residues of Tah1 affected by MEEVD binding can be identified (Fig. 2). Resonance changes with ΔδavgHN = (((ΔδH)2 + (ΔδN/5)2)/5)½ (in ppm) values that are more than one S.D. away from the mean value were selected as being significant. Many residues that exhibit significant changes in chemical shift are located in helix 2A (Gly-40, Asn-43, Lys-44, Ala-45, Ala-47, Leu-48, and Lys-50), which forms the base of the channel where the pentapeptide binds. In addition, there are some residues with high ΔδavgHN values belonging to helix 1A (Asn-12, Leu-14, and Phe-15), helix 1B (Ile-31), helix C (Leu-84), and the unstructured region (Gly-92 and Gln-95). Residues Lys-6 and Lys-8 in helix 1A, Ser-69 and Ala-71 between helix 2B and helix C, and Arg-83 in helix C disappear due to line broadening. None of the residues in helix 2B showed a significant change in chemical shift, consistent with the fact that residues from helix 2B do not contribute to the channel (Fig. 1, B–D). Residues 1–4 do not appear in the HSQC spectra of the free or peptide-bound Tah1, and residues 108 and 109 in the unstructured region do not shift, whereas residues 35, 38, 97, and 105 are prolines.
FIGURE 2.
Mapping the binding of MEEVD to Tah1 by NMR. Shown is a plot of the weighted average chemical shift differences (ΔδavgHN) in the 1H,15N HSQC spectrum between free Tah1 and Tah1 bound to MEEVD (1:10 protein/peptide). The horizontal pink line represents the value of ΔδavgHN that is one S.D. away from the mean value. The secondary structure of Tah1 is shown on top.
Complementary experiments were carried out by titrating 15N-labeled Tah1 with unlabeled Hsp82-MC (residues 260–709 of yeast Hsp82; Tah1 at 200 μm and Hsp82-MC titrated from 0 to 400 μm). The addition of 2 eq of Hsp82-MC to Tah1 caused the complete disappearance of the 1H,15N HSQC signals due to relaxation apart from those belonging to the unstructured C-terminal region of Tah1. The addition of 0.5 eq of Hsp82-MC to Tah1 demonstrated that residues most affected by Hsp82-MC binding to Tah1 are located in the channel created by the three α-helices, as observed with the peptide (data not shown).
In parallel with the NMR titration experiments, binding studies of Tah1 and Tah1 mutants to MEEVD were also carried out using ITC experiments (supplemental Fig. 2) to determine residues critical for mediating this interaction. The mutational studies were based on the NMR titration results of Fig. 2 as well as on the alignment shown in Fig. 1A of other TPR domains whose structures were solved in complex with an EEVD peptide. Based on these structures, it was found that five highly conserved residues are typically involved in mediating electrostatic interactions with the peptide and form a two-carboxylate clamp that tightly interacts with the ultimate Asp-0 residue of the bound peptide. These residues are also conserved in Tah1, namely Lys-8, Asn-12, Asn-43, Lys-79, and Arg-83 (Fig. 1, A, C, and D). Asn-12 and Asn-43 show significant chemical shift changes upon MEEVD binding to Tah1 (Fig. 2), whereas the 1H,15N HSQC signals of Lys-8 and Arg-83 disappear, indicating chemical exchange. No significant chemical shift changes were detected for Lys-79; however, Leu-84, which is directly after Arg-83, exhibits significant chemical shift changes (Fig. 2), indicating that the peptide binds to this region of the protein. The measured dissociation constant (Kd) by ITC for the Tah1-MEEVD interaction is 0.55 ± 0.06 μm, similar to that obtained by NMR above, with a stoichiometry of about 1:1 (Table 3). The interaction is mainly enthalpically driven. The positive entropy of binding might reflect the release of water molecules from the binding site upon the interaction of MEEVD with Tah1. Consistent with the known complex structures and with our NMR titration experiments, mutation of residues that form the two-carboxylate clamp either reduces (K8A and N12A of helix 1A) or abolishes (N43A of helix 2A, K79A and R83A of helix C) binding of MEEVD to Tah1. Furthermore, Mutation of the Lys-50 residue of helix 2A, which exhibited the highest chemical shift change upon MEEVD titration (Fig. 2), was found to reduce peptide binding affinity to Tah1. All of these residues form the positively charged channel of Tah1 (Fig. 1D).
TABLE 3.
Thermodynamic parameters for the interaction of MEEVD peptide with Tah1 and its mutants
| Protein | Kd | ΔG | ΔH | ΔS | n |
|---|---|---|---|---|---|
| μm | kcal mol−1 | kcal mol−1 | cal mol−1K−1 | ||
| Tah1 | 0.55 (0.06)a | −7.9 (0.1) | −6.9 (0.1) | 3.8 (0.4) | 1.14 (0.01) |
| Tah1(K8A) | 36.76 (13.11) | −5.6 (2.0) | −5.3 (1.5) | 1.1 (5.4) | 1.26 (0.24) |
| Tah1(N12A) | 18.83 (3.26) | −6.0 (0.1) | −4.6 (0.7) | 4.9 (2.4) | 0.76 (0.08) |
| Tah1(N43A) | No binding | No binding | No binding | No binding | No binding |
| Tah1(K50A) | 23.20 (6.64) | −5.9 (0.2) | −4.9 (0.8) | 3.6 (3.0) | 1.17 (0.01) |
| Tah1(Q56A) | 0.65 (0.06) | −7.8 (0.1) | −7.0 (0.1) | 3.1 (0.3) | 1.17 (0.01) |
| Tah1(R66A) | 0.83 (0.10) | −7.7 (0.1) | −7.6 (0.1) | 0.3 (0.5) | 1.09 (0.01) |
| Tah1(K79A) | No binding | No binding | No binding | No binding | No binding |
| Tah1(R83A) | No binding | No binding | No binding | No binding | No binding |
| Tah1(1–74) | No binding | No binding | No binding | No binding | No binding |
| Tah1(1–93) | 0.30 (0.06) | −8.3 (0.1) | −3.2 (0.1) | 18.4 (0.4) | 0.73 (0.01) |
a Numbers in parenthesis represent S.D.
Interestingly, Lys-79 and Arg-83 are part of helix C in Tah1 rather than of helix 3A of a third TPR motif as in the case of the other TPR domain proteins (Fig. 1A). Indeed, removal of helix C by truncating Tah1 at residue 74 (Tah1(1–74)) results in a soluble protein that has only two TPR motifs but that cannot bind the MEEVD peptide (Table 3). On the other hand, removal of the unstructured region of Tah1 (Tah1(1–93)) does not significantly affect peptide binding and gives a similar Kd and n as WT protein but a higher ΔS, possibly indicating higher solvent contribution upon peptide binding to Tah1. Finally, consistent with the NMR results of Fig. 2, mutation of residues in helix 2B (Q56A and R66A) has no effect on MEEVD binding to Tah1.
C Termini of Tah1 and Pih1 Are Required for Tah1-Pih1 Interaction
We had proposed and shown earlier that Tah1 forms a ternary complex with Hsp90 and Pih1 (2). This was based on pull-down assays, yeast two-hybrid screens, and functional assays. Pih1 itself was found to be an unstable protein that is stabilized by its interactions with Tah1 and Hsp90. We found that the C terminus of Tah1 (residues 76–111; Figs. 1A and 3A) is required for its interaction with Pih1 (2). To further characterize the interaction between Tah1 and Pih1 and to understand how the stability of Pih1 is modulated by the Hsp90/Tah1 system, we mapped the interaction surface between Tah1 and Pih1 by size exclusion chromatography.
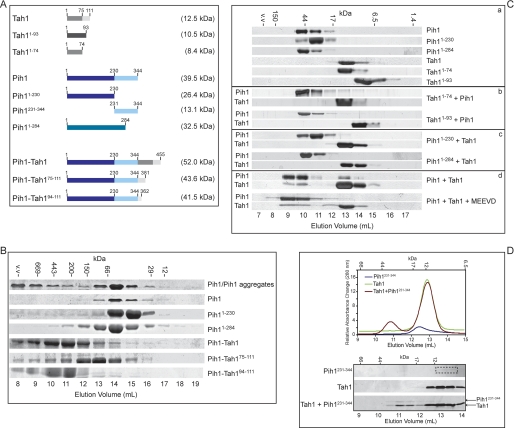
FIGURE 3.
Mapping the interaction between Tah1 and Pih1. A, schematic diagram of the different Tah1 and Pih1 constructs used in the binding experiments. The theoretical molecular weights of the constructs are provided on the right. B, size exclusion chromatography of Pih1 constructs on Superdex 200 column. The elution volume is indicated at the bottom, and the elution positions of the molecular weight standards are indicated at the top of the lanes. C, elution profiles for the different Pih1 (panel a), Tah1 (panel a), and Pih1 + Tah1 (panels b–d) constructs on a Superdex 75 column. D, elution profiles of Tah1 (100 μg), Pih1(231–344) (100 μg), and the mixture of Tah1 (100 μg) and Pih1(231–344) (100 μg) on a Superdex 75 column as observed by absorbance at 280 nm (top) or SDS-PAGE analysis (bottom).
As we demonstrated before (2), full-length Pih1 protein has a tendency to aggregate and to form multiple oligomeric states that spread across the Superdex 200 size exclusion column (Fig. 3B). As shown in the figure, purified Pih1 typically migrates as two peaks on the column; one peak is close to the void volume, above 669 kDa, whereas the other peak is around 50 kDa, close to the molecular mass of a Pih1 monomer (see also Fig. 3C). Upon isolation, this monomeric Pih1 did not readily reaggregate within the time frame of the experiment (Fig. 3, B and C, a). Because Pih1 does not contain any known motifs, we examined the Pih1 sequence and noticed that the Pih1 C-terminal region contains hydrophobic and proline-rich patches. Several C-terminal truncation constructs were then made to find a stable and well behaved fragment of this protein. Pih1(1–230) and Pih1(1–284) had such characteristics. Both, Pih1(1–230) and Pih1(1–284) eluted as monomers on Superdex 200 (Fig. 3B) and Superdex 75 size exclusion columns (Fig. 3C, a). These observations suggest that the aggregation-prone property of Pih1 might result mainly from the Pih1 C-terminal sequence (addressed further below).
In order to stabilize Pih1, we initially naively fused full-length Pih1 to full-length Tah1. The Pih1-Tah1 fusion construct spreads out and runs predominantly as large oligomers and higher order aggregates on a Superdex 200 column (Fig. 3B). However, when Pih1 is fused to the C-terminal 75–111 fragment of Tah1, which contains the C helix and the unstructured region of Tah1 (Fig. 1, A and B), the Pih1-Tah1(75–111) fusion protein migrates as a stable, possibly monomeric protein (Fig. 3B). This is consistent with our earlier observation that Pih1 physically interacts with Tah1 through the Tah1 C terminus (2). When Pih1 is fused to the unstructured C-terminal region of Tah1, the Pih1-Tah1(94–111) fusion protein migrates as higher order oligomers, similar to the Pih1-Tah1 fusion protein. These results suggest that amino acid residues in the C helix and the unstructured region of Tah1 may play an important role in stabilizing the interaction of Pih1 with Tah1. The aggregation of the Pih1-Tah1 fusion might be due to destabilizing steric structural clashes present in such a bigger fusion.
Consistent with the requirement for the C terminus of Tah1 to mediate the Tah1-Pih1 interaction, the deletion of the C helix and the unstructured region of Tah1 (Tah1(1–74)) or only of the unstructured region (Tah1(1–93)) abolished the binding to Pih1 (Fig. 3C, b). Note that, unexpectedly, Tah1(1–93) migrates faster than Tah1 or Tah1(1–74) on the size exclusion column for undetermined reasons. The deletion of the C terminus of Pih1 also abolished the binding of Tah1 to Pih1 (Fig. 3C, c). Hence, the C terminus of Pih1 is required for binding to Tah1. This was further confirmed by observing that the isolated Pih1(231–344) fragment can bind to Tah1 (Fig. 3D). This is in agreement with a recent report showing that residues 199–344 of Pih1 are sufficient for binding to Tah1 (36). Finally, the presence of excess MEEVD peptide did not abolish the Pih1-Tah1 interaction (Fig. 3C, d), suggesting the presence of a Pih1-Tah1-MEEVD ternary complex, which is consistent with our proposal of the presence of Pih1-Tah1-Hsp90 ternary complex (2).
C Terminus of Pih1 Destabilizes the Protein
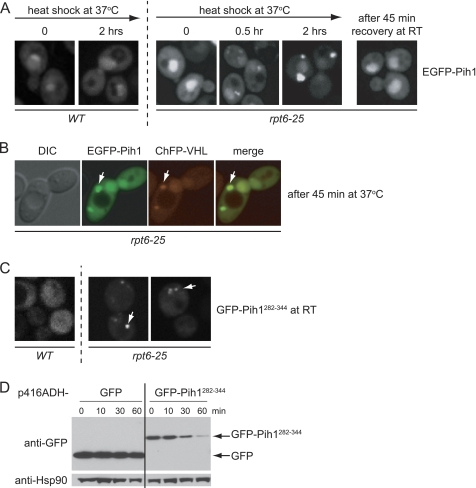
The C terminus of Pih1 seems to play a major role in the destabilization and aggregation of the purified protein in vitro (Fig. 3B). To assess whether this is also true in vivo, initially, WT and rpt6-25 mutant yeast cells (a proteasome ts mutant) expressing full-length EGFP-Pih1 (enhanced GFP fused to the N terminus of Pih1) were grown to midlog phase at room temperature, and then the temperature was shifted to 37 °C for 2 h (Fig. 4A). At the indicated time points, EGFP-Pih1 was visualized in vivo using confocal fluorescence microscopy. At the permissive temperature (room temperature), EGFP-Pih1 was distributed in the cytoplasm and nucleus in both WT and rpt6-25 strains. However, although EGFP-Pih1 localization was unchanged in the WT strain at 37 °C, the inhibition of proteasome-mediated degradation in rpt6-25 strain at this non-permissive temperature induced rapid recruitment of EGFP-Pih1 to discrete foci in the cytoplasm. These cytoplasmic foci became more prominent after 2 h of incubation at 37 °C. The clustering of EGFP-Pih1 in the cytoplasm was fully reversible because shifting the temperature back to room temperature led to the complete dissipation of the foci within 45 min. We hypothesized that these cytoplasmic foci of Pih1 may resemble those formed by misfolded proteins like von Hippel-Lindau (VHL) factor when expressed in yeast. The proper folding of VHL requires the presence of its cofactor elongin BC (37); the absence of elongin BC leads to the misfolding, ubiquitination, and subsequent degradation of VHL in WT yeast cells (38). Therefore, EGFP-Pih1 was co-expressed with ChFP-VHL (cherry fluorescent protein fused to the N terminus of VHL) in rpt6-25 strain to assess whether aggregated Pih1 was with the VHL misfolded protein foci. Complete colocalization of EGFP-Pih1 with ChFP-VHL (Fig. 4B) was observed, suggesting that cytoplasmic foci of EGFP-Pih1 consist of misfolded EGFP-Pih1.
FIGURE 4.
The C terminus of Pih1 destabilizes the protein. A, shown are confocal fluorescence images of S288C WT and rpt6-25 yeast cells expressing EGFP-Pih1. The cells were grown at room temperature (RT) and then exposed to heat shock for 2 h. The rpt6-25 cells were then allowed to recover at room temperature. B, confocal fluorescence images of rpt6-25 cells co-expressing EGFP-Pih1 (green) and ChFP-VHL (red) after exposure to heat shock for 45 min. C, confocal fluorescence images of S288C WT and rpt6-25 cells expressing GFP-Pih1(282–344) at room temperature. D, Western blot, using anti-GFP antibodies, of W303 WT cells expressing GFP or GFP-Pih1(282–344) grown to midlog phase after treatment with cyclohexamide to arrest translation. The Western blot against Hsp90 is shown as a control.
To explicitly test whether the C terminus of Pih1 is responsible for destabilizing the protein, GFP was fused to the 282–344 C-terminal fragment of Pih1, and the localization of this fusion protein, GFP-Pih1(282–344), was assessed in both WT and rpt6-25 mutant cells. We observed GFP-Pih1(282–344) aggregation in 70% of mutant cells but not in WT cells, even at the permissive temperature (Fig. 4C). Furthermore, GFP-Pih1(282–344) was rapidly degraded when translation was arrested by the addition of cyclohexamide in log phase WT cells, whereas GFP was stable (Fig. 4D). This is also consistent with our previous observation that the depletion of endogenous Tah1 in yeast results in the rapid degradation of endogenous Pih1 (2).
These results strongly suggest that the C terminus of Pih1 is responsible for Pih1 instability and aggregation. Furthermore, it is interesting to note that Pih1(282–344) can act as a degradation tag in yeast.
DISCUSSION
The structure of Tah1 is rather unique in that it has only two TPR motifs and a C helix and yet is stable and able to bind the MEEVD peptide. All other known TPR domains that interact with Hsp90/Hsp70 consist of three TPR motifs and a capping helix (Fig. 1A). Hence, the structure of Tah1 was rather unexpected (39). The C helix of Tah1 is not a regular solubility helix as it appears in other TPR proteins, because it hosts two of the five highly conserved residues that establish electrostatic interactions with MEEVD (Figs. 1 and 2 and Table 3). Hence, the C helix serves the role of the third TPR motif in other domains. The dissociation constant of 0.5 μm that we obtained for MEEVD binding to Tah1 by ITC and NMR measurements is consistent with the one obtained for the hepta- and decapeptides described by Millson et al. (39); however, it differs from that for the unprotected peptide, which was reported to have a Kd of about 35 μm. It is also consistent with the Kd obtained for full-length Hsp90 binding obtained by Eckert et al. (36).
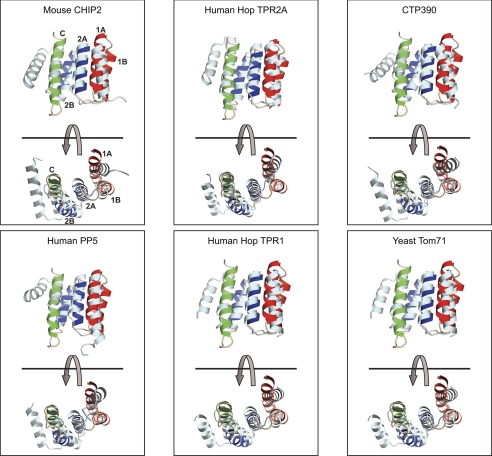
The major overall difference between the structure of Tah1 and that of other TPR domains that bind Hsp90/Hsp70 is that helix 1A and helix C are closer to helix 2A than they are in the other TPR repeat proteins, and, as a result, the binding channel is narrower (Fig. 5). Also, helix 2B appears to be slightly further away from helix 2A in Tah1 compared with other TPR domains. It is reasonable to suggest that this tight packing of helices contributes to the stability of Tah1. The majority of residues that are typically conserved in TPR domains and that are critical for stabilizing the hydrophobic core of these proteins are conserved in Tah1 (40). Hence, Tah1 can be considered to be a minimal TPR domain protein that can bind to Hsp90/Hsp70. The only other minimal stable TPR domain that we know of is that of rat Tom20 (95 residues), which has only one TPR that binds to the mitochondrial targeting presequence but not to Hsp90/Hsp70 (41).
FIGURE 5.
Comparison of Tah1 structure with that of other TPR domains. Shown is the ribbon representation of the lowest energy Tah1 structure without the unstructured region overlaid with the three TPR motifs (in white) of mouse CHIP2 (Protein Data Bank entry 2C2L) (22), human Hop TPR2A (1ELR) (21), the engineered protein CTP390 (3KD7) (16), human G83R protein phosphatase 5 TPR domain (2BUG) (23), human Hop TPR1 (1ELW) (21), and S. cerevisiae Tom71 (3FP2) (24). The structure alignment was carried out in Coot (available on the York Structural Biology Web site) (46).
Helices 1B and 2A of Tah1 are closest in terms of primary sequence and length to the consensus helices found in TPR motifs, which are typically 14 residues long (40). Helix 1A of Tah1 consists of 16 residues and has an additional turn at the C terminus (Fig. 5). In contrast, helix 1A in Tom71 and protein phosphatase 5 is 20 and 19 residues long, respectively, with the additional turns present at the N terminus of the helix. Helix 2B of Tah1 (12 residues) is shorter than that of equivalent helices in the other domains of the family, but helix C (18 residues), which is the equivalent of helix 3A in other domains, is longer. The angle between the two helices within a TPR motif in Tah1 is similar to that measured for other TPR motifs and is about 155–165°.
The NMR measurements of Fig. 2 and the binding experiments of Fig. 3, coupled with the single point mutation ITC results (Table 1 and supplemental Fig. 2), indicate that the amino acid residues involved in binding the Hsp90 C-terminal sequence are Lys-8, Asn-12, Asn-43, Lys-50, Lys-79, and Arg-83. These results are in agreement with our previous (2) and current data showing that neither TPR motifs of Tah1 alone (Tah1(1–75)) nor the C terminus of Tah1(76–111) can bind to Hsp90 on its own. Instead, the five helices of Tah1 are necessary for the binding.
The binding of Tah1 to Pih1 stabilizes Pih1 and is found to be mediated by the C-terminal unstructured regions in the respective proteins. The C helix and the unstructured region of Tah1 are required for this interaction (Fig. 3, B and C). Although we currently have no structural information on the fold of the Pih1 C terminus, it is predicted, using Jpred (42), to predominantly consist of β strands. However, this region of the protein seems to be “problematic” because it leads to Pih1 aggregation in vitro (Fig. 3B) or to its destabilization in vivo (Fig. 4). Furthermore, Pih1(282–344) can lead to the degradation of GFP in vivo (Fig. 4D). It is interesting to note that the pI of Pih1(231–344) and Pih1(282–344) is 9.3 and 9.7, respectively, whereas the pI of Tah1(75–111) and Tah1(94–111) is 4.7 and 3.8 (also see Fig. 1C), respectively. Hence, the stabilizing interaction between these two proteins is expected to be mainly electrostatic.
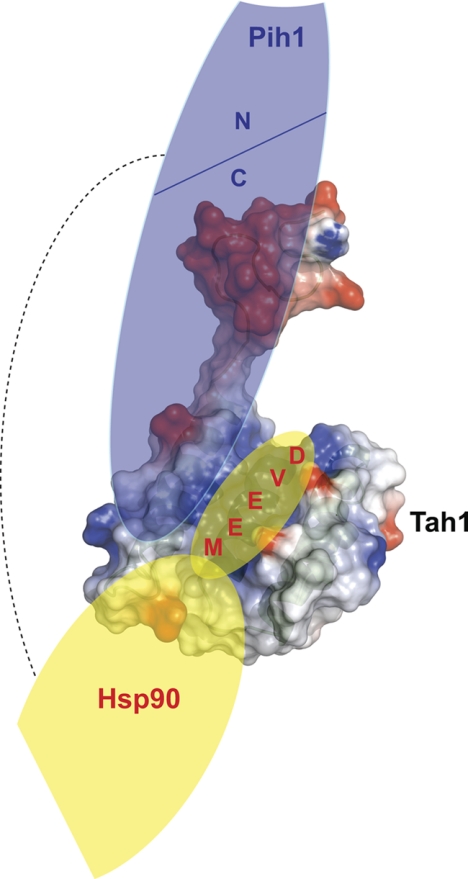
In Fig. 6, we present a schematic of the interactions of Tah1 with Hsp90 and Pih1. Our experiments suggest that the Hsp90 C terminus will bind within the channel formed by Tah1. On the other hand, the C terminus of Tah1 will form strong interactions with the C terminus of Pih1. However, our earlier data (2) as well as those of others (36) suggest that Hsp90 directly binds Pih1 as well. Such a positioning of binding partners would allow this small TPR domain protein to form a ternary complex with Hsp90 and Pih1. Our data collectively suggest that Tah1 is essential for the formation of the Hsp90-Tah1-Pih1 ternary complex that stabilizes Pih1.
FIGURE 6.
Model of the Hsp90-Tah1-Pih1 complex. Model of the complex formed by Pih1 (blue), Tah1 (represented by its electrostatic surface potential), and Hsp90 (yellow) based on the results of our analysis. The C-terminal region of Pih1 interacts with the C helix and the unstructured C terminus of Tah1, whereas the MEEVD peptide at the very C terminus of Hsp90 binds in the positively charged channel of Tah1. Data also indicate that Pih1 and Hsp90 directly interact (dotted line).
Supplementary Material
Acknowledgments
We thank Dr. Francesca Cantini and Prof. Antonio Rosato for help with the structure calculations and software usage. We thank Dr. Hermann Torsten for help with UNIO. We thank Elisa Leung and Usheer Kanjee for help with the ITC experiments and Dr. Majida El Bakkouri for help with the structure figures. A.P.-L. and B. J. acknowledge the WeNMR project (European FP7 e-Infrastructure grant, Contract 261572) for use of Web portals, computing, and storage facilities and CERM (University of Florence) for technical support.
This work was supported by a grant from the Canadian Institutes of Health Research (MOP-93778) to WAH, Spanish Ministerio de Ciencia e Innovación (MICINN, SAF2008-01845) to AP-L.

This article contains supplemental Figs. 1 and 2.
The atomic coordinates and structure factors (code 2L6J) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- TPR
- tetratricopeptide repeat
- ITC
- isothermal titration calorimetry
- VHL
- von Hippel-Lindau
- HSQC
- heteronuclear single quantum coherence.
REFERENCES
- 1. Zhao R., Davey M., Hsu Y. C., Kaplanek P., Tong A., Parsons A. B., Krogan N., Cagney G., Mai D., Greenblatt J., Boone C., Emili A., Houry W. A. (2005) Navigating the chaperone network. An integrative map of physical and genetic interactions mediated by the hsp90 chaperone. Cell 120, 715–727 [DOI] [PubMed] [Google Scholar]
- 2. Zhao R., Kakihara Y., Gribun A., Huen J., Yang G., Khanna M., Costanzo M., Brost R. L., Boone C., Hughes T. R., Yip C. M., Houry W. A. (2008) Molecular chaperone Hsp90 stabilizes Pih1/Nop17 to maintain R2TP complex activity that regulates snoRNA accumulation. J. Cell Biol. 180, 563–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jha S., Dutta A. (2009) RVB1/RVB2. Running rings around molecular biology. Mol. Cell 34, 521–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huen J., Kakihara Y., Ugwu F., Cheung K. L., Ortega J., Houry W. A. (2010) Rvb1-Rvb2. Essential ATP-dependent helicases for critical complexes. Biochem. Cell Biol. 88, 29–40 [DOI] [PubMed] [Google Scholar]
- 5. Boulon S., Marmier-Gourrier N., Pradet-Balade B., Wurth L., Verheggen C., Jády B. E., Rothé B., Pescia C., Robert M. C., Kiss T., Bardoni B., Krol A., Branlant C., Allmang C., Bertrand E., Charpentier B. (2008) The Hsp90 chaperone controls the biogenesis of L7Ae RNPs through conserved machinery. J. Cell Biol. 180, 579–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Thérien C., Bergeron D., Bourassa S., Greenblatt J., Chabot B., Poirier G. G., Hughes T. R., Blanchette M., Price D. H., Coulombe B. (2007) Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell 27, 262–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M. C., Ahmad Y., Neel H., Lamond A. I., Bertrand E. (2010) HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell 39, 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Horejsí Z., Takai H., Adelman C. A., Collis S. J., Flynn H., Maslen S., Skehel J. M., de Lange T., Boulton S. J. (2010) CK2 phospho-dependent binding of R2TP complex to TEL2 is essential for mTOR and SMG1 stability. Mol. Cell 39, 839–850 [DOI] [PubMed] [Google Scholar]
- 9. Ni L., Saeki M., Xu L., Nakahara H., Saijo M., Tanaka K., Kamisaki Y. (2009) RPAP3 interacts with Reptin to regulate UV-induced phosphorylation of H2AX and DNA damage. J. Cell. Biochem. 106, 920–928 [DOI] [PubMed] [Google Scholar]
- 10. Inoue M., Saeki M., Egusa H., Niwa H., Kamisaki Y. (2010) PIH1D1, a subunit of R2TP complex, inhibits doxorubicin-induced apoptosis. Biochem. Biophys. Res. Commun. 403, 340–344 [DOI] [PubMed] [Google Scholar]
- 11. Hirano T., Kinoshita N., Morikawa K., Yanagida M. (1990) Snap helix with knob and hole. Essential repeats in S. pombe nuclear protein nuc2+. Cell 60, 319–328 [DOI] [PubMed] [Google Scholar]
- 12. Sikorski R. S., Boguski M. S., Goebl M., Hieter P. (1990) A repeating amino acid motif in CDC23 defines a family of proteins and a new relationship among genes required for mitosis and RNA synthesis. Cell 60, 307–317 [DOI] [PubMed] [Google Scholar]
- 13. D'Andrea L. D., Regan L. (2003) TPR proteins. The versatile helix. Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 14. Yang J., Roe S. M., Cliff M. J., Williams M. A., Ladbury J. E., Cohen P. T., Barford D. (2005) Molecular basis for TPR domain-mediated regulation of protein phosphatase 5. EMBO J. 24, 1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith D. F. (2004) Tetratricopeptide repeat cochaperones in steroid receptor complexes. Cell Stress Chaperones 9, 109–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cortajarena A. L., Wang J., Regan L. (2010) Crystal structure of a designed tetratricopeptide repeat module in complex with its peptide ligand. FEBS J. 277, 1058–1066 [DOI] [PubMed] [Google Scholar]
- 17. Goebl M., Yanagida M. (1991) The TPR snap helix. A novel protein repeat motif from mitosis to transcription. Trends Biochem. Sci. 16, 173–177 [DOI] [PubMed] [Google Scholar]
- 18. Blatch G. L., Lässle M. (1999) The tetratricopeptide repeat. A structural motif mediating protein-protein interactions. BioEssays 21, 932–939 [DOI] [PubMed] [Google Scholar]
- 19. Wegele H., Müller L., Buchner J. (2004) Hsp70 and Hsp90. A relay team for protein folding. Rev. Physiol. Biochem. Pharmacol. 151, 1–44 [DOI] [PubMed] [Google Scholar]
- 20. Pearl L. H., Prodromou C. (2006) Structure and mechanism of the Hsp90 molecular chaperone machinery. Annu. Rev. Biochem. 75, 271–294 [DOI] [PubMed] [Google Scholar]
- 21. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Structure of TPR domain-peptide complexes. Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 22. Zhang M., Windheim M., Roe S. M., Peggie M., Cohen P., Prodromou C., Pearl L. H. (2005) Chaperoned ubiquitylation: Crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 [DOI] [PubMed] [Google Scholar]
- 23. Cliff M. J., Harris R., Barford D., Ladbury J. E., Williams M. A. (2006) Conformational diversity in the TPR domain-mediated interaction of protein phosphatase 5 with Hsp90. Structure 14, 415–426 [DOI] [PubMed] [Google Scholar]
- 24. Li J., Qian X., Hu J., Sha B. (2009) Molecular chaperone Hsp70/Hsp90 prepares the mitochondrial outer membrane translocon receptor Tom71 for preprotein loading. J. Biol. Chem. 284, 23852–23859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Savchenko A., Yee A., Khachatryan A., Skarina T., Evdokimova E., Pavlova M., Semesi A., Northey J., Beasley S., Lan N., Das R., Gerstein M., Arrowmith C. H., Edwards A. M. (2003) Strategies for structural proteomics of prokaryotes. Quantifying the advantages of studying orthologous proteins and of using both NMR and X-ray crystallography approaches. Proteins 50, 392–399 [DOI] [PubMed] [Google Scholar]
- 26. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 27. Herrmann T., Güntert P., Wüthrich K. (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J. Mol. Biol. 319, 209–227 [DOI] [PubMed] [Google Scholar]
- 28. Cornilescu G., Delaglio F., Bax A. (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR 13, 289–302 [DOI] [PubMed] [Google Scholar]
- 29. Dominguez C., Boelens R., Bonvin A. M. (2003) HADDOCK. A protein-protein docking approach based on biochemical or biophysical information. J. Am. Chem. Soc. 125, 1731–1737 [DOI] [PubMed] [Google Scholar]
- 30. Bertini I., Case D. A., Ferella L., Giachetti A., Rosato A. (2011) Grid-enabled web portal for NMR structure refinement with AMBER. Bioinformatics 27, 2384–2390 [DOI] [PubMed] [Google Scholar]
- 31. Bhattacharya A., Tejero R., Montelione G. T. (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]
- 32. Hu Y., Rolfs A., Bhullar B., Murthy T. V., Zhu C., Berger M. F., Camargo A. A., Kelley F., McCarron S., Jepson D., Richardson A., Raphael J., Moreira D., Taycher E., Zuo D., Mohr S., Kane M. F., Williamson J., Simpson A., Bulyk M. L., Harlow E., Marsischky G., Kolodner R. D., LaBaer J. (2007) Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 17, 536–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Alberti S., Gitler A. D., Lindquist S. (2007) A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast 24, 913–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mumberg D., Müller R., Funk M. (1995) Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 156, 119–122 [DOI] [PubMed] [Google Scholar]
- 35. Cliff M. J., Williams M. A., Brooke-Smith J., Barford D., Ladbury J. E. (2005) Molecular recognition via coupled folding and binding in a TPR domain. J. Mol. Biol. 346, 717–732 [DOI] [PubMed] [Google Scholar]
- 36. Eckert K., Saliou J. M., Monlezun L., Vigouroux A., Atmane N., Caillat C., Quevillon-Chéruel S., Madiona K., Nicaise M., Lazereg S., Van Dorsselaer A., Sanglier-Cianférani S., Meyer P., Moréra S. (2010) The Pih1-Tah1 cochaperone complex inhibits Hsp90 molecular chaperone ATPase activity. J. Biol. Chem. 285, 31304–31312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Feldman D. E., Thulasiraman V., Ferreyra R. G., Frydman J. (1999) Formation of the VHL-elongin BC tumor suppressor complex is mediated by the chaperonin TRiC. Mol. Cell 4, 1051–1061 [DOI] [PubMed] [Google Scholar]
- 38. Kaganovich D., Kopito R., Frydman J. (2008) Misfolded proteins partition between two distinct quality control compartments. Nature 454, 1088–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Millson S. H., Vaughan C. K., Zhai C., Ali M. M., Panaretou B., Piper P. W., Pearl L. H., Prodromou C. (2008) Chaperone ligand-discrimination by the TPR-domain protein Tah1. Biochem. J. 413, 261–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Magliery T. J., Regan L. (2004) Beyond consensus. Statistical free energies reveal hidden interactions in the design of a TPR motif. J. Mol. Biol. 343, 731–745 [DOI] [PubMed] [Google Scholar]
- 41. Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. (2000) Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell 100, 551–560 [DOI] [PubMed] [Google Scholar]
- 42. Cole C., Barber J. D., Barton G. J. (2008) The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36, W197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., Higgins D. G. (2007) ClustalW and ClustalX version 2.0. Bioinformatics 23, 2947–2948 [DOI] [PubMed] [Google Scholar]
- 44. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript. Analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 45. Rocchia W., Sridharan S., Nicholls A., Alexov E., Chiabrera A., Honig B. (2002) Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies. Applications to the molecular systems and geometric objects. J. Comput. Chem. 23, 128–137 [DOI] [PubMed] [Google Scholar]
- 46. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.