Background: Proteases responsible for a CCL15-(25–92) product have not been elucidated.
Results: All 14 CC monocyte chemoattractants, including CCL15, are processed by multiple MMPs.
Conclusion: MMP-processing of CCL15, CCL23, and CCL16 functional activity is altered by MMP processing.
Significance: This is the first study showing MMPs can activate CC chemokines and hence monoycte chemoattraction with potential to propagate inflammation.
Keywords: Arthritis, Chemokines, Chemotaxis, Inflammation, Mass Spectrometry (MS), Matrix Metalloproteinase (MMP), Monocytes, Protease
Abstract
Leukocyte migration and activation is orchestrated by chemokines, the cleavage of which modulates their activity and glycosaminoglycan binding and thus their roles in inflammation and immunity. Early research identified proteolysis as a means of both activating or inactivating CXC chemokines and inactivating CC chemokines. Recent evidence has shown activating cleavages of the monocyte chemoattractants CCL15 and CCL23 by incubation with synovial fluid, although the responsible proteases could not be identified. Herein we show that CCL15 is processed in human synovial fluid by matrix metalloproteinases (MMPs) and serine proteases. Furthermore, a family-wide investigation of MMP processing of all 14 monocyte-directed CC chemokines revealed that each is precisely cleaved by one or more MMPs. By MALDI-TOF-MS, 149 cleavage sites were sequenced including the first reported instance of CCL1, CCL16, and CCL17 proteolysis. Full-length CCL15-(1–92) and CCL23-(1–99) were cleaved within their unique 31 and 32-amino acid residue extended amino termini, respectively. Unlike other CCL chemokines that lose activity and become receptor antagonists upon MMP cleavage, the prominent MMP-processed products CCL15-(25–92, 28–92) and CCL23-(26–99) are stronger agonists in calcium flux and Transwell CC receptor transfectant and monocytic THP-1 migration assays. MMP processing of CCL16-(1–97) in its extended carboxyl terminus yields two products, CCL16-(8–77) and CCL16-(8–85), with both showing unexpected enhanced glycosaminoglycan binding. Hence, our study reveals for the first time that MMPs activate the long amino-terminal chemokines CCL15 and CCL23 to potent forms that have potential to increase monocyte recruitment during inflammation.
Introduction
Chemokines are an important superfamily of chemoattractant cytokines mediating directional leukocyte migration in innate and acquired host defense responses in a concentration-dependent manner. Through interaction of basic amino acid residues in the chemokine carboxyl-terminal α-helix with negatively charged glycosaminoglycans (GAGs)3 of proteoglycans in the extracellular matrix or on cells, chemokines form a haptotactic gradient, the slope of which directs leukocyte migration (1, 2). There are four subfamilies of chemokines based upon the proximity of the conserved amino-terminal cysteine (C) residues, the largest being the CC and CXC subfamilies. Chemokines exert activity by binding to receptors on leukocyte surfaces; CC chemokines bind to CC receptors (CCRs), whereas CXC chemokines bind CXCRs. The predominant CXCRs, namely CXCR1 and CXCR2, are expressed by neutrophils with monocytes expressing CCR1, CCR2, CCR3, CCR5, and CCR8 (3, 4). The flexible amino terminus of a chemokine is involved in binding and activating its cognate receptor (5, 6). Receptor activation causes intracellular signaling, including calcium mobilization, resulting in cell activation that is characterized by cell migration, gene transcription, relocation of receptors to the cell membrane, and the release of further inflammatory mediators. Inappropriate continual recruitment and activation of leukocytes can result in tissue damage in chronic inflammatory diseases such as rheumatoid arthritis. Thus, regulation of cellular recruitment and termination of this intercellular signaling is critical to both initiate and then later dampen inflammatory responses.
Post-translational modifications of both the amino and carboxyl termini of chemokines by proteolytic processing is a versatile mechanism of regulation, trimming CXC chemokines into stronger agonists for recruitment of neutrophils, of both CXC and CC chemokines to receptor antagonists to terminate signaling, to switch receptor use, and to shed CX3CL1 or, alternatively, to modify the GAG binding site to prevent or disrupt haptotactic gradient formation (7–19).
A number of chemokines are processed in vitro by proteases and in particular by serine proteases from neutrophils and by matrix metalloproteinases (MMPs) (8, 12, 13, 20–29). Serine proteases, including cathepsin G and neutrophil elastase, are secreted by activated neutrophils during an inflammatory response; natural inhibitors include serpins. MMPs are an important family of extracellular endopeptidases that are up-regulated in stimulated stromal cells and leukocytes and are pathognomonic of many chronic inflammatory diseases. The activity of MMPs is regulated by tissue inhibitors of metalloproteinases (TIMPs) with the net individual activities of different MMPs being both beneficial and detrimental in disease (30).
In the CXC chemokine subfamily the neutrophil chemoattractants CXCL8 and CXCL5 are processed, in particular by the neutrophil-specific MMP-8 (also known as collagenase-2), to become potent receptor agonists and form a feed-forward mechanism, a critical step for neutrophil recruitment (16, 27). In contrast, all seven neutrophil CXC agonists in man are inactivated by macrophage-derived MMP-12, terminating the recruitment of neutrophils (21). Multiple MMPs generate potent CCR1, CCR2, and CCR5 receptor antagonists by cleaving CCL2, -7, -8, and -13 to terminate monocyte recruitment (12, 13). Notably, proteolysis of human CC chemokines that results in an activating cleavage is limited to serine protease activity on CCL4 (31), CCL14 (32–34), CCL15 (20), and CCL23 (20).
In an in vitro assay, the 92-amino acid residue CCL15-(1–92) and the 99 amino acid residue CCL23-(1–99), neither of which is a potent chemoattractant in the full-length form (35, 36), were processed by synovial fluid from arthritic patients to the products CCL15-(25–92) and CCL23-(19–99) that have enhanced CCR1 agonist activity (20). However, despite the importance of this observation, the specific proteases responsible for these cleavages could not be identified despite considerable effort. Amino-terminally truncated CCL15 and CCL23 were both identified in synovial fluid from arthritic patients at concentrations of 10–100-fold that of CCL3 and CCL5 (20), indicating that these truncated chemokines may contribute to the cellular recruitment that is observed in chronic inflammation.
Herein we utilized inhibitors to identify the protease classes responsible for the activating cleavages of CCL15 in synovial fluid, finding that both serine proteases and MMPs are responsible. In view of the importance of macrophage recruitment, this encouraged us to identify other MMP chemokine substrates. Therefore, we performed a global evaluation of MMP processing of all 14 CC chemokines that are involved in monocyte recruitment. We report that MMP processing of the long amino-terminal CCL15 and CCL23 chemokines and the long carboxyl-terminal CCL16, notably by the monocyte/macrophage specific MMP-12, results in increased receptor activation or GAG binding, respectively. These data thereby point to a critical role for MMPs in the promotion and regulation of monocyte recruitment. Our results implicate new feed-forward mechanisms whereby macrophage and synovial fluid proteases promote the recruitment of monocytes, potentiating the inflammatory response.
EXPERIMENTAL PROCEDURES
Proteinases and Chemokines
Recombinant human MMPs 1, 2, 3, 8, 9, 12, 13, and soluble MMP-14 were expressed and purified (37). MMP-7 was from U. S. Biochemical Corp. All full-length chemokines were chemically synthesized using t-butoxycarbonyl solid phase chemistry, purified by high performance liquid chromatography, and validated for activity as described (38). A 19-mer peptide corresponding to CCL23-(5–23) was chemically synthesized by Sigma. CCL15-(25–92, 28–92), CCL16-(8–77, 8–85), and CCL23-(26–99), utilized for in vitro experiments, were the fully truncated products of MMP-12 cleavage of full-length counterparts; these preparations lack full-length chemokine as determined by matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS). Controls for functional assays, namely full-length chemokine and MMP-12 alone, were prepared at the same time.
Chemokine Cleavage in Synovial Fluid
Cleavage of 2.5 μg of CCL15-(1–92) in 0.25 μg of synovial fluid pooled from eight rheumatoid arthritis patients was performed at 37 °C for 16 h in cleavage assay buffer in the absence or presence of 1 mm PMSF (general serine proteinase inhibitor), 10 μm marimastat (small molecule general metalloproteinase inhibitor) (39), 100 μm TIMP-1 and TIMP-2 (endogenous metalloproteinase inhibitors), 10 μm E64 (cysteine protease inhibitor), 10 μm pepstatin (aspartyl protease inhibitor), and 100 μm leupeptin (inhibitor of specific serine and cysteine proteases) either alone or in combination. Cleavage of CCL15 in the synovial fluid was assessed as described (40) by MALDI-TOF MS on a Voyager-DE STR (Applied Biosystems) or a 4700 tandem mass spectrometer (Applied Biosystems) using the matrices sinapic acid or α-cyano-4-hydroxycinnamic acid, respectively, and confirmed by silver-staining after 15% Tris-Tricine SDS-PAGE.
Chemokine Cleavage Assays
Zymogen forms of MMP-1, -2, -8, -9, -13, and -14 were activated by incubation with 1 mm p-aminophenylmercuric acetate for 45 min at 37 °C. The recombinant MMP-7 and -12 lacked a prodomain and so did not require activation. MMP-3 was activated by incubation with 0.25 μm chymotrypsin for 30 min at 37 °C, and then the chymotrypsin activity was inhibited by the addition of 1 mm PMSF; all experiments with MMP-3 included a chymotrypsin/PMSF only control. The concentrations of active MMPs were determined by active-site titration against TIMP-1 or TIMP-2 in a peptide cleavage assay using the quenched fluorescent peptide Mca-Pro-Leu-Gly-Leu-Dpa-Ala-Arg-NH2(Dpa is N3-(2,4-dinitrophenyl)-l-2–3-diaminopropionyl). In vitro chemokine cleavage assays with recombinant MMPs were performed in 50 mm Tris, 200 mm NaCl, 5 mm CaCl2, pH 7.4, for 16 h at 37 °C. Initially, an enzyme to substrate molar ratio of 1:10 was assessed to screen for cleavage activity followed by lower enzyme concentrations to 1:1000 to suggest biological relevance. Cleavage assay products were analyzed as above by MALDI-TOF MS and confirmed by silver-staining after 15% Tris-Tricine SDS-PAGE. Chemokine cleavage was defined to be positive when the MS spectra showed a cleavage product with a greater than 20% ion intensity of the full-length chemokine.
For kinetic analysis CCL15 was incubated with increasing molar ratios of each of the MMPs. Gels of the reaction products were stained with Coomassie Brilliant Blue R-250, and bands were quantified by densitometry using Alpha-imager software and the results fitted to the equation kcat/Km = (ln 2)/Et½).
Cells
Human CCR1-transfected B300-19 (PreB) cells were kindly provided by Dr. B. Moser (Cardiff, UK) and cultured in RPMI 1640 supplemented with 10% fetal bovine serum, 2 mm glutamine, 50 μm β-mercaptoethanol, and 1.5 μg/ml puromycin. The human monocytic cell line, THP-1 (ATCC), were grown in RPMI 1640 supplemented with 10% fetal bovine serum.
Calcium Mobilization
Calcium mobilization of THP-1- or CCR1-transfected B300-19 cells was evaluated as previously described (40). Briefly, 1 × 107 cells/ml were loaded with 2 μm Fluo-4-acetoxymethyl ester (Molecular Probes) for 30 min at 37 °C, 5% CO2 in RPMI 1640 media supplemented with 1% fetal bovine serum. Cells were washed 3 times and resuspended at 1 × 106 cells/ml in fresh assay buffer (Ca2+/Mg2+-free Hanks' balanced salt solution (Invitrogen), 20 mm HEPES, 2.5 mm probenecid (Sigma), 0.1% bovine serum albumin (BSA)). Assays were performed on 6.5 × 105 cells in 800-μl cuvettes on an LS50B spectrofluorimeter (PerkinElmer Life Sciences) in time-drive mode with excitation and emission at 494 and 515 nm, respectively. Calibration was made by the addition of 5 μm ionomycin (Sigma) followed by 1 mm MnCl2 (FisherBiotech) to determine Fmax and Fmin, respectively. Absolute Ca2+ was calculated as Kd × ((F − Fmin)/(Fmax − F)) where the Kd of Fluo-4 was 345 nm (Molecular Probes). Experiments were carried out in duplicate and repeated a minimum of three times. Statistical significance of cleaved versus full-length chemokines was evaluated by two-tailed t test in Prism (GraphPad).
Transwell Migration
The chemotactic response of THP-1 or CCR1-transfected B300-19 cells was evaluated (40). Chemokine or MMP and buffer controls, diluted in BSA-supplemented RPMI 1640, in the lower chamber were separated from 2 × 105 cells/ml in the upper chamber of a 96-well Boyden chamber (Neuroprobe) by a 5 μm pore filter. Assays performed at 37 °C, 5% CO2 were 90 min for THP-1 and 3 h for CCR1-transfected cells. Non-migrating cells were aspirated, and the upper chamber was washed with 2 mm EDTA. After transfer to a Maxisorb 96-well plate, the DNA content of the lower chamber was determined by CyQuant reagent (Invitrogen). The chemotactic index was calculated by the ratio of the relative fluorescence of samples from cells migrating in response to chemokine compared with the media control. Experiments were carried out in quadruplicate and repeated a minimum of three times. Statistical significance of cleaved versus full-length chemokines were evaluated by 2-way analysis of variance and Bonferroni post-test.
Glycosaminoglycan Binding
HiTrapTM cation exchange Sepharose or heparin-Sepharose (GE healthcare) 1-ml columns were washed and equilibrated before loading 25 μg of full-length or cleaved chemokine in 1 ml of 10 mm potassium phosphate, pH 7.5. Using an Akta Purifier (Amersham Biosciences), columns were washed with 10 × Vt of 10 mm potassium phosphate and then a linear gradient of 0 to 1.5 m NaCl over 30 min at a flow rate of 1 ml/min and monitored by in-line absorbance at 215 nm. ASCII files were imported into Prism (GraphPad) and normalized against base line. The effects of soluble GAGs, namely heparan sulfate and chondroitin sulfate A, B, and C (Seikagaku), on the in vitro cleavage of CCL16 by MMP-12 were assessed with a chemokine to GAG ratio of 1:5 (w/w) at an enzyme to substrate ratio of 1:25 (w/w).
RESULTS
Synovial Fluid Processing of CCL15
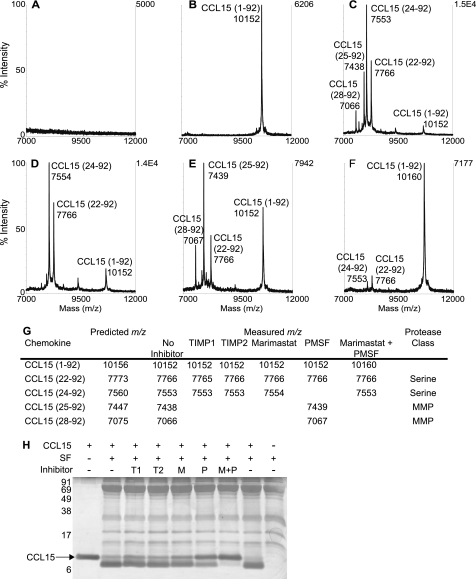
The key role of MMPs in processing chemokines is now well established, including the activation of CXC chemokines critical for neutrophil recruitment. However, an activating cleavage of CC chemokines by MMP activity was hitherto unknown. To evaluate the protease classes responsible for CCL15 processing in vivo (20), a pool of synovial fluids from eight arthritic patients (Fig. 1A) was combined with full-length CCL15 (Fig. 1B) and then incubated in the absence (Fig. 1C) or presence (Fig. 1, D–G) of protease inhibitors for 16 h at 37 °C. Endogenous synovial fluid proteases process CCL15 to products with spectral peaks detected by MALDI-TOF (Fig. 1C) having m/z values of 7066, 7438, 7553, and 7766. In addition to the previously observed product CCL15-(25–92) in synovial fluid (20), we found additional cleavages of CCL15 to produce the products CCL15-(28–92), CCL15-(24–92), and CCL15-(22–92) (Fig. 1G).
FIGURE 1.
Synovial fluid MMP and serine proteases cleave CCL15. Synovial fluid (SF) from eight rheumatoid arthritis patients was pooled, and 25-μg samples were incubated with 2.5 μg of CCL15 in the absence or presence of protease inhibitors for 16 h at 37 °C. Mass spectra are shown of C18 ZipTip prepared samples after 16 h of incubation of SF (A), CCL15-(1–92) (B), SF + CCL15 (C), SF + CCL15 + marimastat (D), SF + CCL15 + PMSF (E), and SF + CCL15 + marimastat + PMSF (F). G, cleavage products were assigned by MALDI-TOF mass by comparison of measured with predicted mass to charge ratios (m/z) with a +1 charge ionization ([M + H]+). H, cleavage was visualized on silver-stained 15% Tris-Tricine gels. T1, TIMP-1; T2, TIMP-2; M, marimastat; P, PMSF.
Two spectral peaks corresponding to CCL15-(28–92) (m/z 7066) and CCL15-(25–92) (m/z 7,738) were eliminated by the metalloproteinase inhibitors TIMP-1, TIMP-2, or marimastat (Fig. 1D). Thus, these cleavage products are due to MMP activity. In the presence of PMSF, the peak corresponding to CCL15-(24–92) (m/z 7553) was eliminated, and that for CCL15-(22–92) (m/z 7766) was significantly reduced (Fig. 1E), showing a role for serine proteases in vivo. CCL15-(24–92) and -(22–92) were previously shown to result from cathepsin G (20) and neutrophil elastase activity (20, 41), respectively. Pepstatin or leupeptin had no effect on processing of CCL15, revealing that neither cysteine nor carboxyproteases cleaved the chemokine (not shown).
In the presence of both marimastat and PMSF, CCL15 remains intact in synovial fluid (Fig. 1F). Together, these results suggest that neutrophil serine proteases and MMPs are responsible for CCL15 processing in vivo. Moreover, the presence of two products when one protease class is inhibited implies that processing by one protease family is not dependent upon another. The proteolytic activity in synovial fluid was impressive as a 10-fold excess of CCL15 on a weight for weight basis was fully cleaved (Fig. 1H).
Selective Processing of CCL15 by MMPs
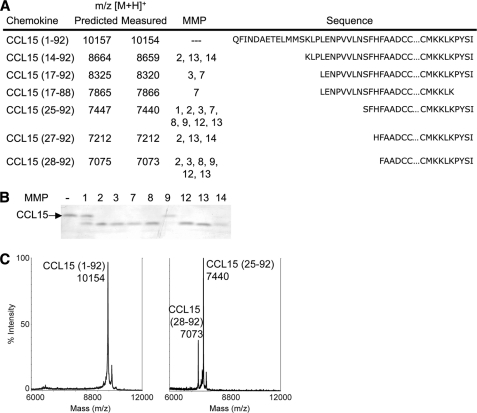
To evaluate which MMPs could be responsible for CCL15 processing observed in synovial fluid, in vitro cleavage assays were performed. CCL15-(1–92) is processed at the amino terminus by all MMPs tested (Fig. 2) and by high concentrations of MMP-7 at the carboxyl terminus. Interestingly, all MMPs processed CCL15-(1–92) to CCL15-(25–92), one of the two forms observed by MMPs present in synovial fluid (Fig. 2A) and previously observed from recombinant production in insect cells (42). The second synovial fluid product, CCL15-(28–92), results from MMP-2, -3, -8, -9, -12, and -13 processing.
FIGURE 2.
All MMPs evaluated precisely cleaved CCL15 at one or more sites. Human CCL15-(1–92) was incubated with recombinant MMP-1, -2, -3, -7, -8, -9, -12, -13, and -14 for 16 h at 37 °C. A, cleavage products were assigned by MALDI-TOF MS by comparison of measured with predicted mass to charge ratios (m/z) with a +1 charge ionization ([M + H]+). Corresponding sequences are shown. B, cleavage assay products were visualized on silver-stained 15% Tris-Tricine gels. C, mass spectra of cleavage assay products of CCL15 in the absence (left) or presence (right) of MMP-12 show that the predominant truncated forms CCL15-(25–92) and -(28–92) are both present in the form used for functional assays.
By silver-stained gels the processing of CCL15 by MMP-9 appears minimal (Fig. 2B), although both CCL15-(25–92) and CCL15-(28–92) cleavage products are observed on MALDI-TOF MS (Fig. 2A). MMP-12 processing of CCL15-(1–92) results in complete truncation to two products as evident by MALDI-TOF MS (Fig. 2C); this preparation was used for in vitro functional assays.
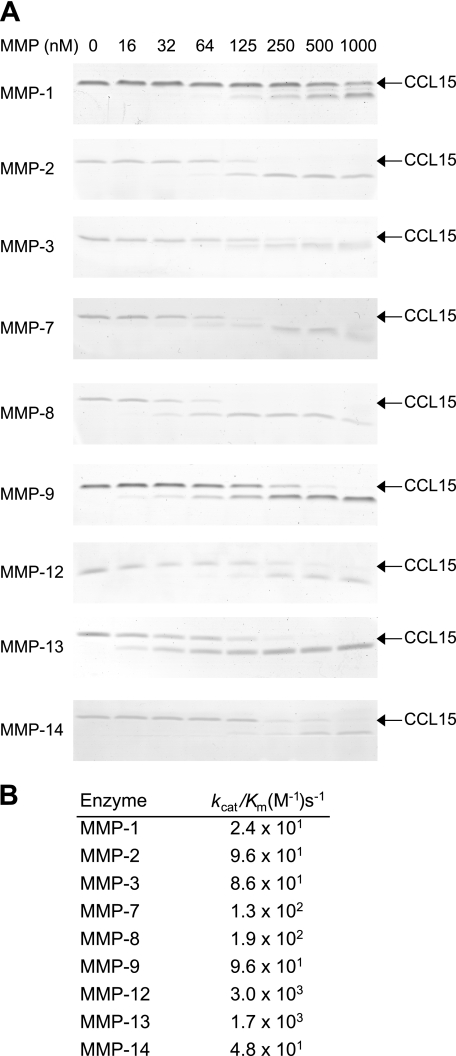
Kinetics Analysis of CCL15 Processing
MMP-12 most efficiently processed CCL15 (Fig. 3), and with a rate of 3000 m−1s−1, this is >100-fold faster than MMP-1, the least efficient MMP. Interestingly, two of the most efficient MMPs with the fastest rates were leukocyte-specific MMPs, macrophage MMP-12, and neutrophil MMP-8.
FIGURE 3.
Kinetics of CCL15 processing by MMPs. CCL15-(1–92) was incubated with increasing concentrations of recombinant MMP-1, -2, -3, -7, -8, -9, -12, -13, and -14 for 16 h and 37 °C at a 1:10 molar ratio. A, cleavage assay products were visualized on Coomassie Brilliant Blue R-250-stained 15% Tris-Tricine gels. B, kcat/Km was determined after densitometry of gels.
All Monocytic-responsive CC Ligands Are Processed by MMPs
To determine the novelty of MMP activation of CCL15, we next evaluated the MMP-processing capacity toward all monocyte chemoattractants in the CC chemokine subfamily, finding that every one of the CCR1, -3, -5, and -8 ligands was cleaved by one or more of the MMPs tested: MMP-1, -2, -3, -7, -8, -9, -12, -13, and -14 (Table 1). Of the 149 cleavage sites we sequenced in the 14 chemokines, 32 are new, being described for the first time, with 28 of these generating previously unidentified amino or carboxyl termini of the chemokines.
TABLE 1.
MMP-truncation products of CC chemokines
Chemokine truncation products after MMP cleavage were deconvoluted from MALDI-TOF MS spectra. The residue numbers of the resultant chemokine cleavage products are shown in parentheses. Sites in roman type represent novel cleavage sites; italics represent truncation sites previously identified. NC, indicates that the full-length chemokine was not cleaved by that MMP. ND, indicates that the cleavage site could not be determined. Activity of MMP-cleaved chemokine is based upon the information provided in footnotes.
| MMP Chemokine | 1 | 2 | 3 | 7 | 8 | 9 | 12 | 13 | 14 | Effect of cleavage |
|---|---|---|---|---|---|---|---|---|---|---|
| CCL1 (1–73) | NC | NC | (8–73) | (7–73) | NC | NC | (7–73) | (6–73) | NC | Unknowna |
| (7–70) | ||||||||||
| CCL2 (1–76) | (5–76) | NC | (5–76) | (5–76) | (5–76) | (5–76) | (5–76) | (5–76) | NC | Loss of agonistb,c |
| (5–67) | ||||||||||
| CCL3 (1–70) | (1–47) | NC | NC | (9–63) | (16–64) | (16–64) | ND | (1–47) | NC | Loss of agonista,d |
| (1–47) | ||||||||||
| CCL4 (1–69) | (16–62) | (6–44) | (6–44) | (1–61) | (7–69) | (14–61) | (7–69) | NC | (6–44) | Monomerizea,b,c |
| (7–69) | (6–69) | (7–69) | ||||||||
| CCL5 (1–69) | NC | NC | NC | (1–65) | NC | NC | NC | (5–69) | NC | Antagonistb |
| CCL7 (1–76) | (5–76) | (5–76) | (5–76) | (7–76) | NC | (5–76) | (5–76) | (5–76) | (9–76) | Antagonistb,c |
| (5–76) | (7–76) | |||||||||
| (5–76) | ||||||||||
| CCL8 (1–76) | (1–73) | (1–73) | (5–76) | (1–73) | (1–73) | NC | (7–76) | (1–73) | (6–76) | Antagonistb,c |
| (5–73) | (5–73) | (5–73) | (1–73) | (5–73) | ||||||
| (5–73) | (1–73) | |||||||||
| CCL11 (1–74) | NC | (10–74) | NC | (10–74) | NC | NC | (10–74) | NC | NC | Loss of agonistd |
| CCL13 (1–75) | (5–72) | (5–75) | (4–75) | (6–75) | (5–72) | (5–72) | (5–72) | (5–72) | (4–75) | Antagonistb,c |
| (5–75) | (4–75) | |||||||||
| (5–72) | ||||||||||
| CCL14 (1–74) | NC | NC | NC | (1–70) | NC | NC | (1–71) | NC | NC | Increased agonista,c |
| (1–66) | (4–72) | |||||||||
| (7–74) | ||||||||||
| CCL15 (1–92) | (25–92) | (14–92) | (17–92) | (17–88) | (25–92) | (25–92) | (25–92) | (25–92) | (14–92) | Increased agonistb,c |
| (25–92) | (25–92) | (17–92) | (28–92) | (28–92) | (28–92) | (27–92) | (27–92) | |||
| (27–92) | (28–92) | (25–92) | (28–92) | |||||||
| (28–92) | ||||||||||
| CCL16 (1–97) | (8–85) | (5–97) | (8–85) | (8–85) | (8–85) | (8–85) | (5–97) | (8–85) | (5–97) | Altered GAG binding |
| (8–77) | (8–77) | (8–85) | (8–77) | (8–85) | ||||||
| (8–77) | ||||||||||
| CCL17 (1–71) | NC | NC | NC | (4–69) | (9–71) | NC | (4–69) | (4–71) | NC | Unknowna |
| CCL23 (1–99) | (11–99) | (11–99) | (11–99) | (11–99) | (11–99) | (11–99) | (11–99) | (28–99) | (14–99) | Increased agonistb,c |
| (17–99) | (17–99) | (21–99) | (17–99) | (17–99) | (28–99) | (17–99) | (1–90) | (30–99) | ||
| (23–99) | (26–99) | (26–99) | (23–99) | (26–99) | (23–99) | |||||
| (26–99) | (28–99) | (1–90) | (26–99) | (26–99) | ||||||
| (28–99) | (1–90) | |||||||||
| (30–99) |
a Unknown effect requiring further characterization.
b Predicted activity from studies reporting truncations at adjacent or nearby sites.
c Published known activity from protein engineering studies or characterization of cleavage at this site by other proteases.
d Predicted activity from reports of cleavages at homologous sites in homologous chemokines.
The broadest specificities observed are for MMP-7 and -12, which processed 14 and 13 chemokines, respectively. Unlike the other MMPs we used, MMP-7 and 12 lack a hemopexin C-domain. Although we have previously shown multiple examples where the hemopexin domain binds specific chemokines and increases cleavage, these results indicate that the hemopexin domain also restricts the substrate repertoire of MMPs. The processing of CCL2, -7, -8, and -13 by MMP-7 is in contrast to the absence of cleavage by MMP-7 we previously reported (13), which we attribute to improved quality control for the only commercially obtained MMP used.
Among the 32 novel truncation sites identified, this is the first report of any proteolysis of three chemokines: CCL1, CCL16, and CCL17. Although not investigated here, based upon the critical role of the amino termini in other CCL chemokines that have only 8–10 residues before the conserved cysteines (7, 10, 12, 13, 15, 18, 43, 44), we predict that the amino-terminal cleavages observed in CCL1 and CCL17 result in a similar loss of agonist activity of these chemokines. Similarly, the CCL5 (5–69) product produced by MMP-13 activity is also most likely to lose agonist activity similar to the CCL5-(3–69) and -(4–69) products previously evaluated (15, 45).
A novel amino-terminal truncation resulting in CCL11-(10–74) is observed after processing by MMP-2, -7, and -12. Unlike previously identified CCL11 truncations of two or three residues, which do not alter the chemoattractant potential (46), this MMP-processing is notable as it occurs between the two conserved cysteines at 6PTTC↓CFNL13, which are components of the two characteristic stabilizing disulfide bonds of the CCL chemokines. Almost certainly then, cleavage here will inactivate the chemokine through structural relaxation. Notably, this is a highly unusual cleavage site specificity unique among the MMPs and reveals that the active site of these MMPs can undergo rearrangement to accommodate the rigid disulfide-linked cysteine residues at P1 and P1′.
Herein, the first identification of both amino- and carboxyl-terminal proteolysis sites in CCL3 is identified. MMP-8 truncation of CCL3 was recently observed in vivo (47), although the site was not sequenced. We show that MMP-8 amino-terminally truncates CCL3-(1–70) to the product CCL3-(16–64), which is likely to be the site of the observed inactive in vivo product, likely through loss of receptor binding mediated via Phe13 (48). The related chemokine CCL4 is known to lose dimer affinity by removal of 5–8 amino-terminal residues (49), whereas the truncation product CCL4-(14–61) lacks activity (48). Thus, both are expected to occur after MMP cleavage of CCL4.
The CCL14-(7–74) product, here shown to result from MMP-12 processing, has been shown to enhance calcium mobilization (50). However, carboxyl-terminal truncations of the chemokine, observed here by MMP activity, have not previously been identified, and thus the functional effects of this require further investigation.
Although there was limited promiscuity of the MMPs toward chemokines, this was not observed with all chemokines. As was found for CCL15, CCL16 and CCL23 are most susceptible to processing, being cleaved by all MMPs evaluated. Moreover, multiple cleavage sites by a given MMP were observed. Therefore, these chemokine processing events were selected for further evaluation.
Selective Processing of CCL23 by MMPs
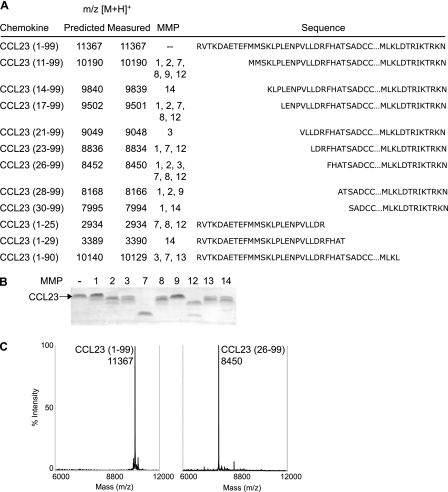
The 97-amino acid residue CCL23 is processed by all MMPs assessed at the amino terminus and at the carboxyl terminus by MMP-3, -7, and -13 (Fig. 4, A and B). The detection of CCL23-(26–99) and the counterpart CCL23-(1–25) by MALDI-TOF MS after MMP-1, 2, 3, 7, 8, and -12 cleavage (Fig. 4A) suggests that although multiple cleavages can occur within the first 25 residues of CCL23-(1–99), CCL23-(26–99) is the predominant stable end product. Similarly, both fragments resulting from cleavage of CCL23 by MMP-14 were observed, namely CCL23-(1–29) and CCL23-(30–99). CCL23-(30–99) is known to result from elastase and activity within synovial fluid (20), but we were unable to identify the protease classes present in synovial fluid responsible for CCL23 cleavage (results not shown). The CCL23-(26–99) preparation we used for in vitro functional assays lacks full-length chemokine as shown by MALDI-TOF MS (Fig. 4C).
FIGURE 4.
All MMPs evaluated precisely cleaved CCL23 at one or more sites. Human CCL23-(1–99) was incubated with recombinant MMP-1, -2, -3, -7, -8, -9, -12, -13, and -14 for 16 h at 37 °C. A, cleavage products were assigned by MALDI-TOF MS by comparison of measured with predicted mass-to-charge ratios (m/z) with a +1 charge ionization ([M + H]+). Corresponding sequences are shown. B, cleavage assay products were visualized on silver-stained 15% Tris-Tricine gels. C, mass spectra of cleavage assay products of CCL23 in the absence (left) or presence (right) of MMP-12 show that the predominant truncated form CCL23-(26–99) is present in the form used for functional assays.
Selective Processing of CCL16 by MMPs
CCL16 is processed by MMPs at both the amino and carboxyl termini (Fig. 5). This is the first identification of CCL16 proteolysis. A CCL16-(5–97) product was observed after MMP-2, -8, and -14 activity, and CCL16-(8–85) or -(8–77) were generated by all MMPs tested (Fig. 5). By silver-stained gels the activity of MMP-1 appears minimal (Fig. 5A), although a CCL16-(8–85) product was observed by MALDI-TOF MS (Fig. 5B); the cleavage sites are shown in Fig. 5C. Conversely, MMP-14 degrades CCL16, yet both the CCL16-(5–97) and (8–85) products represent a >20% intensity in the mass spectra (not shown), indicating that these are likely the initial cleavage products. For in vitro functional assays we used MMP-12 at a 1:10 molar ratio with CCL16 to result in complete truncation of CCL16 to two products by MALDI-TOF MS (Fig. 5D).
FIGURE 5.
All MMPs evaluated precisely cleaved CCL16. Human CCL16-(1–97) was incubated with recombinant MMP-1, -2, -3, -7, -8, -9, -12, -13, and -14 for 16 h at 37 °C. A, cleavage assay products were visualized on silver-stained 15% Tris-Tricine gels. B, cleavage sites were assigned by MALDI-TOF MS by comparison of measured with predicted mass-to-charge ratios (m/z) with +1 charge ionization ([M + H]+). C, cleavage sites of each MMP are shown on the CCL16 sequence. D, mass spectra of cleavage assay products of CCL16 in the absence (left) or presence (right) of MMP-12 show that the predominant truncated forms CCL16-(8–77) and -(8–85) are both present in the form used for functional assays.
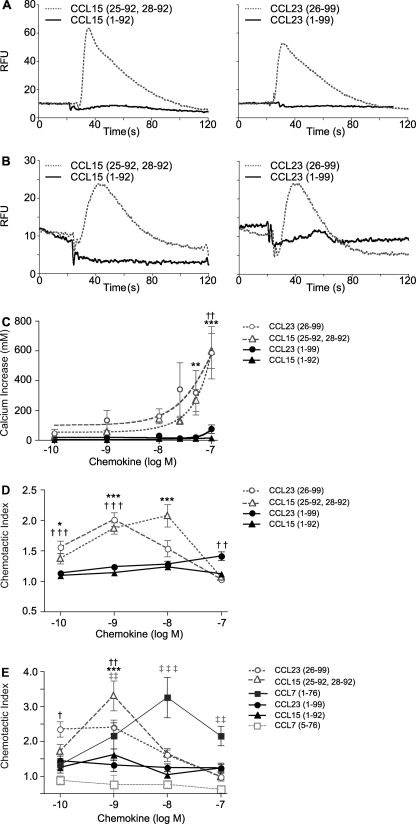
Calcium Mobilization in CCR1-transfected B300-19 and THP-1 Cells in Response to MMP-processed CCL15 and CCL23
MMP-cleaved chemokines induced a significant calcium flux after CCL15-(25–92, 28–92) and CCL23-(26–99) treatment of human CCR1-transfected B300-19 cells (Fig. 6A). With no activity observed by full-length CCL15 and CCL23, this shows that MMPs activate these two chemokines. To confirm this surprising result, similar experiments were performed using THP-1 cells (Fig. 6B); again, MMP-processed chemokines caused a concentration-dependent increase in THP-1 cell intracellular calcium mobilization, whereas a very minor response to CCL15-(1–92) and CCL23-(1–99) only occurred at the highest concentration evaluated (Fig. 6C). Notably, calcium mobilization did not occur after stimulation with the 19-mer peptide corresponding to CCL23-(5–23). Finally, MMP-processing of CCL16 did not alter calcium mobilization (not shown).
FIGURE 6.
MMP-mediated cleavages of CCL15 and CCL23 activates chemokine agonism. Calcium mobilization (A–C) and chemotactic potential (D–E) of CCL15 and CCL23 are enhanced by MMP processing. A and B, representative traces of calcium flux, shown as a burst in relative fluorescence units (RFU) after the addition of 10 nm of full-length or MMP-truncated chemokine in CCR1-transfected B300-19 (A) and THP-1 cells loaded with Fluo-4 calcium indicator reagent (B). C, shown is the calcium mobilization dose response of THP-1 cells wherein calcium concentrations were calculated from relative fluorescence based on calibration with ionomycin and MnCl2. Transwell migration of CCR1-transfected B300-19 for 3 h (D) or THP-1 cells for 90 min (E) toward full-length or truncated chemokine through a 5-μm pore-sized filter is shown. Migrated cells were quantified by CyQUANT assay and displayed as chemotactic index, defined as the ratio of cells migrating in response to stimulus compared with buffer control. Results shown are the mean ± S.E. of three or more biological replicates of experiments completed in quadruplicate. Statistical analysis was by two-way ANOVA with Bonferroni post-test; the levels of significance comparing full-length to MMP-cleaved counterpart are: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Chemotactic Response of CCR1 Transfectants and THP-1 Cells to MMP-processed CCL15 and CCL23
We mechanistically confirmed the calcium mobilization results by chemotaxis assays. Consistent with the Ca2+ flux results, both CCR-1-transfected B300-19 and THP-1 cells had a concentration-dependent response, with the characteristic hyperbolic curve to CCL15-(25–92, 28–92) and CCL23-(26–99) but notably with only a minimal response to intact CCL15 and CCL23 (Fig. 6, D and E). The response of THP-1 cells to CCL15-(25–92, 28–92) was highest at 1 nm and thus was considerably more potent than CCL7 (Fig. 6E). Indeed, equivalent chemotactic potential of cleaved CCL15 was only observed at a 10-fold higher concentration of CCL7, a prodigal monocyte chemoattractant. The 19-mer peptide corresponding to CCL23-(5–23) did not have chemotactic potential for THP-1 cells (supplemental Fig. S1). MMP-processing of CCL16 did not alter the Transwell migration compared with full-length CCL16 (not shown).
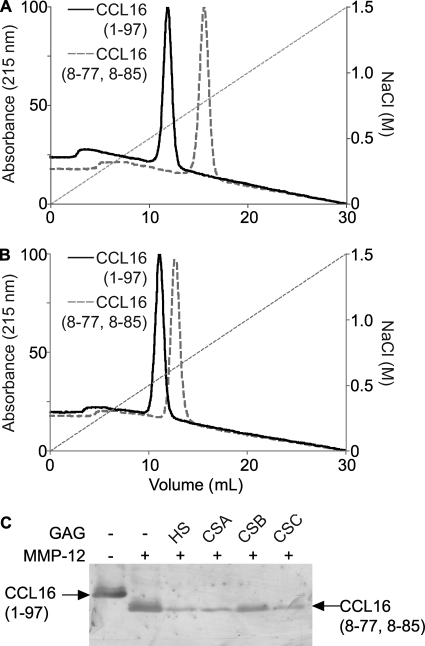
Heparin Binding Affinity of CCL16 Is Increased by MMP Processing
Given the role of chemokine carboxyl termini in GAG binding, we hypothesized that cleavage of the carboxyl terminus of CCL16-(1–97) would reduce GAG binding of the MMP-truncated chemokine compared with the full-length counterpart. The binding of MMP-12-processed chemokine CCL16-(8–77, 8–85) compared with intact CCL16-(1–97) was characterized using heparin-Sepharose chromatography (Fig. 7A). Surprisingly, the truncated CCL16 has greater affinity for heparin-Sepharose, reproducibly eluting at 0.71 m versus 0.52 m NaCl for the intact molecule. To control for charge effects, equivalent experiments were performed with a strong cation exchange Sepharose column (Fig. 7B). CCL16-(1–97) eluted at 0.51 m NaCl, whereas CCL16-(8–77, 8–85) reproducibly eluted at 0.56 m NaCl. The shift in elution on the strong cation exchange column was not as marked as on the heparin column, suggesting heparin specificity and that the interaction of specific amino acid residues or their spatial arrangement and not charge alone is responsible for the increased GAG binding by the chemokine. As previously observed for CXCL11 (8), soluble GAGs had no effect on the processing of CCL16 by MMP-12 (Fig. 7C), showing that cleavage occurs of both soluble and GAG-bound full-length chemokine without masking of the cleavage site.
FIGURE 7.
Heparin binding is enhanced upon CCL16 processing. Shown are elution profiles of CCL16-(1–97) and MMP-cleaved CCL16-(8–77, 8–85) from heparin-Sepharose column (A) or strong cation exchange column (B). The absorbance at 215 nm was measured during a 30-ml gradient that reached 1.5 m NaCl. C, CCL16-(1–97) was incubated with recombinant MMP-12 in the presence of heparan sulfate (HS), chondroitin sulfate A (CSA), CSB, or CSC in 5-fold excess (w/w) for 16 h at 37 °C. Cleavage assay products were visualized on silver-stained 15% Tris-Tricine gels.
DISCUSSION
We report the first instance of MMP-processing of CC chemokines that results in enhanced monocyte recruitment specifically by selective activation cleavages of CCL15, CCL16, and CCL23. Notably, these three chemokines are distinguished from other chemokines by the presence of uniquely long amino- or carboxyl-terminal sequences. Hence, these terminal peptides are responsible for chemokine latency, functioning as propeptides, and requiring their proteolytic removal for signaling that is 10-fold more potent than by CCL7. Together with previous findings for activation cleavages of CXC chemokines for neutrophil chemotaxis (16, 27, 28), it is now apparent that both macrophages and neutrophils have the potential to be recruited by MMP-activating cleavages of select chemokines in the right proteolytic milieu in vivo. Indeed, cleavage of CCL15 occurred ex vivo as determined by MALDI-TOF MS analysis of chemokine cleavage products in pooled synovial fluid from arthritis patients.
In addition to MMPs, serine proteases were also capable of cleaving CCL15 ex vivo as also found by Berahovich et al. (20). Notably, the macrophage-specific MMP-12 was kinetically the most efficient in CCL15 activation (Fig. 3), suggesting it potentially contributes to a feed-forward mechanism of monocyte recruitment. Overall, the two most efficient MMPs with the highest kinetic rates in vitro were leukocyte-specific MMP-12 and neutrophil-specific MMP-8. Finally, we also found that MMP-12 cleaved the classically regarded monocyte chemoattractants, namely CCL2, -7, -8, and -13, to generate products known to be CCR1, -2, and -3 receptor antagonists (12, 13). Together with the cleavage and inactivation by MMP-12 of all seven CXCL chemokines involved in neutrophil chemoattraction (21), this reveals an important role for the macrophage in dampening the cellular response in inflammation.
In showing that MMPs can activate select CCL chemokines, this reveals pleiotropic roles for MMPs in monocyte recruitment. Previous studies have shown the MMP-generated antagonist forms of CCL2, -7, -8, and -13 to be crucial in the resolution of inflammation by the active termination of monocyte recruitment (12, 13). Moreover, we recently found another crucial role for MMPs where genetic ablation of neutrophil MMP-8 delayed neutrophil apoptosis leading to massive and prolonged inflammatory infiltrates in tissue (51). This study reveals both the selectivity of MMP proteolytic roles and further complexity in the exquisite pleiotropic regulation of inflammation.
By the addition of protease inhibitors, we determined that both MMP and serine proteases generate stable CCL15 cleavage products in human arthritic synovial fluid. Neutrophil elastase cleaves CCL15 to a (22–92) product (20) that we also found, whereas cathepsin G processing results in products of (24–92, 27–93, and 29–92) (20, 41), rendering these neutrophil proteases as likely candidates responsible for the additional cleavages we observed. Interestingly, previous studies using PMN supernatant did not identify the CCL15-(25–92) product (20, 41) that was observed here. Although all MMPs are capable of processing CCL15 in vitro, the in vivo response may be specific to one or just a few MMPs due to other factors such as limited expression, interactors, or localization. Indeed, although several MMPs are capable of processing murine CXCL5/LIX in vitro, MMP-8 cleavage of the chemokine is critical in vivo and is not compensated for by elevated neutrophil MMP-9, which also cleaves at the same site as MMP-8 in the Mmp8−/− mouse (16).
These results suggest the utilization of two new mechanisms for monocyte recruitment: enhanced haptotactic gradient formation and the generation of potent receptor agonists with increased chemotactic potential that can now be formally tested in appropriate in vivo models. A distinctive feature of CCL16, CCL15, and CCL23 is proteins size; at 97, 92, and 99 amino acids in length, respectively, they are much longer than typical chemokines that consist of 65–80 amino acids. The carboxyl terminus of CCL16 extends 44 amino acids after the conserved fourth cysteine residue, whereas CCL15 and CCL23 have extended amino termini of 31 and 32 amino acids, respectively. Also, unlike all other chemokines evaluated, these three chemokines were cleaved by all MMPs tested (Table 1), suggesting that the extended termini, most likely showing structural disorder, increases their susceptibility to proteolysis and, hence, possibilities for proteolytic regulation. Indeed, truncated forms of both CCL15 and CCL23 were previously observed during recombinant chemokine protein expression in heterologous cells (35, 52, 53).
Select chemokines also have increased activity after dipeptidyl peptidase, cathepsin, urokinase-type plasminogen activator, and chymase proteolysis (20, 23, 32), and synthetic CCL15-(25–92) increases Ca2+ mobilization (53–55), yet ours is the first study to identify MMP-activation. This was shown for MMP-cleaved CCL15 by calcium mobilization in THP-1 cells (Fig. 6, B and C) through CCR1 activity (Fig. 6A). Similarly, CCL23-(26–99) enhances Ca2+ mobilization in both THP-1- and CCR1-transfected B300-19 cells (Fig. 6, A–C). This was reflected by an elevation in chemotactic response to MMP-processed CCL15 and CCL23 by CCR1-transfectant (Fig. 6D) and THP-1 (Fig. 6E) cells.
Previous studies of another subfamily of chemokines, the neutrophil chemoattractants CXCL5 and CXCL8, have shown that MMP processing of the amino termini results in products with 10-fold higher chemotactic potential than the full-length counterpart (16, 28, 56, 57). Although these neutrophil chemoattractants do not have an extended amino terminus like CCL15 and CCL23, the precise processing and maintenance of the critical ELR sequence enables CXCR2 activation. As for neutrophil recruitment where only two of the seven ELR+ CXCR2 binding human chemokines are activated by proteolysis, a similar limited proteolytic selectivity for just two CCL chemokines suggests the importance of proteolysis in the control of monocyte and macrophage recruitment under certain, but not all circumstances.
The cleavage patterns are complex and differ even between closely related chemokines in the same species and between species due to sequence diversity. Indeed, unlike human CCL7 (12, 13), murine CCL7 is not cleaved to an antagonist at position 4↓5 by MMPs (not shown). Because we have specifically studied human chemokine cleavage by human MMPs and there are no homologous murine chemokines for CCL15, CCL16, and CCL23, confirmatory studies in murine models of inflammation will be difficult to interpret. However, the presence of amino-terminal truncation forms of CCL15 and CCL23 in rheumatoid arthritis was recently shown through the use of antibodies (20); the carboxyl termini, but not amino termini, of truncated CCL15 and CCL23 were identified in synovial fluid. Although neither the exact truncation sites nor the proteases responsible could be identified, Berahovich et al. (20) showed that synovial fluid cleaved CCL15-(1–92) to a (25–92) product in vitro. We now show the broad ability of MMPs in cleaving CCL15 at 22VLN↓SFH27 by MMP-1, -2, -3, -7, -8, -9, -12, and -13 (Fig. 2). Notably, cleavage at this position by the small S1′ pocket MMPs 1 and 7 is unexpected due to the large serine residue at P1′, thereby indicating conformational plasticity in the protease active site upon substrate interaction. This was also indicated by the unusual cleavage of CCL11 at an expected very rigid and structurally complex region of the chemokine at 9C↓C10 by MMPs, which also has not been previously described.
The situation is different for the long carboxyl-terminal chemokine CCL16 where MMP processing neither altered the chemotactic response nor Ca2+ mobilization in THP-1 cells, as compared with full-length chemokine (not shown). This supports the findings of Howard et al. (58), who describe the chemotactic potential of CCL16 as minimal. Indeed, we found that a chemotactic response toward CCL16 was only observed at concentrations >10 nm, but this is obtainable after MMP cleavage leading to enhanced GAG binding in tissues.
Immobilization on the cell surface and presentation to circulating leukocytes through high affinity interactions with GAG side chains of proteoglycans are critical for the in vivo activity of many chemokines (2). By creating a high, localized concentration of chemokine, the haptotactic gradient provides the directionality for leukocytes as well as potentially placing the chemokine in an optimal orientation for chemokine-receptor interactions (59). Disruption of haptotactic gradients by proteolysis of the proteoglycan core protein decreases chemotaxis (60), and MMP-truncation of the Th1 cell chemokine CXCL11 reduces GAG binding efficiency (8). In contrast, we observed increased GAG binding upon MMP processing of CCL16 to CCL16-(8–77, 8–85). Removal of 12 and 20 carboxyl-terminal residues may expose or lead to rearrangement of previously buried residues to form stronger interactions with the heparin used in our analysis. Specifically, the residues 47KRNR50, which correlate with the signature GAG binding motif BBXB (where B represents a basic amino acid) that is found in other CC chemokines (61–63), may be exposed, resulting in increased GAG binding in vitro with a stronger haptotactic gradient formed in vivo to promote monocyte migration.
In addition to the identification of the truncated chemokine forms of CCL23 by MALDI-TOF MS, the cleaved amino termini CCL23-(1–25) and -(1–29) were identified (Fig. 4A). This suggests that the amino terminus is released as a complete segment and, therefore, may have functionality. The amino terminus of CCL15 between residues Asp4 to Leu23 is 89.5% homologous and 94.7% similar to CCL23 (supplemental Fig. S1), suggesting that functionality within this peptide may be shared between these two chemokines. The alternatively spliced chemokine CCL23β contains an insert of 18 amino acids between Asp24↓Phe25, termed SHAAGtide, which is chemotactic for both monocytes and neutrophils through FPRL1 binding (64). In contrast to SHAAGtide, CCL23-(5–23) did not chemoattract THP-1 cells or isolated peripheral blood neutrophils nor did it alter THP-1 cell chemotaxis to CCL7 (supplemental Fig. S1). Furthermore, in three different assays the peptide had no antibacterial properties against Escherichia coli or Staphylococcus aureus, although we were unable to confirm the antimicrobial activities of CCL15-(1–92) previously published (65) (not shown). Finally, CCL23-(5–23) did not activate THP-1 cells or neutrophils as assayed by reactive oxygen species production and degranulation, respectively (not shown). Like SHAAGtide, CCL23-(5–23) was proteolytically processed in vitro, with cleavage sites comparable with those observed for CCL23 (supplemental Fig. S1). The exception is MMP-14 cleavage of CCL23-(5–23) to -(11–23) and not the -(14–99) product observed in full-length chemokine, suggesting that additional sites within the full-length chemokine modify the MMP cleavage preference. This may be due in part to exosite interactions of the MMP and chemokine that sterically hinder access by MMP-14 to Phe10–Met11. Such unexpected negative substrate selectivity has also been found in the selectivity of MMPs in cleaving CCL2 and CCL7 (66).
In identifying the proteases responsible for CCL15 cleavage in synovial fluid, which could not be done before (20), our work adds to the importance of MMPs in regulating chemokine activity. The presence of four CCL15 truncation variants after processing by synovial fluid suggests that serine proteases and MMPs work in concert as part of the protease web (30). Furthermore, processing by one protease family is not dependent upon another given the presence of two products when one class of proteases was inhibited. In homeostasis or acute inflammation, protease activity is tightly regulated by latency as a zymogen, by localization, and by endogenous inhibitors, and so a single protease or class may be responsible for normal function. In synovial fluid used here, the proteolytic activity was significant, completely processing a 10-fold excess of chemokine compared with total protein in the fluid. This suggests that in chronic disease many regulation mechanisms are overcome and, thus, multiple proteases can contribute to disease progression through redundant processing of chemokines. Together with the absence of homologous chemokines in mouse, such redundancy renders specific in vivo validation difficult, and indeed our several attempts to do this in various systems did not provide conclusive in vivo confirmation.
In our previous work we proposed a model whereby macrophages produce MMP-12, which terminates neutrophil recruitment (21). We now build upon that model and suggest that MMPs, with an important but not exclusive contribution by macrophage MMP-12, act in a positive feed-forward mechanism to process CCL15, CCL16, and CCL23, which has potential to promote monocyte recruitment. Our study also suggests that the long amino- and carboxyl-terminal chemokines CCL16, CCL15, and CCL23 are expressed as prochemokines. Upon stimulation of an inflammatory response, the associated increased activity of MMPs together with serine proteases (20, 41) results in precise truncations of the full-length chemokines to promote leukocyte chemoattraction through improved gradient formation or receptor activation. Collectively then, MMPs have the potential to regulate all limbs of the recruitment and dissipation phases of neutrophils and macrophages in inflammation.
Supplementary Material
This work was supported by a Canadian Institutes of Health Research grant, an Infrastructure Grant from the Michael Smith Research Foundation (University of British Columbia Centre for Blood Research), and by the British Columbia Proteomics Network.

This article contains supplemental Fig. S1.
- GAG
- glycosaminoglycan
- MMP
- matrix metalloproteinase
- TIMP
- tissue inhibitor of metalloproteinase
- SF
- synovial fluid
- CCR
- CC receptor
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Laguri C., Arenzana-Seisdedos F., Lortat-Jacob H. (2008) Relationships between glycosaminoglycan and receptor binding sites in chemokines. The CXCL12 example. Carbohydr. Res. 343, 2018–2023 [DOI] [PubMed] [Google Scholar]
- 2. Proudfoot A. E., Handel T. M., Johnson Z., Lau E. K., LiWang P., Clark-Lewis I., Borlat F., Wells T. N., Kosco-Vilbois M. H. (2003) Glycosaminoglycan binding and oligomerization are essential for the in vivo activity of certain chemokines. Proc. Natl. Acad. Sci. U.S.A. 100, 1885–1890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Murphy P. M. (2002) International Union of Pharmacology. XXX. Update on chemokine receptor nomenclature. Pharmacol. Rev. 54, 227–229 [DOI] [PubMed] [Google Scholar]
- 4. Murphy P. M., Baggiolini M., Charo I. F., Hébert C. A., Horuk R., Matsushima K., Miller L. H., Oppenheim J. J., Power C. A. (2000) International union of pharmacology. XXII. Nomenclature for chemokine receptors. Pharmacol. Rev. 52, 145–176 [PubMed] [Google Scholar]
- 5. Baysal C., Atilgan A. R. (2001) Elucidating the structural mechanisms for biological activity of the chemokine family. Proteins 43, 150–160 [DOI] [PubMed] [Google Scholar]
- 6. Blanpain C., Doranz B. J., Bondue A., Govaerts C., De Leener A., Vassart G., Doms R. W., Proudfoot A., Parmentier M. (2003) The core domain of chemokines binds CCR5 extracellular domains. whereas their amino terminus interacts with the transmembrane helix bundle. J. Biol. Chem. 278, 5179–5187 [DOI] [PubMed] [Google Scholar]
- 7. Clark-Lewis I., Schumacher C., Baggiolini M., Moser B. (1991) Structure-activity relationships of interleukin-8 determined using chemically synthesized analogs. Critical role of NH2-terminal residues and evidence for uncoupling of neutrophil chemotaxis, exocytosis, and receptor binding activities. J. Biol. Chem. 266, 23128–23134 [PubMed] [Google Scholar]
- 8. Cox J. H., Dean R. A., Roberts C. R., Overall C. M. (2008) Matrix metalloproteinase processing of CXCL11/I-TAC results in loss of chemoattractant activity and altered glycosaminoglycan binding. J. Biol. Chem. 283, 19389–19399 [DOI] [PubMed] [Google Scholar]
- 9. Dean R. A., Overall C. M. (2007) Proteomics discovery of metalloproteinase substrates in the cellular context by iTRAQ labeling reveals a diverse MMP-2 substrate degradome. Mol. Cell. Proteomics 6, 611–623 [DOI] [PubMed] [Google Scholar]
- 10. Gong J. H., Clark-Lewis I. (1995) Antagonists of monocyte chemoattractant protein 1 identified by modification of functionally critical NH2-terminal residues. J. Exp. Med. 181, 631–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McQuibban G. A., Butler G. S., Gong J. H., Bendall L., Power C., Clark-Lewis I., Overall C. M. (2001) Matrix metalloproteinase activity inactivates the CXC chemokine stromal cell-derived factor-1. J. Biol. Chem. 276, 43503–43508 [DOI] [PubMed] [Google Scholar]
- 12. McQuibban G. A., Gong J. H., Tam E. M., McCulloch C. A., Clark-Lewis I., Overall C. M. (2000) Inflammation dampened by gelatinase A cleavage of monocyte chemoattractant protein-3. Science 289, 1202–1206 [DOI] [PubMed] [Google Scholar]
- 13. McQuibban G. A., Gong J. H., Wong J. P., Wallace J. L., Clark-Lewis I., Overall C. M. (2002) Matrix metalloproteinase processing of monocyte chemoattractant proteins generates CC chemokine receptor antagonists with anti-inflammatory properties in vivo. Blood 100, 1160–1167 [PubMed] [Google Scholar]
- 14. Mortier A., Van Damme J., Proost P. (2008) Regulation of chemokine activity by posttranslational modification. Pharmacol. Ther. 120, 197–217 [DOI] [PubMed] [Google Scholar]
- 15. Struyf S., De Meester I., Scharpé S., Lenaerts J. P., Menten P., Wang J. M., Proost P., Van Damme J. (1998) Natural truncation of RANTES abolishes signaling through the CC chemokine receptors CCR1 and CCR3, impairs its chemotactic potency, and generates a CC chemokine inhibitor. Eur. J. Immunol. 28, 1262–1271 [DOI] [PubMed] [Google Scholar]
- 16. Tester A. M., Cox J. H., Connor A. R., Starr A. E., Dean R. A., Puente X. S., López-Otín C., Overall C. M. (2007) LPS responsiveness and neutrophil chemotaxis in vivo require PMN MMP-8 activity. PLoS One 2, e312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vergote D., Butler G. S., Ooms M., Cox J. H., Silva C., Hollenberg M. D., Jhamandas J. H., Overall C. M., Power C. (2006) Proteolytic processing of SDF-1α reveals a change in receptor specificity mediating HIV-associated neurodegeneration. Proc. Natl. Acad. Sci. U.S.A. 103, 19182–19187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Weber M., Uguccioni M., Baggiolini M., Clark-Lewis I., Dahinden C. A. (1996) Deletion of the NH2-terminal residue converts monocyte chemotactic protein 1 from an activator of basophil mediator release to an eosinophil chemoattractant. J. Exp. Med. 183, 681–685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wolf M., Albrecht S., Märki C. (2008) Proteolytic processing of chemokines. Implications in physiological and pathological conditions. Int. J. Biochem. Cell Biol. 40, 1185–1198 [DOI] [PubMed] [Google Scholar]
- 20. Berahovich R. D., Miao Z., Wang Y., Premack B., Howard M. C., Schall T. J. (2005) Proteolytic activation of alternative CCR1 ligands in inflammation. J. Immunol. 174, 7341–7351 [DOI] [PubMed] [Google Scholar]
- 21. Dean R. A., Cox J. H., Bellac C. L., Doucet A., Starr A. E., Overall C. M. (2008) Macrophage-specific metalloelastase (MMP-12) truncates and inactivates ELR+ CXC chemokines and generates CCL2, -7, -8, and -13 antagonists. Potential role of the macrophage in terminating polymorphonuclear leukocyte influx. Blood 112, 3455–3464 [DOI] [PubMed] [Google Scholar]
- 22. Delgado M. B., Clark-Lewis I., Loetscher P., Langen H., Thelen M., Baggiolini M., Wolf M. (2001) Rapid inactivation of stromal cell-derived factor-1 by cathepsin G associated with lymphocytes. Eur. J. Immunol. 31, 699–707 [DOI] [PubMed] [Google Scholar]
- 23. Proost P., Menten P., Struyf S., Schutyser E., De Meester I., Van Damme J. (2000) Cleavage by CD26/dipeptidyl peptidase IV converts the chemokine LD78β into a most efficient monocyte attractant and CCR1 agonist. Blood 96, 1674–1680 [PubMed] [Google Scholar]
- 24. Proost P., Struyf S., Schols D., Durinx C., Wuyts A., Lenaerts J. P., De Clercq E., De Meester I., Van Damme J. (1998) Processing by CD26/dipeptidyl-peptidase IV reduces the chemotactic and anti-HIV-1 activity of stromal-cell-derived factor-1α. FEBS Lett. 432, 73–76 [DOI] [PubMed] [Google Scholar]
- 25. Proost P., Struyf S., Schols D., Opdenakker G., Sozzani S., Allavena P., Mantovani A., Augustyns K., Bal G., Haemers A., Lambeir A. M., Scharpé S., Van Damme J., De Meester I. (1999) Truncation of macrophage-derived chemokine by CD26/dipeptidyl-peptidase IV beyond its predicted cleavage site affects chemotactic activity and CC chemokine receptor 4 interaction. J. Biol. Chem. 274, 3988–3993 [DOI] [PubMed] [Google Scholar]
- 26. Van den Steen P. E., Husson S. J., Proost P., Van Damme J., Opdenakker G. (2003) Carboxyl-terminal cleavage of the chemokines MIG and IP-10 by gelatinase B and neutrophil collagenase. Biochem. Biophys. Res. Commun. 310, 889–896 [DOI] [PubMed] [Google Scholar]
- 27. Van den Steen P. E., Proost P., Wuyts A., Van Damme J., Opdenakker G. (2000) Neutrophil gelatinase B potentiates interleukin-8 10-fold by amino-terminal processing, whereas it degrades CTAP-III, PF-4, and GRO-α and leaves RANTES and MCP-2 intact. Blood 96, 2673–2681 [PubMed] [Google Scholar]
- 28. Van Den Steen P. E., Wuyts A., Husson S. J., Proost P., Van Damme J., Opdenakker G. (2003) Gelatinase B/MMP-9 and neutrophil collagenase/MMP-8 process the chemokines human GCP-2/CXCL6, ENA-78/CXCL5, and mouse GCP-2/LIX and modulate their physiological activities. Eur. J. Biochem. 270, 3739–3749 [DOI] [PubMed] [Google Scholar]
- 29. Wolf M., Clark-Lewis I., Buri C., Langen H., Lis M., Mazzucchelli L. (2003) Cathepsin D specifically cleaves the chemokines macrophage inflammatory protein-1 α, macrophage inflammatory protein-1β, and SLC that are expressed in human breast cancer. Am. J. Pathol. 162, 1183–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Overall C. M., Kleifeld O. (2006) Tumor microenvironment-opinion. Validating matrix metalloproteinases as drug targets and anti-targets for cancer therapy. Nat. Rev. Cancer 6, 227–239 [DOI] [PubMed] [Google Scholar]
- 31. Lambeir A. M., Proost P., Durinx C., Bal G., Senten K., Augustyns K., Scharpé S., Van Damme J., De Meester I. (2001) Kinetic investigation of chemokine truncation by CD26/dipeptidyl peptidase IV reveals a striking selectivity within the chemokine family. J. Biol. Chem. 276, 29839–29845 [DOI] [PubMed] [Google Scholar]
- 32. Detheux M., Ständker L., Vakili J., Münch J., Forssmann U., Adermann K., Pöhlmann S., Vassart G., Kirchhoff F., Parmentier M., Forssmann W. G. (2000) Natural proteolytic processing of hemofiltrate CC chemokine 1 generates a potent CC chemokine receptor (CCR)1 and CCR5 agonist with anti-HIV properties. J. Exp. Med. 192, 1501–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Stropes M. P., Schneider O. D., Zagorski W. A., Miller J. L., Miller W. E. (2009) The carboxyl-terminal tail of human cytomegalovirus (HCMV) US28 regulates both chemokine-independent and chemokine-dependent signaling in HCMV-infected cells. J. Virol. 83, 10016–10027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vakili J., Ständker L., Detheux M., Vassart G., Forssmann W. G., Parmentier M. (2001) Urokinase plasminogen activator and plasmin efficiently convert hemofiltrate CC chemokine 1 into its active. J. Immunol. 167, 3406–3413 [DOI] [PubMed] [Google Scholar]
- 35. Macphee C. H., Appelbaum E. R., Johanson K., Moores K. E., Imburgia C. S., Fornwald J., Berkhout T., Brawner M., Groot P. H., O'Donnell K., O'Shannessy D., Scott G., White J. R. (1998) Identification of a truncated form of the CC chemokine CK β8 demonstrating greatly enhanced biological activity. J. Immunol. 161, 6273–6279 [PubMed] [Google Scholar]
- 36. Youn B. S., Zhang S. M., Lee E. K., Park D. H., Broxmeyer H. E., Murphy P. M., Locati M., Pease J. E., Kim K. K., Antol K., Kwon B. S. (1997) Molecular cloning of leukotactin-1. A novel human β-chemokine, a chemoattractant for neutrophils, monocytes, and lymphocytes, and a potent agonist at CC chemokine receptors 1 and 3. J. Immunol. 159, 5201–5205 [PubMed] [Google Scholar]
- 37. Butler G. S., Tam E. M., Overall C. M. (2004) The canonical methionine 392 of matrix metalloproteinase 2 (gelatinase A) is not required for catalytic efficiency or structural integrity. Probing the role of the methionine-turn in the metzincin metalloprotease superfamily. J. Biol. Chem. 279, 15615–15620 [DOI] [PubMed] [Google Scholar]
- 38. Clark-Lewis I., Vo L., Owen P., Anderson J. (1997) Chemical synthesis, purification, and folding of C-X-C and C-C chemokines. Methods Enzymol. 287, 233–250 [DOI] [PubMed] [Google Scholar]
- 39. auf dem Keller U., Bellac C. L., Li Y., Lou Y., Lange P. F., Ting R., Harwig C., Kappelhoff R., Dedhar S., Adam M. J., Ruth T. J., Bénard F., Perrin D. M., Overall C. M. (2010) Novel matrix metalloproteinase inhibitor [18F]marimastat-aryltrifluoroborate as a probe for in vivo positron emission tomography imaging in cancer. Cancer Res. 70, 7562–7569 [DOI] [PubMed] [Google Scholar]
- 40. Starr A. E., Overall C. M. (2009) Chapter 13. Characterizing proteolytic processing of chemokines by mass spectrometry, biochemistry, neo-epitope antibodies, and functional assays. Methods Enzymol. 461, 281–307 [DOI] [PubMed] [Google Scholar]
- 41. Richter R., Bistrian R., Escher S., Forssmann W. G., Vakili J., Henschler R., Spodsberg N., Frimpong-Boateng A., Forssmann U. (2005) Quantum proteolytic activation of chemokine CCL15 by neutrophil granulocytes modulates mononuclear cell adhesiveness. J. Immunol. 175, 1599–1608 [DOI] [PubMed] [Google Scholar]
- 42. Forssmann U., Mägert H. J., Adermann K., Escher S. E., Forssmann W. G. (2001) Hemofiltrate CC chemokines with unique biochemical properties. HCC-1/CCL14a and HCC-2/CCL15. J. Leukoc. Biol. 70, 357–366 [PubMed] [Google Scholar]
- 43. Jarnagin K., Grunberger D., Mulkins M., Wong B., Hemmerich S., Paavola C., Bloom A., Bhakta S., Diehl F., Freedman R., McCarley D., Polsky I., Ping-Tsou A., Kosaka A., Handel T. M. (1999) Identification of surface residues of the monocyte chemotactic protein 1 that affect signaling through the receptor CCR2. Biochemistry 38, 16167–16177 [DOI] [PubMed] [Google Scholar]
- 44. Oravecz T., Pall M., Roderiquez G., Gorrell M. D., Ditto M., Nguyen N. Y., Boykins R., Unsworth E., Norcross M. A. (1997) Regulation of the receptor specificity and function of the chemokine RANTES (regulated on activation, normal T cell expressed and secreted) by dipeptidyl peptidase IV (CD26)-mediated cleavage. J. Exp. Med. 186, 1865–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Lim J. K., Burns J. M., Lu W., DeVico A. L. (2005) Multiple pathways of amino-terminal processing produce two truncated variants of RANTES/CCL5. J. Leukoc. Biol. 78, 442–452 [DOI] [PubMed] [Google Scholar]
- 46. Noso N., Bartels J., Mallet A. I., Mochizuki M., Christophers E., Schröder J. M. (1998) Delayed production of biologically active O-glycosylated forms of human eotaxin by tumor necrosis factor-α-stimulated dermal fibroblasts. Eur J Biochem. 253, 114–122 [DOI] [PubMed] [Google Scholar]
- 47. Quintero P. A., Knolle M. D., Cala L. F., Zhuang Y., Owen C. A. (2010) Matrix metalloproteinase-8 inactivates macrophage inflammatory protein-1α to reduce acute lung inflammation and injury in mice. J. Immunol. 184, 1575–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Laurence J. S., Blanpain C., Burgner J. W., Parmentier M., LiWang P. J. (2000) CC chemokine MIP-1 β can function as a monomer and depends on Phe-13 for receptor binding. Biochemistry 39, 3401–3409 [DOI] [PubMed] [Google Scholar]
- 49. Laurence J. S., LiWang A. C., LiWang P. J. (1998) Effect of N-terminal truncation and solution conditions on chemokine dimer stability. Nuclear magnetic resonance structural analysis of macrophage inflammatory protein 1 β mutants. Biochemistry 37, 9346–9354 [DOI] [PubMed] [Google Scholar]
- 50. Richter R., Casarosa P., Ständker L., Münch J., Springael J. Y., Nijmeijer S., Forssmann W. G., Vischer H. F., Vakili J., Detheux M., Parmentier M., Leurs R., Smit M. J. (2009) Significance of N-terminal proteolysis of CCL14a to activity on the chemokine receptors CCR1 and CCR5 and the human cytomegalovirus-encoded chemokine receptor US28. J. Immunol. 183, 1229–1237 [DOI] [PubMed] [Google Scholar]
- 51. Cox J. H., Starr A. E., Kappelhoff R., Yan R., Roberts C. R., Overall C. M. (2010) Matrix metalloproteinase 8 deficiency in mice exacerbates inflammatory arthritis through delayed neutrophil apoptosis and reduced caspase 11 expression. Arthritis Rheum. 62, 3645–3655 [DOI] [PubMed] [Google Scholar]
- 52. Forssmann U., Delgado M. B., Uguccioni M., Loetscher P., Garotta G., Baggiolini M. (1997) CKβ8, a novel CC chemokine that predominantly acts on monocytes. FEBS Lett. 408, 211–216 [DOI] [PubMed] [Google Scholar]
- 53. Pardigol A., Forssmann U., Zucht H. D., Loetscher P., Schulz-Knappe P., Baggiolini M., Forssmann W. G., Mägert H. J. (1998) HCC-2, a human chemokine. Gene structure, expression pattern, and biological activity. Proc. Natl. Acad. Sci. U.S.A. 95, 6308–6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Escher S. E., Forssmann U., Frimpong-Boateng A., Adermann K., Vakili J., Sticht H., Detheux M. (2004) Functional analysis of chemically synthesized derivatives of the human CC chemokine CCL15/HCC-2, a high affinity CCR1 ligand. J. Pept. Res. 63, 36–47 [DOI] [PubMed] [Google Scholar]
- 55. Escher S. E., Sticht H., Forssmann W. G., Rösch P., Adermann K. (1999) Synthesis and characterization of the human CC chemokine HCC-2. J. Pept. Res. 54, 505–513 [DOI] [PubMed] [Google Scholar]
- 56. Nufer O., Corbett M., Walz A. (1999) Amino-terminal processing of chemokine ENA-78 regulates biological activity. Biochemistry 38, 636–642 [DOI] [PubMed] [Google Scholar]
- 57. Wuyts A., D'Haese A., Cremers V., Menten P., Lenaerts J. P., De Loof A., Heremans H., Proost P., Van Damme J. (1999) NH2- and COOH-terminal truncations of murine granulocyte chemotactic protein-2 augment the in vitro and in vivo neutrophil chemotactic potency. J. Immunol. 163, 6155–6163 [PubMed] [Google Scholar]
- 58. Howard O. M., Dong H. F., Shirakawa A. K., Oppenheim J. J. (2000) LEC induces chemotaxis and adhesion by interacting with CCR1 and CCR8. Blood 96, 840–845 [PubMed] [Google Scholar]
- 59. Kuschert G. S., Coulin F., Power C. A., Proudfoot A. E., Hubbard R. E., Hoogewerf A. J., Wells T. N. (1999) Glycosaminoglycans interact selectively with chemokines and modulate receptor binding and cellular responses. Biochemistry 38, 12959–12968 [DOI] [PubMed] [Google Scholar]
- 60. Li Q., Park P. W., Wilson C. L., Parks W. C. (2002) Matrilysin shedding of syndecan-1 regulates chemokine mobilization and transepithelial efflux of neutrophils in acute lung injury. Cell 111, 635–646 [DOI] [PubMed] [Google Scholar]
- 61. Graham G. J., Wilkinson P. C., Nibbs R. J., Lowe S., Kolset S. O., Parker A., Freshney M. G., Tsang M. L., Pragnell I. B. (1996) Uncoupling of stem cell inhibition from monocyte chemoattraction in MIP-1α by mutagenesis of the proteoglycan binding site. EMBO J. 15, 6506–6515 [PMC free article] [PubMed] [Google Scholar]
- 62. Laurence J. S., Blanpain C., De Leener A., Parmentier M., LiWang P. J. (2001) Importance of basic residues and quaternary structure in the function of MIP-1 β. CCR5 binding and cell surface sugar interactions. Biochemistry 40, 4990–4999 [DOI] [PubMed] [Google Scholar]
- 63. Proudfoot A. E., Fritchley S., Borlat F., Shaw J. P., Vilbois F., Zwahlen C., Trkola A., Marchant D., Clapham P. R., Wells T. N. (2001) The BBXB motif of RANTES is the principal site for heparin binding and controls receptor selectivity. J. Biol. Chem. 276, 10620–10626 [DOI] [PubMed] [Google Scholar]
- 64. Miao Z., Premack B. A., Wei Z., Wang Y., Gerard C., Showell H., Howard M., Schall T. J., Berahovich R. (2007) Proinflammatory proteases liberate a discrete high affinity functional FPRL1 (CCR12) ligand from CCL23. J. Immunol. 178, 7395–7404 [DOI] [PubMed] [Google Scholar]
- 65. Kotarsky K., Sitnik K. M., Stenstad H., Kotarsky H., Schmidtchen A., Koslowski M., Wehkamp J., Agace W. W. (2010) A novel role for constitutively expressed epithelial-derived chemokines as antibacterial peptides in the intestinal mucosa. Mucosal Immunol. 3, 40–48 [DOI] [PubMed] [Google Scholar]
- 66. Overall C. M., Tam E. M., Kappelhoff R., Connor A., Ewart T., Morrison C. J., Puente X., López-Otín C., Seth A. (2004) Protease degradomics. Mass spectrometry discovery of protease substrates and the CLIP-CHIP, a dedicated DNA microarray of all human proteases and inhibitors. Biol. Chem. 385, 493–504 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.