SUMMARY
Mitochondrial dysfunction causes poorly understood tissue-specific pathology stemming from primary defects in respiration, coupled with altered reactive oxygen species (ROS), metabolic signaling and apoptosis. The A1555G mtDNA mutation that causes maternally inherited deafness disrupts mitochondrial ribosome function, in part, via increased methylation of the mitochondrial 12S rRNA by the methyltransferase mtTFB1. In patient-derived A1555G cells, we show that 12S rRNA hyper-methylation causes ROS-dependent activation of AMP kinase and the pro-apoptotic nuclear transcription factor E2F1. This retrograde mitochondrial-stress relay is operative in vivo as transgenic-mtTFB1 mice exhibit enhanced 12S rRNA methylation in multiple tissues, increased E2F1 and apoptosis in the stria vascularis and spiral ganglion neurons of the inner ear, and progressive E2F1-dependent hearing loss. This transgenic-mtTFB1 mouse mitochondrial disease model provides a robust platform for deciphering the complex tissue-specificity of human mitochondrial-based disorders, as well as the precise pathogenic mechanism of maternally inherited deafness and its exacerbation by environmental factors.
INTRODUCTION
Mitochondria are multi-functional cellular organelles involved in oxidative metabolism, ion homeostasis, signal transduction, and apoptosis and hence contribute to human disease by a variety of mechanisms. Mitochondrial pathogenesis is complex and involves maternally inherited diseases due to mutations in mtDNA, as well as Mendelian-inherited disease due to mutations in nuclear genes required for mitochondrial function (Dimauro and Schon, 2003). Furthermore, mitochondrial dysfunction is implicated in more common disorders such as diabetes, heart disease, neurodegeneration, and age-related diseases (Wallace, 2005, Shadel 2008), making efforts to better understand of how mitochondria cause and exacerbate human disease pathology imperative and of broad significance.
A major hurdle in understanding mitochondrial disease pathogenesis is the current lack of understanding of the often-extreme tissue specificity involved (DiMauro and Schon, 2003). Because defective oxidative phosphorylation (OXPHOS) per se can be pathogenic, this is often attributed to variable energetic thresholds in different cell types. However, defective OXPHOS is also frequently associated with increased mitochondrial reactive oxygen species (ROS) that are a major cause of pathology because they promote molecular damage, oxidative stress, and cell death. Finally, mitochondria and ROS are involved in signal transduction pathways ({Hamanaka and Chandel, 2010) that are likely an underappreciated downstream cause of pathology due to mitochondrial dysfunction. For example, a major signaling node that reacts to mitochondrial dysfunction is the AMP kinase pathway, which responds to cellular energy decline that can occur when OXPHOS is disrupted (Hardie, 2007), and is regulated by ROS (Emerling et al., 2009; Quintero et al., 2006).
A hallmark case of tissue-specific mitochondrial disease pathology is maternally inherited deafness caused by the A1555G mutation in human mtDNA (Prezant et al., 1993). This relatively common mutation (Vandebona et al., 2009) causes non-syndromic and/or aminoglycoside-induced deafness and occurs in the 12S rRNA gene, encoding the RNA component of the small (28S) mitochondrial ribosome subunit. It is also in close proximity to an evolutionarily conserved stem-loop that contains two tandem adenine residues that are methylated by the site-specific rRNA adenine N6-di-methyltransferase, h-mtTFB1/TFB1M (McCulloch et al., 2002; Seidel-Rogol et al., 2003). This methylation occurs during ribosome biogenesis in bacteria (Pulicherla et al., 2009) and is essential in mice, the lack of which disrupts mitochondrial 28S ribosome subunit assembly (Metodiev et al., 2009).
Hearing loss caused by the A1555G mutation is irreversible (Prezant et al., 1993), implying death of critical, irreplaceable cells in the inner ear by an unknown pathogenic cell-death pathway. The A1555G mutation alters mitochondrial ribosome function and translation (Cotney et al., 2009; Guan et al., 1996, 2000; Hobbie et al., 2008), which in turn causes OXPHOS defects that are thought to contribute to deafness pathology. However, mitochondrial translation and OXPHOS defects observed in patient-derived primary and cytoplasmic hybrid (cybrid) cell lines are mild and highly dependent on the nuclear genetic background (Guan et al., 1996, 2000), suggesting that other aspects of mitochondrial dysfunction may also be operative. Finally, polymorphisms near the TFB1M gene encoding h-mtTFB1 are nuclear modifiers of the A1555G deafness phenotype (Bykhovskaya et al., 2004), suggesting a connection between 12S rRNA methylation and hearing loss (Shadel, 2004b).
We recently showed that patient-derived A1555G cybrids are hyper-methylated at the 12S rRNA stem-loop methylated by h-mtTFB1 (Cotney et al., 2009). These A1555G cybrids exhibit defective mitochondrial biogenesis and membrane potential and heightened sensitivity to stress-induced apoptosis (Cotney et al., 2009), apparently due to disrupted coordination of overall mitochondrial biogenesis with the assembly of mitochondrial ribosomes (Cotney et al., 2007). Remarkably, these phenotypes are shared by HeLa cells with wild-type mtDNA that instead have hyper-methylated 12S rRNA due to over-expression of h-mtTFB1, leading us to conclude that hyper-methylation of mitochondrial ribosomes per se is a key molecular defect driving the apoptotic phenotype (Cotney et al., 2009).
In the current study, we tested the hypothesis that this hyper-methylation of mitochondrial ribosomes instigates a unique form of mitochondrial stress signaling involved in the deafness pathology of the A1555G mutation. We show that increased mitochondrial ROS generated in A1555G cybrids activate the pro-apoptotic nuclear transcription factor E2F1 in an AMP kinase-dependent manner, and that over-expression of mtTFB1 in mice (to model pathogenesis due to increased mitochondrial 12S rRNA methylation) is sufficient to cause progressive hearing loss associated with tissue-specific up-regulation of E2F1 and apoptosis of critical cells of the inner ear.
RESULTS
A mitochondrial stress-response pathway activates nuclear transcription factor E2F1
We hypothesized that mitochondrial defects due to 12S rRNA hyper-methylation elicit a mitochondrial retrograde stress signal (Butow and Avadhani, 2004) that explains the apoptotic susceptibility we documented previously in A1555G and h-mtTFB1 methyltransferase over-expression cell lines (Cotney et al., 2009). Therefore, we performed microarray analysis on these cell lines, which exhibit 12S hyper-methylation for different reasons (i.e. due to the A1555G point mutation or over-expression of the h-mtTFB1), reasoning that this would define a common 12S methylation transcriptional signature, which, in turn, would allow elements of this mitochondrial stress-response pathway to be deciphered.
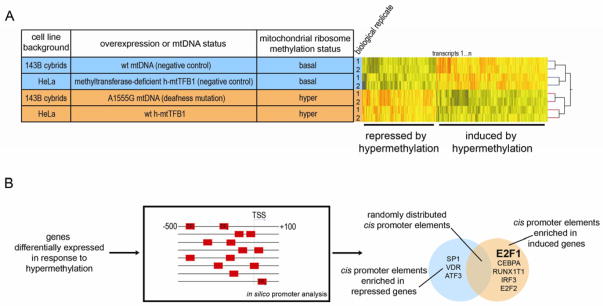
Expression microarrays were performed on A1555G cybrids, using analogous 143B cybrids containing the corresponding wild-type mtDNA (“A” at position 1555) as the negative control, and on HeLa cells over-expressing h-mtTFB1 compared to those expressing a methyltransferase-deficient G65A mutant of h-mtTFB1 as a negative control (Figure 1A). We identified genes that were significantly up-regulated or down-regulated in the hyper-methylation cell lines (Table S1), which we reasoned would include genes that were responding to 12S hyper-methylation specifically. We then analyzed the promoters of this set of hyper-methylation-responsive genes (Figure 1B) for transcription factor binding sites and found that E2F1 sites were the most frequently over-represented in genes up-regulated in 12S hyper-methylation (Table 1). We next crossed our microarray dataset with a published dataset identifying E2F1-responsive genes and found significant overlap (Table S2), again confirming E2F1 induction in our experimental system. We also prepared a list of E2F1 targets by selecting human genes whose promoter is predicted to contain cis-elements for E2F1, and found that there is a significant enrichment of E2F1 targets in the transcripts induced by 12S rRNA hyper-methylation (Table S2), again suggesting an increase in E2F1 transcriptional activity.
Figure 1. Integrative genomic analysis reveals up-regulation of nuclear transcription factor E2F1 in response to mitochondrial stress induced by mitochondrial ribosome hyper-methylation.
(A) Table showing the four cell lines on which we performed microarray analysis to develop a signature of the gene expression response to mitochondrial 12S hyper-methylation: two control lines with “basal” mitochondrial 12S hyper-methylation (blue) and two experimental cell lines with 12S hyper-methylation (orange), with the number of biological replicates indicated. To the right of the table is the representative heat map of all replicates of the cell lines analyzed, with transcripts that are induced or repressed depending on 12S hyper-methylation status delineated. “wt” indicates wild-type with regard to either mtDNA or h-mtTFB1. Transcripts whose expression was significantly different (t-test p-value<0.05) between the two groups (“basal” versus “hyper”-methylated) represented a 12S hyper-methylation signature list that was analyzed further (total of 4333 transcripts, 2492 induced and 1841 repressed).
(B) The promoter regions (−500 to +100 bp of the predicted transcription initiation site) of the genes that were differentially expressed were analyzed for known transcription factor binding sites. The Venn diagram indicates the top transcription factors identified in the repressed (blue) and induced (orange) genes. Since E2F1 was the most over-represented binding site in the genes induced by 12S rRNA hyper-methylation it became a focus of this study.
Table 1.
Transcription factors whose cis-elements are detected in the promoters of transcripts up-regulated in response to 12S rRNA hyper-methylation but not in the promoters of the down-regulated transcripts. The p-value was calculated using Fisher’s exact test.
| Transcription Factor | Up-regulated target genes | Down-regulated target genes | p-value (Fisher’s exact test) |
|---|---|---|---|
| E2F1 | 50 | 0 | 0.00010 |
| CEBPA | 43 | 0 | 0.00012 |
| RUNX1T1 | 15 | 0 | 0.0010 |
| IRF3 | 13 | 0 | 0.0018 |
| E2F2 | 9 | 0 | 0.0128 |
E2F1 mediates pro-apoptotic signaling in cells containing the human deafness-associated A1555G mutation
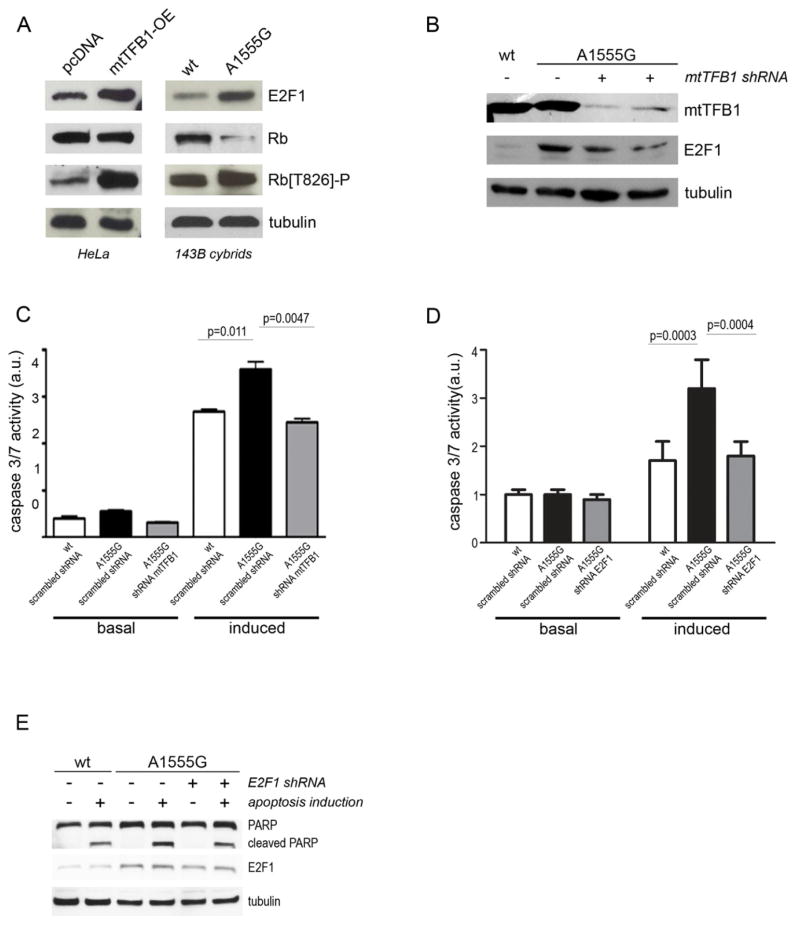
E2F1 is one of a family of eight E2F transcription factors that are involved in cell cycle regulation through their cyclin-dependent interactions with pocket proteins, including the tumor suppressor Rb (DeGregori and Johnson, 2006). Of particular significance, E2F1 is involved in pro-apoptotic signaling (Field et al., 1996; Hou et al., 2000; Polager and Ginsberg, 2009). This connection to cell death, in conjunction with our microarray results, made E2F1 a likely candidate for a stress-responsive protein that is responding to mitochondrial dysfunction caused by the A1555G mutation and perhaps also the causative factor in the observed apoptosis susceptibility in the 12S hyper-methylation cell lines (Cotney et al., 2009). Entirely consistent with this concept, we confirmed E2F1 transcriptional induction in cells with 12S rRNA hyper-methylation by q-PCR (Figure S1A), and found that the steady-state level of E2F1 protein was increased in A1555G cybrids and in HeLa cells that over-express h-mtTFB1 compared to appropriate control lines (Figure 2A and Figure S1B). In addition, we observed a decrease in the steady-state amount of Rb (Figure 2A and S1B) and/or its increased phosphorylation (Figure 2A and S1B), either of which is consistent with increased E2F1 activity in the 12S hyper-methylated cell lines.
Figure 2. Hyper-methylation of mitochondrial 12S rRNA leads to activation of E2F1 and E2F1-dependent apoptosis.
(A) Western blot analysis of whole cell extracts for E2F1, Rb and phosphorylated-Rb, (Rb[T268]-P) in HeLa cells that over-express h-mtTFB1 (h-mtTFB1-OE) compared to cell containing an empty-vector negative control (pcDNA), and in human 143B cybrids containing wild-type (wt) or A1555G mtDNA. Tubulin was probed as a loading control. Quantification is shown in Figure S1B.
(B) Effect of h-mtTFB1 knock-down (to reduce 12S hyper-methylation) on E2F1 levels in A1555G cybrids by Western blot using tubulin as a loading control. Two independent knock-down constructs were used (+), and a scrambled shRNA was used as a negative control (−). 143B cybrids with wild-type (wt) mtDNA treated with the negative control shRNA are shown in lane 1. Quantification is shown in Figure S1C.
(C) Quantitation of apoptosis (caspase 3/7 activity) in 143B cybrids containing wild-type (wt) or A1555G mtDNA without (−) or with (+) h-mtTFB1 knocked down by shRNA (see Figure S2A). Basal and induced apoptosis refer to the presence and absence of staurosporine. The values plotted represent mean ± standard deviation (n=12), with t-test p-values indicated.
(D) Quantitation of apoptosis (caspase 3/7 activity) in 143B cybrids containing wild-type (wt) or A1555G mtDNA without (−) or with (+) E2F1 knocked down by shRNA. Basal and induced apoptosis refer to the presence and absence of staurosporine. The values plotted represent mean ± standard deviation (n=12), with t-test p-values indicated.
(E) Western blot analysis of full-length and cleaved PARP and of E2F1 in wild-type (wt) and A1555G treated with scrambled (−) or E2F1 (+) shRNA without (−) or with (+) induction of apoptosis by starosporine. E2F1, full-length (PARP) and caspase-3-cleaved forms of PARP are shown, with tubulin probed as a loading control. Quantification is shown in Figure S1D.
Given the association of active E2F1 with apoptosis, we next tested whether the up-regulation of E2F1 per se in A1555G cybrids is involved in their heightened sensitivity to apoptotic cell death. In strong support of this concept, we found that suppressing the hyper-methylation of the 12S rRNA via knock-down of h-mtTFB1 abolished E2F1 induction (Figure 2B and S1C) and reduced the susceptibility of A1555G cybids to apoptosis (Figure 2C). Furthermore, reduction of E2F1 to near wild-type levels by shRNA in the A1555G cybrids abrogated the increase in apoptosis susceptibility, as measured by caspase 3/7 activity (Figure 2D) and accumulation of the cleaved form of PARP (Figure 2E and S1D). Altogether, these results indicate that the Rb-E2F1 apoptotic pathway responds to mitochondrial stress induced by hyper-methylation of 12S rRNA in mitochondrial ribosomes.
ROS-dependent AMPK activation underlies E2F1 induction in A1555G cells
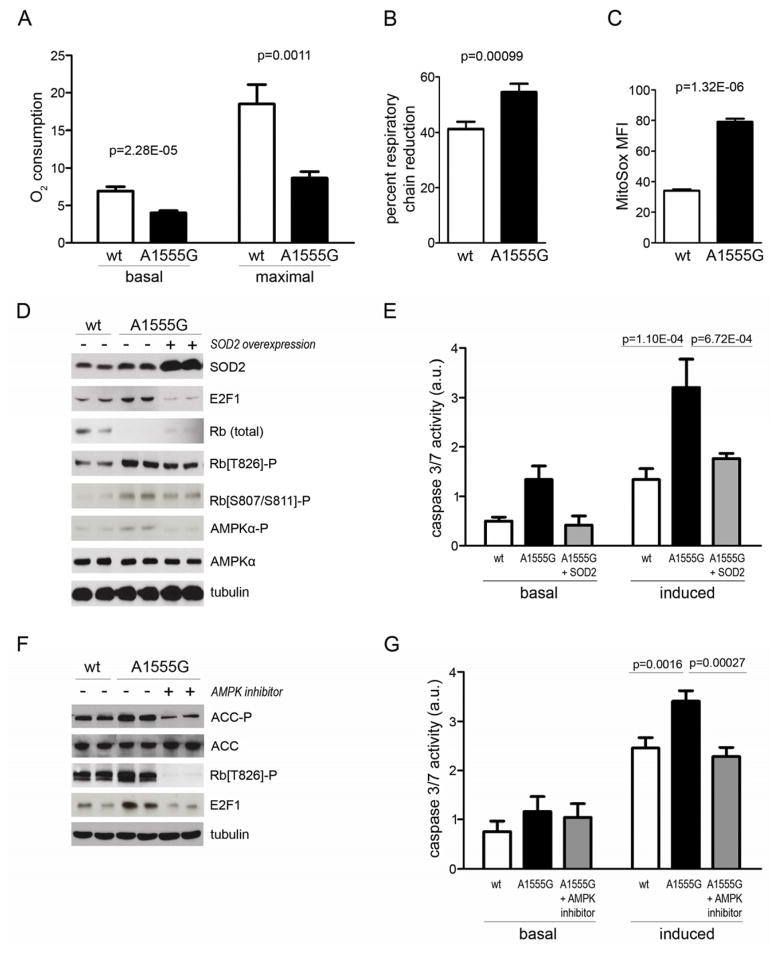
To determine the proximal signal leading to E2F1 activation we examined respiratory function in A1555G and corresponding wild-type cybrids and found that the former have decreased basal and maximal mitochondrial O2 consumption rates (Figure 3A). Furthermore, the respiratory chain in the A1555G cells is reduced (Figure 3B), a condition conducive to increased ROS production (e.g. superoxide) (Jones et al., 2005). Accordingly, we found that A1555G cells have increased mitochondrial superoxide, as measured by MitoSox (Figure 3C). To determine if this increased superoxide was underlying pro-apoptotic signaling to E2F1 in A1555G cybrids, we overexpressed mitochondrial superoxide dismutase 2 (SOD2) and found that this eliminated E2F1 induction, increased the total amount of Rb, decreased Rb hyper-phosphorylation and eliminated the apoptosis susceptibility in A1555G cells (Figure 3D and E).
Figure 3. A pro-apoptotic mitochondrial stress-signaling pathway that involves mitochondrial ROS-dependent activation of E2F1 by AMP kinase.
(A) Basal and maximal (+ FCCP uncoupler) mitochondrial O2 consumption rates in 143B cybrids containing wild-type (wt) or A1555G mtDNA are shown. The mean ± standard deviation (n=24) is plotted, with t-test p-values indicated.
(B) Degree of respiratory chain reduction in the same cells as in (A). This is the ratio of basal to maximal respiration, the latter of which was considered 100% reduction (i.e. the chain handling the maximal amount of electrons). The mean ± standard deviation (n=12) is plotted, with t-test p-values indicated.
(C) Mitochondrial ROS in 143B cybrids containing wild-type (wt) or A1555G mtDNA as measured by FACS analysis of MitoSox staining. The mean fluorescence intensity ± standard deviation is plotted (n=3), with t-test p-values indicated.
(D) Western blot analysis of A1555G cybrids that overexpress SOD2. Immunoblotting of E2F1, Rb, phospho-Rb T826 and S807/811, AMPKα and phospho-AMPKα T172 are shown in A1555G cybrids transfected with the SOD2 over-expression vector (lanes 5–6), and in control cybrids with wild-type mtDNA (“wt”, lanes 1–2) and A1555G mtDNA (lanes 3–4) transfected with empty vector. Lanes 1–2, 3–4 and 5–6 represent independent biological replicates. Tubulin was probed as a loading control.
(E) Quantitation of apoptosis (caspase 3/7 activity) in 143B cybrids containing wild-type (wt) mtDNA (white bars) or A1555G mtDNA without (black bars) or with SOD2 over-expression (grey bars). Basal and induced apoptosis refer to the presence and absence of staurosporine. The values plotted represent mean ± standard deviation (n=12), with t-test p-values indicated.
(F) Effect of pharmacological AMPK inhibition on the levels of acetyl-CoA carboxylase (ACC), phospho-ACC S79 (ACC-P), phospho-Rb T826 and E2F1 in A1555G cybrids (lanes 5–6), determined by western blot of whole cell extracts using tubulin as a loading control. Lanes 1–2 (wild-type cybrids) and 3–4 (A1555G cybrids) are the vehicle controls (−). The lanes 1–2, 3–4 and 5–6 represent independent biological replicates.
(G) Quantitation of apoptosis (caspase 3/7 activity) in 143B cybrids containing wild-type (wt) mtDNA (white bars), or A1555G mtDNA without (black bars) or with AMPK inhibition (grey bars). Basal and induced apoptosis refer to the presence and absence of staurosporine. The values plotted represent mean ± standard deviation (n=12), with t-test p-values indicated.
We next sought to determine how mitochondrial superoxide is sensed and promotes E2F1 activation. AMP-dependent kinases (AMPK) are established sensors of mitochondrial dysfunction. Furthermore, AMPK activity is activated by superoxide (Quintero et al., 2006) and regulates E2F1 by phosphorylating Rb (Dasgupta and Milbrandt, 2009) making it a potential candidate in the stress pathway we have elucidated. Accordingly, we found that A1555G cybrids have increased active AMPKα (phospho-AMPKα) compared to wild-type controls, and that AMPKα activity was restored close to normal upon over-expression of SOD2 (Figure 3D). To determine if the increased AMPK activity was linked to E2F1 induction in A1555G cells, we treated A1555G cells with AMPK inhibitor compound C, which was confirmed by reduced phosphorylation of AMPK target acetyl-CoA carboxylase (ACC). Pharmacological inactivation of AMPK in A1555G cybrids reduced Rb phosphorylation and E2F1 levels (Figure 3F), and lowered apoptosis susceptibility of the A1555G cells (Figure 3G). Taken together, these results strongly implicate AMPK as a ROS-sensitive component of the mitochondrial stress-response pathway that leads to pro-apoptotic E2F1 signaling in cells containing the deafness-causing A1555G mutation.
Transgenic mice that over-express the mitochondrial rRNA methyltransferase mtTFB1 exhibit increased mitochondrial 12S rRNA methylation in multiple tissues and progressive hearing loss
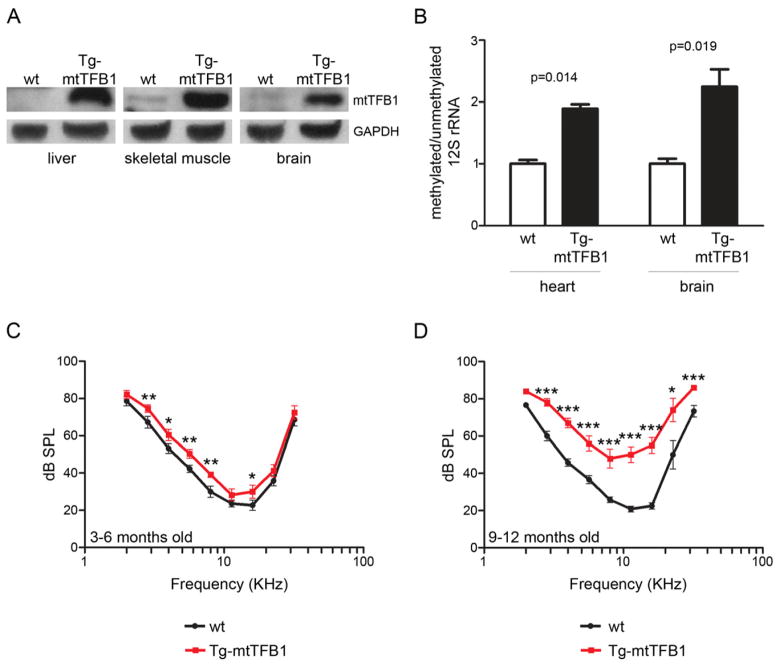
The cell culture studies above strongly implicate an E2F1-dependent, pro-apoptotic mitochondrial stress signaling pathway downstream of mitochondrial 12S hyper-methylation as a potential pathogenic component of deafness caused by the A1555G mtDNA mutation. To test this hypothesis directly, we generated transgenic mice that globally over-express the mouse mtTFB1 rRNA methyltransferase (Tg-mtTFB1) responsible for the specific mitochondrial 12S methylation event implicated (Seidel-Rogol et al., 2003). These animals are viable and fertile, and have increased mtTFB1 protein in multiple tissues (Figure 4A) that corresponds with increased 12S rRNA methylation (Figure 4B). When we measured hearing function in Tg-mtTFB1 mice by the auditory brainstem response (ABR) and compared them to age-matched, non-transgenic littermate controls, we found that at 3–6 months of age Tg-mtTFB1 mice have higher ABR thresholds across the entire range of frequencies, indicating significant hearing loss (Figure 4C). Similar hearing-threshold defects are observed in patients with the A1555G mutation (Noguchi et al., 2004). This hearing loss in the Tg-mtTFB1 animals progresses dramatically with age, with a much greater difference in ABR thresholds in animals aged to 9–12 months (Figure 4D). It is important to note that, even though the C57BL/6 genetic background used here exhibits premature age-related hearing loss (Someya et al., 2009), significant decreases in ABR in these mice are not observed until 12–15 month of age, which is beyond the timeframe we analyzed in this study.
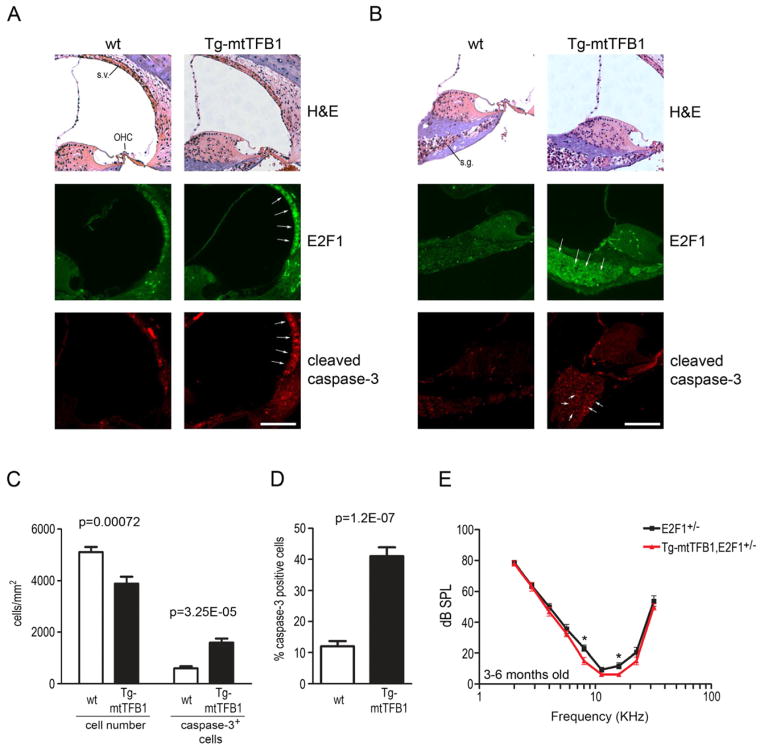
Figure 4. Transgenic mice that over-express the mtTFB1 rRNA methyltransferase exhibit 12S hyper-methylation and progressive hearing loss.
(A) Western blot of mtTFB1 in wild-type (wt) and transgenic mtTFB1 (Tg-mtTFB1) mouse tissues using GAPDH as loading control.
(B) Quantification of the degree of 12S rRNA methylation in Tg-mtTFB1 mouse heart and brain tissues using a methylation-sensitive mitochondrial 12S rRNA primer-extension assay (Cotney et al., 2009). The values plotted represent mean of methylated/unmethylated ± standard deviation (n=3), with t-test p-values indicated.
(C) Hearing thresholds determined by ABR analysis of Tg-mtTFB1 (red points) and control wild-type (wt), non-transgenic littermate control mice (black points) are shown in the audiogram (frequency vs. threshold curve). The threshold (in sound pressure level, dB SPL) was tested with 5-dB resolution and frequencies in half octave steps from 32 to 2 kHz. The values plotted represent mean ± standard error of the mean, with t-test p-values indicated (*, p<0.05; **, p<0.01; ***, p<0.001). Mice were tested at ages 3–6 months (9 wt mice and 12 Tg-mtTFB1 mice were used).
(D) same as in (C) except mice were tested at ages 9–12 months (5 wild-type mice and 6 Tg-mtTFB1 mice were used).
E2F1 induction and apoptosis in critical inner ear cells of Tg-mtTFB1 mice
The cochlea of the Tg-mtTFB1 mice display a largely normal structure, with no obvious loss of the hair cells in the organ of Corti (Figure 5A, upper panel) or obvious disruption of the spiral ganglion (Figure 5B, upper panel). Not observing significant hair cell dysfunction or apoptosis even in one-year-old animals with severe hearing loss, we turned our attention to other critical inner ear cell types. We found that E2F1 protein was increased in the stria vascularis (Figure 5A, middle panel) and in the spiral ganglion neurons (Figure 5B, middle panel) of Tg-mtTFB1 mice, but not in the organ of Corti. This tissue-specific up-regulation of E2F1 correlated with in increased caspase 3/7 staining in the stria vascularis (Figure 5A, bottom panel) and spiral ganglion neurons (Figure 5B, bottom panel), consistent with E2F1 enacting a pro-apoptotic signal in critical cells involved in inner ear homeostasis and hearing. To confirm the caspase-3 activation in the spiral ganglion of Tg-mtTFB1 mice, we measured the number of spiral ganglion neurons and the number of caspase-3-positive neurons, and found that Tg-mtTFB1 mice have ~30% fewer spiral ganglion neurons and a 3-fold increase in the number of caspase-3-positive cells (Figure 5C). Consequently, the percentage of caspase-3-positive cells is 4-fold higher in Tg-mtTFB1 spiral ganglion (Figure 5D).
Figure 5. Deafness pathology in Tg-mtTFB1 mice involves up-regulation of E2F1 and apoptosis in the stria vascularis and spinal ganglion neurons of the inner ear.
(A) H&E staining of the organ of Corti and stria vascularis (top panels) from representative 1-year old wild-type (wt, left column) and Tg-mtTFB1 mice (right column). The middle panels show immunohistochemistry staining for E2F1 and the lower panels show immunohistochemistry for cleaved caspase-3 in similar representative sections. White arrows indicate areas where significant changes in signal are observed. Stria vascularis (s.v.), outer hair cells (OHC), scale bar, 100μm. See also Figure S2.
(B) same as in (A) but sections that highlight staining in the spiral ganglion neurons are shown. Spiral ganglion (s.g.), scale bar, 100μm.
(C) quantification of cellular density in the spiral ganglion (the left side of the plot) and of caspase-3 positive spiral ganglion neurons (right side). The sections were obtained from 3 wild-type and 3 Tg-mtTFB1 mice at 1 year of age, and six independent sections were counted for each animal. The values plotted represent mean ± standard deviation, with t-test p-values indicated.
(D) same as in (C), but expressing the percentage of caspase-3 positive cells in the spiral ganglion. The values plotted represent mean ± standard deviation, with t-test p-values indicated.
(E) ABR analysis of E2F1+/− mice (black points) and E2F1+/−/Tg-mtTFB1 mice (red points) performed as described in Figure 4C (n=). Mice were tested at ages 3–6 months (8 E2F1+/− mice and 10 E2F1+/−/Tg-mtTFB1 mice were used). The values plotted represent mean ± standard error of the mean, with t-test p-values indicated (*, p<0.05).
In support of the cell-type specificity of the mitochondrial-stress pathway to E2F1 in vivo, we found that E2F1 is not induced in other tissues of Tg-mtTFB1 mice (Figure S2A). Interestingly, activation of AMPK, measured by phosphorylation of both AMPKα and ACC, is detectable in some tissues (skeletal muscle and heart) but not in the liver. To further confirm the tissue-specificity of E2F1 activation, we measured the transcript levels of E2F1, and found an increase in the stria vascularis, but not in the organ of Corti or in the brain (Figure S2B). Furthermore, there is increased phospho-AMPK in the stria vascularis of Tg-mtTFB1 mice (Figure S2C). Finally, analysis of available microarray datasets from the mtDNA-mutator mouse (Trifunovic et al., 2004), which exhibits age-related pathology including deafness (Someya et al., 2008), revealed that E2F1 transcripts and its targets are up-regulated in the cochlea, but not in skeletal muscle (Figure S2D and Suppl. Table 3), further supporting the role of E2F1 in cell death in the inner ear.
E2F1 mediates hearing loss in Tg-mtTFB1 mice
In order to determine if E2F1 induction per se has a causal role in the observed hearing loss in this new mouse model of deafness, we crossed the Tg-mtTFB1 mice with E2F1 knock-out mice, to generate mice heterozygous for E2F1 that either over-express Tg-mtTFB1 (E2F1+/−,Tg-mtTFB1) or not (E2F1+/−). Evaluation of hearing function in these animals by ABR revealed that over-expression of mtTFB1 no longer promoted significant hearing loss in the E2F1+/− genetic background (Figure 5E), which has reduced expression of E2F1. These results confirm a causal role for activation of E2F1 above a certain threshold level in the hearing loss observed in Tg-mtTFB1 mice.
DISCUSSION
In this study we endeavored to gain new insight into the complexity and tissue specificity of mitochondrial-based diseases by studying the common human mtDNA mutation A1555G that has one primary outcome, deafness, and modeling its pathogenic mechanism in mice. Based on our results, we conclude that a major pathogenic driving force is cell death due to activation of the pro-apoptotic nuclear transcription factor E2F1 by a cell-type-specific mitochondrial stress pathway. Below we will discuss the results that led us to this conclusion and the potential broader implications of this study with regard to the documented environmental exacerbation of this form of deafness (Warchol, 2010), age-related hearing loss (presbycusis), and other diseases where mitochondria are implicated in pathogenesis.
We propose that mitochondrial stress due the hyper-methylation of the mtDNA-encoded 12S rRNA is a critical component of the inner ear pathology associated with deafness caused by the human mtDNA A1555G mutation. This is perhaps best demonstrated by our results in the Tg-mtTFB1 transgenic mice, which exhibit 12S rRNA hyper-methylation in multiple tissues and activation of the pro-apoptotic transcription factor E2F1 that causes progressive hearing loss similar to that observed in human A1555G patients. Deafness is the primary effect of the A1555G mutation even though most patients are homoplasmic (i.e. have 100% mutant mtDNA) in all cells and tissues (Fischel-Ghodsian, 1999; Prezant et al., 1993). This implies a cell-type specific response to mitochondrial dysfunction is at play. Our results showing that up-regulation of E2F1 and increased cell death occurs in the stria vascularis and in the spiral ganglion neurons, but not in the organ of Corti, is consistent with activation of E2F1 by mitochondrial stress occurring only in certain critical cells in the inner ear and their subsequent apoptosis via this pathway.
E2F1-dependent cell death in the stria vascularis and spiral ganglion neurons is consistent with the pattern of hearing loss across all frequencies that we observe in the Tg-mtTFB1 mice. With regard to the irreversible nature of the deafness, neurodegeneration in spiral ganglion itself could itself be responsible, as has been proposed for age-related hearing loss (Someya et al., 2009). Alternatively, or in addition, dysfunction of the stria vascularis and spiral ganglion neurons could ultimately result in a higher susceptibility of irreplaceable inner hair cells to undergo cell death (Fetoni et al., 2011). Furthermore, since activation of the cell cycle in post-mitotic neuronal cells is an established trigger for apoptosis (Chen et al., 2003; Hou et al., 2000; Shadel, 2004a) the mitochondrial stress pathway to E2F1 identified herein may also may make some cells of the inner ear inherently more susceptible to cell death due to being forced into the cell cycle, as opposed to activation the E2F1 apoptosis pathway per se. Based on these considerations, we propose that A1555G patients would likewise be more prone to eventual loss of critical inner ear cells via this mitochondrial stress pathway to E2F1, explaining their irreversible hearing loss either spontaneously, as a function of age, or in response to environmental stimuli such as noise or aminoglycosides. Even though our results strongly implicate 12S hyper-methylation per se in the deafness phenotype in mice, it is likely that defects in mitochondrial translation caused by the A1555G mutation, such as infidelity (Hobbie et al., 2008), conspire with hyper-methylation to produce the precise deafness pathology in humans and/or mediate its exacerbation by aminoglycoside antibiotics. Therefore, it remains important to determine precisely which mechanisms and cell types within the inner ear instigate deafness pathology in this new mouse model and in human A1555G patients. A full understanding of these events may be therapeutic in this regard and may also help in advancing efforts to regenerate hearing function.
Our results in A1555G cybrids also provide key insight into the nature of the pathogenic mitochondrial stress-signaling pathway. Specifically, hyper-methylation of mitochondrial ribosomes, which are needed for translational and assembly of the respiratory chain (Bonawitz et al., 2006), disrupts mitochondrial respiration in a manner that increases ROS production (Figure 3). We propose that increased mitochondrial superoxide is sensed by AMPK (Emerling et al., 2009; Quintero et al., 2006), activation of which relays the stress signal to E2F1. Since E2F1 activity is associated with pro-apoptotic signaling and is necessary for the enhanced cell death on A1555G cybrids (Figure 2D) and hearing loss in Tg-mtTFB1 mice (Figure 5E), we speculate ROS- and AMPK-dependent activation of E2F1 is the major mitochondrial stress-signaling pathway involved under these circumstances. Going forward it will be important to determine precisely how E2F1 is regulated by ROS and AMPK.
The marked tissue-specificity of E2F1 induction and, to a lesser degree, of AMPK activation (Figure S2A), provides insight into the complex mechanisms underlying tissue-specificity of mitochondrial diseases. The requirement for multiple sequential steps to fully activate the mitochondrial ROS-AMPK-E2F1 apoptotic pathway provides a variety of opportunities for different tissues to suppress a pathogenic mechanism. For example, tissues with lower OXPHOS activity or better redox buffering could prevent initial activation of the pathogenic mechanism. Alternatively, in other tissues, AMPK may not respond to ROS, E2F1 may not be activated by AMPK, or E2F1 may not be a potent trigger of apoptosis, any of which would presumably prevent a pathogenic outcome. These observations may be generalizable, in that other pathogenic mitochondrial retrograde signaling pathways likely exist with their differential activation or readout in tissues contributing to the complex tissue specificity observed in mitochondrial diseases.
Finally, the induction of E2F1 in response to a mitochondrial malfunction may represent a paradigm in mitochondrial pathogenesis where the cause of the disease is not the immediate OXPHOS dysfunction, but instead the misinterpretation of resulting retrograde signals generated by mitochondria. The pivotal role of AMPK as a sensor of energy charge and now as a ROS-dependent regulator of E2F1 activity highlights the complexity of mitochondrial stress responses and the need for additional research to uncover other such pathways. Finally, it is tempting to speculate that aberrant mitochondrial stress signaling may also be of general significance in diseases and circumstances where mitochondrial dysfunction and apoptosis are implicated, such as heart disease, cancer, neurodegenerative diseases and aging.
EXPERIMENTAL PROCEDURES
Cell culture, shRNA-mediated gene knock-down, FACS analysis and 12S methylation assays
143B osteosarcroma cybrids containing either wild-type (A1555A) or mutant (A1555G) mtDNA (Guan et al., 2001) were grown in DMEM high glucose (Sigma D5648) plus 10% fetal calf serum at 37°C and 5% CO 2. Prior to all experiments, cells were plated at 5000 cells/cm2 and grown for 96 hours to 70–80% confluence. When puromycin was used as for selection, it was removed at least 24 hours prior to harvesting cells. HeLa cells that over-express h-mtTFB1 have been described (Cotney et al., 2009; Cotney et al., 2007).
Retroviral-mediated shRNA knock-down of mtTFB1 and E2F1 by was carried as described (Cotney et al., 2009) with the following modifications. Wild-type or mutant 143B cybrids were transfected and plated at 5000/cm2 in DMEM with 10% fetal calf serum and 600 ng/ml puromycin and used for experiments at 70–80% confluence. Puromycin was removed 48 hours prior to analysis. Target sequences ligated into the pSiren-RetroQ vector (Clonetech) for mtTFB1 and E2F1 knock-down, as well the scrambled negative-control shRNA used were described previously (Cotney et al., 2009, Goto et al., 2006). To overexpress SOD2, cybrids were transfected with pCMV-SPORT6-SOD2 (Open Biosystems MMM4769-99609684) or pcDNA3.0 as a negative control as described (Cotney et al., 2009). To inhibit AMPK, cybrids were treated for 16h with 10μM of compound C (Calbiochem) and then used immediately for experiments.
To analyze mitochondrial superoxide by FACS, wild-type and A1555G cybrids were treated with 50 nM MitoSOX (Invitrogen) for 20 minutes at 37°C, rinsed and detached with trypsin. Fluorescence was measured using a FACScan (BD Biotechnology) and analyzed using FlowJo software. Primer-extension analysis of mitochondrial 12S rRNA adenine dimethylation was performed on 1–5 μg of total cellular or tissue RNA prepared using the RNEasy Kit (Qiagen) as described (Cotney et al., 2007; Seidel-Rogol et al., 2003), except 2 μM each of dATP, dCTP, dTTP and 0.5 μM ddGTP were used.
Western blotting
Whole cell extracts of cultured human cells were prepared in 1.5% n-dodecylmaltoside in PBS as described (Raimundo et al., 2009). Gels were loaded with 50 μg of total protein per well, and the following antibodies were used for immunoblotting: E2F1 (Santa Cruz, sc193), mtTFB1/TFBM1 (McCulloch and Shadel, 2003) or that provided by Dr. Craig Cameron, tubulin (Thermo, DM1A), Rb (BD Pharmingen, 554136), phospho-Rb (T826) (Abcam, ab4779-50), phospho-Rb (S807/811) (Cell Signaling, 9308S), AMPKα (Cell Signaling, 2603), phospho-AMPKα (T172) (Cell Signaling, 2535S), acetyl-CoA carboxylase (ACC) (Cell Signaling, 3676), phospho-ACC (S79) (Cell Signaling, 3661), SOD2 (Assay Designs, SOD-111), PARP (Cell Signaling, 9542) and GAPDH (Ambion, AM4300). Western blots were performed on 3–4 biological replicates and at least three technical replicates.
Apoptosis Assays
To induce E2F1-dependent apoptosis, cybrids were treated with 200 nM staurosporine for 3 hours in the presence of 10 mM wortmannin as described (Hallstrom et al., 2008). Cells were then rinsed and used for two different apoptosis assays. Caspase 3/7 activity was determined using a plate reader one-hour after treatment with Caspase-Glo reagent (Promega) as described by the manufacturer, and full-length and cleaved PARP were assayed by Western blot.
Oxygen consumption analysis
Cells were plated in XF96 plates (SeaHorse Biosciences) at 10000 cells/well and the next day cellular O2 consumption was determined in a SeaHorse Bioscience XF96 extracellular flux analyzer according to manufacturer instructions. Cells were maintained at 37°C in normal growth medium without serum. In some wells 8μM of the mitochondrial uncoupler FCCP was added.
Microarrays and data analysis
Total cellular RNA from the cultured cells was prepared using an RNeasy mini RNA extraction kit (Qiagen GmbH, Hilden, Germany) and used for the expression microarray procedure in conjunction with the Yale University W.M. Keck Foundation Biotechnology Resource Laboratory. RNA integrity was first verified by an Agilent Bioanalyzer and then amplified, labelled and hybridized onto GeneChip HG-U133 Plus 2.0 arrays (Affymetrix, Agilent, Santa Clara, U.S.A.) using standard protocols recommended by the manufacturer, starting from 5 μg of total RNA. Data were normalized by the RMA method (Irizarry et al., 2003), using commercial software (GeneSpring, Agilent, Santa Clara, California, U.S.A.). For each biological sample, two RNA samples were prepared from two independently collected cell pellets. To analyze the expression data, samples were grouped according to their 12S rRNA methylation status (Figure 1A). The four samples in the hyper-methylation group were compared to the four samples with basal methylation to control for non-specific effects of over-expression of mtTFB1 independent of increased 12S rRNA methylation and in an attempt to subtract effects due to cell nuclear genetic background. Transcripts whose expression was significantly changed between the two groups formed the 12S rRNA hyper-methylation list (4333 transcripts; Table S1). The student’s t-test was used to determine statistically significant changes in expression, with a cut-off p-value of 0.05 (Raimundo et al., 2009). The false-positive rate was determined to be 3.8% as described (Hovatta et al., 2005). The correlation of the Q-PCR with the microarray results for six genes tested was R2=0.77 (Raimundo et al., 2009).
To determine which cis-elements were present in the promoters of the genes up- or down-regulated in 12S rRNA hyper-methylated samples, we used the Bibliosphere software (Genomatix Software GmbH, Munich, Germany). To minimize false positives, a cut-off of at least two cis element-gene co-citations was implemented. Enrichment in cis-elements of the genes up- or down-regulated was determined by Fisher’s exact test.
Generation and analysis of transgenic mice that over-express mtTFB1
A mouse mtTFB1 cDNA was amplified with primers containing EcoR1 and EcoRV restriction sites and ligated into the vector pCAGGS that contains a CMV immediate early-enhancer/chicken β-actin/rabbit β-globin (CAG) promoter (Niwa et al., 1991) and confirmed by sequencing. Purified pCAGGS/mtTFB1 plasmid was digested with SalI and PstI, to liberate a fragment containing the enhancer, promoter, mtTFB1 ORF, and poly-A tail. This linear DNA fragment was then used by the Yale Animal Genomic Services to microinject pro-nuclear, C57BL/6J X SJL/J embryos. Seventy-four animals were born from the six foster mothers. After genotyping, one female (F1) founder was identified that contained the transgene. This female was then mated with a wild-type C57BL/6J male to generate the second generation (F2) and then serially to create subsequent generations. Female F3 to F5 transgenics and control non-transgenic littermates were used for all of the experiments.
For analysis of mouse samples, tissues from 9-month old mice were harvested, immediately frozen in liquid nitrogen and stored at -80C. The tissues were ground using a mortar and pestle previously frozen at -80C, kept on dry ice during the procedure. Whole tissue homogenates were prepared as described (Cotney et al., 2009). Primer extension analysis and q-PCR were performed on RNA purified from these same mouse tissues using the RNAeasy kit using the method already described for human cells, but with the following mouse 12S-specific primer: 5′-ATTATTCCAAGC-3′. To generate Tg-mtTFB1/E2F1+/− mice, a C57BL/6J male E2F1−/− was obtained from the Jackson Laboratory (Maine, USA) and mated with F4 Tg-mtTFB1 females, yielding E2F1+/− progeny with and without the mtTFB1 transgene.
Auditory Brainstem Response and Cochlear Histology
Detailed methods for the ABR analysis can be found in the Supplemental Materials. A total of 21 animals (9 Tg-mtTFB1 and 12 wild-type littermates) between the ages of 3 and 6 months, and 11 animals between 9–12 months (5 wild-type, 6 Tg-mtTFB1), were used for auditory brainstem response (ABR) testing. The E2F1+/− animals were tested at 3–6 months of age (total of 20, 8 wild-type and 12 Tg-mtTFB1). In all experiments, age-matched littermates were compared directly on the same day and by the same experienced specialist blinded to the genotype of the mice. The mice were subsequently combined into 3–6 and 9–12 groups for data presentation and statistical analysis. Animals were anesthetized with chloral hydrate (480 mg/kg IP) and all recordings were conducted in a sound-attenuating chamber (Industrial Acoustics Corp) using a customized TDT3 system (Tucker-Davis Technologies, Inc.). A t-test was used to determine the statistical significance of the threshold difference at each frequency.
The inner ear was prepared for histology by removing the temporal bone and exposing and rupturing the bulla. The cochlea was perfused with 4% paraformaldehyde (in PBS) using a syringe outfitted with small diameter tubing through the oval and round windows. The tissue was incubated overnight, decalcified in 10% EDTA in PBS for 3 days, and then paraffin embedded, sectioned in 5 mm samples and mounted onto glass slides by the Yale Histology section. Following deparaffinization and rehydration, sections were used for immunohistochemistry and hematoxylin staining. Images were obtained using an Olympus IX71 microscope and Metamorph image analysis software. Nine independent cochlear sections were analyzed from three one-year-old wild-type and three Tg-mtTFB1 mice. Spiral ganglion cells were counted using the DAPI counterstain in an area of 100×100 pixels and normalized to area (mm2). Cells displaying cytoplasmic foci of caspase-3 were considered caspase-3+.
Supplementary Material
Research Highlights.
Mitochondrial stress promotes a pro-apoptotic E2F1 signaling cascade
E2F1 activation is mediated by mitochondrial ROS-dependent AMP kinase signaling
Hearing loss in Tg-mtTFB1 mice models deafness caused by a human mtDNA mutation
Deafness involves tissue-specific E2F1-driven apoptosis in critical inner ear cells
Acknowledgments
This study was supported by grants R01 HL-059655 to G.S.S. and R01 DC000273 to J.S-S from the U.S. NIH and a UMDF fellowship to T.E.S. The authors thank Zimei Zhang for animal husbandry, Yale Animal Genomic Services for help in generating the Tg-mtTFB1 mice, and Dr. Susan Kaech for comments on the manuscript, helpful discussions and access to the SeaHorse.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Bykhovskaya Y, Mengesha E, Wang D, Yang H, Estivill X, Shohat M, Fischel-Ghodsian N. Human mitochondrial transcription factor B1 as a modifier gene for hearing loss associated with the mitochondrial A1555G mutation. Mol Genet Metab. 2004;82:27–32. doi: 10.1016/j.ymgme.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Chen P, Zindy F, Abdala C, Liu F, Li X, Roussel MF, Segil N. Progressive hearing loss in mice lacking the cyclin-dependent kinase inhibitor Ink4d. Nat Cell Biol. 2003;5:422–426. doi: 10.1038/ncb976. [DOI] [PubMed] [Google Scholar]
- Cotney J, McKay SE, Shadel GS. Elucidation of separate, but collaborative functions of the rRNA methyltransferase-related human mitochondrial transcription factors B1 and B2 in mitochondrial biogenesis reveals new insight into maternally inherited deafness. Hum Mol Genet. 2009;18:2670–2682. doi: 10.1093/hmg/ddp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Wang Z, Shadel GS. Relative abundance of the human mitochondrial transcription system and distinct roles for h-mtTFB1 and h-mtTFB2 in mitochondrial biogenesis and gene expression. Nucleic Acids Res. 2007;35:4042–4054. doi: 10.1093/nar/gkm424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta B, Milbrandt J. AMP-activated protein kinase phosphorylates retinoblastoma protein to control mammalian brain development. Dev Cell. 2009;16:256–270. doi: 10.1016/j.devcel.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGregori J, Johnson DG. Distinct and Overlapping Roles for E2F Family Members in Transcription, Proliferation and Apoptosis. Curr Mol Med. 2006;6:739–748. doi: 10.2174/1566524010606070739. [DOI] [PubMed] [Google Scholar]
- DiMauro S, Schon EA. Mitochondrial respiratory-chain diseases. N Engl J Med. 2003;348:2656–2668. doi: 10.1056/NEJMra022567. [DOI] [PubMed] [Google Scholar]
- Emerling BM, Weinberg F, Snyder C, Burgess Z, Mutlu GM, Viollet B, Budinger GR, Chandel NS. Hypoxic activation of AMPK is dependent on mitochondrial ROS but independent of an increase in AMP/ATP ratio. Free Radic Biol Med. 2009;46:1386–1391. doi: 10.1016/j.freeradbiomed.2009.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fetoni AR, Picciotti PM, Paludetti G, Troiani D. Pathogenesis of presbycusis in animal models: A review. Exp Gerontol. 2011 doi: 10.1016/j.exger.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Field SJ, Tsai FY, Kuo F, Zubiaga AM, Kaelin WG, Jr, Livingston DM, Orkin SH, Greenberg ME. E2F-1 functions in mice to promote apoptosis and suppress proliferation. Cell. 1996;85:549–561. doi: 10.1016/s0092-8674(00)81255-6. [DOI] [PubMed] [Google Scholar]
- Fischel-Ghodsian N. Mitochondrial deafness mutations reviewed. Hum Mutat. 1999;13:261–270. doi: 10.1002/(SICI)1098-1004(1999)13:4<261::AID-HUMU1>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Goto Y, Hayashi R, Kang D, Yoshida K. Acute loss of transcription factor E2F1 induces mitochondrial biogenesis in HeLa cells. J Cell Physiol. 2006;209:923–934. doi: 10.1002/jcp.20802. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Biochemical evidence for nuclear gene involvement in phenotype of non-syndromic deafness associated with mitochondrial 12S rRNA mutation. Hum Mol Genet. 1996;5:963–971. doi: 10.1093/hmg/5.7.963. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. A biochemical basis for the inherited susceptibility to aminoglycoside ototoxicity. Hum Mol Genet. 2000;9:1787–1793. doi: 10.1093/hmg/9.12.1787. [DOI] [PubMed] [Google Scholar]
- Guan MX, Fischel-Ghodsian N, Attardi G. Nuclear background determines biochemical phenotype in the deafness-associated mitochondrial 12S rRNA mutation. Hum Mol Genet. 2001;10:573–580. doi: 10.1093/hmg/10.6.573. [DOI] [PubMed] [Google Scholar]
- Hallstrom TC, Mori S, Nevins JR. An E2F1-dependent gene expression program that determines the balance between proliferation and cell death. Cancer Cell. 2008;13:11–22. doi: 10.1016/j.ccr.2007.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanaka RB, Chandel NS. Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem Sci. 2010;35:505–513. doi: 10.1016/j.tibs.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie DG. AMP-activated/SNF1 protein kinases: conserved guardians of cellular energy. Nat Rev Mol Cell Biol. 2007;8:774–785. doi: 10.1038/nrm2249. [DOI] [PubMed] [Google Scholar]
- Hobbie SN, Bruell CM, Akshay S, Kalapala SK, Shcherbakov D, Bottger EC. Mitochondrial deafness alleles confer misreading of the genetic code. Proc Natl Acad Sci U S A. 2008;105:3244–3249. doi: 10.1073/pnas.0707265105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou ST, Callaghan D, Fournier MC, Hill I, Kang L, Massie B, Morley P, Murray C, Rasquinha I, Slack R, et al. The transcription factor E2F1 modulates apoptosis of neurons. J Neurochem. 2000;75:91–100. doi: 10.1046/j.1471-4159.2000.0750091.x. [DOI] [PubMed] [Google Scholar]
- Hovatta I, Tennant RS, Helton R, Marr RA, Singer O, Redwine JM, Ellison JA, Schadt EE, Verma IM, Lockhart DJ, et al. Glyoxalase 1 and glutathione reductase 1 regulate anxiety in mice. Nature. 2005;438:662–666. doi: 10.1038/nature04250. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones RG, Plas DR, Kubek S, Buzzai M, Mu J, Xu Y, Birnbaum MJ, Thompson CB. AMP-activated protein kinase induces a p53-dependent metabolic checkpoint. Mol Cell. 2005;18:283–293. doi: 10.1016/j.molcel.2005.03.027. [DOI] [PubMed] [Google Scholar]
- McCulloch V, Seidel-Rogol BL, Shadel GS. A human mitochondrial transcription factor is related to RNA adenine methyltransferases and binds S-adenosylmethionine. Mol Cell Biol. 2002;22:1116–1125. doi: 10.1128/MCB.22.4.1116-1125.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 interacts with the C-terminal activation region of h-mtTFA and stimulates transcription independently of its RNA methyltransferase activity. Mol Cell Biol. 2003;23:5816–5824. doi: 10.1128/MCB.23.16.5816-5824.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metodiev MD, Lesko N, Park CB, Camara Y, Shi Y, Wibom R, Hultenby K, Gustafsson CM, Larsson NG. Methylation of 12S rRNA is necessary for in vivo stability of the small subunit of the mammalian mitochondrial ribosome. Cell Metab. 2009;9:386–397. doi: 10.1016/j.cmet.2009.03.001. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Polager S, Ginsberg D. p53 and E2f: partners in life and death. Nat Rev Cancer. 2009;9:738–748. doi: 10.1038/nrc2718. [DOI] [PubMed] [Google Scholar]
- Prezant TR, Agapian JV, Bohlman MC, Bu X, Oztas S, Qiu WQ, Arnos KS, Cortopassi GA, Jaber L, Rotter JI, et al. Mitochondrial ribosomal RNA mutation associated with both antibiotic-induced and non-syndromic deafness. Nat Genet. 1993;4:289–294. doi: 10.1038/ng0793-289. [DOI] [PubMed] [Google Scholar]
- Pulicherla N, Pogorzala LA, Xu Z, HCOF, Musayev FN, Scarsdale JN, Sia EA, Culver GM, Rife JP. Structural and functional divergence within the Dim1/KsgA family of rRNA methyltransferases. J Mol Biol. 2009;391:884–893. doi: 10.1016/j.jmb.2009.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero M, Colombo SL, Godfrey A, Moncada S. Mitochondria as signaling organelles in the vascular endothelium. Proc Natl Acad Sci U S A. 2006;103:5379–5384. doi: 10.1073/pnas.0601026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raimundo N, Vanharanta S, Aaltonen LA, Hovatta I, Suomalainen A. Downregulation of SRF-FOS-JUNB pathway in fumarate hydratase deficiency and in uterine leiomyomas. Oncogene. 2009;28:1261–1273. doi: 10.1038/onc.2008.472. [DOI] [PubMed] [Google Scholar]
- Seidel-Rogol BL, McCulloch V, Shadel GS. Human mitochondrial transcription factor B1 methylates ribosomal RNA at a conserved stem-loop. Nat Genet. 2003;33:23–24. doi: 10.1038/ng1064. [DOI] [PubMed] [Google Scholar]
- Shadel GS. Coupling the mitochondrial transcription machinery to human disease. Trends Genet. 2004a;20:513–519. doi: 10.1016/j.tig.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Shadel GS. A dual-function mitochondrial transcription factor tunes out deafness. Mol Genet Metab. 2004b;82:1–3. doi: 10.1016/j.ymgme.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Shadel GS. Expression and maintenance of mitochondrial DNA: new insights into human disease pathology. Am J Pathol. 2008;172:1445–1456. doi: 10.2353/ajpath.2008.071163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Xu J, Kondo K, Ding D, Salvi RJ, Yamasoba T, Rabinovitch PS, Weindruch R, Leeuwenburgh C, Tanokura M, et al. Age-related hearing loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial apoptosis. Proc Natl Acad Sci U S A. 2009;106:19432–19437. doi: 10.1073/pnas.0908786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yamasoba T, Kujoth GC, Pugh TD, Weindruch R, Tanokura M, Prolla TA. The role of mtDNA mutations in the pathogenesis of age-related hearing loss in mice carrying a mutator DNA polymerase gamma. Neurobiol Aging. 2008;29:1080–1092. doi: 10.1016/j.neurobiolaging.2007.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, et al. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature. 2004;429:417–423. doi: 10.1038/nature02517. [DOI] [PubMed] [Google Scholar]
- Vandebona H, Mitchell P, Manwaring N, Griffiths K, Gopinath B, Wang JJ, Sue CM. Prevalence of mitochondrial 1555A-->G mutation in adults of European descent. N Engl J Med. 2009;360:642–644. doi: 10.1056/NEJMc0806397. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warchol ME. Cellular mechanisms of aminoglycoside ototoxicity. Curr Opin Otolaryngol Head Neck Surg. 2010;18:454–458. doi: 10.1097/MOO.0b013e32833e05ec. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.