Abstract
We recently reported that hydrogen peroxide-inducible clone-5 (Hic-5, also named ARA55, androgen receptor-associated protein 55) can bind to the TGF-β signaling regulator Smad3, thereby inhibiting certain Smad3-dependent TGF-β responses. We now show that Hic-5 can also control TGF-β responses through an alternative mechanism involving Smad7, a key negative regulator of TGF-β signaling. Hic-5 binds directly to Smad7. This interaction requires the LIM3 domain of Hic-5, and enhances TGF-β signaling through causing loss of Smad7 protein but not mRNA. Enforced expression of Hic-5 reverses the ability of Smad7 to suppress TGF-β-induced phosphorylation of Smads 2 and 3 and activation of the plasminogen activator inhibitor-1 promoter (in NRP-154 and PC3 prostate carcinoma and WPMY-1 prostate myofibroblast cell lines). Lentiviral-mediated shRNA silencing of endogenous Hic-5 reduced TGF-β responses in PC3 and WPMY-1 cells. Further work suggests that the level of Smad7 is modulated by its physical interaction with Hic-5 and targeted to a degradation pathway not likely to be proteasomal. Our findings support that Hic-5 functions as a cell type-specific activator of TGF-β signaling through its ability to physically interact with and neutralize Smad7.
Keywords: Smad, TGF-β, prostate, Smad3, Smad2, Smad7, PAI-1, Hic-5
Introduction
Transforming growth factor-β (TGF-β1) is a multifunctional cytokine that plays important roles in cell proliferation, cell cycle arrest, differentiation, apoptosis, formation of ECM and immunosuppression (Massague et al., 2000), and functions as a tumor suppressor in many tissues including the prostate (Danielpour, 2005; Song et al., 2003a; Tang et al., 1999). However, a number of studies more recently show that TGF-β also functions as a tumor promoter, particularly in late stage cancers (Wakefield and Roberts, 2002). The mechanisms by which TGF-β suppresses or promotes tumor growth and the molecular switches controlling such differential responses remain poorly understood.
Well-defined transducers of TGF-β signals include two trans-membrane serine/threonine kinase receptors, TβRI and TβRII (Massague et al., 1992; Wrana et al., 1992), receptor Smads (Smad2 and Smad3) and co-Smad (Smad4) (Wrana and Attisano, 1996; Zhang et al., 1996). Smad7 negatively regulates TGF-β signaling (Hayashi et al., 1997; Nakao et al., 1997) through binding to both TβRI and HECT family E3 ubiquitin ligases (i.e., Smad ubiquitination regulatory factors (Smurfs) 1 or 2 (Ebisawa et al., 2001; Kavsak et al., 2000) and WW-domain containing protein 1 (Komuro et al., 2004)). Smad7 recruits such E3 ligases to promote proteasomal degradation of TβRI (Kavsak et al., 2000). Smad7 was also found to interact with the deubiquitinating enzyme UCH37 and alternatively stabilize TβRI (Wicks et al., 2005), implying that Smad7 functions as a multifunctional adaptor protein to either stabilize or destabilize TβRI. Additionally, Smad7 has been reported to interact with Smad-binding element, thus interfering with formation of regulatory Smad-DNA complex in the nucleus (Zhang et al., 2007).
Smad7 is transcriptionally induced by TGF-β through Smads 3 and 4 (Nakao et al., 1997; von Gersdorff et al., 2000), providing a negative-feedback mechanism for TGF-β signaling. The suppressive effects of Smad7 are extinguished by its proteasomal degradation pathway involving Smurfs 1 or 2 or other ubiquitin ligases such as RING finger family Arkadia (Koinuma et al., 2003) and HECT family AIP4 (Lallemand et al., 2005). The amino-terminal lysine residues 64 and 70 of Smad7 have been identified as the ubiquitination sites, and acetylation of these two residues stabilizes Smad7 (Gronroos et al., 2002; Simonsson et al., 2005).
Hic-5, also called ARA55, was originally identified as a TGF-β or hydrogen peroxide-inducible protein in mouse osteoblast MC3T3-E1 cells (Shibanuma et al., 1994). Hic-5 belongs to the LIM domain protein superfamily and has high homology to the focal adhesion adaptor protein paxillin (Thomas et al., 1999). Although Hic-5 can shuttle between the nucleus and the cytosol (Mori et al., 2006; Shibanuma et al., 2003), it mainly localizes to focal adhesions in competition with paxillin for binding to focal adhesion kinase (Fujita et al., 1998) and proline-rich tyrosine kinase 2 (PYK2) (Matsuya et al., 1998), thus modulating integrin-mediated signals that control cell proliferation and migration (Nishiya et al., 2001) or causing cellular senescence (Shibanuma et al., 1994; Shibanuma et al., 1997). In the nuclear compartment Hic-5 functions as a steroid hormone-dependent coactivator for nuclear receptors such as AR, glucocorticoid receptor and progesterone receptor (Fujimoto et al., 1999). This role is well supported by a recent report showing that Hic-5 functions as an adaptor protein for recruitment of such coactivators as TIF2 and p300 in the complexes assembled at glucocorticoid receptor-responsive elements (Heitzer and DeFranco, 2006b). The seemingly discrepant functions of Hic-5, either suppressing cell growth through interrupting focal adhesions or promoting cell growth through interaction with steroid hormone receptors, could be reconciled by its phosphorylation state. PYK2 phosphorylates Hic-5 at tyrosine 43, which prevents its function as a coactivator of AR (Dong et al., 2002) and possibly other steroid hormone receptors (Fujimoto et al., 1999).
Hic-5 was previously reported to block certain Smad3-mediated transcriptional responses of TGF-β through a direct physical interaction involving the MH2 domain of Smad3 and the LIM3 domain of Hic-5 (Wang et al., 2005). We now show that the binding of Smad7 to Hic-5 requires the LIM3 domain of Hic-5, and this binding reverses the ability of Smad7 to suppress TGF-β signaling by promoting depletion of Smad7. Our data support that the overall suppressive activity of Hic-5 of on both Smad3 and Smad7 enhances Smad2 activity, while suppressing Smad3-dependent responses. We suggest that the overall effects of Hic-5 on TGF-β responses depend on the balance of Smad3- and Smad2-dependent signals.
Results
Hic-5 can bind to Smad7
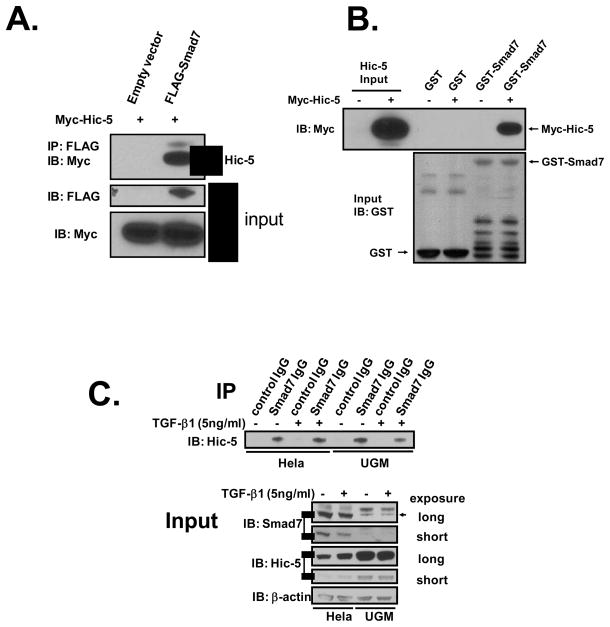
We previously reported that Hic-5 physically associates to Smad3 and inhibits certain Smad3-mediated transcriptional responses (Wang et al., 2005). Further screening of the interaction of Hic-5 with other Smads revealed a rather robust physical interaction with Smad7, as shown by co-IP experiments in HEK293 cells 24 h after transfection with FLAG-Smad7 and Myc-Hic-5. Anti-FLAG M2 MoAb pulled down Myc-Hic-5 only in cells that expressed FLAG-Smad7 (Figure 1A). Likewise, anti-Myc IgG pulled down Smad7 only when Myc-Hic-5 was expressed (data not shown). To test whether the physical interaction of Hic-5 with Smad7 was direct, we conducted a cell-free GST pull-down experiment using purified bacterially expressed GST-Smad7 and in vitro transcribed and translated Myc-Hic-5. We show that GST-Smad7, but not GST, pulled down Myc-Hic-5 (Figure 1B), supporting a direct interaction between Smad7 and Hic-5. We also examined whether endogenous Smad7 interacts with endogenous Hic-5 in Hela and rat urogenital sinus mesenchymal cells, which unlike HEK293 cells express both proteins at detectable levels. Anti-Smad7 rabbit IgG but not control rabbit IgG pulled down endogenous Hic-5 in both of these cells, as detected with a Hic-5-specific monoclonal antibody (Figure 1C). These results support that Hic-5 rather robustly binds to Smad7 at the endogenous level.
Figure 1. Binding of Hic-5 to Smad7 through Hic-5 LIM3 domain.
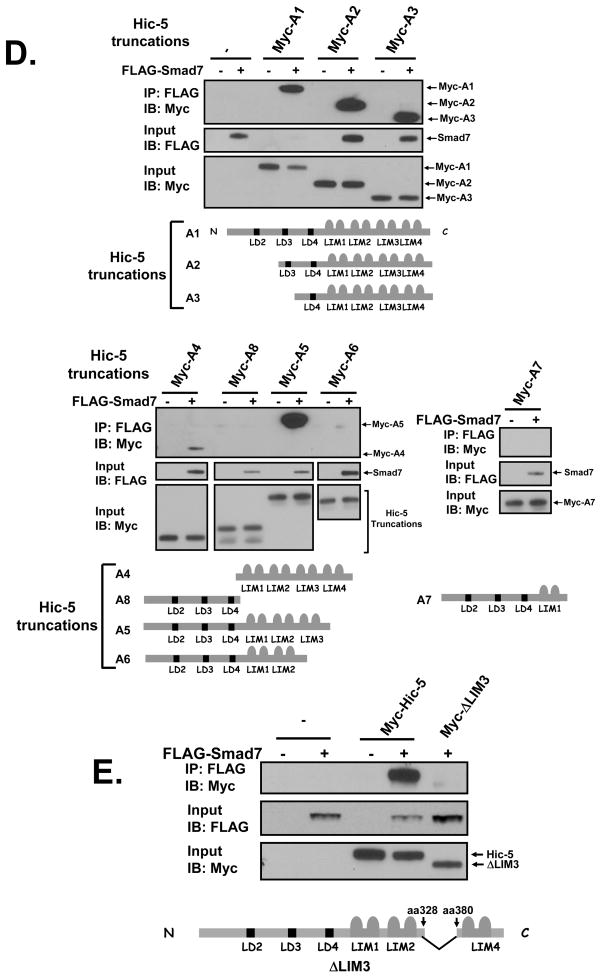
A, Interaction between Hic-5 and Smad7. HEK293 cells were co-transfected with 1 μg/well of Myc-Hic-5 and 1μg/well of FLAG-Smad7 or the empty vector. Cell lysates were immunoprecipitated with anti-FLAG M2 monoclonal antibody, and both the immunoprecipitated proteins and the input cells lysates were immunoblotted with anti-Myc antibody A-14. B, the binding of Smad7 to Hic-5 is direct. In vitro transcribed and translated Myc-Hic-5 was incubated with GST or GST-Smad7 fusion protein overnight at 4°C in the presence of glutathione-Sepharose™ 4B beads. Elution of GST from the beads by treatment (50° C, 5 min) with SDS-PAGE sample loading buffer released both GST-Smad7 and Myc-Hic-5 as detected following immunoblotting with anti-GST and anti-Myc A-14 antibodies, respectively. C, Hela or rat urogenital sinus mesenchymal (UGM) cells were plated in 100-mm dishes with 5% FBS/DMEM/F-12 medium. Twenty-four hours before harvesting, TGF-β1 (5 ng/ml) or vehicle was added to cells. Cells were lysed in cold RIPA buffer and then the endogenous Smad7 was immunoprecipitated with control rabbit IgG or anti-Smad7 rabbit IgG. The resultant eluates were immunoblotted with anti-Hic-5 mouse antibody. D and E, Hic-5 LIM3 domain is the binding region. HEK293 cells were co-transfected with 1 μg/well of full-length Myc-Hic-5 (Hic-5 A1), Myc-Hic-5 truncation vectors (Hic-5 A2 – Hic-5 A8, see map in figure), or Myc-Hic-5 without the LIM3 domain (Hic-5ΔLIM3) and 1 μg/well of FLAG-Smad7 or the empty vector. Immunoprecipitation and immunoblotting were completed in the same way as described in A. All of the data shown are representative of two to three independent experiments.
To define the domains of Hic-5 involved in this interaction, we conducted co-IP assays using a series of Hic-5 truncations co-expressed with Smad7 in HEK293 cells. We found that all the Hic-5 truncations containing the LIM3 domain were able to bind to Smad7, while those without this domain (A6, A7, A8 and ΔLIM3) failed to bind (Figure 1D and 1E). This suggests that the LIM3 domain of Hic-5 is critical for its binding to Smad7.
Hic-5 can induce Smad7 protein loss
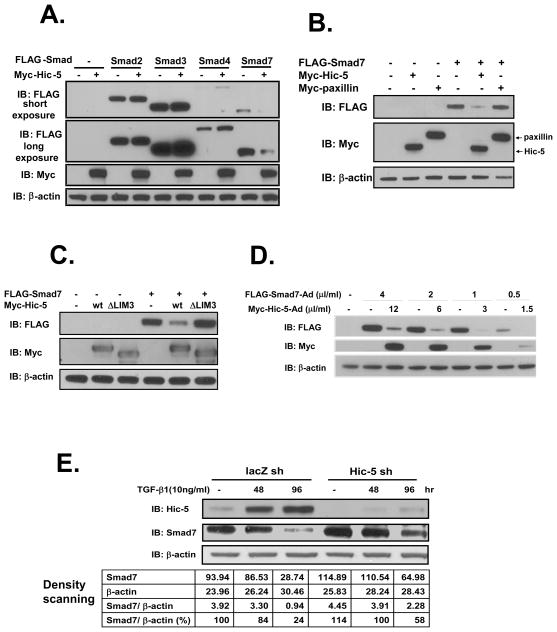
Interestingly, in the above co-IP assays we consistently observed that Smad7 protein levels were depressed by Hic-5 in HEK293 cells. This Hic-5-induced protein loss seems selective for Smad7, as it did not occur with other FLAG-Smads transfected in parallel (Figure 2A). Moreover, the Hic-5 homologue paxillin, which does not bind to Smad7 (data not shown2), did not downregulate Smad7 levels (Figure 2B). These results suggest that loss of Smad7 expression by Hic-5 may depend on their physical interaction. To test this, the ΔLIM3 construct of Hic-5, which does not bind Smad7 (Figure 1E), was co-transfected with Smad7 in HEK293T cells, and changes in the expression levels of Smad7 were observed 24 h after transfection. As expected, Hic-5 ΔLIM3 was unable to repress Smad7 protein levels, in contrast to wild-type Hic-5 (Figure 2C). Therefore, our results suggest that the LIM3 domain of Hic-5 or/and the binding of Hic-5 to Smad7 is necessary for the ability of Hic-5 to downregulate Smad7.
Figure 2. Binding of Hic-5 to Smad7 leads to Smad7 protein loss.
A, Hic-5 selectively causes Smad7 protein loss. HEK293 cells were co-transfected with 1 μg/well of vectors bearing FLAG-Smad cDNA fragments and 1 μg/well of Myc-Hic-5 or the empty vector. The same amount of cell lysate was immunoblotted to detect tagged proteins. B and C, Binding is necessary for Smad7 protein reduction by Hic-5. HEK293 (B) or HEK293T (C) cells were co-transfected with 1 μg/well of the empty vector or Myc-tagged plasmids (Hic-5, paxillin or Hic-5 ΔLIM3) and 1 μg/well of FLAG-Smad7. Tagged proteins were detected as described in A. D, Downregulation of Smad7 protein by Hic-5 in NRP-154 cells. NRP-154 cells were co-infected with different doses (v/v dilution) of the adenoviruses Myc-Hic-5-AdMax (Ad) and FLAG-Smad7-Ad overnight. After 36 h, cells were harvested for immunoblotting to detect the expression of exogenous proteins. E, Silencing Hic-5 in DU145 cells. DU145 cells infected with lentivirus bearing lacZ shRNA or Hic-5 shRNA were treated with vehicle or TGF-β1 (10 ng/ml, recombinant human TGF-β1, R&D Systems, Inc.) for 48 h or 96 h. Band intensities were evaluated with Quantity One 4.5.1 (Bio-Rad). Cell lysates were immunoblotted for Hic-5 and Smad7, respectively. All of the data shown are representative of three independent experiments.
We next tested whether Hic-5 could suppress Smad7 levels in other cells lines, such as the NRP-154 rat prostatic tumorigenic cell line, by adenoviral transduction (Admax) to efficiently deliver both Hic-5 and Smad7. We found that essentially 100% of these cells were transduced by AdMax(Ad)-β-galactosidase, assessed by following conversion of the substrate X-gal to blue product. Overexpression of Hic-5 in NRP-154 cells by adenoviral transduction resulted in a robust loss of Smad7 protein (Figure 2D). We next determined whether such regulation also occurred at the endogenous level. For this we used the DU145 human prostate carcinoma cell line with which we showed expression of both endogenous Hic-5 and Smad7, where levels of Hic-5 were also >10-fold induced by a 96 h treatment with TGF-β1 (Figure 2E). Such induction occurred with a concomitant ~ 3-fold decrease in the expression of Smad7. DU145 cells in which Hic-5 was stably silenced showed elevation of Smad7 expression, and TGF-β1 only modestly decreased Smad7 in Hic-5-silenced cells (Figure 2E). The slight loss of Smad7 levels in the silenced cells reflects a slight elevation in Hic-5 expression in the Hic-5-silenced cells following 96 h induction with TGF-β. These results support that Hic-5 can downregulate Smad7 at the endogenous level.
Structural analysis of Smad7 for interaction with Hic-5
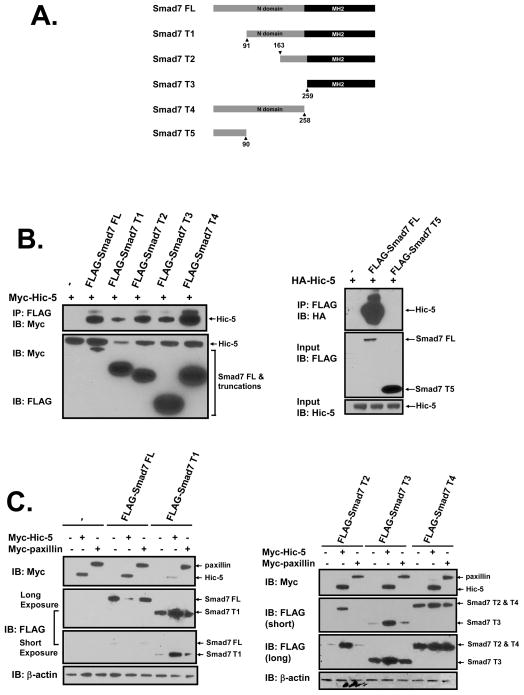
To better define how the physical interaction of Smad7 with Hic-5 suppresses the expression of Smad7, we developed various FLAG-Smad7 truncations (Smad7 T1-T5, Figure 3A), which were co-transfected with full-length Myc-Hic-5 in HEK293 cells. Cell lysates were immunoprecipitated with anti-FLAG IgG and analyzed for expression of Myc-Hic-5 by Western blot analysis. We showed the Smad7 truncations except for T5 physically interacted with Hic-5. Our results demonstrate that at least two sites of Smad7 associated with Hic-5, with a site of interaction in the MH2 domain (T3) and another in the remaining N-terminal portion of Smad7 (T4) (Figure 3B). The first 90 amino acids (aa) of Smad7 (Smad7 T5) did not pull down Hic-5, suggesting that this region does not include an interaction domain, although when missing (Smad7 T1), the amount of Hic-5 pulled down or the level of Hic-5 in the input were substantially less compared to full-length Smad7, and there was no loss of Smad7 T1 (Figure 3B and 3C). In contrast to full-length Smad7, the truncations with an intact C-terminus, but lacking in the N-terminal 90 aa, were stabilized by Hic-5 (Figure 3C). Therefore, this small N-terminal sequence seems to be critical for Smad7 protein loss by Hic-5. Taken together, our data support that two binding sites stabilize the complex of Hic-5 with Smad7, and a third region (amino acids 1–90) promotes loss of Smad7.
Figure 3. Interaction of Smad7 truncations with Hic-5.
A, Smad7 truncation constructs. B, Binding of Smad7 truncations to Hic-5. HEK293 cells were co-transfected with 1 μg/well of Myc-Hic-5 and 1 μg/well of FLAG-Smad7 truncations or the empty vector. Cell lysates were immunoprecipitated with anti-FLAG M2 monoclonal antibody, and both the immunoprecipitated proteins and input lysates were immunoblotted with anti-Myc antibody A-14. C, Effects of Hic-5 on Smad7 truncation protein levels. HEK293 cells were co-transfected with 2 μg/well vectors bearing 1 μg Myc-Hic-5 and 1 μg of each of the FLAG-Smad7 truncations or the empty vector. Cell lysate was immunoblotted to detect tagged proteins. All of the data shown are representative of two independent experiments.
Effects of Hic-5-Smad7 interaction on TGF-β signaling
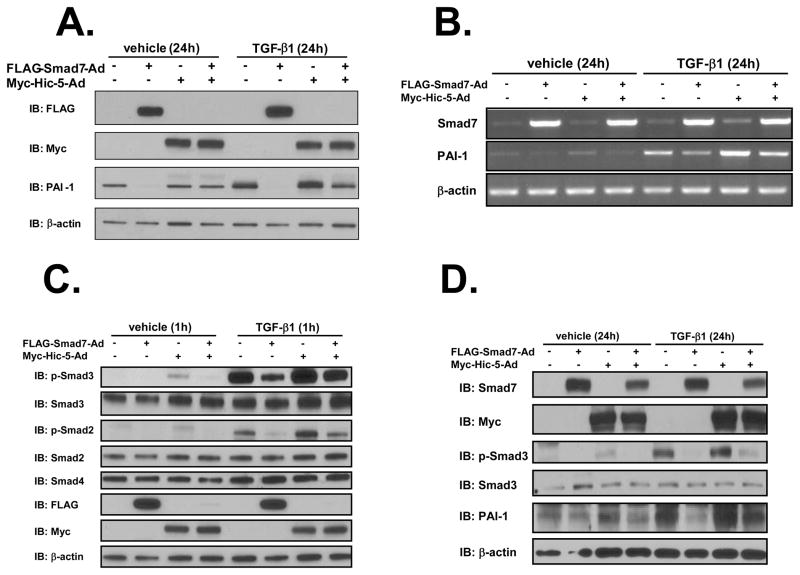
We further explored the biological impact of Hic-5 on Smad7’s ability to control PAI-1 levels. NRP-154 cells were infected with Hic-5-Ad, Smad7-Ad, both viral constructs, or empty vector control Ad. Twenty-four h after transduction, cells were treated for 1 h or 24 h with either TGF-β1 or vehicle and subjected to Western blot for PAI-1. Overexpression of Hic-5 depleted essentially all exogenously-introduced Smad7 (Figure 4A). Moreover, Hic-5 protected both basal and TGF-β-induced PAI-1 protein and mRNA from Smad7-mediated repression (Figure 4A and 4B). The level of Hic-5 in NRP-154 cells expressed with this adenovirus was comparable to that expressed in normal rat prostate fibroblasts (Supplementary Figure 1A), and it was thereby within a physiological range.
Figure 4. Effects of Hic-5-Smad7 interaction on TGF-β-induced R-Smad activation and PAI-1 expression.
A and D, NRP-154 cells (A) or PC3 cells (D) were co-infected with ±FLAG-Smad7-Ad (1:1000 v/v dilution) and ±Myc-Hic-5-Ad (3:1000 v/v dilution) overnight. Then cells were added with vehicle or 10 ng/ml TGF-β1. Twenty-four h later, cells were harvested for immunoblotting to detect protein level of Hic-5 or Smad7 and endogenous protein level of PAI-1 and β-actin (loading control). B, NRP-154 cells were treated in the same way as described in A but mRNA levels of Smad7 and endogenous PAI-1 were measured. C, NRP-154 cells were treated as described in A. However, TGF-β1 (10 ng/ml) was added 1 h before harvesting. All of the data shown are representative of three independent experiments.
A parallel RT-PCR analysis was conducted to determine whether changes in exogenous or endogenous Smad7 mRNA reflected that of protein levels. This analysis revealed that neither exogenous nor endogenous Smad7 mRNA levels were suppressed by Hic-5 (Figure 4B), suggesting that loss of exogenous Smad7 by Hic-5 occurred specifically at the protein level. In contrast, PAI-1 mRNA levels were influenced by TGF-β, Smad7 and Hic-5 with a pattern that was similar to changes in PAI-1 protein levels.
Next, we examined the effects of Hic-5-Smad7 interaction on downstream effectors of TGF-β signaling (Smads) in NRP-154 cells. Hic-5 reversed the ability of Smad7 to suppress TGF-β-induced (1 h) phosphorylation of Smads 2 and 3. These changes did not reflect the total levels of Smads 2, 3 or 4, indicating that Hic-5 reverses the ability of Smad7 to block TGF-β-induced activation of Smads 2 and 3 (Figure 4C). Interestingly, Hic-5 overexpression alone appeared to slightly activate Smads 2 and 3 without exogenous TGF-β and slightly enhance the ability of TGF-β to phosphorylate Smads 2 and 3, consistent with the ability of Hic-5 to promote loss of endogenous Smad7 and relieve its inhibitory activity on Smad activation by autocrine TGF-β.
We also examined effects of Hic-5 on Smad7’s influence on TGF-β responses and Smad activation in the PC3 human prostate carcinoma cell line. In these cells, Hic-5 suppressed the level of exogenous Smad7 protein, and Smad7 activity with respect to TGF-β-induced PAI-1 expression and Smad3 phosphorylation (Figure 4D).
Hic-5-induced Smad7 protein loss is not likely through a proteasomal pathway
Because Hic-5 did not repress Smad7 mRNA levels (Figure 4B), it is reasonable to hypothesize that this Smad7 protein loss occurs through a protein degradation pathway. We examined the possibility that Hic-5 promotes the proteasomal degradation of Smad7 by testing whether Hic-5 enhances the ubiquitination of Smad7. HEK293 cells were first co-transfected with HA-ubiquitin, FLAG-Smad7 or/and Myc-Hic-5, and immunoprecipitated FLAG-Smad7 to detect its ubiquitination by Western blot for HA. Hic-5 significantly increased the HA-ubiquitin pulled down by Smad7, but there was no difference in the amount of ubiquitinated protein in Myc-Hic-5 immunoprecipitates whether or not Smad7 was co-expressed (Supplementary Figure 2A and 2B). Consistent with our findings, HA-ubiquitin pulled down both Smad7 and Hic-5 (Supplementary Figure 2C), and coexpression of Hic-5 slightly enhanced the amount of Smad7 pulled down despite a drop in total Smad7 levels (input), suggesting that Hic-5 enhances the ubiquitination of Smad7. However, it is unlikely that such ubiquitination promotes proteasomal degradation pathway of Smad7, as 26S proteasomal inhibitors (lactacystin, MG132 and epoxomicin) did not block loss of Smad7 by Hic-5 (Supplementary Figure 2D). Moreover, mutating the classical ubiquitination sites on Smad7 (lysine residues 64 and 70 (Gronroos et al., 2002)) did not abate the loss of Smad7 by Hic-5 (Supplementary Figure 2E). Deletion of the Smurf E3-ligase binding domain of Smad7 also failed to prevent loss of Smad7 by Hic-5 (Supplementary Figure 2E). These results suggest that degradation of Smad7 by Hic-5 does not occur through a proteasomal pathway.
Functional Inactivation of Smad7 by Hic-5
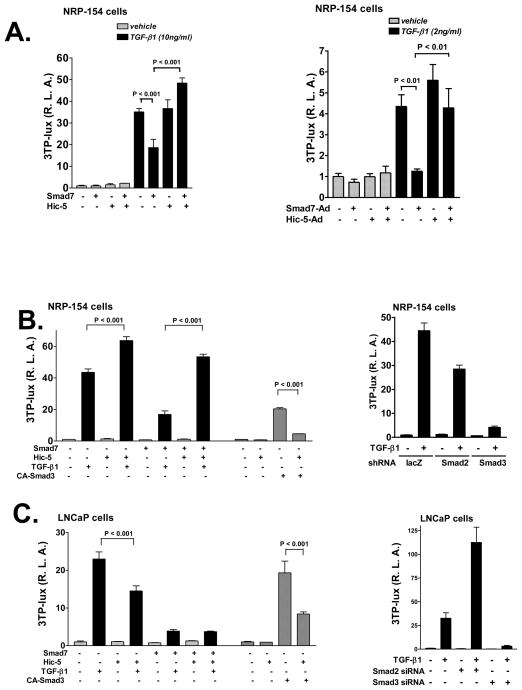
Our results led us to hypothesize that Hic-5 can reverse the ability of Smad7 to suppress TGF-β responses. We tested this hypothesis by first using a highly TGF-β-responsive promoter construct, 3TP-lux, which contains the PAI-1 promoter upstream of the firefly luciferase reporter. TGF-β activates this promoter through Smads 2 and 3 and MEK signaling (Song et al., 2006a; Yue et al., 1999). NRP-154 cells were co-transfected with 3TP-lux, along with CMV-Renilla and expression plasmids for Smad7, or/and Hic-5 or empty vector only. These cells were then treated with or without TGF-β1, and 24 h later relative luciferase activity (firefly/renilla luciferase ratios) was assessed. Transfection of Smad7 led to about 50% to 80% inhibition of TGF-β-induced reporter activity in NRP-154 cells, which was reversed in cells co-expressing Hic-5 (Figure 5A). Similar results were obtained in the WPMY-1 human prostate myofibroblast cell line which expresses endogenous Hic-5 (Supplementary Figure 3). Thus, Hic-5 reverses the inhibitory effect of Smad7 on TGF-β-induced promoter activity in both stromal and epithelial cell lines. Moreover, expression of Hic-5 alone enhanced TGF-β-induced 3TP-lux, consistent with the suppression of endogenous Smad7 by Hic-5 (Figure 5B and Supplementary Figure 3).
Figure 5. Inactivation of Smad7 by Hic-5.
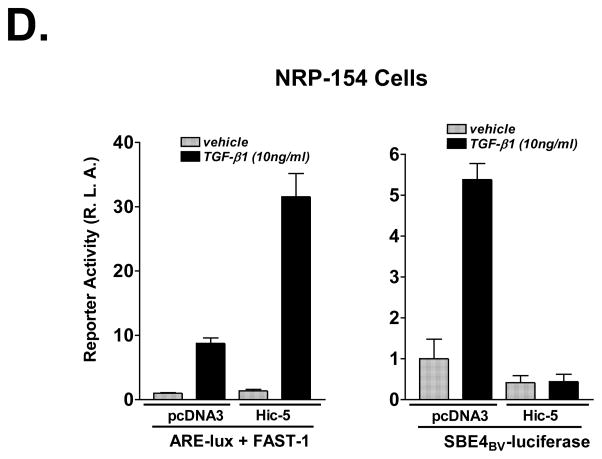
A, Left panel, NRP-154 cells were co-transfected (calcium phosphate method (Wang et al., 2005)) with 0.2 μg/well of 3TP-lux reporter, 12.5 ng/well of pRL-CMV internal control, ±0.1 μg/well of FLAG-Smad7 and ±0.7 μg/well of Myc-Hic-5. The next day, cells were treated with 10 ng/ml TGF-β1 or vehicle for 24 h before harvesting. Right panel, NRP-154 cells were infected with FLAG-Smad7-Ad (1:1000 v/v dilution) and Myc-Hic-5-Ad (3:1000 v/v dilution) constructs and then were transfected with the same amount of 3TP-lux reporter and pRL-CMV internal control plasmid as used in A. The next day, 2 ng/ml TGF-β1 or vehicle was used to treat cells for 24 h before harvesting. B, NRP-154 cells were co-transfected with the same amount of various plasmids as described in A, except that ±0.65 μg/well of Myc-Hic-5 was used in this experiment. The next day, 10 ng/ml TGF-β1 or vehicle was added and luciferase activity was measured 24 h later. CA-Smad3 (0.1 μg/well) was used as an inducer in a parallel experiment, where cells were harvested 24 h after transfection. Smad2 and Smad3 were silenced with shRNA according to our recent published protocol (Yang et al., 2008). C, LNCaP cells were treated in the same way as described in B. Smad2 and Smad3 were silenced with siRNA (Supplementary Figure 5) according to another publication form our laboratory (Song et al., 2006b). D, NRP-154 cells were co-transfected with 0.2 μg/well of reporter (ARE-lux/FAST-1 or SBE4BV-luciferase), 20 ng/well of pRL-CMV internal control and ±0.8 μg/well of Myc-Hic-5. The next day, cells were treated with 10 ng/ml TGF-β1 or vehicle for 24 h before harvesting for luciferase test. In all reporter assays, dual luciferase tests were performed as described before (Wang et al., 2005). Luciferase values represent the average of triplicate determinations ±SEM. All of the data shown are representative of three independent experiments.
In our previous report (Wang et al., 2005), we showed that Hic-5 can suppress TGF-β signaling through intercepting Smad3. Our current report suggests that this suppressive effect of Hic-5 on Smad3-dependent TGF-β response is counter-balanced by activation of TGF-β signals through reversing the inhibitory effects (negative feedback) of Smad7 on TGF-β receptors. We predicted that the net result of suppression of Smad3 and degradation of Smad7 will be suppression of Smad3-dependent TGF-β signals and activation of Smad2-dependent and Smad3-independent TGF-β signals. In our previous report, Hic-5 was shown to inhibit TGF-β-induced 3TP-lux and PAI-1 expression in LNCaP human prostate carcinoma cells, constitutively active (CA)-Smad3-induced 3TP-lux and Smad binding element reporter construct SBE4BV-luciferase activities in both LNCaP and NRP-154 cells, and TGF-β-induced SBE4BV-luciferase activity in both LNCaP and NRP-154 cells. We reconfirmed these findings side-by-side with and without overexpression of Smad7, and showed that Smad7 reverses TGF-β responses in both cell lines; however, Hic-5 reverses the suppressive effect of Smad7 on 3TP-lux activity in NRP-154 cells but not in LNCaP cells (Figure 5B and 5C). Initially, these data suggested that Hic-5 does not cause degradation of Smad7 in LNCaP cells. However, Western blot analysis revealed that Hic-5 also caused loss of Smad7 in LNCaP cells (Supplementary Figure 4), although not as robustly as in NRP-154 cells, suggesting an alternative mechanism for cell line differences in the effects of Hic-5 on TGF-β-induced 3TP-lux activity. We provide data suggesting that the above cell line differences are at least partly due to cell line differences in the relative effects of Smad3 and Smad2 on TGF-β-induced 3TP-lux. In NRP-154 cells, silencing Smad2 and Smad3 blocked about 30% and 80%, respectively, of the TGF-β-induced 3TP-lux activity (Figure 5B). In LNCaP cells, however, silencing Smad3 blocks TGF-β-induced 3TP-lux activity, while silencing Smad2 actually enhances it (Figure 5C, Supplementary Figure 5). Thus, to better test our model that Hic-5 suppresses Smad3 while activating Smad2 responses, we examined the effects of Hic-5 in NRP-154 cells on TGF-β-induced Smad2 and Smad3 activities using a promoter construct more specific for Smad3 (SBE4BV-luciferase) and one more specific for Smad2 (ARE-lux/FAST-1) (Chen et al., 1997). Consistent with our model, Hic-5 suppressed TGF-β-induced SBE4BV-luciferase activity, whereas it enhanced TGF-β-induced ARE-lux/FAST-1 activity (Figure 5D). These results suggest that Hic-5 differentially alters the overall activities of Smad2 and Smad3, thus suppressing Smad3 transcriptional responses while enhancing Smad2 transcriptional responses (Figure 6).
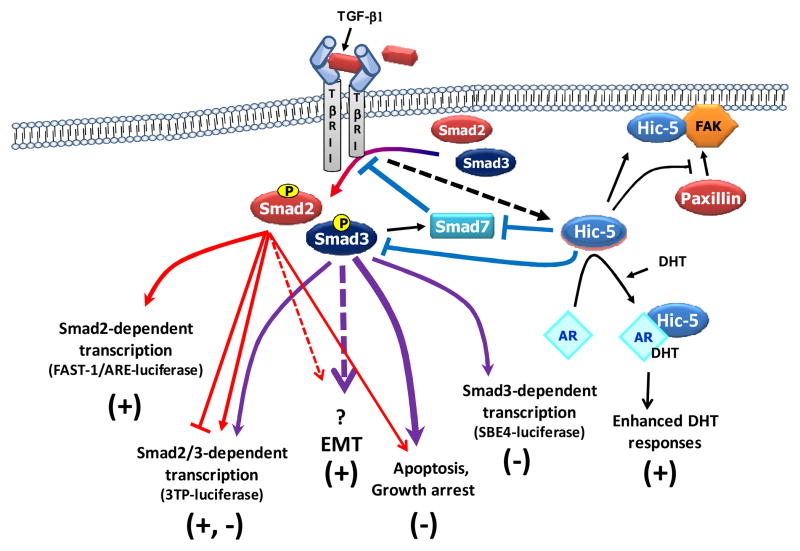
Figure 6. Signal transduction diagram depicting how Hic-5 affects TGF-β signaling.
Hic-5, which is induced by TGF-β through a mechanism that is yet to be resolved, binds to and inactivates both Smad3 and Smad7. Inactivation of Smad7 through its degradation relieves the inhibitory action of Smad7 on TGF-β receptors, which leads to phosphorylation and thus activation of Smads 2 and 3. Depletion of Smad7 frees up Hic-5 and is expected to enable further binding to and suppression of Smad3. We thus propose that the overall effect of Hic-5 on TGF-β signaling is inhibition of Smad3-dependent pathway and enhancement of Smad2-dependent or Smad-independent pathway.
Function of endogenous Hic-5 on Smad3-independent TGF-β responses
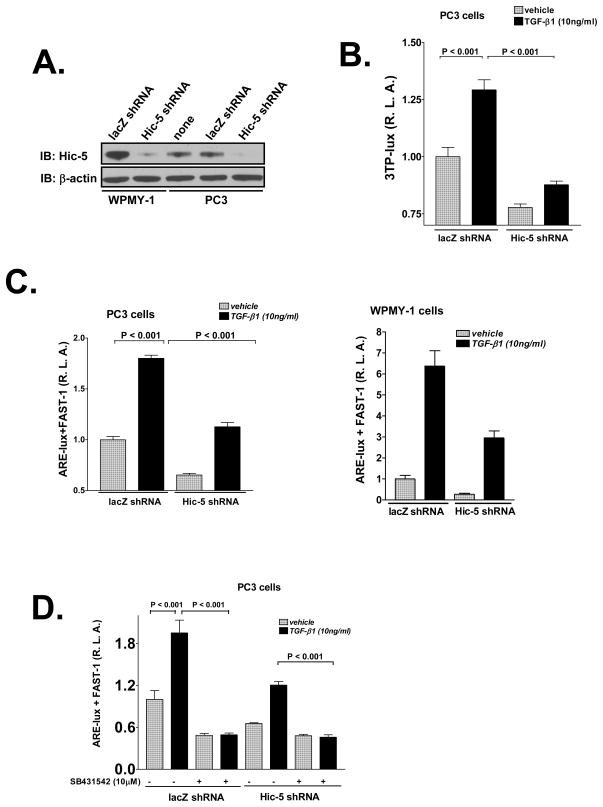
Whole cell lysates from various cell lines were tested for levels of endogenous Hic-5. WPMY-1 and PC3 cells express high levels of Hic-5 relative to other prostate cell lines (Supplementary Figure 1A and 1B). We thus explored the role of endogenous Hic-5 on TGF-β responses in these two cell lines by silencing expression of Hic-5 with a lentiviral construct bearing shRNA. This construct efficiently knocked down endogenous Hic-5, while the control lacZ shRNA did not (Figure 7A). As expected, silencing Hic-5 repressed TGF-β-induced 3TP-lux reporter activity (Figure 7B). Similarly, loss of endogenous Hic-5 by Hic-5 shRNA reduced TGF-β-induced Smad2 activity as shown byARE-lux/FAST-1 activity in PC3 and WPMY-1 cells (Figure 7C), consistent with Hic-5 being an inhibitor of Smad7. Basal luciferase activity for both of the above promoter constructs was repressed by silencing Hic-5 with shRNA, which was possibly due to suppression of autocrine TGF-β signaling triggered by elevated levels endogenous Smad7 resulting from loss of Hic-5. We tested this possibility by examining whether shutting off autocrine TGF-β signaling with a specific TβRI kinase inhibitor, SB431542, would diminish the ability of Hic-5 to repress ARE-lux activity PC3 cells. Our results (Figure 7D) suggest that the ability of endogenous Hic-5 to enhance basalARE-lux activity occurs through enhanced autocrine TGF-β activity. Taken together, these data support that Hic-5 activates the biological activity of Smad2, consistent with loss of Smad7 as predicted from our model (Figure 6). We provide supplemental data with the DU145 cell line that endogenous Hic-5 enhances TGF-β-induced EMT, as assessed by loss of E-cadherin staining and increased expression of vimentin (Supplementary Figure 6A and 5B). We speculate that the permissive effect of Hic-5 on TGF-β-induced EMT may involve the ability of Hic-5 to differentially regulate the activities of Smad2 and Smad3 (Figure 6).
Figure 7. Role of Endogenous Hic-5 on TGF-β responses.
A, Silencing Hic-5 in WPMY-1 and PC3 cells. WPMY-1 or PC3 cells were infected with lentiviral lacZ shRNA or Hic-5 shRNA construct overnight and then maintained in culture medium. After 1 day, cells were harvested and endogenous Hic-5 protein was detected with anti-Hic-5 antibody. B, PC3 cells bearing lacZ shRNA or Hic-5 shRNA were transfected with 0.2 μg/well of 3TP-lux reporter and 20 ng/well of pRL-CMV internal control. The next day, cells were treated with 10 ng/ml TGF-β1 or vehicle for 24 h and then harvested for luciferase test. C, PC3 or WPMY-1 cells bearing lacZ shRNA or Hic-5 shRNA were treated as described in B except that ARE-lux co-expressed with FAST-1 was used as the reporter. D, PC3 cells bearing lacZ shRNA or Hic-5 shRNA were treated as described in C. However, after transfection, cells were added with 10 μM TβRI kinase inhibitor SB431542 or vehicle. Luciferase values in B, C and D represent the average of triplicate determinations ±SEM. All of the data shown are representative of two to three independent experiments.
Discussion
Here we report the first evidence that Hic-5 forms a direct physical complex with Smad7, leading to loss of Smad7 protein levels and thus a functional inactivation of Smad7. Although Smad7 is a TGF-β-inducible protein whose function was first proposed to be feedback inhibition of TGF-β signaling, subsequent studies showed that its effects are cell-type dependent and could be independent of suppressing TGF-β activity (Hong et al., 2007; Landstrom et al., 2000; Mazars et al., 2001). Smad7 has been shown to induce apoptosis through activating p38 kinase (Edlund et al., 2003) or inhibiting nuclear factor-κB (Lallemand et al., 2001). The ability of Hic-5 to reverse these new roles of Smad7 awaits further work. However, consistent with Smad7’s function as an inhibitor of TGF-β receptor signaling, our study shows that Hic-5 can enhance TGF-β-induced phosphorylation of both Smads 2 and 3, leading to induced expression of PAI-1 (Figure 4Aand 4C) and 3TP-lux activity (Figure 5Aand 5B). In fact, in the absence of exogenous TGF-β1, Hic-5 can induce phosphorylation of both Smads 2 and 3 (Figure 4C and 4D), likely through enhancing autocrine TGF-β activity and consistent with its suppression by Smad7 (Figure 4C and 4D) or by SB431542 (Figure 7D). Activation of autocrine TGF-β activity by Hic-5 in PC3 cells is consistent with the recent report that Hic-5 induces activation of autocrine TGF-β1 in hypertrophic scar fibroblasts (Dabiri et al., 2008). The activation of TGF-β signaling by Hic-5 through loss of Smad7 is balanced by a more specific suppression of the activity of Smad3 down-stream of its activation by TGF-β receptors. This is supported by the selective association of Hic-5 to CA-Smad3 (not CA-Smad2) and the suppression of Smad3-dependent promoter activity (Wang et al., 2005). Use of mammalian two-hybrid systems in that study suggested that Hic-5 inhibits the transcriptional activity of Smad3 by blocking transcriptional co-activators in the nucleus. Thus, by interacting with both Smad7 and Smad3, we predict that Hic-5 will have a net effect on enhancing Smad3-independent TGF-β responses, depending on the relative level of Smad7 or activity of Smad7 in suppressing TGF-β receptor signaling.
In our model, Hic-5 reduces the relative ratio of the biological activity of Smad3 to Smad2 or to non-Smad TGF-β signaling (Figure 6). Although activated by the same receptors, Smad2, Smad3, and non-Smad signaling play differential roles in gene expression and cell function (Moustakas and Heldin, 2005). In stromal cells, TGF-β induces collagen types I and III, and fibronectin expression through a Smad-independent mechanism (Wilkes and Leof, 2006). Importantly, a high ratio of Smad3 to Smad2 was reported as critical to the ability of TGF-β to promote cytostatic responses on epithelial cells (Kim et al., 2005). In contrast to the cytostatic effect of TGF-β in normal epithelial cells, TGF-β is a survival factor/growth promoter in normal stromal cells (Rahimi and Leof, 2007), which is the cell type that expresses high levels of Hic-5 at least in the prostate (Heitzer and DeFranco, 2006a). Thus, the selective expression of Hic-5 in stromal cells is likely to contribute loss of cytostatic activity of TGF-β by suppressing the activity of Smad3 while enhancing that of Smad2. TGF-β-induced survival of stromal cells correlates with a stromal-specific TGF-β-induced activation of c-Abl, PI3K and PAK2, which occur through a Smad2- and Smad3-independent mechanism (Wilkes and Leof, 2006). Hic-5 may thus enhance or promote the Smad-independent activation of those kinases.
Hic-5 has recently been reported to play a key role in TGF-β-induced EMT, leading to downregulation or disruption of cell-cell contact mediator E-cadherin in response to TGF-β, as shown in the MCF10A human epithelial cell line (Tumbarello and Turner, 2007). Consistent with that study, we show that silencing Hic-5 in DU145 cells repressed TGF-β-induced EMT (Supplementary Figure 6A and 5B). In epithelial cells undergoing EMT and in stromal cells, TGF-β has been reported to induce the expression of Hic-5, which likely functions to inhibit Smad3-dependent responses, such as apoptosis (data not shown), while activating Smad3-independent responses. Hic-5 may further facilitate cancer cell migration through enhancing TGF-β-induced expression of PAI-1, a protein associated with poor cancer prognosis (Durand et al., 2004). Ectopic expression of Hic-5 in prostate carcinoma may stimulate tumor cell migration, invasion and metastasis (Horrevoets, 2004). Interestingly, TGF-β-induced EMT has been reported to be Smad3-dependent in many systems (Moustakas and Heldin, 2005), which may be potentially antagonized by Hic-5. If so, other signals are likely to override this antagonistic activity.
At least two regions of Smad7, localized in both N- and C-termini, are involved in Smad7-Hic-5 binding (Figure 3B). Correspondingly, although the LIM3 domain of Hic-5 is required for its interaction with Smad7, this domain may not be the only site interacting with Smad7. Our data suggest that binding of Hic-5 to Smad7 is necessary but not sufficient for the ability of Hic-5 to cause Smad7 loss, and a region within that the first 90 aa of Smad7 is critical to its degradation by Hic-5. However, the complex lacking this region leads to loss of Hic-5 instead of Smad7 (Figure 3B and 3C). Therefore, the first 90 aa of Smad7 are critical for the specificity of Smad7 loss versus that of Hic-5. It is thus likely that the interface of Smad7 and Hic-5 serves as a docking site for an enzyme involved in the degradation of Smad7, requiring elements within the first 90 aa of Smad7 to confer specificity.
In summary, Hic-5 has dual roles on TGF-β signaling as defined by its interactions with both Smad3 and Smad7, with an overall generalized effect of enhancing Smad3-independent transcriptional responses (Figure 6). We propose that high levels of Hic-5 found in stromal cells and associated with certain epithelial tumors may function as part of the intriguingly complex cellular circuitry that governs the pleiotropic responses of TGF-β between normal differentiated epithelium and normal mesenchymal cells or invasive carcinomas. We thus hypothesize that Hic-5 is part of the functional switch of TGF-β from a tumor suppressor to an oncogene in during tumor progression (Wakefield and Roberts, 2002).
Materials and Methods
Plasmids
See supplementary section.
Cell Passaging
HEK293 cells (passage 40–55) were cultured in DMEM/F-12 (Invitrogen) supplemented with 5% FBS (Hyclone). NRP-154 rat prostatic tumorigenic cell line (Danielpour et al., 1994) (passage number 18–30) was cultured in GM2.1 medium (DMEM/F-12 containing 5% FBS, 5 μg/ml insulin and 0.1 μM dexamethasone). Both PC3 and DU145 human prostatic carcinoma cell lines and WPMY-1 human prostatic myofibroblast cell line were cultured in 5% FBS/DMEM/F-12 medium. LNCaP human prostatic carcinoma cell line was cultured in 10% FBS/DMEM/F-12 medium in flasks coated with poly-D-lysine.
Co-IP and Western blot
HEK293 cells were plated for 48 h in 5% FBS/DMEM/F-12 medium in 6-well dishes at 2.5 × 105 cells/2ml/well. Cells were transfected with 2 μg plasmid DNA by calcium phosphate precipitation, and 36 h later the tagged proteins were co-immunoprecipitated as described before (Chipuk et al., 2002a; Chipuk et al., 2002b).
Antibodies
Antibodies for HA tag (Y-11, sc805), Myc tag (A-14, sc-789), Smad3 (FL-425, sc-8332), Smad4 (B-8, sc-7966), Smad7 (H-79, sc-11392) and PAI-1 (H-135, sc-8979) were from Santa Cruz Biotechnology. Antibodies for phospho-Smad2 (Ser465/467, #3101) and phospho-Smad3 (Ser433/435, #9514) were from Cell Signaling Technology, Inc. FLAG M1 (F-3040) and M2 (F3165) antibodies were from Sigma. Hic-5 antibody (Cat. No. 611165) was from BD Transduction Laboratories.
Adenovirus Vectors and Infection
see supplementary section.
Transfection and Luciferase Assay
NRP-154, PC3 and WPMY-1 cells (2 × 105) plated overnight in 12-well dishes with GM2.1 (NRP-154) or 5% FBS/DMEM/F-12 (PC3, WPMY-1) were transfected for 3 h with DNA (1 μg) and 2.5 μg of polyethylenimine (~25 kDa) in 0.5 ml of DMEM/F-12 + 15 mM HEPES (pH 7.4). Cells were washed once with PBS and then cultured in GM3 (NRP-154) or in 1% FBS/DMEM/F-12 medium (PC3, WYMP-1). LNCaP cells were cultured and transfected as described previously (Song et al., 2006a). Dual luciferase activity was assessed as before (Song et al., 2003b).
RNA Preparation and RT-PCR
see supplementary section.
Lentiviral Hic-5 shRNA Construct Preparation
All lentiviral vectors (Wiznerowicz and Trono, 2003) were from Addgene. The shRNA sequence targeting Hic-5 nucleic acid 317–345 (Rahman et al., 2003) was used to silence Hic-5. The dimerized Hic-5 oligo was inserted to pLVTHM vector between MluI and ClaI restriction enzyme sites. Lentiviral construct generation was then performed according to the published protocol in HEK293T cells (Wiznerowicz and Trono, 2003). The lacZ control shRNA sequence was: 5′ GTG ACC AGC GAA TAC CTG T 3′ (Qin et al., 2003).
GST Pull-Down Assay
see supplementary section.
Supplementary Material
Acknowledgments
We thank Drs. Joan Massagué for 3TP-lux plasmid, Bert Vogelstein for SBE4BV-luciferase plasmid, Kohei Miyazono for full-length mouse Flag-Smad7-pcDNA3 plasmid, Bing-Cheng Wang for the pcDNA3-Myc plasmid, Ryosuke Takahashi and Yu-Chung Yang for HA-ubiquitin plasmid, Malcolm Whitman for ARE-lux and Myc-FAST-1. This work was supported by NCI grants 1R01CA102074 and 1R01CA092102.
Footnotes
Abbreviations: TGF-β, transforming growth factor-β; ECM, extracellular matrix; Hic-5, hydrogen peroxide-inducible clone-5; AR, androgen receptor; ARA55, androgen receptor-associated protein 55; co-IP, co-immunoprecipitation; HEK293, human embryonic kidney cell line 293; TβRI, TGF-β type I receptor; TβRII, TGF-β type II receptor; DMEM/F-12, Dulbecco’s modified Eagle’s medium/Ham’s F-12; FBS, fetal bovine serum; R. L. A., relative luciferase activity; PAI-1, plasminogen activator inhibitor-1; 3TP-lux, a TGF-β-responsive reporter containing PAI-1 promoter region; ARE-lux, a Smad2-responsive reporter containing activin response element; FAST-1, forkhead activin signal transducer-1; Ad, adenovirus vector; GST, Glutathione-S-transferase; PBS, phosphate buffered saline; shRNA, small hairpin RNA; EMT: epithelial-mesenchymal transition.
H. Wang, K. Song, T. L. Krebs, D. Danielpour, unpublished data.
References
- Chen X, Weisberg E, Fridmacher V, Watanabe M, Naco G, Whitman M. Smad4 and FAST-1 in the assembly of activin-responsive factor. Nature. 1997;389:85–9. doi: 10.1038/38008. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Cornelius SC, Pultz NJ, Jorgensen JS, Bonham MJ, Kim SJ, et al. The androgen receptor represses transforming growth factor-beta signaling through interaction with Smad3. J Biol Chem. 2002a;277:1240–8. doi: 10.1074/jbc.M108855200. [DOI] [PubMed] [Google Scholar]
- Chipuk JE, Stewart LV, Ranieri A, Song K, Danielpour D. Identification and characterization of a novel rat ov-serpin family member, trespin. J Biol Chem. 2002b;277:26412–21. doi: 10.1074/jbc.M201244200. [DOI] [PubMed] [Google Scholar]
- Dabiri G, Tumbarello DA, Turner CE, Van De Water L. Hic-5 Promotes the Hypertrophic Scar Myofibroblast Phenotype by Regulating the TGF-beta1 Autocrine Loop. J Invest Dermatol. 2008 doi: 10.1038/jid.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielpour D. Functions and regulation of transforming growth factor-beta (TGF-beta) in the prostate. Eur J Cancer. 2005;41:846–57. doi: 10.1016/j.ejca.2004.12.027. [DOI] [PubMed] [Google Scholar]
- Danielpour D, Kadomatsu K, Anzano MA, Smith JM, Sporn MB. Development and characterization of nontumorigenic and tumorigenic epithelial cell lines from rat dorsal-lateral prostate. Cancer Res. 1994;54:3413–21. [PubMed] [Google Scholar]
- Dong C, Zhu S, Wang T, Yoon W, Li Z, Alvarez RJ, et al. Deficient Smad7 expression: a putative molecular defect in scleroderma. Proc Natl Acad Sci U S A. 2002;99:3908–13. doi: 10.1073/pnas.062010399. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Durand MK, Bodker JS, Christensen A, Dupont DM, Hansen M, Jensen JK, et al. Plasminogen activator inhibitor-I and tumour growth, invasion, and metastasis. Thromb Haemost. 2004;91:438–49. doi: 10.1160/TH03-12-0784. [DOI] [PubMed] [Google Scholar]
- Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 2001;276:12477–80. doi: 10.1074/jbc.C100008200. [DOI] [PubMed] [Google Scholar]
- Edlund S, Bu S, Schuster N, Aspenstrom P, Heuchel R, Heldin NE, et al. Transforming growth factor-beta1 (TGF-beta)-induced apoptosis of prostate cancer cells involves Smad7-dependent activation of p38 by TGF-beta-activated kinase 1 and mitogen-activated protein kinase kinase 3. Mol Biol Cell. 2003;14:529–44. doi: 10.1091/mbc.02-03-0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto N, Yeh S, Kang HY, Inui S, Chang HC, Mizokami A, et al. Cloning and characterization of androgen receptor coactivator, ARA55, in human prostate. J Biol Chem. 1999;274:8316–21. doi: 10.1074/jbc.274.12.8316. [DOI] [PubMed] [Google Scholar]
- Fujita H, Kamiguchi K, Cho D, Shibanuma M, Morimoto C, Tachibana K. Interaction of Hic-5, A senescence-related protein, with focal adhesion kinase. J Biol Chem. 1998;273:26516–21. doi: 10.1074/jbc.273.41.26516. [DOI] [PubMed] [Google Scholar]
- Gronroos E, Hellman U, Heldin CH, Ericsson J. Control of Smad7 stability by competition between acetylation and ubiquitination. Mol Cell. 2002;10:483–93. doi: 10.1016/s1097-2765(02)00639-1. [DOI] [PubMed] [Google Scholar]
- Hayashi H, Abdollah S, Qiu Y, Cai J, Xu YY, Grinnell BW, et al. The MAD-related protein Smad7 associates with the TGFbeta receptor and functions as an antagonist of TGFbeta signaling. Cell. 1997;89:1165–73. doi: 10.1016/s0092-8674(00)80303-7. [DOI] [PubMed] [Google Scholar]
- Heitzer MD, DeFranco DB. Hic-5/ARA55, a LIM domain-containing nuclear receptor coactivator expressed in prostate stromal cells. Cancer Res. 2006a;66:7326–33. doi: 10.1158/0008-5472.CAN-05-2379. [DOI] [PubMed] [Google Scholar]
- Heitzer MD, DeFranco DB. Mechanism of action of Hic-5/androgen receptor activator 55, a LIM domain-containing nuclear receptor coactivator. Mol Endocrinol. 2006b;20:56–64. doi: 10.1210/me.2005-0065. [DOI] [PubMed] [Google Scholar]
- Hong S, Lee C, Kim SJ. Smad7 sensitizes tumor necrosis factor induced apoptosis through the inhibition of antiapoptotic gene expression by suppressing activation of the nuclear factor-kappaB pathway. Cancer Res. 2007;67:9577–83. doi: 10.1158/0008-5472.CAN-07-1179. [DOI] [PubMed] [Google Scholar]
- Horrevoets AJ. Plasminogen activator inhibitor 1 (PAI-1): in vitro activities and clinical relevance. Br J Haematol. 2004;125:12–23. doi: 10.1111/j.1365-2141.2004.04844.x. [DOI] [PubMed] [Google Scholar]
- Kavsak P, Rasmussen RK, Causing CG, Bonni S, Zhu H, Thomsen GH, et al. Smad7 binds to Smurf2 to form an E3 ubiquitin ligase that targets the TGF beta receptor for degradation. Mol Cell. 2000;6:1365–75. doi: 10.1016/s1097-2765(00)00134-9. [DOI] [PubMed] [Google Scholar]
- Kim SG, Kim HA, Jong HS, Park JH, Kim NK, Hong SH, et al. The endogenous ratio of Smad2 and Smad3 influences the cytostatic function of Smad3. Mol Biol Cell. 2005;16:4672–83. doi: 10.1091/mbc.E05-01-0054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koinuma D, Shinozaki M, Komuro A, Goto K, Saitoh M, Hanyu A, et al. Arkadia amplifies TGF-beta superfamily signalling through degradation of Smad7. Embo J. 2003;22:6458–70. doi: 10.1093/emboj/cdg632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komuro A, Imamura T, Saitoh M, Yoshida Y, Yamori T, Miyazono K, et al. Negative regulation of transforming growth factor-beta (TGF-beta) signaling by WW domain-containing protein 1 (WWP1) Oncogene. 2004;23:6914–23. doi: 10.1038/sj.onc.1207885. [DOI] [PubMed] [Google Scholar]
- Lallemand F, Mazars A, Prunier C, Bertrand F, Kornprost M, Gallea S, et al. Smad7 inhibits the survival nuclear factor kappaB and potentiates apoptosis in epithelial cells. Oncogene. 2001;20:879–84. doi: 10.1038/sj.onc.1204167. [DOI] [PubMed] [Google Scholar]
- Lallemand F, Seo SR, Ferrand N, Pessah M, L’Hoste S, Rawadi G, et al. AIP4 restricts transforming growth factor-beta signaling through a ubiquitination-independent mechanism. J Biol Chem. 2005;280:27645–53. doi: 10.1074/jbc.M500188200. [DOI] [PubMed] [Google Scholar]
- Landstrom M, Heldin NE, Bu S, Hermansson A, Itoh S, ten Dijke P, et al. Smad7 mediates apoptosis induced by transforming growth factor beta in prostatic carcinoma cells. Curr Biol. 2000;10:535–8. doi: 10.1016/s0960-9822(00)00470-x. [DOI] [PubMed] [Google Scholar]
- Massague J, Andres J, Attisano L, Cheifetz S, Lopez-Casillas F, Ohtsuki M, et al. TGF-beta receptors. Mol Reprod Dev. 1992;32:99–104. doi: 10.1002/mrd.1080320204. [DOI] [PubMed] [Google Scholar]
- Massague J, Blain SW, Lo RS. TGFbeta signaling in growth control, cancer, and heritable disorders. Cell. 2000;103:295–309. doi: 10.1016/s0092-8674(00)00121-5. [DOI] [PubMed] [Google Scholar]
- Matsuya M, Sasaki H, Aoto H, Mitaka T, Nagura K, Ohba T, et al. Cell adhesion kinase beta forms a complex with a new member, Hic-5, of proteins localized at focal adhesions. J Biol Chem. 1998;273:1003–14. doi: 10.1074/jbc.273.2.1003. [DOI] [PubMed] [Google Scholar]
- Mazars A, Lallemand F, Prunier C, Marais J, Ferrand N, Pessah M, et al. Evidence for a role of the JNK cascade in Smad7-mediated apoptosis. J Biol Chem. 2001;276:36797–803. doi: 10.1074/jbc.M101672200. [DOI] [PubMed] [Google Scholar]
- Mori K, Asakawa M, Hayashi M, Imura M, Ohki T, Hirao E, et al. Oligomerizing potential of a focal adhesion LIM protein Hic-5 organizing a nuclear-cytoplasmic shuttling complex. J Biol Chem. 2006;281:22048–61. doi: 10.1074/jbc.M513111200. [DOI] [PubMed] [Google Scholar]
- Moustakas A, Heldin CH. Non-Smad TGF-beta signals. J Cell Sci. 2005;118:3573–84. doi: 10.1242/jcs.02554. [DOI] [PubMed] [Google Scholar]
- Nakao A, Afrakhte M, Moren A, Nakayama T, Christian JL, Heuchel R, et al. Identification of Smad7, a TGFbeta-inducible antagonist of TGF-beta signalling. Nature. 1997;389:631–5. doi: 10.1038/39369. [DOI] [PubMed] [Google Scholar]
- Nishiya N, Tachibana K, Shibanuma M, Mashimo JI, Nose K. Hic-5-reduced cell spreading on fibronectin: competitive effects between paxillin and Hic-5 through interaction with focal adhesion kinase. Mol Cell Biol. 2001;21:5332–45. doi: 10.1128/MCB.21.16.5332-5345.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin XF, An DS, Chen IS, Baltimore D. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100:183–8. doi: 10.1073/pnas.232688199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi RA, Leof EB. TGF-beta signaling: a tale of two responses. J Cell Biochem. 2007;102:593–608. doi: 10.1002/jcb.21501. [DOI] [PubMed] [Google Scholar]
- Rahman MM, Miyamoto H, Lardy H, Chang C. Inactivation of androgen receptor coregulator ARA55 inhibits androgen receptor activity and agonist effect of antiandrogens in prostate cancer cells. Proc Natl Acad Sci U S A. 2003;100:5124–9. doi: 10.1073/pnas.0530097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibanuma M, Kim-Kaneyama JR, Ishino K, Sakamoto N, Hishiki T, Yamaguchi K, et al. Hic-5 communicates between focal adhesions and the nucleus through oxidant-sensitive nuclear export signal. Mol Biol Cell. 2003;14:1158–71. doi: 10.1091/mbc.02-06-0099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibanuma M, Mashimo J, Kuroki T, Nose K. Characterization of the TGF beta 1-inducible hic-5 gene that encodes a putative novel zinc finger protein and its possible involvement in cellular senescence. J Biol Chem. 1994;269:26767–74. [PubMed] [Google Scholar]
- Shibanuma M, Mochizuki E, Maniwa R, Mashimo J, Nishiya N, Imai S, et al. Induction of senescence-like phenotypes by forced expression of hic-5, which encodes a novel LIM motif protein, in immortalized human fibroblasts. Mol Cell Biol. 1997;17:1224–35. doi: 10.1128/mcb.17.3.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsson M, Heldin CH, Ericsson J, Gronroos E. The balance between acetylation and deacetylation controls Smad7 stability. J Biol Chem. 2005;280:21797–803. doi: 10.1074/jbc.M503134200. [DOI] [PubMed] [Google Scholar]
- Song K, Cornelius SC, Danielpour D. Development and characterization of DP-153, a nontumorigenic prostatic cell line that undergoes malignant transformation by expression of dominant-negative transforming growth factor beta receptor type II. Cancer Res. 2003a;63:4358–67. [PubMed] [Google Scholar]
- Song K, Cornelius SC, Reiss M, Danielpour D. Insulin-like growth factor-I inhibits transcriptional responses of transforming growth factor-beta by phosphatidylinositol 3-kinase/Akt-dependent suppression of the activation of Smad3 but not Smad2. J Biol Chem. 2003b;278:38342–51. doi: 10.1074/jbc.M304583200. [DOI] [PubMed] [Google Scholar]
- Song K, Krebs TL, Danielpour D. Novel permissive role of epidermal growth factor in transforming growth factor beta (TGF-beta) signaling and growth suppression. Mediation by stabilization of TGF-beta receptor type II. J Biol Chem. 2006a;281:7765–74. doi: 10.1074/jbc.M511781200. [DOI] [PubMed] [Google Scholar]
- Song K, Wang H, Krebs TL, Danielpour D. Novel roles of Akt and mTOR in suppressing TGF-beta/ALK5-mediated Smad3 activation. Embo J. 2006b;25:58–69. doi: 10.1038/sj.emboj.7600917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang B, de Castro K, Barnes HE, Parks WT, Stewart L, Bottinger EP, et al. Loss of responsiveness to transforming growth factor beta induces malignant transformation of nontumorigenic rat prostate epithelial cells. Cancer Res. 1999;59:4834–42. [PubMed] [Google Scholar]
- Thomas SM, Hagel M, Turner CE. Characterization of a focal adhesion protein, Hic-5, that shares extensive homology with paxillin. J Cell Sci. 1999;112 ( Pt 2):181–90. doi: 10.1242/jcs.112.2.181. [DOI] [PubMed] [Google Scholar]
- Tumbarello DA, Turner CE. Hic-5 contributes to epithelial-mesenchymal transformation through a RhoA/ROCK-dependent pathway. J Cell Physiol. 2007;211:736–47. doi: 10.1002/jcp.20991. [DOI] [PubMed] [Google Scholar]
- von Gersdorff G, Susztak K, Rezvani F, Bitzer M, Liang D, Bottinger EP. Smad3 and Smad4 mediate transcriptional activation of the human Smad7 promoter by transforming growth factor beta. J Biol Chem. 2000;275:11320–6. doi: 10.1074/jbc.275.15.11320. [DOI] [PubMed] [Google Scholar]
- Wakefield LM, Roberts AB. TGF-beta signaling: positive and negative effects on tumorigenesis. Curr Opin Genet Dev. 2002;12:22–9. doi: 10.1016/s0959-437x(01)00259-3. [DOI] [PubMed] [Google Scholar]
- Wang H, Song K, Sponseller TL, Danielpour D. Novel function of androgen receptor-associated protein 55/Hic-5 as a negative regulator of Smad3 signaling. J Biol Chem. 2005;280:5154–62. doi: 10.1074/jbc.M411575200. [DOI] [PubMed] [Google Scholar]
- Wicks SJ, Haros K, Maillard M, Song L, Cohen RE, Dijke PT, et al. The deubiquitinating enzyme UCH37 interacts with Smads and regulates TGF-beta signalling. Oncogene. 2005;24:8080–4. doi: 10.1038/sj.onc.1208944. [DOI] [PubMed] [Google Scholar]
- Wilkes MC, Leof EB. Transforming growth factor beta activation of c-Abl is independent of receptor internalization and regulated by phosphatidylinositol 3-kinase and PAK2 in mesenchymal cultures. J Biol Chem. 2006;281:27846–54. doi: 10.1074/jbc.M603721200. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–61. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrana JL, Attisano L. MAD-related proteins in TGF-beta signalling. Trends Genet. 1996;12:493–6. doi: 10.1016/s0168-9525(96)30109-1. [DOI] [PubMed] [Google Scholar]
- Wrana JL, Carcamo J, Attisano L, Cheifetz S, Zentella A, Lopez-Casillas F, et al. The type II TGF-beta receptor signals diverse responses in cooperation with the type I receptor. Cold Spring Harb Symp Quant Biol. 1992;57:81–6. doi: 10.1101/sqb.1992.057.01.011. [DOI] [PubMed] [Google Scholar]
- Yang J, Song K, Krebs TL, Jackson MW, Danielpour D. Rb/E2F4 and Smad2/3 link survivin to TGF-beta-induced apoptosis and tumor progression. Oncogene. 2008 doi: 10.1038/onc.2008.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue J, Frey RS, Mulder KM. Cross-talk between the Smad1 and Ras/MEK signaling pathways for TGFbeta. Oncogene. 1999;18:2033–7. doi: 10.1038/sj.onc.1202521. [DOI] [PubMed] [Google Scholar]
- Zhang S, Fei T, Zhang L, Zhang R, Chen F, Ning Y, et al. Smad7 antagonizes transforming growth factor beta signaling in the nucleus by interfering with functional Smad-DNA complex formation. Mol Cell Biol. 2007;27:4488–99. doi: 10.1128/MCB.01636-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng X, We R, Derynck R. Receptor-associated Mad homologues synergize as effectors of the TGF-beta response. Nature. 1996;383:168–72. doi: 10.1038/383168a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.