Background: Fc receptor-mediated macrophage activation is a major cause of glomerular damage in crescentic glomerulonephritis.
Results: We investigated the role of a novel rat Fc receptor, Fcgr3-rs, in human and rat macrophage activation.
Conclusion: We showed that this receptor prevents the cell signaling of its paralogue (Fcgr3).
Significance: These results provide a novel way to inhibit Fc receptor-mediated cell activation in macrophages.
Keywords: Gene Regulation, Macrophages, Phagocytosis, Rat, Receptor Regulation, Signal Transduction, Fc receptor-mediated Signaling
Abstract
Crescentic glomerulonephritis (Crgn) is a complex disease where the initial insult is often the glomerular deposition of antibodies against intrinsic or deposited antigens in the glomerulus. The role of Fc receptors in the induction and progression of Crgn is increasingly recognized, and our previous studies have shown that copy number variation in Fcgr3 partially explains the genetic susceptibility of the Wistar-Kyoto (WKY) rat to nephrotoxic nephritis, a rat model of Crgn. The Fcgr3-related sequence (Fcgr3-rs) is a novel rat-specific Fc receptor with a cytoplasmic domain 6 amino acids longer than its paralogue, Fcgr3. The Fcgr3-rs gene is deleted from the WKY rat genome, and this deletion is associated with enhanced macrophage activity in this strain. Here, we investigated the mechanism by which the deletion of Fcgr3-rs in the WKY strain leads to increased macrophage activation. By lentivirus-mediated gene delivery, we generated stably transduced U937 cells expressing either Fcgr3-rs or Fcgr3. In these cells, which lack endogenous Fcgr3 receptors, we show that Fcgr3-rs interacts with the common Fc-γ chain but that Fc receptor-mediated phagocytosis and signaling are defective. Furthermore, in primary macrophages, expression of Fcgr3-rs inhibits Fc receptor-mediated functions, because WKY bone marrow-derived macrophages transduced with Fcgr3-rs had significantly reduced phagocytic activity. This inhibitory effect on phagocytosis was mediated by the novel cytoplasmic domain of Fcgr3-rs. These results suggest that Fcgr3-rs may act to inhibit Fcgr3-mediated signaling and phagocytosis and could be considered as a novel mechanism in the modulation of Fc receptor-mediated cell activation in autoimmune diseases.
Introduction
In crescentic glomerulonephritis, leukocytes, especially monocytes/macrophages, infiltrating the glomerulus play a critical role in the development of the disease by recognizing immunoglobulins and complement factors via cell surface receptors (1). One of the checkpoints regulating the immune response in glomerular inflammation is through leukocyte cell surface receptors for the Fc (fragment crystallizable) portion of IgG. The family of Fc receptors can regulate the fine balance between pro-inflammatory versus anti-inflammatory states of cell activation, and there is increasing evidence suggesting that genetic susceptibility to autoimmune diseases is partly dependent on Fc receptor-mediated activating and inhibitory signaling pathways determining the net signal (pro- or anti-inflammatory) in the progression of the disease (2, 3). In systemic lupus erythematosus, aberrant expression or the presence of allelic variants of FcγRs with altered functionality have been reported to contribute to the pathogenesis of the disease (4).
There are two distinct classes of Fc receptors: the activatory and the inhibitory receptors. Most activatory receptors serve as the ligand-binding component of a receptor that also includes a signal transducing molecule containing an immunoreceptor tyrosine-based activation motif (ITAM)2 (5). In monocytes and macrophages, this role is fulfilled by the common γ chain. Signaling via its ITAM motif triggers the activation of multiple downstream signaling pathways including the activation of Src and Syk kinases, calcium mobilization, and NF-κB activation, leading ultimately to cellular responses such as phagocytosis and cytokine production. The importance of activatory Fcγ receptors in glomerulonephritis has been shown using mice engineered to lack the ITAM-bearing Fc receptor γ chain. We and others have shown that these mice are protected from glomerulonephritis induced by antibodies to glomerular basement membrane (6–8). In contrast, the inhibitory Fc receptor FcγRIIB has an immunoreceptor tyrosine-based inhibitory motif and does not use the common γ chain (3). Activatory and inhibitory Fcγ receptors are often expressed on the same cell, and when co-aggregated by IgG antibody, the net signal to the cell depends on the sum total of the activator and inhibitor signals, thus setting thresholds for effector cell responses (3).
In our previous studies of nephrotoxic nephritis (NTN), a model of crescentic glomerulonephritis in the Wistar-Kyoto (WKY) rat, we performed genome-wide linkage analysis for glomerular crescents, macrophage infiltration, and proteinuria in an F2 population derived from NTN-susceptible WKY and NTN-resistant Lewis rats (9). The most significant linkage (logarithm of odds > 8) was obtained with a quantitative trait locus mapping to chromosome 13 (Crgn1) accounting, respectively, for 21.8, 16.7, and 12.9% of the genetic variance in crescent formation, proteinuria, and macrophage infiltration. Positional cloning of Crgn1 led to the identification and functional characterization of the Fcgr3-related sequence (Fcgr3-rs), a novel rat-specific activatory Fc receptor arising from copy number variation in the Fcgr3 gene (9). The genomic rearrangement in the Fcgr3 gene is such that there is a duplication of exon 5 in the NTN-resistant Lewis genomic DNA with the presence of a shorter exon lacking a sequence of 226 bp in its 3′-untranslated region. This shorter exon 5 was deleted in the WKY genome, suggesting a copy number variation in the Fcgr3 gene. Sequence analysis of this shorter copy of exon 5 revealed deletion of a single guanine nucleotide at position 129 (ΔG129) in the coding sequence of the cytoplasmic domain, resulting in a frameshift and generating a novel cytoplasmic domain 6 amino acids longer than that encoded by Fcgr3. We designated this copy number variant Fcgr3-related sequence (Fcgr3-rs) and showed that this novel variant is transcribed and translated in the Lewis strain, whereas it is deleted from the WKY genome (9).
Given the increased macrophage activity and associated NTN susceptibility in the WKY rat that lacks the Fcgr3-rs molecule, it is important to understand how the presence of this molecule might be influencing macrophage activity. Thus, we have studied the role of Fcgr3-rs in macrophage activation. We first investigated whether the genomic duplication/deletion event giving rise to Fcgr3-rs occurred during the derivation of inbred Wistar-related colonies. We have then generated U937 cells stably expressing either Fcgr3 or Fcgr3-rs and have studied the role of Fcgr3-rs in macrophage activation both in U937 cells and WKY bone marrow-derived macrophages (BMDMs) transduced with Fcgr3-rs. Our results show that the absence of Fcgr3-rs is not specific to the WKY strain but is widely distributed throughout the rat phylogenetic tree. Furthermore, in both human monocyte cell lines and primary rat macrophages, expression of Fcgr3-rs inhibits Fcgr3-mediated signaling and phagocytosis, suggesting that the rat possesses a unique mechanism by which Fc receptor-mediated cell activation can be modulated.
EXPERIMENTAL PROCEDURES
Animals
WKY (WKY/NCrl) rats were purchased from Charles River. All of the procedures were performed in accordance with the United Kingdom Animals (Scientific Procedures) Act.
Reagents
RPMI 1640 containing 25 mm l-glutamine was purchased from Biowhittaker (Berkshire, UK). FCS was from PAA (Yeovil, UK) or Labtech (East Sussex, UK). All of the growth media were supplemented with penicillin (100 units/ml) and streptomycin (100 μg/ml), supplied as a 100× sterile solution from Biowhittaker. LPS was obtained from Sigma. Antibodies were from Millipore (anti-Myc), Santa Cruz (F(ab′)2, anti-Syk, anti-CD16), GE Healthcare (HRP-conjugated antibodies for Western blot), or Upstate Biotech (anti-γ chain). pCSGW vector was a gift from Richard Jenner (University College London, London, UK).
Fcgr3 Exon 5 Genotyping
Fcgr3 exon 5 genotype was determined by PCR and agarose gel electrophoresis by using primers to amplify exon 5 genomic DNA from 134 laboratory rat strains and substrains available in the National BioResource project for the rat in Japan. The primer sequence used is as follows: 5′-GTCCCTAAATTCTGAATTTC-3′ and 5′-AAAGAAGTCACAGAAAGGAG-3′.
Lentiviral Constructs
Fcgr3 and Fcgr3-rs were amplified from Lewis spleen cDNA with forward primer 5′-AAGCTTGCCACCATGACTTTGGAG-3′ and reverse primer 5′-TCTAGATTATTAGCCATACGATGGGAT-3′. Clones were reamplified with forward primer 5′-CGGAAGCTTGCCACCATGGAGCAGAAACTCATCTCTGAAGAGGATCTGACTTTGGAGACCCAGATGTTTCAG-3′ and reverse primer 5′-CCTTGAGCACCTGGATCCATGGGG-3′ and recloned. Site-directed mutagenesis was performed on the Fcgr3 construct for ΔG129 with primers 5′-GAGAAATCTTCAAACCTCGGGGAGGACTGGAGGAAATCCC-3′ (forward) and 5′-GGGATTTCCTCCAGTCCTCCCCGAGGTTTGAAGATTTCTC-3′(reverse). All α subunit constructs were cloned into the pcDNA3.1/Hygro(+) expression vector (Invitrogen). For inserting an N-terminal Myc tag into the Fcgr3 constructs, 1.0 μg of the Fcgr3, Fcgr3-rs, and Fcgr3-ΔG constructs were amplified with forward primer 5′-CGGAAGCTTGCCACCATGGAGCAGAAACTCATCTCTGAAGAGGATCTGACTTTGGAGACCCAGATGTTTCAG-3′ and reverse primer 5′-CCTTGAGCACCTGGATCCATGGGG-3′. Constructs and PCR products were cut with HindIII and BamHI and ligated to each other. Lentivirus constructs were prepared as follows: DNA was excised from pCDNA3.1 by digestion with HindIII/XbaI, and blunt ends were created using Pfu DNA polymerase. This was ligated into a SmaI cut pENTR4.3F. This “in house” modified version of the pENTR4 gateway vector contains a multiple cloning site downstream of a CMV promoter and upstream of an internal ribosome entry sequence, which mediates expression of an enhanced GFP sequence. Inserts in the correct orientation were identified by diagnostic restriction enzyme digests. Subsequent digestion of this vector with SalI and NotI released a DNA “cassette” containing the “gene of interest” internal ribosome entry sequence-EGP sequences. This was then cloned into the pCSGW lentiviral transfer vector (10) following removal of the vectors own GFP sequence.
Lentiviral Production and Titration
Transfer vector (pCSGW), lentiviral mammalian expression vector (envelope, pMD2.G), and packaging vector (psPAX2) were mixed at a ratio of 4:3:1 and used to infect confluent monolayers of 293T17 cells by calcium phosphate precipitation. The medium was replaced after 16 h. Media containing virus were collected after a subsequent 24 and 48 h of culture. Insoluble debris was removed by centrifugation and sterilized by filtration through 0.2-μ filters. This crude viral supernatant was stored at −80 °C until use. Titers of viral preparations produced in this way were established by infection of U937 cells as follows. Briefly, U937 cells were mixed with varying dilutions of viral supernatant (typically 1/5 to 1/500) in the presence of 8 mg/ml polybrene (hexadimethrine bromide; Sigma). The cells were “spinoculated” by spinning at 2200 rpm for 2 h at 30 °C. After 72 h, the percentage of GFP-positive cells (FITC channel) was determined by FACS analysis (using a LSRII FACS). The lowest dilution factor with GFP+ score of <20% was used to calculate viral titer.
U937 Lentiviral Line Production
Human monoblastic leukemia U937 cells were infected with lentivirus carrying the gene of interest at a ratio of 1:1 (cells:virus) as described above. The cells were then cultured overnight before removal of virus containing media. Cell surface Myc-positive cells (Myc-Fcgr3 or Myc-Fcgr3RS) were isolated by panning on 10-cm Petri dishes coated with anti-Myc antibody (Millipore). Adherent cells were expanded in culture and used in subsequent experiments. GFP-positive cells expressing pCSGW (no surface Myc) were isolated by cell sorting based on FITC fluorescence. Subsequent analysis of cell lines by FACS analysis confirmed 90–100% of cells to be FITC-positive.
FcR Stimulation
The cells were resuspended in ice-cold serum-free RPMI and anti-Myc antibody (Millipore) added to a final concentration of 130 μg/ml. After 60 min of incubation on ice, the cells were washed three times and resuspended in serum free RPMI with 8 mg/ml goat anti-mouse IgGF(ab′)2. The cells were then incubated at 37 °C for the indicated times. The reaction was stopped by the addition of gel sample buffer (2% SDS, 10% glycerol, 0.5% β-mercaptoethanol, 50 mm Tris, pH 6.8).
Immunoprecipitations and Western Blot Analysis
The cells were lysed in ice-cold lysis buffer (25 mm Tris, pH 7.6, 150 mm NaCl, 1 mm EDTA, 0.1% Triton X-100, pH 7.8, 1% digitonin), containing protease inhibitor mixture (Sigma) with 1 mm DTT, 10 mm NaF, 100 mm sodium orthovanadate). The nuclei and cell debris were removed by centrifugation, and supernatants were precleared with protein G-Sepharose. Myc-labeled proteins were then precipitated with anti-Myc antibody (Millipore) or isotype matched control antibody and protein G-Sepharose for 1.5 h. Immunoprecipitated complexes were washed in lysis buffer and were resolved on 4–20% SDS-PAGE gels (Criterion).
After electrotransfer on to nitrocellulose membrane, the membranes were blocked in 2% BSA/TBS/Tween (0.05%) and probed with the appropriate antibodies at 4 °C overnight followed by HRP-conjugated secondary antibody. Blots were developed using ECL (Amersham Biosciences).
ELISAs
At 24 h after stimulation, the supernatants were harvested. The concentrations of TNFα and IL-10 were determined by ELISA (PharMingen) according to the manufacturer's instructions. Absorbance was read and analyzed at 450 nm on a spectrophotometric ELISA plate reader (Labsystems Multiskan Biochromic) using the Ascent software program.
BMDM Preparation and Phagocytosis Assay
Femurs from adult WKY rats were isolated and flushed with Hanks' balanced salt solution. Total BM-derived cells were plated and infected with lentivirus carrying the gene of interest at a ratio of 5:1 (cells: virus). 48 h following the infection, the medium was replaced with DMEM that contained 25 mm HEPES (Sigma), 25% L929 conditioned medium, 25% decomplemented FBS (Biosera), penicillin (100 units/ml; Invitrogen), streptomycin (100 μg/ml; Invitrogen), and cultured for 5 days. These cells were characterized as macrophages by ED-1 staining. To assess phagocytosis, latex polystyrene 6.0-μm microspheres (50 beads/macrophage; Polysciences Inc.) were opsonized with BSA-anti-BSA IgG (Sigma) and added to transduced WKY macrophages cultured in 8-well glass chamber slides. Following staining with Diff-Quick fix (Dade Behring), 200 macrophages were counted to determine the number of beads ingested per cell.
Expression Analysis by qRT-PCR
Stably transduced U937 cells were differentiated in macrophages after adding PMA (100 nm) for 48 h and stimulated with LPS (Sigma; 1 μg/ml) for 3 h. Total RNA was extracted using TRIzol (Invitrogen), and one-step qRT-PCR was performed using 100 ng of total RNA and Brilliant II SYBR® Green QRT-PCR Master Mix Kit (Agilent) with specific primers for Tnfa and IL1b1 in an ABI 7900 sequence detection system (Applied Biosystems, Warrington, UK). In rat BMDMs, Fcgr3 levels relative to total Fcgr3 (Fcgr3 + Fcgr3-rs) were assessed by selecting a forward primer where Fcgr3 differs from Fcgr3-rs by two single nucleotide polymorphisms at the end of the 3′-end. The selected primers were Fcgr3-Forward: 5′TCTAGTGTGGTTCCATGCCG-3′ and Fcgr3-Reverse: 5′-TGTCCTGTGGAGCCTTGTACT-3′; total Fcgr3 (Fcgr3+Fcgr3-rs)-Forward: 5′-CTCCAGACCCCTCAACTGGT-3′ and total Fcgr3 (Fcgr3+Fcgr3-rs)-Reverse: 5′-GCAGTAGTAGTTCCCACTGT-3′.
Statistical Analysis
Statistical differences in mean values were compared using one-way analysis of variance followed by Dunnett's multiple comparison post-test; p < 0.05 was considered statistically significant.
RESULTS
The Phylogeny of Genomic Deletion of Fcgr3-rs in Laboratory Rat
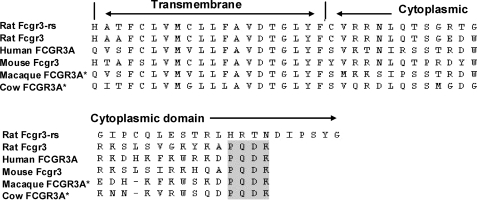
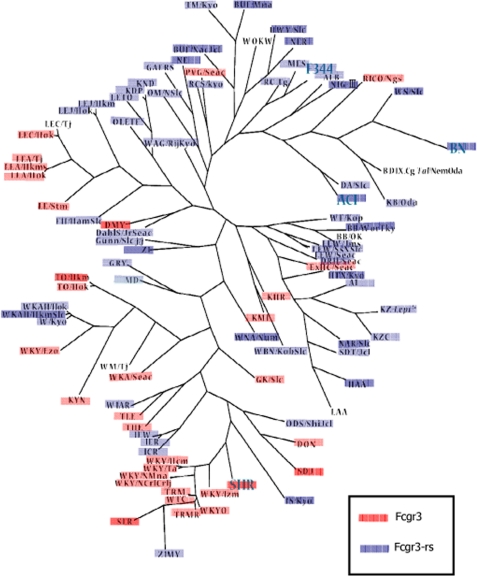
Exon 5 of Fcgr3-rs contains a deletion of a single guanine nucleotide at position 129 (ΔG129) in the coding sequence of the cytoplasmic domain (9). When compared with Fcgr3, this ΔG129 variant results in a frameshift in the Fcgr3-rs coding sequence and generates a novel cytoplasmic domain 6 amino acids longer than that encoded by Fcgr3 that was not found in any other species (Fig. 1). We have previously shown that Fcgr3-rs was deleted from NTN-susceptible Kyoto-derived strains (9), but to investigate the distribution of this deletion throughout the rat genealogical tree, we carried out a phylogenetic tree analysis of 94 inbred laboratory strains and 40 of their substrains (Fig. 2). The results show that the deletion of Fcgr3-rs is widespread throughout the rat phylogeny (32 of 94 strains), suggesting that the origin of the deletion event is earlier then the derivation of these strains. We also investigated the relative levels of Fcgr3 and Fcgr3-rs mRNA levels in Lewis rat BMDMs by qRT-PCR and showed that although there is relatively increased Fcgr3 expression when compared with Fcgr3-rs, LPS stimulation did not change the relative levels of both receptors (Table 1).
FIGURE 1.
Rat-specific Fcgr3-rs is a novel Fc receptor with a unique cytoplasmic domain 6 amino acids longer then Fcgr3. Shown is a comparison of amino acid sequences of transmembrane and cytoplasmic domains of rat Fcgr3 and Fcgr3-rs, human Fcgr3, and FCGR3 in other species. Fcgr3-rs has lost the terminal PQDK sequence (highlighted) that is highly conserved across species. *, amino acid prediction based on human FCGR3A using Ensembl Genome Browser.
FIGURE 2.
Phylogenetic tree of 134 laboratory rat strains according to the presence or absence of Fcgr3-rs in the genome. A phylogenetic tree of 94 strains is shown. Strains having numerous substrains (i.e. SHR, F344) are shown in capital letters, and all of the substrains showed the same genotype as the founder strain. The tree was developed through a heuristic search for maximum parsimony implemented in PAUP 4.0b10. TreeView was used to display the radial tree. For BD IX, LEC/Tj, and WM/Tj, genotypes could not be determined.
TABLE 1.
Relative mRNA levels of Fcgr3 and Fcgr3-rs in Lewis rat BMDMs
Expression was assessed by qRT-PCR with specific primers that differentiate between Fcgr3 and Fcgr3-rs in basal and LPS-stimulated (100 ng/ml, for 5 h) BMDMs. Fcgr3 and Fcgr3-rs expression levels were normalized to total Fcgr3 (Fcgr3 + Fcgr3-rs) levels.
| Expression | BMDM |
|
|---|---|---|
| Basal | LPS | |
| Fcgr3/(Fcgr3 + Fcgr3-rs) | 0.73 ± 0.02 | 0.67 ± 0.01 |
| Fcgr3-rs/(Fcgr3 + Fcgr3-rs) | 0.27 ± 0.02 | 0.33 ± 0.01 |
U937 Cells Expressing Either Fcgr3 or Fcgr3-rs: Surface Expression and Interaction with γ Chain
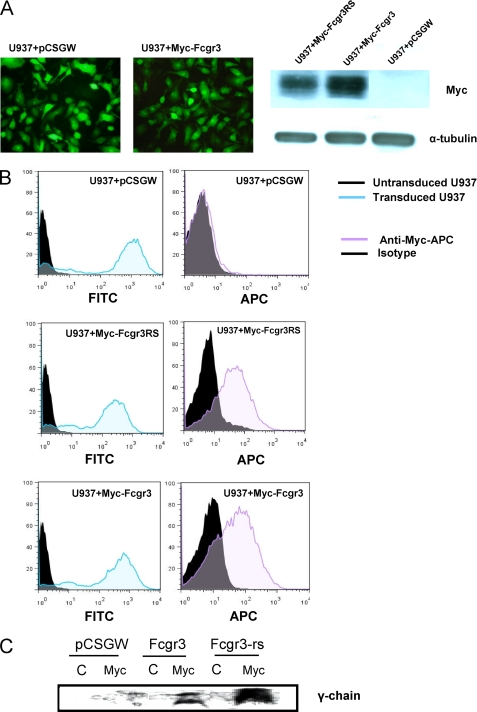
The U937 cell line does not express any endogenous FcgRIIIA (11) and is therefore an ideal tool for the analysis of FcgRIII receptors, allowing direct functional comparison between the rat orthologues Fcgr3 and Fcgr3-rs. Thus, to study the role of Fcgr3-rs in macrophage activation, we generated U937 cells stably transduced with lentivirus constructs expressing the protein of interest. U937 cells were infected with either Myc-tagged Fcgr3 (Myc-Fcgr3), Myc tagged-Fcgr3-rs (Myc-Fcgr3RS), or empty (pCSGW) lentiviral vectors. All of the lentiviral vectors contained GFP. Expression of Myc-Fcgr3 and Myc-Fcgr3-rs was confirmed by fluorescence microscopy, detecting GFP+ cells and also by a Myc Western blot on lysates from stably transduced U937 cells (Fig. 3A). We then investigated whether Myc-Fcgr3 and Myc-Fcgr3-rs are expressed on the surface of U937 cells. Flow cytometry analysis using an anti-Myc-APC mouse antibody (mAb) showed that both Myc-Fcgr3 and Myc-Fcgr3-rs are expressed on the cell surface at similar levels in these cells (Fig. 3B). Because Fcgr3 transport to and expression at the cell surface is known to be dependent on its association with the common γ chain (12), these findings implied that both the Fcgr3 and Fcgr3-rs molecules were associated with γ chain at the cell surface. Co-immunoprecipitation analysis of U937 cells expressing either Fcgr3 or Fcgr3-rs demonstrated that this was indeed the case (Fig. 3C). Thus, although Fcgr3-rs has an additional cytoplasmic domain 6 amino acids longer than its paralogue Fcgr3, it binds the γ-chain on the surface of stably transduced and PMA-differentiated U937 macrophages.
FIGURE 3.
Surface expression of Fcgr3-rs and Fcgr3 and assessment of their interaction with the γ chain in stably transduced U937 cells. A, illustration of GFP expression in U937 + pCSGW and U937 + Myc-Fcgr3 by fluorescent microscopy (×40). Myc and α-tubulin (loading control) Western blots of whole cell lysates from stably transduced U937 cells are shown. B, the human monoblastic leukemia cell line U937 was stably transduced to express either Myc-Fcgr3 (U937 + Myc-Fcgr3) or Myc-Fcgr3RS (U937 + Myc-Fcgr3RS) lentiviral constructs co-expressing GFP together with transgene. Untransduced cells (U937) and cells transduced with lentiviral construct expressing GFP alone (without any transgene, U937 + pCSGW) were used as controls. Surface expression was measured by using anti-Myc-APC mAb. All of the transduced cells were GFP+ as shown by FITC emission. C, stably transduced U937 cells were differentiated into macrophages after adding PMA (100 nm) for 48 h, and cell lysates were precipitated with anti-Myc antibody. Immunoprecipitated proteins were blotted with anti-γ chain antibody.
Fcgr3-rs-mediated Signaling and Phagocytosis in U937 Cells
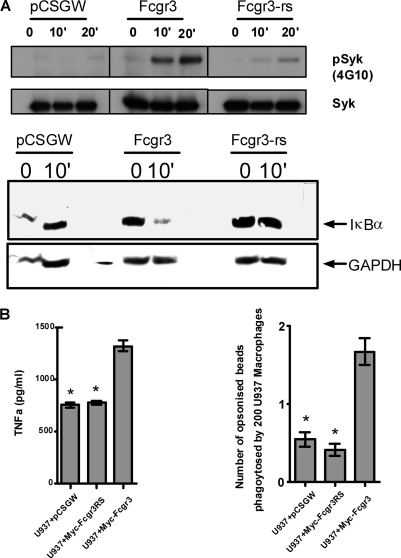
In most macrophage Fc receptor complexes (with the exception of FcgrII), the ITAM-containing common γ-chain is necessary for downstream signaling (13, 14). Thus, the dimeric γ-chain forms multichain complexes with the ligand binding α-chain of Fcgr3 to allow signal transduction (3). This signaling starts with tyrosine phosphorylation of the ITAMs by kinases of the SRC family (15, 16) and leads to the recruitment and subsequent phosphorylation of the Syk kinase, followed by the activation of various downstream targets such as PI3K and NF-κB. Subesquent changes in gene expression led to the induction of inflammatory cytokines such as TNFα. We were therefore keen to understand whether the Fcgr3-rs is able to transduce signals similar to those described for Fcgr3. However, despite its interaction with the γ-chain, cross-linking of Fcgr3-rs with anti-Myc antibody does not lead to phosphorylation of Syk or the degradation of IκBα (Fig. 4A). This was not the case for the U937 macrophages stably transduced with Fcgr3 (Fig. 4A).
FIGURE 4.
Fc receptor-mediated signaling and phagocytosis in U937 cells stably transduced with Myc-Fcgr3RS. A, Fc receptor stimulation after cross-linking with anti-Myc Ab for a maximum of 20 min and analysis of Syk phosphorylation and IκBα degradation in stably transduced U937-derived macrophages. B, TNFα levels were assessed by sandwich ELISA after cross-linking stably transduced U937 macrophages by anti-Myc mAb. Supernatants were collected after 48 h of incubation. Fc receptor-mediated bead phagocytosis was assayed by incubating macrophages with IgG opsonized latex beads. Nonopsonized beads did not show any phagocytosis. *, p < 0.001 when compared with U937 + Myc-Fcgr3.
Furthermore, Fc receptor-mediated TNFα production and IgG-opsonized bead phagocytosis were also significantly less in U937 macrophages expressing Fcgr3-rs when compared with U937 macrophages stably transduced with Fcgr3 (Fig. 4B). Taken together, these results suggest that the rat-specific Fcgr3-rs is unable to mediate the same intracellular signals as the wild type receptor.
Fcgr3-rs-and TLR4-mediated TNFα and IL-1β Expression Levels
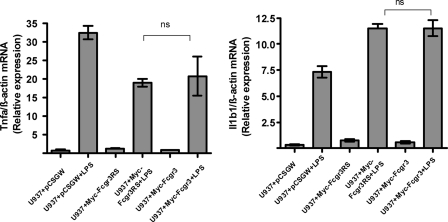
To test whether the relative reduction in TNFα production in U937 cells transduced with Myc-Fcgr3RS is specific to receptor cross-linking, we have stimulated the stably transduced PMA-differentiated U937 cells with LPS for 3 h and measured TNFα and IL-1β expression levels by qRT-PCR (Fig. 5). Although the results have confirmed the previously shown up-regulation of these genes in U937 cells (17), there was no significant difference in the up-regulation of TNFα and IL-1β between U937 + Myc-Fcgr3 and U937 + Myc-Fcgr3RS, demonstrating that the inability of these cells to produce TNFα is not an intrinsic property of these cells and is specific to Fcgr3 stimulation (Fig. 5).
FIGURE 5.
Fcgr3-rs does not affect TLR4-mediated Tnfa and Il1b1 expressions in stably transduced U937 cells. Stably transduced U937 cells were differentiated in macrophages after adding PMA (100 nm) for 48 h and stimulated with LPS (1 μg/ml) for 3 h. Tnfa and Il1b1 gene expression were measured by qRT-PCR. ns, not significant.
The Role of the Novel Cytoplasmic Domain of Fcgr3-rs in Rat and Human Macrophage Activation
To study the effect of Fcgr3-rs on the phagocytic activity of primary WKY BMDMs where other endogenous, FcRs (Fcgr3 and others) are expressed, we transduced these cells with lentivirus encoding either Myc-Fcgr3rs or Myc-Fcgr3 and used cells transduced with the empty vector (pCSGW) as controls.
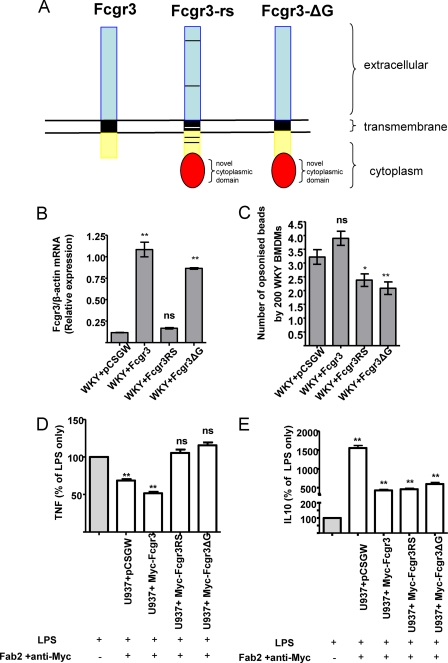
Because the Fcgr3-rs also contains a number of single-base pair substitutions in its extracellular domain, we were also keen to establish whether these might explain the differences between the related sequence and wild type Fcgr3 molecules, possibly as a result of altered FcR binding. Thus, we also generated an additional lentiviral construct encoding a chimeric protein to specifically test the effect of the novel cytoplasmic domain of Fcgr3-rs on Fc receptor-mediated phagocytosis. Fcgr3-ΔG contains the wild type Fcgr3 extracellular, transmembrane, and cytoplasmic domains with the presence of the Fcgr3-rs-specific novel cytoplasmic domain (9) (Fig. 6A).
FIGURE 6.
Fcgr3-rs inhibits Fc receptor-mediated phagocytosis in WKY BMDMs. A, schematic representation of the functional differences between Fcgr3 and Fcgr3-rs. Although there are amino acid changes in the Fcgr3-rs extracellular, transmembrane, and cytoplasmic domains (indicated by bars) when compared with Fcgr3, the major structural difference is the presence of a novel cytoplasmic domain. The chimeric Fcgr3-ΔG has the extracellular transmembrane and cytoplasmic domains identical to Fcgr3, the only difference being the presence of the novel cytoplasmic domain. B, Fcgr3 expression was assessed by qRT-PCR in stably transduced U937 cells by using specific primers designed based on polymorphisms in the transmembrane domain differentiating between Fcgr3 and Fcgr3-rs. The expression of Myc-Fcgr3 ΔG was also assessed and showed similar expression levels as Fcgr3. This is because Fcgr3 and Fcgr3-ΔG share the same sequence in the transmembrane domain. C, WKY bone marrow-derived cells were transduced with lentiviral constructs co-expressing GFP with either Myc-tagged Fcgr3-rs (Myc-Fcgr3RS) or Myc-tagged wild type Fcgr3 (Myc-Fcgr3) or Myc-tagged Fcgr3-ΔG. The lentiviral construct expressing GFP alone (without any transgene) was used as control (pCSGW). The cells were then differentiated into BM-derived macrophages by culturing in DMEM containing L929, and Fc receptor-mediated phagocytosis was performed. **, p < 0.01; *, p < 0.05; ns, not significant when compared with pCSGW. D and E, LPS-induced TNFα (D) and IL-10 (E) levels were assessed by sandwich ELISA after cross-linking stably transduced U937 macrophages by anti-Myc mAb and F(ab′)2. Following cross-linking, the cells were washed, and supernatants were collected after 5 h of incubation with LPS (1 μg/ml). TNFα and IL-10 production were expressed as a percentage of that produced in LPS-stimulated cells without FcR cross-linking (100% of cytokine production). **, p < 0.001 when compared with LPS-stimulated cells without FcR cross-linking (100% of cytokine production; histogram in gray)
We first measured the expression of Fcgr3 by using primers specific for the transmembrane domain, thereby allowing differential amplification between Fcgr3 and Fcgr3-rs transcripts (Fig. 6B). We then measured the number of opsonized beads phagocytosed by 200 WKY BMDMs transduced with each different lentiviral construct.
Phagocytosis was significantly reduced in Fcgr3-rs transduced WKY BMDMs when compared with those transduced with pCSGW (Fig. 6C). This inhibitory effect is clearly mediated by the novel cytoplasmic domain of Fcgr3-rs, because similar reductions in phagocytic activity were seen in WKY BMDMs transduced either with Fcgr3-ΔG or with Fcgr3-rs (Fig. 6C). Taken together, these data suggest that the presence of FcRs containing the related sequence cytoplasmic domain is able to reduce phagocytosis mediated by endogenous FcRs in the WKY BMDMs.
To further understand the mechanism of action of the Fcgr3-rs, we then investigated how the Fcgr3 and Fcgr3-rs interact with TLR4-mediated TNFα production following LPS stimulation. U937 cells transduced with FcRs (U937 + Myc-Fcgr3, U937 + MycFcgr3RS, and U937 + Myc-Fcgr3ΔG) were cross-linked with anti-Myc antibodies and IgG F(ab′)2. Following washing, the cells were stimulated with LPS for 5 h. TNFα and IL-10 protein levels were measured by sandwich ELISA.
In the absence of any additional FcR expression in these cells, treatment with Ab·Fab complexes resulted in a 38% decrease in TNF production. This was further decreased in the presence of the Fcgr3 molecule to 50% of that seen in cells that are not cross-linked, revealing that signals from Fcgr3 act to reduce TLR4-induced TNF production in U937 cells (Fig. 6D). In contrast, the presence of either the Fcgr3-rs or Fcgr3ΔG did not result in any changes in TNFα production compared with cells that are not cross-linked. Levels of IL-10 produced in these cells did not differ significantly between Fcgr3 and FcgR3-rs or FcgR3-ΔG cells (Fig. 6E), revealing that the Fcgr3-rs is not functioning in these cells by increasing levels of this anti-inflammatory cytokine.
DISCUSSION
There is considerable evidence from experimental models of crescentic glomerulonephritis that Fcγ receptors play a central role in disease progression. In mice, the FcR chain-deficient mice (FcRγ−/−) lack surface expression of both murine activator Fc receptors I, III, and IV, and the high affinity IgE receptor (FcRI) (18). Several studies have shown that FcRγ−/− mice are protected from disease in accelerated nephrotoxic nephritis (6, 7, 19), and we have shown that the disease is mediated by Fcγ receptors on circulating leukocytes and not intrinsic renal cells (8). Our previous studies in the NTN model in the Wistar Kyoto rat showed that the disease architecture is complex and highly heritable, with seven quantitative trait loci (Crgn1–7) linked to the percentage of glomerular crescents and controlling mainly macrophage activation through distinct transcriptome profiles (9, 20–22). In this model, we showed that copy number variation in rat Fcgr3 locus led to the deletion of Fcgr3-rs in the WKY rat, predisposing this strain to enhanced macrophage activity (9).
The purpose of the current study was to understand the mechanism by which the genomic deletion of Fcgr3-rs leads to macrophage overactivation and whether the novel cytoplasmic domain binds to the common γ chain to trigger downstream signaling pathways. We first investigated the distribution of Fcgr3-rs deletion throughout the rat phylogeny and found that the deletion was widespread. It was previously reported that five outbred Wistar and one Long-Evans stocks have Fcgr3-rs (23), suggesting that the deletion has originated from the ancestor outbred Wistar colony in the Wistar institute in the United States. We now confirm and extend these findings because all of the laboratory rat strains that we genotyped for Fcgr3 copy number variation are derived from outbred Wistar and Long-Evans stocks.
Previously, to investigate how Fcgr3-rs exerts its inhibitory effect on Fcγ receptor signaling, we have transfected COS7 cells and have shown that although Fcgr3-rs is expressed on the cell surface, it is less efficient than Fcgr3 in mediating phagocytosis (9). It also had an inhibitory effect on Fcgr3-mediated phagocytosis when both receptors were co-transfected in COS7 cells (9). However, the COS7 cells are not primarily phagocytic cells and do not endogenously express the γ chain, making it difficult to draw mechanistic conclusions concerning the role of Fcgr3-rs in Fc receptor-mediated signaling and activation. Here we have examined the signaling function of Fcgr3-rs in a human monocyte cell line U937, a well characterized cell line previously used to study Fc receptor function (11). U937 cells constitutively express only Fcgr1 and Fcgr2a but not Fcgr3 (11). We have used lentivirus-mediated gene transfer to generate stably transduced U937 cell lines. Our results showed that Fcgr3-rs associates with the common γ-chain in these cells, but Fc receptor-mediated signaling and phagocytosis are defective. Importantly, WKY BMDMs transduced with the Fcgr3-rs also showed reduced bead phagocytosis, and this was mediated through the rat-specific novel cytoplasmic domain.
It is well established that the transmembrane domain is required for association with the γ chain and consequently cell surface expression of Fcgr3 (24). Because of the amino acid changes between Fcgr3 and Fcgr3-rs in the transmembrane domains, it could therefore be argued that the amino acid changes simply prevented Fcgr3-rs expression. This, however, was not the case, and in both U937 cells and primary rat macrophages, Fcgr3-rs was expressed at the cell surface at similar levels to Fcgr3. The use of the ΔG chimeric molecule, which contains the extracellular and transmembrane domains of Fcgr3 combined with the cytoplasmic domain of Fcgr3-rs, also demonstrates that the differences seen between Fcgr3 and Fcgr3-rs functions are mediated only by the novel cytoplasmic domain of Fcgr3-rs and cannot be explained by point mutations in the extracellular domain of Fcgr3-rs, which might simply have prevented Fc binding of IgG.
The majority of signaling cascades emanating from the Fcgr3 receptor complex are thought to be derived from the γ chain via its ITAM motifs. Thus, removal of the γ chain cytoplasmic domain or mutations of tyrosine residues within the ITAM motifs affects intracellular Ca+ levels, PI3K activity and receptor-induced phosphorylation events (25). In particular there is compelling evidence that activation of Syk is essential for subsequent events such as receptor-induced phagocytosis (26, 27). However, the cytoplasmic tail of the Fcgr3 molecule is unlikely to be completely redundant in this process, and Hou et al. (28) have suggested that sequences within the cytoplasmic domain of Fcgr3, in particular those close to the transmembrane domain, are required for signaling. Our findings demonstrate that signaling from the Fcgr3-rs is clearly different from that mediated by Fcgr3, because Fcgr3-rs·γ chain complexes do not generate phosphorylated Syk molecules or activate the NF-κB transcription factor that is required for transcription of Fc-responsive genes such as TNFα. Accordingly, IgG-induced TNFα production and phagocytosis are both significantly lower in U937 cells expressing Fcgr3-rs compared with Fcgr3. Moreover, expressing Fcgr3-rs in primary rat WKY BMDMs resulted in lower levels of phagocytosis than cells expressing Fcgr3 alone, suggesting that Fcgr3-rs has an inhibitory effect on Fcgr3-mediated phagocytosis in these cells.
Although the Fcgr3-rs cytoplasmic domain includes a novel sequence with a single tyrosine molecule, this is not an immunoreceptor tyrosine-based inhibitory motif in the same way as that found in the FcgrIIb receptor. Elegant studies by Kim et al. (29) have shown that specific regions of the transmembrane region of Fcγ receptors are important for signaling leading to phagocytosis. The same group has provided further evidence on differences in the signaling properties of FcγR1/γ and FcγRIIA such as their interaction with Syk and Src-related tyrosine kinases (30). The region of the cytoplasmic domain shown by Hou et al. (28) to be important in signaling is present in the Fcgr3-rs; however, it is possible that the altered sequence of the remainder of the cytoplasmic domain may render this region inaccessible to essential downstream signaling molecules, thereby rendering it inactive. The question of whether the Fcgr3-rs exerts its effects by acting as a decoy receptor, binding immunoglobulins, but not signaling, or whether its novel cytoplasmic domain induces distinct signaling pathways that act to inhibit those from the wt Fcgr3 is therefore an interesting one.
Our demonstration herein that the -rs does not induce Syk or NF-κB activation suggests that this molecule may not signal, and this is underscored by the demonstration that LPS-induced TNF and IL-10 production is unaffected in U937 cells by the presence of the -rs. Taken together, these data add further weight to the argument that Fcgr3-rs is a decoy receptor that is able to bind Fc but unable to signal such that its expression inhibits co-expressed Fc receptor-mediated signaling and cell activation. However, we cannot completely rule out a potential additional role of this receptor, which will lead to inhibition by a novel negative signaling pathway. Investigations into this question are continuing in this laboratory.
In conclusion, our results suggest that the rat-specific Fcgr3-rs acts to inhibit Fcgr3-mediated signaling and phagocytosis in both rodent and human cells and could be considered as a novel therapeutic mechanism for the modulation of Fc receptor-mediated cell activation in macrophage-mediated diseases such as crescentic glomerulonephritis.
Acknowledgments
We thank Sunil Modi for technical assistance. We are thankful to the National BioResource Project for providing rat strains.
This work was supported by intramural funding from the Wellcome Trust, Clinical Sciences Centre, and the United Kingdom Medical Research Council. Recipient of an Imperial College Junior Research Fellowship.
- ITAM
- immunoreceptor tyrosine-based activation motif
- FcR
- Fc receptor
- BMDM
- bone marrow-derived macrophage
- NTN
- nephrotoxic nephritis
- Crgn
- Crescentic glomerulonephritis
- WKY
- Wistar-Kyoto
- qRT
- quantitative RT
- PMA
- phorbol 12-myristate 13-acetate.
REFERENCES
- 1. Mason J. C., Pusey C. D. (2008) The Kidney in Systemic Autoimmune Diseases, 1st Ed., Elsevier, Amsterdam [Google Scholar]
- 2. Nimmerjahn F., Ravetch J. V. (2007) Fc-receptors as regulators of immunity. Adv. Immunol. 96, 179–204 [DOI] [PubMed] [Google Scholar]
- 3. Nimmerjahn F., Ravetch J. V. (2008) Fcγ receptors as regulators of immune responses. Nat. Rev. Immunol. 8, 34–47 [DOI] [PubMed] [Google Scholar]
- 4. Brown E. E., Edberg J. C., Kimberly R. P. (2007) Fc receptor genes and the systemic lupus erythematosus diathesis. Autoimmunity 40, 567–581 [DOI] [PubMed] [Google Scholar]
- 5. Strzelecka A., Kwiatkowska K., Sobota A. (1997) Tyrosine phosphorylation and Fcγ receptor-mediated phagocytosis. FEBS Lett. 400, 11–14 [DOI] [PubMed] [Google Scholar]
- 6. Park S. Y., Ueda S., Ohno H., Hamano Y., Tanaka M., Shiratori T., Yamazaki T., Arase H., Arase N., Karasawa A., Sato S., Ledermann B., Kondo Y., Okumura K., Ra C., Saito T. (1998) Resistance of Fc receptor-deficient mice to fatal glomerulonephritis. J. Clin. Invest. 102, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Suzuki Y., Shirato I., Okumura K., Ravetch J. V., Takai T., Tomino Y., Ra C. (1998) Distinct contribution of Fc receptors and angiotensin II-dependent pathways in anti-GBM glomerulonephritis. Kidney Int. 54, 1166–1174 [DOI] [PubMed] [Google Scholar]
- 8. Tarzi R. M., Davies K. A., Robson M. G., Fossati-Jimack L., Saito T., Walport M. J., Cook H. T. (2002) Nephrotoxic nephritis is mediated by Fcγ receptors on circulating leukocytes and not intrinsic renal cells. Kidney Int. 62, 2087–2096 [DOI] [PubMed] [Google Scholar]
- 9. Aitman T. J., Dong R., Vyse T. J., Norsworthy P. J., Johnson M. D., Smith J., Mangion J., Roberton-Lowe C., Marshall A. J., Petretto E., Hodges M. D., Bhangal G., Patel S. G., Sheehan-Rooney K., Duda M., Cook P. R., Evans D. J., Domin J., Flint J., Boyle J. J., Pusey C. D., Cook H. T. (2006) Copy number polymorphism in Fcgr3 predisposes to glomerulonephritis in rats and humans. Nature 439, 851–855 [DOI] [PubMed] [Google Scholar]
- 10. Bainbridge J. W., Stephens C., Parsley K., Demaison C., Halfyard A., Thrasher A. J., Ali R. R. (2001) In vivo gene transfer to the mouse eye using an HIV-based lentiviral vector. Efficient long-term transduction of corneal endothelium and retinal pigment epithelium. Gene Ther. 8, 1665–1668 [DOI] [PubMed] [Google Scholar]
- 11. Floto R. A., Clatworthy M. R., Heilbronn K. R., Rosner D. R., MacAry P. A., Rankin A., Lehner P. J., Ouwehand W. H., Allen J. M., Watkins N. A., Smith K. G. (2005) Loss of function of a lupus-associated FcγRIIb polymorphism through exclusion from lipid rafts. Nat. Med. 11, 1056–1058 [DOI] [PubMed] [Google Scholar]
- 12. Park J. G., Isaacs R. E., Chien P., Schreiber A. D. (1993) In the absence of other Fc receptors, Fc γ RIIIA transmits a phagocytic signal that requires the cytoplasmic domain of its γ subunit. J. Clin. Invest. 92, 1967–1973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lowry M. B., Duchemin A. M., Robinson J. M., Anderson C. L. (1998) Functional separation of pseudopod extension and particle internalization during Fc γ receptor-mediated phagocytosis. J. Exp. Med. 187, 161–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Davis W., Harrison P. T., Hutchinson M. J., Allen J. M. (1995) Two distinct regions of FC γ RI initiate separate signalling pathways involved in endocytosis and phagocytosis. EMBO J. 14, 432–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ghazizadeh S., Bolen J. B., Fleit H. B. (1994) Physical and functional association of Src-related protein tyrosine kinases with Fc γ RII in monocytic THP-1 cells. J. Biol. Chem. 269, 8878–8884 [PubMed] [Google Scholar]
- 16. Wang A. V., Scholl P. R., Geha R. S. (1994) Physical and functional association of the high affinity immunoglobulin G receptor (Fc γ RI) with the kinases Hck and Lyn. J. Exp. Med. 180, 1165–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sharif O., Bolshakov V. N., Raines S., Newham P., Perkins N. D. (2007) Transcriptional profiling of the LPS induced NF-κB response in macrophages. BMC Immunol. 8, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Takai T., Li M., Sylvestre D., Clynes R., Ravetch J. V. (1994) FcR γ chain deletion results in pleiotrophic effector cell defects. Cell 76, 519–529 [DOI] [PubMed] [Google Scholar]
- 19. Wakayama H., Hasegawa Y., Kawabe T., Hara T., Matsuo S., Mizuno M., Takai T., Kikutani H., Shimokata K. (2000) Abolition of anti-glomerular basement membrane antibody-mediated glomerulonephritis in FcRγ-deficient mice. Eur. J. Immunol. 30, 1182–1190 [DOI] [PubMed] [Google Scholar]
- 20. Behmoaras J., Bhangal G., Smith J., McDonald K., Mutch B., Lai P. C., Domin J., Game L., Salama A., Foxwell B. M., Pusey C. D., Cook H. T., Aitman T. J. (2008) Jund is a determinant of macrophage activation and is associated with glomerulonephritis susceptibility. Nat. Genet. 40, 553–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Behmoaras J., Smith J., D'Souza Z., Bhangal G., Chawanasuntoropoj R., Tam F. W., Pusey C. D., Aitman T. J., Cook H. T. (2010) Genetic loci modulate macrophage activity and glomerular damage in experimental glomerulonephritis. J. Am. Soc. Nephrol. 21, 1136–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Maratou K., Behmoaras J., Fewings C., Srivastava P., D'Souza Z., Smith J., Game L., Cook T., Aitman T. (2011) Characterization of the macrophage transcriptome in glomerulonephritis-susceptible and -resistant rat strains. Genes Immun. 12, 78–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuramoto T., Nakanishi S., Serikawa T. (2008) Functional polymorphisms in inbred rat strains and their allele frequencies in commercially available outbred stocks. Physiol. Genomics 33, 205–211 [DOI] [PubMed] [Google Scholar]
- 24. Lanier L. L., Yu G., Phillips J. H. (1991) Analysis of Fc γ RIII (CD16) membrane expression and association with CD3 ζ and Fc epsilon RI-γ by site-directed mutation. J. Immunol. 146, 1571–1576 [PubMed] [Google Scholar]
- 25. Indik Z. K., Park J. G., Hunter S., Schreiber A. D. (1995) The molecular dissection of Fc γ receptor mediated phagocytosis. Blood 86, 4389–4399 [PubMed] [Google Scholar]
- 26. Crowley M. T., Costello P. S., Fitzer-Attas C. J., Turner M., Meng F., Lowell C., Tybulewicz V. L., DeFranco A. L. (1997) A critical role for Syk in signal transduction and phagocytosis mediated by Fcγ receptors on macrophages. J. Exp. Med. 186, 1027–1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wirthmueller U., Kurosaki T., Murakami M. S., Ravetch J. V. (1992) Signal transduction by Fc γ RIII (CD16) is mediated through the γ chain. J. Exp. Med. 175, 1381–1390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hou X., Dietrich J., Geisler N. O. (1996) The cytoplasmic tail of FcγRIIIAα is involved in signaling by the low affinity receptor for immunoglobulin G. J. Biol. Chem. 271, 22815–22822 [DOI] [PubMed] [Google Scholar]
- 29. Kim M. K., Huang Z. Y., Hwang P. H., Jones B. A., Sato N., Hunter S., Kim-Han T. H., Worth R. G., Indik Z. K., Schreiber A. D. (2003) Fcγ receptor transmembrane domains: role in cell surface expression, γ chain interaction, and phagocytosis. Blood 101, 4479–4484 [DOI] [PubMed] [Google Scholar]
- 30. Huang Z. Y., Hunter S., Kim M. K., Chien P., Worth R. G., Indik Z. K., Schreiber A. D. (2004) The monocyte Fcγ receptors FcγRI/γ and FcγRIIA differ in their interaction with Syk and with Src-related tyrosine kinases. J. Leukoc. Biol. 76, 491–499 [DOI] [PubMed] [Google Scholar]