Full text
PDF


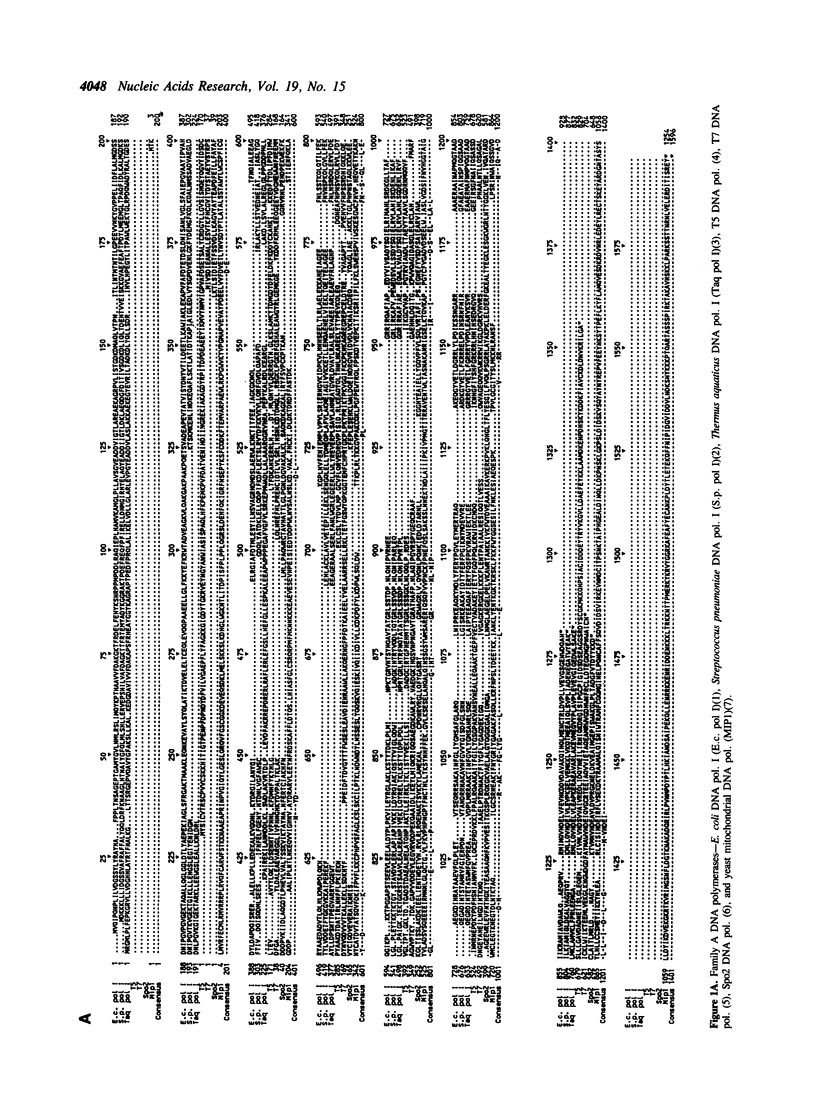


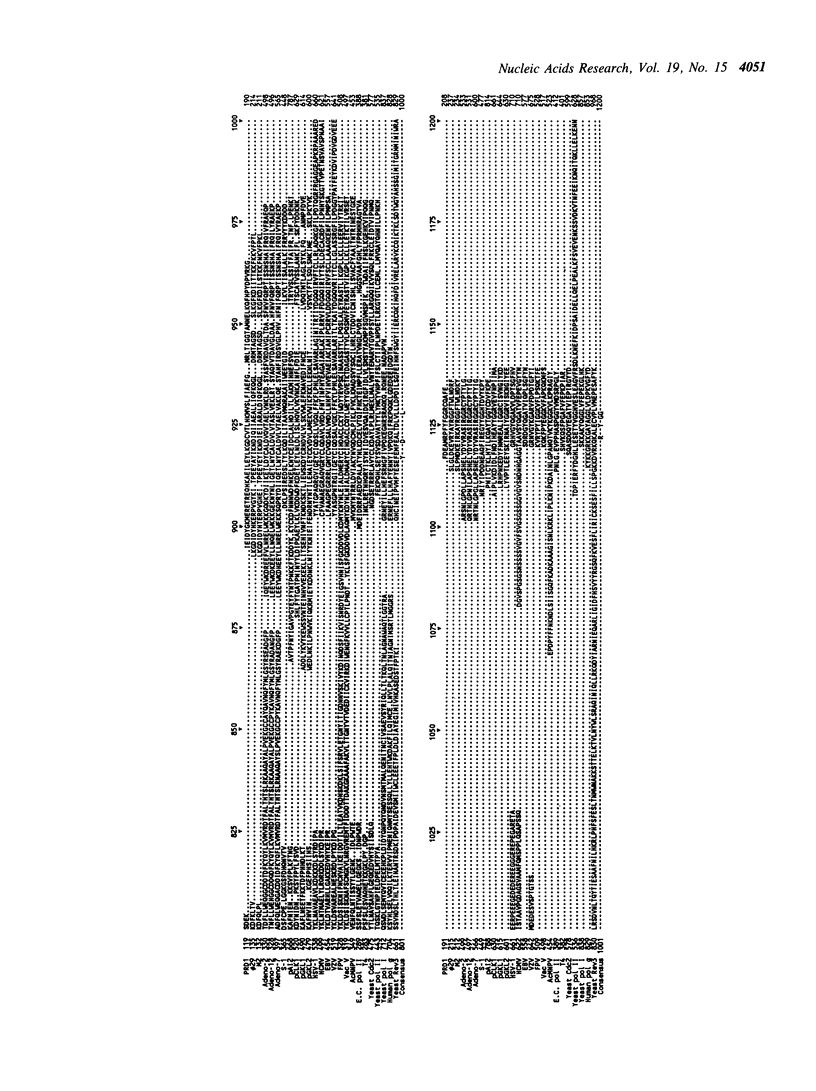

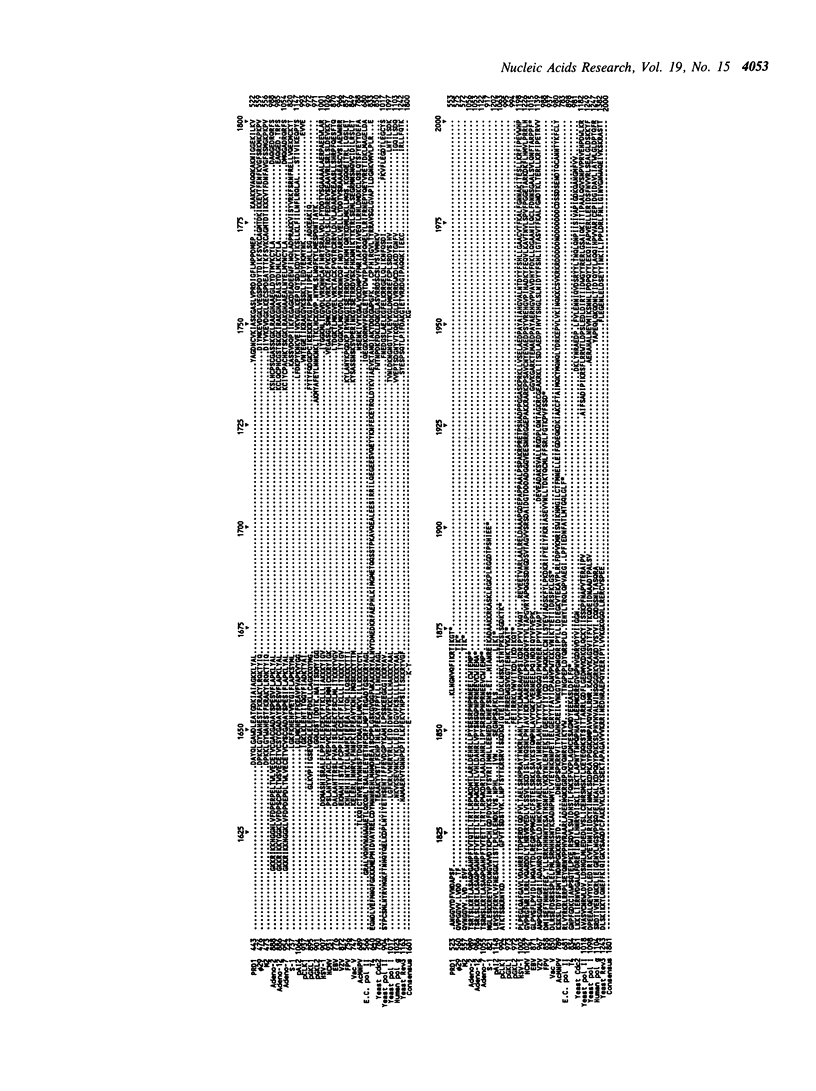
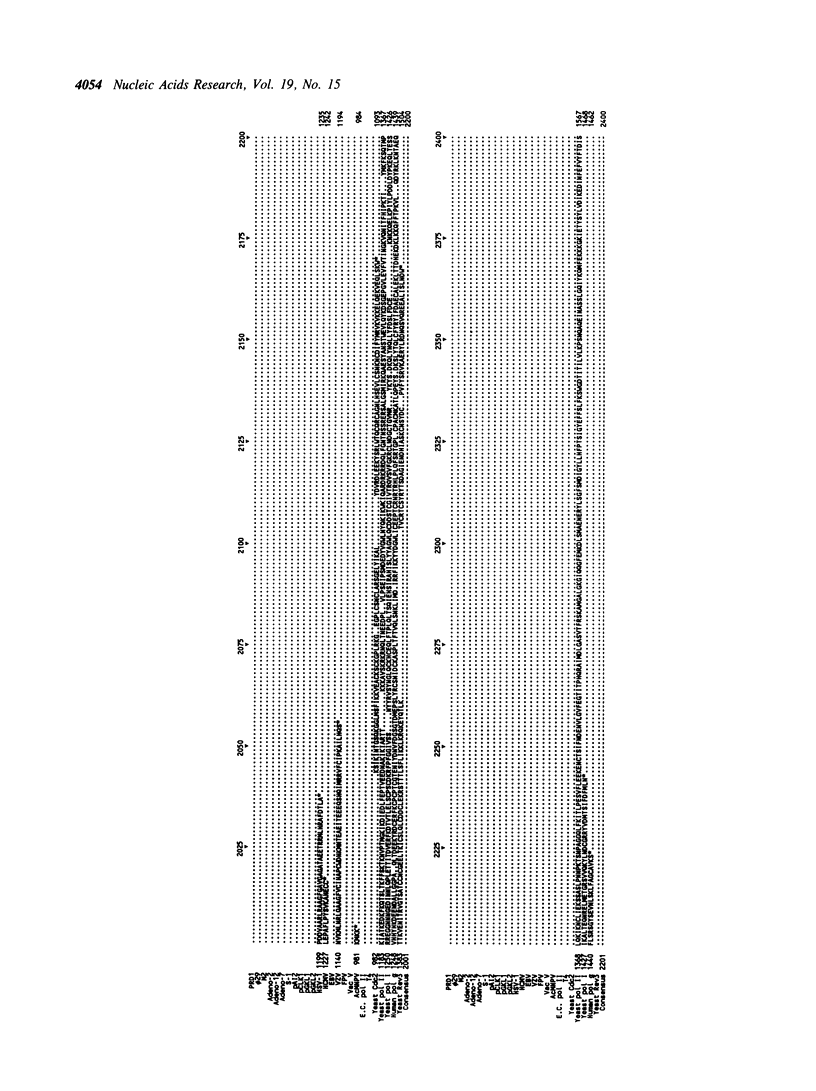

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abbotts J., SenGupta D. N., Zmudzka B., Widen S. G., Notario V., Wilson S. H. Expression of human DNA polymerase beta in Escherichia coli and characterization of the recombinant enzyme. Biochemistry. 1988 Feb 9;27(3):901–909. doi: 10.1021/bi00403a010. [DOI] [PubMed] [Google Scholar]
- Argos P., Tucker A. D., Philipson L. Primary structural relationships may reflect similar DNA replication strategies. Virology. 1986 Mar;149(2):208–216. doi: 10.1016/0042-6822(86)90122-4. [DOI] [PubMed] [Google Scholar]
- Baer R., Bankier A. T., Biggin M. D., Deininger P. L., Farrell P. J., Gibson T. J., Hatfull G., Hudson G. S., Satchwell S. C., Séguin C. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984 Jul 19;310(5974):207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- Beck P. J., Gonzalez S., Ward C. L., Molineux I. J. Sequence of bacteriophage T3 DNA from gene 2.5 through gene 9. J Mol Biol. 1989 Dec 20;210(4):687–701. doi: 10.1016/0022-2836(89)90102-2. [DOI] [PubMed] [Google Scholar]
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Bernad A., Lázaro J. M., Salas M., Blanco L. The highly conserved amino acid sequence motif Tyr-Gly-Asp-Thr-Asp-Ser in alpha-like DNA polymerases is required by phage phi 29 DNA polymerase for protein-primed initiation and polymerization. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernad A., Zaballos A., Salas M., Blanco L. Structural and functional relationships between prokaryotic and eukaryotic DNA polymerases. EMBO J. 1987 Dec 20;6(13):4219–4225. doi: 10.1002/j.1460-2075.1987.tb02770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binns M. M., Stenzler L., Tomley F. M., Campbell J., Boursnell M. E. Identification by a random sequencing strategy of the fowlpoxvirus DNA polymerase gene, its nucleotide sequence and comparison with other viral DNA polymerases. Nucleic Acids Res. 1987 Aug 25;15(16):6563–6573. doi: 10.1093/nar/15.16.6563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco L., Bernad A., Salas M. MIP1 DNA polymerase of S. cerevisiae: structural similarity with the E. coli DNA polymerase I-type enzymes. Nucleic Acids Res. 1991 Feb 25;19(4):955–955. doi: 10.1093/nar/19.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner C. A., Hays S., McEntee K., Goodman M. F. DNA polymerase II is encoded by the DNA damage-inducible dinA gene of Escherichia coli. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7663–7667. doi: 10.1073/pnas.87.19.7663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H., Lawrence C. B., Bryan S. K., Moses R. E. Aphidicolin inhibits DNA polymerase II of Escherichia coli, an alpha-like DNA polymerase. Nucleic Acids Res. 1990 Dec 11;18(23):7185–7186. doi: 10.1093/nar/18.23.7185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J., Scott J. E. The complete DNA sequence of varicella-zoster virus. J Gen Virol. 1986 Sep;67(Pt 9):1759–1816. doi: 10.1099/0022-1317-67-9-1759. [DOI] [PubMed] [Google Scholar]
- Dekker B. M., van Ormondt H. The nucleotide sequence of fragment HindIII-C of human adenovirus type 5 DNA (map positions 17.1-31.7). Gene. 1984 Jan;27(1):115–120. doi: 10.1016/0378-1119(84)90244-0. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky D. I., Crumpacker C. S. Site-specific mutagenesis of a highly conserved region of the herpes simplex virus type 1 DNA polymerase gene. J Virol. 1990 Mar;64(3):1394–1397. doi: 10.1128/jvi.64.3.1394-1397.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Earl P. L., Jones E. V., Moss B. Homology between DNA polymerases of poxviruses, herpesviruses, and adenoviruses: nucleotide sequence of the vaccinia virus DNA polymerase gene. Proc Natl Acad Sci U S A. 1986 Jun;83(11):3659–3663. doi: 10.1073/pnas.83.11.3659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler J. A., Hoppe M. S., van Bree M. P. The nucleotide sequence of the genes encoded in early region 2b of human adenovirus type 7. Gene. 1983 Jan-Feb;21(1-2):145–159. doi: 10.1016/0378-1119(83)90156-7. [DOI] [PubMed] [Google Scholar]
- Foury F. Cloning and sequencing of the nuclear gene MIP1 encoding the catalytic subunit of the yeast mitochondrial DNA polymerase. J Biol Chem. 1989 Dec 5;264(34):20552–20560. [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Hall J. D., Mount D. W., Retondo M. J., Weller S. K., Coen D. M. Sequence and mapping analyses of the herpes simplex virus DNA polymerase gene predict a C-terminal substrate binding domain. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7969–7973. doi: 10.1073/pnas.82.23.7969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingeras T. R., Sciaky D., Gelinas R. E., Bing-Dong J., Yen C. E., Kelly M. M., Bullock P. A., Parsons B. L., O'Neill K. E., Roberts R. J. Nucleotide sequences from the adenovirus-2 genome. J Biol Chem. 1982 Nov 25;257(22):13475–13491. [PubMed] [Google Scholar]
- Hall J. D. Modeling functional sites in DNA polymerases. Trends Genet. 1988 Feb;4(2):42–46. doi: 10.1016/0168-9525(88)90065-0. [DOI] [PubMed] [Google Scholar]
- Hammond R. A., Barnes M. H., Mack S. L., Mitchener J. A., Brown N. C. Bacillus subtilis DNA polymerase III: complete sequence, overexpression, and characterization of the polC gene. Gene. 1991 Feb 1;98(1):29–36. doi: 10.1016/0378-1119(91)90100-p. [DOI] [PubMed] [Google Scholar]
- Ito J., Braithwaite D. K. Yeast mitochondrial DNA polymerase is related to the family A DNA polymerases. Nucleic Acids Res. 1990 Nov 25;18(22):6716–6716. doi: 10.1093/nar/18.22.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Ishino Y., Toh H., Nakata A., Shinagawa H. Escherichia coli DNA polymerase II is homologous to alpha-like DNA polymerases. Mol Gen Genet. 1991 Apr;226(1-2):24–33. doi: 10.1007/BF00273583. [DOI] [PubMed] [Google Scholar]
- Joyce C. M., Kelley W. S., Grindley N. D. Nucleotide sequence of the Escherichia coli polA gene and primary structure of DNA polymerase I. J Biol Chem. 1982 Feb 25;257(4):1958–1964. [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Hsieh J. C., Ito J. Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8287–8291. doi: 10.1073/pnas.84.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Schultz M., Ito J. Site-specific mutagenesis of PRD1 DNA polymerase: mutations in highly conserved regions of the family B DNA polymerase. Biochem Biophys Res Commun. 1990 Aug 16;170(3):1294–1300. doi: 10.1016/0006-291x(90)90534-t. [DOI] [PubMed] [Google Scholar]
- Kempken F., Meinhardt F., Esser K. In organello replication and viral affinity of linear, extrachromosomal DNA of the ascomycete Ascobolus immersus. Mol Gen Genet. 1989 Sep;218(3):523–530. doi: 10.1007/BF00332419. [DOI] [PubMed] [Google Scholar]
- Koiwai O., Yokota T., Kageyama T., Hirose T., Yoshida S., Arai K. Isolation and characterization of bovine and mouse terminal deoxynucleotidyltransferase cDNAs expressible in mammalian cells. Nucleic Acids Res. 1986 Jul 25;14(14):5777–5792. doi: 10.1093/nar/14.14.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouzarides T., Bankier A. T., Satchwell S. C., Weston K., Tomlinson P., Barrell B. G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987 Jan;61(1):125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancy E. D., Lifsics M. R., Munson P., Maurer R. Nucleotide sequences of dnaE, the gene for the polymerase subunit of DNA polymerase III in Salmonella typhimurium, and a variant that facilitates growth in the absence of another polymerase subunit. J Bacteriol. 1989 Oct;171(10):5581–5586. doi: 10.1128/jb.171.10.5581-5586.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larder B. A., Kemp S. D., Darby G. Related functional domains in virus DNA polymerases. EMBO J. 1987 Jan;6(1):169–175. doi: 10.1002/j.1460-2075.1987.tb04735.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawyer F. C., Stoffel S., Saiki R. K., Myambo K., Drummond R., Gelfand D. H. Isolation, characterization, and expression in Escherichia coli of the DNA polymerase gene from Thermus aquaticus. J Biol Chem. 1989 Apr 15;264(11):6427–6437. [PubMed] [Google Scholar]
- Leavitt M. C., Ito J. T5 DNA polymerase: structural--functional relationships to other DNA polymerases. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4465–4469. doi: 10.1073/pnas.86.12.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez P., Martinez S., Diaz A., Espinosa M., Lacks S. A. Characterization of the polA gene of Streptococcus pneumoniae and comparison of the DNA polymerase I it encodes to homologous enzymes from Escherichia coli and phage T7. J Biol Chem. 1989 Mar 5;264(7):4255–4263. [PubMed] [Google Scholar]
- Matsukage A., Nishikawa K., Ooi T., Seto Y., Yamaguchi M. Homology between mammalian DNA polymerase beta and terminal deoxynucleotidyltransferase. J Biol Chem. 1987 Jul 5;262(19):8960–8962. [PubMed] [Google Scholar]
- Matsumoto K., Takano H., Kim C. I., Hirokawa H. Primary structure of bacteriophage M2 DNA polymerase: conserved segments within protein-priming DNA polymerases and DNA polymerase I of Escherichia coli. Gene. 1989 Dec 14;84(2):247–255. doi: 10.1016/0378-1119(89)90498-8. [DOI] [PubMed] [Google Scholar]
- McHenry C. S. DNA polymerase III holoenzyme of Escherichia coli: components and function of a true replicative complex. Mol Cell Biochem. 1985 Feb;66(1):71–85. doi: 10.1007/BF00231826. [DOI] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Morrison A., Christensen R. B., Alley J., Beck A. K., Bernstine E. G., Lemontt J. F., Lawrence C. W. REV3, a Saccharomyces cerevisiae gene whose function is required for induced mutagenesis, is predicted to encode a nonessential DNA polymerase. J Bacteriol. 1989 Oct;171(10):5659–5667. doi: 10.1128/jb.171.10.5659-5667.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M. M., Brown N. C. Inhibition of a discrete bacterial DNA polymerase by 6-(p-hydroxyphenylazo)-uracil and 6-(p-hydroxyphenylazo-)-isocytosine. Nat New Biol. 1972 Nov 15;240(98):80–82. doi: 10.1038/newbio240080a0. [DOI] [PubMed] [Google Scholar]
- Oeser B., Tudzynski P. The linear mitochondrial plasmid pClK1 of the phytopathogenic fungus Claviceps purpurea may code for a DNA polymerase and an RNA polymerase. Mol Gen Genet. 1989 May;217(1):132–140. doi: 10.1007/BF00330952. [DOI] [PubMed] [Google Scholar]
- Paces V., Vlcek C., Urbánek P., Hostomský Z. Nucleotide sequence of the major early region of Bacillus subtilis phage PZA, a close relative of phi 29. Gene. 1985;38(1-3):45–56. doi: 10.1016/0378-1119(85)90202-1. [DOI] [PubMed] [Google Scholar]
- Paillard M., Sederoff R. R., Levings C. S. Nucleotide sequence of the S-1 mitochondrial DNA from the S cytoplasm of maize. EMBO J. 1985 May;4(5):1125–1128. doi: 10.1002/j.1460-2075.1985.tb03749.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson R. C., Cheung L. C., Mattaliano R. J., White S. T., Chang L. M., Bollum F. J. Expression of human terminal deoxynucleotidyl transferase in Escherichia coli. J Biol Chem. 1985 Sep 5;260(19):10495–10502. [PubMed] [Google Scholar]
- Pizzagalli A., Valsasnini P., Plevani P., Lucchini G. DNA polymerase I gene of Saccharomyces cerevisiae: nucleotide sequence, mapping of a temperature-sensitive mutation, and protein homology with other DNA polymerases. Proc Natl Acad Sci U S A. 1988 Jun;85(11):3772–3776. doi: 10.1073/pnas.85.11.3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polesky A. H., Steitz T. A., Grindley N. D., Joyce C. M. Identification of residues critical for the polymerase activity of the Klenow fragment of DNA polymerase I from Escherichia coli. J Biol Chem. 1990 Aug 25;265(24):14579–14591. [PubMed] [Google Scholar]
- Reha-Krantz L. J. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J Mol Biol. 1988 Aug 20;202(4):711–724. doi: 10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- Rådén B., Rutberg L. Nucleotide sequence of the temperate Bacillus subtilis bacteriophage SPO2 DNA polymerase gene L. J Virol. 1984 Oct;52(1):9–15. doi: 10.1128/jvi.52.1.9-15.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savilahti H., Bamford D. H. The complete nucleotide sequence of the left very early region of Escherichia coli bacteriophage PRD1 coding for the terminal protein and the DNA polymerase. Gene. 1987;57(1):121–130. doi: 10.1016/0378-1119(87)90183-1. [DOI] [PubMed] [Google Scholar]
- Scheuermann R., Tam S., Burgers P. M., Lu C., Echols H. Identification of the epsilon-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci U S A. 1983 Dec;80(23):7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SenGupta D. N., Zmudzka B. Z., Kumar P., Cobianchi F., Skowronski J., Wilson S. H. Sequence of human DNA polymerase beta mRNA obtained through cDNA cloning. Biochem Biophys Res Commun. 1986 Apr 14;136(1):341–347. doi: 10.1016/0006-291x(86)90916-2. [DOI] [PubMed] [Google Scholar]
- Shepard D., Oberfelder R. W., Welch M. M., McHenry C. S. Determination of the precise location and orientation of the Escherichia coli dnaE gene. J Bacteriol. 1984 May;158(2):455–459. doi: 10.1128/jb.158.2.455-459.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu L. M., Hong J. S., Wei Y. F., Engler J. A. Nucleotide sequence of the genes encoded in early region 2b of human adenovirus type 12. Gene. 1986;46(2-3):187–195. doi: 10.1016/0378-1119(86)90403-8. [DOI] [PubMed] [Google Scholar]
- Smith H. O., Annau T. M., Chandrasegaran S. Finding sequence motifs in groups of functionally related proteins. Proc Natl Acad Sci U S A. 1990 Jan;87(2):826–830. doi: 10.1073/pnas.87.2.826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer E. K., Rush J., Fung C., Reha-Krantz L. J., Karam J. D., Konigsberg W. H. Primary structure of T4 DNA polymerase. Evolutionary relatedness to eucaryotic and other procaryotic DNA polymerases. J Biol Chem. 1988 Jun 5;263(16):7478–7486. [PubMed] [Google Scholar]
- Stark M. J., Mileham A. J., Romanos M. A., Boyd A. Nucleotide sequence and transcription analysis of a linear DNA plasmid associated with the killer character of the yeast Kluyveromyces lactis. Nucleic Acids Res. 1984 Aug 10;12(15):6011–6030. doi: 10.1093/nar/12.15.6011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomalski M. D., Wu J. G., Miller L. K. The location, sequence, transcription, and regulation of a baculovirus DNA polymerase gene. Virology. 1988 Dec;167(2):591–600. [PubMed] [Google Scholar]
- Tomasiewicz H. G., McHenry C. S. Sequence analysis of the Escherichia coli dnaE gene. J Bacteriol. 1987 Dec;169(12):5735–5744. doi: 10.1128/jb.169.12.5735-5744.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tommasino M., Ricci S., Galeotti C. L. Genome organization of the killer plasmid pGK12 from Kluyveromyces lactis. Nucleic Acids Res. 1988 Jul 11;16(13):5863–5878. doi: 10.1093/nar/16.13.5863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsurumi T., Maeno K., Nishiyama Y. Nucleotide sequence of the DNA polymerase gene of herpes simplex virus type 2 and comparison with the type 1 counterpart. Gene. 1987;52(2-3):129–137. doi: 10.1016/0378-1119(87)90039-4. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Wong S. W., Wahl A. F., Yuan P. M., Arai N., Pearson B. E., Arai K., Korn D., Hunkapiller M. W., Wang T. S. Human DNA polymerase alpha gene expression is cell proliferation dependent and its primary structure is similar to both prokaryotic and eukaryotic replicative DNA polymerases. EMBO J. 1988 Jan;7(1):37–47. doi: 10.1002/j.1460-2075.1988.tb02781.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H., Ito J. Nucleotide sequence of the major early region of bacteriophage phi 29. Gene. 1982 Mar;17(3):323–335. doi: 10.1016/0378-1119(82)90149-4. [DOI] [PubMed] [Google Scholar]



