Background: Activated FVIII (FVIIIa) stability is critical for cofactor function.
Results: Mass spectrometry analysis reveals that Lys1967 and/or Lys1968 are buried in factor VIII and surface-exposed in dissociated FVIIIa. Lys1967/Lys1968 variants differentially affect FVIIIa activity over time.
Conclusion: Lys1967 and Lys1968 have an opposite contribution to FVIIIa stability.
Significance: Insight is increased about how FVIIIa activity is controlled to prevent thrombosis or bleeding.
Keywords: Enzyme Kinetics, Factor VIII, Mass Spectrometry (MS), Protein Stability, Protein Structure, Surface Plasmon Resonance
Abstract
The A2 domain rapidly dissociates from activated factor VIII (FVIIIa) resulting in a dampening of the activity of the activated factor X-generating complex. The amino acid residues that affect A2 domain dissociation are therefore critical for FVIII cofactor function. We have now employed chemical footprinting in conjunction with mass spectrometry to identify lysine residues that contribute to the stability of activated FVIII. We hypothesized that lysine residues, which are buried in FVIII and surface-exposed in dissociated activated FVIII (dis-FVIIIa), may contribute to interdomain interactions. Mass spectrometry analysis revealed that residues Lys1967 and Lys1968 of region Thr1964-Tyr1971 are buried in FVIII and exposed to the surface in dis-FVIIIa. This result, combined with the observation that the FVIII variant K1967I is associated with hemophilia A, suggests that these residues contribute to the stability of activated FVIII. Kinetic analysis revealed that the FVIII variants K1967A and K1967I exhibit an almost normal cofactor activity. However, these variants also showed an increased loss in cofactor activity over time compared with that of FVIII WT. Remarkably, the cofactor activity of a K1968A variant was enhanced and sustained for a prolonged time relative to that of FVIII WT. Surface plasmon resonance analysis demonstrated that A2 domain dissociation from activated FVIII was reduced for K1968A and enhanced for K1967A. In conclusion, mass spectrometry analysis combined with site-directed mutagenesis studies revealed that the lysine couple Lys1967-Lys1968 within region Thr1964-Tyr1971 has an opposite contribution to the stability of FVIIIa.
Introduction
Factor VIII (FVIII)2 serves its role in the coagulation cascade as a cofactor for activated factor IX (FIXa) during the proteolytic conversion of factor X (FX) into activated FX (1). To perform this role, FVIII comprises multiple domains that are grouped in a heavy chain (domains A1-a1-A2-a2-B) and a light chain (domains a3-A3-C1-C2) (2). The A domains are homologous to the A domains of ceruloplasmin, the C domains to those of discoidin, and the B domain is unique to FVIII. a1, a2, and a3 are spacer regions that are rich in acidic amino acid residues (3). Because of limited proteolysis of the B domain, FVIII circulates in plasma as a heterogeneous protein of which the light chain is noncovalently linked to the heavy chain (1).
FVIII is found in plasma in a tight complex with its carrier protein von Willebrand factor (4). The role of von Willebrand factor is to protect FVIII from rapid clearance from the circulation and to prevent premature binding of FVIII to its ligands (5, 6). Proteolytic cleavage between a3 and the A3 domain by thrombin results in the dissociation of the FVIII-von Willebrand factor complex (4). Additional cleavages by thrombin between a1 and the A2 domain, and between a2 and the B domain, are required to convert FVIII into the activated heterotrimeric protein (FVIIIa) (7, 8). FVIIIa subsequently binds via its C1 domain and C2 domain to phosphatidylserine-containing procoagulant surfaces thereby forming a platform for high affinity interaction with FIXa (1, 9, 10).
FVIIIa is rapidly inactivated to prevent the unlimited production of activated FX (FXa). Several mechanisms have been proposed that may drive this inactivation. One mechanism involves proteolytic cleavage of FVIIIa by activated protein C. It has been proposed that activated protein C inactivates FVIIIa through proteolytic cleavages in the A1 and A2 domain (11–13). It has also been shown that FIXa, FXa, and plasmin are able to cleave FVIIIa inducing inactivation of the cofactor (13–16).
Apart from protease-assisted inactivation of the cofactor, spontaneous dissociation of the A2-a2 domain from the A1-a1/A3-C1-C2 dimer also leads to dampening of the activity of the FXa-generating complex (17–19). It has been demonstrated that residues that control the dissociation rate of the A2-a2 domain from A1-a1/A3-C1-C2 dimer, and as such affect the stability of FVIIIa, are of critical importance for cofactor function (20, 21). For a number of hemophilia A variants carrying single amino acid substitutions, it has been suggested that a decreased stability of FVIIIa is the cause for the bleeding disorder (21, 22). Employing molecular modeling studies on the available FVIII structures in combination with site-directed mutagenesis, Fay and co-workers have now successfully identified several amino acid residues that enhance or decrease the stability of FVIII (21, 23–25).
The recently solved crystal structures of FVIII have also assisted the biochemical studies addressing FVIIIa stability (26, 27). They have, for instance, served as a basis for molecular dynamics studies from which conclusions have been drawn about the critical contacts between the individual FVIII domains (28). Yet, a limitation of the crystal structures is their relatively low resolution.
We have now employed chemical footprinting in conjunction with mass spectrometry to identify lysine residues that contribute to FVIIIa stability. We hypothesized that those lysine residues, which are buried in FVIII and not in dissociated FVIIIa (dis-FVIIIa), may contribute to the interaction between the A2-a2 domain and the A1-a1/A3-C1-C2 dimer. To identify these residues, we took advantage of the notion that buried lysine residues are less prone to chemical modification than those that are exposed to the protein surface (29). The lysines with the largest change in surface exposure upon FVIIIa dissociation were therefore evaluated for their contribution to the stability of FVIIIa.
EXPERIMENTAL PROCEDURES
Materials
HEPES was from SERVA (Heidelberg, Germany), NaCl was from FAGRON (Rotterdam, The Netherlands) and Tris-HCl from Invitrogen. All other fine chemicals were from Merck.
Proteins
Monoclonal antibodies EL14, KM33, and CLB-CAg 9 have been described previously (30–32). B domain-deleted FVIII and the FVIII variants thereof have been constructed and purified as described (10, 33) with the exception that FVIII was stored in 50 mm HEPES (pH 7.4), 0.8 m NaCl, 5 mm CaCl2, and 50% glycerol. Plasma-derived FVIII was purified from Aafact (Sanquin, Amsterdam) according to the same procedure. To this end, Aafact was taken up in 50 mm imidazole (pH 6.7), 50 mm CaCl2, and 0.8 m NaCl prior to purification. The purification of FX, FIXa, and thrombin is described by Mertens and Bertina (34). FXa was from Enzyme Research (South Bend, IN).
Chemical Modification of FVIII and FVIIIa
70 nm plasma-derived FVIII was incubated with 2 nm thrombin in 50 mm HEPES (pH 7.4), 150 mm NaCl, and 5 mm CaCl2 for 2 h at 37 °C to ensure dissociation. Thrombin activation was terminated by the addition of 1.6 units/ml hirudin. FVIII was chemically modified by tandem mass tag (TMT)-127 for 2 h at 37 °C (Thermo Fisher Scientific) according to instructions of the manufacturer for labeling whole proteins. 100 mm lysine dissolved in 140 mm Tris-HCl was employed to stop the reaction. To label nonactivated FVIII with TMT-126, FVIII was first incubated with hirudin and then with thrombin prior to the addition of TMT-126. The TMT-labeled proteins were pooled in a one-to-one molar ratio, and the free cysteines were alkylated employing 2.5 mm dithiothreitol for 15 min at 50 °C followed by a 30-min incubation at 37 °C with 15 mm iodoacetamide in a buffer containing 50 mm ammonium bicarbonate and 2 mm CaCl2. Finally, the sample was cleaved by the addition of 0.1 μg chymotrypsin for an overnight incubation at 37 °C. Obtained peptides were concentrated and washed employing a C18 Ziptip (Millipore Corporation) according to the instructions of the manufacturer.
Mass Spectrometry Analysis
FVIII peptides were separated by reverse-phase chromatography and sprayed in a LTQ OrbitrapXL mass spectrometer (Thermo Fisher Scientific) essentially as described (35). During reverse-phase chromatography, we employed a 40-min gradient from 0 to 35% (v/v) acetonitrile with 0.05% (v/v) acetic acid. Collision-induced dissociation (CID) spectra and higher energy collision-induced dissociation (HCD) spectra were acquired as described by Dayon et al. (36). The three most intense precursor ions in the full scan (300–2000 m/z, resolving power 30,000) with a charge state of 2+ or higher were selected for CID using an isolation width of 2 Da, a 35% normalized collision energy, and an activation time of 30 ms. The same precursor ions were subjected to HCD with a normalized collision energy of 60%, which allows for the identification of the reporter group from the TMT label.
Peptide Identification and Selection
The identity of the peptides, including TMT labeled lysine residues, and the TMT-127/TMT-126 ratio thereof were assessed employing Proteome discoverer 1.1 software (Thermo Fisher Scientific). The SEQUEST search algorithm and protein data base 25.H_sapiens.fasta were used to identify the peptides. The presence of the TMT label on peptides generates additional ion fragments in the tandem MS spectra that negatively influence the SEQUEST Xcorr score (37). We therefore employed the following peptide selection criteria: (i) the peptide is identified in three of four independent experiments; (ii) the peptide is identified from both the CID and the HCD spectrum; (iii) lysine residues within the peptide are all modified by a TMT label (+225.1558 Da) and the cysteines are alkylated; (iv) peptides that included (or are close to) a thrombin cleavage site were excluded from the analysis. TMT ratios were normalized to the average ratio obtained for all peptides within one experiment.
FXa-generating Assay
Factor Xase activity measurement and the preparation of phospholipid vesicles were performed as described before (30). Briefly, 25 μm sonicated phospholipid vesicles comprising 15% phosphatidylserine, 20% phosphatidylethanolamine, and 65% phosphatidylcholine were mixed with 0.3 nm FVIII, 200 nm FX, and 0–16 nm FIXa in a buffer containing 40 mm Tris-HCl (pH 7.8), 150 mm NaCl, 0.2% (w/v) bovine serum albumin (BSA) (Merck). The reaction was started with the addition of 1.5 mm CaCl2 and 1 nm thrombin. The amount of FXa generated per minute was subsequently assessed as described (30). For the FVIIIa stability analysis, the FVIII variants were first incubated at varying time intervals with 2 nm thrombin and 1.5 mm CaCl2 in the presence and absence of increasing concentrations of FIXa (0–16 nm) at 25 °C in 40 mm Tris-HCl (pH 7.8), 150 mm NaCl, 0.2% (w/v) BSA, and 25 μm phospholipid vesicles. The generated amount of FXa was assessed as described (30). Residual FVIII activity was derived from the initial rate of FXa formation as a function of the FIXa concentration. This residual FVIIIa activity was assessed 2, 5, 7.5, 12.5, 20, and 40 min after thrombin activation of at least three independent experiments. The inactivation rate constant, kinact, was estimated by fitting the mean data to a one-phase exponential decay equation (24) by nonlinear least square regression analysis using Prism 5 (GraphPad Software).
Surface Plasmon Resonance Analysis
Surface plasmon resonance analysis was performed on a BIAcoreTM3000 system (Uppsala, Sweden) essentially as described (10, 33). EL14 was first coupled to a CM5 chip (GE Healthcare) to a density of 5000 response units (RU) employing the amide coupling kit as prescribed by the manufacturer. FVIII WT, K1967A, and K1968A were bound to immobilized EL14 by passing the proteins over EL14 at a flow rate of 20 μl/min 20 mm HEPES (pH 7.4), 150 mm NaCl, 5 mm CaCl2 and 0.005% Tween 20. After thrombin activation on the chip, the remaining FVIII light chain and/or A2 domain was assessed by passing either 100 nm KM33 or 100 nm CLB-CAg 9 over the immobilized proteins at a flow rate of 20 μl/min in the same buffer as described above.
RESULTS
Lysine Residues within Region Thr1964-Tyr1971 Are Buried in FVIII and Not in Dissociated FVIIIa
To identify FVIII lysine residues that are less prone to chemical modification in FVIII than in dis-FVIIIa, we labeled the lysine residues in plasma-derived FVIII with TMT-126 and those in dis-FVIIIa with TMT-127. Labeled FVIII and dis-FVIIIa were subsequently pooled and cleaved into peptides by chymotrypsin. A unique property of the TMT labels is that the same peptide ions carrying a TMT-126 or a TMT-127 exhibit an identical mass. However, fragmentation of these ions employing HCD generates a reporter ion from the TMT label as well as a mass spectrum of the peptide. The intensity of the signals of the reporter ions within each mass spectrum allows for calculating the TMT-127/TMT-126 ratio of the lysine-containing peptides (36, 38). As such, it can be assessed whether a lysine residue is more readily labeled in dis-FVIIIa or in FVIII (supplemental Fig. S1). To facilitate peptide identification, we also performed CID of the peptide ions.
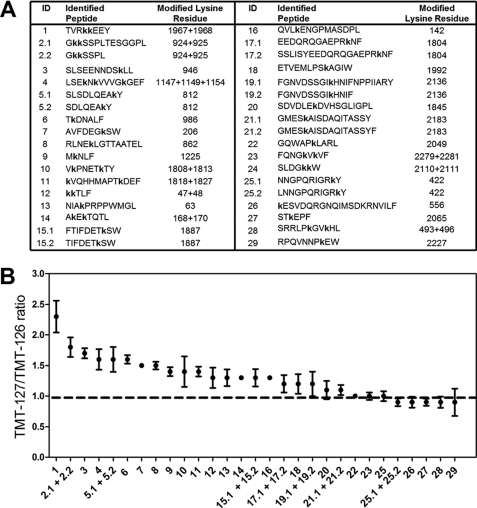
The above-mentioned approach was employed on plasma-derived FVIII. Fig. 1A shows the identified peptides that comprise TMT-modified lysine residues. Peptide identification scores are shown in supplemental Table S1. The average TMT-127/TMT-126 ratio obtained from peptides comprising the same TMT-modified lysine residues are displayed in Fig. 1B. The result reveals that most of the identified peptides exhibit an increased TMT-127/TMT-126 ratio. However, region Thr1964-Tyr1971 comprising the residues Lys1967 and Lys1968 was most prone to chemical modification in dis-FVIIIa (supplemental Fig. S2). This finding implies that Lys1967 and Lys1968 are buried within the protein core in FVIII and not in dis-FVIIIa.
FIGURE 1.
Surface exposure of amino acid regions upon dissociation of FVIIIa. A, TMT-labeled FVIII peptides that were identified by mass spectrometry as described under “Experimental Procedures.” Indicated are the identification number of the peptide (ID), the peptide sequence, and the identity of the modified lysine residue. The lowercase letter k indicates that this residue was modified with a TMT label. B, average TMT-127/TMT-126 ratio of peptides that include the same labeled lysine residues. The ID number of the peptides from which the TMT ratio is calculated is indicated on the x axis. TMT ratios that deviate from 1 (dashed line) suggest that the involved lysine residues within the peptide are more exposed to the protein surface in dis-FVIIIa. Data represent the mean ± S.D. (error bars) of the TMT-127/TMT-126 ratios obtained from all the HCD spectra that belong to the indicated peptides.
K1968A Exhibits Enhanced Cofactor Function
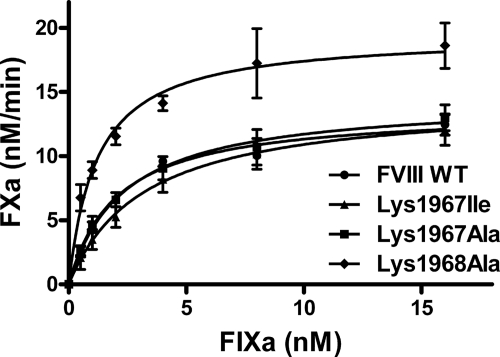
The mass spectrometry results suggest that the lysine residues 1967 and 1968 may contribute to the stability of FVIIIa. Intriguingly, replacement of Lys1967 with an isoleucine has been associated with mild hemophilia A, implying that this lysine may also directly contribute to cofactor activity (22). Replacement of Lys1967 by an alanine in FVIII, however, did not result in a marked change in the specific one-stage clotting activity and the specific two-stage chromogenic activity. In addition, the relative specific activities of a K1967I FVIII variant were still 70% of FVIII WT. Surprisingly, a K1968A variant revealed a moderate (if any) increase in the specific chromogenic activity and a 2-fold increase in specific clotting activity (supplemental Table S2). To assess FVIII activity under more defined experimental conditions, we next evaluated the FVIII variants for their ability to enhance the activity of the FXa-generating complex employing purified proteins. To this end, FXa generation was assessed at increasing concentrations of FIXa in the presence of the activated FVIII variants (Fig. 2). The results showed that there was only a small difference in the efficiency of FXa generation for K1967I and K1967A compared with that of FVIII WT. However, K1968A displayed a markedly more effective FXa generation relative to FVIII WT. This was reflected by an almost 2-fold decrease in the apparent affinity constant (KD) for FIXa binding (KD = 2 nm for FVIII WT and 1.2 nm for K1968A) and an increased maximum rate of FXa generation.
FIGURE 2.
FX activation by FIXa in the presence of recombinant FVIII variants. Activation of FX was assessed by increasing concentration of FIXa (0–16 nm) in the presence of phospholipids, Ca2+ ions, and 0.3 nm FVIII WT and variants thereof as described under “Experimental Procedures.” Shown is the amount of FXa generated per minute for FVIII WT (circles), K1967I (triangles), K1967A (squares), and K1968A (diamonds). Data represent mean ± S.D. (error bars) of at least three independent experiments.
Lys1967 and Lys1968 Exhibit Opposite Effect on FVIIIa Activity Over Time
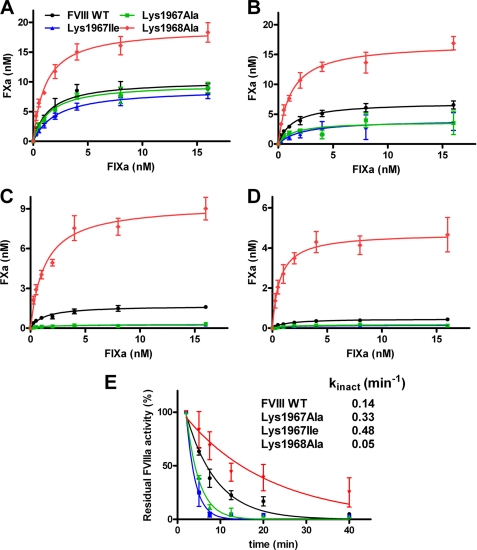
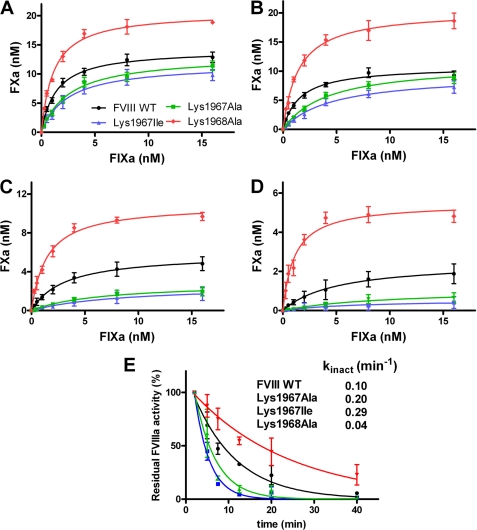
The finding that the hemophilic K1967I variant displays almost normal cofactor function suggests that Lys1967 may indeed contribute to the stability of FVIIIa. Because the A2 domain dissociation from FVIIIa inactivates the cofactor, we addressed whether or not mutation of the residues 1967 and 1968 affects the activity of FVIIIa over time. To this end, we activated FVIII WT and variants thereof with thrombin, and we measured the time-dependent decrease in cofactor activity at increasing FIXa concentrations. As expected, the results showed that there was a decline in cofactor activity over time for FVIII WT (Fig. 3). However, the rate of inactivation was about 3-fold increased for K1967I and 2-fold for K1967A. Strikingly, K1968A showed an about 3-fold decrease in the rate of inactivation relative to that of FVIIIa WT (Fig. 3E). This finding suggests that there is a decreased stability of activated K1967I and K1967A. In contrast, activated K1968A has an enhanced stability compared with FVIIIa WT.
FIGURE 3.
Lysine residues 1967 and 1968 have a differential effect on activity of FVIIIa over time. A–D, 0.3 nm FVIII WT and the variants thereof activated by thrombin as described under “Experimental Procedures.” FXa generation was allowed for 1 min in the presence of increasing concentrations of FIXa (0–16 nm), phospholipids, and Ca2+ ions 2 min (A) 5 min (B) 20 min (C), or 40 min (D) after thrombin activation. FVIII WT is displayed in black circles, K1967I in blue triangles, K1967A in green squares, and K1968A in red diamonds. Data represent mean ± S.D. (error bars) of at least three independent experiments. E, normalized decrease in residual FVIIIa activity over time for FVIII WT and FVIII variants. The inactivation rate constants, kinact, were assessed as described under “Experimental Procedures.” The S.D. values were within 15% of the obtain values for kinact.
FVIIIa Stability Assessed by Surface Plasmon Resonance Analysis
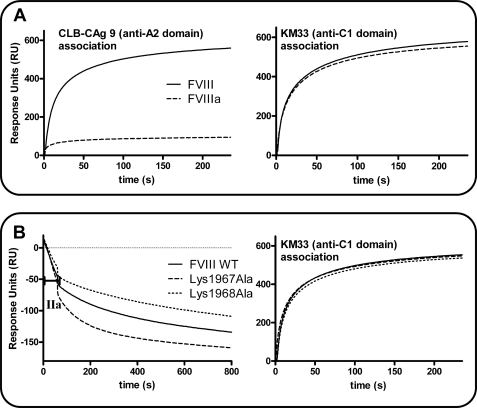
Replacement of the lysine residues 1967 and 1968 may exhibit a differential effect on A2-a2 domain dissociation from the A1-a1/A3-C1-C2 dimer. To this end, we assessed dissociation of the A2 domain from FVIIIa employing surface plasmon resonance analysis essentially as employed by Pipe et al. (39). We first perfused 200 nm thrombin or buffer over FVIII that was bound via its C2 domain to the immobilized monoclonal antibody EL14. A rapid decline in RU was observed, indicative of A2-a2 domain dissociation from the A1-a1/A3-C1-C2 dimer (data not shown). We subsequently passed anti-A2 domain antibody CLB-CAg 9 and anti-C1 domain antibody KM33 over thrombin-activated FVIII and over nonactivated FVIII. The result shows that there was hardly any association of CLB-CAg 9 to FVIIIa whereas this antibody readily bound nonactivated FVIII. In contrast, KM33 was equally effective in binding FVIIIa and FVIII (Fig. 4A). This finding suggests that the A2 domain is indeed rapidly lost from FVIIIa. We next perfused 2 nm thrombin over immobilized FVIII WT, K1967A, and K1968A (Fig. 4B). The result revealed that there is an increased decay in RU for K1967A relative to that of FVIII WT. However, this decrease in RU was reduced for the K1968A variant. When we subsequently perfused KM33 over the immobilized proteins, an almost identical increase in RU was observed for all the variants. This suggests that the differential decrease in RU is caused by an altered dissociation rate of the A2-a2 domain from the A1-a1/A3-C1-C2 dimer. Taken together, the data imply that the lysine residues 1967 and 1968 play an opposite role in the interaction between the A2-a2 domain and A1-a1/A3-C1-C2 dimer.
FIGURE 4.
Replacement of Lys1967 or Lys1968 exhibits a differential effect on A2-a2 domain dissociation from the A1-a1/A3-C1-C2 dimer. A, 200 nm thrombin or buffer was perfused for 60 s over FVIII WT that was immobilized to a density of 1500 RU on the surface of a CM5 sensor chip via anti-C2 domain antibody EL14. Left panel shows the association of 100 nm anti-A2 domain antibody CLB-CAg 9 to FVIII (solid line) or thrombin-activated FVIII (dashed line). Right panel shows the association of 100 nm anti-C1 domain antibody KM33 to FVIII (solid line) or thrombin-activated FVIII (dashed line). B, FVIII WT, K1967A, and K1968A were immobilized via EL14 to the surface of a CM5 sensor chip to a density of 1500 RU. Left panel, 2 nm thrombin (IIa) was perfused for 60 s over the immobilized FVIII variants. The decay in RU was subsequently followed in time for FVIII WT (solid line), K1967A (dashed line), and K1968A (dotted line). Right panel shows the association of KM33 to thrombin-activated FVIII WT (solid line), K1967A (dashed line), and K1968A (dotted line).
FIXa Enhances Stability of Activated FVIII WT and Variants Thereof
It has been proposed that binding of FVIIIa to FIXa leads to a decreased rate of A2 domain dissociation from FVIIIa (40, 41). We now assessed the effect of FIXa on the time-dependent activity of the FVIII variants. To this end, we activated FVIII in the presence of increasing concentrations of FIXa, and the time-dependent residual activity of the FVIII variants was established. The data revealed a decreased cofactor function over time for FVIII WT and its variants (Fig. 5). Both K1967A and K1967I displayed again a 2-fold and a 3-fold increase in the rate of inactivation relative to that of activated FVIII WT. Furthermore, the K1968A variant showed again a prolonged activity over time (Fig. 5E). However, comparison of the observations in Figs. 3E and 5E also reveals that the rate of FVIIIa inactivation of the FVIII variants is decreased when these variants are activated in the presence of FIXa. The activated K1968A variant was the least (if at all) affected by the presence and absence of FIXa. These findings together imply that FIXa can still reduce the rate of A2 domain dissociation from the FVIII variants. This suggests that the lysine residues 1967 and 1968 do not directly contribute to factor IXa binding. Collectively, the results of our study show that Lys1967 and Lys1968 have an opposite contribution to FVIIIa stability.
FIGURE 5.
FIXa enhances stability of FVIII WT and variants thereof. A–D, 0.3 nm FVIII WT and the variants thereof were activated by 2 nm thombin in the presence of increasing concentrations of FIXa (0–16 nm) as described under “Experimental Procedures.” FXa generation was allowed for 1 min in the presence of phospholipids and Ca2+ ions 2 min (A), 5 min (B), 20 min (C), and 40 min (D) after thrombin activation. FXa generation in the presence of FVIII WT is displayed in black circles, K1967I in blue triangles, K1967A in green squares, and K1968A in red diamonds. Data represent mean ± S.D. (error bars) of at least three independent experiments. E, normalized decrease in residual FVIIIa activity over time for FVIII WT and the FVIII variants is shown. The inactivation rate constants, kinact, were assessed as described under “Experimental Procedures.” The S.D. values were within 15% of the obtain values for kinact.
DISCUSSION
Lysine amino acid residues that are buried within the protein core of FVIII are expected to be completely protected from chemical modification by TMT-126. The same is true for lysine residues that form an interdomain salt bridge with a negatively charged amino acid residue. When these regions are only exposed to the protein surface in dis-FVIIIa, these regions should then only carry the TMT-127 modification. However, the high internal dynamics of the structural core of FVIII in solution will allow for penetrations of the TMT-126 labels inside FVIII. Because of this “molecular breathing,” part of the inaccessible lysine residues will be chemically modified by TMT-126 after all. This explains why there are no FVIII regions identified that completely lack TMT-126. An increased TMT-127/TMT-126 ratio does, however, imply that at least the lysines of the involved FVIII regions are buried in FVIII and surface-exposed in dis-FVIIIa.
The highest TMT-127/TMT-126 ratio is observed for peptide Thr1964-Tyr1971 which includes the residues Lys1967 and Lys1968 (Fig. 1). In agreement with our initial hypothesis, this observation is compatible with our finding that these residues contribute to the stability of FVIIIa (Figs. 3–5). Intriguingly, the crystal structure of Ngo et al. shows that Lys1887 and Lys1804 are directed with their side chain toward the A2 domain (26). Our data show that these lysine residues are also partially protected from chemical modification in FVIII (Fig. 1). This suggests that these lysine residues may contribute to the interaction between the A1-a1/A3-C1-C2 dimer and the A2 domain as well.
Peptides with a relatively high incorporation level of TMT-127 are almost all derived from the first 500 residues of the B domain of FVIII (Fig. 1). However, the B domain is removed from FVIII upon activation and therefore plays no role in maintaining the stability of FVIIIa. This result does suggest that the identified lysines of these B domain regions are buried in FVIII and not in the released B domain. The most likely explanation for this observation is that the B domain changes its conformation upon its release from FVIII. However, this finding also opens the possibility that the identified lysines are involved in shielding functional sites on FVIII similar to what has been proposed for a positively charged region within the B domain of factor V (8, 42). Yet, B domain-deleted FVIII shows, unlike B domain-deleted factor V, no cofactor activity (43). On the other hand, FVIII requires additional cleavages by thrombin to obtain full cofactor function. This suggests that additional changes in FVIII are necessary to enhance the activity of the FXa-generating complex (1, 7, 8). It can therefore not be fully excluded that the B domain contributes to shielding functional regions in FVIII after all.
Other identified peptides from the A1 and A3 domains show a modest but decreased protection from chemical modification of their lysine residues after dissociation of FVIIIa (Fig. 1). The significance thereof remains to be established. Some of these lysine residues may contribute to the stability of the A1-a1/A3-C1-C2 dimer itself. It is of interest to mention that the side chain of Lys206 is situated close to the side chain of Glu2004 in both reported structures of FVIII (26, 27). This suggests that these residues may form an interdomain salt bridge between the A3 and A1 domains. Our finding that lysine 206 is partially protected from chemical modification in FVIII agrees with this observation (Fig. 1). A similar level of protection from chemical modification was observed for the FVIII peptide, including the residues Lys1808 and Lys1813. These lysine residues are, however, not in close proximity to residues from the A2 domain or the A1 domain. Previously, we have demonstrated that FVIII peptides comprising these residues (i.e. Lys1804-Lys1818 and Glu1811-Gln1820) inhibit FIXa binding to the FVIII light chain (6). From this observation, we have concluded that these peptides include a FIXa binding region. The findings of the present study suggest that this FIXa binding region is more surface-exposed in the A3 domain of dis-FVIIIa. It is tempting to speculate that this is also the case for intact FVIIIa. If so, this implies that the FIXa binding region is only optimally exposed to the protein surface after activation of FVIII. This may then, in part, explain the observation that only FVIIIa can enhance the activity FIXa (7). This aspect is, therefore, a subject for further investigation.
The available crystal structures of FVIII reveal that A2 domain region 661–665 is in close proximity to region Thr1964-Tyr1971 (26, 27). Notably, in the crystal structure by Shen et al., Val663 is positioned between the lysine residues 1967 and 1968. In both structures, the side chain of Lys1967 is directed toward the inside of the FVIII molecule, whereas Lys1968 is exposed to the surface (supplemental Fig. S3). Employing molecular dynamics studies on these structures, Venkateswarlu proposed a refined structure of FVIII in which the side chain of Lys1967 has a direct interaction with the side chain of Glu665 of the A2 domain (28). This suggestion is compatible with our observation that replacement of Lys1967 by an alanine increases the dissociation rate of the A2-a2 domain from the A1-a1/A3-C1-C2 dimer (Fig. 5). However, Fay and co-workers demonstrated that replacement of residue Glu665 for a valine or an alanine increases the stability of FVIIIa (24). This finding is in disagreement with the suggestion that interaction between Lys1967 and Glu665 contributes to the binding of the A2-a2 domain to the A1-a1/A3-C1-C2 dimer. Although it is speculation, Glu665 may in fact interact with Lys1968 instead of Lys1967, resulting into destabilization of FVIIIa. Replacement of either Lys1968 or Glu665 by a hydrophobic amino acid residue would then prevent this interaction, thereby increasing the stability of FVIIIa. Even though it seems likely that there is an interaction between the A2 domain residues 661–665 and the A3 domain residues 1964–1971, the above-mentioned observations demonstrate that a high resolution structure of FVIII, and if possible at all of FVIIIa, is required to gain insight into the interactions within FVIII at the level of individual amino acid residues.
The physiological relevance of Lys1967 and Lys1968 for cofactor functions remains to be established. However, Lys1967 seems to be required for stabilizing FVIIIa sufficiently long to mediate effective blood coagulation. This provides an explanation for the observation that K1967I is associated with hemophilia A. The same explanation has previously been proposed for other hemophilia A FVIII variants (21, 22). Lys1968 may, on the other hand, be critical to prevent prolonged cofactor function. Patients with an elevated risk for thrombosis who are a carrier of a Lys1968 mutation have, however, not yet been identified. In conclusion, employing mass spectrometry analysis combined with functional studies, we have identified a lysine couple in FVIII with a remarkable opposite function in the biology of FVIII. Our findings provide novel insight about how FVIIIa activity is controlled to prevent thrombosis or bleeding.
Supplementary Material

This article contains supplemental Figs. S1–S3 and Tables S1 and S2.
- FVIII
- factor VIII
- FVIIIa
- activated FVIII
- CID
- collision-induced dissociation
- dis-FVIIIa
- dissociated FVIIIa
- FIXa
- activated factor IX
- FX
- factor X
- FXa
- activated FX
- HCD
- higher energy CID
- RU
- response units
- TMT
- tandem mass tag.
REFERENCES
- 1. Fay P. J. (2006) Factor VIII structure and function. Int. J. Hematol. 83, 103–108 [DOI] [PubMed] [Google Scholar]
- 2. Vehar G. A., Keyt B., Eaton D., Rodriguez H., O'Brien D. P., Rotblat F., Oppermann H., Keck R., Wood W. I., Harkins R. N. (1984) Structure of human factor VIII. Nature 312, 337–342 [DOI] [PubMed] [Google Scholar]
- 3. Church W. R., Jernigan R. L., Toole J., Hewick R. M., Knopf J., Knutson G. J., Nesheim M. E., Mann K. G., Fass D. N. (1984) Coagulation factors V and VIII and ceruloplasmin constitute a family of structurally related proteins. Proc. Natl. Acad. Sci. U.S.A. 81, 6934–6937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lollar P., Hill-Eubanks D. C., Parker C. G. (1988) Association of the factor VIII light chain with von Willebrand factor. J. Biol. Chem. 263, 10451–10455 [PubMed] [Google Scholar]
- 5. Lillicrap D. (2008) Extending half-life in coagulation factors: where do we stand? Thromb. Res. 122, S2–8 [DOI] [PubMed] [Google Scholar]
- 6. Lenting P. J., Donath M. J., van Mourik J. A., Mertens K. (1994) Identification of a binding site for blood coagulation factor IXa on the light chain of human factor VIII. J. Biol. Chem. 269, 7150–7155 [PubMed] [Google Scholar]
- 7. Duffy E. J., Parker E. T., Mutucumarana V. P., Johnson A. E., Lollar P. (1992) Binding of factor VIIIa and factor VIII to factor IXa on phospholipid vesicles. J. Biol. Chem. 267, 17006–17011 [PubMed] [Google Scholar]
- 8. Camire R. M., Bos M. H. (2009) The molecular basis of factor V and VIII procofactor activation. J. Thromb. Haemost. 7, 1951–1961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lü J., Pipe S. W., Miao H., Jacquemin M., Gilbert G. E. (2011) A membrane-interactive surface on the factor VIII C1 domain cooperates with the C2 domain for cofactor function. Blood 117, 3181–3189 [DOI] [PubMed] [Google Scholar]
- 10. Meems H., van den Biggelaar M., Rondaij M., van der Zwaan C., Mertens K., Meijer A. B. (2011) C1 domain residues Lys2092 and Phe2093 are of major importance for the endocytic uptake of coagulation factor VIII. Int. J. Biochem. Cell Biol. 43, 1114–1121 [DOI] [PubMed] [Google Scholar]
- 11. Fay P. J., Smudzin T. M., Walker F. J. (1991) Activated protein C-catalyzed inactivation of human factor VIII and factor VIIIa: identification of cleavage sites and correlation of proteolysis with cofactor activity. J. Biol. Chem. 266, 20139–20145 [PubMed] [Google Scholar]
- 12. Fay P. J., Haidaris P. J., Huggins C. F. (1993) Role of the COOH-terminal acidic region of A1 subunit in A2 subunit retention in human factor VIIIa. J. Biol. Chem. 268, 17861–17866 [PubMed] [Google Scholar]
- 13. Eaton D., Rodriguez H., Vehar G. A. (1986) Proteolytic processing of human factor VIII: correlation of specific cleavages by thrombin, factor Xa, and activated protein C with activation and inactivation of factor VIII coagulant activity. Biochemistry 25, 505–512 [DOI] [PubMed] [Google Scholar]
- 14. Lamphear B. J., Fay P. J. (1992) Proteolytic interactions of factor IXa with human factor VIII and factor VIIIa. Blood 80, 3120–3126 [PubMed] [Google Scholar]
- 15. O'Brien D. P., Johnson D., Byfield P., Tuddenham E. G. (1992) Inactivation of factor VIII by factor IXa. Biochemistry 31, 2805–2812 [DOI] [PubMed] [Google Scholar]
- 16. Nogami K., Shima M., Matsumoto T., Nishiya K., Tanaka I., Yoshioka A. (2007) Mechanisms of plasmin-catalyzed inactivation of factor VIII: a crucial role for proteolytic cleavage at Arg336 responsible for plasmin-catalyzed factor VIII inactivation. J. Biol. Chem. 282, 5287–5295 [DOI] [PubMed] [Google Scholar]
- 17. Lollar P., Parker E. T. (1991) Structural basis for the decreased procoagulant activity of human factor VIII compared with the porcine homolog. J. Biol. Chem. 266, 12481–12486 [PubMed] [Google Scholar]
- 18. Lollar P., Parker C. G. (1990) pH-dependent denaturation of thrombin-activated porcine factor VIII. J. Biol. Chem. 265, 1688–1692 [PubMed] [Google Scholar]
- 19. Fay P. J., Haidaris P. J., Smudzin T. M. (1991) Human factor VIIIa subunit structure: reconstruction of factor VIIIa from the isolated A1/A3-C1-C2 dimer and A2 subunit. J. Biol. Chem. 266, 8957–8962 [PubMed] [Google Scholar]
- 20. Gale A. J., Radtke K. P., Cunningham M. A., Chamberlain D., Pellequer J. L., Griffin J. H. (2006) Intrinsic stability and functional properties of disulfide bond-stabilized coagulation factor VIIIa variants. J. Thromb. Haemost. 4, 1315–1322 [DOI] [PubMed] [Google Scholar]
- 21. Wakabayashi H., Fay P. J. (2008) Identification of residues contributing to A2 domain-dependent structural stability in factor VIII and factor VIIIa. J. Biol. Chem. 283, 11645–11651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kemball-Cook G., Tuddenham E. G., Wacey A. I. (1998) The factor VIII Structure and Mutation Resource Site: HAMSTeRS version 4. Nucleic Acids Res. 26, 216–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wakabayashi H., Griffiths A. E., Fay P. J. (2009) Combining mutations of charged residues at the A2 domain interface enhances factor VIII stability over single point mutations. J. Thromb. Haemost. 7, 438–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wakabayashi H., Varfaj F., Deangelis J., Fay P. J. (2008) Generation of enhanced stability factor VIII variants by replacement of charged residues at the A2 domain interface. Blood 112, 2761–2769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wakabayashi H., Griffiths A. E., Fay P. J. (2011) Increasing hydrophobicity or disulfide bridging at the factor VIII A1 and C2 domain interface enhances procofactor stability. J. Biol. Chem. 286, 25748–25755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ngo J. C., Huang M., Roth D. A., Furie B. C., Furie B. (2008) Crystal structure of human factor VIII: implications for the formation of the factor IXa-factor VIIIa complex. Structure 16, 597–606 [DOI] [PubMed] [Google Scholar]
- 27. Shen B. W., Spiegel P. C., Chang C. H., Huh J. W., Lee J. S., Kim J., Kim Y. H., Stoddard B. L. (2008) The tertiary structure and domain organization of coagulation factor VIII. Blood 111, 1240–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Venkateswarlu D. (2010) Structural investigation of zymogenic and activated forms of human blood coagulation factor VIII: a computational molecular dynamics study. BMC Struct. Biol. 10, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Shkriabai N., Datta S. A., Zhao Z., Hess S., Rein A., Kvaratskhelia M. (2006) Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45, 4077–4083 [DOI] [PubMed] [Google Scholar]
- 30. Meems H., Meijer A. B., Cullinan D. B., Mertens K., Gilbert G. E. (2009) Factor VIII C1 domain residues Lys2092 and Phe2093 contribute to membrane binding and cofactor activity. Blood 114, 3938–3946 [DOI] [PubMed] [Google Scholar]
- 31. van Helden P. M., van den Berg H. M., Gouw S. C., Kaijen P. H., Zuurveld M. G., Mauser-Bunschoten E. P., Aalberse R. C., Vidarsson G., Voorberg J. (2008) IgG subclasses of anti-FVIII antibodies during immune tolerance induction in patients with hemophilia A. Br. J. Haematol. 142, 644–652 [DOI] [PubMed] [Google Scholar]
- 32. Stel H. V. (1984) Monoclonal antibodies against factor VIII-von Willebrand factor. Ph.D. thesis, University of Amsterdam, 1984 [Google Scholar]
- 33. van den Biggelaar M., Meijer A. B., Voorberg J., Mertens K. (2009) Intracellular cotrafficking of factor VIII and von Willebrand factor type 2N variants to storage organelles. Blood 113, 3102–3109 [DOI] [PubMed] [Google Scholar]
- 34. Mertens K., Bertina R. M. (1980) Pathways in the activation of human coagulation factor X. Biochem. J. 185, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Haren S. D., Herczenik E., ten Brinke A., Mertens K., Voorberg J., Meijer A. B. (2011) HLA-DR-presented peptide repertoires derived from human monocyte-derived dendritic cells pulsed with blood coagulation factor VIII. Mol. Cell Proteomics 10, M110.002246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dayon L., Pasquarello C., Hoogland C., Sanchez J. C., Scherl A. (2010) Combining low- and high-energy tandem mass spectra for optimized peptide quantification with isobaric tags. J. Proteomics 73, 769–777 [DOI] [PubMed] [Google Scholar]
- 37. Thingholm T. E., Palmisano G., Kjeldsen F., Larsen M. R. (2010) Undesirable charge-enhancement of isobaric tagged phosphopeptides leads to reduced identification efficiency. J. Proteome Res. 9, 4045–4052 [DOI] [PubMed] [Google Scholar]
- 38. Thompson A., Schäfer J., Kuhn K., Kienle S., Schwarz J., Schmidt G., Neumann T., Johnstone R., Mohammed A. K., Hamon C. (2003) Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904 [DOI] [PubMed] [Google Scholar]
- 39. Pipe S. W., Eickhorst A. N., McKinley S. H., Saenko E. L., Kaufman R. J. (1999) Mild hemophilia A caused by increased rate of factor VIII A2 subunit dissociation: evidence for nonproteolytic inactivation of factor VIIIa in vivo. Blood 93, 176–183 [PubMed] [Google Scholar]
- 40. Lamphear B. J., Fay P. J. (1992) Factor IXa enhances reconstitution of factor VIIIa from isolated A2 subunit and A1/A3-C1-C2 dimer. J. Biol. Chem. 267, 3725–3730 [PubMed] [Google Scholar]
- 41. Fay P. J., Beattie T. L., Regan L. M., O'Brien L. M., Kaufman R. J. (1996) Model for the factor VIIIa-dependent decay of the intrinsic factor Xase: role of subunit dissociation and factor IXa-catalyzed proteolysis. J. Biol. Chem. 271, 6027–6032 [DOI] [PubMed] [Google Scholar]
- 42. Toso R., Camire R. M. (2004) Removal of B-domain sequences from factor V rather than specific proteolysis underlies the mechanism by which cofactor function is realized. J. Biol. Chem. 279, 21643–21650 [DOI] [PubMed] [Google Scholar]
- 43. Lind P., Larsson K., Spira J., Sydow-Bäckman M., Almstedt A., Gray E., Sandberg H. (1995) Novel forms of B-domain-deleted recombinant factor VIII molecules: construction and biochemical characterization. Eur. J. Biochem. 232, 19–27 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.