Background: The N terminus is required for localization and functions of Mps1, Bub1, and BubR1 kinases.
Results: A novel Bub1/BubR1-related TPR motif is identified in Mps1 and is required for kinase activity.
Conclusion: TPR domain of Mps1 regulates kinase activity, Mps1 chromosome alignment, and checkpoint functions.
Significance: Identification of a novel domain in Mps1 enhances our understanding of its contribution to maintaining genome integrity.
Keywords: Cell Cycle, Checkpoint Control, Kinetochore, Mitosis, Mitotic Spindle, Chromosome Congression, Mps1, Spindle Checkpoint, TPR Domain
Abstract
Kinetochore targeting of the mitotic kinases Bub1, BubR1, and Mps1 has been implicated in efficient execution of their functions in the spindle checkpoint, the self-monitoring system of the eukaryotic cell cycle that ensures chromosome segregation occurs with high fidelity. In all three kinases, kinetochore docking is mediated by the N-terminal region of the protein. Deletions within this region result in checkpoint failure and chromosome segregation defects. Here, we use an interdisciplinary approach that includes biophysical, biochemical, cell biological, and bioinformatics methods to study the N-terminal region of human Mps1. We report the identification of a tandem repeat of the tetratricopeptide repeat (TPR) motif in the N-terminal kinetochore binding region of Mps1, with close homology to the tandem TPR motif of Bub1 and BubR1. Phylogenetic analysis indicates that TPR Mps1 was acquired after the split between deutorostomes and protostomes, as it is distinguishable in chordates and echinoderms. Overexpression of TPR Mps1 resulted in decreased efficiency of both chromosome alignment and mitotic arrest, likely through displacement of endogenous Mps1 from the kinetochore and decreased Mps1 catalytic activity. Taken together, our multidisciplinary strategy provides new insights into the evolution, structural organization, and function of Mps1 N-terminal region.
Introduction
Mitosis equally distributes the duplicated genome to each of the nascent daughter cells. Defects in chromosome segregation can lead to aneuploidy, which in turn is implicated in tumorigenesis (1, 2). Attachment of mitotic chromosomes to spindle microtubules is mediated by the kinetochore (KT),5 a protein-rich framework that assembles onto the centromeric region of DNA molecules (3, 4). Notably, the KT functions not only as a structural platform but also as a signaling hub to coordinate chromosome attachment, spindle assembly checkpoint (SAC) activity, and the metaphase to anaphase transition (5). The SAC is a signaling cascade that prolongs mitosis until all chromosomes form stable attachments. The target of the checkpoint is Cdc20, a substrate-specific subunit of the anaphase-promoting complex/cyclosome that catalyzes the polyubiquitination of the key mitotic proteins cyclin B and Securin, targeting them for eventual degradation. The delay imposed on mitotic exit is not permanent, and cells that cannot satisfy the checkpoint ultimately die or exit mitosis as a result of cyclin B degradation or inactivation and enter the next G1 as single tetraploids (6).
The core components of the spindle checkpoint are highly conserved and include a number of serine/threonine kinases such as Bub1, BubR1, and the dual specificity kinase Mps1. Mps1 was originally discovered in a yeast genetic screen for mutants producing monopolar spindles (7). Its role in SAC signaling was subsequently identified in yeasts and confirmed in higher eukaryotes (7–10). During mitosis, Mps1 kinase activity is crucial for Mad1 and Mad2 kinetochore recruitment (10–14). Recent inhibitor and chemical genetics studies confirmed these observations and indicated that Mps1 activity facilitates the conformational activation of Mad2 to a form capable of Cdc20 binding and inhibition (14, 15).
In addition to the Mps1 C-terminal kinase domain, the N-terminal region is critical for KT localization of Mps1 and the recruitment of core checkpoint components to unattached kinetochores in mammalian cells (9, 11, 12, 15–17). In budding yeast, distinct regions of N-terminal Mps1 are required for spindle pole body duplication and biorientation (18). In mammalian mitoses, deletion of the N-terminal 100 amino acids prevented kinetochore recruitment and resulted in chromosome alignment defects as well as inefficient mitotic arrest in response to microtubule poisons (15). More recently, studies of Mps1 in meiosis I of mammalian oocytes demonstrated that N-terminal Mps1 is required for spindle checkpoint control, for correct timing of prometaphase I, and for chromosome alignment (19). Indeed, mice expressing an Mps1 fragment lacking residues 47–154 exhibited severely reduced fertility confirming the essential role of Mps1 for embryonic development (19).
The TPR is a degenerate tandem repeat of 34 amino acid residues encoding an α-helix-turn-α-helix motif that is present in proteins of diverse biological functions in diverse organisms, ranging from bacteria to humans (20). In Bub1 and BubR1, the TPR adopts an overall fold that closely resembles that of many other triple TPR folds despite the high amino acid sequence divergence (21). The uniform arrangement of neighboring α-helices gives rise to the formation of a right-handed superhelical structure that creates a regular, elongated amphipathic (i.e. one side hydrophobic and the other side hydrophilic) groove. This topology also creates a continuous concave surface on one side with a contrasting convex surface on the other side. At the same time, TPR Bub1 and TPR BubR1 exhibit unique features, including a shallow groove in the first TPR unit, the insertion of a 310-helix between the second and third TPR tandem repeats, and the noncanonical packing interactions established between the α-helices of the second TPR unit (21).
Because the functions thus far attributed to the KT localization domain of Mps1 are reminiscent of the role of the N-terminal regions of Bub1 and BubR1, we set out to investigate the biophysical, functional, and evolutionary characteristics of the N-terminal region of Mps1. Using a interdisciplinary strategy, we demonstrate that the N-terminal domain of Mps1 is globular, predominantly α-helical, stable in a wide range of pH, and likely to be organized as a triple tandem repeat of the TPR motif. We also show that overexpression of the TPR-containing fragment of Mps1 in cells results in mislocalization of endogenous Mps1, chromosome congression defects, and a weakened spindle checkpoint response. Evolutionary analysis of the putative Mps1 TPR regions reveals its presence in chordates and echinoderms and indicates that it likely evolved from the TPR domain of Bub1 or BubR1 at or after the emergence of the deuterostomes.
EXPERIMENTAL PROCEDURES
Structural Bioinformatics Analyses
The sequence of human N-terminal Mps1 was compared with all the protein sequences deposited in Swiss-Prot by using BLAST. A PSI-BLAST search produced an alignment between N-terminal Mps1 and close homologues and highlighted conserved residues in N-terminal Mps1 family. Homologous proteins with known structure were identified by using the sequence-structure homology (fold)-recognition program FUGUE (22), which initially searches for homologues in the structural profile library derived from structure-based alignments in the HOMSTRAD (23) database. The alignment produced by FUGUE for the highest scoring hit (TPR BubR1) was formatted with JOY (24) and analyzed visually to highlight the conservation of structurally and functionally important residues. The model of N-terminal Mps1 was constructed with MODELLER (25) and validated with PROCHECK (26), VERIFY3D (27), JOY, and visual inspection by using three-dimensional graphics software. All of these programs revealed that the model needed no further modifications.
Protein Expression and Purification
N-terminal Mps1 fragments (1–239, 55–239, 58–210, 51–210, 58–175, 51–175, and 55–210; numbering according to human Mps1) were amplified and cloned into pGEX-6P3 and expressed in Escherichia coli BL21(DE3) at 20 °C, 250 rpm in 2× YT broth. Expression was induced with isopropyl 1-thio-β-d-galactopyranoside for 3 h. Cell lysis was performed using BugBuster (Merck) in TBS buffer (40 mm Tris, 200 mm NaCl, 1 mm DTT, pH 8.0). The lysate was cleared by centrifugation, and the soluble fraction was loaded onto a chromatographic column packed with TBS-equilibrated GST-Sepharose. After washing, recombinant Mps1 fragments were eluted in TBS buffer containing 20 mm reduced glutathione and concentrated. The cleaved GST tag was removed by passing the digested sample through a GST-Sepharose column. As the final purification step, Mps1 fragments were loaded onto a gel filtration column (Superdex 75, HR 26/60) and eluted in 10 mm Tris buffer, 200 mm NaCl, pH 8.0, at 1 ml/min. After analysis by SDS-PAGE and measurement of UV absorption spectra (200–300 nm), fractions containing pure Mps1 were collected, concentrated, and stored at −20 °C. N-terminal sequencing and mass spectrometry were carried out to confirm protein identity and purity.
Monolayers, Null Ellipsometry, and Surface Pressure Measurements
Protein monolayers were prepared on a circular trough (surface = 20 cm2), and the surface pressure was measured with a sensor (Nima Technology Ltd., Coventry, UK) using a Wilhelmy plate with a precision of ± 0.5 mN/m. All measurements were performed in 20 mm phosphate buffer, pH 7 (0.01 mm Na2HPO4, 0.01 mm NaH2PO4, ultrapure water), and a temperature of 20 ± 2 °C.
Ellipsometry measurements were carried out with an in-house automated ellipsometer (28) in a “null ellipsometer” configuration (29). He-Ne laser beam (λ = 632.8 nm, Melles Griot) is polarized with a Glan-Thompson polarizer and reflected on the surface of the trough (incidence angle of 52.12°). After reflection on the water surface, the laser light passed through a λ/4 retardation plate, a Glan-Thompson analyzer, and a photomultiplier. The analyzer angle, multiplied by two, yielded the value of the ellipsometric angle (Δ), i.e. the phase difference between parallel and perpendicular polarization of the reflected light. The ellipsometric angle, Δ0, is proportional to the quantity of protein adsorbed at the interface in case of a monolayer. Hence, the variation of the ellipsometric angle is a relevant probe for changes occurring at the interface. Using the measured ellipsometric angle, Δ0, and estimating the refractive index increment of the protein to 0.2 ml/g, the surface concentration, Γ0 of adsorbed protein was calculated using the relationship between Δ and Γ reported by De Feijter et al. (30), Γ (mg/m2) = (0.2) Δ0. The laser beam probed a surface of 1 mm2 and a depth on the order of 1 μm. Initial values of the ellipsometric angle (Δ0) and surface tension of pure buffer solutions were recorded on the subphase for at least half an hour. These values have been subtracted from all data presented below. Values of Δ were stable and recorded every 4 s with a precision of ±0.5°.
Determination of the Shear Elastic Constant
The principles and implementation of our experimental setup for the measurement of the lateral rigidity of monolayers and the procedure for data analysis have been extensively described before (31). Briefly, at the center of a 48-mm diameter Teflon trough, a 10-mm diameter paraffin-coated aluminum disc floats at the air/water interface, in contact with the monolayer, whose rigidity is measured. The subphase is 5 mm deep. The float carries a small magnet and is kept centered by a permanent magnetic field, B0 = 6 × 10−5 tesla, parallel to the earth's field and created by a small solenoid located just above the float. Sensitive angular detection of the float rotation is achieved by using a mirror fixed on the magnet to reflect a laser beam onto a differential photodiode. A sinusoidal torque excitation is applied to the float in the 0.01–100 Hz frequency range by an oscillating field perpendicular to the permanent solenoid field. The latter field acts as a restoring torque equivalent to a monolayer with a rigidity of 0.16 mN/m. This number set the sensitivity limit of the rheometer. The device behaves like a simple harmonic oscillator. The resistance that the monolayer opposes to the rotation of the float is directly measured. An important advantage of this setup is the absence of a physical link between the outside and the float torsion (i.e. no torsion wire). This allows high sensitivities such that the applied deformation is very small, below uxy ∼10−7, where uxy is the horizontal component of the deformation tensor. This device introduces very small excitation strains (from 10−3 down to 10−6) to the system. Because in previous experiments we have shown that pure shear elastic response spectra exhibit a linear stress-strain relationship over this range (31), we concluded that the rotation coupling between the float and the contacting monolayer is satisfactory. Moreover, we have showed that such small strains do not create plastic deformations on fragile surface objects (32). For the experimental procedure, the amplitude and phase of the mechanical response of the pure subphase was first analyzed in the frequency range 0.01–100 Hz to assess that no rigidity was detected. This measurement takes approximately 1 h. Then the protein solution was directly poured into the trough, and the mechanical response of the layer formed at the interface was recorded at the fixed frequency of 5 Hz. At the end of the kinetics, when the shear elastic constant, μ (expressed in mN/m), reached a constant value, a new measurement between 0.01 and 100 Hz was recorded to determine whether the system behaves as an elastic layer. Rigidity measurements were performed at 20 °C in parallel to ellipsometry.
Far-UV Circular Dichroism
Spectra were recorded on an AVIV 62-S spectropolarimeter (AVIV) previously calibrated with camphorsulfonic acid and equipped with a temperature control unit. In all experiments, spectra were recorded at 25 °C in a 0.1-cm quartz cell using an average time of 1.0 s, a step size of 0.5 nm, 1-nm bandwidth, and averaged over 20 scans. After subtraction of the buffer base line, the CD data were normalized to calculate the mean molar ellipticity. For studies on protein stability as a function of temperature, five unfolding curves were recorded upon heating from 25 to 95 °C at a rate of 1 °C/min and 50 s accumulation time. The apparent melting temperature, Tm, was determined from differential melting curves of the function d[θ222](T)/dt. The concentration of protein solutions was determined from amino acid composition analysis at the PNAC facility (Department of Biochemistry, University of Cambridge).
Analytical Gel Permeation Chromatography and Protein Analysis
Analytical and preparative size-exclusion chromatography was performed in a HiLoad 26/60 Superdex 75 preparative grade column previously equilibrated in TBS buffer. For determination of the protein oligomerization state, the following proteins standards were used: BSA (67 kDa), ovalbumin (43 kDa), chymotrypsinogen A (25 kDa), thaumatin I (22 kDa), and ribonuclease A (13 kDa). MALDI-TOF, amino acid composition analysis, and N-terminal sequencing by the Edman degradation method were conducted at the PNAC facility.
Evolutionary Bioinformatic Analysis
To study the evolutionary history of N-terminal Mps1, we generated 160 amino acids overlapping (by 10 amino acids) sequence fragments encompassing the entire human Mps1 protein, which corresponds to the approximate length of a triple tandem repeat of the TPR domain. This allowed us to examine whether the different regions of the protein may have different evolutionary histories. Each fragment was then blasted (PSI-BLAST) against all sequences in the nonredundant protein sequences database (NCBI). For each fragment, all matches were identified and classified by organism and significance of match (E-value), which estimates the number of hits “expected” by chance. An interactive electronic version of the matrix is available on line. Secondary structure alignment of the TPR region in Mps1 orthologs was performed using PSIPRED (33).
Cell Culture and Transfections
HeLaS3 and 293T cells were routinely maintained in DMEM (Hyclone) containing 10% fetal bovine serum (FBS, PAA Laboratories) at 37 °C and 5% CO2. Transfections and siRNA-mediated depletions in HeLa cells were performed using TransIT-LT1 (Mirus) and Oligofectamine (Invitrogen) reagents, respectively, according to the manufacturers' instructions. Transfections in 293T cells were performed using polyethyleneimine (PEI) at a 15:2 (PEI/DNA) ratio. Cell extracts for pulldowns, immunoprecipitations, and Western blots were prepared in RIPA lysis buffer (10 mm Tris, pH 7.5, 150 mm NaCl, 0.5% Triton X-100, 1% sodium deoxycholate, 1 mm sodium vanadate, 10 μg/ml leupeptin, 1 μg/ml pepstatin A, 10 μg/ml aprotinin, and 1 mg/ml Pefabloc). All Western blots and immunoprecipitations were performed with lysates equalized for protein content using the BCA assay (Pierce). Drug treatments were performed at the following concentrations and durations unless otherwise indicated: thymidine (Acros Organics, 2 mm for 16 h); S-trityl-l-cysteine (STLC, Sigma, 0.5 μm for 16 h); MG132 (Calbiochem, 20 μm for 1.5 h); reversine (Sigma, 0.5 μm for 30 min); nocodazole (Sigma, 3.3 μm, 16 h).
Immunofluorescence, Microscopy, Image Processing, and Quantification
Cells grown on coverslips were fixed and permeabilized simultaneously for 10 min at room temperature and processed for indirect immunofluorescence microscopy as described previously (34), For Mps1 staining, cells were fixed in 1% formaldehyde, quenched with glycine, and permeabilized using 0.5% Triton X-100 before being stained (12). Cells were imaged by confocal microscopy on a Leica DMI6000B inverted microscope equipped with a WaveFX spinning disc and an Orca-ER camera (Quorum Technologies). Image acquisition was performed using Volocity software (PerkinElmer Life Sciences). Optical sections were acquired with identical exposure times for each channel within an experiment and then projected into a single picture using ImageJ (rsb.info.nih.gov). Image processing was performed in Photoshop, and images shown in the same figure have been identically scaled. Quantification of kinetochore intensities was performed essentially as described previously (34). Antibodies against the following were used in this study: Mps1 (Sigma, clone N1. Note that this antibody did not recognize recombinant or overexpressed Mps1(1–239)), Bub1 (35), BubR1 (34), CREST (Immunovision), Mad2 (36), α-tubulin (DM1A, Sigma), MYC (9E10 and A-14, Santa Cruz Biotechnology), Sgo1 (Abnova), Plk1 (37), and mCherry (GTX59788, GeneTex). Antibodies against Blinkin and CenpE were kind gifts from Iain Cheeseman and Tim Yen, respectively. Anti-Mps1 phosphospecific antibodies (anti-Thr(P)-676, anti-Thr(P)-686, and anti-Ser(P)-821) were a generous gift from Patrick Eyers. Hoechst 33342 (Sigma) was used to stain the chromosomes.
RESULTS
Structure Model
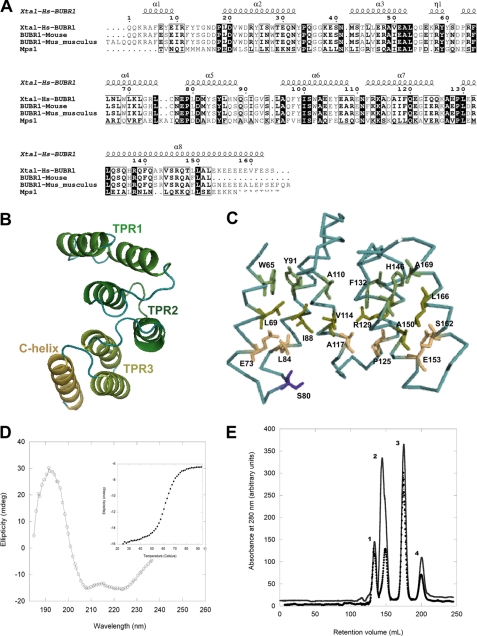
The observation that the multidomain protein kinases Bub1, BubR1, and Mps1 require kinetochore attachment to perform efficiently their essential roles in the SAC prompted us to investigate to what extent the similarity of functions is the result of evolutionary conservation among these checkpoint kinases. Sequence alignments of N-terminal Mps1 (residues 61–210) against Bub1 and BubR1 from various species indicate that the highest similarity of N-terminal Mps1 is with the N-terminal region of BubR1 (20.3% sequence identity, Fig. 1A) followed by N-terminal Bub1 (14.2% sequence identity). Analysis using FUGUE software shows a Z-score for N-terminal Mps1 of 24.36 thus indicating (with 99% confidence) that an evolutionary relationship and common fold exist between the TPR domains of Bub1 and BubR1 and N-terminal Mps1. Because of the higher amino acid sequence identity between human Mps1 and human BubR1, we modeled the N-terminal region of Mps1 using the crystal structure of human BubR1 (38) as template (Fig. 1, B and C). Some conserved features of a canonical TPR can be recognized in the Mps1 structure model, including a pattern of large residues (WYFL) in TPR helix A and smaller ones (ESAG) in TPR helix B at equivalent positions to those described for canonical TPRs (Fig. 1B). Such residue distribution is expected to be important for the establishment of interactions between TPR-forming helices, as these interactions should confer stability to the N-terminal domain (Fig. 1C). Furthermore, the model of the three-dimensional structure includes an additional α-helix immediately downstream of the third TPR unit of N-terminal Mps1, which might function as a C-terminal “capping” helix, as observed in certain TPR structures (39, 40). The model allows the mapping of residues that are fully conserved and predicted to be exposed at the surface. For instance, serine 80, a residue that can be phosphorylated in vivo (41), is mapped onto the flexible loop region that links helices A and B of TPR1 (Fig. 1C). The predicted spatial location of this residue is consistent with its accessibility to protein kinases. These observations prompted us to investigate further the predicted structural similarity between N-terminal Mps1, Bub1, and BubR1 using a multidisciplinary experimental strategy involving biochemical, biophysical, and cellular methods.
FIGURE 1.
Identification of a TPR motif in the N-terminal region of Mps1. A, amino acid sequence alignment of human N-terminal Mps1 and BUB1 and BUBR1 from different species. Secondary structure elements are mapped onto the crystal structure of N-terminal BUBR1 (Protein Data Bank code 2WVI). Figure was generated with ESPript (68). B, structure model of N-terminal Mps1 predicts this region is organized as a triple tandem of the TPR motif. C, model highlighting the Mps1 residues that are conserved and located in positions that define a canonical TPR motif. Most of these conserved residues are predicted to be engaged in stabilizing stacking interactions. Purple indicates the phosphorylation site serine 80 is highlighted. D, far-UV CD confirms N-terminal Mps1 is organized as a predominantly α-helical region. Inset, the thermal denaturation of this domain is highly cooperative and follows a two-state unfolding process. E, size-exclusion chromatogram of molecular mass markers only (●): peak 1, bovine serum albumin; peak 2, ovalbumin; peak 3, chymotrypsinogen A; peak 4, ribonuclease A. For the second chromatogram (−), the same molecular mass markers were combined with Mps1(1–239) prior to gel filtration. Mps1(1–239) retention time was closed to that of ovalbumin (43 kDa) thus revealing the former self-associates to form stable dimers.
N-terminal Mps1 Is Homodimeric and Predominantly α-Helical
Based on our secondary structure analysis for the possible domain boundaries of the putative Mps1 TPR region using diverse secondary structure prediction programs (JPred, PsiPred, and PredictProtein), several N-terminal fragments encompassing the N-terminal Mps1 region were generated (see under “Experimental Procedures”), of which fragment 1–239 was the most stable after GST cleavage and removal. Far-UV CD confirms that Mps1(1–239) is a predominantly α-helical protein (Fig. 1D) with ∼62% α-helix content as estimated from 2,2,2,-trifluoroethanol titration experiments (data not shown). The thermal unfolding of Mps1(1–239) resembles that of Bub1 and BubR1 TPRs and follows a highly cooperative two-state transition process (Tm = 62 °C; Fig. 1D, inset), indicating that Mps1(1–239) functions as a single independently folded domain. As Mps1 dimerization has been suggested to be critical for Mps1 functions in mitotic checkpoint signaling (14, 42), we investigated the oligomerization state of Mps1(1–239) in aqueous solutions. Analytical gel filtration chromatography revealed that Mps1(1–239) associates to form homodimers (Fig. 1E), thus lending support to this notion.
Some of the seven predicted α-helices of Mps1(1–239) are also predicted to be of an amphipathic nature. Because the affinity of this class of α-helices for water/air interfaces is importantly influenced by the magnitude of the hydrophobic moment (μH), this parameter was estimated for each of the predicted α-helices of this domain. The calculated μH values were as follows: helix 1 (61–75), 0.43; helix 2 (84–94), 0.59; helix 3 (107–119), 0.62; helix 4 (126–135), 0.02; helix 5 (143–155), 0.09; helix 6 (159–172), 0.54; and helix 7 (177–192), 0.34. The average hydrophobic moment (μHavg) of Mps1(1–239) was 0.37 kcal/mol per residue, which is very close to that calculated for TPR Bub1 and TPR BubR1 (μHavg = 0.38 and μHavg = 0.32 kcal/mol per residue, respectively) (43) indicating similarity in tertiary structure. Because the amphipathic character that results from the spatial arrangement of tandem repeats of the TPR motif confers surface activity to the protein when deposited onto monolayers, we set out to investigate in more detail the physiochemical properties of the putative TPR tandem repeat of N-terminal Mps1 at the air/water interface.
Mps1(1–239) as a Surface-active Domain
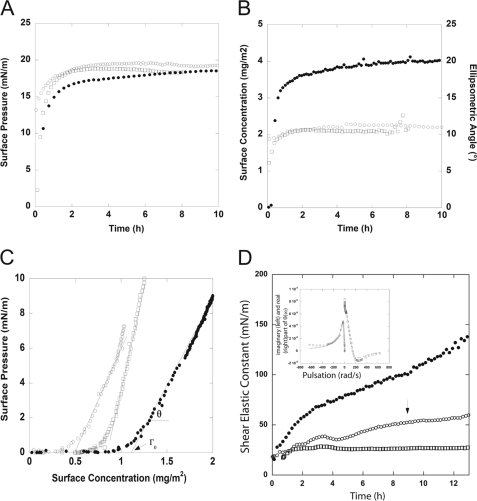
A typical feature of amphiphilic molecules (such as TPR folds) is the gradual migration of molecules toward the air/water interface. This property leads to an increase of the surface pressure until the surface is saturated and can be characterized by monitoring the evolution of surface pressure as a function of protein subphase concentration. The ellipsometric angle (Δ) at equilibrium (20 ± 1°), which is proportional to the quantity of protein adsorbed at the interface, as well as the maximum surface pressure (18 mN/m) confirmed that N-terminal Mps1(1–239) is highly surface-active (Fig. 2, A and B). After 6 h, the surface concentration remains virtually constant at 2.1 mg/m2. The maximal surface pressure of Mps1(1–239) as measured at the bulk concentration of 30 μg/ml is very similar to that of TPR domains of Bub1, BubR1, and PP5 (i.e. between 18 and 20 mN/m), again suggesting a similar tertiary structure (43).
FIGURE 2.
Biophysical characterization of the N-terminal fragment Mps1(1–239). Interfacial properties of Mps1(1–239), TPR Bub1, and TPR BubR1 are shown. Surface pressure (A) and surface concentration (Γ = 0.2 Δ) (B) together with the corresponding ellipsometric angle at air/water interfaces were determined by null ellipsometry measurements. Solutions of Mps1(1–239) (□), TPR Bub1 (○), and TPR BubR1 (●) are shown. Protein solutions were prepared at 30 μg/ml in 20 mm phosphate buffer, pH 7. C, surface pressure versus surface concentration of Mps1(1–239) (□), TPR Bub1 (○), and TPR BubR1 (●). θ corresponds to the slope dπ/dΓ, and Γ0 is the surface concentration at which the surface pressure becomes different from zero. Γ0 is calculated from the intersect of the slope π versus Γ. Mps1(1–239) (□), TPR Bub1 (○), and TPR BubR1 (●) at 1 μg/ml in 20 mm phosphate buffer, pH 7, are shown. D, rheology measurements of Mps1(1–239) (□), TPR Bub1 (○), and TPR BubR1 (●). The graph shows the evolution of the shear elastic constant, μ, versus time measured at the fixed frequency of 5 Hz, during protein adsorption at the interface. Protein solutions were prepared at 30 μg/ml in 20 mm phosphate buffer, pH 7. The error bar on μ is ± 5 mN/m. Inset, at the end of the kinetic (around 9 h, indicated by the arrow in the graph) the angular deviation θ(ω) versus the pulsation was measured. The curves correspond to Mps1(1–239). An elastic layer model (harmonic oscillator) fit the imaginary and real part of the response. For clarity, the imaginary part has been plotted versus −ω.
The adsorbed surface concentration (2.1 mg/m2) and surface pressure (1.3 mN/m) of Mps1(1–239) monitored at the low bulk concentration of 5 μg/ml made it possible to record initial adsorption events, which in turn allowed extraction of several parameters that are relevant to surface activity such as Γ0, the surface concentration at which the surface pressure becomes different from zero, and θ, which corresponds to the increase of surface pressure relative to the increase of surface concentration. The Γ0 value of Mps1(1–239) was 0.84 mg/m2, and θ value was 25 mN·m/mg, similar to those of Bub1 and BubR1 (Fig. 2C).
In the first steps of Mps1(1–239) adsorption at this low bulk concentration, the transport of protein molecules from the subphase to the interface is assumed to be a diffusion-controlled process. Thus, estimation of the diffusion coefficient allows a comparison of the transport rate of protein molecules from the subphase to the interface. The magnitude of the diffusion coefficient of Mps1(1–239), 0.2 × 10–10 m2· s−1, was of the same order of magnitude as that of TPR Bub1 (0.7 × 10–10 m2·s−1), TPR BubR1 (4.8 × 10–10 m2·s−1), and TPR PP5 (3 × 10–10 m2·s−1).
Consistent with null ellipsometry, Brewster angle microscopy (BAM), which is very sensitive to the amount of matter adsorbed at the interface, allowed the direct observation of the rapid migration of Mps1(1–239) from the bulk toward the surface. At the low concentration of 5 μg/ml, protein adsorption could be noticed as early as 1 min after deposition (Fig. 2D). After 6 h, the protein interfacial layer thus formed remained stable, showed a high contrast with respect to pure buffer solution, and was homogeneous at the micrometer scale (Fig. 2D). Such behavior closely resembles BAM observations of TPR Bub1 and TPR BubR1 monolayers (43, 46).
The study of the evolution of the shear elastic constant as a function of time provides clues about the cohesiveness and possible self-organization of the molecules reaching the interface to form a layer. The shear elastic constant of Mps1(1–239) monolayers measured at a fixed frequency of 5 Hz reveals an elastic monolayer thus indicating that medium and long range interactions play a major role in the cohesion of the layer (supplemental Fig. S1). This is consistent with the observation that extensive medium and long range interactions are one important functional characteristic of the TPR motif (47) and constitute the interactions most frequently observed in all-α-helix proteins compared with the classes all-β, α+β, and α/β (48). Collectively, our bioinformatics, biochemical, and biophysical data indicate that Mps1(1–239) most likely adopts a TPR fold that is highly similar to that of the mitotic kinases Bub1 and BubR1.
Evolutionary Analysis of the Mps1 N-terminal Region
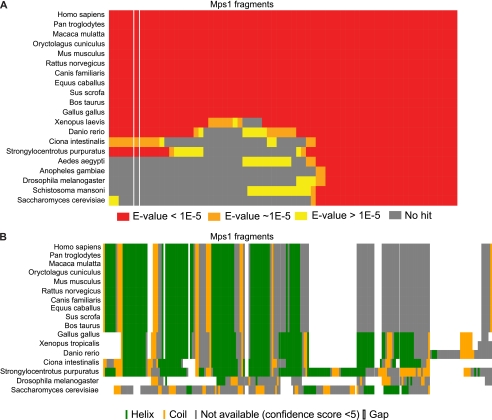
To study the evolutionary history of the Mps1 TPR domain, we blasted overlapping regions of the hMps1 protein sequence (see under “Experimental Procedures”) against all sequences in the nonredundant protein sequences database. Our bioinformatic analysis indicates that the MPS1 TPR appeared in the lineage leading to the deuterostomes (Fig. 3A) as no significant PSI-BLAST hits for the N-terminal region were detected outside the group, except for few exceptions such as Albugo (oomycetes, data not shown) that may result from either contamination in the sequenced genomic DNA or horizontal gene transfer from animals. We identified fragments homologous to the human MPS1 TPR region in some Urochordates (Ciona intestinalis) and Echinoderms (Strongylocentrotus purpuratus) but in no arthropods or other animal lineages (see full sequence analysis on Landry lab homepage). Secondary structure analysis of the hMps1 TPR and the corresponding regions in Mps1 orthologues corroborate these results and support the idea that the TPR domain appeared with the emergence of the deuterostome lineage, as deuterostomes share the great majority of the secondary structures present in that region, in particular the pattern of α-helix-turn-α-helix (Fig. 3B), although organisms like Drosophila and Saccharomyces cerevisiae do not.
FIGURE 3.
Mps1 TPR domain appeared after the emergence of deuterostomes. A, human Mps1 overlapping fragments (length, 160 amino acids; offset, 10 amino acids) were blasted (PSI-BLAST) against the entire NCBI sequence database (nonredundant). For each listed organism for which we could find a hit for one or more fragments, we show if the hit was highly significant (red rectangles, E-value <1e-5), close to the threshold (orange rectangles, ∼1e-5 < E-value < 1), or not significant (yellow rectangles, E-value ≥1). If we could not find a hit for a specific fragment, the corresponding rectangle is gray. The column highlighted between white lines corresponds to the exact hMps1 TPR region. B, secondary structure prediction of the TPR region of hMps1 and the corresponding region in Mps1 orthologues. Alignment and prediction were performed using PSIPRED (33).
Mps1(1–239) Localization
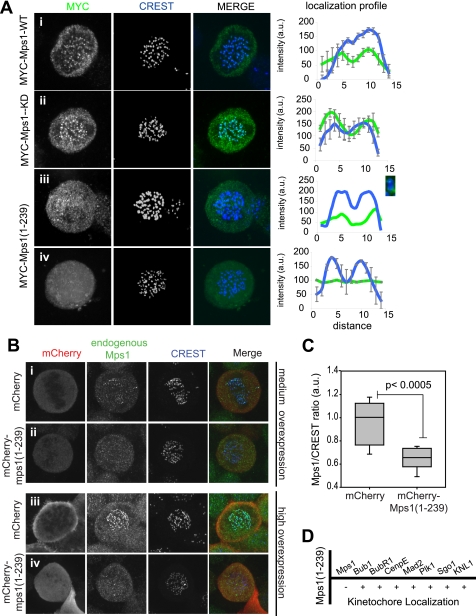
The identification of a presumptive TPR fold in the N-terminal region of Mps1 that is related to and behaves similarly to the TPR domains of Bub1 and BubR1 led us to postulate that these regions may have similar functions in the cell. As the Mps1 N-terminal region and the TPR domain of Bub1 and BubR1 are critical for KT localization of these proteins, we investigated the role of Mps1(1–239) in Mps1 KT recruitment. We first overexpressed triple MYC (3×MYC)-tagged Mps1 wild-type (WT), kinase-dead (KD), and Mps1(1–239) and compared the localization of each of these constructs with the KT marker CREST. As expected, 3×MYC-Mps1-WT and 3×MYC-Mps1-KD localization overlapped that of CREST, and both were clearly visible at KTs (Fig. 4A, rows i and ii), as confirmed by colocalization profiles across sister KT pairs. We also noted that Mps1-KD showed stronger KT staining as reported previously (11, 14, 42). In contrast, we found that 3×MYC-Mps1(1–239) was largely cytoplasmic (Fig. 4A, row iii). However, we occasionally observed low level kinetochore recruitment as demonstrated by partial overlap with CREST (Fig. 4A, row iv), in good agreement with previous observations (16).
FIGURE 4.
Localization of Mps1(1–239). A, localization of 3×MYC-Mps1 constructs. HeLa cells were transfected with 3×MYC-Mps1 WT (row i), -Mps1-KD (row ii) or Mps1(1–239) (rows iii and iv) and synchronized in mitosis by release from a single thymidine arrest before being fixed and stained for immunofluorescence with antibodies against MYC (green) and CREST (blue). The panel on the right shows the colocalization profile between MYC and CREST signal across sister kinetochore pairs (3–4 pairs). A, row iii, colocalization of one sister kinetochore pair is included, and the pair is detailed in the inset. B, immunofluorescence of cells expressing mCherry alone (row i, medium; row iii, highly overexpressing) or mCherry-Mps1(1–239) (row ii, medium; row iv, highly overexpressing) were synchronized in mitosis and fixed for immunofluorescence as in A. Cells were stained with anti-Mps1 antibodies (green) and CREST autoimmune serum (blue). C, endogenous Mps1/CREST signal ratio (arbitrary units; a.u.) in cells highly overexpressing mCherry or mCherry-Mps1(1–239), n = 9 cells each. D, cells were transfected, synchronized, and fixed as in A and were then stained for the indicated proteins. Kinetochore localization was scored for these proteins in cells overexpressing Mps1(1–239) relative to control cells.
While studying the localization of the Mps1 constructs, we noticed that in cells strongly overexpressing Mps1(1–239), endogenous Mps1 was often absent from or difficult to detect at KTs. To address this in more detail, we compared endogenous Mps1 KT recruitment in cells expressing low, medium, and high levels of Mps1(1–239). Because Mps1 kinase activity negatively regulates its KT localization (11, 14, 42), we performed these experiments with or without treatment of cells with the Mps1 inhibitor reversine prior to fixation. The same results were obtained under both conditions. Whereas low mCherry-Mps1(1–239) expression did not visibly interfere with KT docking of endogenous Mps1 (data not shown), we observed a decrease in KT Mps1 levels in cells expressing medium levels of mCherry-Mps1(1–239) compared with cells expressing similar levels of the mCherry-tag alone (Fig 4B, row ii versus i). This effect was further enhanced in cells expressing high levels of mCherry-Mps1(1–239) where endogenous Mps1 was almost undetectable at KTs, suggesting that overexpression of this Mps1 N-terminal fragment interferes with KT targeting of endogenous Mps1. Quantification of the levels of endogenous Mps1 relative to the CREST autoimmune antigen in cells highly expressing mCherry-Mps1(1–239) confirmed these observations (Fig. 4C). These results argue that Mps(1–239) on its own is not sufficient for KT docking but likely plays an important role in the context of the full-length Mps1 protein as its overexpression interferes with docking of endogenous Mps1. As the Mps1(1–239) purified fragment behaved as a dimer in solution, we interpret these results to mean that Mps1(1–239) interferes with endogenous Mps1 by recruiting it away from KTs. In agreement with this, we found that a GST-Mps1(1–239) fragment is able to pull down endogenous Mps1 from mitotic cell extracts (supplemental Fig. S2).
Mps1 kinase activity is required for the KT docking of a number of checkpoint proteins, including Mad1, Mad2, Bub1, BubR1, CenpE, Plk1, and Sgo1 (10, 15, 16, 49, 50). However, in most cases whether the N-terminal region plays a role is not clear. To investigate this, we overexpressed the empty 3×MYC vector or 3×MYC-Mps1(1–239) in HeLa cells and determined whether overexpression of Mps1(1–239) disrupted KT recruitment of these proteins in cells synchronized in mitosis by release from G1/S arrest. Our results indicate that whereas endogenous Mps1 is clearly reduced in cells expressing Mps1(1–239), Bub1, Mad2, BubR1, CenpE, Plk1, Sgo1, and the KNL-1/Mis12/Ndc80 (KMN) subunit Blinkin (also known as hKNL1, AF15q14, D40, and CASC5), are all normally localized to the KT (Fig. 4D).
Overexpression of Mps1(1–239) Disrupts Chromosome Congression and Weakens the Spindle Checkpoint
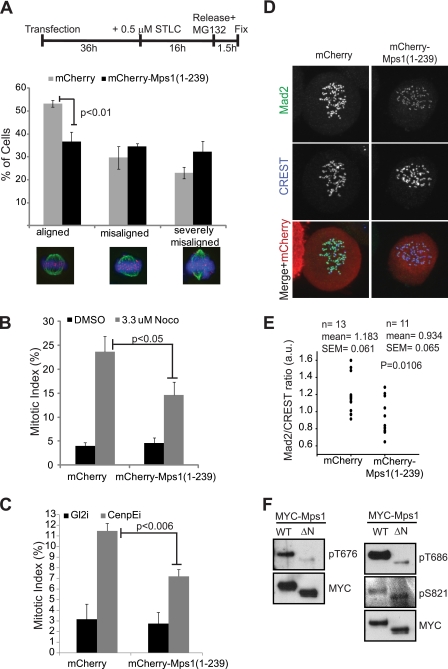
It has been demonstrated that Mps1 kinase activity is required for both spindle checkpoint function and efficient chromosome congression (13, 51). In addition, overexpression of an extended N-terminal kinetochore-binding fragment of Mps1(1–301) or an Mps1 allele lacking the first 100 amino acids (Mps1ΔN) decreased mitotic arrest efficiency in cells treated with nocodazole (15, 16). Similar to the case of TPR Bub1 and TPR BubR1, a series of bioinformatics analysis of the Mps1 region following the TPR domain, residues 240–301, consistently predicted this region to be of low structural complexity (data not shown). In oocyte meiosis I, the absence of Mps1 from KTs severely impairs chromosome segregation and fertility in mice (19). To test whether the TPR domain of Mps1 is involved in regulating these processes, we overexpressed Mps1(1–239) in HeLa cells and determined the efficiency of chromosome alignment in these cells. Although most Mps1(1–239)-expressing cells were able to align chromosomes to a metaphase formation after a 1-h treatment with the proteosomal inhibitor MG132, about 10% (compared with 5% of control cells) displayed mild alignment defects, including 1–2 lagging chromosomes and broad metaphase plates (data not shown). We reasoned that Mps1(1–239) overexpression may reveal a more severe phenotype in cells undergoing recovery from spindle stress. To test this, we treated Mps1(1–239)-overexpressing cells with STLC, an inhibitor for the Eg5 kinesin motor, which results in monopolar spindle formation and mitotic delay because of the inability of the centrosomes to separate. STLC washout reverses this phenotype and allows for bipolar spindle formation, although the rate of KT-MT attachment errors is elevated. We found that significantly fewer Mps1(1–239)-expressing cells aligned at metaphase plates in the presence of MG132 1.5 h after release from STLC compared with control cells (Fig. 5A). This indicates that Mps1(1–239) overexpression disrupts efficient chromosome congression and metaphase alignment during the recovery from spindle stress.
FIGURE 5.
Functional analysis of Mps1(1–239). A, HeLa cells were transfected with mCherry or mCherry-Mps1A and then synchronized in STLC followed by release into MG132 according to the schematic. After 1.5 h of MG132 treatment, cells were fixed and processed for immunofluorescence with antibodies against α-tubulin (green) and CREST (Cy5, illustrated here in magenta). Hoechst 33342 was used to visualize the DNA (blue). Chromosome alignment at metaphase plates was examined in cells expressing either mCherry or mCherry-Mps1(1–239) and grouped into the three indicated categories based on the extent of the alignment. Only cells in which both spindle poles were visible in the same plane were counted (n = 3, ≥100 cells/experiment). B, cells transfected with either mCherry or mCherry-Mps1 were treated for 16 h either with DMSO as control or 3.3 μm nocodazole (Noco), fixed, and stained with Hoechst 33342 to visualize the DNA. The mitotic index of transfected cells was counted (n = 5, ≥100 cells/experiment). C, cells were transfected as in B but were also simultaneously transfected with siRNA oligonucleotides targeting Gl2 (as control) or CenpE. 48 h after transfection, the cells were fixed and stained as in B before mitotic index counts were performed (n = 3, ≥100 cells/experiment). D, cells were transfected with either mCherry or mCherry-Mps1(1–239) and treated with 3.3 μm nocodazole for 16 h before being fixed and stained for Mad2 (green) and CREST (Cy5, shown here in blue). E, quantification of the Mad2/CREST ratio from cells in D. F, 293T cells were transfected with 3×MYC Mps1-WT or 3×MYC Mps1ΔN and treated with nocodazole for 16 h before being harvested. Immunoprecipitated Mps1 fragments were resolved by SDS-PAGE and immunoblotted with anti-Thr(P)-676, anti-Thr(P)-686, and anti-Ser(P)-S821 Mps1 antibodies. The membranes were stripped and reprobed for MYC to demonstrate equal input.
Mps1 is required for SAC function, but the role of the N-terminal domain remains controversial (15, 16, 19). To test whether overexpression of TPR Mps1 interferes with efficient checkpoint activity, we took two independent approaches. Recent reports have indicated that rigorous assessment of checkpoint signaling requires microtubules to be completely depolymerized, as occurs at very high concentrations of nocodazole (3.2 μm) (52). The spindle checkpoint by definition cannot be satisfied under these conditions, thus enabling the dissection of checkpoint function from chromosome congression. We therefore treated cells expressing mCherry or mCherry-Mps1(1–239) with DMSO or 3.3 μm nocodazole and quantified the percentage of mitotic cells under these conditions. DMSO-treated mCherry and mCherry-Mps1(1–239) cells demonstrated no significant difference in the percentage of mitotically arrested cells (Fig. 5B). However, treatment of Mps1(1–239)-overexpressing cells with 3.3 μm nocodazole resulted in an attenuated checkpoint response with a significantly smaller percentage of cells remaining in mitosis compared with cells expressing mCherry alone 16 h after drug treatment (Fig. 5B). These observations indicate that Mps1(1–239) overexpression disrupts SAC signaling independent of its role on chromosome alignment.
As a second approach, we tested the effect of Mps1(1–239) overexpression on CenpE-mediated mitotic delay. In mammalian cells, disruption of CenpE function prolongs mitotic timing in a checkpoint-dependent manner (53). Cells in which CenpE is depleted or inhibited exhibit elevated levels of checkpoint proteins at unaligned kinetochores, including Mps1, and co-injection of antibodies targeting both CenpE and Mps1 abrogated the mitotic arrest observed upon inactivation of CenpE alone (16). To test whether overexpression of TPR Mps1 attenuates the checkpoint response in CenpE-depleted cells, we expressed either mCherry or mCherry-Mps1(1–239) in cells treated with control siRNA or in cells depleted of CenpE. Overexpression of mCherry-Mps1(1–239) did not increase the percentage of mitotic cells compared with the expression of mCherry alone in control cells as indicated above (Fig. 5C). As expected, depletion of CenpE resulted in an increase in the mitotic index of cells expressing mCherry. This increase was significantly attenuated in cells transfected with mCherry-Mps1(1–239) indicating that this fragment disrupted the SAC response induced by CenpE inactivation (Fig. 5C). Collectively, our data indicate that Mps1(1–239) overexpression attenuates the checkpoint response under conditions of both partial and complete microtubule disruption.
Mps1 kinase activity is thought to mediate checkpoint function by facilitating the KT recruitment of Mad2 and its conversion from open and inactive (O-Mad2) to closed and activated (C-Mad2) capable of Cdc20 binding and inhibition (14). We therefore reasoned that the increased slippage observed in nocodazole-treated cells overexpressing Mps1(1–239) may be due to a decrease in KT recruitment of Mad2. To test this, we took advantage of a recently described Mad2 monoclonal antibody that specifically recognizes the Cdc20 inhibitory form of Mad2, C-Mad2, to determine KT recruitment of “active” Mad2 in cells overexpressing Mps1(1–239) (54). Using this antibody, we observed that although Mad2 was not completely lost at KTs in Mps1(1–239)-expressing cells treated with 3.3 μm nocodazole, C-Mad2 levels at KTs were visibly diminished (Fig. 5D). Quantification of Mad2/CREST ratios in these cells indeed revealed a significant trend toward lower Mad2 levels at KTs compared with control cells (Fig. 5E). As tight control of Mad2 is crucial for optimal checkpoint function and even a small reduction of the protein attenuated checkpoint signaling (see for example Refs. 55, 56), our observations suggest that Mps1(1–239) overexpression may allow increased slippage by reducing the levels of active KT-bound C-Mad2.
The conversion of O-Mad2 to checkpoint-proficient C-Mad2 is dependent on Mps1 kinase activity (14, 15), implying that the decrease in C-Mad2 at KTs in cells overexpressing Mps1(1–239) may be due to decreased Mps1 catalytic activity. Mps1 dimerization, which is likely mediated by the TPR domain, has been shown to enhance its catalytic activity (57); we therefore reasoned that mutation of the Mps1 TPR domain may inhibit Mps1 function through attenuation of kinase activity. Mps1 is known to autophosphorylate both in vitro and in vivo at Thr-676 and Thr-686 in the activation loop resulting in increased catalytic activity (57–60), whereas phosphorylation at Ser-821 is likely due to MAPK (58, 61). To test the contribution of the Mps1 TPR to kinase function, we generated Mps1 lacking the N-terminal 100 amino acids (Mps1ΔN (15)) and thus a significant portion of the TPR domain, and we tested the effect of this mutation on kinase function using phosphospecific antibodies against Mps1 (58). Immunoprecipitation of Mps1-WT and Mps1ΔN followed by Western blotting clearly revealed that autophosphorylation of Mps1 at Thr-676 and Thr-686 is severely compromised in Mps1ΔN compared with Mps1-WT, whereas phosphorylation of the proline-directed, MAPK site Ser-821 remains unchanged. Together, these results indicate that Mps1 TPR is required for efficient functioning of this checkpoint protein, and mutations in this region reduce the checkpoint response, likely through attenuation of kinase activity.
DISCUSSION
Biophysical Characteristics of the Mps1 N Terminus
Although the requirement of the N-terminal region of Mps1 for both its localization at KTs and efficient functionality has been reported by several groups (9, 15, 16, 18, 19), a more detailed understanding of the functional significance and structural organization of this region was a pending assignment in the field. To this purpose, we combined a bioinformatics approach with biochemical and biophysical analyses and provided novel evidence strongly suggesting that Mps1 residues 1–239 adopt a stable TPR fold that remarkably mimics the N-terminal TPR-containing region of Bub1 and BubR1. For instance, far-UV circular dichroism confirmed that Mps1(1–239) is a predominantly α-helical protein domain, whereas analytical gel filtration shows that the protein self-associates to form stable dimers. Moreover, ellipsometry, polarized fluorescence microscopy, and BAM experiments revealed that like TPR Bub1 and TPR BubR1 domains, when deposited onto monolayers, Mps1(1–239) functions as a surface-active domain that forms stable, rigid monolayers at the air/water interface. Definition of protein physicochemical properties and behavior at interfaces can yield relevant information such as the molecular orientation of protein domains in phospholipid films (62), the differential affinity for the substrate across lipid-binding proteins (63), and the relative surface activity and interfacial stability of proteins sharing the TPR motif, including Bub1, BubR1, and protein phosphatase 5 (43). The latter case is of particular importance as it provided a framework to investigate the interfacial behavior of Mps1(1–239) at the air/water interface. Indeed, the biophysical characterization of the N-terminal fragment Mps1(1–239) revealed striking similarities with TPR Bub1 and TPR BubR1. For instance, Langmuir-Blodget films coupled to null ellipsometry measurements showed a similar maximal surface pressure and surface concentration at air/water interfaces for the three proteins, whereas BAM observations revealed the rapid migration of Mps1(1–239) from the bulk toward the surface in a fashion that closely resembles the behavior of TPR Bub1 and TPR BubR1 monolayers. Moreover, the evolution of the shear elastic constant of Mps1(1–239) interfacial layers measured at the end of the adsorption kinetics (i.e. around 4 h) also reveals a monolayer of a rigidity comparable with that of TPR Bub1 and TPR BubR1 films.
Evolution of the Mps1 TPR Domain
Through extensive protein database searches, we found that the MPS1 TPR most likely appeared at the split between the protostomes and deuterostomes because it is found in the chordates as well as in echinoderms. We favor this scenario over domain loss in other animal lineages such as the insects, as the domain also appears to be absent in fungi. This conclusion is supported by secondary structure analysis that identified the analogous pattern of α-helices and turns predicted in hMps1 in the Mps1 homologues of other deuterostomes. Our results also imply that the MPS1 TPR region has a more limited phylogenetic distribution than the BUB1 or BUBR1 TPR, suggesting that the MPS1 TPR could have been acquired from one of these two genes. Indeed alignment generated by FUGUE indicated that the highest scoring hit was TPR BubR1, and a search in the SMART database using the human Mps1 TPR fragment as query retrieved the Mad3_BUB1_I domain (SMART accession number SM00777, 5.02e+01) as the most significant hit. The reciprocal SMART search with S. cerevisiae and Drosophila Mps1 N-terminal regions, neither of which is predicted to form a TPR domain, returned no significant hit. Together, these observations suggest two possible scenarios for the emergence of MPS1 TPR. If BUB1 and BUBR1 duplicated in the deuterostome ancestor, the MPS1 TPR was acquired at about the same time as this duplication, making it difficult to assign its origin to either BUB1 or BUBR1 because little divergence would have accumulated between the two genes. The alternative scenario holds that duplication occurred after the chordate divergence from the echinoderms. In this case, the MPS1 TPR would have derived from the BUB1 and BUBR1 ancestor. Because of the limited set of genomes from echinoderms available to date, it is difficult to favor one scenario over the other.
Mps1 TPR Functions
The demonstration that Mps1(1–239) behaves as an independently folded domain prompted us to test its function(s) in mitotic cells. Our data show that Mps1(1–239) is only partially competent in KT localization, but its overexpression nevertheless causes mislocalization of endogenous Mps1, likely through dimerization with endogenous Mps1 and its titration away from KTs. This idea is supported by our gel filtration and GST pulldown experiments (Fig. 1E and supplemental Fig. S2). Also in agreement with our observations, deletion of N-terminal residues that are predicted to form part of TPR Mps1 preclude Mps1 KT binding and interfere with full Mps1 functionality in both mitotic and meiotic systems (15, 19). This behavior is remarkably reminiscent of the Bub1 TPR domain. Mutations in the Bub1 TPR that eliminated the interaction with the KMN subunit Blinkin and thus abrogated KT recruitment resulted in checkpoint deficiency and caused severe defects in chromosome alignment and segregation (64, 65). These data are in full agreement with the shared evolutionary history between the TPR domains of the Bub kinases and Mps1 and argue that at least transient KT docking of TPR Mps1 is required for optimal activity.
The direct binding partners of Mps1 responsible for its KT recruitment are not known, although both Aurora B and Hec1 play an important role in this process (9, 50, 66). Bub1 and BubR1 TPR domains bind directly to Blinkin, and disruption of this interaction by depletion of Blinkin or mutation of the TPR domains precludes KT recruitment of Bub1 and BubR1 resulting in loss of checkpoint control and severe errors in chromosome segregation during mitosis (64, 67). However, despite the close relationship between the TPR domains of Bub1, BubR1, and Mps1, we found no interaction between Mps1(1–239) and Blinkin, and depletion of Blinkin did not significantly affect KT docking of endogenous Mps1, nor did Mps1(1–239) noticeably interfere with Blinkin KT docking (data not shown and Fig. 4D), suggesting that Mps1 may bind to other core KT proteins to facilitate its recruitment. Because TPR Mps1 overexpression may affect not only the function of Mps1 but also that of Bub1 and BubR1, we decided to investigate whether N-terminal Mps1 physically interacts with N-terminal Bub1 and/or N-terminal BubR1. For this, we used recombinant proteins and studied the possibility of heterodimer complex formation between TPR Bub1 and TPR Mps1 as well as between TPR BubR1 and TPR Mps1 using analytical size-exclusion chromatography and dynamic light scattering. The formation of stable binary complexes between N-terminal Mps1 with N-terminal Bub1 or BubR1 was not detected with the aforementioned techniques suggesting that these TPRs do not physically interact with each other at least under our in vitro conditions (data not shown).
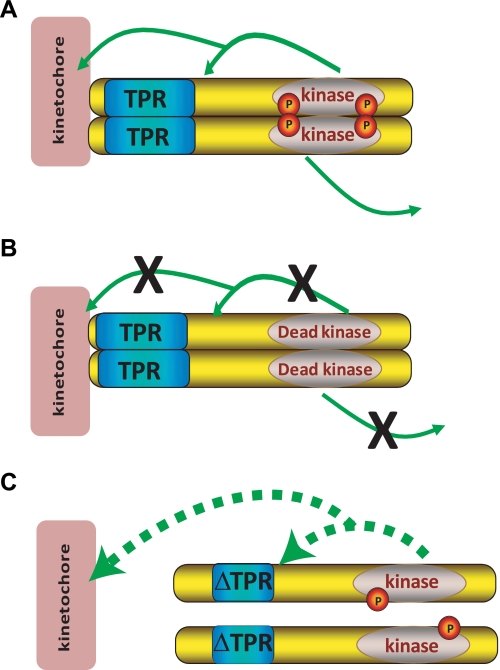
Mps1 KT localization is also regulated by its own catalytic activity (11, 42), and recent studies identified a number of autophosphorylation sites that map to the N-terminal domain, which may regulate Mps1 KT recruitment (17, 41, 58, 59). Importantly, we demonstrate here that the Mps1 TPR enhances kinase activity as N-terminal truncation of Mps1 resulted in a marked decrease of Mps1 autophosphorylation at two sites in the activation loop critical for optimal kinase function (Fig. 5F). We imagine that the TPR contributes to Mps1 activation and function through both dimerization and KT enrichment, both of which increase the local concentration of Mps1 (Fig. 6A). Because kinase-inactive Mps1 exhibits longer residency at KTs, autophosphorylation or phosphorylation of a KT target may promote release of Mps1 into the cytoplasm (Fig. 6B). Abolishing TPR function (Fig. 6C) results in inefficient dimerization and KT binding of Mps1, effectively diluting the monomeric enzyme in the cytoplasm and reducing kinase activity.
FIGURE 6.
Model for Mps1 TPR function. A, Mps1-WT can both dimerize and dock at KTs, thus increasing its local concentration and activity. Mps1 autophosphorylation or phosphorylation of a KT protein allows for its release into the cytoplasm. B, inactive Mps1 can dimerize and localize but cannot phosphorylate itself or other targets and is thus not released from the KT as efficiently as the WT protein. C, Mps1 lacking a functional TPR cannot localize to the KT and cannot dimerize resulting in reduced concentration of Mps1 protein and reduced kinase activity. This is manifest as reduced autophosphorylation and reduced phosphorylation of potential targets. Ultimately this results in suboptimal Mps1 activity.
Our interdisciplinary approach demonstrates that the N-terminal region of Mps1 likely forms a TPR domain that is evolutionarily, structurally, and biophysically related to the TPR domains of Bub1 and BubR1. Functionally, this region in Mps1 is critical for KT binding and for optimal Mps1 kinase activation in cells. Although the Mps1 TPR awaits direct structural confirmation, the data presented here provide novel evidence for the close relationship between these three checkpoint kinases and prompt a closer examination of the role of KT binding and the TPR domains in regulating their catalytic activity during mitosis.
Supplementary Material
Acknowledgments
We thank Drs. Maria J. Fernandes and Masahiko Sato for use of their microscopes and Iain Cheeseman, Tim Yen, and Patrick Eyers for their kind gifts of reagents.
This work was supported by a “Fondation des Etoiles” starting grant (to S. E.), Wellcome Trust Programme Grant RG44650 (Structural Biology of Cell Signalling and Regulation: Multiprotein Systems and the Achievement of High Signal-to-Noise Ratios) (to T. L. B.), and Canadian Institutes of Health Research Grant GMX-191597 (to C. R. L.).

This article contains supplemental Figs. S1 and S2.
- KT
- kinetochore
- TPR
- tetratricopeptide repeat
- SAC
- spindle assembly checkpoint
- STLC
- S-trityl-l-cysteine
- BAM
- Brewster angle microscopy
- mN
- millinewton.
REFERENCES
- 1. Holland A. J., Cleveland D. W. (2009) Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat. Rev. Mol. Cell Biol. 10, 478–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thompson S. L., Compton D. A. (2008) Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180, 665–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cheeseman I. M., Desai A. (2008) Molecular architecture of the kinetochore-microtubule interface. Nat. Rev. Mol. Cell Biol. 9, 33–46 [DOI] [PubMed] [Google Scholar]
- 4. Santaguida S., Musacchio A. (2009) The life and miracles of kinetochores. EMBO J. 28, 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Welburn J. P., Cheeseman I. M. (2008) Toward a molecular structure of the eukaryotic kinetochore. Dev. Cell 15, 645–655 [DOI] [PubMed] [Google Scholar]
- 6. Rieder C. L., Maiato H. (2004) Stuck in division or passing through. What happens when cells cannot satisfy the spindle assembly checkpoint. Dev. Cell 7, 637–651 [DOI] [PubMed] [Google Scholar]
- 7. Weiss E., Winey M. (1996) The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 132, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. He X., Jones M. H., Winey M., Sazer S. (1998) Mph1, a member of the Mps1-like family of dual specificity protein kinases, is required for the spindle checkpoint in S. pombe. J. Cell Sci. 111, 1635–1647 [DOI] [PubMed] [Google Scholar]
- 9. Stucke V. M., Baumann C., Nigg E. A. (2004) Kinetochore localization and microtubule interaction of the human spindle checkpoint kinase Mps1. Chromosoma 113, 1–15 [DOI] [PubMed] [Google Scholar]
- 10. Abrieu A., Magnaghi-Jaulin L., Kahana J. A., Peter M., Castro A., Vigneron S., Lorca T., Cleveland D. W., Labbé J. C. (2001) Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell 106, 83–93 [DOI] [PubMed] [Google Scholar]
- 11. Santaguida S., Tighe A., D'Alise A. M., Taylor S. S., Musacchio A. (2010) Dissecting the role of MPS1 in chromosome biorientation and the spindle checkpoint through the small molecule inhibitor reversine. J. Cell Biol. 190, 73–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Tighe A., Staples O., Taylor S. (2008) Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores. J. Cell Biol. 181, 893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones M. H., Huneycutt B. J., Pearson C. G., Zhang C., Morgan G., Shokat K., Bloom K., Winey M. (2005) Chemical genetics reveals a role for Mps1 kinase in kinetochore attachment during mitosis. Curr. Biol. 15, 160–165 [DOI] [PubMed] [Google Scholar]
- 14. Hewitt L., Tighe A., Santaguida S., White A. M., Jones C. D., Musacchio A., Green S., Taylor S. S. (2010) Sustained Mps1 activity is required in mitosis to recruit O-Mad2 to the Mad1-C-Mad2 core complex. J. Cell Biol. 190, 25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Maciejowski J., George K. A., Terret M. E., Zhang C., Shokat K. M., Jallepalli P. V. (2010) Mps1 directs the assembly of Cdc20 inhibitory complexes during interphase and mitosis to control M phase timing and spindle checkpoint signaling. J. Cell Biol. 190, 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Liu S. T., Chan G. K., Hittle J. C., Fujii G., Lees E., Yen T. J. (2003) Human MPS1 kinase is required for mitotic arrest induced by the loss of CENP-E from kinetochores. Mol. Biol. Cell 14, 1638–1651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Xu Q., Zhu S., Wang W., Zhang X., Old W., Ahn N., Liu X. (2009) Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by Mps1 phosphorylation. Mol. Biol. Cell 20, 10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Araki Y., Gombos L., Migueleti S. P., Sivashanmugam L., Antony C., Schiebel E. (2010) N-terminal regions of Mps1 kinase determine functional bifurcation. J. Cell Biol. 189, 41–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hached K., Xie S. Z., Buffin E., Cladière D., Rachez C., Sacras M., Sorger P. K., Wassmann K. (2011) Mps1 at kinetochores is essential for female mouse meiosis I. Development 138, 2261–2271 [DOI] [PubMed] [Google Scholar]
- 20. D'Andrea L. D., Regan L. (2003) TPR proteins. The versatile helix. Trends Biochem. Sci. 28, 655–662 [DOI] [PubMed] [Google Scholar]
- 21. Bolanos-Garcia V. M., Blundell T. L. (2011) BUB1 and BUBR1. Multifaceted kinases of the cell cycle. Trends Biochem. Sci. 36, 141–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shi J., Blundell T. L., Mizuguchi K. (2001) FUGUE. Sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J. Mol. Biol. 310, 243–257 [DOI] [PubMed] [Google Scholar]
- 23. de Bakker P. I., Bateman A., Burke D. F., Miguel R. N., Mizuguchi K., Shi J., Shirai H., Blundell T. L. (2001) HOMSTRAD. Adding sequence information to structure-based alignments of homologous protein families. Bioinformatics 17, 748–749 [DOI] [PubMed] [Google Scholar]
- 24. Mizuguchi K., Deane C. M., Blundell T. L., Johnson M. S., Overington J. P. (1998) JOY. Protein sequence-structure representation and analysis. Bioinformatics 14, 617–623 [DOI] [PubMed] [Google Scholar]
- 25. Sali A., Blundell T. L. (1993) Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 234, 779–815 [DOI] [PubMed] [Google Scholar]
- 26. Laskowski R. A., MacArthur M. W., Thornton J. M. (1998) Validation of protein models derived from experiment. Curr. Opin. Struct. Biol. 8, 631–639 [DOI] [PubMed] [Google Scholar]
- 27. Lüthy R., Bowie J. U., Eisenberg D. (1992) Assessment of protein models with three-dimensional profiles. Nature 356, 83–85 [DOI] [PubMed] [Google Scholar]
- 28. Berge B., Renault A. (1993) Ellipsometry Study of 2D crystallization of 1-alcohol monolayers at the water surface. Europhys. Let. 21, 773–777 [Google Scholar]
- 29. Azzam R. M., Bashara N. M. (1977) Ellipsometry and Polarized Light. North-Holland Personal Library, Amsterdam [Google Scholar]
- 30. De Feijter J. A., Benjamins J., Veer F. A. ( 1978) Ellipsometry as a tool to study the adsorption behavior of synthetic and biopolymers at the air–water interface. Biopolymers 17, 1759–1772 [Google Scholar]
- 31. Vénien-Bryan C., Lenne P. F., Zakri C., Renault A., Brisson A., Legrand J. F., Berge B. (1998) Characterization of the growth of 2D protein crystals on a lipid monolayer by ellipsometry and rigidity measurements coupled to electron microscopy. Biophys. J. 74, 2649–2657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Renault A., Lenne P. F., Zakri C., Aradian A., Vénien-Bryan C., Amblard F. (1999) Surface-induced polymerization of actin. Biophys. J. 76, 1580–1590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jones D. T. (1999) Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292, 195–202 [DOI] [PubMed] [Google Scholar]
- 34. Elowe S., Hümmer S., Uldschmid A., Li X., Nigg E. A. (2007) Tension-sensitive Plk1 phosphorylation on BubR1 regulates the stability of kinetochore microtubule interactions. Genes Dev. 21, 2205–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 36. Chan Y. W., Fava L. L., Uldschmid A., Schmitz M. H., Gerlich D. W., Nigg E. A., Santamaria A. (2009) Mitotic control of kinetochore-associated dynein and spindle orientation by human Spindly. J. Cell Biol. 185, 859–874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Baumann C., Körner R., Hofmann K., Nigg E. A. (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128, 101–114 [DOI] [PubMed] [Google Scholar]
- 38. D'Arcy S., Davies O. R., Blundell T. L., Bolanos-Garcia V. M. (2010) Defining the molecular basis of BubR1 kinetochore interactions and APC/C-CDC20 inhibition. J. Biol. Chem. 285, 14764–14776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Das A. K., Cohen P. W., Barford D. (1998) The structure of the tetratricopeptide repeats of protein phosphatase 5. Implications for TPR-mediated protein-protein interactions. EMBO J. 17, 1192–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F. U., Moarefi I. (2000) Structure of TPR domain-peptide complexes. Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell 101, 199–210 [DOI] [PubMed] [Google Scholar]
- 41. Dou Z., von Schubert C., Körner R., Santamaria A., Elowe S., Nigg E. A. (2011) Quantitative mass spectrometry analysis reveals similar substrate consensus motif for human Mps1 kinase and Plk1. PloS one 6, e18793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jelluma N., Dansen T. B., Sliedrecht T., Kwiatkowski N. P., Kops G. J. (2010) Release of Mps1 from kinetochores is crucial for timely anaphase onset. J. Cell Biol. 191, 281–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Beaufils S., Grossmann J. G., Renault A., Bolanos-Garcia V. M. (2008) Characterization of the tetratricopeptide-containing domain of BUB1, BUBR1, and PP5 proves that domain amphiphilicity over amino acid sequence specificity governs protein adsorption and interfacial activity. J. Phys. Chem. B 112, 7984–7991 [DOI] [PubMed] [Google Scholar]
- 44. Deleted in proof.
- 45. Deleted in proof.
- 46. Bolanos-Garcia V. M., Beaufils S., Renault A., Grossmann J. G., Brewerton S., Lee M., Venkitaraman A., Blundell T. L. (2005) The conserved N-terminal region of the mitotic checkpoint protein BUBR1. A putative TPR motif of high surface activity. Biophys. J. 89, 2640–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Beddoe T., Bushell S. R., Perugini M. A., Lithgow T., Mulhern T. D., Bottomley S. P., Rossjohn J. (2004) A biophysical analysis of the tetratricopeptide repeat-rich mitochondrial import receptor, Tom70, reveals an elongated monomer that is inherently flexible, unstable, and unfolds via a multistate pathway. J. Biol. Chem. 279, 46448–46454 [DOI] [PubMed] [Google Scholar]
- 48. Gromiha M. M., Selvaraj S. (2004) Inter-residue interactions in protein folding and stability. Prog. Biophys. Mol. Biol. 86, 235–277 [DOI] [PubMed] [Google Scholar]
- 49. Vigneron S., Prieto S., Bernis C., Labbé J. C., Castro A., Lorca T. (2004) Kinetochore localization of spindle checkpoint proteins. Who controls whom? Mol. Biol. Cell 15, 4584–4596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Martin-Lluesma S., Stucke V. M., Nigg E. A. (2002) Role of Hec1 in spindle checkpoint signaling and kinetochore recruitment of Mad1/Mad2. Science 297, 2267–2270 [DOI] [PubMed] [Google Scholar]
- 51. Jelluma N., Brenkman A. B., van den Broek N. J., Cruijsen C. W., van Osch M. H., Lens S. M., Medema R. H., Kops G. J. (2008) Mps1 phosphorylates Borealin to control Aurora B activity and chromosome alignment. Cell 132, 233–246 [DOI] [PubMed] [Google Scholar]
- 52. Yang Z., Kenny A. E., Brito D. A., Rieder C. L. (2009) Cells satisfy the mitotic checkpoint in Taxol and do so faster in concentrations that stabilize syntelic attachments. J. Cell Biol. 186, 675–684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tanudji M., Shoemaker J., L'Italien L., Russell L., Chin G., Schebye X. M. (2004) Gene silencing of CENP-E by small interfering RNA in HeLa cells leads to missegregation of chromosomes after a mitotic delay. Mol. Biol. Cell 15, 3771–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fava L., Kaulich M., Nigg E., Santamaria A. (2011) Probing the in vivo function of Mad1:C-Mad2 in the spindle assembly checkpoint. EMBO J. 30, 3322–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hübner N. C., Wang L. H., Kaulich M., Descombes P., Poser I., Nigg E. A. (2010) Re-examination of siRNA specificity questions role of PICH and Tao1 in the spindle checkpoint and identifies Mad2 as a sensitive target for small RNAs. Chromosoma 119, 149–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Niault T., Hached K., Sotillo R., Sorger P. K., Maro B., Benezra R., Wassmann K. (2007) Changing Mad2 levels affects chromosome segregation and spindle assembly checkpoint control in female mouse meiosis I. PloS One 2, e1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kang J., Chen Y., Zhao Y., Yu H. (2007) Autophosphorylation-dependent activation of human Mps1 is required for the spindle checkpoint. Proc. Natl. Acad. Sci. U.S.A. 104, 20232–20237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tyler R. K., Chu M. L., Johnson H., McKenzie E. A., Gaskell S. J., Eyers P. A. (2009) Phosphoregulation of human Mps1 kinase. Biochem. J. 417, 173–181 [DOI] [PubMed] [Google Scholar]
- 59. Jelluma N., Brenkman A. B., McLeod I., Yates J. R., 3rd, Cleveland D. W., Medema R. H., Kops G. J. (2008) Chromosomal instability by inefficient Mps1 auto-activation due to a weakened mitotic checkpoint and lagging chromosomes. PloS One 3, e2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mattison C. P., Old W. M., Steiner E., Huneycutt B. J., Resing K. A., Ahn N. G., Winey M. (2007) Mps1 activation loop autophosphorylation enhances kinase activity. J. Biol. Chem. 282, 30553–30561 [DOI] [PubMed] [Google Scholar]
- 61. Zhao Y., Chen R. H. (2006) Mps1 phosphorylation by MAP kinase is required for kinetochore localization of spindle-checkpoint proteins. Curr. Biol. 16, 1764–1769 [DOI] [PubMed] [Google Scholar]
- 62. Vié V., Legardinier S., Chieze L., Le Bihan O., Qin Y., Sarkis J., Hubert J. F., Renault A., Desbat B., Le Rumeur E. (2010) Specific anchoring modes of two distinct dystrophin rod subdomains interacting in phospholipid Langmuir films studied by atomic force microscopy and PM-IRRAS. Biochim. Biophys. Acta 1798, 1503–1511 [DOI] [PubMed] [Google Scholar]
- 63. Dubreil L., Vié V., Beaufils S., Marion D., Renault A. (2003) Aggregation of puroindoline in phospholipid monolayers spread at the air-liquid interface. Biophys. J. 85, 2650–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kiyomitsu T., Obuse C., Yanagida M. (2007) Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev. Cell 13, 663–676 [DOI] [PubMed] [Google Scholar]
- 65. Klebig C., Korinth D., Meraldi P. (2009) Bub1 regulates chromosome segregation in a kinetochore-independent manner. J. Cell Biol. 185, 841–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Saurin A. T., van der Waal M. S., Medema R. H., Lens S. M., Kops G. J. (2011) Aurora B potentiates Mps1 activation to ensure rapid checkpoint establishment at the onset of mitosis. Nat. Commun. 2, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kiyomitsu T., Murakami H., Yanagida M. (2011) Protein interaction domain mapping of human kinetochore protein Blinkin reveals a consensus motif for binding of spindle assembly checkpoint proteins Bub1 and BubR1. Mol. Cell. Biol. 31, 998–1011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.