Abstract
Streptomyces coelicolor is the genetically best characterized species of a populous genus belonging to the Gram-positive Actinobacteria. Streptomycetes are filamentous soil organisms, well known for the production of a plethora of biologically active secondary metabolic compounds. The Streptomyces developmental life cycle is uniquely complex, and involves coordinated multicellular development with both physiological and morphological differentiation of several cell types, culminating in production of secondary metabolites and dispersal of mature spores. This review presents a current appreciation of the signaling mechanisms used to orchestrate the decision to undergo morphological differentiation, and the regulators and regulatory networks that direct the intriguing development of multigenomic hyphae, first to form specialized aerial hyphae, and then to convert them into chains of dormant spores. This current view of S. coelicolor development is destined for rapid evolution as data from “-omics” studies shed light on gene regulatory networks, new genetic screens identify hitherto unknown players, and the resolution of our insights into the underlying cell biological processes steadily improve.
Keywords: Differentiation, Development, Sporulation, Cell division, Chromosome segregation
Introduction
Streptomycetes belong to nature’s most competent chemists and produce a stunning multitude and diversity of secondary metabolites. These soil bacteria continue to provide a goldmine of antibiotics and other biologically active compounds that have been and will be explored by generations of microbiologists (Hopwood, 2007). Streptomycetes also exhibit a complex developmental biology, involving mycelial growth, multicellular behaviour, intercellular communication, and morphological differentiation coordinated with various physiological processes. The developmental life cycle culminates in production of dormant spores that allow the otherwise sessile mycelial organisms to spread to new locations.
Streptomyces development has been the subject of intense genetic and molecular biology research since the isolation of the first mutants specifically blocked in the process (Hopwood, et al., 1970). Much progress has been made with the excellent model organism S. coelicolor, but there is also variation in the developmental life cycles and the strategies used to control them among streptomycetes. Most notably, extensive studies of the streptomycin-producer S. griseus have revealed both conserved features and important and intriguing variations compared to S. coelicolor (reviewed in e.g., Chater & Horinouchi, 2003, Horinouchi, 2007). Streptomycetes are members of the phylum Actinobacteria, comprising Gram-positive bacteria with high GC content in their DNA. This is a large and ancient (deeply branching) group of bacteria with many interesting features. Various members represent a gradient of morphological and developmental complexity, from simple coccoid cells like the Micrococcus, and rod-shaped or pleiomorphic organisms like the industrially important Corynebacterium, and pathogens like Mycobacterium tuberculosis, to the highly complex mycelium of Streptomyces and related genera. An informative dimension has now been added by the rapidly growing genome sequence information, which opens fantastic possibilities for comparative and evolutionary studies, both within Streptomyces and among the Actinobacteria (see for example Chater & Chandra, 2006, Ventura, et al., 2007). Nonetheless, our basic understanding of Streptomyces developmental life cycle will continue to depend heavily on genetic, molecular, and cell biological analyses of good experimental model organisms like S. coelicolor.
Our aim is to review the current understanding of how Streptomyces morphological differentiation is regulated, strongly biased towards what has been learned from experimental work on S. coelicolor. We will focus specifically on the regulatory networks that direct formation of spore-bearing aerial hyphae and the production of dormant spores. This is followed by an overview of stimuli and signaling mechanisms coordinating the decision to initiate morphological development. Finally, we discuss links between the developmental regulators and key cell biological processes like cell division, chromosome segregation, cell wall assembly and nucleoid structure. However, we will begin this review with the outsider’s perspective in mind, and will start with a brief overview of Streptomyces development in order to help disseminate the knowledge to a broader audience. After setting the stage, we continue with more detailed descriptions of developmental regulation in Streptomyces, focusing mainly on recent progress. Physiological differentiation and secondary metabolism occur at similar time as morphological differentiation; however, we will not comment much on that subject except where the information supports the discussion on development. A range of excellent previous reviews on different aspects of Streptomyces development are available, and are referred to when appropriate in the following text.
Streptomyces development for the uninitiated
The polarized growth of Streptomyces hyphae
The vegetative phase of growth starts by germination of a dormant spore when it encounters a suitable environment. Germination involves swelling of the spore, followed by establishment of cell polarity and apical growth, leading to outgrowth of one or more hyphae from the spore. The hyphae grow by tip extension and branching and give rise to mycelial networks, which are more reminiscent of filamentous fungi than of other bacteria. The pronounced apical assembly of the peptidoglycan wall contrasts sharply with growth of well-known rod-shaped bacteria like Escherichia coli and Bacillus subtilis, but, this unique mode of directing cell wall synthesis is shared by other members of the Actinobacteria such as corynebacteria and mycobacteria (Daniel & Errington, 2003, Margolin, 2009). While the actin-like MreB cytoskeleton has a key role in orchestration of lateral cell wall synthesis in most other rod-shaped bacteria, Streptomyces and its actinobacterial relatives instead use the coiled-coil protein DivIVA for directing apical cell wall synthesis (Flärdh, 2003, Hempel, et al., 2008, Letek, et al., 2008). Remarkably, neither cell division, which gives rise to the widely-spaced crosswalls that delimit the syncytial hyphal cells, nor genes known to be involved in chromosome segregation are required for vegetative hyphal growth in Streptomyces (McCormick, et al., 1994, Dedrick, et al., 2009, McCormick, 2009). Instead tip extension, hyphal branching, and presumably pole-ward transport of nucleoids, are crucial for efficient growth. We refer the reader to recent reviews for further discussion of apical growth in streptomycetes (Flärdh & Buttner, 2009, Margolin, 2009, Flärdh, 2010).
Multicellular development
The growing vegetative (or substrate) mycelium explores the environment for available nutrients, degrading encountered polymeric substrates by secretion of hydrolytic enzymes. On solid laboratory medium, Streptomyces colonies develop over the course of a few days into a complex multicellular consortium of different cell types (see e.g., Chater, 1998). While peripheral regions of the colony may still be growing, non-growing parts of the colony start secondary metabolism and synthesis of antibiotics. At the same time, specialized aerial hyphae emerge on the colony surface, many of which go on to produce spores. The lysis of substrate mycelium in central parts of the colony, perhaps through a form of programmed cell death (Miguelez, et al., 1999), is thought to release nutrients that can be recycled for developmental processes. Thus antibiotic production might be for protection of this endogenous source of nutrients from exploitation by other neighboring organisms (Chater, 2011). Finally, the fact that both physiological and morphological differentiation involve several types of extracellular communication and signaling (as will be discussed below) further contributes to the concept of multicellular development in a Streptomyces colony (Chater & Losick, 1997, Elliot, et al., 2008, Chater, et al., 2010).
Formation of an aerial mycelium involves assembly of a hydrophobic sheath on the cell surface
The vegetative mycelium is nonmotile, and streptomycetes use dormant unicellular spores as the means for dispersal in the environment. In the majority of species, spores are formed on reproductive aerial hyphae, which form a fuzzy layer on the colony surface called the aerial mycelium (Fig. 1)(Wildermuth, 1970). The aerial hyphae and mature spores, but not the vegetative hyphae, are surrounded by a superficial fibrous sheath that give them a hydrophobic cell surface (Fig. 2AB) (Hopwood, et al., 1970). At least 3 different classes of proteins – the lantibiotic-like SapB peptide, the chaplins, and the rodlins - are parts of the sheath that enables aerial hyphae to escape the aqueous substrate and extend into the air. These components have been thoroughly reviewed elsewhere (see e.g., Willey, et al., 2006, Flärdh & Buttner, 2009, Chater, et al., 2010), and are only briefly introduced here.
Figure 1.
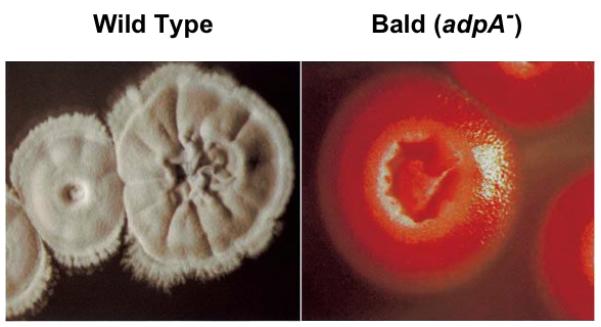
Visual appearance of the surfaces of wild type and bald mutant colonies. Wild type S. coelicolor colonies differentiate and form an aerial mycelium with a fuzzy gray appearance due to the conversion of aerial hyphae into chains of gray-pigmented spores. In contrast, the bald mutant, with a mutation in the adpA (bldH) gene, produces smooth and lustrous colonies lacking an aerial mycelium. For this particular bld mutant, the red-pigmented antibiotic undecylprodigiosin is still produced. Colonies were grown for 5 days on rich medium (R2YE) at 30°C. This figure was adapted from (Nguyen, et al., 2003) with permission from American Society for Microbiology.
Figure 2.
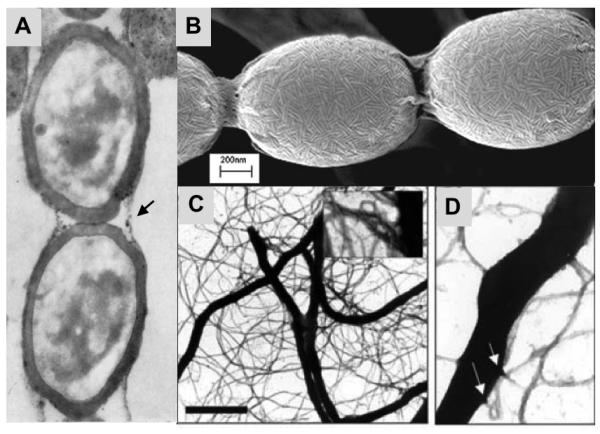
Cell surface of spores derived from aerial hyphae and the extracellular matrix of vegetative hyphae involved in attachment to hydrophobic surfaces. Panel A, A TEM image of a portion of a chain of mature spores from a wild type strain is shown. The thickened spore walls are surrounded by vestiges of a superficial fibrous sheath (black arrow). Image reprinted from (Hopwood, et al., 1970) with permission from Society for General Microbiology. Panel B, The SEM image shows that chaplin fibers assemble into paired rodlet structures on the surface of the spore. The assembly is dependent on the rodlin proteins, but the rodlins are not structural components of the structures. Image reproduced from (Di Berardo, et al., 2008) with permission from American Society for Microbiology. Panels C and D, Formation of fimbriae in extracellular matrix of vegetative hyphae are revealed by TEM with negative staining. Fimbriae are formed in the extracellular matrix in a minimal medium with mannitol, but not glucose, as carbon source. Arrows in D highlight that fibers emanate from spiked protrusions. Scale bar in C is 5 μm for the image in panel C, 100 nm for inset panel of C and 1.25 μm for panel D. Images reproduced from (de Jong, et al., 2009a) with permission from the authors.
SapB was the first hydrophobic morphogenetic protein discovered in S. coelicolor (Willey, et al., 1991). It derives from the C-terminal half of a precursor protein called RamS, and is the result of proteolytic processing and posttranslational modification in a fashion similar to the processing of lantibiotics (Kodani, et al., 2004), a class of peptide antibiotics. However, SapB is unusual in that it lacks antibiotic activity and has a structural role. RamS is encoded in the rapid aerial mycelium operon (ramCSAB) (Nguyen, et al., 2002, O’Connor, et al., 2002). The other coding regions of the ram locus are believed to be responsible for at least some of the posttranslational modification (RamC) and as components of an ABC type transporter (RamAB) for the export of the finished product (Willey, et al., 2006). SapB exists both in the medium surrounding the colony and tightly associated with the surface of the spore, but it is still unclear if the final processing of RamS (preSapB) occurs intracellularly, extracellularly or during export. SapB is required for aerial mycelium formation only on certain media (Willey, et al., 1991, Capstick, et al., 2007).
The chaplins (Chp) are a family of secreted hydrophobic proteins that are necessary for constructing the fibrous sheath on aerial hyphae (Claessen, et al., 2003, Elliot, et al., 2003b). These proteins are synthesized under all tested growth conditions, and are responsible for the SapB-independent pathway for aerial mycelium formation that is observed on some media (Capstick, et al., 2007). Characterized by a common hydrophobic chaplin domain, these proteins sort into two groups: long chaplins ChpA-C (ca. 225 aa) containing two copies of the chaplin domain, and short chaplins ChpD-H (ca. 55 aa) consisting of only one domain. The long chaplins are believed to be attached to the cell wall, and are required to organize the remainder of the shorter chaplin variants into paired rodlet fibers decorating the surface of the mature spores (see SEM in Fig. 2B). Recent data show that all of the small chaplins are capable of forming amyloid fibrils in vitro, apparently similar to the structures found on the spore surface, and that the chaplin domain, as studied in ChpH, contains two amyloidogenic regions that contribute differentially to the assembly of the chaplin fibrils (Capstick, et al., 2011, Sawyer, et al., 2011). It has recently been shown that chaplins are also structural components of amyloid-like fimbriae that are involved in surface attachment of S coelicolor hyphae grown in the presence of hydrophobic surfaces (Fig. 2CD)(de Jong, et al., 2009a). These fimbriae are organized and attached to the cells using cellulose-like fibres that emanate from protrusions on the hyphal surface. Analysis of a mutant lacking the cellulose synthase encoded by cslA suggest that the cellulose-like compound is also involved in assembly of the hydrophobic sheath of aerial hyphae (Xu, et al., 2008), suggesting parsimonial roles of both chaplins and cellulose in formation of different types of cell surface structures.
Rodlins are the third known class of proteins participating in the formation of the surface structure of aerial hyphae in S. coelicolor (Claessen, et al., 2002). Rodlins influence the organization of the chaplins into rodlet structures, but are not themselves components of the distinct rodlet layer observed on the spore surface (Fig. 2B), and are not required for the ability to form aerial mycelium or acquisition of surface hydrophobicity (Claessen, et al., 2002, Claessen, et al., 2004, Sawyer, et al., 2011). Homologues of the rodlin-encoding genes are only present in some members of the Streptomycetaceae family, and consistent with their role in organizing the rodlet layer, an organism that lacks rodlins (S. avermitilis) has a smooth spore surface (Claessen, et al., 2004).
Differentiation of the aerial mycelium
It is the apical cell of an aerial hypha that is converted to spores, and we will refer to this as the sporogenic cell (Fig. 3). The delimitation of an apical sporogenic cell from the subapical stem by a crosswall is referred to as a basal septum (arrows in Fig. 3), although it is only well characterized in S. griseus (Wildermuth & Hopwood, 1970, Kwak, et al., 2001b). Formation of the basal septum is likely to be a key event in the developmental process, and allows some developmental genes to be specifically expressed in either the apical sporogenic cell or in the subapical stem compartment (discussed in e.g., Dalton, et al., 2007, Flärdh & Buttner, 2009). Each long apical cell is subdivided synchronously by evenly-spaced sporulation septa into a string of unicellular prespores (Fig. 3), and these septa eventually permit the separation and release of individual spores. Closely coordinated with septation, copies of the linear genome are actively segregated and evenly distributed into prespores.
Figure 3.
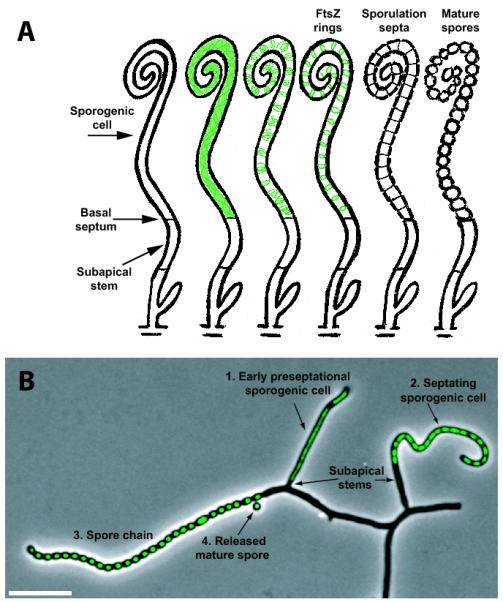
Stages in the differentiation of aerial hyphae into chains of spores. (A) Schematic representation of the conversion of the apical sporogenic cell of an aerial hypha into a chain of spores. A basal septum delimits the sporogenic cell from the underlying cell (referred to as a “subapical stem”), and provides the ability to have compartment-specific gene expression. The developmental upregulation of FtsZ production in the sporogenic cell, the assembly of FtsZ into helical filaments, and then shorter spirals coalesce into regularly positioned Z rings are indicated (green). Synchronous division and metamorphosis of the resulting string of single-celled compartments results in a chain of spores. Image reproduced from (Grantcharova, et al., 2005) with permission from American Society for Microbiology. (B) The production and nucleoid-localization of a HupS-EGFP fusion protein is used as a molecular marker of the sporogenic apical cells. The hypha labeled 1 shows an aerial hyphal branch in which a clear distinction is seen between the sporogenic apical cell in which hupS is expressed and a short subapical stem. In hypha 2, sporulation septation is in progress in the sporogenic cell, as indicated by the separation of individual nucleoids. In hypha 3, constrictions indicate that septation has been completed and spores are rounding up while laying down a thick spore wall. A released mature spore with a condensed nucleoid is also indicated (4). HupS-EGFP fluorescence is shown in green overlay on a phase-contrast image. Scale bar, 8 μm. Image reproduced and amended from (Salerno, et al., 2009) with permission from American Society for Microbiology.
Properties of mature Streptomyces spores
The differentiation of Streptomyces prespores into mature spores prepares them for dispersal and survival in a quiescent state for extended periods of time, able to withstand desiccation and other physical or chemical challenges. However, Streptomyces spores are significantly less resistant to adverse conditions than endospores produced by Bacillus and other genera of the Firmicutes (low G+C Gram-positives), and differ ultrastructurally in several ways. After sporulation septation has been completed, Streptomyces spore maturation involves the formation of a thick spore wall (Fig. 2A) (Wildermuth & Hopwood, 1970, Potúcková, et al., 1995), which has to be assembled from the inside of the prespore itself. In sharp contrast, the deposition of the cortex and coat layers of Bacillus endospores is accomplished from the mother cell, i.e., from the outside of the developing prespore (Henriques & Moran, 2007). Further, in order to maintain genetic integrity, spore DNA should be protected to prevent damage. In Bacillus, a class of small, acid soluble, DNA-binding proteins (SASPs) and the characteristic accumulation of dipicolinic acid (DPA) complexed with Ca2+ strongly contribute to the stress resistance and longevity of the endospores (Setlow, 2007), but DPA and SASP homologues are absent from Streptomyces spores (Ensign, 1978). Instead, other developmentally controlled nucleoid-associated proteins contribute to nucleoid structure and protection in Streptomyces spores (Fig. 3B)(see below and Chater, 2011). Finally, during maturation of the prespores, a pigment is produced, which in S. coelicolor is a grey-pigmented compound synthesized by enzymes encoded in the whiE gene cluster (Davis & Chater, 1990).
Key developmental regulators identified by classical genetics
Classical genetic screens led to isolation of developmentally defective mutants, being of two main phenotypic categories. Mutants with the Bld phenotype (Fig. 1) fail to form an aerial mycelium and therefore have a “bald” and lustrous appearance compared to the hirsute wild type colonies (Hopwood, et al., 1970). Other mutants can make aerial mycelium but fail to produce the spore pigment, resulting in a characteristic white colony surface – the Whi phenotype (Hopwood, et al., 1970, Chater, 1972). Such Whi mutants are blocked at different stages of spore formation, but are in a few cases only defective in synthesis of the pigment. Studies of Bld and Whi mutants have led to the identification a range of genes required at different steps of development of aerial hyphae to mature spores. Many of these genes encode various types of developmental regulators. The following section describes the developmental regulatory networks in S. coelicolor in which several of the bld and whi genes have central positions.
Regulatory pathways in S. coelicolor sporulation
Here we aim to give a current view of the regulatory pathways governing morphological differentiation in Streptomyces. We focus mainly on established direct interactions between regulators in S. coelicolor, recent developments in the field, and some of the intriguing key questions that we suggest require attention.
AdpA - an important early regulator of development
From studies in S. griseus, AdpA (A-factor-dependent protein) has been identified as a major regulator of both secondary metabolism and morphological differentiation (reviewed by Horinouchi, 2007). This AraC/XylS family protein mediates the regulatory response to the γ-butyrolactone signal molecule A-factor in S. griseus. The A-factor-specific receptor (ArpA) has only one known specific target in S. griseus – it represses the adpA promoter. In the presence of A-factor, ArpA dissociates from DNA, allowing induction of adpA expression (Ohnishi, et al., 1999). The AdpA protein then transcriptionally activates a large regulon of genes involved in sporulation or antibiotic production, including the gene for the pathway-specific regulator of streptomycin biosynthesis (strR); other secondary metabolite genes; amfR regulating genes for formation of the S. griseus SapB orthologue; adsA encoding the RNA polymerase σ factor AdsA/BldN; ssgA required for control of sporulation septation; sgiA encoding a protease inhibitor protein (all are discussed elsewhere in this paper); and several genes for secreted proteases and other extracellular proteins (see references in Horinouchi, 2007). The large effects of adpA on A- factor- and stationary phase-induced gene expression have been further substantiated and extended by transcriptomic and proteomic analyses (Akanuma, et al., 2009, Hara, et al., 2009).
AdpA has a key role also in development of S. coelicolor, but the picture of control is not yet as complete. One of the classical bld loci in S. coelicolor, bldH (mutant phenotype shown in Fig. 1) (Champness, 1988), encodes the S. coelicolor AdpA orthologue (Nguyen, et al., 2003, Takano, et al., 2003a). Henceforth, we will refer to the S. coelicolor gene as adpA. Microarray analyses revealed decreased expression of several sporulation genes in a S. coelicolor adpA deletion mutant (Xu, et al., 2009b). Transcription of the developmentally regulated protease inhibitor gene sti1 (orthologous to the S. griseus AdpA-target gene sgiA) is adpA-dependent (Kim, et al., 2005, Xu, et al., 2009b), and AdpA binds directly to and activates the sti1 promoter (Fig. 4) (Wolanski, et al., 2011). In addition, AdpA binds to the promoter of ramR, and expression of this gene is strongly reduced in an adpA mutant (Wolanski, et al., 2011). RamR is the regulator of ramCSAB (Fig. 4) and therefore directly controls production of the surface-active SapB peptide (see above)(Nguyen, et al., 2002, O’Connor, et al., 2002).
Figure 4.
Summary of an early part of the developmental regulatory pathways in S. coelicolor, centered on the regulator AdpA and leading to aerial mycelium formation. Demonstrated direct interactions at transcriptional, translational, or post-translational levels are shown as solid lines. Filled arrows indicate positive or activating effects, perpendicular lines indicate negative or repressing effects, and lines ending in bullet indicate that direct interaction has been demonstrated, but the effect on gene expression is not clear. In the case of STI, it is known that expression of sti is strongly decreased in a bldD mutant, but it has not been clarified whether this is mediated by the direct binding of BldD to the promoter region of sti (open arrowhead), or via an indirect effect. One BldD target that is not mentioned in the text is bldC, encoding a MerR-family transcription factor that is conditionally required for aerial mycelium formation (Hunt, et al., 2005). Dashed lines indicate hypothetical interactions. The effects of AdpA on STI and σBldN are based on genetic data and extrapolations from data for S. griseus. Dotted arrows not pointing to symbols are used to indicate unknown but anticipated target genes. The question mark indicates unknown mechanisms for counteracting BldD activity.
Another likely AdpA target is bldN, encoding the ECF family sigma factor σBldN that is unconditionally required for aerial mycelium formation in S. coelicolor (Bibb, et al., 2000). The direct orthologue of σBldN in S. griseus, σAdsA, is part of the AdpA regulon (Yamazaki, et al., 2000, Yamazaki, et al., 2004). Expression from the bldN promoter in S. coelicolor requires adpA (Fig. 4), but contradicting data have been reported on whether this is reflected in the abundance of σBldN/AdsA protein (Bibb, et al., 2000, Bibb & Buttner, 2003, Takano, et al., 2003a, Xu, et al., 2009b). One of the approaches that led to discovery of the chaplin proteins (discussed above) was a transcriptomic comparison between the bldN mutant and its wildtype parent, and all the eight chaplin genes fail to be upregulated in the mutant (Elliot, et al., 2003b). However, it is still unclear whether chp genes are directly controlled by σBldN. Hitherto, the only known direct target for σBldN in S. coelicolor is bldM (Fig. 4), and the bldM mutant phenotype is similar to that of bldN (Bibb, et al., 2000, Molle & Buttner, 2000). BldM is an atypical response regulator that may not need to be phosphorylated to carry out its biological function (Molle & Buttner, 2000). So far, no genes controlled by BldM have been identified.
Overall, AdpA emerges as a major regulator of morphological development also in S. coelicolor, and it appears to control both an extracellular proteolytic cascade (via sti1) and production of key hydrophobic sheath components SapB (via ramR) and chaplins (potentially via bldN), all having crucial roles in aerial mycelium formation. Intriguingly, both adpA and SapB-deficient mutants have only conditional Bld phenotypes, and it would be interesting to know whether bldN can be expressed independently of adpA on mannitol-containing medium on which adpA mutants are phenotypically suppressed but bldN and chaplin-free mutants are still bald (Champness, 1988, Bibb, et al., 2000, Capstick, et al., 2007).
Regulation of adpA involves a feedforward loop with the bldA tRNA
A fundamental difference from the S. griseus case is that S. coelicolor adpA does not respond to γ-butyrolactone signaling, consistent with the lack of effect of these signaling compounds on morphological differentiation in S. coelicolor (Takano, et al., 2003a, Takano, 2006). Then, how is adpA developmentally regulated in S. coelicolor? The cellular content of AdpA protein changes during development, peaking during the time of aerial mycelium formation, and decreasing late during development (Wolanski, et al., 2011). At the transcriptional level, adpA is autoregulated (Wolanski, et al., 2011), directly controlled by bldD (Takano, et al., 2003a, den Hengst, et al., 2010), and the transcripts are a substrate for RNase III (encoded by absB) (Huang, et al., 2005, Xu, et al., 2009b). However, respective contributions are not obvious since S. coelicolor adpA is transcribed throughout development in surface cultures (Takano, et al., 2003a).
At the translational level, adpA depends on the bldA tRNA - the only tRNA that reads the leucine codon TTA. The TTA codon is significantly underrepresented in Streptomyces genomes, and viability of bldA mutants shows that it is not present in any essential gene (for a thorough review of the roles of bldA and TTA codons in Streptomyces development and secondary metabolism, see Chater & Chandra, 2008). A survey of four sequenced Streptomyces genomes revealed around 150-250 genes with TTA codons per organism, but only three are present and have TTA codons in all four genomes, two of which are of unknown function and the third being adpA (Chater & Chandra, 2008). We extended the correlation of previous analyses to 23 adpA genes currently available in GenBank and Streptomyces genome sequences and found that there is a single TTA codon at the same position in all but two adpA genes. Mutation of the TTA codon in S. coelicolor adpA to the synonymous TTG or CTC codons makes morphological development and aerial mycelium formation largely independent of bldA (Nguyen, et al., 2003, Takano, et al., 2003a). Thus, most of the effect of bldA mutations on morphological differentiation in S. coelicolor is due to restricted expression of adpA, and this can be bypassed by simply removing the TTA codon in adpA.
Importantly, a positive feedback loop between AdpA and bldA has been demonstrated for S. griseus and S. coelicolor (Higo, et al., 2011). In both organisms, AdpA binds to two sites in the promoter region for bldA, and adpA is required for bldA transcription (Fig. 4). Together with the bldA-dependence of adpA translation, this provides an elegant feedforward mechanism to control the production of the key regulator AdpA (Higo, et al., 2011). This finding strongly supports the suggestion that the bldA tRNA is used as a regulatory device for developmental control of gene expression in Streptomyces, allowing efficient translation of TTA-containing genes only at the right stage of growth or development (see discussion and references in Chater & Chandra, 2008). Further evidence for developmental regulation of the bldA tRNA is that BldD (described below) has one of its strongest binding sites in the S. coelicolor genome within the bldA gene (den Hengst, et al., 2010). However, the binding site is located immediately downstream of the sequence encoding to the 3′-end of the mature tRNA, and it is not clear how bldA expression is affected by BldD binding to this site far from the promoter. Perhaps BldD could alter production of an antisense RNA that is reported to initiate within bldA (Leskiw, et al., 1993), or it could function via effects on transcriptional termination or elongation possibly influencing maturation of bldA tRNA (den Hengst, et al., 2010).
BldD – a transition state regulator with wide-ranging effects on Streptomyces development
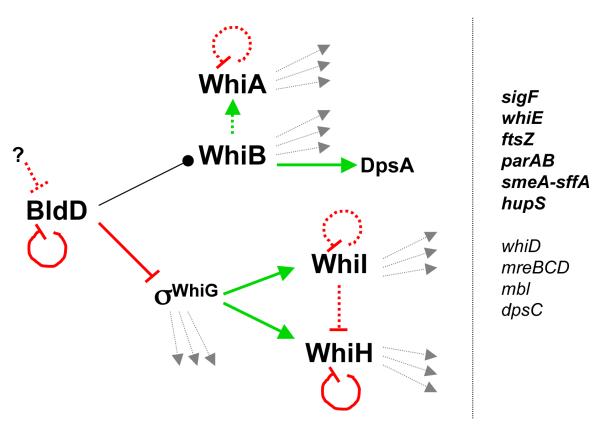
BldD belongs to the XRE family of transcription factors and carries a helix-turn-helix DNA-binding motif similar to the lambda repressor (Kim, et al., 2006). bldD mutants are blocked in aerial mycelium formation on rich medium, but like many other bld mutants, including those mutant for bldA and adpA mentioned above, development is restored on minimal medium with mannitol as carbon source (Merrick, 1976, Champness, 1988, Elliot, et al., 2003a). Initial studies showed that BldD acts as a repressor of some known developmental regulatory genes (Figs. 4-6): bldN, encoding the sigma factor σBldN/AdsA; bldM, encoding a response-regulator-type protein; whiG, encoding the sporulation sigma factor σWhiG; and sigH, encoding the stress-related σH (Elliot, et al., 2001, Kelemen, et al., 2001). These observations led to the suggestion that BldD may work in an analogous way to the so called transition state regulators AbrB and SinR in B. subtilis by repressing developmental genes during exponential growth (Strauch & Hoch, 1993, Elliot, et al., 2001).
Figure 6.
Schematic summary of key regulators required for further differentiation of aerial hyphae into spores in S. coelicolor. Demonstrated direct interactions at transcriptional, translational, or post-translational levels are shown as solid lines. Filled arrows indicate positive or activating effects, perpendicular lines indicate negative or repressing effects, and lines ending in bullet indicate that direct interaction has been demonstrated, but the effect on gene expression is not clear. Dashed lines indicate hypothetical interactions. The question marks indicate unknown mechanisms for counteracting BldD activity. Dotted arrows not pointing to symbols are used to indicate unknown but anticipated target genes. The right-hand part of the figure lists in bold genes that are transcriptionally upregulated during sporulation in a way that requires one or more of the indicated regulators encoded by whiA, whiB, whiG, whiI, or whiH. Other listed genes are some examples that are upregulated during sporulation, but for which there is no data available regarding the mechanism of their dependence on early whi genes.
A recent report reveals the full extent of the vast BldD regulon in the S. coelicolor. Since BldD is mainly produced during vegetative growth, it lends itself very well to chromatin immunoprecipitation-microarray analysis (ChIP-chip), and at least 167 putative BldD targets were identified using this method (den Hengst, et al., 2010). Around 80% of those identified contained a readily recognized 15 bp palindromic sequence motif, and for several genes, it was demonstrated that BldD binds to this motif. This analysis was complemented by a transcriptomic survey of the differences in gene expression between a bldD mutant and the wild type parent during growth in liquid culture. Overall, the results show that BldD controls a very large regulon that includes bldC, whiB, ftsZ, ssgA, ssgB, smeA-sffA, as well as the targets mentioned above and several other genes known to influence morphological differentiation or secondary metabolism (den Hengst, et al., 2010). The BldD binding motif was also found upstream of some developmental genes in other streptomycete genomes (den Hengst, et al., 2010), and BldD is a direct regulator of the erythromycin biosynthesis genes and required to form aerial mycelium in Saccharopolyspora erythraea (Chng, et al., 2008), and represses the amfTSAB operon involved in making the SapB-like peptide AmfS in S. griseus (Ueda, et al., 2005). Clearly, BldD has a conserved and important role in developmental biology of Streptomyces and their relatives.
If the role of BldD is to repress key developmental genes during growth, it is an apparent paradox that bldD loss-of-function mutants are blocked in sporulation. Unless this phenotype is not simply due to the premature expression of developmental regulators, there should be critical genes that directly or indirectly depend on bldD for developmental upregulation. A candidate may be the pathway controlled by AdpA (described above). One of the genes showing the strongest dependence on bldD in the transcriptomic data is sti1, which encodes the extracellular protease inhibitor STI and is directly controlled by AdpA (Kim, et al., 2005, Xu, et al., 2009b, den Hengst, et al., 2010, Wolanski, et al., 2011). In addition, BldD binds to promoters of multiple members of the putative AdpA regulon, including adpA itself, sti1, bldN, as well as an internal site in the bldA tRNA gene (Fig. 4)(den Hengst, et al., 2010). With this in mind, it could be important to determine how bldD affects adpA expression, whether a TTA-free allele of adpA would be expressed in a bldD mutant background, and whether engineering of an adpA allele that is expressed independently of bldD would suppress any of the phenotypic effects of a bldD deletion.
How is BldD-mediated repression of developmental genes relieved?
The significance of BldD-mediated repression of developmental genes is illustrated by a study of sigH indicating that negative control by BldD both temporally and spatially restricts the activity of the promoter sigHp2 to the apical part of sporulating hyphae (Kelemen, et al., 2001). The question then arises how BldD is controlled so that BldD-mediated repression of critical sporulation genes is relieved at the right time and in the right cell type. Transcripts of bldD decrease in abundance during development, but it is not known how bldD is transcriptionally regulated, except that it acts as an autorepressor (Elliot, et al., 1998, Elliot & Leskiw, 1999, Kelemen, et al., 2001). The highest accumulation of BldD protein is during early stages of growth and it decreases during development in surface cultures (den Hengst, et al., 2010). Similar to its relative the lambda repressor, BldD may be subject to proteolytic control and BldD proteolysis activity was highest in cell extracts from spore-forming stages of development of S. coelicolor compared to extracts from vegetative hyphae (Lee, et al., 2007). A mutation inactivating two of the four clpP genes for the proteolytic subunit of the ClpXP protease gives rise to a Bld phenotype in S. lividans (de Crécy-Lagard, et al., 1999), and one of them, clpP1, is in S. coelicolor directly controlled by AdpA (Wolanski, et al., 2011). In addition, mutation of clpX also affects the ability to form aerial mycelium (Viala & Mazodier, 2003). It should be interesting to identify the developmental ClpXP targets and whether BldD may be one of them.
Studies of the whiJ locus give hints about bldB and bldD regulation
Like bldD, whiJ encodes an XRE family protein. whiJ and its two immediate neighbors have multiple paralogs in S. coelicolor, many of which form gene clusters similar to that of whiJ (Gehring, et al., 2000, Aínsa, et al., 2010). Intriguingly, all of the investigated whiJ mutant alleles that give rise to Whi phenotypes allow synthesis of a truncated WhiJ with an intact N-terminal DNA-binding domain, but a complete deletion of whiJ has no detectable effect on development (Aínsa, et al., 2010). Further, deletion of the gene (designated SCO4542 and encoding a small acidic protein of unknown function) immediately downstream of whiJ abolished aerial mycelium formation or sporulation, depending on the medium, but this phenotype was completely suppressed by deletion of whiJ (Aínsa, et al., 2010). This result led to the suggestion that WhiJ is able to repress differentiation (although this role is normally dispensable), that the SCO4542 gene product normally is counter-acting the effect of whiJ, and that mutations that cause constitutive activity of WhiJ (deletion of its C-terminal part or inactivation of SCO4542) therefore lead to arrested development (Aínsa, et al., 2010). Interestingly, bldB encodes an orphan paralogue of SCO4542 not connected to any other whiJ-like genes (Pope, et al., 1998, Eccleston, et al., 2002). The bldB mutants are defective in both aerial mycelium formation and antibiotic production (Merrick, 1976, Champness, 1988, Eccleston, et al., 2002). BldB forms oligomers, probably dimers, and a genetic investigation led to the suggestion that BldB operates by interacting with some other protein (Eccleston, et al., 2002, Eccleston, et al., 2006). If BldB works in a way similar to that of SCO4542 and acts by counteracting an XRE-family repressor protein, it appears that a screen for suppressors of the bldB phenotype may be a meaningful way forward. An obvious candidate to test for interplay with BldB is BldD, which belongs to the XRE family and is inactivated during development in an unknown way (see above).
New complexity in developmental sigma factor regulation – BldG controls σH activity
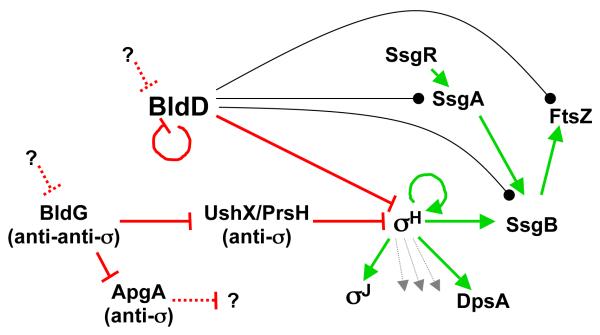
The classical bld gene bldG (Champness, 1988) encodes a protein similar to anti-sigma factor antagonists like the B. subtilis SpoIIAA controlling the forespore-specific σF (Bignell, et al., 2000). bldG is co-transcribed with the downstream gene apgA (antagonistic partner of BldG), which encodes an anti-sigma factor that indeed interacts with BldG and counteracts its activity (Parashar, et al., 2009). It is still not clear which developmental sigma factor is controlled by ApgA, but the investigators made a number of observations that led them to suggest that BldG may act via other anti-sigma factors in addition to ApgA (Parashar, et al., 2009). This turned out to be correct with the recent identification of BldG as the regulator of the stress-response and developmental sigma factor σH in S. coelicolor and S. griseus (Sevcikova, et al., 2011, Takano, et al., 2011). In both species, the activity of σH is modulated in response to osmotic shock, but the expression is also developmentally regulated from one of its promoters, the BldD-repressed and autoregulated sigHp2 (Fig. 5)(Kormanec, et al., 2000, Kelemen, et al., 2001, Takano, et al., 2003b, Viollier, et al., 2003). The co-transcribed gene immediately upstream of sigH encodes its anti-sigma factor named RshA in S. griseus and UshX or PrsH in S. coelicolor (Sevcikova & Kormanec, 2002, Takano, et al., 2003b). Using different approaches, two groups identified BldG as the direct antagonist of this anti-σH factor (Sevcikova, et al., 2011, Takano, et al., 2011), and we note that this regulatory interaction between homologs of BldG and UshX/PrsH is conserved in M. tuberculosis (Beaucher, et al., 2002). The expression of ssgB, a regulator of sporulation septation (discussed below), depends on sigH (Kormanec & Sevcikova, 2002). σH also seems to control the expression of the ECF sigma factor gene sigJ and dpsA encoding a nucleoid-associated protein (see below)(Mazurakova, et al., 2006, Facey, et al., 2011). In total, these findings indicate an important developmental role for sigH (Fig. 5), even though reports on sigH mutant phenotypes are contradictory (Sevcikova, et al., 2001, Takano, et al., 2003b, Viollier, et al., 2003). Deletion of the entire prsH-sigH transcription unit in S. coelicolor caused a bald phenotype on all tested media, possibly suggesting that the anti-σH gene has more roles than controlling σH (Viollier, et al., 2003).
Figure 5.
Summary of the regulatory connectivity in the intersection of aerial mycelium formation and early events in the differentiation of aerial hyphae in S. coelicolor. Demonstrated direct interactions at transcriptional, translational, or post-translational levels are shown as solid lines. Filled arrows indicate positive or activating effects, perpendicular lines indicate negative or repressing effects, and lines ending in bullet indicate that direct interaction has been demonstrated, but the effect on gene expression is not clear. Dashed lines indicate hypothetical interactions. The question marks indicate unknown mechanisms for counteracting BldD and BldG activity, and the still hypothetical sigma factor controlled by anti-sigma factor ApgA. Dotted arrows not pointing to symbols are used to indicate unknown but anticipated target genes.
The key regulators of spore formation in aerial hyphae
From analyses of Whi phenotype mutants, five central regulatory genes have emerged that are strictly and non-conditionally required for early stages of the conversion of aerial hyphae to spores - whiA, whiB, whiG, whiH, and whiI (reviewed in e.g., Chater & Chandra, 2006, Elliot, et al., 2008, Chater, 2011). All five encode transcription factors (see below and Fig. 6), and a crucial future task is to map the largely unknown regulons controlled by these key regulators.
The sigma factor σWhiG is an important regulator required at the earliest known stage of aerial hyphal differentiation, but not formation of the aerial mycelium (Chater, et al., 1989). Two key developmental genes, whiI and whiH, are directly controlled by σWhiG (Fig. 6): an atypical response regulator of the FixJ subfamily and a member of the GntR family, respectively (Ryding, et al., 1998, Aínsa, et al., 1999). whiI and whiH mutants show distinctive blocks in development, neither of which reiterate the whiG phenotype, and additional σWhiG-regulated genes are likely to contribute to development (see Chater, 2011). The exact role of BldD in the regulation of whiG remains unclear (Fig. 6), and whiG transcripts were detected throughout colony development in the wildtype, suggesting that a post-transcriptional regulatory switch may affect the onset of whiG-dependent transcription (Kelemen, et al., 1996, Elliot, et al., 2001). Both whiI and whiH appear to be autorepressors and whiI negatively influences expression of whiH (Ryding, et al., 1998, Aínsa, et al., 1999), suggesting that negative feedback loops may also contribute to the regulation (Fig. 6). In addition, it has been suggested these factors are sensitive to yet unidentified physiological signals (see e.g., Chater & Chandra, 2006, Chater, 2011).
WhiI has a critical function because whiI-null mutants are blocked for sporulation septation and chromosome condensation (Aínsa, et al., 1999). A transcriptomic survey has identified genes dependent on whiI, and although it is unclear whether they are directly controlled by WhiI, at least three of the identified genes are required for normal development of spores (Tian, et al., 2007, Zhang, et al., 2010). An interesting feature of WhiI (shared with BldM and RamR), is that the phosphorylation pocket is atypical and there is no cognate histidine kinase connected to WhiI (Aínsa, et al., 1999, Hutchings, et al., 2004). Conserved phosphorylation pocket residues are not required for WhiI function at the early stage of differentiation, and mutation of these residues primarily affected spore maturation. This led to the suggestion that WhiI may switch between two different states, one required for formation of prespore compartments by septation, and one needed later during spore maturation (Tian, et al., 2007).
whiA and whiB are connected by their essentially identical mutant phenotypes with very long, tightly coiled, but poorly septated aerial hyphae (Chater, 1972, Flärdh, et al., 1999, Aínsa, et al., 2000). Both genes are transcribed individually from developmentally regulated promoters, with contributions in both cases from at least one constitutive promoter. The developmentally regulated whiA promoter is active specifically in aerial hyphae in S. ansochromogenes (Xie, et al., 2007). Transcription of whiA and whiB is independent of whiG, leading to models that whiA/whiB and whiG/whiH/whiI form two parallel but converging pathways required for sporulation (Fig. 6) (Soliveri, et al., 1992, Chater, 1999, Aínsa, et al., 2000). The binding of BldD to the whiB promoter region suggests one possibility for developmental control of the whiA/whiB pathway (den Hengst, et al., 2010). We assume that whiB expression is repressed by BldD (and therefore derepressed during development), and note that upregulation of the developmental promoter of whiA depends on whiB (in addition to whiA itself) (Jakimowicz, et al., 2006). Thus, it is conceivable that BldD-mediated derepression of whiB also indirectly contributes to developmental expression of whiA (Fig. 6).
whiA is the third gene in a putative operon that is conserved in many Gram-positive bacteria (e.g., yvcJKL in B. subtilis), and WhiA homologues are also found among Fusobacteria and Thermotogae (Aínsa, et al., 2000, Görke, et al., 2005; Knizewski & Ginalski, 2007). Structural analyses show that WhiA from Thermotoga maritima consists of a duplicated N-terminal domain with folds similar to a specific family of homing endonucleases, and that WhiA-like proteins may bind DNA in a way similar to these enzymes but they have lost important catalytic residues for endonuclease activity (Knizewski & Ginalski, 2007, Kaiser, et al., 2009). Further, WhiA-like proteins have a C-terminal domain with similarity to domain 4 in σ70-type sigma factors (including a predicted helix-turn-helix motif) that may also contribute to DNA binding. These studies predict that WhiA is a DNA-binding protein that interacts with an interesting asymmetric and possibly bipartite sequence motif, but this remains to be verified experimentally.
WhiB-like transcription factors act at different stages of development
Homologues of WhiB constitute a large family of WhiB-like (Wbl) proteins, which are small proteins characterized by four conserved cysteine residues and found exclusively among Actinobacteria (Soliveri, et al., 2000, Gao, et al., 2006). Many Actinobacteria encode multiple Wbl proteins, including S. coelicolor, which encodes 11 chromosomal Wbls (den Hengst & Buttner, 2008). Previous observations suggested that Wbl proteins act as transcription factors in Streptomyces (reviewed in Chater & Chandra, 2006, den Hengst & Buttner, 2008, Chater, 2011). The first demonstration that WhiB in S. coelicolor binds DNA has now been published; WhiB directly controls dpsA (Fig. 6), encoding a nucleoid-associated protein that is further discussed below (Facey, et al., 2011). Recent papers show that four mycobacterial Wbls are also DNA-binding proteins that interact with certain regulatory sequences in promoters (Guo, et al., 2009, Singh, et al., 2009, Rybniker, et al., 2010, Smith, et al., 2010). Several Wbls bind an iron-sulphur cluster (for review and references, see den Hengst & Buttner, 2008), and, consistent with that observation, DNA-binding of two mycobacterial Wbls is responsive to nitric oxide or redox state (Singh, et al., 2009, Smith, et al., 2010).
In addition to whiB itself, two further members of the wbl gene family are required for morphological differentiation in S. coelicolor, but affect different developmental stages. whiB is specifically required at an early pre-septational stage of aerial hyphal sporulation (see above). whiD is required for correct placement of septa and spore maturation (Molle, et al., 2000). A recent systematic study of the remaining chromosomal wbl genes in S. coelicolor revealed that wblA acts pleiotropically on both secondary metabolism and morphological differentiation, and seems to influence an early and previously unrecognized developmental transition of aerial hyphal cells (Fowler-Goldsworthy, et al., 2011). Interestingly, wblA is a target for AdpA in S. griseus (Hara, et al., 2009), and there is a sequence matching the AdpA binding site consensus in the S. coelicolor wblA promoter region as well. The S. coelicolor wblA mutant is characterized by a strongly reduced production of spores, absence of the normal grey spore pigment, and instead has an unusual and conspicuous pink aerial mycelium, apparently because the mutant aerial hyphae uncharacteristically produce both of the pigmented antibiotics in S. coelicolor. wblA has previously been implicated as a negative regulator of antibiotic biosynthesis (Kang, et al., 2007, Noh, et al., 2010). Transcriptome analysis shows that it affects an interesting set of genes during colony development, including many with known functions related to differentiation, primary and secondary metabolism (Fowler-Goldsworthy, et al., 2011). This provides a clear illustration of the complex interrelations between physiological and morphological differentiation in a Streptomyces colony.
Control by σF during spore development
The sigF gene (encoding σF) is expressed late during differentiation of aerial hyphae, mainly in the apical sporogenic cell (Kelemen, et al., 1996, Sun, et al., 1999). The sigF mutant phenotype is restricted to defects in spore maturation, including production of thin-walled spores with less condensed nucleoids and reduced spore pigmentation (Potúcková, et al., 1995). The spore pigmentation defect can be ascribed to the σF-dependence of the whiEp2 promoter, which drives expression of a gene involved in a late step of synthesis of the spore coloration (Kelemen, et al., 1998), but additional direct targets for σF remain to be identified. The control of σF involves transcriptional dependence on the sporulation-specific regulators encoded by whiG, whiA, whiB, and whiI (Kelemen, et al., 1996), and may also include the action of anti-sigma factors. σF interacts in yeast two-hybrid assays with the anti-sigma factor homologue SCO4677, and screening for interaction partners to this protein yielded two hits to predicted anti-sigma factor antagonist-like proteins (Kim, et al., 2008b). Consistent with a role of SCO4677 as anti-σF factor, a ΔSCO4677 mutant showed enhanced or accelerated development, but also enhanced actinorhodin production in liquid culture, which suggests SCO4677 regulation of at least one additional sigma factor (Kim, et al., 2008b). No phenotypic effect of the proposed anti-σF antagonists were detected in constructed deletion mutants, and their functions remain unclear.
σN, subapical stem compartments, and a new spore surface protein
Immediately adjacent to sigF is sigN, which is also developmentally regulated but expressed independently of sigF (Dalton, et al., 2007). (Note that sigN should not be confused with bldN, encoding another sigma factor.) One of the two promoters for sigN is specifically active in sporulating cultures, dependent on bldC, bldD, and bldG, and autoregulated (Dalton, et al., 2007). This sigN promoter and the other known σN-target nepAp become active in the aerial hyphal cell that is immediately below what will become the spore chain, leading to definition of this compartment as the “subapical stem” (Fig. 2)(Dalton, et al., 2007), although there are conflicting observations of nepA expression also in developing spore chains (de Jong, et al., 2009b). The expression of nepA in the subapical stem does not depend on formation of the spore chain, and it remains to be elucidated how this cell type-specific control of nepA is exerted (Dalton, et al., 2007). NepA is predicted to be secreted as a short 49 amino acid protein, and like chaplins and rodlins, NepA is extractable from spores only with trifluoroacetic acid, suggesting a tight association with spore surfaces (de Jong, et al., 2009b). Interestingly, nepA and the rodlin genes may be examples of gene expression coupled to aerial hyphae formation as such, since they are neither expressed in a multiple chaplin deletion mutant nor a bldN mutant that fails to express chaplin genes (Elliot, et al., 2003b, Claessen, et al., 2006, de Jong, et al., 2009b). In one report, the nepA mutant showed a similar phenotype to the sigN mutant with a medium-dependent defect in aerial mycelium formation (Dalton, et al., 2007). On the other hand, a second study reported different phenotypes and ascribed those to more efficient spore germination in the nepA mutant compared to its parent, suggesting a role for NepA in maintenance of spore dormancy (de Jong, et al., 2009b). Additional work is needed to understand the role of nepA, which is broadly conserved in streptomycetes.
Cues and signals feeding into the decision to begin aerial mycelium formation
A central developmental decision in Streptomyces is if and when to initiate aerial mycelium formation. At least as observed on laboratory media, aerial mycelium formation is a coordinated behavior of large numbers of cells on the colony surface, involving both extensive extracellular signaling and various physiological and nutritional stimuli. However, the details of the sensing mechanisms and the connections to the regulatory pathways outlined in the previous section are in most cases still obscure. We believe that information generated by the newer “-omics” methods will aide our understanding of how morphological differentiation is initiated.
Nutritional downshift as a cue for sporulation
It is a common conception that nutrient starvation stimulates morphological development in Streptomyces, but convincing demonstration of this and possible sensing mechanisms remains elusive. The potential link between morphological development and the depletion of nutritional sources is best illustrated by some Streptomyces species that undergo sporulation in submerged liquid culture, and for which a simple nutritional down shift (nutrient deprivation) can be used to induce liquid culture sporulation, for example in S. griseus (Kendrick & Ensign, 1983, Daza, et al., 1989). On solid medium, cessation of primary growth in the vegetative mycelium, presumably due in part to nutrient depletion, is followed by a transition phase during which major metabolic resources are reorganized, before a second growth phase occurs in aerial mycelium (Granozzi, et al., 1990). A similar transition phase is observed for growth in liquid culture (e.g., Huang, et al., 2001, Novotna, et al., 2003). Current studies are being done using post-genomic resources to analyze the dynamic transition in detail, mainly in liquid culture and with a focus on secondary metabolism (Rodriguez-Garcia, et al., 2007, Lian, et al., 2008, Nieselt, et al., 2010, Lewis, et al., 2011), but events during the transition phase should also be important for understanding the onset of morphological development. The following section discusses intracellular signaling molecules related to general nutrient depletion.
ppGpp and GTP pools
The ubiquitous intracellular alarmone guanosine tetraphosphate (ppGpp) regulates global and specific gene expression in response to decreasing nutrient availability in many bacteria (Potrykus & Cashel, 2008). In S. coelicolor, ppGpp is directly or indirectly affecting morphological differentiation and a relA null mutant is blocked for differentiation, at least under nitrogen limitation (Chakraburtty & Bibb, 1997, Hesketh, et al., 2007). However, most attention in S. coelicolor has been given to the influence of ppGpp on regulation of antibiotic production because overproduction of or failure to make the modified nucleotide seem to have more of an influence on certain aspects of antibiotic production than on morphological differentiation (Chakraburtty, et al., 1996, Chakraburtty & Bibb, 1997, Hesketh, et al., 2001, Sun, et al., 2001).
A drop in the GTP pool or the GTP:GDP ratio may also be an important signal for morphological differentiation in Streptomyces. Decoyinine, an inhibitor of GMP synthase that modifies GTP/GDP pools, can induce aerial mycelium formation and sporulation in several Streptomyces species (Ochi, 1986, Glazebrook, et al., 1990). The Obg family of GTP-binding proteins are potential sensors of the intracellular guanine nucleotide pools, although their exact cellular functions are unknown (Czyz & Wegrzyn, 2005). Obg has been characterized for S. coelicolor and S. griseus, and it was proposed that this essential protein participates as a regulator of morphological differentiation by sensing the intracellular GTP pool (Okamoto, et al., 1997, Okamoto & Ochi, 1998). This is analogous to the requirement of Obg for transition from stage 0 to stage II in B. subtilis sporulation (Kok, et al., 1994). No additional work has appeared on the function of this potential transition sensor and it would be useful to investigate an Obg depletion strain by microarray analysis. In addition, Obg could be a link between stress response and development as Obg is required for the activation of the stress sigma factor σB in B. subtilis (Scott & Haldenwang, 1999) and binds to a stress response anti-sigma factor UsfX in the actinomycete Mycobacterium tuberculosis (Sasindran, et al., 2011). We note that the closest homolog of UsfX in S. coelicolor is the anti-sigma factor UshX, which regulates σH (see above and Fig. 5), and one putative function of Obg might therefore be in the regulation of σH activity in response to the intracellular GTP pool.
Effects of preferred natural carbon sources
Cellulose and chitin are the most abundant natural polymers, and are important sources of nutrients for streptomycetes, which are major agents for their recycling in the soil (Schrempf, 2001). The S. coelicolor genome encodes 13 chitinases/chitosanases (Bentley, et al., 2002), and unusually among bacteria, the phosphotransferase system (PTS) of S. coelicolor is biased for the utilization of N-acetylglucosamine (GlcNAc), the monomer for chitin (Nothaft, et al., 2003). Further emphasizing the importance of chitin-derived substrates, inclusion of GlcNAc in the medium strongly affects both morphological development and pigmented antibiotic production for S. coelicolor, although in a complex and medium-dependent fashion (Rigali, et al., 2008). On poor medium, GlcNAc stimulates development, but on rich media, GlcNAc inhibits aerial mycelium formation, and this inhibitory effect depends on the major PTS permease-encoding gene, nagE2 (Rigali, et al., 2006, Nothaft, et al., 2010). The genes for PTS, chitinases, and other components of GlcNAc catabolism are repressed by the GntR-family transcription factor DasR, which integrates the detection and metabolism of GlcNAc; interaction with an intermediate of degradation (glucoseamine-6-phosphate) reduces the binding of DasR to its target promoters (Rigali, et al., 2006, Rigali, et al., 2008, van Wezel & McDowall, 2011). DasR acts as a global regulator that directly controls a range of other genes, including that for the pathway-specific regulator of actinorhodin biosynthesis, actII-ORF4 (Rigali, et al., 2008, van Wezel & McDowall, 2011). Further, dasR mutants have a Bld phenotype on glucose-containing media, but the basis for this developmental defect is not understood (Rigali, et al., 2006). As another piece of the story, the chitobiose-binding protein DasA, part of an ABC-type transporter, also acts as a link between chitin utilization and morphogenesis, and dasA mutants are, similar to dasR, bald on rich sporulation medium even in the absence of GlcNAc (Colson, et al., 2008). It is not known how these effects on development are mediated, but this very interesting connection between the sensing of a major carbon source and initiation of development certainly deserves further attention.
CebR is the regulator for cellulose utilization in S. griseus, the other highly abundant carbon source in the soil (Marushima, et al., 2009). Initial analysis suggests that there are some similarities between CebR and utilization of cellulose (for S. griseus) to the importance of DasR in integrating signals from chitin utilization. There appears to be an important link between the natural carbon sources and morphogenesis for both S. coelicolor (see above) and S. griseus (Seo, et al., 2002). While there is one locus in S. griseus, there are two copies of the entire ceb locus in S. coelicolor, including two paralogs of cebR (Marushima, et al., 2009). Genetic analyses of the presumably redundant loci for cellulose utilization have not been done for S. coelicolor.
The screens for classic developmental bld mutants defective in aerial mycelium formation were carried out on glucose-containing (Merrick, 1976, Champness, 1988, Nodwell, et al., 1999) or maltose-containing media (Champness, 1988). Hence, the bld mutants deriving from these screens have a bald phenotype on glucose-containing media but, with the exception of bldBmutants, the phenotypes can be partially suppressed by growth on carbon sources such as arabinose, glycerol, galactose or mannitol (Merrick, 1976, Champness, 1988, Nodwell, et al., 1999). The full significance of this conditionality is still unclear. It may relate to catabolite repression effects of glucose, but it is also worth being reminded that growth on glucose is acidogenic while growth on mannitol is not (see below). However, with the relatively recent appreciation of the importance of signaling molecules emanating from chitin and cellulose metabolism in Streptomyces, it appears that new insights might be gleaned if screens for bld mutants are repeated but carried out on well-buffered media with such ecologically relevant carbon sources as chitooligosaccharide- (chitin catabolic intermediates) and/or cellooligosaccharide- (cellulose catabolic intermediates). This might lead to the identification of new developmental regulatory mutants unconditionally blocked on all carbon sources or to a parallel alternative pathway regulating aerial mycelium formation.
Streptomyces development involves cyclic-di-GMP and other cyclic nucleotides
Recent analyses have established a role for cyclic-di-GMP in the control of morphological differentiation of S. coelicolor. Cyclic-di-GMP is a common signaling molecule in a variety of microorganisms (Jenal & Malone, 2006). Genes encoding three c-di-GMP binding proteins were identified as being under the control of bldD in S. coelicolor, suggesting a potential involvement in Streptomyces development (den Hengst, et al., 2010). Overexpression of either cdgA or cdgB and the deletion of cdgB affects both morphological and physiological differentiation, suggesting that c-di-GMP concentrations play a role in both processes (den Hengst, et al., 2010, Tran, et al., 2011). Furthermore, overexpression of engineered versions of cdgA and cdgB encoding enzymes with an incapacitated active sites for synthesizing c-di-GMP did not interfere with the production of aerial hyphae (den Hengst, et al., 2010, Tran, et al., 2011). The role of c-di-GMP in Streptomyces development certainly deserves further attention. There are also reports of cyclic AMP (cAMP), the cAMP-binding protein Crp, and other prediceted cyclic nucleotide-binding proteins affecting development, but their exact influence on the life cycle are still ill-defined (Saito, et al., 2006, Susstrunk, et al., 1998, Derouaux, et al., 2004, Tian, et al., 2007).
Quorum sensing and other forms of extracellular signaling
One form of cell-to-cell communication in Streptomyces is reminiscent of bacterial quorum sensing and involves the use of γ-butyrolactones instead of the more well-known N-acyl-homoserine lactones in proteobacteria (Takano, 2006). These hormone molecules are used by different Streptomyces species to signal and coordinate transitions from primary metabolism to antibiotic production and sometimes development. The γ-butyrolactone compound produced by S. griseus is known as A-factor and has a profound and well-investigated effect on antibiotic biosynthesis and morphological differentiation (reviewed by Horinouchi, 2007). A group of related signaling molecules in S. coelicolor are called SCBs for S. coelicolor butanolides, but development is not as reliant on SCB signaling in S. coelicolor as it is on A-factor for S. griseus (Chater & Horinouchi, 2003, Takano, 2006). An up-to-date picture of gene regulation for S. coelicolor by SCB γ-butyrolactones from a genome-wide study has recently been published (D’Alia, et al., 2011). This study, comparing an SCB synthase mutant to the wild type, helps provide molecular details to explain previous antibiotic expression phenotypes. It also showed that expression of some genes that are important for morphological differentiation is shifted earlier in time in the mutant.
The existence of other putative extracellular signals have been inferred from the extracellular complementation behavior between different bld mutants. On rich (osmotically-enhanced) sporulation medium, aerial growth can be restored for certain bld developmental mutants by growing colonies near other mutants or the wild type strain (Willey, et al., 1993). While one bldstrain is still competent to produce a potential diffusible signal, the other mutant cannot produce but can still respond to the putative signal. The linear and hierarchical relation between multiple complementation groups suggested an extracellular cascade of several signal molecules, leading eventually to the production of SapB, and presumably other targets (Chater, 1998). The bldK gene encodes an oligopeptide importer, suggesting that one signal is a small polypeptide (Nodwell, et al., 1996), and the dependence on bldK oligopeptide permease for differentiation is shared by S. griseus, suggesting that the mechanism is conserved (Akanuma, et al., 2011). At least one oligopeptide signal with a molecular weight of 655 Da was partially purified (Nodwell & Losick, 1998). However, the simple model is complicated by the facts that several mutants did not fit into the linear extracellular complementation hierarchy (Nodwell, et al., 1999), and that in a few cases the Bld phenotype and observed extracellular complementation might be attributable to acidification and neutralization, respectively, of the surrounding solid medium (Susstrunk, et al., 1998, Viollier, et al., 2001). These observations complicate but do not necessarily negate the prior accumulated observations. Thus, additional putative extracellular signal molecules may await purification and identification.
As recently reviewed (Chater, et al., 2010), evidence has been accumulating to indicate the importance of extracellular proteolytic signaling and processing during morphological differentiation. A subtilisin-like protease inhibitor called STI (homolog of S. griseus SgiA; Hirano, et al., 2006) and several proteases were investigated in S. coelicolor (Kim, et al., 2005, Kim, et al., 2008a). STI is inactivated by extracellular proteolysis coinciding with the time of aerial mycelium formation and at least one protease that participates in its inactivation has been identified. At least two protease targets for inhibition by STI are known from two-hybrid, immunoprecipitation and biochemical analyses. As discussed by Chater et al. (2010), there is the possibility that some of the regulatory events in a cascade of extracellular proteolysis may intersect with the extracellular signaling cascade defined for the cross complementation of the bald developmental mutants (described above). We agree and expect that products of protease activity may form some of the elusive diffusible signals underlying the extracellular complementation.
Possible links between ectopic sporulation, catabolite repression, and developmental signaling
Except for those Streptomyces species that can sporulate in liquid, there is spatial restriction of development, and spores are not formed in the vegetative mycelium. However, vegetative hyphae have the capacity to form chains of spores (Ohnishi, et al., 2002). For S. coelicolor, overexpression of the key developmental whiG sigma factor is sufficient to induce some ectopic sporulation in vegetative hyphae (Chater, et al., 1989). In addition, deletion of 7.4 kb of DNA near glkA (encoding glucose kinase) results in a similar ectopic sporulation (Esp) phenotype (Kelemen, et al., 1995). glkA mutants are defective in carbon catabolite repression (Angell, et al., 1994, Kwakman & Postma, 1994), and a previous paper has indicated a connection, albeit not well understood, between some bld mutant phenotypes and catabolite repression (Pope, et al., 1996). While the Esp phenotype is not due to deletion of glkA itself (Kelemen, et al., 1995), the deletion responsible for Esp also encompasses another gene involved in catabolite repression, SCO2127 (Chavez, et al., 2009). Importantly, SCO2127 is required for the glucose-mediated repression of sporulation seen in the wildtype (Chavez, et al., 2011). Unfortunately, the possibility of ectopic sporulation in the SCO2127 mutant was not investigated. In an overlay blot, SCO2127 interacts with two proteins, extracellular BldKB subunit of the oligopeptide importer important for aerial mycelium formation and a predicted membrane metalloprotease SCO2582 (Chavez, et al., 2011). This led the authors to raise the question whether cotranslational (presecretion) interaction between SCO2127 and BldKB could function to arrest assembly of the oligopeptide importer as a developmental timing mechanism, and whether SCO2582 could generate the signal imported by BldK. Additional experiments need to be done to validate and further evaluate the significance of the reported results for SCO2127.
Signaling during competition and predation
Under laboratory conditions, S. coelicolor proceeds through its life cycle in solitude. In its natural environment, it must interact and compete with other organisms. Some recent studies of signaling in co-culture should be more similar to events in the natural environment. For example, predation of S. coelicolor by Myxococcus xanthus induces aerial mycelium formation and differentiation into spores, as well as antibiotic production (Perez, et al., 2011). Such co-culture effects and cross-species communication (see other references therein) should be a fertile area for future studies and provide insight into the ways these sessile mycelial species respond to casual contacts, invasions and predation in the natural environment.
Intersection of developmental regulation and cell biology
As our understanding of the regulatory pathways involved in controlling morphological differentiation in S. coelicolor improves, a main challenge is to unravel their direct connections to genes and proteins at the business end of cell differentiation, i.e. those directly involved in cell wall synthesis, morphogenesis, cell cycle processes, nucleoid structure, and various other aspects of cell biology. Developmentally regulated modulations of such processes are crucial for the differentiation of aerial hyphae into mature spores, and interesting mechanisms have evolved for this purpose. Streptomyces development offers a powerful experimental system for investigation of fundamental cellular processes, illustrated for example by cell division and chromosome segregation, as described below. We conclude this paper with a discussion about developmental control of cell cycle-related processes, cell wall assembly, and chromatin structure.
Cell division
Spore formation in aerial hyphae obviously requires temporal and spatial control of cell division. In bacteria, cell division is directed by FtsZ, which assembles into a ring-like cytoskeletal structure termed the Z ring that both provides force for constriction of the cell and acts as a scaffold for recruitment of other components of the divisome – the multiprotein complex that connects the Z ring to synthesis of septal peptidoglycan (Erickson, et al., 2010). Streptomycetes are no exception; ftsZ is required for both the vegetative hyphal crosswalls and the sporulation septa (McCormick, et al., 1994), and several of the typical divisome components are conserved in Streptomyces (for review, see McCormick, 2009). Developmental regulation at the transcriptional level has only been explored for ftsZ, which is developmentally upregulated from the ftsZp2 promoter (Flärdh, et al., 2000, Kwak, et al., 2001b). The striking similarity between a mutant lacking ftsZp2 and a whiH mutant implicated whiH in control of cell division (Flärdh, et al., 2000, Chater, 2001), but WhiH does not appear to bind to this promoter region and likely affects division by some other means (Larsson. J, Chater, K. F., Flärdh, K., manuscript in preparation). Based on available evidence, whiA and whiB may be the best candidates for direct control of ftsZ (Flärdh, et al., 2000). Interestingly, ftsZ is targeted by BldD, but the BldD binding site overlaps with the apparently constitutive ftsZp3 promoter rather than the developmentally controlled ftsZp2 and the effect on ftsZ expression remains unknown (den Hengst, et al., 2010).
After the increase in ftsZ expression, FtsZ assembles into helical structures that can be observed along the length of this sporogenic apical cell (Fig. 3A). The remodeling of these FtsZ filaments into regularly spaced Z rings seems to occur via shorter helical structures coalescing into regular rings (Grantcharova, et al., 2005). It has been unclear how this developmental control of Z ring assembly is brought about since streptomycetes and other Actinobacteria lack clear homologues of most of the proteins that in other bacteria are involved in control of FtsZ assembly, stabilization of the Z ring, and anchoring of the ring to the cytoplasmic membrane (see discussions in Flärdh & Buttner, 2009, McCormick, 2009). The small transmembrane protein CrgA was implicated in the control of cell division for S. coelicolor (Del Sol, et al., 2003, Del Sol, et al., 2006), and this is supported by a report showing septal targeting and interaction of CrgA with FtsZ and other division proteins in M. tuberculosis (Plocinski, et al., 2011).
SsgA-like proteins (SALPs) have been implicated in a range of cellular processes during Streptomyces development, including division (Traag & van Wezel, 2008). The SALPs are small proteins only found among mycelial or sporulating Actinobacteria, and they are present as multiple paralogues in Streptomyces and related genomes (e.g., seven SALPs in S. coelicolor and twelve in the recently sequenced Kitasatospora setae; Noens, et al., 2007, Traag & van Wezel, 2008, Ichikawa, et al., 2010). The structure of one SALP (a trimer of the SsgB orthologue from Thermobifida fusca) shows similarity to a class of ssDNA/RNA-binding proteins, but SALPs are thought to act via protein-protein interactions rather than by binding nucleic acids (Xu, et al., 2009a). The ssgA gene was originally discovered and named based on its ability to suppress sporulation in a hyper-sporulating mutant of S. griseus (Kawamoto & Ensign, 1995). It strongly stimulates septation and mycelium fragmentation when overexpressed, and is required for normal sporulation septation in both S. coelicolor and S. griseus (Kawamoto, et al., 1997, Jiang & Kendrick, 2000, van Wezel, et al., 2000). Likewise, the paralog ssgB is also required for spore formation (Keijser, et al., 2003). Both ssgA and ssgB are developmentally regulated independently of the early regulatory whi genes (Kormanec & Sevcikova, 2002, Traag, et al., 2004). As discussed above, both genes are direct targets for BldD, and in the case of ssgB, there is a dual bldD effect since ssgB is controlled by the BldD-repressed sigH (Fig. 5) (Kelemen, et al., 2001, Kormanec & Sevcikova, 2002, den Hengst, et al., 2010). In contrast to the situation in S. griseus, transcription of ssgA in S. coelicolor is not strongly affected by adpA and instead directly controlled by the IclR-family regulator SsgR encoded by a gene immediately upstream of ssgA (Fig. 5)(Traag, et al., 2004, Wolanski, et al., 2011), but it is not clear if this mediates the developmental regulation.
Recently, SsgA and SsgB were proposed to determine the future sites of cell division in the sporogenic cell and recruit FtsZ to those sites (Willemse, et al., 2011). SsgB from T. fusca was found to interact with both FtsZ and SsgA (from S. coelicolor) in a two-hybrid assay and stimulate GTP-induced polymerization of T. fusca FtsZ in vitro when added in equimolar amounts (Willemse, et al., 2011). In combination with in vivo monitoring of protein-protein interactions and co-localization, this led to a model for cell division control during S. coelicolor sporulation proposing that SsgA forms foci along the sporogenic cell, that SsgB is recruited by these foci, and that SsgA then leaves these future division sites before SsgB recruits FtsZ. The model also suggests that the Z rings are built or nucleated from such SsgB-FtsZ foci (Willemse, et al., 2011), rather than being formed by coalescing helical structures as previously proposed for S. coelicolor and other organisms (Grantcharova, et al., 2005, Erickson, et al., 2010). The molecular details of how SsgA and SsgB contribute to controlling Z ring assembly obviously require further investigation.
Chromosome segregation
One of the striking accomplishments of streptomycetes is the faithful simultaneous segregation of genomic copies into the nascent prespore compartments while dozens of synchronous sporulation septation events are taking place. The ParAB system has a key role in this process. ParA ATPase assembles into structures along the entire length of the long axis of the sporogenic aerial hyphal cell, and ParB forms into nucleoprotein complexes on the centromere-like sequences known as parS sites that are present in multiple copies in the environs of oriC on the chromosome(Jakimowicz, et al., 2002, Jakimowicz, et al., 2007). The ParA structures somehow impart positional information, allowing the ParB:parS complexes (and therefore chromosomes) to be fairly evenly distributed in the multigenomic syncytial aerial hyphae (Jakimowicz, et al., 2007).
Recently, an additional protein named ParJ (SCO1662) was identified as an important participant in developmental chromosome segregation by regulating ParA polymerization activity (Ditkowski, et al., 2010). ParJ, found exclusively in Actinobacteria (Gao, et al., 2006), was identified in a screen for proteins that directly interact with ParA. Consistent with the known involvement of ParA in segregation, a parJ null mutant has a segregation defect. Spacing of septation was also affected in the parJ mutant, resulting in a substantial percentage of minispore compartments. The biochemical interaction between ParJ and ParA was confirmed in vitro, and ParJ induces the depolymerization of ParA filaments (Ditkowski, et al., 2010). Finally, the genetic and biochemical analysis of a paralogue of ParJ (SCO1997) will also be of interest to see if there are some additional interactions between ParA, ParJ and SCO1997 during developmental genome segregation.
Transcriptional analyses of parJ have not yet been reported, but one of the two promoters upstream of the parAB operon is upregulated at the time of development (Kim, et al., 2000), and this is completely dependent on whiA and whiB, while at least some transcript is observed in whiI and whiH mutants (Jakimowicz, et al., 2006). In addition to parAB, smc (encoding a condensation protein) and ftsK (spoIIIE; encoding a septum-localized DNA motor protein) play roles in developmental chromosome segregation without overtly affecting septation and spore development (Wang, et al., 2007, Dedrick, et al., 2009, Kois, et al., 2009). One can infer some degree of compartment specific transcription of ftsK(spoIIIE) and smc because FtsK-EGFP and SMC-EGFP accumulate and localize in the aerial hyphae at the expected stage for chromosome segregation and condensation (Ausmees, et al., 2007, Wang, et al., 2007, Dedrick, et al., 2009, Kois, et al., 2009).
The smeA-sffA operon encodes two proteins that together form a FtsK/SpoIIIE-like architecture; SmeA is a small membrane protein and SffA contain only a single membrane spanning segment and an FtsK DNA motor-like domain (Ausmees, et al., 2007). These proteins appear to interact and SmeA helps target SffA to the sporulation septa. Developmental transcriptional regulation of smeA-sffA is completely dependent on whiG, whiA and whiI (Ausmees, et al., 2007). A smeA-sffA null mutant has slight defects in genome segregation, nucleoid condensation, and spore cell wall appearance, but the exact function is not obvious.
Assembly of the spore wall
S. coelicolor grows vegetatively by a mechanism that does not require the actin-like MreB-type of proteins. It was therefore surprising that the streptomycete genomes, in contrast to those of many other Actinobacteria, invariably carry mreB genes. Recent studies have established that the main function of the S. coelicolor MreB proteins is in spore wall assembly. Two bona fide MreB proteins, MreB and Mbl, appear first at the sporulation septa, and then spread along the walls of the maturing spore (Mazza, et al., 2006, Heichlinger, et al., 2011). By extrapolation from studies in other rod-shaped bacteria (reviewed in e.g., Shaevitz & Gitai, 2011), these actin-homologues are expected to form cytoskeletal filaments under the cytoplasmic membrane, although this has not been confirmed experimentally in Streptomyces prespores. However, consistent with the situation in other bacteria, MreB and Mbl are part of a large Streptomyces spore wall synthesizing complex (SSSC), which includes MreC, MreD, Pbp2, and the RodA/FtsW-like protein Sfr (Heichlinger, et al., 2011, Kleinschnitz, et al., 2011b). Extensive protein interactions have been detected within the complex, and they also extend to the S. coelicolor homologues of RodZ and FtsI, as well as two additional penicillin binding proteins (Kleinschnitz, et al., 2011b). Further interaction partners and possible members of the SSSC complex were found by a two-hybrid screen, including a membrane protein encoded by an interesting gene cluster (cmdABCDEF) required for normal sporulation (Xie, et al., 2009, Kleinschnitz, et al., 2011b), and another membrane protein (SCO2584) that may be related to wall teichoic acid biosynthesis, which was recently reported to be required for normal spore wall assembly (Kleinschnitz, et al., 2011a).
The core components of SSSC are encoded by adjacent operons (mreBCD and pbp2-sfr). The mreBCD operon is subject to developmental regulation from a promoter that is upregulated during sporulation, but it is also expressed from two constitutive promoters (Burger, et al., 2000). It is not clear what significance the expression of mreB may have during vegetative growth phase (Heichlinger, et al., 2011). Transcription of the unlinked mbl gene is strictly developmentally regulated and is only detected late during sporulation (Heichlinger, et al., 2011). A third mreB-like gene is present in some Streptomyces genomes, including S. coelicolor (SCO6166), but it is expressed during vegetative growth and a deletion mutant has no detectable phenotype (Heichlinger, et al., 2011).
Inactivation of any of the genes for the SSSC core components results in highly similar developmental phenotypes, with the production of defective spores with thin spore walls, aberrant shapes, and increased stress sensitivity (Mazza, et al., 2006, Heichlinger, et al., 2011, Kleinschnitz, et al., 2011b). Interestingly, deletion of any one of four genes for different cell wall hydrolases (rpfA, swlA, swlB, swlC) also leads to thin-walled and heat-sensitive spores (Haiser, et al., 2009). Thus, extensive peptidoglycan turnover is required for proper assembly of the spore wall. This may be one consequence of the fact that the Streptomyces spore wall can not be assembled from outside of the existing cell wall (as is the case for the cortex in Bacillusendospores), but rather is being made from the inside, and the original peptidoglycan layer of the aerial hypha is therefore likely to be gradually degraded or at least remodeled so that it can be stretched to accommodate the new and thicker spore wall. The four investigated hydrolase genes do not show clear-cut developmental regulation, and at least two of them have functions also during vegetative growth. Interestingly, their transcripts share a common 5′ untranslated sequence that is broadly conserved among Gram-positives, and is predicted to form a riboswitch structure, suggesting potentially very interesting regulation (Haiser, et al., 2009).
Preservation of the genome
In E. coli, Dps is a nucleoid-associated protein that strongly increases in abundance during entry into stationary phase, forms large crystalline-like nucleoprotein complexes with DNA, and is involved in protecting stationary phase cells against multiple stresses (Nair & Finkel, 2004). S. coelicolor has three dps genes that all influence spore nucleoids and positioning of sporulation septa, but also seem to counteract each other’s effects in a complex way (Facey, et al., 2009). Both dpsA and dpsC are upregulated in sporogenic cells (no data are available for dpsB), but the dps genes are also expressed in response to specific stress conditions (Facey, et al., 2009). Both the stress-induced and developmental upregulations are to a large extent dependent on the SigB-like sigma factor σH (Fig. 5), but also influenced by other sigma factors of the same subfamily, and overlapping regulation of both σH and σB at the single dpsA promoter was reported (Facey, et al., 2011), This illustrates the complex connections between stress and development in Streptomyces (see references and discussion in Chater, 2011). Importantly, the developmental control of dpsA involves direct binding of WhiB to the promoter region, making dpsA the first identified target for the WhiB transcription factor (Facey, et al., 2011).
Another protein specifically associated with spore nucleoids is HupS, which is one of the two Streptomyces homologues of the ubiquitous HU proteins in bacteria (Yokoyama, et al., 2001, Salerno, et al., 2009). In S. coelicolor, hupA and hupS are differentially regulated, with hupA expressed in both vegetative and aerial hyphae, and hupS being specifically expressed in sporogenic compartments (Fig. 3B) and dependent on whiA, whiG, and whiI (Salerno, et al., 2009). HupS contributes to spore nucleoid compaction and is required for development of spore heat resistance. HupS belongs to an actinobacteria-specific subgroup of HU proteins that carry long and unusual, lysine and alanine-rich C-terminal extensions, very similar to the C-terminal parts of eukaryotic linker histones. The the mycobacterial orthologue of HupS, Hlp, is upregulated during cold-shock and anaerobiosis-induced dormancy in M. smegmatis, and has been implicated in repair and recombination processes like non-homologous end-joining (see references in Salerno, et al., 2009). In addition, Hlp/HupS-like proteins are overrepresented in the highly radiation- and desiccation-resistant actinobacterium Kineococcus radiotolerans (four paralogues, compared to zero or one in other Actinobacteria) (Salerno, et al., 2009). This suggests that HupS may not only be involved in compacting spore DNA, but could also facilitate the action of DNA repair mechanisms, presumably acting during spore germination. However, little is known about developmental control of DNA repair and its role in Streptomyces spore survival.
Conclusion
We began this review with broad strokes to provide information to indoctrinate the uninitiated and proceeded to describe the state of the field for the Streptomyces enthusiasts. Our goal was to show that the research on Streptomyces development is vigorous and continuing progress will give a much more coherent view of the fascinating life cycles of these organisms, including anticipated novel and intriguing molecular and cell biological mechanisms. New important resources, such as the dozen genome sequences of diverse species that are being determined by the Broad Institute (Broad Institute, 2011), will support the vibrant cell biology and molecular genetic analyses as many laboratories in numerous countries collaborate and contribute to study various aspects of Streptomyces development. An additional tool in the arsenal is anticipated to accelerate the research progress: S. venezuelae is promoted as a new model organism with several advantages (Flärdh & Buttner, 2009). It will have a completed genome sequence, and its ability to sporulate in submerged culture is a great advantage for many types of analyses, e.g., making developmental microarray analyses and proteomics more feasible. The availability of a generalized transducing phage and a useful transposon mutagenesis system also promise to facilitate and revitalize powerful genetic approaches. It is imperative that the research community continues its mission because there is an important need to understand the basic biology of these unique organisms that produce crucial compounds that human and veterinary medicine rely upon.
References
- Akanuma G, Hara H, Ohnishi Y, Horinouchi S. Dynamic changes in the extracellular proteome caused by absence of a pleiotropic regulator AdpA in Streptomyces griseus. Molecular Microbiology. 2009;73:898–912. doi: 10.1111/j.1365-2958.2009.06814.x. [DOI] [PubMed] [Google Scholar]
- Akanuma G, Ueki M, Ishizuka M, Ohnishi Y, Horinouchi S. Control of aerial mycelium formation by the BldK oligopeptide ABC transporter in Streptomyces griseus. FEMS Microbiology Letters. 2011;315:54–62. doi: 10.1111/j.1574-6968.2010.02177.x. [DOI] [PubMed] [Google Scholar]
- Angell S, Lewis CG, Buttner MJ, Bibb MJ. Glucose repression in Streptomyces coelicolor A3(2): a likely regulatory role for glucose kinase. Molecular and General Genetics. 1994;244:135–143. doi: 10.1007/BF00283514. [DOI] [PubMed] [Google Scholar]
- Ausmees N, Wahlstedt H, Bagchi S, Elliot MA, Buttner MJ, Flärdh K. SmeA, a small membrane protein with multiple functions in Streptomyces sporulation including targeting of a SpoIIIE/FtsK-like protein to cell division septa. Molecular Microbiology. 2007;65:1458–1473. doi: 10.1111/j.1365-2958.2007.05877.x. [DOI] [PubMed] [Google Scholar]
- Aínsa JA, Parry HD, Chater KF. A response regulator-like protein that functions at an intermediate stage of sporulation in Streptomyces coelicolor A3(2) Molecular Microbiology. 1999;34:607–619. doi: 10.1046/j.1365-2958.1999.01630.x. [DOI] [PubMed] [Google Scholar]
- Aínsa JA, Bird N, Ryding NJ, Findlay KC, Chater KF. The complex whiJ locus mediates environmentally sensitive repression of development of Streptomyces coelicolor A3(2) Antonie Van Leeuwenhoek. 2010;98:225–236. doi: 10.1007/s10482-010-9443-3. [DOI] [PubMed] [Google Scholar]
- Aínsa JA, Ryding NJ, Hartley N, Findlay KC, Bruton CJ, Chater KF. WhiA, a protein of unknown function conserved among Gram-positive bacteria, is essential for sporulation in Streptomyces coelicolor A3(2) Journal of Bacteriology. 2000;182:5470–5478. doi: 10.1128/jb.182.19.5470-5478.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaucher J, Rodrigue S, Jacques PE, Smith I, Brzezinski R, Gaudreau L. Novel Mycobacterium tuberculosis anti-ſ factor antagonists control ſF activity by distinct mechanisms. Molecular Microbiology. 2002;45:1527–1540. doi: 10.1046/j.1365-2958.2002.03135.x. [DOI] [PubMed] [Google Scholar]
- Bentley SD, Chater KF, Cerdeno-Tarraga AM, et al. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2) Nature. 2002;417:141–147. doi: 10.1038/417141a. [DOI] [PubMed] [Google Scholar]
- Bibb MJ, Buttner MJ. The Streptomyces coelicolor developmental transcription factor σBldN is synthesized as a proprotein. Journal of Bacteriology. 2003;185:2338–2345. doi: 10.1128/JB.185.7.2338-2345.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb MJ, Molle V, Buttner MJ. σBldN, an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2) Journal of Bacteriology. 2000;182:4606–4616. doi: 10.1128/jb.182.16.4606-4616.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bignell DR, Warawa JL, Strap JL, Chater KF, Leskiw BK. Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in Streptomyces coelicolor antibiotic production and sporulation. Microbiology. 2000;146:2161–2173. doi: 10.1099/00221287-146-9-2161. [DOI] [PubMed] [Google Scholar]
- Broad_Institute . Actinomycetales group Database. Broad Institute; 2011. http://www.broadinstitute.org/annotation/genome/streptomyces_group/MultiHome.html. [Google Scholar]
- Burger A, Sichler K, Kelemen G, Buttner M, Wohlleben W. Identification and characterization of the mre gene region of Streptomyces coelicolor A3(2) Molecular and General Genetics. 2000;263:1053–1060. doi: 10.1007/s004380050034. [DOI] [PubMed] [Google Scholar]
- Capstick DS, Willey JM, Buttner MJ, Elliot MA. SapB and the chaplins: connections between morphogenetic proteins in Streptomyces coelicolor. Molecular Microbiology. 2007;64:602–613. doi: 10.1111/j.1365-2958.2007.05674.x. [DOI] [PubMed] [Google Scholar]
- Capstick DS, Jomaa A, Hanke C, Ortega J, Elliot MA. Dual amyloid domains promote differential functioning of the chaplin proteins during Streptomyces aerial morphogenesis. Proceedings of the National Academy of Science, USA. 2011;108:9821–9826. doi: 10.1073/pnas.1018715108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty R, Bibb M. The ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) plays a conditional role in antibiotic production and morphological differentiation. Journal of Bacteriology. 1997;179:5854–5861. doi: 10.1128/jb.179.18.5854-5861.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraburtty R, White J, Takano E, Bibb M. Cloning, characterization and disruption of a (p)ppGpp synthetase gene (relA) of Streptomyces coelicolor A3(2) Molecular Microbiology. 1996;19:357–368. doi: 10.1046/j.1365-2958.1996.390919.x. [DOI] [PubMed] [Google Scholar]
- Champness WC. New loci required for Streptomyces coelicolor morphological and physiological differentiation. Journal of Bacteriology. 1988;170:1168–1174. doi: 10.1128/jb.170.3.1168-1174.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor A3(2) Journal of General Microbiology. 1972;72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- Chater KF. Taking a genetic scalpel to the Streptomyces colony. Microbiology. 1998;144:1465–1478. doi: 10.1099/00221287-144-6-1465. [DOI] [PubMed] [Google Scholar]
- Chater KF. Developmental decisions during sporulation in the aerial mycelium in Streptomyces. In: Brun YV, Shimkets LJ, editors. Prokaryotic Development. ASM Press; Washington, DC: 1999. pp. 33–48. [Google Scholar]
- Chater KF. Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Current Opinion in Microbiology. 2001;4:667–673. doi: 10.1016/s1369-5274(01)00267-3. [DOI] [PubMed] [Google Scholar]
- Chater KF. Differentiation in Streptomyces: the properties and programming of diverse cell-types. In: Dyson P, editor. Streptomyces: Molecular Biology and Biotechnology. Caister Academic Press; Norfolk, UK: 2011. pp. 43–86. [Google Scholar]
- Chater KF, Losick R. Mycelial life style of Streptomyces coelicolor A3(2) and its relatives. In: Shapiro JA, Dworkin M, editors. Bacteria as Multicellular Organisms. Oxford University Press; New York: 1997. pp. 149–182. [Google Scholar]
- Chater KF, Horinouchi S. Signalling early developmental events in two highly diverged Streptomyces species. Molecular Microbiology. 2003;48:9–15. doi: 10.1046/j.1365-2958.2003.03476.x. [DOI] [PubMed] [Google Scholar]
- Chater KF, Chandra G. The evolution of development in Streptomyces analysed by genome comparisons. FEMS Microbiology Reviews. 2006;30:651–672. doi: 10.1111/j.1574-6976.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- Chater KF, Chandra G. The use of the rare UUA codon to define "expression space" for genes involved in secondary metabolism, development and environmental adaptation in Streptomyces. The Journal of Microbiology. 2008;46:1–11. doi: 10.1007/s12275-007-0233-1. [DOI] [PubMed] [Google Scholar]
- Chater KF, Biro S, Lee KJ, Palmer T, Schrempf H. The complex extracellular biology of Streptomyces. FEMS Microbiology Reviews. 2010;34:171–198. doi: 10.1111/j.1574-6976.2009.00206.x. [DOI] [PubMed] [Google Scholar]
- Chater KF, Bruton CJ, Plaskitt KA, Buttner MJ, Méndez C, Helmann JD. The developmental fate of S. coelicolor hyphae depends on a gene product homologous with the motility s factor of B. subtilis. Cell. 1989;59:133–143. doi: 10.1016/0092-8674(89)90876-3. [DOI] [PubMed] [Google Scholar]
- Chavez A, Garcia-Huante Y, Ruiz B, Langley E, Rodriguez-Sanoja R, Sanchez S. Cloning and expression of the sco2127 gene from Streptomyces coelicolor M145. Journal of Industrial Microbiology and Biotechnology. 2009;36:649–654. doi: 10.1007/s10295-009-0533-z. [DOI] [PubMed] [Google Scholar]
- Chavez A, Forero A, Sanchez M, et al. Interaction of SCO2127 with BldKB and its possible connection to carbon catabolite regulation of morphological differentiation in Streptomyces coelicolor. Applied Microbiology and Biotechnology. 2011;89:799–806. doi: 10.1007/s00253-010-2905-8. [DOI] [PubMed] [Google Scholar]
- Chng C, Lum AM, Vroom JA, Kao CM. A key developmental regulator controls the synthesis of the antibiotic erythromycin in Saccharopolyspora erythraea. Proceedings of the National Academy of Science, USA. 2008;105:11346–11351. doi: 10.1073/pnas.0803622105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, de Jong W, Dijkhuizen L, Wosten HA. Regulation of Streptomyces development: reach for the sky! Trends in Microbiology. 2006;14:313–319. doi: 10.1016/j.tim.2006.05.008. [DOI] [PubMed] [Google Scholar]
- Claessen D, Wösten HA, van Keulen G, Faber OG, Alves AM, Meijer WG, Dijkhuizen L. Two novel homologous proteins of Streptomyces coelicolor and Streptomyces lividans are involved in the formation of the rodlet layer and mediate attachment to a hydrophobic surface. Molecular Microbiology. 2002;44:1483–1492. doi: 10.1046/j.1365-2958.2002.02980.x. [DOI] [PubMed] [Google Scholar]
- Claessen D, Rink R, de Jong W, Siebring J, de Vreugd P, Boersma FG, Dijkhuizen L, Wösten HA. A novel class of secreted hydrophobic proteins is involved in aerial hyphae formation in Streptomyces coelicolor by forming amyloid-like fibrils. Genes & Development. 2003;17:1714–1726. doi: 10.1101/gad.264303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claessen D, Stokroos I, Deelstra HJ, Penninga NA, Bormann C, Salas JA, Dijkhuizen L, Wosten HA. The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Molecular Microbiology. 2004;53:433–443. doi: 10.1111/j.1365-2958.2004.04143.x. [DOI] [PubMed] [Google Scholar]
- Colson S, van Wezel GP, Craig M, Noens EE, Nothaft H, Mommaas AM, Titgemeyer F, Joris B, Rigali S. The chitobiose-binding protein, DasA, acts as a link between chitin utilization and morphogenesis in Streptomyces coelicolor. Microbiology. 2008;154:373–382. doi: 10.1099/mic.0.2007/011940-0. [DOI] [PubMed] [Google Scholar]
- Czyz A, Wegrzyn G. The Obg subfamily of bacterial GTP-binding proteins: essential proteins of largely unknown functions that are evolutionarily conserved from bacteria to humans. Acta Biochimica Polonica. 2005;52:35–43. [PubMed] [Google Scholar]
- D’Alia D, Eggle D, Nieselt K, Hu WS, Breitling R, Takano E. Deletion of the signalling molecule synthase ScbA has pleiotropic effects on secondary metabolite biosynthesis, morphological differentiation and primary metabolism in Streptomyces coelicolor A3(2) Microbial Biotechnology. 2011;4:239–251. doi: 10.1111/j.1751-7915.2010.00232.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton KA, Thibessard A, Hunter JI, Kelemen GH. A novel compartment, the ‘subapical stem’ of the aerial hyphae, is the location of a sigN-dependent, developmentally distinct transcription in Streptomyces coelicolor. Molecular Microbiology. 2007;64:719–737. doi: 10.1111/j.1365-2958.2007.05684.x. [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J. Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell. 2003;113:767–776. doi: 10.1016/s0092-8674(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Davis NK, Chater KF. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Molecular Microbiology. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- Daza A, Martin JF, Dominguez A, Gil JA. Sporulation of several species of Streptomyces in submerged cultures after nutritional downshift. Journal of General Microbiology. 1989;135:2483–2491. doi: 10.1099/00221287-135-9-2483. [DOI] [PubMed] [Google Scholar]
- de Crécy-Lagard V, Servant-Moisson P, Viala J, Grandvalet C, Mazodier P. Alteration of the synthesis of the Clp ATP-dependent protease affects morphological and physiological differentiation in Streptomyces. Molecular Microbiology. 1999;32:505–517. doi: 10.1046/j.1365-2958.1999.01364.x. [DOI] [PubMed] [Google Scholar]
- de Jong W, Wösten HA, Dijkhuizen L, Claessen D. Attachment of Streptomyces coelicolor is mediated by amyloidal fimbriae that are anchored to the cell surface via cellulose. Molecular Microbiology. 2009a;73:1128–1140. doi: 10.1111/j.1365-2958.2009.06838.x. [DOI] [PubMed] [Google Scholar]
- de Jong W, Manteca A, Sanchez J, Bucca G, Smith CP, Dijkhuizen L, Claessen D, Wosten HA. NepA is a structural cell wall protein involved in maintenance of spore dormancy in Streptomyces coelicolor. Molecular Microbiology. 2009b;71:1591–1603. doi: 10.1111/j.1365-2958.2009.06633.x. [DOI] [PubMed] [Google Scholar]
- Dedrick RM, Wildschutte H, McCormick JR. Genetic interactions of smc, ftsK, and parB genes in Streptomyces coelicolor and their developmental genome segregation phenotypes. Journal of Bacteriology. 2009;191:320–332. doi: 10.1128/JB.00858-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sol R, Pitman A, Herron P, Dyson P. The product of a developmental gene, crgA, that coordinates reproductive growth in Streptomyces belongs to a novel family of small actinomycete-specific proteins. Journal of Bacteriology. 2003;185:6678–6685. doi: 10.1128/JB.185.22.6678-6685.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Sol R, Mullins JG, Grantcharova N, Flärdh K, Dyson P. Influence of CrgA on assembly of the cell division protein FtsZ during development of Streptomyces coelicolor. Journal of Bacteriology. 2006;188:1540–1550. doi: 10.1128/JB.188.4.1540-1550.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hengst CD, Buttner MJ. Redox control in actinobacteria. Biochimica et Biophysica Acta. 2008;1780:1201–1216. doi: 10.1016/j.bbagen.2008.01.008. [DOI] [PubMed] [Google Scholar]
- den Hengst CD, Tran NT, Bibb MJ, Chandra G, Leskiw BK, Buttner MJ. Genes essential for morphological development and antibiotic production in Streptomyces coelicolor are targets of BldD during vegetative growth. Molecular Microbiology. 2010;78:361–379. doi: 10.1111/j.1365-2958.2010.07338.x. [DOI] [PubMed] [Google Scholar]
- Derouaux A, Halici S, Nothaft H, Neutelings T, Moutzourelis G, Dusart J, Titgemeyer F, Rigali S. Deletion of a cyclic AMP receptor protein homologue diminishes germination and affects morphological development of Streptomyces coelicolor. Journal of Bacteriology. 2004;186:1893–1897. doi: 10.1128/JB.186.6.1893-1897.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Berardo C, Capstick DS, Bibb MJ, Findlay KC, Buttner MJ, Elliot MA. Function and redundancy of the chaplin cell surface proteins in aerial hypha formation, rodlet assembly, and viability in Streptomyces coelicolor. Journal of Bacteriology. 2008;190:5879–5889. doi: 10.1128/JB.00685-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditkowski B, Troc P, Ginda K, Donczew M, Chater KF, Zakrzewska-Czerwinska J, Jakimowicz D. The actinobacterial signature protein ParJ (SCO1662) regulates ParA polymerization and affects chromosome segregation and cell division during Streptomyces sporulation. Molecular Microbiology. 2010;78:1403–1415. doi: 10.1111/j.1365-2958.2010.07409.x. [DOI] [PubMed] [Google Scholar]
- Eccleston M, Willems A, Beveridge A, Nodwell JR. Critical residues and novel effects of overexpression of the Streptomyces coelicolor developmental protein BldB: evidence for a critical interacting partner. Journal of Bacteriology. 2006;188:8189–8195. doi: 10.1128/JB.01119-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eccleston M, Ahmed Ali R, Seyler R, Westpheling J, Nodwell J. Structural and genetic analysis of the BldB protein of Streptomyces coelicolor. Journal of Bacteriology. 2002;184:4270–4276. doi: 10.1128/JB.184.15.4270-4276.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot M, Damji F, Passantino R, Chater K, Leskiw B. The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. Journal of Bacteriology. 1998;180:1549–1555. doi: 10.1128/jb.180.6.1549-1555.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot MA, Leskiw BK. The BldD protein from Streptomyces coelicolor is a DNA-binding protein. Journal of Bacteriology. 1999;181:6832–6835. doi: 10.1128/jb.181.21.6832-6835.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliot MA, Buttner MJ, Nodwell JR. Multicellular development of Streptomyces. In: Whitworth DE, editor. Myxobacteria: Multicellularity and Differentiation. ASM Press; Herndon, VA, USA: 2008. pp. 419–438. [Google Scholar]
- Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK. BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2) Molecular Microbiology. 2001;40:257–269. doi: 10.1046/j.1365-2958.2001.02387.x. [DOI] [PubMed] [Google Scholar]
- Elliot MA, Locke TR, Galibois CM, Leskiw BK. BldD from Streptomyces coelicolor is a non-essential global regulator that binds its own promoter as a dimer. FEMS Microbiology Letters. 2003a;225:35–40. doi: 10.1016/S0378-1097(03)00474-9. [DOI] [PubMed] [Google Scholar]
- Elliot MA, Karoonuthaisiri N, Huang J, Bibb MJ, Cohen SN, Kao CM, Buttner MJ. The chaplins: a family of hydrophobic cell-surface proteins involved in aerial mycelium formation in Streptomyces coelicolor. Genes & Development. 2003b;17:1727–1740. doi: 10.1101/gad.264403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ensign JC. Formation, properties, and germination of actinomycete spores. Annual Reviews of Microbiology. 1978;32:185–219. doi: 10.1146/annurev.mi.32.100178.001153. [DOI] [PubMed] [Google Scholar]
- Erickson HP, Anderson DE, Osawa M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiology and Molecular Biology Reviews. 2010;74:504–528. doi: 10.1128/MMBR.00021-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facey PD, Hitchings MD, Saavedra-Garcia P, Fernandez-Martinez L, Dyson PJ, Del Sol R. Streptomyces coelicolor Dps-like proteins: differential dual roles in response to stress during vegetative growth and in nucleoid condensation during reproductive cell division. Molecular Microbiology. 2009;73:1186–1202. doi: 10.1111/j.1365-2958.2009.06848.x. [DOI] [PubMed] [Google Scholar]
- Facey PD, Sevcikova B, Novakova R, Hitchings MD, Crack JC, Kormanec J, Dyson PJ, Del Sol R. The dpsA gene of Streptomyces coelicolor: Induction of expression from a single promoter in response to environmental stress or during development. PLoS One. 2011;6:e25593. doi: 10.1371/journal.pone.0025593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flärdh K. Essential role of DivIVA in polar growth and morphogenesis in Streptomyces coelicolor A3(2) Molecular Microbiology. 2003;49:1523–1536. doi: 10.1046/j.1365-2958.2003.03660.x. [DOI] [PubMed] [Google Scholar]
- Flärdh K. Cell polarity and the control of apical growth in Streptomyces. Current Opinion in Microbiology. 2010;13:758–765. doi: 10.1016/j.mib.2010.10.002. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Buttner MJ. Streptomyces morphogenetics: Dissecting differentiation in a filamentous bacterium. Nature Reviews Microbiology. 2009;7:36–49. doi: 10.1038/nrmicro1968. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Findlay KC, Chater KF. Association of early sporulation genes with suggested developmental decision points in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2229–2243. doi: 10.1099/00221287-145-9-2229. [DOI] [PubMed] [Google Scholar]
- Flärdh K, Leibovitz E, Buttner MJ, Chater KF. Generation of a non-sporulating strain of Streptomyces coelicolor A3(2) by the manipulation of a developmentally controlled ftsZ promoter. Molecular Microbiology. 2000;38:737–749. doi: 10.1046/j.1365-2958.2000.02177.x. [DOI] [PubMed] [Google Scholar]
- Fowler-Goldsworthy K, Gust B, Mouz S, Chandra G, Findlay KC, Chater KF. The actinobacteria-specific gene wblA controls major developmental transitions in Streptomyces coelicolor A3(2) Microbiology. 2011 doi: 10.1099/mic.0.047555-0. (in press) [DOI] [PubMed] [Google Scholar]
- Gao B, Paramanathan R, Gupta RS. Signature proteins that are distinctive characteristics of Actinobacteria and their subgroups. Antonie Van Leeuwenhoek. 2006;90:69–91. doi: 10.1007/s10482-006-9061-2. [DOI] [PubMed] [Google Scholar]
- Gehring AM, Nodwell JR, Beverley SM, Losick R. Genomewide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. Proceedings of the National Academy of Science, USA. 2000;97:9642–9647. doi: 10.1073/pnas.170059797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazebrook MA, Doull JL, Stuttard C, Vining LC. Sporulation of Streptomyces venezuelae in submerged cultures. Journal of General Microbiology. 1990;136:581–588. doi: 10.1099/00221287-136-3-581. [DOI] [PubMed] [Google Scholar]
- Granozzi C, Billetta R, Passantino R, Sollazzo M, Puglia AM. A breakdown in macromolecular synthesis preceding differentiation in Streptomyces coelicolor A3(2) Journal of General Microbiology. 1990;136:713–716. doi: 10.1099/00221287-136-4-713. [DOI] [PubMed] [Google Scholar]
- Grantcharova N, Lustig U, Flärdh K. Dynamics of FtsZ assembly during sporulation in Streptomyces coelicolor A3(2) Journal of Bacteriology. 2005;187:3227–3237. doi: 10.1128/JB.187.9.3227-3237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Feng H, Zhang J, et al. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Research. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Görke B, Foulquier E, Galinier A. YvcK of Bacillus subtilis is required for a normal cell shape and for growth on Krebs cycle intermediates and substrates of the pentose phosphate pathway. Microbiology. 2005;151:3777–3791. doi: 10.1099/mic.0.28172-0. [DOI] [PubMed] [Google Scholar]
- Haiser HJ, Yousef MR, Elliot MA. Cell wall hydrolases affect germination, vegetative growth, and sporulation in Streptomyces coelicolor. Journal of Bacteriology. 2009;191:6501–6512. doi: 10.1128/JB.00767-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara H, Ohnishi Y, Horinouchi S. DNA microarray analysis of global gene regulation by A-factor in Streptomyces griseus. Microbiology. 2009;155:2197–2210. doi: 10.1099/mic.0.027862-0. [DOI] [PubMed] [Google Scholar]
- Heichlinger A, Ammelburg M, Kleinschnitz EM, Latus A, Maldener I, Flärdh K, Wohlleben W, Muth G. The MreB-like protein Mbl of S. coelicolor A3(2)depends on MreB for proper localization and contributes to spore wall synthesis. Journal of Bacteriology. 2011;193:1533–1542. doi: 10.1128/JB.01100-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hempel AM, Wang S, Letek M, A GJ, Flärdh K. Assemblies of DivIVA mark sites for hyphal branching and can establish new zones of cell wall growth in Streptomyces coelicolor. Journal of Bacteriology. 2008;90:7579–7583. doi: 10.1128/JB.00839-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques AO, Moran CP., Jr. Structure, assembly, and function of the spore surface layers. Annual Reviews of Microbiology. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- Hesketh A, Sun J, Bibb M. Induction of ppGpp synthesis in Streptomyces coelicolor A3(2) grown under conditions of nutritional sufficiency elicits actII-ORF4 transcription and actinorhodin biosynthesis. Molecular Microbiology. 2001;39:136–144. doi: 10.1046/j.1365-2958.2001.02221.x. [DOI] [PubMed] [Google Scholar]
- Hesketh A, Chen WJ, Ryding J, Chang S, Bibb M. The global role of ppGpp synthesis in morphological differentiation and antibiotic production in Streptomyces coelicolor A3(2) Genome Biology. 2007;8:R161. doi: 10.1186/gb-2007-8-8-r161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higo A, Horinouchi S, Ohnishi Y. Strict regulation of morphological differentiation and secondary metabolism by a positive feedback loop between two global regulators AdpA and BldA in Streptomyces griseus. Molecular Microbiology. 2011;81:1607–1622. doi: 10.1111/j.1365-2958.2011.07795.x. [DOI] [PubMed] [Google Scholar]
- Hirano S, Kato JY, Ohnishi Y, Horinouchi S. Control of the Streptomyces Subtilisin inhibitor gene by AdpA in the A-factor regulatory cascade in Streptomyces griseus. Journal of Bacteriology. 2006;188:6207–6216. doi: 10.1128/JB.00662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood DA. Streptomyces in Nature and Medicine. The Antibiotic Makers. Oxford University Press; New York: 2007. [Google Scholar]
- Hopwood DA, Wildermuth H, Palmer HM. Mutants of Streptomyces coelicolor defective in sporulation. Journal of General Microbiology. 1970;61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- Horinouchi S. Mining and polishing of the treasure trove in the bacterial genus Streptomyces. Bioscience, Biotechnology, and Biochemistry. 2007;71:283–299. doi: 10.1271/bbb.60627. [DOI] [PubMed] [Google Scholar]
- Huang J, Lih CJ, Pan KH, Cohen SN. Global analysis of growth phase responsive gene expression and regulation of antibiotic biosynthetic pathways in Streptomyces coelicolor using DNA microarrays. Genes & Development. 2001;15:3183–3192. doi: 10.1101/gad.943401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, Shi J, Molle V, et al. Cross-regulation among disparate antibiotic biosynthetic pathways of Streptomyces coelicolor. Molecular Microbiology. 2005;58:1276–1287. doi: 10.1111/j.1365-2958.2005.04879.x. [DOI] [PubMed] [Google Scholar]
- Hunt AC, Servin-Gonzalez L, Kelemen GH, Buttner MJ. The bldC developmental locus of Streptomyces coelicolor encodes a member of a family of small DNA-binding proteins related to the DNA-binding domains of the MerR family. Journal of Bacteriology. 2005;187:716–728. doi: 10.1128/JB.187.2.716-728.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchings MI, Hoskisson PA, Chandra G, Buttner MJ. Sensing and responding to diverse extracellular signals? Analysis of the sensor kinases and response regulators of Streptomyces coelicolor A3(2) Microbiology. 2004;150:2795–2806. doi: 10.1099/mic.0.27181-0. [DOI] [PubMed] [Google Scholar]
- Ichikawa N, Oguchi A, Ikeda H, et al. Genome sequence of Kitasatospora setae NBRC 14216T: an evolutionary snapshot of the family Streptomycetaceae. DNA Research. 2010;17:393–406. doi: 10.1093/dnares/dsq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, Chater K, Zakrzewska-Czerwinska J. The ParB protein of Streptomyces coelicolor A3(2) recognizes a cluster of parS sequences within the origin-proximal region of the linear chromosome. Molecular Microbiology. 2002;45:1365–1377. doi: 10.1046/j.1365-2958.2002.03102.x. [DOI] [PubMed] [Google Scholar]
- Jakimowicz D, Mouz S, Zakrzewska-Czerwinska J, Chater KF. Developmental control of a parAB promoter leads to formation of sporulation-associated ParB complexes in Streptomyces coelicolor. Journal of Bacteriology. 2006;188:1710–1720. doi: 10.1128/JB.188.5.1710-1720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakimowicz D, Zydek P, Kois A, Zakrzewska-Czerwinska J, Chater KF. Alignment of multiple chromosomes along helical ParA scaffolding in sporulating Streptomyces hyphae. Molecular Microbiology. 2007;65:625–641. doi: 10.1111/j.1365-2958.2007.05815.x. [DOI] [PubMed] [Google Scholar]
- Jenal U, Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annual Reviews of Genetics. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Jiang H, Kendrick KE. Characterization of ssfR and ssgA, two genes involved in sporulation of S. griseus. Journal of Bacteriology. 2000;182:5521–5529. doi: 10.1128/jb.182.19.5521-5529.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser BK, Clifton MC, Shen BW, Stoddard BL. The structure of a bacterial DUF199/WhiA protein: domestication of an invasive endonuclease. Structure. 2009;17:1368–1376. doi: 10.1016/j.str.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang SH, Huang J, Lee HN, Hur YA, Cohen SN, Kim ES. Interspecies DNA microarray analysis identifies WblA as a pleiotropic down-regulator of antibiotic biosynthesis in Streptomyces. Journal of Bacteriology. 2007;189:4315–4319. doi: 10.1128/JB.01789-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamoto S, Ensign JC. Cloning and characterization of a gene involved in regulation of sporulation and cell division of Streptomyces griseus. Actinomycetologica. 1995;9:136–151. [Google Scholar]
- Kawamoto S, Watanabe H, Hesketh A, Ensign JC, Ochi K. Expression analysis of the ssgA gene product, associated with sporulation and cell division in Streptomyces griseus. Microbiology. 1997;143:1077–1086. doi: 10.1099/00221287-143-4-1077. [DOI] [PubMed] [Google Scholar]
- Keijser BJF, Noens EEE, Kraal B, Koerten HK, van Wezel GP. The Streptomyces coelicolor ssgB gene is required for early stages of sporulation. FEMS Microbiology Letters. 2003;225:59–67. doi: 10.1016/S0378-1097(03)00481-6. [DOI] [PubMed] [Google Scholar]
- Kelemen GH, Plaskitt KA, Lewis CG, Findlay KC, Buttner MJ. Deletion of DNA lying close to the glkA locus induces ectopic sporulation in Streptomyces coelicolor A3(2) Molecular Microbiology. 1995;17:221–230. doi: 10.1111/j.1365-2958.1995.mmi_17020221.x. [DOI] [PubMed] [Google Scholar]
- Kelemen GH, Brown GL, Kormanec J, Potúcková L, Chater KF, Buttner MJ. The positions of the sigma factor genes whiG and sigF in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Molecular Microbiology. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- Kelemen GH, Brian P, Flärdh K, Chamberlin LC, Chater KF, Buttner MJ. Developmental regulation of transcription of whiE, a locus specifying the polyketide spore pigment in Streptomyces coelicolor A3(2) Journal of Bacteriology. 1998;180:2515–2521. doi: 10.1128/jb.180.9.2515-2521.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelemen GH, Viollier PH, Tenor JL, Marri L, Buttner MJ, Thompson CJ. A connection between stress and development in the multicellular prokaryote Streptomyces coelicolor A3(2) Molecular Microbiology. 2001;40:804–814. doi: 10.1046/j.1365-2958.2001.02417.x. [DOI] [PubMed] [Google Scholar]
- Kendrick KE, Ensign JC. Sporulation of Streptomyces griseus in submerged culture. Journal of Bacteriology. 1983;155:357–366. doi: 10.1128/jb.155.1.357-366.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Chater K, Lee KJ, Hesketh A. Changes in the extracellular proteome caused by the absence of the bldA gene product, a developmentally significant tRNA, reveal a new target for the pleiotropic regulator AdpA in Streptomyces coelicolor. Journal of Bacteriology. 2005;187:2957–2966. doi: 10.1128/JB.187.9.2957-2966.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DW, Hesketh A, Kim ES, Song JY, Lee DH, Kim IS, Chater KF, Lee KJ. Complex extracellular interactions of proteases and a protease inhibitor influence multicellular development of Streptomyces coelicolor. Molecular Microbiology. 2008a;70:1180–1193. doi: 10.1111/j.1365-2958.2008.06471.x. [DOI] [PubMed] [Google Scholar]
- Kim ES, Song JY, Kim DW, Chater KF, Lee KJ. A possible extended family of regulators of sigma factor activity in Streptomyces coelicolor. Journal of Bacteriology. 2008b;190:7559–7566. doi: 10.1128/JB.00470-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Calcutt MJ, Schmidt FJ, Chater KF. Partitioning of the linear chromosome during sporulation of Streptomyces coelicolor A3(2) involves an oriC-linked parAB locus. Journal of Bacteriology. 2000;182:1313–1320. doi: 10.1128/jb.182.5.1313-1320.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim IK, Lee CJ, Kim MK, Kim JM, Kim JH, Yim HS, Cha SS, Kang SO. Crystal structure of the DNA-binding domain of BldD, a central regulator of aerial mycelium formation in Streptomyces coelicolor A3(2) Molecular Microbiology. 2006;60:1179–1193. doi: 10.1111/j.1365-2958.2006.05176.x. [DOI] [PubMed] [Google Scholar]
- Kleinschnitz EM, Latus A, Sigle S, Maldener I, Wohlleben W, Muth G. Genetic analysis of SCO2997, encoding a TagF homologue, indicates a role for wall teichoic acids in sporulation of Streptomyces coelicolor A3(2) Journal of Bacteriology. 2011a;193:6080–6085. doi: 10.1128/JB.05782-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleinschnitz EM, Heichlinger A, Schirner K, Winkler J, Latus A, Maldener I, Wohlleben W, Muth G. Proteins encoded by the mre gene cluster in Streptomyces coelicolor A3(2) cooperate in spore wall synthesis. Molecular Microbiology. 2011b;79:1367–1379. doi: 10.1111/j.1365-2958.2010.07529.x. [DOI] [PubMed] [Google Scholar]
- Knizewski L, Ginalski K. Bacterial DUF199/COG1481 proteins including sporulation regulator WhiA are distant homologs of LAGLIDADG homing endonucleases that retained only DNA binding. Cell Cycle. 2007;6:1666–1670. doi: 10.4161/cc.6.13.4471. [DOI] [PubMed] [Google Scholar]
- Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM. The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proceedings of the National Academy of Science, USA. 2004;101:11448–11453. doi: 10.1073/pnas.0404220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kois A, Swiatek M, Jakimowicz D, Zakrzewska-Czerwinska J. SMC protein-dependent chromosome condensation during aerial hyphal development in Streptomyces. Journal of Bacteriology. 2009;191:310–319. doi: 10.1128/JB.00513-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok J, Trach KA, Hoch JA. Effects on Bacillus subtilis of a conditional lethal mutation in the essential GTP-binding protein Obg. Journal of Bacteriology. 1994;176:7155–7160. doi: 10.1128/jb.176.23.7155-7160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kormanec J, Sevcikova B. The stress-response sigma factor ſH controls the expression of ssgB, a homologue of the sporulation-specific cell division gene ssgA, in Streptomyces coelicolor A3(2) Molecular Genetics and Genomics. 2002;267:536–543. doi: 10.1007/s00438-002-0687-0. [DOI] [PubMed] [Google Scholar]
- Kormanec J, Sevcikova B, Halgasova N, Knirschova R, Rezuchova B. Identification and transcriptional characterization of the gene encoding the stress-response sigma factor ſH in Streptomyces coelicolor A3(2) FEMS Microbiology Letters. 2000;189:31–38. doi: 10.1111/j.1574-6968.2000.tb09202.x. [DOI] [PubMed] [Google Scholar]
- Kwak J, Dharmatilake AJ, Jiang H, Kendrick KE. Differential regulation of ftsZ transcription during septation of Streptomyces griseus. Journal of Bacteriology. 2001b;183:5092–5101. doi: 10.1128/JB.183.17.5092-5101.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakman JH, Postma PW. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. Journal of Bacteriology. 1994;176:2694–2698. doi: 10.1128/jb.176.9.2694-2698.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CJ, Won HS, Kim JM, Lee BJ, Kang SO. Molecular domain organization of BldD, an essential transcriptional regulator for developmental process of Streptomyces coelicolor A3(2) Proteins. 2007;68:344–352. doi: 10.1002/prot.21338. [DOI] [PubMed] [Google Scholar]
- Leskiw BK, Mah R, Lawlor EJ, Chater KF. Accumulation of bldA-specified tRNA is temporally regulated in Streptomyces coelicolor A3(2) Journal of Bacteriology. 1993;175:1995–2005. doi: 10.1128/jb.175.7.1995-2005.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letek M, Ordónez E, Vaquera J, Margolin W, Flärdh K, Mateos LM, Gil JA. DivIVA is required for polar growth in the MreB-lacking rod-shaped actinomycete Corynebacterium glutamicum. Journal of Bacteriology. 2008;190:3283–3292. doi: 10.1128/JB.01934-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis RA, Shahi SK, Laing E, Bucca G, Efthimiou G, Bushell M, Smith CP. Genome-wide transcriptomic analysis of the response to nitrogen limitation in Streptomyces coelicolor A3(2) BMC Research Notes. 2011;4:78. doi: 10.1186/1756-0500-4-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian W, Jayapal KP, Charaniya S, Mehra S, Glod F, Kyung YS, Sherman DH, Hu WS. Genome-wide transcriptome analysis reveals that a pleiotropic antibiotic regulator, AfsS, modulates nutritional stress response in Streptomyces coelicolor A3(2) BMC Genomics. 2008;9:56. doi: 10.1186/1471-2164-9-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolin W. Sculpting the bacterial cell. Current Biology. 2009;19:R812–822. doi: 10.1016/j.cub.2009.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marushima K, Ohnishi Y, Horinouchi S. CebR as a master regulator for cellulose/cellooligosaccharide catabolism affects morphological development in Streptomyces griseus. Journal of Bacteriology. 2009;191:5930–5940. doi: 10.1128/JB.00703-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurakova V, Sevcikova B, Rezuchova B, Kormanec J. Cascade of sigma factors in streptomycetes: identification of a new extracytoplasmic function sigma factor σJ that is under the control of the stress-response sigma factor σH in Streptomyces coelicolor A3(2) Archives of Microbiology. 2006;186:435–446. doi: 10.1007/s00203-006-0158-9. [DOI] [PubMed] [Google Scholar]
- Mazza P, Noens EE, Schirner K, et al. MreB of Streptomyces coelicolor is not essential for vegetative growth but is required for the integrity of aerial hyphae and spores. Molecular Microbiology. 2006;60:838–852. doi: 10.1111/j.1365-2958.2006.05134.x. [DOI] [PubMed] [Google Scholar]
- McCormick JR. Cell division is dispensable but not irrelevant in Streptomyces. Current Opinion in Microbiology. 2009;12:689–698. doi: 10.1016/j.mib.2009.10.004. [DOI] [PubMed] [Google Scholar]
- McCormick JR, Su EP, Driks A, Losick R. Growth and viability of Streptomyces coelicolor mutant for the cell division gene ftsZ. Molecular Microbiology. 1994;14:243–254. doi: 10.1111/j.1365-2958.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- Merrick MJ. A morphological and genetic mapping study of bald colony mutants of Streptomyces coelicolor. Journal of General Microbiology. 1976;96:299–315. doi: 10.1099/00221287-96-2-299. [DOI] [PubMed] [Google Scholar]
- Miguelez EM, Hardisson C, Manzanal MB. Hyphal death during colony development in Streptomyces antibioticus: morphological evidence for the existence of a process of cell deletion in a multicellular prokaryote. The Journal of Cell Biology. 1999;145:515–525. doi: 10.1083/jcb.145.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molle V, Buttner MJ. Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at different stages. Molecular Microbiology. 2000;36:1265–1278. doi: 10.1046/j.1365-2958.2000.01977.x. [DOI] [PubMed] [Google Scholar]
- Molle V, Palframan WJ, Findlay KC, Buttner MJ. WhiD and WhiB, homologous proteins required for different stages of sporulation in Steptomyces coelicolor A3(2) Journal of Bacteriology. 2000;182:1286–1295. doi: 10.1128/jb.182.5.1286-1295.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair S, Finkel SE. Dps protects cells against multiple stresses during stationary phase. Journal of Bacteriology. 2004;186:4192–4198. doi: 10.1128/JB.186.13.4192-4198.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen KT, Willey JM, Nguyen LD, Nguyen LT, Viollier PH, Thompson CJ. A central regulator of morphological differentiation in the multicellular bacterium Streptomyces coelicolor. Molecular Microbiology. 2002;46:1223–1238. doi: 10.1046/j.1365-2958.2002.03255.x. [DOI] [PubMed] [Google Scholar]
- Nguyen KT, Tenor J, Stettler H, Nguyen LT, Nguyen LD, Thompson CJ. Colonial differentiation in Streptomyces coelicolor depends on translation of a specific codon within the adpA gene. Journal of Bacteriology. 2003;185:7291–7296. doi: 10.1128/JB.185.24.7291-7296.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieselt K, Battke F, Herbig A, et al. The dynamic architecture of the metabolic switch in Streptomyces coelicolor. BMC Genomics. 2010;11:10. doi: 10.1186/1471-2164-11-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodwell JR, Losick R. Purification of an extracellular signaling molecule involved in production of aerial mycelium by Streptomyces coelicolor. Journal of Bacteriology. 1998;180:1334–1337. doi: 10.1128/jb.180.5.1334-1337.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nodwell JR, McGovern K, Losick R. An oligopeptide permease responsible for the import of an extracellular signal governing aerial mycelium formation in Streptomyces coelicolor. Molecular Microbiology. 1996;22:881–893. doi: 10.1046/j.1365-2958.1996.01540.x. [DOI] [PubMed] [Google Scholar]
- Nodwell JR, Yang M, Kuo D, Losick R. Extracellular complementation and the identification of additional genes involved in aerial mycelium formation in Streptomyces coelicolor. Genetics. 1999;151:569–584. doi: 10.1093/genetics/151.2.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noens EE, Mersinias V, Willemse J, Traag BA, Laing E, Chater KF, Smith CP, Koerten HK, van Wezel GP. Loss of the controlled localization of growth stage-specific cell-wall synthesis pleiotropically affects developmental gene expression in an ssgA mutant of Streptomyces coelicolor. Molecular Microbiology. 2007;64:1244–1259. doi: 10.1111/j.1365-2958.2007.05732.x. [DOI] [PubMed] [Google Scholar]
- Noh JH, Kim SH, Lee HN, Lee SY, Kim ES. Isolation and genetic manipulation of the antibiotic down-regulatory gene, wblA ortholog for doxorubicin-producing Streptomyces strain improvement. Applied Microbiology and Biotechnology. 2010;86:1145–1153. doi: 10.1007/s00253-009-2391-z. [DOI] [PubMed] [Google Scholar]
- Nothaft H, Dresel D, Willimek A, Mahr K, Niederweis M, Titgemeyer F. The phosphotransferase system of Streptomyces coelicolor is biased for N-acetylglucosamine metabolism. Journal of Bacteriology. 2003;185:7019–7023. doi: 10.1128/JB.185.23.7019-7023.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nothaft H, Rigali S, Boomsma B, Swiatek M, McDowall KJ, van Wezel GP, Titgemeyer F. The permease gene nagE2 is the key to N-acetylglucosamine sensing and utilization in Streptomyces coelicolor and is subject to multi-level control. Molecular Microbiology. 2010;75:1133–1144. doi: 10.1111/j.1365-2958.2009.07020.x. [DOI] [PubMed] [Google Scholar]
- Novotna J, Vohradsky J, Berndt P, et al. Proteomic studies of diauxic lag in the differentiating prokaryote Streptomyces coelicolor reveal a regulatory network of stress-induced proteins and central metabolic enzymes. Molecular Microbiology. 2003;48:1289–1303. doi: 10.1046/j.1365-2958.2003.03529.x. [DOI] [PubMed] [Google Scholar]
- Ochi K. A decrease in GTP content is associated with aerial mycelium formation in Streptomyces MA406-A-1. Journal of General Microbiology. 1986;132:299–305. doi: 10.1099/00221287-132-2-299. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Seo J-W, Horinouchi S. Deprogrammed sporulation in Streptomyces. FEMS Microbiology Letters. 2002;216:1–7. doi: 10.1111/j.1574-6968.2002.tb11406.x. [DOI] [PubMed] [Google Scholar]
- Ohnishi Y, Kameyama S, Onaka H, Horinouchi S. The A-factor regulatory cascade leading to streptomycin biosynthesis in Streptomyces griseus: identification of a target gene of the A-factor receptor. Molecular Microbiology. 1999;34:102–111. doi: 10.1046/j.1365-2958.1999.01579.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Ochi K. An essential GTP-binding protein functions as a regulator for differentiation in Streptomyces coelicolor. Molecular Microbiology. 1998;30:107–119. doi: 10.1046/j.1365-2958.1998.01042.x. [DOI] [PubMed] [Google Scholar]
- Okamoto S, Itoh M, Ochi K. Molecular cloning and characterization of the obg gene of Streptomyces griseus in relation to the onset of morphological differentiation. Journal of Bacteriology. 1997;179:170–179. doi: 10.1128/jb.179.1.170-179.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor TJ, Kanellis P, Nodwell JR. The ramC gene is required for morphogenesis in Streptomyces coelicolor and expressed in a cell type-specific manner under direct control of RamR. Molecular Microbiology. 2002;45:45–57. doi: 10.1046/j.1365-2958.2002.03004.x. [DOI] [PubMed] [Google Scholar]
- Parashar A, Colvin KR, Bignell DR, Leskiw BK. BldG and SCO3548 interact antagonistically to control key developmental processes in Streptomyces coelicolor. Journal of Bacteriology. 2009;191:2541–2550. doi: 10.1128/JB.01695-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez J, Munoz-Dorado J, Brana AF, Shimkets LJ, Sevillano L, Santamaria RI. Myxococcus xanthus induces actinorhodin overproduction and aerial mycelium formation by Streptomyces coelicolor. Microbial Biotechnology. 2011;4:175–183. doi: 10.1111/j.1751-7915.2010.00208.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plocinski P, Ziolkiewicz M, Kiran M, et al. Characterization of CrgA, a new partner of the Mycobacterium tuberculosis peptidoglycan polymerization complexes. Journal of Bacteriology. 2011;193:3246–3256. doi: 10.1128/JB.00188-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pope MK, Green BD, Westpheling J. The bld mutants of Streptomyces coelicolor are defective in the regulation of carbon utilization, morphogenesis and cell cell signalling. Molecular Microbiology. 1996;19:747–756. doi: 10.1046/j.1365-2958.1996.414933.x. [DOI] [PubMed] [Google Scholar]
- Pope MK, Green B, Westpheling J. The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. Journal of Bacteriology. 1998;180:1556–1562. doi: 10.1128/jb.180.6.1556-1562.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potrykus K, Cashel M. (p)ppGpp: still magical? Annual Reviews of Microbiology. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Potúcková L, Kelemen GH, Findlay KC, Lonetto MA, Buttner MJ, Kormanec J. A new RNA polymerase sigma factor, σF, is required for the late stages of morphological differentiation in Streptomyces spp. Molecular Microbiology. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- Rigali S, Titgemeyer F, Barends S, Mulder S, Thomae AW, Hopwood DA, van Wezel GP. Feast or famine: the global regulator DasR links nutrient stress to antibiotic production by Streptomyces. EMBO Reports. 2008;9:670–675. doi: 10.1038/embor.2008.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigali S, Nothaft H, Noens EE, et al. The sugar phosphotransferase system of Streptomyces coelicolor is regulated by the GntR-family regulator DasR and links N-acetylglucosamine metabolism to the control of development. Molecular Microbiology. 2006;61:1237–1251. doi: 10.1111/j.1365-2958.2006.05319.x. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Garcia A, Barreiro C, Santos-Beneit F, Sola-Landa A, Martin JF. Genome-wide transcriptomic and proteomic analysis of the primary response to phosphate limitation in Streptomyces coelicolor M145 and in a ΔphoP mutant. Proteomics. 2007;7:2410–2429. doi: 10.1002/pmic.200600883. [DOI] [PubMed] [Google Scholar]
- Rybniker J, Nowag A, van Gumpel E, Nissen N, Robinson N, Plum G, Hartmann P. Insights into the function of the WhiB-like protein of mycobacteriophage TM4--a transcriptional inhibitor of WhiB2. Molecular Microbiology. 2010;77:642–657. doi: 10.1111/j.1365-2958.2010.07235.x. [DOI] [PubMed] [Google Scholar]
- Ryding NJ, Kelemen GH, Whatling CA, Flärdh K, Buttner MJ, Chater KF. A developmentally regulated gene encoding a repressor-like protein is essential for sporulation in Streptomyces coelicolor A3(2) Molecular Microbiology. 1998;29:343–357. doi: 10.1046/j.1365-2958.1998.00939.x. [DOI] [PubMed] [Google Scholar]
- Saito N, Xu J, Hosaka T, Okamoto S, Aoki H, Bibb MJ, Ochi K. EshA accentuates ppGpp accumulation and is conditionally required for antibiotic production in Streptomyces coelicolor A3(2) Journal of Bacteriology. 2006;188:4952–4961. doi: 10.1128/JB.00343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salerno P, Larsson J, Bucca G, Laing E, Smith CP, Flärdh K. One of the two genes encoding nucleoid-associated HU proteins in Streptomyces coelicolor is developmentally regulated and specifically involved in spore maturation. Journal of Bacteriology. 2009;191:6489–6500. doi: 10.1128/JB.00709-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasindran SJ, Saikolappan S, Scofield VL, Dhandayuthapani S. Biochemical and physiological characterization of the GTP-binding protein Obg of Mycobacterium tuberculosis. BMC Microbiology. 2011;11:43. doi: 10.1186/1471-2180-11-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer EB, Claessen D, Haas M, Hurgobin B, Gras SL. The assembly of individual chaplin peptides from Streptomyces coelicolor into functional amyloid fibrils. PLoS One. 2011;6:e18839. doi: 10.1371/journal.pone.0018839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrempf H. Recognition and degradation of chitin by streptomycetes. Antonie Van Leeuwenhoek. 2001;79:285–289. doi: 10.1023/a:1012058205158. [DOI] [PubMed] [Google Scholar]
- Scott JM, Haldenwang WG. Obg, an essential GTP binding protein of Bacillus subtilis, is necessary for stress activation of transcription factor σB. Journal of Bacteriology. 1999;181:4653–4660. doi: 10.1128/jb.181.15.4653-4660.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo J-W, Ohnishi Y, Hirata A, Horinouchi S. ATP-binding cassette transport system involved in regulation of morphological differentiation in response to glucose in Streptomyces griseus. Journal of Bacteriology. 2002;184:91–103. doi: 10.1128/JB.184.1.91-103.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setlow P. I will survive: DNA protection in bacterial spores. Trends in Microbiology. 2007;15:172–180. doi: 10.1016/j.tim.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Sevcikova B, Kormanec J. Activity of the Streptomyces coelicolor stress-response sigma factor ſH is regulated by an anti-sigma factor. FEMS Microbiology Letters. 2002;209:229–235. doi: 10.1111/j.1574-6968.2002.tb11136.x. [DOI] [PubMed] [Google Scholar]
- Sevcikova B, Benada O, Kofronova O, Kormanec J. Stress-response sigma factor ſH is essential for morphological differentiation of Streptomyces coelicolor A3(2) Archives of Microbiology. 2001;177:98–106. doi: 10.1007/s00203-001-0367-1. [DOI] [PubMed] [Google Scholar]
- Sevcikova B, Rezuchova B, Homerova D, Kormanec J. The anti-anti-sigma factor BldG is involved in activation of the stress response sigma factor ſH in Streptomyces coelicolor A3(2) Journal of Bacteriology. 2011;192:5674–5681. doi: 10.1128/JB.00828-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaevitz JW, Gitai Z. The structure and function of bacterial actin homologs. Cold Spring Harbor Perspectives in Biology. 2011;2:a000364. doi: 10.1101/cshperspect.a000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A, Crossman DK, Mai D, Guidry L, Voskuil MI, Renfrow MB, Steyn AJ. Mycobacterium tuberculosis WhiB3 maintains redox homeostasis by regulating virulence lipid anabolism to modulate macrophage response. PLoS Pathogens. 2009;5:e1000545. doi: 10.1371/journal.ppat.1000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LJ, Stapleton MR, Fullstone GJ, et al. Mycobacterium tuberculosis WhiB1 is an essential DNA-binding protein with a nitric oxide-sensitive iron-sulfur cluster. Biochemical Journal. 2010;432:417–427. doi: 10.1042/BJ20101440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliveri J, Brown KL, Buttner MJ, Chater KF. Two promoters for the whiB sporulation gene of Streptomyces coelicolor A3(2) and their activities in relation to development. Journal of Bacteriology. 1992;174:6215–6220. doi: 10.1128/jb.174.19.6215-6220.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliveri JA, Gomez J, Bishai WR. Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology. 2000;146:333–343. doi: 10.1099/00221287-146-2-333. [DOI] [PubMed] [Google Scholar]
- Strauch MA, Hoch JA. Transition-state regulators: sentinels of Bacillus subtilis post-exponential gene expression. Molecular Microbiology. 1993;7:337–342. doi: 10.1111/j.1365-2958.1993.tb01125.x. [DOI] [PubMed] [Google Scholar]
- Sun J, Hesketh A, Bibb M. Functional analysis of relA and rshA, two relA/spoT homologues of Streptomyces coelicolor A3(2) Journal of Bacteriology. 2001;183:3488–3498. doi: 10.1128/JB.183.11.3488-3498.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Kelemen GH, Fernandez-Abalos JM, Bibb MJ. Green fluorescent protein as a reporter for spatial and temporal gene expression in Streptomyces coelicolor A3(2) Microbiology. 1999;145:2221–2227. doi: 10.1099/00221287-145-9-2221. [DOI] [PubMed] [Google Scholar]
- Susstrunk U, Pidoux J, Taubert S, Ullmann A, Thompson CJ. Pleiotropic effects of cAMP on germination, antibiotic biosynthesis and morphological development in Streptomyces coelicolor. Molecular Microbiology. 1998;30:33–46. doi: 10.1046/j.1365-2958.1998.01033.x. [DOI] [PubMed] [Google Scholar]
- Takano E. Gamma-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Current Opinion in Microbiology. 2006;9:287–294. doi: 10.1016/j.mib.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Takano E, Tao M, Long F, et al. A rare leucine codon in adpA is implicated in the morphological defect of bldA mutants of Streptomyces coelicolor. Molecular Microbiology. 2003a;50:475–486. doi: 10.1046/j.1365-2958.2003.03728.x. [DOI] [PubMed] [Google Scholar]
- Takano H, Hosono K, Beppu T, Ueda K. Involvement of ſH and related sigma factors in glucose-dependent initiation of morphological and physiological development of Streptomyces griseus. Gene. 2003b;320:127–135. doi: 10.1016/s0378-1119(03)00818-7. [DOI] [PubMed] [Google Scholar]
- Takano H, Fujimoto M, Urano H, Beppu T, Ueda K. Cross-interaction of anti-ſH factor RshA with BldG, an anti-sigma factor antagonist in Streptomyces griseus. FEMS Microbiology Letters. 2011;314:158–163. doi: 10.1111/j.1574-6968.2010.02155.x. [DOI] [PubMed] [Google Scholar]
- Tian Y, Fowler K, Findlay K, Tan H, Chater KF. An unusual response regulator influences sporulation at early and late stages in Streptomyces coelicolor. Journal of Bacteriology. 2007;189:2873–2885. doi: 10.1128/JB.01615-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag BA, van Wezel GP. The SsgA-like proteins in actinomycetes: small proteins up to a big task. Antonie Van Leeuwenhoek. 2008;94:85–97. doi: 10.1007/s10482-008-9225-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traag BA, Kelemen GH, van Wezel GP. Transcription of the sporulation gene ssgA is activated by the IclR-type regulator SsgR in a whi-independent manner in Streptomyces coelicolor A3(2) Molecular Microbiology. 2004;53:985–1000. doi: 10.1111/j.1365-2958.2004.04186.x. [DOI] [PubMed] [Google Scholar]
- Tran NT, Den Hengst CD, Gomez-Escribano JP, Buttner MJ. Identification and characterization of CdgB, a diguanylate cyclase involved in developmental processes in Streptomyces coelicolor. Journal of Bacteriology. 2011;193:3100–3108. doi: 10.1128/JB.01460-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda K, Takano H, Nishimoto M, Inaba H, Beppu T. Dual transcriptional control of amfTSBA, which regulates the onset of cellular differentiation in Streptomyces griseus. Journal of Bacteriology. 2005;187:135–142. doi: 10.1128/JB.187.1.135-142.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Wezel GP, McDowall KJ. The regulation of the secondary metabolism of Streptomyces: new links and experimental advances. Natural Products Report. 2011;28:1311–1333. doi: 10.1039/c1np00003a. [DOI] [PubMed] [Google Scholar]
- van Wezel GP, van der Meulen J, Kawamoto S, Luiten RG, Koerten HK, Kraal B. ssgA is essential for sporulation of Streptomyces coelicolor A3(2) and affects hyphal development by stimulating septum formation. Journal of Bacteriology. 2000;182:5653–5662. doi: 10.1128/jb.182.20.5653-5662.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventura M, Canchaya C, Tauch A, Chandra G, Fitzgerald GF, Chater KF, van Sinderen D. Genomics of Actinobacteria: tracing the evolutionary history of an ancient phylum. Microbiology and Molecular Biology Reviews. 2007;71:495–548. doi: 10.1128/MMBR.00005-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viala J, Mazodier P. The ATPase ClpX is conditionally involved in the morphological differentiation of Streptomyces lividans. Molecular and General Genetics. 2003;268:563–569. doi: 10.1007/s00438-002-0783-1. [DOI] [PubMed] [Google Scholar]
- Viollier PH, Minas W, Dale GE, Folcher M, Thompson CJ. Role of acid metabolism in Streptomyces coelicolor morphological differentiation and antibiotic biosynthesis. Journal of Bacteriology. 2001;183:3184–3192. doi: 10.1128/JB.183.10.3184-3192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viollier PH, Kelemen GH, Dale GE, Nguyen KT, Buttner MJ, Thompson CJ. Specialized osmotic stress response systems involve multiple SigB-like sigma factors in Streptomyces coelicolor. Molecular Microbiology. 2003;47:699–714. doi: 10.1046/j.1365-2958.2003.03302.x. [DOI] [PubMed] [Google Scholar]
- Wang L, Yu Y, He X, Zhou X, Deng Z, Chater KF, Tao M. Role of an FtsK-like protein in genetic stability in Streptomyces coelicolor A3(2) Journal of Bacteriology. 2007;189:2310–2318. doi: 10.1128/JB.01660-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wildermuth H. Development and organisation of the aerial mycelium in Streptomyces. Journal of General Microbiology. 1970;60:43–50. doi: 10.1099/00221287-60-1-43. [DOI] [PubMed] [Google Scholar]
- Wildermuth H, Hopwood DA. Septation during sporulation in Streptomyces coelicolor. Journal of General Microbiology. 1970;60:57–59. doi: 10.1099/00221287-60-1-51. [DOI] [PubMed] [Google Scholar]
- Willemse J, Borst JW, de Waal E, Bisseling T, van Wezel GP. Positive control of cell division: FtsZ is recruited by SsgB during sporulation of Streptomyces. Genes & Development. 2011;25:89–99. doi: 10.1101/gad.600211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willey J, Schwedock J, Losick R. Multiple extracellular signals govern the production of a morphogenetic protein involved in aerial mycelium formation by Streptomyces coelicolor. Genes & Development. 1993;7:895–903. doi: 10.1101/gad.7.5.895. [DOI] [PubMed] [Google Scholar]
- Willey J, Santamaria R, Guijarro J, Geistlich M, Losick R. Extracellular complementation of a developmental mutation implicates a small sporulation protein in aerial mycelium formation by S. coelicolor. Cell. 1991;65:641–650. doi: 10.1016/0092-8674(91)90096-h. [DOI] [PubMed] [Google Scholar]
- Willey JM, Willems A, Kodani S, Nodwell JR. Morphogenetic surfactants and their role in the formation of aerial hyphae in Streptomyces coelicolor. Molecular Microbiology. 2006;59:731–742. doi: 10.1111/j.1365-2958.2005.05018.x. [DOI] [PubMed] [Google Scholar]
- Wolanski M, Donczew R, Kois-Ostrowska A, Masiewicz P, Jakimowicz D, Zakrzewska-Czerwinska J. The level of AdpA directly affects expression of developmental genes in Streptomyces coelicolor. Journal of Bacteriology. 2011;193:6358–6365. doi: 10.1128/JB.05734-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie P, Zeng A, Qin Z. cmdABCDEF, a cluster of genes encoding membrane proteins for differentiation and antibiotic production in Streptomyces coelicolor A3(2) BMC Microbiology. 2009;9:157. doi: 10.1186/1471-2180-9-157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Li W, Tian Y, Liu G, Tan H. Identification and characterization of sawC, a whiA-like gene, essential for sporulation in Streptomyces ansochromogenes. Archives of Microbiology. 2007;188:575–582. doi: 10.1007/s00203-007-0278-x. [DOI] [PubMed] [Google Scholar]
- Xu H, Chater KF, Deng Z, Tao M. A cellulose synthase-like protein involved in hyphal tip growth and morphological differentiation in Streptomyces. Journal of Bacteriology. 2008;190:4971–4978. doi: 10.1128/JB.01849-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Q, Traag BA, Willemse J, et al. Structural and functional characterizations of SsgB, a conserved activator of developmental cell division in morphologically complex actinomycetes. The Journal of Biological Chemistry. 2009a;284:25268–25279. doi: 10.1074/jbc.M109.018564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Huang J, Lin R, Shi J, Cohen SN. Regulation of morphological differentiation in S. coelicolor by RNase III (AbsB) cleavage of mRNA encoding the AdpA transcription factor. Molecular Microbiology. 2009b;75:781–791. doi: 10.1111/j.1365-2958.2009.07023.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Ohnishi Y, Horinouchi S. An A-factor-dependent extracytoplasmic function sigma factor (σAdsA) that is essential for morphological development in Streptomyces griseus. Journal of Bacteriology. 2000;182:4596–4605. doi: 10.1128/jb.182.16.4596-4605.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki H, Tomono A, Ohnishi Y, Horinouchi S. DNA-binding specificity of AdpA, a transcriptional activator in the A-factor regulatory cascade in Streptomyces griseus. Molecular Microbiology. 2004;53:555–572. doi: 10.1111/j.1365-2958.2004.04153.x. [DOI] [PubMed] [Google Scholar]
- Yokoyama E, Doi K, Kimura M, Ogata S. Disruption of the hup gene encoding a histone-like protein HS1 and detection of HS12 of Streptomyces lividans. Research in Microbiology. 2001;152:717–723. doi: 10.1016/s0923-2508(01)01252-9. [DOI] [PubMed] [Google Scholar]
- Zhang G, Tian Y, Hu K, Feng C. SCO3900, co-transcripted with three downstream genes, is involved in the differentiation of Streptomyces coelicolor. Current Microbiology. 2010;60:268–273. doi: 10.1007/s00284-009-9536-2. [DOI] [PubMed] [Google Scholar]