Background: Colorectal cancer (CRC) to metastatic disease may involve the epithelial-mesenchymal transition (EMT).
Results: STAT3 may regulate N-cadherin, vimentin, and ZEB1 expressions. STAT3-induced cell invasion and down-regulation of E-cadherin may depend on ZEB1.
Conclusion: STAT3 may mediate CRC EMT progression and ZEB1 expression. Activation of STAT3 and ZEB1 proteins may contribute to worse prognosis in CRC patients.
Significance: Our data may provide potential targets to prevent and/or treat CRC invasion.
Keywords: Cell Signaling, Metastasis, STAT3, Tumor, Tumor Therapy, STAT3, ZEB1, Colorectal Cancer, Epithelial-Mesenchymal Transition, Metastasis
Abstract
The progression of colorectal carcinoma (CRC) to invasive and metastatic disease may involve localized occurrences of epithelial-mesenchymal transition (EMT). However, mechanisms of the EMT process in CRC progression are not fully understood. We previously showed that knockdown of signal transducer and activator of transcription 3 (STAT3) up-regulated E-cadherin (a key component in EMT progression) in CRC. In this study, we examined the roles of STAT3 in CRC EMT and ZEB1, an EMT inducer, in STAT3-induced down-regulation of E-cadherin. Knockdown of STAT3 significantly increased E-cadherin and decreased N-cadherin and vimentin expressions in highly invasive LoVo CRC cells. Meanwhile, overexpression of STAT3 significantly reduced E-cadherin and enhanced N-cadherin and vimentin expressions in weakly invasive SW1116 CRC cells. Activation of STAT3 significantly increased CRC cell invasiveness and resistance to apoptosis. Knockdown of STAT3 dramatically enhanced chemosensitivity of CRC cells to fluorouracil. STAT3 regulated ZEB1 expression in CRC cells, and the STAT3-induced decrease in E-cadherin and cell invasion depended on activation of ZEB1 in CRC cells. Additionally, pSTAT3Tyr-705 and ZEB1 expressions were significantly correlated with TNM (tumor, lymph node, and metastasis stages) (p < 0.01). In conclusion, STAT3 may directly mediate EMT progression and regulate ZEB1 expression in CRC. ZEB1 may participate in STAT3-induced cell invasion and E-cadherin down-regulation in CRC cells. The expressions of pSTAT3Tyr-705 and ZEB1 may be positively associated with CRC metastasis. Our data may provide potential targets to prevent and/or treat CRC invasion and metastasis.
Introduction
Despite welcome declines in mortality rates over the past decade, colorectal cancer (CRC)4 remains a common malignancy and one of the leading causes of morbidity and death in the world (1, 2). Epithelial-mesenchymal transition (EMT) is a crucial process in the initiation of the metastatic spread of tumor cells to distal organs (3). EMT may promote epithelial cells to escape from the rigid structural constraints provided by the tissue architecture and adopt a phenotype more amenable to cell migration and movement (4–6). In this progression, epithelial cells may lose adhesion and cell-to-cell contacts (7, 8). Therefore, EMT can be regarded as a pathological process that contributes to cancer progression, particularly in tumor cell invasion and metastasis (9).
The initiation and progression of EMT require transduction of cell signals. The transforming growth factor-β (TGF-β) signaling and activated Ras pathways have been implicated as key EMT inducers in CRC cancer (10–11). The Wnt, PI3K/AKT, and other signaling pathways may also play an important role in the EMT process in CRC progression (12–15). Recently, accumulating evidence has indicated that abnormalities in the Janus kinase/signal transducer and activator of transcription (JAK/STAT) pathway are involved in CRC oncogenesis (16, 17). As a key component of the JAK/STAT pathway, STAT3 is constitutively activated in CRC (18), and several lines of evidence have supported its role in mediating cell motility and migration. STAT3 is important for the migration of sheets of cells in zebrafish embryo development (19), and conditional depletion of this molecule blocks wound healing in mouse keratinocytes (20). Additionally, gastrin may induce EMT in CRC through the JAK2/STAT3 pathway (21). EGF receptor is overexpressed in ovarian carcinoma, whereas EGF-induced EMT in ovarian cancer cells has been shown to depend on IL-6R and the JAK2/STAT3 pathway (22). However, the role of STAT3 in the EMT process of CRC progression is not fully understood.
Loss of E-cadherin expression is a crucial step and fundamental event of EMT in cancer progression (3). As a key component of adherens junctions, E-cadherin plays a crucial role in the maintenance of epithelial integrity (23). Many studies have reported on the regulation of E-cadherin during cancer progression (3, 24, 25), and several proteins, including Snail, ZEB1, and ZEB2, have been identified that may down-regulate E-cadherin in various cancers (3). In our previous studies, we found that knockdown of STAT3 by RNA interference (RNAi) significantly increased E-cadherin expression in CRC cells (18). Whether STAT3 contributes to the EMT process of CRC progression and the mechanisms of STAT3-induced E-cadherin down-regulation are not known. We now show that STAT3 may directly induce cell invasion and participate in resistance to chemotherapy drugs and apoptosis during EMT of CRC progression. To our knowledge, this is the first study to report that STAT3 may directly mediate the EMT process and ZEB1 expression of CRC progression. STAT3-induced cell invasion and decrease in E-cadherin expression depend on activation of ZEB1 in CRC cells. The combination of pSTAT3Tyr-705/ZEB1 may be a novel predictor of CRC metastasis and a potential therapeutic target.
EXPERIMENTAL PROCEDURES
Cell Culture and AG490 Treatment
Two human CRC cell lines SW1116 and LoVo (ATCC, Manassas, VA) were cultured in RPMI 1640 medium (Invitrogen), supplemented with 10% fetal bovine serum (FBS) at 37 °C in humidified 5% CO2 atmosphere. For AG490 (pharmacological JAK2 inhibitor; Sigma) treatment, CRC cells were incubated with 100 μm AG490 for 24 h (18) before harvesting for measurements.
Construction of Plasmids
The DNA fragment encoding the STAT3 gene (GenBank® accession number NM_003150) was amplified from human cDNA with the primers STAT3-F (5′-GCTAAGCTTTATGGCCCAATGGAATCAGCTACAG-3′ and STAT3-R (5′-GCTCTCGAGTCATGGGGGAGGTAGCGCACTCCG-3′), which introduced the cloning sites HindIII and XhoI (underlined), respectively. The cDNA fragment obtained above was verified by sequencing and finally cloned into pCDNA3.1 between the HindIII and XhoI sites to obtain pCDNA3.1-STAT3.
The wild type DNA fragment containing part of the promoter region (−520 to +70 from transcriptional initiation site) of the E-cadherin gene (GenBank® accession number NM_004360) and the wild type DNA fragment containing part of the promoter region (−500 to +100 from the transcriptional initiation site) of the ZEB1 gene (GenBank® accession number NM_001174094) were amplified from human genomic DNA with the following primers, respectively: E-cadherinP-F (5′-GGGGTACCTGTCTCTCTACAAAAAGGCA-3′) and E-cadherinP-R (5′-GGAAGATCTGGGCTGGAGCGGGCTGGAGT-3′); ZEB1 P-F (5′- GGGGTACCAAAGACGTTTCCTTATTCGA-3′) and ZEB1 P-R (5′- GAAGATCTAGAAAGGCGACGGGCTGACC-3′), which introduced the cloning sites KpnI and BglII (underlined), respectively. The DNA fragment obtained above was directly cloned into pGL3-basic (Promega, Madison, WI) between the KpnI and BglII sites to obtain pGL3-E-cadherinPWT and pGL3-ZEB1PWT.
The mutant DNA sequences of the ZEB1 promoter region encompassing both of the two putative binding sites of STAT3 (−500 to +100 from the transcriptional initiation site) or the mutant DNA sequences of the E-cadherin promoter region encompassing both of the two putative binding sites of STAT3 and four putative binding sites of ZEB1 (−520 to +70 from transcriptional initiation site) were synthesized and inserted into pGL3-basic vector. The mutant type constructs were designated as pGL3-basic-ZEB1P MT, pGL3-basic-E-cadherinP STAT3B MT, pGL3-basic-E-cadherinP ZEB1B MT, and pGL3-basic-E-cadherinP STAT3B and ZEB1B MT, respectively. T was replaced with G in each STAT3 binding site of pGL3-basic-ZEB1P MT, pGL3-basic-E-cadherinP STAT3BMT, and pGL3-basic-E-cadherinP STAT3B and ZEB1B MT constructs. CT and CTG was replaced with AA and AAA in each ZEB1 binding site of pGL3-basic-E-cadherinP ZEB1B MT and pGL3-basic-E-cadherinP STAT3B and ZEB1B MT constructs, respectively.
Small Interfering RNA (siRNA) Plasmid Transfections and Lentiviral Transduction
The siRNA against ZEB1 (TCF8; catalog no. L-006564-01-0005), the siRNA against STAT3 (catalog no. L-003544-00-0005), and the control siRNA were purchased from Dharmacon RNA Technology (Lafayette, CO). Twenty-four h before transfection at 30–40% confluence, CRC cells were transferred to 6-well plates. Transfection of siRNAs was carried out with DharmaFECT 1 siRNA transfection reagent (Dharmacon) according to the manufacturer's instructions. Cells were collected for analysis 48 h after transfection.
For plasmid transfections, CRC cells (70% confluence, ∼5 × 106 cells) were transfected with 2 μg of pCDNA3.1-STAT3 or pCDNA3.1 using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were collected for measurements 48 h after transfection.
To stably knock down ZEB1, we infected SW1116 and LoVo cells with MISSION shRNA lentivirus particles (with the puromycin resistance gene) containing a U6 promoter driving shRNA targeting human ZEB1 or scramble negative control (Sigma-Aldrich). Methods used for lentivirus production and infection were performed as described by Gire et al. (26).
Reverse Transcription-PCR (RT-PCR)
Total RNA was extracted by TRIzol reagent (Invitrogen), according to the protocol of the manufacturer, and 1.5 μg of total RNA from cultured cells was reverse transcribed using the PrimeScriptPTMP RT reagent kit (Perfect Real Time) for RT-PCR (Takara, Shiga, Japan).
Quantitative Real-time PCR
Quantitative real-time PCR was carried out on an Applied Biosystems 7900 quantitative PCR system. The primers used were as follows: ZEB1-F (5′-GCCAATAAGCAAACGATTCTG-3′), ZEB1-R (5′-TTTGGCTGGATCACTTTCAAG-3′), ZEB2-F (5′-CGGTGCAAGAGGCGCAAACA-3′), ZEB2-R (5′-GGAGGACTCATGGTTGGGCA-3′), Snail1-F (5′-CACTATGCCGCGCTCTTTC-3′), Snail1-R (5′-GGTCGTAGGGCTGCTGGAA-3′), Snail2-F (5′-AAACTACAGCGAACTGGACACA-3′), Snail2-R (5′-GCCCCAAAGATGAGGAGTATC-3′), Twist1-F (5′-AGTCCGCAGTCTTACGAGGA-3′), Twist1-R (5′- GCCAGCTTGAGGGTCTGAAT-3′), Twist2-F (5′-CAAGCTGAGCAAGATCCAGAC-3′), Twist2-R (5′-GGTCATCTTATTGTCCATCTCG-3′), E12/E47-F (5′- TCAAGCAATAACTTCTCGTCCA-3′), E12/E47-R (5′-CGTCCAGGTGGTCTTCTATCTT-3′)18S-F (5′-CGGACAGGATTGACAGATTGATAGC-3′), and 18S-R (5′-TGCCAGAGTCTCGTTCGTTATCG-3′). All reactions were performed in triplicate in a 10-μl total volume containing Brilliant® SYBR® Green QPCR Master Mix (Takara, Shiga, Japan). The amplified transcript level of each specific gene was normalized to that of 18S.
Western Blot and Antibodies
Western blot analysis was performed using standard techniques as described previously (27). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Kangchen, Shanghai, China) was detected as a loading control. Antibodies used in this study were purchased from Cell Signaling Technology Inc. (Beverly, MA). All primary antibodies were used at a 1:1000 dilution.
Drug Sensitivity Assays to Fluorouracil
Cell proliferation assay was assessed by a tetrazolium salt (WST-8)–based colorimetric assay in Cell Counting Kit 8 (Dojindo, Kumamoto, Japan) (28, 29). Briefly, control and CRC cells treated with different doses of fluorouracil were seeded onto 96-well plates at an initial density of 5 × 103 cells/well. At specified time points, 10 μl of Cell Counting Kit 8 solution were added to each well of the plate, which was then incubated for 2 h. Cell viability was determined by scanning with a microplate reader at 450 nm. Data are expressed as the percentage of viable cells calculated as follows: cell survival rate (%) = (A450(treated) − A450(blank))/(A450(control) − A450(blank)) × 100%.
Detection of Apoptosis
For flow cytometric analysis, an annexin-V fluorescein isothiocyanate/PI double stain assay was performed in accordance with the manufacturer's protocol (BioVision, Mountain View, CA). Analysis was performed using a flow cytometer.
In Vitro Invasion Assay
Cell invasion assays were performed as described by Hecht et al. (30). In brief, chambers with 8-μm pore polycarbonate membranes, coated with Matrigel on the upper side, were used (BD Biosciences). CRC cells or stable CRC cell lines with ZEB1 gene knockdown were transfected with STAT3 siRNA, control siRNA, pCDNA3.1-STAT3, or pCDNA3.1 for 24 h. Transfected cells were then harvested, and 1 × 105 cells were seeded in serum-free medium into the upper chamber, whereas medium supplemented with 15% FBS was applied to the lower chamber as a chemoattractant to induce invasion. After incubation for 48 h, migrated cells on the bottom surface of the filter were fixed, stained, and counted.
Luciferase Assay
Designated combinations of pGL3-E-cadherinPWT, pGL3-ZEB1PWT, and other mutant constructs with other siRNA or plasmids at 1.0 μg and 100 ng of phRL (Renilla luciferase) TK plasmid (Promega) for monitoring transfection efficiency were transiently transfected in triplicate with Lipofectamine 2000 (Invitrogen) or DharmaFECT 1 siRNA transfection reagent (Dharmacon) according to the manufacturer's directions. Twenty-four h after transfection, the cells were collected to detect luciferase activity using the Dual-Luciferase reporter assay system (Promega). Luciferase activity was measured by using a BD Monolight 3010 luminometer (BD Biosciences). Variation in transfection efficiency was normalized by dividing the luciferase activity of the construct by the corresponding Renilla luciferase activity. Promoter activity is reported as the mean ± S.E.
Chromatin Immunoprecipitation (ChIP) Assay
Chromatin immunoprecipitation assays were performed using the ChIP assay kit (Upstate, Charlottesville, VA) following the manufacturer's protocol. ChIP analysis was performed as described previously (31). Antibodies against ZEB1 (Cell Signal Technology), STAT3 (Cell Signal Technology), and normal rabbit IgG (Upstate) were used.
Real-time PCR Quantification of Genomic DNA ChIP
Real-time PCR was performed in triplicate using an Applied Biosystems 7900 quantitative PCR system. Each PCR was carried out in a 10-μl reaction volume by using 3 μl of the eluted immunoprecipitated DNA. The amount of genomic DNA co-precipitated with the specific antibody was calculated in comparison with the total input DNA used for each immunoprecipitation as follows: CBTB = CBTB(genomic input) − CBTB(specific antibody), where CBTB(genomic input) and CBTB(specific antibody) are the mean threshold cycles of PCR performed in triplicate on DNA samples from the genomic input samples and the specific antibody samples, respectively.
Immunohistochemical Staining
All specimens were from patients (35 primary colorectal adenocarcinomas) who underwent surgery in Shanghai Renji Hospital from July 2009 to December 2010. The protocol was approved by the ethics committee of Shanghai Jiao-Tong University School of Medicine Renji Hospital, and the research was carried out according to the provisions of the Helsinki Declaration of 1975. None of the patients received preoperative treatments, such as radiotherapy or chemotherapy. Meanwhile, 21 specimens of normal colonic epithelium, taken from patients without colorectal cancer, were used as negative controls. The expressions of STAT3, pSTAT3Tyr-705, ZEB1, and E-cadherin were examined with primary antibodies (STAT3, pSTAT3Tyr-705, ZEB1, and E-cadherin; dilution 1:100) in consecutive tissue sections using the LSAB+ kit (DakoCytomation, Copenhagen, Denmark) according to the manufacturer's instructions.
The slides were examined independently by two investigators blinded to both clinical and pathologic data. Protein expression was quantified using a visual grading system based on the extent of staining (percentage of positive tumor cells graded on a scale of 0–4: 0, none; 1, 1–25%; 2, 26–50%; 3, 51–75%; 4, 475%) and the intensity of staining (graded on a scale of 0–3: 0, no staining; 1, weak staining; 2, moderate staining; 3, strong staining). For further analysis, an index value was calculated as a product of grades of the extent and intensity of staining to define the cut-off value for high expression of the proteins, and the protein expression was classified into two categories: high (grades 4–12) and low (grades 0–3).
Statistical Analysis
Statistical analysis was performed with SPSS 13.0 software. Data are expressed as means ± S.E. Statistical differences between two groups were determined by Student's t test. Differences between multiple groups were tested using analysis of variance (ANOVA) and checked for significance using Fisher's protected least significant difference test. Analyses comparing the expressions of STAT3, pSTAT3Tyr-705, ZEB1, and E-cadherin were performed using χ2 analysis and Fisher's exact test. Results were considered significant if the p value was less than 0.05. Correlation analysis was performed between pSTAT3Tyr-705 and ZEB1.
RESULTS
Effect of STAT3 on E-cadherin, N-cadherin, and Vimentin Expressions in CRC Cells
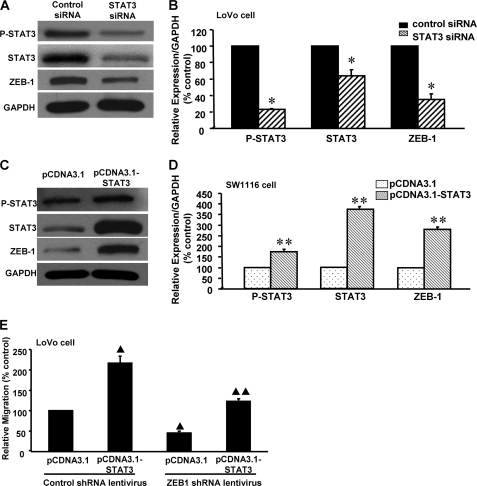
We previously showed that activated STAT3 is constitutively expressed in CRC and mediates cell proliferation, whereas knockdown of STAT3 significantly restores E-cadherin expression (18). Down-regulation of E-cadherin is one of the EMT phenotypes in cancer progression (3). To determine whether STAT3 mediates EMT initiation in CRC cells, the effect of STAT3 siRNA was evaluated in highly invasive LoVo CRC cells. Western blot analysis showed that STAT3 siRNA significantly decreased STAT3 expression and phosphorylation in these cells (Fig. 1, A and B), indicating that the STAT3 was knocked down effectively. Knockdown of STAT3 significantly increased E-cadherin and decreased N-cadherin and vimentin expressions (Fig. 1, A and B), suggesting that STAT3 may contribute to EMT progression in LoVo cells.
FIGURE 1.
Effect of STAT3 on expression of epithelial and mesenchymal markers in LoVo and SW1116 cells. A representative Western blot (A) and the summarized data (B) show that knockdown of STAT3 expression significantly decreased the phosphorylation of STAT3 and the expressions of STAT3, N-cadherin, and vimentin in LoVo cells, whereas E-cadherin expression was dramatically increased, indicating that STAT3 may participate in the regulation of epithelial and mesenchymal markers in colon cancer. Another representative Western blot (C) and the summarized data (D) show that STAT3 overexpression significantly increased the phosphorylation of STAT3 and expressions of STAT3, N-cadherin, and vimentin in SW1116 cells, whereas E-cadherin expression was dramatically reduced. Seventy-five pmol of siRNA duplexes of STAT3, control siRNA, or plasmids complexed with liposomes were applied to each well. After 72 h of transfection, the cells were collected for analysis. n = 3, t test; *, p < 0.01; **, p < 0.05, compared with the pCDNA3.1 or control siRNA groups. Error bars, S.E.
To further confirm the role of STAT3 in down-regulation of E-cadherin and up-regulation of N-cadherin and vimentin in CRC cells, we constructed and transfected the recombinant pCDNA3.1-STAT3 plasmid into low invasion SW1116 cells. Transfection of the pCDNA3.1-STAT3 plasmid significantly increased STAT3 expression and phosphorylation in SW1116 cells, when compared with the pCDNA3.1 control (Fig. 1, C and D), indicating that STAT3 was successfully overexpressed. Overexpression of STAT3 significantly reduced E-cadherin and enhanced N-cadherin and vimentin expressions in SW1116 cells, compared with pCDNA3.1 transfection. These results further indicate that STAT3 may mediate EMT initiation and progression in CRC cells.
Role of STAT3 in Cell Invasion, Cell Colony Formation, Chemosensitivity, and Resistance to Apoptosis in CRC Cells
It has been reported that EMT may induce cell migration, alter invasion properties, promote chemotherapy drug resistance, and prevent apoptosis (3). Aberrant cell survival and resistance to apoptosis are also hallmarks of tumor EMT progression in epithelial carcinoma (32). Therefore, we examined whether STAT3 participates in these aspects of EMT progression in CRC cells. Transwell cell invasion assays showed that knockdown of STAT3 expression significantly reduced the invasion ability of LoVo cells normally characterized as highly invasive (Fig. 2A). Meanwhile, overexpression of STAT3 dramatically increased the invasiveness of SW1116 cells (Fig. 2B), indicating that STAT3 may have significant effects on cell migration and invasion in CRC cells. In cell colony-forming assays, the number of colonies of LoVo cells was reduced by nearly 80% when transfected with STAT3 siRNA as compared with control siRNA (Fig. 2C). Up-regulation of STAT3 significantly increased the number of SW1116 cell colonies, compared with transfection of pCDNA3.1 (Fig. 2D). The data suggest that STAT3 may mediate cell colony formation in CRC cells. In addition, the CRC cell survival rate was lower in the STAT3 siRNA-transfected group than in the control siRNA group after treatment with different doses of fluorouracil (Fig. 2E), indicating that knockdown of STAT3 significantly enhanced chemosensitivity of CRC to this drug. Furthermore, overexpression of STAT3 significantly decreased cell apoptosis in response to different doses of etopside (Fig. 2F), indicating that STAT3 may play an important role in resistance to fluorouracil treatment and etopside-induced apoptosis during the EMT process of CRC progression.
FIGURE 2.
Role of STAT3 in CRC cell EMT phenotypes. Transwell Matrigel invasion assays were performed in LoVo cells transfected with control siRNA or STAT3 siRNA (A) and in SW1116 cells transfected with pCDNA3.1 or pCDNA3.1-STAT3 (B). Twenty-four h after transfection, the cells were plated on transwell inserts, and invasion was assessed after incubation for 72 h. Cells were observed under a microscope and photographed. Cells were counted from five random microscopic fields (200×) per insert in triplicate. The migrated cell numbers were normalized to that of the control group. Data are shown as mean ± S.D. (error bars) from three separate experiments (*, p < 0.05 compared with control siRNA group; **, p < 0.01 compared with the pCDNA3.1 group). Representative colony-forming assays and relative quantitations show inhibition of growth in LoVo cells transfected with STAT3 siRNA compared with control siRNA (C) and increase of growth of SW1116 cells transfected with pCDNA3.1-STAT3 compared with pCDNA3.1 (D). Colony numbers following transfection with STAT3 siRNA or pCDNA3.1-STAT3 are expressed as the relative percentages of colonies compared with the corresponding control groups. Data are means ± S.E. (error bars) of five randomly selected microscopic fields from three independent wells in each group. *, p < 0.05 compared with the control siRNA group; **, p < 0.01 compared with the pCDNA3.1 group. E, dose-response curve of a representative experiment showing relative fluorouracil sensitivity determined by Cell Counting Kit 8 cell proliferation. LoVo cells were treated with fluorouracil after transfection with control or STAT3 siRNA. n = 3, t test; *, p < 0.05, compared with control siRNA group. F, apoptosis of SW116 cells were analyzed by flow cytometric analysis after transfection with pCDNA3.1-STAT3 or pCDNA3.1 following treatment with different doses of etopside. n = 3, t test; ▴, p < 0.01, compared with the pCDNA3.1 group.
JAK/STAT3 Pathway May Regulate ZEB1 Expression in CRC Cells
It has been confirmed that ZEB1, ZEB2, Snail1, Snail2, Twist1, Twist2, and E12/E47 are critical transcription factors that repress E-cadherin expression in EMT of cancer progression (3). After having found that STAT3 may regulate E-cadherin expression, we next detected the expressions of ZEB1, ZEB2, Snail1, Snail2, Twist1, Twist2, and E12/E47 in CRC cells. Although treatment with the JAK/STAT3 pathway inhibitor AG490 significantly down-regulated ZEB1 expression in SW1116 (Fig. 3A) and LoVo (Fig. 3B) cells, it did not dramatically affect the expressions of any of the other molecules detected at the mRNA level in CRC cells, indicating that the JAK/STAT3 pathway may specifically mediate ZEB1 expression in CRC cells. As shown in Fig. 3, C and D, knockdown of STAT3 significantly down-regulated ZEB1 expression in both SW1116 and LoVo cells. The data further indicate that STAT3 may participate in regulating ZEB1 expression in CRC cells.
FIGURE 3.
Role of JAK/STAT3 pathway in ZEB1 expression in CRC cells. Real-time RT-PCR showed that the JAK2 inhibitor AG490 significantly decreased the expression of ZEB1 in SW1116 (A) and LoVo (B) cells, when compared with the control cells. AG490 treatment did not dramatically affect the expression of ZEB2, Snail1, Snail2, Twist1, Twist2, or E12/E47. C, knockdown of STAT3 significantly decreased the ZEB1 mRNA level in SW1116 cells. D, knockdown of STAT3 significantly decreased the ZEB1 mRNA level in LoVo cells. Cells were collected for analysis after treatment with 100 μm AG490 for 24 h. Seventy-five pmol of siRNA duplex of STAT3 or control siRNA complexed with liposomes were applied in each well. After 48 h of transfection, the cells were collected for analysis. n = 3, ANOVA; *, p < 0.05, compared with control; n = 3, t test; **, p < 0.05; ▴, p < 0.01, compared with control siRNA. Error bars, S.E.
Next, we examined whether ZEB1 expression depended on STAT3 in CRC cells. After transfection of STAT3 siRNA into LoVo cells for 48 h, Western blot analysis showed that knockdown of STAT3 significantly down-regulated ZEB1 expression compared with the control (Fig. 4, A and B), indicating that STAT3 may mediate ZEB1 expression in CRC cells. By contrast, when pCDNA3.1-STAT3 was transfected into SW1116 cells for 48 h, the overexpression of STAT3 significantly increased ZEB1 expression, compared with pCDNA3.1 (Fig. 4, C and D). These results further indicate that STAT3 may participate in regulation of ZEB1 expression in CRC cells. Moreover, stable knockdown of ZEB1 dramatically decreased cell invasion in basal conditions. STAT3-induced CRC cell invasion was significantly decreased by stable knockdown of ZEB1 expression in CRC cells (Fig. 4E), indicating that STAT3-induced CRC cell invasion may depend on ZEB1.
FIGURE 4.
Effect of STAT3 on ZEB1 expression and function in CRC cells. A representative Western blot (A) and the summarized data (B) show that transfection with STAT3 siRNA significantly decreased STAT3 phosphorylation and expressions of STAT3 and ZEB1 in LoVo cells. Another Western blot (C) and the summarized data (D) show that STAT3 phosphorylation and expressions of STAT3 and ZEB1 were boosted in STAT3-overexpressing SW1116 cells. E, stable knockdown of ZEB1 dramatically decreased cell invasion in basal condition and blocked STAT3-induced cell invasion in LoVo cells. n = 3, t test; *, p < 0.05, compared with control siRNA. n = 3, t test; **, p < 0.05, compared with pCDNA3.1. n = 3, ANOVA; ▴, p < 0.01, compared with pCDNA3.1+ control shRNA lentivirus; ▴▴, p < 0.05, compared with pCDNA3.1-STAT3+ control shRNA lentivirus. Error bars, S.E.
Furthermore, DNA sequence analysis of the ZEB1 promoter regions (nt −500 to 100) revealed two putative STAT3 binding sites (Fig. 5A). Thus, in the ChIP assay, we designed one primer set (nt −310 to −130, containing two putative STAT3 binding sites), with product sizes of ∼200 bp, to amplify part of the ZEB1 promoter regions. As shown in Fig. 5B, this primer set showed amplifiable products. In contrast, no detectable amplification was observed in cell lysates incubated with non-relevant rabbit IgG or cell lysate without antibody incubation (negative controls). In the dual luciferase assay, mutation of STAT3 binding sites of ZEB1 promoter exhibited almost a 60% decrease in relative luciferase activity compared with that of wild type construct (Fig. 5E), suggesting that STAT3 may directly bind to the ZEB1 promoter and regulate the transcriptional activity of ZEB1. We further found that overexpression of STAT3 significantly up-regulated transcriptional activity of the ZEB1 promoter in CRC cells (Fig. 5C). Knockdown of STAT3 significantly down-regulated transcriptional activity of the ZEB1 promoter (Fig. 5D). The data suggest that STAT3-induced up-regulation of ZEB1 may depend on activation of ZEB1 transcription in CRC cells.
FIGURE 5.
Role of STAT3 in the regulation of ZEB1 expression in CRC cells. A, bioinformatic analysis of STAT3 transcriptional factor binding site in part of the ZEB1 gene promoter region. The numbers on the left side indicate the locations upstream of the first base of the initial transcription site. STAT3-binding sites are highlighted, and the DNA sequence encompassed by two arrows was amplified in the ChIP assay. B, representative results from three experiments showed that ZEB1 DNA was detectable in the chromatin sample immunoprecipitated from SW1116 cells using an antibody against STAT3, suggesting that STAT3 binds to the ZEB1 promoter. Input DNA was used as a positive control; rabbit IgG and cell lysates without antibody were used as negative controls. C, overexpression of STAT3 caused a nearly 40% increase in ZEB1 promoter luciferase activity in SW1116 cells. D, knockdown of STAT3 significantly decreased ZEB1 promoter luciferase activity in SW1116 cells. E, two putative STAT3 binding sites were located between nt −500 and +100. White and black rhombuses indicate wild type and mutant sequences for STAT3 binding sites, respectively. WT, wild type; MT, mutant type. Mutation of STAT3 binding sites significantly decreased transcriptional activity of the ZEB1 promoter in a luciferase assay. n = 3, t test; **, p < 0.05, compared with pCDNA3.1+pGL3-basic-ZEB1PWT. n = 3, t test; *, p < 0.05, compared with control siRNA+pGL3-basic-ZEB1PWT; ▴, p < 0.01, compared with pGL3-basic-ZEB1PWT. Error bars, S.E.
Effects of ZEB1 on STAT3-induced E-cadherin Down-regulation
In order to determine whether ZEB1 mediates STAT3-induced E-cadherin down-regulation in CRC cells, pCDNA3.1-STAT3 and control plasmid were introduced into CRC cells after ZEB1 siRNA and control siRNA transfection 48 h later. Western blot analysis showed that knockdown of ZEB1 expression significantly increased the expression of E-cadherin in CRC cells. Furthermore, STAT3-induced E-cadherin down-regulation was dramatically blocked by down-regulation of ZEB1 (Fig. 6, B and C), indicating that ZEB1 may participate in STAT3-induced E-cadherin down-regulation in CRC cells.
FIGURE 6.
Role of ZEB1 in STAT3-regulated E-cadherin expression in CRC cells. A, a representative Western blot analysis showed that transfection of ZEB1 siRNA significantly reduced the expression of ZEB1. A representative Western blot (B) and the summarized data (C) showed that STAT3 overexpression did not down-regulate E-cadherin expression after ZEB1 knockdown. D, knockdown of STAT3 or ZEB1 expression caused a nearly 2-fold increase in E-cadherin promoter luciferase activity in SW1116 cells. E, overexpression of STAT3 significantly decreased E-cadherin promoter luciferase activity in SW1116 cells. ZEB1 siRNA significantly blocked STAT3-induced down-regulation of E-cadherin promoter luciferase activity in SW1116 cells; n = 3, ANOVA; ▴, p < 0.05, compared with pCDNA3.1+control siRNA; ▴▴, p < 0.05, compared with pCDNA3.1-STAT3+control siRNA; n = 3, ANOVA; *, p < 0.05, compared with control siRNA+pGL3-basic-E-cadherinPWT; **, p < 0.05, compared with pCDNA3.1+control siRNA+pGL3-basic-E-cadherinPWT; ***, p < 0.05, compared with pCDNA3.1-STAT3+control siRNA+pGL3-basic-E-cadherinPWT. Error bars, S.E.
Luciferase assays were then performed to further confirm whether STAT3 repressed E-cadherin expression via altering ZEB1 regulation of E-cadherin transcription. We found that knockdown of STAT3 or ZEB1 significantly up-regulated E-cadherin promoter activity in CRC cells (Fig. 6D). Knockdown of ZEB1 significantly abolished the STAT3-induced decrease in E-cadherin promoter activity (Fig. 6E). Furthermore, DNA sequence analysis of the E-cadherin promoter regions (nt −520 to +70) revealed two putative STAT3 binding sites and four putative ZEB1 binding sites (Fig. 7A). Thus, in the ChIP assay, we designed one primer set (E-cadherin-a, nt −206 to 40, containing three putative ZEB1 binding sites), with product sizes of ∼250 bp, to amplify part of the E-cadherin promoter regions. As shown in Fig. 7, B and C, this primer set showed amplifiable products. In contrast, no detectable amplification was observed in cell lysates incubated with non-relevant rabbit IgG or cell lysate without antibody incubation (negative controls). These results indicate that ZEB1 may directly bind to the E-cadherin promoter. Furthermore, results of a ChIP assay and real-time PCR analysis demonstrated that overexpression of STAT3 increased ZEB1 recruitment to the E-cadherin promoter (Fig. 7, D and E), whereas it was decreased by knockdown of STAT3 (Fig. 7, F and G). The data suggest that STAT3 may play an important role in ZEB1 binding to the E-cadherin promoter in SW1116 and LoVo cells. Moreover, the transcriptional activity of the E-cadherin promoter was significantly increased after mutation of ZEB1 binding sites or dual mutation of STAT3 and ZEB1 binding sites in the E-cadherin promoter (Fig. 8). The data further suggests that ZEB1 may directly bind to the E-cadherin promoter and regulate the transcriptional activity of E-cadherin.
FIGURE 7.
A, bioinformatic analysis of STAT3 and ZEB1 transcriptional factor binding site in part of the E-cadherin gene promoter region. The numbers on the left indicate the locations upstream of the first base of the initial transcription site. STAT3-binding sites and ZEB1-binding sites are highlighted, and the DNA sequence surrounded by two arrows was for ChIP. Representative results from three experiments show that E-cadherin DNA was detected in the chromatin sample immunoprecipitated from SW1116 (B) and LoVo (C) cells using an antibody against ZEB1, respectively, suggesting that ZEB1 binds to the E-cadherin promoter. Input DNA was used as a positive control; rabbit IgG and cell lysates without antibody were used as negative controls. Real-time PCR of the ChIP samples showed that overexpression of STAT3 dramatically increased the binding efficiency of ZEB1 to the E-cadherin promoter in SW1116 (D) and LoVo (E) cells. Knockdown of STAT3 significantly decreased the binding efficiency of ZEB1 to the E-cadherin promoter in SW1116 (F) and LoVo (G) cells. n = 3, t test; *, p < 0.01, compared with pCDNA3.1. n = 3, t test; **, p < 0.05, compared with control siRNA. Error bars, S.E.
FIGURE 8.
Two putative STAT3 binding sites and four putative ZEB1 binding sites were located between nt −520 and +70 of the E-cadherin 5′-flanking region. White and black rhombuses indicate a wild or mutant sequence for STAT3 binding sites, respectively. White and black triangles indicate a wild type or mutant sequence for ZEB1 binding sites, respectively. WT, wild type; STAT3B MT, mutant type of each STAT3 mutation binding site; ZEB1B MT, mutant type of each ZEB1 mutation binding site; ZEB1B and STAT3B MT, mutant type of each ZEB1 and STAT3 mutation binding site. Mutation of ZEB1 binding sites or dual mutation of STAT3 and ZEB1 binding sites significantly increased the transcriptional activity of E-cadherin promoter in the luciferase assay. Mutation of STAT3 binding sites had no obvious effect on the transcriptional activity of the E-cadherin promoter. n = 3, ANOVA; *, p < 0.05, compared with pGL3-basic-E-cadherinPWT. Error bars, S.E.
In addition, in order to examine whether STAT3 may directly bind to the E-cadherin promoter, we designed two primer sets (E-cadherin-b, nt −416 to −224, containing one putative STAT3 binding site; E-cadherin-c, nt −83 to 56, containing one putative STAT3 binding site), with product sizes of ∼200 bp to amplify parts of the E-cadherin promoter regions. However, no E-cadherin DNA was detectable in the immunoprecipitated chromatin sample of SW1116 or LoVo cell lysates by using an antibody against STAT3 (data not shown). Further dual luciferase assay showed that the transcriptional activity of E-cadherin promoter was not dramatically changed after mutation of STAT3 binding sites in E-cadherin promoter (Fig. 8). The data suggest that STAT3 may not directly bind to the E-cadherin promoter.
Expressions of pSTAT3, STAT3, ZEB1, and E-cadherin in CRC Tissues
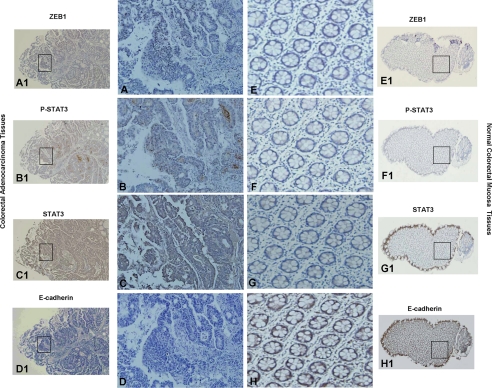
Table 1 showed the frequencies of STAT3, pSTAT3Tyr-705, ZEB1, and E-cadherin expression in normal and tumor colon tissues by immunohistochemical staining. In consecutive tissue sections, pSTAT3Tyr-705, mostly present in the nucleus, was found with higher expression in 16.7% of the normal colon epithelium samples and 65.7% of the colon adenocarcinoma samples (Fig. 9, B and F). ZEB1 showed predominantly nuclear localization, with occasional cytoplasmic staining (Fig. 9, A and E). Immunostaining with an anti-ZEB1 antibody showed higher expression in 19 and 60% of normal colon epithelium samples and colon adenocarcinoma samples, respectively. E-cadherin was observed at higher expression in 100% of the normal colon epithelium samples and in 20% of the adenocarcinoma samples (Fig. 9, D and H). Our data suggest that the expressions of pSTAT3Tyr-705 and ZEB1 were significantly up-regulated (χ2 test, p < 0.001), whereas the expression of E-cadherin was significantly decreased (χ2 test, p < 0.001) in colorectal adenocarcinoma.
TABLE 1.
Frequencies of STAT3, pSTAT3, ZEB1, and E-cadherin expression in normal and tumor colon samples
pSTAT3Tyr-705 and ZEB1 were mostly present in adenocarcinoma, whereas the expression of E-cadherin was significantly decreased in adenocarcinoma (χ2 test). There was no significant difference in STAT3 expression between normal epithelia and adenocarcinoma.
| n | STAT3 |
p value | pSTAT3 |
p value | ZEB1 |
p value | E-cadherin |
p value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| High | Low | High | Low | High | Low | High | Low | ||||||
| Normal epithelium | 21 | 19 | 2 | 0.14 | 3 | 18 | <0.001a | 4 | 17 | <0.01a | 21 | 0 | <0.001a |
| Adenocarcinoma | 35 | 26 | 9 | 23 | 12 | 21 | 14 | 7 | 28 | ||||
a p < 0.001.
FIGURE 9.
Expressions of STAT3, ZEB1, and E-cadherin in CRC. Shown is immunohistochemical analysis of consecutive tissue sections for ZEB1, pSTAT3Tyr-705, STAT3, and E-cadherin in normal colorectal mucosa and high grade CRC. A and B, nuclear staining of ZEB1 and pSTAT3Tyr-705 were mostly presented in adenocarcinoma. Absent expression of ZEB1 (E) or pSTAT3Tyr-705 (F) was evident in the stroma or epithelium of the normal colorectal mucosa. C and G, cytoplasmic and nuclear staining of STAT3 was frequently detected in normal mucosa and adenocarcinoma. H, cytoplasmic staining of E-cadherin was predominantly detected in normal colorectal mucosa. D, staining of E-cadherin was dramatically decreased in adenocarcinoma, compared with normal colorectal epithelium. A–H (original magnification, ×200), representative areas from A1–H1 (original magnification, ×40), respectively.
Moreover, Table 2 showed that the high expressions of pSTAT3Tyr-705 and ZEB1 were significantly more frequent in poorly differentiated CRC tissue than in well differentiated CRC tissue (Fisher's exact test, p < 0.001). Furthermore, high expression of pSTAT3Tyr-705 was significantly correlated with high expression of ZEB1 (r = 0.7385, p < 0.001). There was no significant difference in high STAT3 expressions between the CRC tissues of different differentiation levels. In addition, other clinical characteristics, including age and gender, were not directly related to the expressions of STAT3, pSTAT3Tyr-705, and ZEB1. The results indicate that the high expressions of pSTAT3Tyr-705 and ZEB1 may play important roles in CRC metastasis.
TABLE 2.
STAT3, pSTAT3, and ZEB1 expressions in CRC tissues with different levels of differentiation
Correlation analysis of expressions of STAT3, pSTAT3Tyr-705, and ZEB1 and clinicopathologic characteristics in CRC patients. Statistical analysis was conducted with Fisher's exact test. p values less than 0.05 were considered statistically significant. pSTAT3Tyr-705 and ZEB1 expressions were significantly related to TNM stage (Fisher exact test), suggesting that constitutive activation of STAT3 and elevation of ZEB1 may contribute to poor prognosis in CRC patients.
| STAT3 |
p value | pSTAT3 |
p value | ZEB1 |
p value | ||||
|---|---|---|---|---|---|---|---|---|---|
| High | Low | High | Low | High | Low | ||||
| Sex | |||||||||
| Male | 16 | 5 | 1 | 13 | 8 | 0.72 | 12 | 9 | 0.68 |
| Female | 10 | 4 | 10 | 4 | 7 | 7 | |||
| Gender | |||||||||
| ≤65 | 16 | 3 | 0.25 | 14 | 5 | 0.3 | 10 | 9 | 0.83 |
| >65 | 10 | 6 | 9 | 7 | 9 | 7 | |||
| TNM stage | |||||||||
| I/II | 10 | 6 | 0.25 | 4 | 12 | <0.001a | 5 | 11 | <0.001a |
| III/IV | 16 | 3 | 19 | 0 | 16 | 3 | |||
a p < 0.001.
DISCUSSION
The JAK/STAT signaling pathway plays a significant role in immune function, cell growth, and differentiation (33). Accumulating evidence has indicated that STAT3 correlates with cell proliferation in breast carcinoma (34) and non-small cell lung cancer (35). In a previous study, we found that activation of STAT3 may mediate human CRC tumorigenesis and progression, and knockdown of STAT3 significantly increases the expression of E-cadherin (18).
As a calcium-dependent cell adhesion molecule, E-cadherin is expressed predominantly in epithelial tissues (36). It is an important tumor suppressor gene, which may lose expression and function in tumor progression and invasion (37–40). The loss of E-cadherin expression is a crucial step in the initiation of tumor metastasis and a fundamental event in EMT (3). EMT plays important roles in tumor formation and progression through enhancing cell proliferation, migration, and drug resistance and preventing apoptosis (3). It has been reported that the TGFβ, Wnt, Notch, and EGF signaling pathways may induce EMT in the progression of different cancers (41–44). Although IL-6 may induce an EMT phenotype in human breast cancer cells (45) and TGF-β1 induces EMT in mouse hepatocytes (46) via activation of STAT3, the role of STAT3 is not yet well understood in human CRC cancer EMT progression.
In our present study, we found that activation of STAT3 may induce changes in epithelial cells resembling that of EMT progression for the following reasons. 1) Knockdown of STAT3 significantly decreased N-cadherin and vimentin expressions (Fig. 1, A and B). 2) Overexpression of STAT3 significantly reduced E-cadherin and enhanced N-cadherin and vimentin expressions (Fig. 1, C and D). 3) Down-regulation of STAT3 significantly reduced cell invasion ability (Fig. 2A) and cell colony numbers (Fig. 2C) of CRC cells. 4) Overexpression of STAT3 dramatically increased the invasion ability (Fig. 2B) and cell colony numbers (Fig. 2D) in CRC cells. Furthermore, Thiery et al. (3) have demonstrated that EMT may be involved in more than invasion in cancer; it may be involved as well in resistance to chemotherapy and cell death. Therefore, we also explored whether STAT3 participates in the resistance to fluorouracil (a chemotherapy drug) treatment and promotes CRC cell resistance to etopside-induced cell apoptosis. Our data (Fig. 2, E and F) indicate that STAT3-induced EMT may cause resistance to conventional chemotherapy and cell death in colon cancer.
Because down-regulation of E-cadherin expression is a key initiating event in EMT, transcription factors that repress E-cadherin have been defined as inducers of EMT. The major transcriptional repressors that are known to down-regulate E-cadherin expression include those in the Snail and ZEB families (6, 47). In our study, ZEB1 expression was significantly reduced after treatment with the JAK/STAT pathway inhibitor AG490 and STAT3 siRNA transfection (Fig. 3, A–D). However, ZEB2, Snail1, Snail2, Twist1, Twist2, and E12/E47 expressions were not obviously altered in response to AG490. The data suggest that ZEB1 but not ZEB2, Snail1, Snail2, Twist1, Twist2, or E12/E47 may mediate STAT3-induced colon cancer progression. Results of the cell invasion assays further supported this conclusion. Stable knockdown of ZEB1 significantly reduced cell invasion at the basal conditions and blocked STAT3-induced cell invasion in CRC cells (Fig. 4E). This result is consistent with another report showing that ZEB1 expression is important during colon cancer progression (48). In addition, we further found that ZEB1 expression was significantly decreased by knockdown of STAT3 (Fig. 4, A and B), whereas it was dramatically increased by overexpression of STAT3 (Fig. 4, C and D). These results further indicate that the activation of STAT3 is highly important for ZEB1 expression in colon carcinoma. Further analysis revealed two putative STAT3 binding sites (Fig. 5A) in the ZEB1 promoter region. Results of the ChIP assay (Fig. 5B) and luciferase assay (Fig. 5, C–E) confirmed that STAT3 may directly bind to the ZEB1 promoter. STAT3-induced up-regulation of ZEB1 may depend on activation of ZEB1 transcription in CRC cells.
The mechanisms of STAT3-induced E-cadherin down-regulation in EMT of CRC progression are poorly understood. We examined whether STAT3 may directly bind to the E-cadherin promoter. Although we found two putative STAT3 binding sites in the E-cadherin promoter region, no E-cadherin DNA was detected in the immunoprecipitated chromatin sample of SW1116 or LoVo cell lysates by using an antibody against STAT3 (data not shown). In addition, the transcriptional activity of E-cadherin promoter was not dramatically changed after mutation of putative STAT3 binding sites in the E-cadherin promoter (Fig. 8). The data suggest that STAT3 may not directly bind to the E-cadherin promoter. Based on the data described above, we hypothesized that ZEB1 may mediate STAT3-induced E-cadherin down-regulation in CRC cells. This hypothesis was further supported by our finding that STAT3-induced decrease of E-cadherin expression (Fig. 6, B and C) and promoter transcription activity (Fig. 6E) was significantly blocked by knockdown of ZEB1 in CRC cells. In the real-time ChIP PCR assay, overexpression of STAT3 increased ZEB1 recruitment to the E-cadherin promoter (Fig. 7, D and E), whereas it was decreased by knockdown of STAT3 (Fig. 7, F and G). The transcriptional activity of the E-cadherin promoter was significantly increased after mutation of ZEB1 binding sites or dual mutation of STAT3 and ZEB1 binding sites in the E-cadherin promoter in the dual luciferase assay (Fig. 8). The data suggest that ZEB1 may contribute to the STAT3-induced decrease in E-cadherin expression via increasing its binding efficiency to the E-cadherin promoter region.
Our detection of pSTAT3Tyr-705 and ZEB1 expressions by immunohistochemistry further showed that they were remarkably increased in colon adenocarcinoma, compared with normal colon epithelium (Fig. 9). However, E-cadherin expression was significantly decreased in high grade colon adenocarcinoma (Fig. 9). The data are consistent with other reports showing that pSTAT3Tyr-705 is constitutively activated and correlated with CRC dedifferentiation (49). ZEB1 expression is absent in normal colon and breast tissues, whereas ZEB1-positive tumor tissues express low levels of cytokeratin (50). Furthermore, our study also demonstrates that high tumor, lymph node, and metastasis (TNM) staging of CRC was significantly more frequent in patients with high expressions of pSTAT3Tyr-705 and ZEB1 than in those with low expression (Tables 1 and 2). Moreover, a very strong positive correlation between pSTAT3Tyr-705 and ZEB1 expression was identified in cancerous tissues. These observations imply that pSTAT3Tyr-705 and ZEB1 expression may be powerful indicators of CRC metastasis.
In conclusion, STAT3 may play an important role in the EMT process of CRC progression. As an EMT inducer, ZEB1 expression may be regulated by STAT3. ZEB1 may participate in STAT3-induced cell invasion in CRC cells. Moreover, ZEB1 may mediate STAT3-induced down-regulation of E-cadherin via directly inhibiting transcriptional activity of the E-cadherin promoter in CRC. Constitutive activation of STAT3 and elevation of ZEB1 may contribute to poor prognosis in CRC patients.
This work was supported by National Natural Science Foundation of China Grant 30900757 (to F. J. Y.), National Natural Science Foundation of China Grant 91129724 (to J. H.), and “Chen Guang” Project Grant 09CG13 (to J. H.).
- CRC
- colorectal cancer
- EMT
- epithelial-mesenchymal transition
- ANOVA
- analysis of variance
- nt
- nucleotide(s)
- TNM
- tumor, lymph node, and metastasis.
REFERENCES
- 1. Bates R. C., Mercurio A. M. (2005) The epithelial-mesenchymal transition (EMT) and colorectal cancer progression. Cancer Biol. Ther. 4, 365–370 [DOI] [PubMed] [Google Scholar]
- 2. Markowitz S. D., Dawson D. M., Willis J., Willson J. K. (2002) Focus on colon cancer. Cancer Cell 1, 233–236 [DOI] [PubMed] [Google Scholar]
- 3. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 4. Hay E. D. (1995) An overview of epithelio-mesenchymal transformation. Acta Anat. 154, 8–20 [DOI] [PubMed] [Google Scholar]
- 5. Savagner P. (2001) Leaving the neighborhood. Molecular mechanisms involved during epithelial-mesenchymal transition. BioEssays 23, 912–923 [DOI] [PubMed] [Google Scholar]
- 6. Thiery J. P. (2003) Epithelial-mesenchymal transitions in development and pathologies. Curr. Opin. Cell Biol. 15, 740–746 [DOI] [PubMed] [Google Scholar]
- 7. Arima Y., Inoue Y., Shibata T., Hayashi H., Nagano O., Saya H., Taya Y. (2008) Rb depletion results in deregulation of E-cadherin and induction of cellular phenotypic changes that are characteristic of the epithelial-to-mesenchymal transition. Cancer Res. 68, 5104–5112 [DOI] [PubMed] [Google Scholar]
- 8. Papageorgis P., Lambert A. W., Ozturk S., Gao F., Pan H., Manne U., Alekseyev Y. O., Thiagalingam A., Abdolmaleky H. M., Lenburg M., Thiagalingam S. (2010) Smad signaling is required to maintain epigenetic silencing during breast cancer progression. Cancer Res. 70, 968–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arias A. M. (2001) Epithelial mesenchymal interactions in cancer and development. Cell 105, 425–431 [DOI] [PubMed] [Google Scholar]
- 10. Bhowmick N. A., Ghiassi M., Bakin A., Aakre M., Lundquist C. A., Engel M. E., Arteaga C. L., Moses H. L. (2001) Transforming growth factor-β1 mediates epithelial to mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 12, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ellenrieder V., Hendler S. F., Boeck W., Seufferlein T., Menke A., Ruhland C., Adler G., Gress T. M. (2001) Transforming growth factor beta1 treatment leads to an epithelial-mesenchymal transdifferentiation of pancreatic cancer cells requiring extracellular signal-regulated kinase 2 activation. Cancer Res. 61, 4222–4228 [PubMed] [Google Scholar]
- 12. Joyce T., Cantarella D., Isella C., Medico E., Pintzas A. (2009) A molecular signature for Epithelial to Mesenchymal transition in a human colon cancer cell system is revealed by large-scale microarray analysis. Clin. Exp. Metastasis 26, 569–587 [DOI] [PubMed] [Google Scholar]
- 13. Gulhati P., Bowen K. A., Liu J., Stevens P. D., Rychahou P. G., Chen M., Lee E. Y., Weiss H. L., O'Connor K. L., Gao T., Evers B. M. (2011) mTORC1 and mTORC2 regulate EMT, motility, and metastasis of colorectal cancer via RhoA and Rac1 signaling pathways. Cancer Res. 71, 3246–3256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cottonham C. L., Kaneko S., Xu L. (2010) miR-21 and miR-31 converge on TIAM1 to regulate migration and invasion of colon carcinoma cells. J. Biol. Chem. 285, 35293–35302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kalogeropoulou M., Voulgari A., Kostourou V., Sandaltzopoulos R., Dikstein R., Davidson I., Tora L., Pintzas A. (2010) TAF4b and Jun/activating protein-1 collaborate to regulate the expression of integrin α6 and cancer cell migration properties. Mol. Cancer Res. 8, 554–568 [DOI] [PubMed] [Google Scholar]
- 16. Spano J. P., Milano G., Rixe C., Fagard R. (2006) JAK/STAT signaling pathway in colorectal cancer. A new biological target with therapeutic implications. Eur. J. Cancer 42, 2668–2670 [DOI] [PubMed] [Google Scholar]
- 17. Lassmann S., Schuster I., Walch A., Göbel H., Jütting U., Makowiec F., Hopt U., Werner M. (2007) STAT3 mRNA and protein expression in colorectal cancer. Effects on STAT3-inducible targets linked to cell survival and proliferation. J. Clin. Pathol. 60, 173–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xiong H., Zhang Z. G., Tian X. Q., Sun D. F., Liang Q. C., Zhang Y. J., Lu R., Chen Y. X., Fang J. Y. (2008) Inhibition of JAK1,2/STAT3 signaling induces apoptosis, cell cycle arrest, and reduces tumor cell invasion in colorectal cancer cells. Neoplasia 10, 287–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yamashita S., Miyagi C., Carmany-Rampey A., Shimizu T., Fujii R., Schier A. F., Hirano T. (2002) Stat3 controls cell movements during zebrafish gastrulation. Dev. Cell 2, 363–375 [DOI] [PubMed] [Google Scholar]
- 20. Sano S., Itami S., Takeda K., Tarutani M., Yamaguchi Y., Miura H., Yoshikawa K., Akira S., Takeda J. (1999) Keratinocyte-specific ablation of Stat3 exhibits impaired skin remodeling, but does not affect skin morphogenesis. EMBO J. 18, 4657–4668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ferrand A., Kowalski-Chauvel A., Bertrand C., Pradayrol L., Fourmy D., Dufresne M., Seva C. (2004) Involvement of JAK2 upstream of the PI 3-kinase in cell-cell adhesion regulation by gastrin. Exp. Cell Res. 301, 128–138 [DOI] [PubMed] [Google Scholar]
- 22. Colomiere M., Ward A. C., Riley C., Trenerry M. K., Cameron-Smith D., Findlay J., Ackland L., Ahmed N. (2009) Cross-talk of signals between EGFR and IL-6R through JAK2/STAT3 mediate epithelial-mesenchymal transition in ovarian carcinomas. Br. J. Cancer 100, 134–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Perez-Moreno M., Jamora C., Fuchs E. (2003) Sticky business. Orchestrating cellular signals at adherens junctions. Cell 112, 535–548 [DOI] [PubMed] [Google Scholar]
- 24. Qiao Y., Jiang X., Lee S. T., Karuturi R. K., Hooi S. C., Yu Q. (2011) FOXQ1 regulates epithelial-mesenchymal transition in human cancers. Cancer Res. 71, 3076–3086 [DOI] [PubMed] [Google Scholar]
- 25. Ren D., Minami Y., Nishita M. (2011) Critical role of Wnt5a-Ror2 signaling in motility and invasiveness of carcinoma cells following Snail-mediated epithelial-mesenchymal transition. Genes Cells 16, 304–315 [DOI] [PubMed] [Google Scholar]
- 26. Gire V., Roux P., Wynford-Thomas D., Brondello J. M., Dulic V. (2004) DNA damage checkpoint kinase Chk2 triggers replicative senescence. EMBO J. 23, 2554–2563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lu R., Wang X., Chen Z. F., Sun D. F., Tian X. Q., Fang J. Y. (2007) Inhibition of the extracellular signal-regulated kinase/mitogen-activated protein kinase pathway decreases DNA methylation in colon cancer cells. J. Biol. Chem. 282, 12249–12259 [DOI] [PubMed] [Google Scholar]
- 28. Morita Y., Naka T., Kawazoe Y., Fujimoto M., Narazaki M., Nakagawa R., Fukuyama H., Nagata S., Kishimoto T. (2000) Signals transducers and activators of transcription (STAT)-induced STAT inhibitor-1 (SSI-1)/suppressor of cytokine signaling-1 (SOCS-1) suppresses tumor necrosis factor α-induced cell death in fibroblasts. Proc. Natl. Acad. Sci. U.S.A. 97, 5405–5410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang Y. Y., Zhou G. B., Yin T., Chen B., Shi J. Y., Liang W. X., Jin X. L., You J. H., Yang G., Shen Z. X., Chen J., Xiong S. M., Chen G. Q., Xu F., Liu Y. W., Chen Z., Chen S. J. (2005) AML1-ETO and C-KIT mutation/overexpression in t(8;21) leukemia. Implication in stepwise leukemogenesis and response to Gleevec. Proc. Natl. Acad. Sci. U.S.A. 102, 1104–1109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hecht M., Papoutsi M., Tran H. D., Wilting J., Schweigerer L. (2004) Hepatocyte growth factor/c-Met signaling promotes the progression of experimental human neuroblastomas. Cancer Res. 64, 6109–6118 [DOI] [PubMed] [Google Scholar]
- 31. Fu X., Beer D. G., Behar J., Wands J., Lambeth D., Cao W. (2006) cAMP-response element-binding protein mediates acid-induced NADPH oxidase NOX5-S expression in Barrett esophageal adenocarcinoma cells. J. Biol. Chem. 281, 20368–20382 [DOI] [PubMed] [Google Scholar]
- 32. Jäättelä M. (1999) Escaping cell death. Survival proteins in cancer. Exp. Cell Res. 248, 30–43 [DOI] [PubMed] [Google Scholar]
- 33. Niwa Y., Kanda H., Shikauchi Y., Saiura A., Matsubara K., Kitagawa T., Yamamoto J., Kubo T., Yoshikawa H. (2005) Methylation silencing of SOCS-3 promotes cell growth and migration by enhancing JAK/STAT and FAK signalings in human hepatocellular carcinoma. Oncogene 24, 6406–6417 [DOI] [PubMed] [Google Scholar]
- 34. Zhang F., Li C., Halfter H., Liu J. (2003) Delineating an oncostatin M-activated STAT3 signaling pathway that coordinates the expression of genes involved in cell cycle regulation and extracellular matrix deposition of MCF-7 cells. Oncogene 22, 894–905 [DOI] [PubMed] [Google Scholar]
- 35. Alvarez J. V., Greulich H., Sellers W. R., Meyerson M., Frank D. A. (2006) Signal transducer and activator of transcription 3 is required for the oncogenic effects of non-small-cell lung cancer-associated mutations of the epidermal growth factor receptor. Cancer Res. 66, 3162–3168 [DOI] [PubMed] [Google Scholar]
- 36. Cavallaro U., Liebner S., Dejana E. (2006) Endothelial cadherins and tumor angiogenesis. Exp. Cell Res. 312, 659–667 [DOI] [PubMed] [Google Scholar]
- 37. Blaschuk O. W., Devemy E. (2009) Cadherins as novel targets for anti-cancer therapy. Eur. J. Pharmacol. 625, 195–198 [DOI] [PubMed] [Google Scholar]
- 38. Jeanes A., Gottardi C. J., Yap A. S. (2008) Cadherins and cancer. How does cadherin dysfunction promote tumor progression? Oncogene 27, 6920–6929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Makrilia N., Kollias A., Manolopoulos L., Syrigos K. (2009) Cell adhesion molecules. Role and clinical significance in cancer. Cancer Invest. 27, 1023–1037 [DOI] [PubMed] [Google Scholar]
- 40. Paschos K. A., Canovas D., Bird N. C. (2009) The role of cell adhesion molecules in the progression of colorectal cancer and the development of liver metastasis. Cell. Signal. 21, 665–674 [DOI] [PubMed] [Google Scholar]
- 41. Gan Y., Shi C., Inge L., Hibner M., Balducci J., Huang Y. (2010) Differential roles of ERK and Akt pathways in regulation of EGFR-mediated signaling and motility in prostate cancer cells. Oncogene 29, 4947–4958 [DOI] [PubMed] [Google Scholar]
- 42. Luna-Zurita L., Prados B., Grego-Bessa J., Luxán G., del Monte G., Benguría A., Adams R. H., Pérez-Pomares J. M., de la Pompa J. L. (2010) Integration of a Notch-dependent mesenchymal gene program and Bmp2-driven cell invasiveness regulates murine cardiac valve formation. J. Clin. Invest. 120, 3493–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stemmer V., de Craene B., Berx G., Behrens J. (2008) Snail promotes Wnt target gene expression and interacts with β-catenin. Oncogene 27, 5075–5080 [DOI] [PubMed] [Google Scholar]
- 44. Vincent T., Neve E. P., Johnson J. R., Kukalev A., Rojo F., Albanell J., Pietras K., Virtanen I., Philipson L., Leopold P. L., Crystal R. G., de Herreros A. G., Moustakas A., Pettersson R. F., Fuxe J. (2009) A SNAIL1-SMAD3/4 transcriptional repressor complex promotes TGF-β-mediated epithelial-mesenchymal transition. Nat. Cell Biol. 11, 943–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sullivan N. J., Sasser A. K., Axel A. E., Vesuna F., Raman V., Ramirez N., Oberyszyn T. M., Hall B. M. (2009) Interleukin-6 induces an epithelial-mesenchymal transition phenotype in human breast cancer cells. Oncogene 28, 2940–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chen Y. L., Lv J., Ye X. L., Sun M. Y., Xu Q., Liu C. H., Min L. H., Li H. P., Liu P., Ding X. (2011) Sorafenib inhibits transforming growth factor β1-mediated epithelial-mesenchymal transition and apoptosis in mouse hepatocytes. Hepatology 53, 1708–1718 [DOI] [PubMed] [Google Scholar]
- 47. Nieto M. A. (2002) The snail superfamily of zinc finger transcription factors. Nat. Rev. Mol. Cell Biol. 3, 155–166 [DOI] [PubMed] [Google Scholar]
- 48. Peinado H., Olmeda D., Cano A. (2007) Snail, Zeb, and bHLH factors in tumor progression. An alliance against the epithelial phenotype? Nat. Rev. Cancer 7, 415–428 [DOI] [PubMed] [Google Scholar]
- 49. Kusaba T., Nakayama T., Yamazumi K., Yakata Y., Yoshizaki A., Inoue K., Nagayasu T., Sekine I. (2006) Activation of STAT3 is a marker of poor prognosis in human colorectal cancer. Oncol. Rep. 15, 1445–1451 [PubMed] [Google Scholar]
- 50. Aigner K., Dampier B., Descovich L., Mikula M., Sultan A., Schreiber M., Mikulits W., Brabletz T., Strand D., Obrist P., Sommergruber W., Schweifer N., Wernitznig A., Beug H., Foisner R., Eger A. (2007) The transcription factor ZEB1 (δEF1) promotes tumor cell dedifferentiation by repressing master regulators of epithelial polarity. Oncogene 26, 6979–6988 [DOI] [PMC free article] [PubMed] [Google Scholar]