Abstract
Specific interactions between proteins and DNA are fundamental to many biological processes. In this review, we provide a revised view of protein-DNA interactions that emphasizes the importance of the three-dimensional structures of both macromolecules. We divide protein-DNA interactions into two categories: those where the protein recognizes the unique chemical signatures of the DNA bases (base readout) and those where the protein recognizes a sequence-dependent DNA shape (shape readout). We further divide base readout into those interactions that occur in the major groove from those that occur in the minor groove. Analogously, the readout of DNA shape is subdivided into global shape recognition, for example when the DNA helix exhibits an overall bend, and local shape recognition, for example when a base pair step is kinked or when a region of the minor groove is narrow. Based on the >1500 structures of protein-DNA complexes now available in the Protein Data Base, we argue that individual DNA binding proteins combine multiple readout mechanisms to achieve DNA binding specificity. Specificity that distinguishes between families frequently involves base readout in the major groove while shape readout is often exploited for higher resolution specificity, to distinguish between members within the same DNA-binding protein family.
Keywords: Protein-DNA binding, Direct readout, Indirect readout, DNA base recognition, DNA shape recognition, Narrow minor groove, DNA kinks, DNA bending, B-DNA, A-DNA, Z-DNA
II. Introduction
II.a General comments
More than 50 years after the structure of DNA was first proposed by Watson and Crick [1], biologists are still working to achieve a complete understanding of how proteins interact with genomes. One of the most important questions that remain is one of specificity – how do the large and diverse number of DNA binding proteins encoded by eukaryotic genomes recognize their specific binding sites? Moreover, most DNA binding proteins are part of large families that share DNA binding domains with very similar biochemical properties. How do proteins with closely related DNA binding domains carry out their unique functions in vivo? Providing answers to these questions is especially timely given the need to accurately annotate the many complete genome sequences that are now available, an endeavor that is still a major unsolved challenge.
The size and complexity of this problem has recently been underscored by several publications that use high throughput approaches, such as protein binding microarrays or the bacterial one-hybrid system, to generate an unprecedented database of the DNA sequence preferences for a large number of DNA binding proteins [2–5]. In one such recent report, Bulyk and colleagues describe the binding site preferences for 104 mouse transcription factors, in many cases analyzing multiple members from the same transcription factor family [6]. To highlight just one example, the DNA binding site preferences for 21 members of the Sox (SRY-related high mobility group box)/TCF (T cell factor) family of transcriptional regulators were compared. Remarkably, although each factor executes unique functions, 14 of the 21 prefer to bind the sequence ACAAT. Moreover, although small differences in sequence preference were identified, these did not always correlate with the extent of sequence identity of the DNA binding domains. For example, Sox1 preferred the sequence ATTTAAAT, while its two most closely related relatives (Sox14 and Sox21), as well as a much more distantly related family member, Sex-determining Region Y (SRY), preferred the sequence ACAAT. This study also revealed that many transcription factors have the capacity to recognize two distinct binding sites (so-called primary and secondary binding sites) and that there is a previously underappreciated interdependence between neighboring base pairs within a binding site.
Observations such as these raise a number of fundamental questions regarding protein-DNA recognition whose answers will require a better understanding of the rules that govern how proteins bind to DNA sequences. We suggest that the linear sequence of base pairs in a binding site is only a small part of the story, and that the three-dimensional structures of both macromolecules must be taken into account to fully understand protein-DNA recognition. In particular, local variations in DNA structure – DNA topography – may be as important as protein structure. A recent study that examined the evolutionary constraints on DNA topology strongly supports this point of view [7]. Remarkably, the authors found that DNA shape in the human genome, as measured by hydroxyl radical cleavage patterns, is evolutionarily constrained. Moreover, these cleavage patterns, which are correlated with the solvent accessibility of the DNA helix [8], were found to be a much better predictor of functional DNA elements than the linear DNA sequence [7]. Thus, to more fully understand the rules that govern protein-DNA recognition, we must consider the structures of DNA and protein as equal partners.
II.b. Previous definitions: direct versus indirect readout mechanisms
Understanding how proteins recognize their DNA binding sites has a long history. Initially, based on early low-resolution X-ray structures of nucleic acid duplexes [9], it was realized that the major groove of the DNA helix offered a set of base-specific hydrogen bond donors and acceptors and non-polar groups that could be recognized by a complementary set of donors and acceptors presented by amino acid side chains [10]. Accordingly, the idea soon evolved that short DNA sequences could serve as binding sites that were specifically read by a complementary sequence of amino acid side chains [11]. This mechanism of protein-DNA recognition, now commonly referred to as direct readout, is evident in nearly all of the >1500 structures of protein-DNA complexes that have been solved and deposited in the Protein Data Base (PDB). Nevertheless, as was realized many years ago [12], there is not a simple recognition code or one-to-one correspondence between DNA and protein sequences. Thus, direct readout, by itself, cannot be sufficient to account for the specificities of protein-DNA interactions.
Although elements of direct readout contribute to nearly all protein-DNA complexes, these structures also reveal that bound DNA frequently deviates from a standard B-form double-helix. In some cases, deviations from a B-form helix are large and clearly contribute to DNA binding specificity (e.g. papillomavirus E2 protein and TATA binding protein (TBP); [13–15]). In these cases, a bend or some other deformation of the DNA helix is required to establish a set of hydrogen bonds or non-polar interactions between the protein and DNA that are much less likely to occur in the absence of the deformation. From such observations the term indirect readout was coined [12]. In the strictest definition, indirect readout is defined as protein-DNA interactions that depend on base pairs that are not directly contacted by the protein [16]; in other words, base pairs that create or facilitate a specific DNA structure that is subsequently recognized by a protein. However, in many cases, this strict definition is not always satisfied and the term indirect readout has been taken to mean any interaction between DNA and protein where the DNA is not a B-form helix. This looser definition has less value because it simply encompasses all interactions that are not direct.
II.c Goals for this review
In this review, we reevaluate the mechanisms that underlie protein-DNA recognition in light of new and previous structures of protein-DNA complexes. We suggest that the terms direct and indirect readout both describe idealized extremes that rarely exist in isolation in real protein-DNA complexes and therefore have limited value. For example, rarely are direct hydrogen bonds formed between protein side chains and DNA in the complete absence of any deviation from a B-form helix. Conversely, rarely are protein-DNA interactions purely indirect. As will be detailed below, this reevaluation suggests that protein-DNA recognition utilizes a continuum of readout mechanisms that depend on the structural features and flexibility of both macromolecules including the sequence-dependent propensity of DNA to assume conformations that deviate from ideal B-DNA. This more nuanced view suggests that protein-DNA and protein–protein recognition are in many ways analogous phenomena.
In order to reassess protein-DNA readout mechanisms, we divide this review into three main sections. In the first, we briefly discuss the range of protein structures that bind DNA. Because there are excellent recent reviews that already cover this topic [17–19], we simply summarize the major protein superfamiles that are observed in DNA binding proteins. Second, because interactions between proteins and DNA depend on the interplay between both macromolecules, we review how DNA structures vary and the relationships between these structures and DNA sequence. Finally, with these structural considerations as background, we review the range of interactions that are observed at protein-DNA interfaces, identifying common themes that are used both across and within individual families of DNA binding proteins. In our revised view, we propose to replace the terms direct readout and indirect readout with the more informative terms, base readout and shape readout, which we further subdivide to reflect the way proteins recognize DNA sequences. Our goal is to present a richer and more subtle view of protein-DNA recognition that more accurately reflects the way in which evolution has fine-tuned these essential interactions.
Because the perspective offered here is structural in its origins, we do not review thermodynamics measurements of protein-DNA interactions nor do we summarize the many insights available from the application of simulation methodologies to the recognition problem [20]. Rather, our goal is to review recent structural evidence regarding readout mechanisms of DNA sequences, recognizing that the a deeper understanding of the underlying forces and their interactions will require the application of a variety of experimental and computational approaches both to specific systems and on a genome-wide scale. It is our hope that the presentation and integration of structural data presented in this review will serve to facilitate and to focus such studies.
III. Structure of DNA binding proteins
The first protein-DNA complexes solved by X-ray crystallography were the catabolite gene activator protein (CAP) [21], Cro repressor [22], and λ repressor [23] bound to their binding sites. Since then, more than 1,500 structures of protein-DNA complexes have been deposited in the Protein Data Bank.
Proteins utilize a wide range of DNA-binding structural motifs, such as the helix-turn-helix (HTH) motif of homeodomains, to recognize DNA. Many proteins also contain flexible segments outside a globular core that mediate important specific and nonspecific interactions. For example, λ repressor has an N-terminal arm that contacts bases in the major groove [24], the phage Φ29 transcriptional regulator p4 uses N-terminal β-turn substructures to make base specific contacts in the major groove [25], while homeodomain proteins have N-terminal arms and linker regions that dock in the minor groove of the DNA [26–29]. These flexible regions, which are sometimes not included in the strict definition of these DNA binding domains, can have profound and essential roles in binding specificity.
According to SCOP classifications [30], DNA binding proteins, whose structures are currently available in complexes with DNA, are grouped into more than 70 superfamilies (Table 1). Due to this large number, it is not possible to discuss each superfamily here, and thus we focus only on a few representative examples. In Table 1 we group DNA binding proteins into the following categories based on the overall secondary structure content of the DNA binding domains: (1) mainly α, (2) mainly β, (3) mixed α/β, and (4) multi-domain proteins that have more than one of the aforementioned three domains. It is evident from the table that certain local motifs, such as the HTH motif, are used repeatedly and can be found within different global domain architectures. Moreover, depending on the protein and DNA binding site, any one type of motif can be used in multiple ways to interact with DNA. These observations support one of the main points of this review: that protein-DNA interactions depend on the interplay between two equal partners, the DNA and the protein; both macromolecules have their own characteristic three-dimensional structures that must accommodate to the other to achieve specificity.
Table 1.
Architecture of DNA binding proteins
| SCOP superfamily | Number of PDB entries |
Architecture of DNA binding domains |
DNA binding motif |
|---|---|---|---|
| DNA/RNA polymerases | 159 | multidomain, mixed α/β | |
| nucleotidyltransferase | 127 | multidomain, mixed α/β | |
| Ribonuclease H-like | 104 | multidomain, mixed α/β | |
| restriction endonuclease-like | 89 | mixed α/β | |
| Homeodomain-like | 75 | mainly alpha | Helix-turn-helix |
| `Winged helix` DNA-binding domain | 75 | mainly α with a small β-ribbon (wing) | winged Helix-turn-helix |
| Lesion bypass DNA Polymerase | 60 | multidomain, mixed α/β | |
| lambda repressor-like DNA-binding domains | 57 | mainly α | Helix-turn-helix |
| Glucocorticoid receptor-like | 53 | mixed α/β | zinc finger |
| p53-like transcription factors | 53 | mainly β | Immunoglobulin-like β-sandwich |
| DNA breaking-rejoining enzymes | 45 | multidomain, mixed α/β | |
| DNA-glycosylase | 40 | mixed α/β | |
| S-adenosyl-L-methionine-dependent methyltransferases | 40 | mixed α/β | |
| Histone-fole | 29 | mainly α | |
| Leucine zipper domain | 27 | mainly α | helix-loop-helix |
| DNA/RNA polymerases | 27 | multidomain, mixed α/β | |
| TATA-box binding protein-like | 24 | mainly β | TBP β-sheet |
| Homing endonucleases | 24 | mixed α/β | |
| C2H2 and C2HC zinc fingers | 22 | mixed α/β | zinc finger |
| E set domains | 21 | mainly β | Immunoglobulin-like β-sandwich |
| Chromo domain-like | 19 | mainly β | β-barrel |
| DNA repair protein MutS | 18 | multidomain, mixed α/β | |
| Ribbon-helix-helix | 16 | mixed α/β | ribbon-helix-helix |
| Uracil-DNA glycosylase-like | 16 | mixed α/β | |
| His-Me finger endonucleases | 14 | mixed α/β | |
| HMG-box | 13 | mainly α | helix-turn-helix |
| Origin of replication-binding domain, RBD-like | 13 | mixed α/β | |
| P-loop containing nucleoside triphosphate hydrolases | 12 | multidomain, mixed α/β | |
| Putative DNA-binding domain | 12 | mainly α | |
| Zn2/Cys6 DNA-binding domain | 11 | mixed α/β | zinc finger |
| IHF-like DNA-binding proteins | 10 | mixed α/β | |
| RNase A-like | 9 | mixed α/β | |
| HLH, helix-loop-helix DNA-binding domain | 8 | mainly α | helix-loop-helix |
| SRF-like | 8 | mixed α/β | |
| Zn2/Cys4 DNA-binding domain | 8 | mixed α/β | zinc finger |
| C-terminal effector domain of the bipartite response regulators | 7 | mainly α | helix-turn-helix |
| DNase I-like | 5 | mixed α/β | |
| Retrovirus zinc finger-like domains | 5 | mixed α/β | zinc finger |
| TrpR-lke | 5 | mainly α | helix-turn-helix |
| Viral DNA-binding domain | 5 | mixed α/β | |
| PIN domain-like | 5 | mixed α/β | ribbon-helix-helix |
| Zinc finger design | 4 | mixed α/β | zinc finger |
| DNA-binding domains of HMG-I(Y) | 4 | peptide | AT-hook |
| Transcription factor IIA (TFIIA) | 4 | mainly β | β-barrel |
| replication terminator protein (Tus) | 4 | multidomain, mixed α/β | |
| UDP-Glycosyltransferase/glycogen phosphorylase | 4 | mixed α/β | |
| Replication modulator SeqA, C-terminal DNA-binding domain | 4 | mainly α | |
| DNA binding domain | 4 | mixed α/β | β-sheet |
| FMT C-terminal domain-like | 4 | mixed α/β | |
| Sigma3 and sigma4 domains of RNA polymerase sigma factors | 3 | mainly α | helix-turn-helix |
| Methylated DNA-protein cysteine methyltransferase domain | 3 | mixed α/β | |
| DNA-binding domain of intron-encoded endonucleases | 3 | mixed α/β | |
| Cryptochrome/photolyase FAD-binding domain | 3 | mixed α/β | |
| T4 endonuclease V | 2 | mainly α | helix-turn-helix |
| SMAD MH1 domain | 2 | mixed α/β | |
| KorB DNA-binding domain-like | 2 | mainly α | helix-turn-helix |
| DNA topoisomerase IV, α subunit | 2 | multidomain, mixed α/β | |
| SMAD MH1 domain | 2 | mixed α/β | |
| 5' to 3' exonuclease catalytic domain | 2 | mixed α/β | |
| Metallo-dependent phosphatases | 2 | multidomain, mixed α/β | |
| WD40 repeat-like | 2 | mainly β | |
| Xylose isomerase-like | 1 | mixed α/β | |
| RNA polymerase | 1 | multidomain, mixed α/β | |
| GCM domain | 1 | mixed α/β | β-sheet |
| ATP-dependent DNA ligase DNA-binding domain | 1 | multidomain, mixed α/β | |
| Transposase IS200-like | 1 | mixed α/β | |
| Thioredoxin-like | 1 | multidomain, mixed α/β | |
| Holliday junction resolvase RusA | 1 | mixed α/β | |
| Skn-1 | 1 | mainly α | |
| ARID-like | 1 | mainly α | helix-turn-helix |
| GCM domain | 1 | mixed α/β | β-sheet |
| Phage replication organizer domain | 1 | mainly α | |
| Bet v1-like | 1 | mixed α/β | |
| AbrB/MazE/MraZ-like | 1 | mainly β |
This table lists DNA binding protein domains in different SCOP superfamilies, whose structures in complexes with DNAs are available in the PDB as of August 2009. When they are well defined, the DNA binding motifs used by these SCOP superfamilies are listed in the 4th column.
III.a. Mainly α
Proteins in 16 SCOP superfamilies have DNA binding domains with mainly α helical architecture, for example, homeodomains, leucine zipper proteins, and λ repressor-like proteins. The α-helix is the most frequently used secondary structure element for specific DNA recognition in the major groove. The positioning of the helix in the major groove can vary between different protein families and also among different proteins within the same family, as reviewed previously [17]. The Lac repressor [31, 32] and intron endonucleases [33–35] demonstrate that α-helices can also be used to interact with DNA in the minor groove. Based on the structural context in which the α-helices are found, the mainly α-class of proteins uses a number of local structural motifs for DNA binding.
Helix-turn-helix
The HTH motif is seen in many proteins in different SCOP families, and is one of the most frequently represented structural motifs in DNA binding proteins. The recognition helix of the HTH motif binds DNA through a series of hydrogen bonds and hydrophobic interactions with exposed bases, while the other helix stabilizes the interaction between the protein and DNA, but does not play a particularly strong role in recognition. Although the HTH motif is highly conserved, its structural context and precise orientation relative to the DNA binding sites it recognizes can vary between different proteins, and the structures outside the HTH core region can be very different in different proteins. For example, in homeodomains, the 2nd and 3rd helices of the three-helix bundle comprise the HTH motif with the 3rd helix (recognition helix) contacting the major groove, in an orientation that is nearly parallel to the flanking DNA backbones. The motility gene repressor (MogR) DNA binding domain contains seven α-helices connected by short loops: the first three helices form a three-helix bundle, the 4th helix forms a small dimerization interface, and helices 5–7 form a three-helix bundle DNA binding domain that contains a HTH motif (α6 and α7), in which α7 is the recognition helix [36]. Although the HTH motif is used most often in the major groove, some proteins use this motif to interact with the minor groove, for example, O6-alkylguanine-DNA alkyltransferase (AGT) [37].
A large class of HTH motif-containing proteins have an additional anti-parallel β-sheet, hence its name “winged helix-turn-helix” (wHTH) motif [38]. Proteins in more than 80 SCOP families contain the wHTH motif, including the hepatocyte nuclear factors-3 (HNF-3)/forkhead family of transcription factors [39], Ets domain [40] and multiple antibiotic resistance (MarR)-like transcription factors [41]. The “wing” typically sits over the minor groove to make additional DNA contacts. However, in some cases, the wings rather than the HTH motif contact the DNA in the major groove, as seen in regulatory factor X1 (RFX1) [42]. Many proteins also contain a second wing, which makes additional DNA contacts.
Helix-loop-helix and leucine zipper motifs
The helix-loop-helix motif consists of a short α-helix connected by a loop to a longer α-helix. Part of this motif is a dimerization domain that interacts with other helix-loop-helix proteins to form homo- or heterodimers; the dimerization partner often determines DNA binding affinity and specificity since two α-helices, one from each monomer, bind to the major groove of the target DNA [43–46].
III.b. Mainly β
Although less common than α-helices, β-strands and intervening loops embedded in the mainly β-domain structures are used by proteins in 7 SCOP superfamilies to recognize specific DNA sequences.
TATA box binding protein
TATA binding proteins (TBP) use a large β-sheet surface to recognize DNA by binding in the minor groove [14, 15]. Insertion of the concave, ten-stranded β-sheet of TBP into the groove requires profound DNA distortion. As will be discussed in the following sections, the TATA box DNA undergoes dramatic unwinding and bending that allows for contacts between the protein's concave surface and the edges of the base pairs in the otherwise recessed minor groove.
Immunoglobulin-like β-sandwich
Immunoglobulin-like structural domains are used for DNA binding in diverse families of proteins, such as p53-like transcription factors [47], E-set domains [48], and Runt domains [49]. The sequence conservation of the immunoglobulin-like domains in different families is low and the structures outside the domain diverge significantly. Although the overall fold is a β-sandwich, DNA recognition is achieved mainly by intervening loops. Like mainly α-helical DNA binding domains, the orientation of the β-sandwich domains relative to the DNA varies among different proteins and different families of proteins.
β-trefoil
The beta-trefoil is a capped β-barrel with an approximate three-fold symmetry, i.e., four strands are repeated in a three-fold arrangement, where strands 1 and 4 form the walls of the β-barrel and strands 2 and 3 contribute to the cap structure to give a 12-stranded structure. The β-trefoil domain of CSL, the nuclear effector of Notch signaling, contacts DNA via the loop between strands βA1 and βA2 [50].
β-β-β
The structure of AgrAC [51] reveals a novel topology, having 10 β-strands arranged into three antiparallel β-sheets, which are arranged roughly parallel to each other in an elongated β-β-β sandwich, and a small two-turn α-helix that is not involved in DNA binding. Base specific contacts are made with residues from intervening loops at both the major and minor grooves.
III.c. Mixed α/β
A large number of proteins, which belong to 48 SCOP superfamilies, use mixed α/β domains to bind DNA, although the major secondary structure elements used for recognition can be any one or any combination of α-helix, β-strand, or loop.
Zinc finger proteins
The zinc finger is a compact, ~30-amino acid DNA binding domain. Zinc fingers are the most minimal of DNA binding domains, with a relatively short α-helix, a two-stranded antiparallel β-sheet, and a Zn2+ ion coordinated by cysteine and histidine residues [52]. Zinc fingers are classified by the type and order of the zinc coordinating residues, e.g. Cys2His2, Cys4, and Cys6. Zinc fingers often occur as tandem repeats with two, three or more fingers that can bind in the major groove typically spaced at 3-base pair intervals. The α-helix of each domain (the "recognition helix") makes sequence-specific contacts to DNA bases in the major groove; residues from a single recognition helix can contact four or more bases to yield an overlapping pattern of contacts with adjacent zinc fingers.
Ribbon-helix-helix
A family of transcription factors from bacteria contain the ribbon-helix-helix (RHH) motif [53] that consists of a two-stranded anti-parallel β-ribbon followed by two α-helices. DNA recognition is achieved by insertion of the β-ribbon into the major groove, whereas the two helices comprise most of the hydrophobic core and are involved in dimerization. The prototypical examples are Met repressor MetJ [54] and Arc repressor [55].
Other mixed α/β domains
Structural studies of seemingly dissimilar restriction endonucleases with remarkable DNA sequence specificity demonstrated that they all share a common structural core with a mixed α/β architecture [56]. A large amount of structural data also reveal that DNA polymerases, DNA lesion repair enzymes and DNA modifying enzymes all have mixed α/β domain structures (Table 1).
III.d. Multi-domain proteins
Many DNA binding proteins contain multiple DNA binding domains, which can work together to recognize different regions of a target sequence, achieving high affinity and recognition specificity. For example, POU domain proteins, such as Oct-1 [57] and Brn-5 [58], contain a homeodomain (POUHD) and POU specific domain (POUS) that are connected by a flexible linker, and MarA consists of two HTH motifs that contact two successive major grooves [59]. Another example is the Rel-homology domain proteins such as NF-κB p50 that have two immunoglobulin-like domains in each monomer: the N-terminal domain mediates DNA contacts primarily in the major groove, while the C-terminal domain mediates homo- and heterodimer interactions in addition to contacting DNA [48, 60]. The side chains involved in dimer interactions lie along one face of the β-sandwich, leaving the loops free to contact the DNA. The E. coli transcription factor Rob, which belongs to the AraC/XylS family has two HTH domains: one binds specifically to DNA, whereas the other only forms a single salt bridge with the DNA backbone [61, 62]. TCF binds to specific DNA sequences through a high mobility group (HMG) domain. Recent data suggest that DNA recognition by Drosophila TCF occurs through a bipartite mechanism, involving both the HMG domain and the C-clamp, which enables TCF to locate and activate wingless-regulated enhancers in the nucleus [63].
IV. Sequence-dependent variations of DNA structure
Most current analyses of the information content in a nucleotide sequence view DNA as a one-dimensional string of letters based on an alphabet consisting of only four characters A, C, G, and T. Yet these bases are chemical entities that, along with the inclusion of the backbone sugar and phosphate groups, create a three-dimensional double-stranded structure in which each base pair has a specific chemical and conformational signature [10]. Although this textbook view of the double-helix is well-known, what is much less appreciated is that DNA structure varies in a sequence-dependent manner [20, 64], and that structural variations are used by proteins to recognize DNA sequences [65].
In this section we review the main ways in which DNA structures are known to deviate from idealized B-DNA. We distinguish between effects that vary the geometry of the helix in a localized manner (local shape, e.g. minor groove width and DNA kinks) from those that deform the overall cylindrical shape of the double-helix (global shape, e.g. DNA bending, A-DNA, and Z-DNA). In addition, although some DNA sequences do not produce a well-defined structure per se, they may be highly flexible, and therefore have a strong propensity to assume a non-B-like structure when bound to a protein. This property, commonly referred to as deformability, is another sequence-dependent feature that is used by proteins to recognize specific DNA sequences. To help make the connection between DNA sequence and DNA structure, Table 2 lists DNA sequences that have a tendency to assume a particular DNA structure.
Table 2.
Tendency of DNA sequence elements to form specific structural motifs.
| Sequence element | Structural motif | References |
|---|---|---|
| AT-rich | B-DNA | [71, 114] |
| GC-rich | A-DNA at low humidity | [75, 76, 186] |
| A-tract | B’-DNA, narrow minor groove, bending, rigid for ≥ 4 bp | [80–82, 85, 216] |
| TATA box | high deformability, A-DNA, TA-DNA upon TBP binding | [77, 187] |
| RY alternating (especially GC alternating) | Z-DNA at high salt concentration, upon cytosine methylation or supercoiling | [78, 79, 195] |
| YpR step (especially TpA step) | compresses major groove, high deformability, ‘hinge’ step, kinking | [83, 87–89] |
| RpY step | compresses minor groove, low deformability | [83, 87, 88] |
The table reflects general tendencies for some sequences to have particular structural characteristics. It is important to stress, however, that DNA conformation depends on environmental conditions (e.g., humidity and salt concentration) and the larger sequence context [64, 75]. For example, although AT-rich DNA is usually observed in B-form, TATA box containing oligonucleotides were crystallized in A-form [187], which is the basis for TBP specificity. In addition, due to their high deformability, the structure of TATA boxes is affected by long-range sequence effects [217], and by supercoiling. A TATA box flanked by GC alternating regions can also assume a Z-DNA conformation [218].
Differences in DNA shape can produce electrostatic potentials of varying magnitudes, a characteristic that can be read by proteins. For example, narrow minor grooves locally enhance the negative electrostatic potential of DNA through electrostatic focusing [65], which describes the deformation of field lines due to the shape of the dielectric boundary between solute and solvent [66]. This phenomenon was first described for a cavity of superoxide dismutase [67] but has also been shown to play a role in codon-anticodon recognition in transfer RNAs [68], in shaping electrostatic potentials around diverse RNA structures [69], and in shifting pKas in RNA catalytic sites [70]. As will be discussed further below, the effect appears to play an important role in protein-DNA recognition. In the following sections we therefore refer to electrostatic potential surfaces shown in Figure 1, which illustrate the close connection between shape and electrostatic potential in different DNA structures.
Figure 1. Molecular shape and electrostatic potential of A-DNA, B-DNA, and Z-DNA.
The upper panels show the molecular shape in GRASP2 images (convex surfaces in green and concave surfaces in grey/black) [219] of the three helical forms of DNA constructed with 3DNA [91] from fiber diffraction data [71, 79]. Each DNA helix is comprised of 14-mers. The width and depth stated below were calculated with Curves [220, 221]. The lower panels show how the electrostatic potential at the molecular surface varies due to shape and atomic charges. The electrostatic potentials were calculated by solving the Poisson-Boltzmann equation with DelPhi [66, 222] at a salt concentration of 0.145 M (other parameters as described in Methods of [65]). Negative electrostatic potentials are shown in red and positive electrostatic potentials in blue.
a. A-DNA with a narrow and deep major groove (2.2 Å wide; 9.5 Å deep) and a wide and shallow minor groove (10.9 Å wide; no defined depth). The model is of the alternating sequence d(GC)7.
b. B-DNA (alternating sequence d(GC)7) with a wide and shallow major groove (5.9 Å wide; 5.5 Å deep) and a narrow and deep minor groove (11.4 Å wide; 4.0 Å deep).
c. B-DNA (alternating sequence d(AT)7). Since the models are built based on fiber diffraction data, the shape of GC and AT alternating B-DNA does not reflect a sequence dependence.
d. Z-DNA which lacks a major groove (13.2 Å wide; no defined depth), while the minor groove is narrow and deep (2.4 Å wide, 5.0 Å deep). The model is of the alternating sequence d(GC)7.
e. A-DNA exhibits a strongly negative major groove but a hydrophobic minor groove surface, which is partially due to its exposed C3'-endo sugar moieties.
f. B-DNA (alternating sequence d(GC)7) exhibits a negative minor groove and less negative major groove.
g. B-DNA (alternating sequence d(AT)7). Variations in electrostatic potential between GC and AT alternating B-DNA reflect the different functional groups of the base pairs (e.g., positive guanine amino group in GC minor groove, neutral thymine methyl group in AT major groove).
h. Z-DNA exhibits a negative minor groove and a positive surface on opposing edges of the bases.
IV.a Global shape variations
IV.a.i. Polymorphisms of the double-helix
B-DNA
The most common form of double-stranded DNA is B-DNA, which is generally favored in aqueous solution similar to the environment in cells [71]. Most DNA-binding proteins recognize B-DNA and its structural variants. B-DNA is a right-handed double-helix with base pairs oriented perpendicular to the helix axis. Ideal B-DNA exhibits a wide and shallow major groove and a narrow and deep minor groove [64] (Figures 1b and 1c). As is evident from Figures 1f and 1g, the minor groove of B-DNA generally exhibits a more electronegative potential than the major groove. The differences in the potential in either groove between AT- and GC-rich sequences is due to the disposition of polar groups at the base edges; specifically AT-rich sequences display more negative electrostatic potentials in the minor groove than GC-rich sequences [72, 73] (Figures 1f and 1g). These effects are further enhanced by sequence-dependent effects on groove width, as discussed below.
A-DNA
A-DNA is observed under dehydrated conditions and in some protein-DNA complexes [74]. GC-rich sequences have an increased tendency to assume A-DNA or A/B intermediate conformations [75]. This property is at least in part due to the fact that GC base pairs have three hydrogen bonds, while AT base pairs have only two. This property makes GC base pairs more planar, allowing consecutive GC base pair steps to slide relative to each other, which promotes the A/B transition [76]. Although less pronounced, such a tendency is also observed for TATA boxes, in part because the TpA step counters propeller twisting. A-DNA is also a right-handed double-helix with the base pairs shifted towards the minor groove and, compared to B-DNA, tilted with respect to the helix axis by about 20°. This results in a narrow and very deep major groove and wide and very shallow minor groove [64] (Figure 1a). Based on this geometry, the A-DNA major groove resembles the shape of the B-DNA minor groove, which explains why, in contrast to B-DNA, the A-form major groove has a more negative electrostatic potential then its shallow minor groove [69] (Figure 1e).
TA-DNA
TA-DNA is a variant of A-DNA observed in TATA boxes, which are specifically recognized by TBP. It differs from A-DNA mainly by a larger base pair inclination of around 50° relative to the helix axis. This feature led to the description of TA-DNA as tilted A-DNA [77]. The TA-DNA geometry exhibits a positive roll (rotation between adjacent base pairs with respect to the base pairing axis), which explains the opening of the TATA box minor groove observed in TATA-TBP complexes [14, 15].
Z-DNA
Alternating purine-pyrimidine sequences were observed to form a left-handed double-helix under high salt concentrations [78, 79]. Due to the zig-zag conformation of its backbone, this topology was coined Z-DNA. Thought to form when B-DNA is deformed by supercoiling, Z-DNA does not have a pronounced major groove; instead, the base edges form a convex surface. The minor groove, however, resembles the dimensions of the B-DNA minor groove, but with a zig-zag trajectory of the backbone (Figure 1d) and a uniform negative electrostatic potential (Figure 1h).
IV.a.ii. DNA bending
We define DNA bending as curvature distributed over a stretch of several base pairs that leads to a different orientation of the regions on both sides of the curvature (Figure 2a). Bending has frequently been observed for sequences that contain A-tracts, which are stretches of A:T base pairs that include ApA (TpT) and ApT, but not TpA steps [80–82]. Various models have been established to explain the molecular origin of bending [83, 84] but all associate bending with wedge angles between adjacent base pairs, that can involve both roll and tilt. For example, Crothers and Shakked have suggested that bending is caused by contributions both from negative roll in A-tracts and positive roll in regions adjacent to A-tracts [85].
Figure 2. Comparison of readout mechanisms based on local shape, bending, and kinking in protein-DNA complexes.
a. HPV-18 E2 bound to DNA (PDB ID 1jj4) shows bending over a large stretch of the helix. The smooth curvature is visualized by the helix axis (blue) calculated with Curves [220].
b. The Lac repressor kinks the DNA at a central CpG base pair step stabilized by the partial intercalation of leucines (PDB ID 2kei). The helix axes calculated for both sides of the kink (blue) show an abrupt change in the helix trajectory caused by the kink.
c. Phage 434 repressor recognizes local shape deformations of its operator with arginine residues (PDB ID 2or1) [65]. The narrow region of the minor groove that is contacted by arginines is highlighted in blue.
d. For the same structure shown in c, the electrostatic potential of the operator calculated in the absence of the repressor is plotted on the molecular surface. In comparison with Figures 1f and 1g, the bottom of the minor groove is only red indicating enhanced negative electrostatic potential [65].
It is likely that the phasing of wedge angles is the critical factor for overall curvature. If short A-tracts (regions with negative roll) are phased by half a helical turn, the overall curvature cancels due to bending towards opposite sides of the helix. In a sequence where regions with negative roll are phased by a helical turn, the overall curvature is enhanced. The effect is further enhanced if regions with negative roll are in phase of half a helical turn with regions of positive roll as both regions would bend the double-helix in the same direction. Such a pattern has been reported for the nucleosome [83] and the papillomavirus E2 binding site [86]. Ultimately, the source of sequence-dependent bending can be traced to the conformational properties of individual dinucleotide steps [87, 88], their tendency to form wedge angles, and the composition of these dinucleotide steps in a DNA sequence.
IV.b. Local shape variations
IV.b.i. DNA kinks
We distinguish a kink from a DNA bend by defining a kink as a local disruption of an otherwise linear helix (Figure 2b). DNA kinks result from the complete or partial loss in stacking at a single base pair step. The pyrimidine-purine (YpR) steps TpA, CpA (TpG), and CpG are least stabilized through base stacking interactions and, of these, the TpA step has the weakest stacking interactions [64, 89] (Table 2). Therefore, it is the most flexible of the 10 unique dinucleotides and is referred to as a ”hinge” step [85, 88]. Because kinks occur at individual base pair steps, regions adjacent to a kink can remain in a straight B-form conformation or be curved. Bending and kinking can enhance each other as is the case for CpA steps adjacent to an A-tract [90]. Kinks are often stabilized by protein binding in cases where the loss of stacking interactions is compensated by the intercalation of hydrophobic side chains, which usually further deforms the kinked dinucleotide.
IV.b.ii. Minor groove narrowing
Minor groove width is another feature that varies locally in DNA structures [65] (Figure 2c). Differences in minor groove width arise from differences in the hydrogen bonding pattern of each base pair and from differing stacking interactions for each dinucleotide step. To optimize both types of interactions, DNA structures vary with respect to three rotational parameters: roll (relative rotation between adjacent base pairs with respect to the base pairing axis), helix twist (relative rotation between adjacent base pairs with respect to the helix axis), and propeller twist (relative rotation between bases within a base pair with respect to the base pairing axis) [91, 92]. ApT base pair steps usually have negative roll angles, which leads to a compression of the minor groove [83] (Table 2). In an A-tract sequence, ApT and ApA (TpT) steps exhibit a negative roll, and the bifurcated hydrogen bonds of A:T base pairs lead to propeller twisting, both enhancing minor groove narrowing [82, 86]. In addition, several A:T base pairs in a row enhance propeller twisting by allowing the formation of inter-base pair hydrogen bonds in the major groove [80]. In contrast to ApA (TpT) and ApT, propeller twisted TpA steps would lead to a steric clash of the cross-strand adenines [85]. Therefore, TpA ”hinge” steps tend to locally widen the minor groove and break rigid A-tract structures [88] (Table 2).
V. Mechanisms of protein-DNA recognition
General comments
Protein-DNA interfaces involve on average 24 protein residues and 12 nucleotides [93], making it likely that each interface is composed of many different types of interactions. While all interactions contribute to binding affinity, specificity can be viewed as resulting from a subset of interactions that are sequence-specific. It is these specificity-determining contacts that we are most concerned with here.
Given our focus on specificity, it is important to define what we mean by this term and to point out that DNA binding proteins generally exhibit multiple tiers of specificity. All homeodomains, for example, have an asparagine at position 51, which is important for the specific binding of these proteins to AT-rich sequences, such as TAAT (e.g., Engrailed and Antennapedia; [26, 94, 95]). Thus, Asn51 can be considered to be a critical determinant of homeodomain DNA binding specificity. However, as all homeodomains have Asn51, this residue cannot contribute to specificity within this superfamily. On a finer level, position 50 of the homeodomain partially fulfills this role: when it is a glutamine, preferred binding sites are TAATTG or TAATTA (where the Gln contacts are underlined) but when it is a lysine, the preferred binding site is TAATCC [96–99]. However, the subset of homeodomain proteins that have a glutamine at position 50 is still very large, and includes all of the Hox homeodomains, of which there are 39 in humans alone. Therefore, Gln50 cannot contribute to specificity within this subset of homeodomain proteins. In addition to Asn51 and Gln50, which are presented from a HTH recognition helix in the major groove, Hox proteins also bind to the minor groove, where DNA shape, in particular minor groove width, can be read [29]. As will be discussed below, this mode of protein-DNA recognition contributes to the specificity within the Hox family. From this one example, we see that DNA binding proteins use multiple readout mechanisms and that specificity is ultimately achieved by combinations of these mechanisms that successively fine-tune the selection of binding sites.
Although contacts between proteins and the DNA backbone are thought to have little impact on specificity [100], backbone contacts may play a role in specificity through the positioning of protein recognition elements in orientations that allow them to make other, more specific, contacts, such as hydrogen bonds to the bases [101, 102]. Indeed, protein families often contain conserved backbone-contacting residues that preserve the interface orientation for an entire family [102]. In addition, specificity may depend on contacts to the DNA backbone if these contacts can only be made when the DNA assumes a sequence-dependent structure that deviates from ideal B-DNA (below referred to as “non-ideal B-DNA”). An example is the readout of narrow minor groove regions, where the phosphates are located in positions that differ from ideal B-DNA. The Arg repressor from Mycobacterium tuberculosis, for instance, specifically recognizes a narrow minor groove region via extensive phosphate contacts from a four-stranded β-sheet that lies above the groove without inserting any side chain into the groove [103].
Protein-DNA recognition is also more complex than a simple docking process of two structurally preformed macromolecules. Some proteins fold only in the presence of DNA. For example, the leucine zippers of Fos and Jun are helical only when they form a heterodimer, and the basic regions are helical only when the dimer binds DNA [104, 105]. Moreover, other domains in both proteins appear to be unstructured until bound by cofactors such as CREB binding protein (CBP)/p300 [106]. Lymphoid enhancer factor-1 (LEF-1) also transitions from a relatively unstructured state to a well-folded domain upon DNA binding [107]. The sequence-specific binding of Cys2His2 zinc finger proteins to DNA causes their linker regions to fold, cap and thereby stabilize the preceding helix, which helps to orient the next zinc finger correctly for binding in the major groove [108]. Finally, binding of the zinc finger domain of retinoid X receptor (RXR) to DNA leads to folding of the dimerization region, which is disordered in the unbound protein [109]. DNA binding can also induce conformational changes in the bound protein that can change its properties. For example, the binding of the Glucocorticoid Receptor (GR) to its response elements induces conformational changes that expose transcriptional activation surfaces [110]. Moreover, different GR binding sites result in distinct GR activities, that, based on X-ray data, could be explained by changes in the orientation of a GR loop induced by a modification of DNA backbone contacts [111].
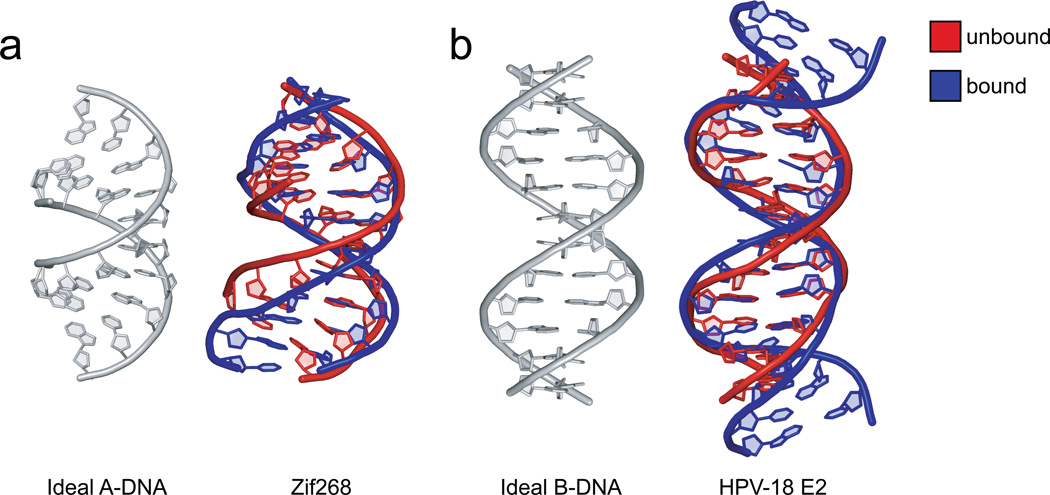
DNA can also change conformation and pre-existing sequence-dependent conformations can be stabilized or enhanced upon protein binding (Figure 3). For example, in specifically designed non-cognate GR complexes, the DNA is able to distort so as to maximize the number of cognate interface interactions, even if these are only maintained by a single strand [102, 112]. Such effects make it difficult to unambiguously determine if non-ideal B-DNA structures observed in protein-DNA complexes are intrinsic to the DNA sequence, induced by the protein, or some combination of the two. The relative impact of intrinsic vs. induced effects on DNA structure can only be assessed with certainty by comparing the structure of the free DNA binding site with its protein-bound form. Such structural information is currently restricted to the binding sites of only a handful of proteins, including the EcoRI endounclease [113, 114], Trp repressor [12, 115], Met repressor [54, 116], purine repressor [31, 116], NF-κB [48, 117], Zif286 zinc fingers [116, 118, 119] (Figure 3a), papillomavirus E2 protein [13, 81, 120] (Figure 3b), and Runt domain [49, 121, 122],. The limited size of this group is largely due to the lack of free DNA structures [20]. In their place, theoretical approaches have been developed to estimate the impact of intrinsic vs. induced effects when only the bound form is available [123, 124] or to predict the structure of the unbound DNA binding site [20, 29, 86].
Figure 3. DNAs bound to proteins have features already present in unbound DNAs.
a. The structure of the unbound FIN-B sequence (PDB ID 2b1c) is similar to ideal A-DNA (grey), while the bound structure of the Zif286-DNA complex (PDB ID 1a1f) has some A-DNA characteristics, notably a wider minor groove than normally found in B-DNA.
b. The specific HPV-18 E2 site (PDB ID 1ilc) contains an A-tract AATT in the central region of the helix, which, although not contacted by the protein, bends the free-DNA structure (red) in a similar manner as seen in the bound structure (blue) of the HPV-18 E2-DNA complex (PDB ID 1jj4). In comparison to ideal B-DNA (grey), the bending is reflected by a minor groove narrowing in the center of the free and bound DNA.
With this background in mind, below we discuss the various mechanisms proteins use to recognize their binding sites, attempting to organize them from a structure-based perspective. Note that we only have space in this review to support each readout mechanism with a small number of examples. Further, because any one DNA binding protein typically uses a variety of readout mechanisms, the same example may be used multiple times.
V.a. Base readout
One well-established way for proteins to achieve DNA binding specificity is through contacts with the bases in either the major or minor groove that recognize the chemical signature of the base or base pair. This type of recognition is generally mediated by the formation of hydrogen bonds between amino acids and bases, which convey the highest degree of specificity, and, in some cases, by water-mediated hydrogen bonds, or hydrophobic contacts.
V.a.i. Base-specific interactions in the major groove
Hydrogen bonds with bases
Hydrogen bonds with bases can confer greater specificity in the major groove than in the minor groove because the four possible base pairs have a unique pattern of hydrogen bond donors and acceptors in the major but not in the minor groove [10, 125] (Figure 4). Proteins that form hydrogen bonds with bases in the major groove use HTH domains (e.g., homeodomains, 434 repressor, λ repressor, Trp repressor, Myb domain), zinc finger domains (e.g., TFIIIA), immunoglobin fold domains (e.g., p53, NF-κB, STAT, and NFAT), and the N-terminal end of basic Leucine zipper (bZip) domains or the Max transcription factor [17–19].
Figure 4. Base recognition in the major and minor groove.
Sequence specific patterns on the edges of the bases in the major groove underlie the ability for proteins to readout base pairs through hydrogen bonds and hydrophobic contacts (hydrogen bond acceptors in red, donors in blue, thymine methyl group in yellow, and base carbon hydrogens in white). In contrast, A:T versus T:A and C:G versus G:C are indistinguishable in the minor groove. The three panels show successive rotations of 90° around the helix axis. The dodecamer d(GACT)3 was built based on fiber diffraction data with 3DNA [91].
As noted above, the orientation of the recognition helix in the major groove is similar for homeodomain-DNA interfaces [126] but can vary among different families [17] and even within a given family, as between the Trp and λ repressors [100]. In some cases, as observed for the KorA repressor, the recognition helix induces a widening of the major groove [127]. Besides α-helices, hydrogen bonds between β-sheets and bases can be used as well in specific recognition. Hydrogen bonds between bases in the major groove with the convex side of a β-sheet are observed in the binding of the MetJ and Arc repressors to DNA [128]. The width of the major groove adjusts to the size of the β-sheet (widened in Arc repressor and narrowed in MetJ repressor), and the side of the β-sheet interacting with DNA generally exhibits more positive electrostatic potentials [128].
Specificity conveyed through hydrogen bonds in either groove depends on the number of contacts formed between protein residues and DNA bases but also on the uniqueness of the hydrogen bonding geometry. Bidentate hydrogen bonds (two hydrogen bonds with different donor and acceptor atoms) have the highest degree of specificity followed by bifurcated hydrogen bonds (two hydrogen bonds that share the donor) and single hydrogen bonds. Whereas single hydrogen bonds usually do not contribute to specificity, bidentate hydrogen bonds are a source of remarkable selectivity [129]. Bidentate hydrogen bonds can be formed with one base, two bases in a base pair, two adjacent bases in one strand, or two bases diagonally in different base pairs and opposite strands.
As discussed above, the specificity achieved through hydrogen bonds with bases depends on the pattern of donors and acceptors at the base edges in both grooves (Figure 4). Since DNA usually occurs in Watson-Crick geometry [1], this pattern is specific for each of the four base pairs in the major groove. However, base pair geometry can vary. For instance, Hoogsteen base pairs [130] have been observed in structures with deformed DNA sequences such as the TBP/TATA box complex [131] and at the ends of oligonucleotides where the helical structure is preserved through stacking interactions (e.g., in a p53 tetramer complex, [122]). To date, a Hoogsteen base pair not present at the end of an oligonucleotide has only been observed in one complex with undistorted B-DNA, of the MATα2 homeodomain bound to a specific binding site [132]. Interestingly, the Hoogsteen base pair (underlined) occurs in the center of the binding site CATGTAATT and was seen in crystals generated under various conditions [132]. A transition from a Watson-Crick to Hoogsteen geometry alters the pattern of hydrogen bond donors and acceptors in both grooves and the conformation of the double-helix. Although this single example should be interpreted with caution, it raises the possibility that non-Watson-Crick base pairs may contribute in important ways to binding specificity. As high-resolution structures are required to visualize such geometries, they may be present at a greater frequency than is evident in existing structures.
In many structures, hydrogen bonds between protein and DNA are mediated by intervening water molecules. The bridging of hydrogen bonds by water molecules has frequently been observed for enzymes [133], and most hydrogen bonds in the Trp repressor-DNA interface are water-mediated [12, 125]. Mutagenesis experiments have shown that the CTAG tracts in both half sites of the Trp repressor’s binding site are most critical for its sequence specificity [134]. Highly ordered water molecules also mediate the specific readout of bases in the RXR/retinoid acid receptor (RAR)-DNA complex involving several arginine and lysine residues [135]. Interestingly for the Lac repressor, the protein-DNA interface retains a significant portion of its hydration when it binds non-specifically but not in the specific complex [136].
These data suggest that water-mediated hydrogen bonds in the major groove can be used for specific readout because they often reflect the positions of hydrogen bond donors and acceptors at the base edges. This is not the case for water molecules in the minor groove where the donor-acceptor patterns become unrecognizable.
Hydrophobic contacts with bases
Whereas hydrogen bonds with bases are highly specific in recognizing purines, hydrophobic contacts with bases are mainly used to read pyrimidines. Protein side chains employ hydrophobic interactions to differentiate thymine from cytosine [125] as in bacteriophage 434 repressor and 434 Cro binding to their operator sites [137, 138]. Four thymine methyl groups form a cleft that is specifically recognized by a valine in the lambdoid bacteriophage P22 c2 repressor-operator complex [139].
Hydrophobic contacts with bases also play a key role in the sequence-specific recognition of single-stranded DNA by bacterial cold shock proteins, which recognize poly-thymine strands through stacking interactions with phenylalanines and histidines and distinguish thymine from cytosine through hydrogen bonding [140, 141].
V.a.ii. Base-specific interactions in the minor groove
Proteins can also form hydrogen bonds with bases in the minor groove, although, as discussed above, the pattern of donors and acceptors in the minor groove does not distinguish AT from TA or GC from CG base pairs [10] (Figure 4). Some proteins, such as zinc finger proteins with Cys2Cys2 GATA-like domains, that form hydrogen bonds in the major groove also bind in the minor groove [19]. High mobility group (HMG) proteins form hydrogen bonds in the minor groove [19] but rely on the recognition of DNA shape and flexibility discussed below to achieve specificity. This is also apparent for the binding of TBP to the minor groove as the six observed hydrogen bonds with the TATA box are not sufficient for the protein to attain specificity [14, 15, 142].
In some cases a spine of hydration in narrow minor groove regions is contacted by proteins, as observed in the DNA complexes formed by the IFN-β enhanceosome [143] and the integration host factor (IHF) [142]. In other cases only individual water molecules are displaced from narrow minor groove regions when amino acids intrude into the groove (e.g., α2-Arg7 in the MATa1/MATα2-DNA complex [144]). The displacement of water molecules from the narrow minor groove has been shown to provide a strong thermodynamic driving force for DNA binding [145–147].
Hydrophobic contacts with bases
Architectural proteins only contact the minor groove, which is often associated with a dramatic widening and extensive hydrophobic contacts [142]. This mechanism is employed by TBP, SRY, and LEF-1. The TBP/TATA box interface is completely dehydrated and the abundance of hydrophobic contacts in the interface [148] suggests that they contribute to specificity. While 12 of the 16 hydrogen bond acceptors in the minor groove remain unsatisfied upon TBP binding, these base atoms mainly engage in hydrophobic contacts with non-polar side chains [14, 15, 142].
V.b. Shape readout
For most DNA binding proteins, the readout of base pairs through hydrogen bonds or hydrophobic contacts is not sufficient to explain specificity. Other factors that have been proposed to contribute to specificity are sequence-dependent DNA structure and deformability [20, 149]. These readouts, which all depend on deviations from ideal B-DNA, comprise a diverse set of mechanisms that all fall under the general heading of binding a non-ideal B-DNA shape. As such, we collectively refer to them as shape readout. Further, we distinguish between local shape readout mechanisms, in which the DNA helix deviates from ideal B-DNA in a localized manner, and global shape readout mechanisms, in which most of the DNA binding site is either deformed or in a non-ideal B-form conformation.
Both local and global shape readouts can contribute to DNA binding specificity. For local shape readout, such as minor groove narrowing, recent results suggest that the shape of the minor groove within a binding site can be "read" by a complementary set of basic side chains, most typically arginines, when presented in the correct conformation [65]. In contrast, global shape readout, such as a gradual bend in the DNA helix, may position elements of the DNA backbone such that these otherwise non-specific contacts can become highly specific. Below, we discuss each of these types of readouts, providing specific examples to illustrate them.
V.b.i. Local shape readout
As described in the DNA structure section, the two predominant local shape deviations from ideal B-DNA are 1) small regions of 3–8 base pairs where the minor groove is narrow and 2) DNA kinks, which are caused by the unstacking of a single base pair.
V.b.i.1. Minor groove shape
The N-terminal arms of homeodomain proteins have been observed in the minor grooves of several structures, but only recently have they been shown to play a role in DNA binding specificity. In particular, the binding of the Hox protein Sex combs reduced (Scr) and its co-factor Extradenticle (Exd) to a Scr specific (fkh250) and a Hox consensus (fkh250con) site shows how N-terminal arm arginines use minor groove shape to achieve specificity [29]. Whereas both Arg3 and Arg5 of Scr are ordered in the minor groove of the specific binding site (Figure 5c), Arg3 is disordered when presented with the Hox consensus site (Figure 5d). Arg3 does not form direct base contacts but instead forms a hydrogen bond with His-12, which in turn contacts the bases through a water-mediated hydrogen bond. Mutagenesis studies have shown that Arg3 plays a critical role in Scr in vivo specificity [29].
Figure 5. Hox DNA binding specificity mediated by local shape recognition.
All panels show either the fkh250 binding site or the fkh250con binding site. fkh250, but not fkh250con, has two minor groove minima, which creates a more negative electrostatic potential (minus signs). “W” refers to the Hox YPWM motif that makes a direct contact with the cofactor Exd. See [29] for details.
a. In the absence of Exd, Scr does not bind with high affinity to fkh250 because the arginines on the N-terminal arm and linker of Scr are not positioned correctly.
b. Other Hox proteins do not bind well to fkh250 even in the presence of Exd because their Nterminal arms and linker regions do not have the correct residues.
c. The Scr/Exd heterodimer binds well to fkh250 because the Scr N-terminal arm and linker region have the correct residues, and Exd positions them correctly by binding the YPWM motif (W).
d. Other Hox/Exd heterodimers bind well to fkh250con. This binding site is not as selective because it has a less negative electrostatic potential. Thus, the sequences of the Hox N-terminal arms and linker regions are not as important for binding.
The Scr specific and Hox consensus sites differ in minor groove shape, a structural feature that appears to be intrinsic to these sequences. These local variations in shape result in the enhancement of negative electrostatic potential at distinct positions that attract arginines into the minor groove [20, 29] (Figures 2c and 2d). The Scr N-terminal arm uses these sequence-dependent variations in shape and electrostatic potential to achieve DNA binding specificity [150] (Figures 5c and 5d). Since narrow minor grooves are often associated with AT-rich sequences (Table 2), enhancement of negative electrostatic potential in the minor groove, which in turn is recognized by arginines, offers a general mechanism for sequence-specific recognition of DNA shape [65].
In addition to the results for Scr, mutagenesis studies on the Hox protein Ultrabithorax (Ubx) also suggest a role for linker and N-terminal arm residues in DNA binding specificity, even when Ubx binds as a monomer [151, 152]. Although no crystal structures are yet available to visualize these interactions, an intriguing possibility is that these residues may be reading differences in minor groove shape.
The use of arginines to bind to narrow regions of the minor groove is widespread among DNA binding proteins [65]. However, the manner in which the arginines are presented to the minor groove can differ (Figure 6). In the case of Scr/Exd, heterodimer formation between these two homeodomain proteins is necessary to position Arg3 and His-12, which are normally on an unstructured part of the Hox protein, so that these side chains can insert into the minor groove (Figure 6a). In the case of the POU domain protein Brn-5 binding to its element CRH-II, the arginines that insert into a narrow region of the minor groove come from the linker region that separates the POUHD from the POUS domain [58]. Thus, as with Scr/Exd, two DNA binding domains are required to position the Brn-5 arginines, but in this case both domains are in the same protein (Figure 6b). Not all POU proteins use this method to position the relevant arginines [153]. For example, the Oct-1/PORE complex uses the Arg2 and Arg5 side chains of two Oct-1 monomers to bind to two short A-tracts, ATTT and AAAT [154], and a Pit-1 dimer binds to DNA in a similar fashion as the Oct-1 dimer but uses Arg49 of the POU specific domain to distinguish its ATAC site from the ATGC site of the Oct-1 dimer [153].
Figure 6. Examples of minor groove shape recognition.
Each panel shows a different example in which basic residues bind to minor grooves.
a. Arginine residues present on Scr’s N-terminal arm and linker region requires heterodimerzation with Exd to be positioned correctly to insert into a narrow minor groove region of fkh250. (PDB ID 2r5z)
b. Arginine residues present on the linker region that separates POUHD from POUS of Brn-5 insert into a narrow minor groove of the CRH-II binding site. (PDB ID 3d1n)
c. Arginine residues present on a C-terminal extension of a MogR homodimer insert into narrow regions of the flaA binding site. (PDB ID 3fdq)
d. An N-terminal extension from the γδ resolvase has an arginine that inserts into a narrow minor groove and a second arginine that inserts into the major groove of its binding site. (PDB ID 1gdt)
e. MEF2A recognizes a narrow minor groove of the MEF2A binding site via an arginine and glycine present on an N-terminal strand and via a lysine present on alpha helix α1. (PDB ID 1egw)
f. A histidine residue of IRF-3 inserts into a narrow minor groove region of the IFN-β enhancer. (PDB ID 1t2k)
Proteins from families other than homeodomains also use the mechanism of local minor groove shape readout [65]. MogR, for example, binds as a homodimer in which arginines present on a C-terminal extension from both monomers contact a narrow minor groove composed of two anti-parallel A-tracts that are separated by a TpA ”hinge” step [36] (Figure 6c). The γδ resolvase forms an arginine contact to a narrow minor groove with its N-terminal extension and uses another N-terminal arginine to contact the major groove [155] (Figure 6d).
In all of the above examples, the arginines that insert into the minor groove come from otherwise unstructured strands that must be positioned due to heterodimerization (Scr/Exd), homodimerization (Oct-1, MogR), or via two adjacent DNA binding domains in the same protein (Brn-5). Arginines that insert into minor grooves can also be integral to DNA binding domains. For example, MEF2A, from the myocyte enhancer factor-2 family, uses its α1 helix, which is positioned on top of the minor groove, to contact the MEF2A minor groove [156] (Figure 6e).
Minor groove-interacting arginines are often presented as part of short sequence motifs that include more than one arginine, such as RQR in Scr [29], RPR in Engrailed [26], RKKR in POU homeodomains [157], RGHR in MATα2 [144], and PGR in MogR [36]. The observation that arginine-rich motifs bind to the minor groove was also made for the phage 434 repressor (KRPR) (Figures 2c and 2d) and the Hin recombinase (GRPR), for which arginine mutations were shown to have a dramatic effect on binding affinity [158]. The RQR motif of Scr introduces its arginines like a fork into the minor grove with the glutamine pointing away from the DNA like the fork’s handle [29]. Other arginine-rich motifs orient the arginine side chains differently, allowing them to recognize distinct minor groove shapes.
Unlike homeodomain proteins, that rely on both major and minor groove interactions to achieve specificity, the architectural proteins TBP, SRY, LEF-1, IHF, and HMG-I(Y) only contact the minor groove. For example, the N-terminal arm of IHF inserts two arginines deep into a narrow region of the minor groove complemented by a third arginine that contacts a different narrow region [142]. HMG-I(Y) proteins bind to the AT-rich minor grooves but, in contrast to IHF, stabilize essentially straight instead of deformed DNA [142].
Although arginine is the most abundant residue that inserts into minor grooves, lysines can also be observed in such regions, although at a much lower frequency [65]. The difference between these two basic amino acids is due, at least in part, to the higher free energy associated with removing lysines, which has a less delocalized positive charge distribution, from the aqueous phase [65]. The importance of solvation effects is illustrated by the IFN-β enhanceosome structure, which exhibits a number of lysines in the proximity of the minor groove, clearly solvated rather than intruding into the groove [143, 159]. However, the enhanceosome uses histidines (from IRF-3 and IRF-7) to penetrate narrow minor groove regions formed by A-tracts [143, 159]. His 40 of IRF-1, which is conserved across the IRF family, also inserts into narrow minor groove regions [160, 161] (Figure 6f). Histidine is also observed to insert into the minor groove in the Scr/Exd-fkh250 structure [29].
V.b.i.2. Major groove shape
There are indications that sequence-dependent major groove shape is used in readout mechanisms. Indeed, minor and major groove geometry are correlated with each other [162]. The human regulatory factor hRFX1 is a wHTH protein, which recognizes the DNA major groove with its β–hairpin wing in place of the recognition helix used by other wHTH proteins [42]. In turn, hRFX1 protein places its H3 helix over the minor groove, from which a single lysine contacts the groove [42]. The minor groove widens, resulting in a narrowing of the major groove that in turn improves major groove shape complementarity [38]. In another example, domain 4 of the E. coli extracytoplasmic function σ factor σE specifically recognizes the GGAACTT element based on major groove shape complementarity, which is achieved by narrowing the minor groove [163]. The AT base pairs in the σE binding site (underlined), which are highly conserved despite a lack of strong base contacts, are located in the center of a narrow minor groove [163] and were shown in genetic screening experiments to inhibit transcription when mutated [164].
V.b.i.3. Kinks
As discussed above, DNA kinks occur when the linearity of the helix is abruptly broken, most often due to the unstacking of a flexible base pair step such as TpA (Table 2). Kinks can contribute to binding specificity by promoting conformations that optimize protein-DNA and protein-protein contacts. As an example, the conformational flexibility of the ATA region allows the Tramtrack binding site to adjust to the contacting zinc finger [165]. DNA recognition by endonuclease EcoRV also depends upon the deformability of a TpA step [166]. The binding site of the γδ resolvase comprises a central TATA element and exhibits kinks at both TpA steps, [155]. The flexibility intrinsic to TpA steps also plays a role in the specific binding of the RevErb nuclear hormone receptor as it binds to a site that contains two TpA steps [167]. Although neither of these steps engage in base-specific contacts with RevErb, they show different degrees of deformation, indicating the importance of their flexibility.
The DNA binding site of the catabolite activator protein (CAP) protein shows dramatic kinks at two CpA (TpG) steps [16, 168], which cause, along with two additional smaller kinks, an overall bending of the DNA of about 90° around the protein [169, 170]. The kink at the CpA (TpG) step makes it possible for an arginine residue to engage in partial stacking interactions with the guanine [125]. The HincII endonuclease recognizes its cognate site GTYRAC based on the deformability of its central YpR step and shows the highest affinity when this step is CpG [171]. Similarly, the binding of the EcoRI endonuclease to the Dickerson dodecamer involves a kink at the center of its binding site [172]. Kinking and deformability likely explain the previously reported correlation between base stacking and enzymatic activity [173].
Intercalation
Due to weaker stacking interactions, kinks are often stabilized through the intercalation of protein side chains, that in turn causes further deformation of the DNA helix. The specific DNA binding site of the Lac repressor adjusts to the protein by forming a kink of about 36° at its central CpG step, which widens the minor groove where two leucine residues interact with the kinked base pair step through partial intercalation [136] (Figure 2b). In contrast, a nonspecific DNA sequence, which has been designed to be different in all positions from the Lac operator, does not form a kink upon binding to the Lac repressor, but the protein rearranges its backbone and side chain conformations to engage in phosphate contacts [174]. When the purine repressor is bound to its cognate site GCAAACGTTTGC, a similar kink is observed at its central CpG step (underlined) and stabilized by the partial intercalation of two leucine residues from the minor groove side [31]. Although the conformation of the flanking A-tract regions is very similar in the structure of free and PurR-bound DNA, a kink is not observed in the unbound site [116]. This observation argues that, in this case, it is not DNA structure per se but its deformability that is recognized by PurR.
The yeast TBP structure shows phenylalanine intercalations in the first and last base pair step (underlined) of its TATATAAA binding site [14]. Whereas the first intercalation site is a flexible TpA step, the second site is likely determined by spacing [142, 148]. Architectural proteins that intercalate hydrophobic amino acids between base pairs from the minor groove are the HMG box proteins SRY and LEF-1 [142]. These intercalating hydrophobic residues are conserved in HMG domains, and are usually flanked by basic amino acids [175]. SRY and LEF-1 both use Asn10 to convey specificity through tripartite polar contacts with base pairs preceding the intercalation pocket. Closely related to SRY, DNA bending SOX domains represent another subgroup of HMG boxes [176]. The SOX2/Oct-4/DNA ternary complex is characterized by the intercalation of methionine and phenylalanine residues into an ApA (TpT) step inducing a kink [154]. The SOX17 protein also uses its HMG domain to cause a drastic kink of a ApA (TpT) step through the intercalation of a phenylalanine-methionine dipeptide [177].
V.b.ii. Global shape readout
We include in this category the recognition of DNA sequences where the entire binding site is not in a classic B-form helix. Examples are the recognition of bent DNA, where the curvature is distributed along the entire helix, A-DNA, sequences that have elements of both A- and B-DNA, and Z-DNA.
V.b.ii.1. Bent DNA
The papillomavirus E2 protein provides a clear example of DNA bending playing a role in protein-DNA recognition. The E2 protein binds as a dimer to two half sites separated by a linker of four base pairs [86, 178]. Although only the underlined half sites of the ACCGN4CGGT consensus binding site are contacted by the protein, the variable linker optimizes these contacts through bending, which in turn enhances interactions between the protomers of the E2 dimer [13, 81]. The DNA is similarly bent in complex with the E2 proteins of the bovine papillomavirus BPV-1 [13] and the human papillomavirus HPV-18 [179] (Figure 2b). However, whereas the BPV-1 E2 protein binds with similar binding affinity to consensus sites with various linker sequences, the HPV-18 E2 protein shows a strong preference for AATT linkers [180], and the HPV-16 E2 protein for AATT and AAAA linkers [178]. X-ray crystallographic studies and Monte Carlo predictions stressed that the E2 binding site with AATT linker is also bent when not bound to the protein whereas the site with ACGT linker is essentially straight [81, 86, 120] (Figure 3b). A correlation of the structural data with binding studies suggests that high-affinity sites are pre-bent as seen in the E2-DNA complex whereas low affinity sites require the protein to induce the site to bend [178, 179].
Bending was also suggested to play a role in the specificity of homeodomains by facilitating contacts with the recognition helix [181]. The specific DNA recognition by the phage 434 repressor is associated with bending of its operator [149], which decreases with the number of G-C base pairs in its operator sequence [182]. Long A-tracts are associated with bending and are present, for instance, in the binding sites of the MATa1/MATα2 heterodimer [144] and the NF-κB protein [48]. The conformation of the NF-κB binding site in its bound state is similar to the bending already present in its free state [117, 183]. The RXR-RAR heterodimer recognizes the same half sites as the RXR homodimer. However, the smooth bending of the AAA region between both half sites in place of the kink induced by the RXR homodimer contributes to RXR-RAR specificity [135]. The restriction endonucleases BglII and BamHI recognize DNA sites, AGATCT and GGATCC, with an identical core region (underlined) but bending differentiates both binding sites [184]. In contrast, the similar binding sites of the endonucleases MunI and EcoRI, CAATTG and GAATTC, cannot be distinguished through bending and require an arginine contact to read the outer C:G base pair [184].
V.b.ii.2. A-DNA
Whereas sugars are usually buried in the minor groove of B-DNA, they are much more exposed in A-DNA and provide about 50% of the protein-DNA interface in the TBP–DNA complex, where the DNA is in an A-form conformation [14]. Although arginine and lysine frequently interact with nucleotides in B-DNA conformations, non-polar amino acids such as alanine, leucine, phenylalanine, and valine contact nucleotides in A-DNA conformations [185]. These types of contacts are thus associated with GC-rich sequences [75, 76, 186] and with TATA boxes [187] (Table 2). The higher accessibility of C3’-endo sugars of A-DNA in comparison to buried C2’-endo sugars of B-DNA [185] also contributes to the specificity of zinc finger proteins for GC-rich sequences [116] and of TBP for TATA boxes [77].
The B- to A-transition that transforms the sugar conformations and widens the minor groove is often associated with the intrusion of hydrophobic residues into the minor groove [188]. B- to A-transitions are often observed in complexes with endonucleases since A-DNA makes the phosphate oxygen of the bond that is cleaved more accessible [74]. Other proteins that recognize A/B-intermediate conformations are the Trp repressor and the C. elegans Tc3 transposase [74]. The transcription factor for polymerase IIIA (TFIIIA) also binds to an A-DNA-like binding site [189]. In general, zinc finger proteins tend to bind A/B-intermediates in major grooves that are deep like A-DNA and wide like B-DNA [119] and that have the increased helix diameter typical for A-DNA [190]. Zinc fingers from the human glioblastoma protein (GLI) show the base pair inclination that is distinct for A-DNA [191]. In other complexes, only a limited number of base pairs exhibit A-DNA conformations while the remaining site resembles B-DNA, as seen in two regions of the I-PolI binding site [74].
Interestingly, binding sites of the mouse Cys2His2 zinc finger protein Zif268 crystallize in A-like conformations when both unbound and bound by the protein [116] [118, 119] (Figure 3a). These observations suggest that this DNA sequence has an intrinsic tendency to assume an A-like conformation, and that exposed hydrophobic surfaces of A-like sugars may be generally recognized by zinc fingers [189]. Another example of the recognition of a DNA that has an A/B intermediate structure is the Runt domain and its binding site [49]. In this case, the unbound binding site was observed both in A- [192] and B-DNA [121] conformations. Perhaps related to such observations is that some transcription factors, like TFIIIA, Bicoid, and p53, bind to both DNA and RNA, which almost exclusively exhibits A-form topology [193].
V.b.ii.3. Z-DNA
The zig-zag positioning of phosphates along a left-handed Z-DNA helix is specifically recognized by the double-stranded RNA adenosine deaminase (ADAR1), which is an RNA editing enzyme with a wHTH motif [194]. Z-DNA structures have only been observed to form with purine-pyrimidine alternating sequences that can adopt a left-handed helix [78, 79, 195]. The Zα-domain of ADAR1 has a conformation tailored to recognize a row of five phosphates in one zig-zag shaped backbone of Z-DNA. Since the tumor-associated DLM-1 protein also recognizes Z-DNA via five phosphates along a zig-zag shaped left-handed strand, phosphate positions seem to be the signature code recognized by Z-DNA binding proteins [196] (Figures 1d and 1h).
VI. Examples of higher order protein-DNA interactions
The above discussion highlights examples that illustrate specific readout mechanisms, and thus provides a reductionist perspective on DNA recognition. However, individual DNA binding proteins combine many, if not most, of these readout mechanisms to achieve the correct affinity and specificity required for function. To illustrate this, below we discuss a few examples of protein-DNA recognition in which combinations of readout mechanisms are clearly used.
VI.a. The nucleosome
The presence of nucleosomes in eukaryotic genomes profoundly affects the activity of transcription factors and other DNA binding proteins [197–200]. Although some factors can bind to nucleosomal DNA, others can only bind nucleosome-free DNA. For instance, the packaging of DNA in nucleosomes is expected to narrow the minor groove of TATA boxes, thus precluding TBP binding [148]. In contrast, the bending of nucleosomal DNA was suggested to assist p53 binding at the DNA surface facing away from the histone core [90]. Due to the intimate relationship between protein-DNA recognition and nucleosome binding, attempts to predict nucleosome positions in genomic DNA have received a great deal of attention [201–204]. Because DNA deformability (kinks), DNA bending, and local shape recognition all contribute to nucleosome positioning, these mechanisms need to be considered in any prediction algorithm.
The bendability of short sequences accommodates the wrapping of DNA around the histone core in the nucleosome [148, 149]. The presence of short A-tracts of only three A:T base pairs stabilizes the deformation required for regions of the nucleosomal DNA facing the histones, where the minor groove is compressed [65, 205]. Consequently, the distribution of short A-tracts in yeast in vivo sequences reflects the periodicity of a helical turn in congruence with the structural periodicity caused by the wrapping of nucleosomal DNA around the histone core [65]. In addition, kinks of CpA steps adjacent to short A-tracts can enhance the overall curvature in regions where the minor groove faces the histones [90] while, due to their flexibility, kinks of TpA steps are also used to help wrap the DNA around the histone core. Taking both observations together, the deformability of short A-tracts and YpR steps provides more information about the periodicity than was originally observed for dinucleotides [201, 206–208].
The periodicity of short A-tracts in nucleosomal DNA also results in a periodic narrowing of the minor groove, which is in turn read by arginines present at the histone–DNA interface [65]. Nucleosome-bound DNA contains, on average, 10 of these intrinsically narrow minor groove regions, most of which are likely to be contacted by arginines. Thus, in addition to DNA kinks and bends, nucleosome-DNA interactions also rely on the recognition of local variations in DNA shape [65].
VI.b. E. coli IHF
A combination of kinking, bending, and intercalation is used to achieve DNA binding specificity for the E. coli nucleoid protein IHF, which also functions as a transcriptional activator [209]. The IHF α/β heterodimer sharply bends DNA by about 160° in order to bring distant binding sites of the λ repressor into close proximity [210]. IHF recognizes three DNA sites, TATCAA in the central region of its binding site, and a six base pair A-tract and TTG region at its flanking regions [209]. The large bending is partially induced by the A-tract with its intrinsically narrow minor groove at one side of the IHF-DNA complex [211]. On the other side of the complex, the TpG (CpA) step in the TTG element narrows the minor groove through kinking, which is recognized through the insertion of βArg46 [212]. The TTG to TAG mutation, which shifts the YpR step 5’ by one base pair, indicates that the IHF protein discriminates between A:T and T:A base pairs in this region, based on the flexibility of the YpR step [212]. The α-arm of the protein contacts the minor groove of the central consensus element with three arginine residues. Two large kinks at ApA (TpT) steps caused by proline intercalations are the main contributors to the U-form shape of the IHF-bound DNA [210].
VI.c Cooperativity
DNA binding proteins often bind DNA cooperatively to create higher-order nucleoprotein complexes that reflect the combinatorial control of gene expression. DNA binding cooperativity is most typically attributed to direct protein-protein interactions between adjacent DNA binding factors that significantly promote the assembly of higher order complexes. Notable examples are Hox-Exd/Pbx heterodimers [28, 29, 213], the MATa1-MATα2 heterodimer [144], and the NFAT-Fos-Jun heterotrimer [214]. Whereas cofactors in all previous examples directly bind to DNA, the cofactor CBFβ enhances the binding of the Drosophila Runt domain to DNA without forming any DNA contact [49].
In addition to this classical form of cooperativity, sequence-dependent DNA structure may also promote the cooperative binding of multiple factors. One particularly striking example is the assembly of the IFN-β enhanceosome, which is composed of at least eight DNA binding proteins: a heterodimer of ATF-2/c-Jun, a heterodimer of p50/Rel, and four IRF monomers, all bound to a highly conserved ~55 base pair element [143]. In addition, the architectural protein HMGA1 binds, perhaps transiently, in the minor groove to at least two positions, inducing DNA bends that facilitate the assembly of the enhanceosome [215]. Remarkably, despite the binding of eight transcription factors, a paucity of protein-protein interactions is observed, arguing that cooperativity is likely to be achieved in some other manner [143]. One appealing suggestion is that the final DNA structure, which is optimized for enhanceosome assembly, depends on the intrinsic deformability of the DNA [159]. According to this view, the binding of each factor improves the binding of the other factors through an affect on DNA structure. This idea follows logically from many of the other examples described above where DNA shape and deformability contribute to specificity on a smaller scale. Thus, if correct, sequence-dependent DNA structure may be a critical component in the binding not only of individual factors to their binding sites, but also in the assembly of higher order, multi-protein complexes. This idea fits well with another recent observation that was also pointed out at the beginning of this review, namely, that DNA shape is under evolutionary selection and provides a better indicator of functional elements than conservation of the primary DNA sequence [7].
VII. Summary points
DNA binding proteins use a wide range of mechanisms to bind specifically to binding sites.
The three-dimensional structure of the binding site must be taken into consideration when understanding binding specificity.
The main readout mechanisms are 1) the recognition of bases and 2) the recognition of DNA shape.
The recognition of bases can be further subdivided into those interactions that occur in the major groove, which provides the greatest potential for specificity, and those that occur in the minor groove.
The recognition of DNA shape can be further subdivided into the recognition of local shape variation (e.g. minor groove width) and the recognition of global shape variation (e.g. bent DNA).
Any one DNA binding protein is likely to use a combination of readout mechanisms.
Readout mechanisms are often interrelated (e.g., bending towards the minor groove also narrows it).
The formation of higher-order protein-DNA complexes may depend on sequence-dependent DNA structures that are optimized to promote assembly.
VIII. Future issues
The annotation of genomes must take into account DNA structure.
The rules governing the relationships between DNA sequence and DNA structure need to be better understood.
Understanding intrinsic versus induced effects on DNA structure is an important goal and would benefit from additional structural analyses of free DNAs.
Understanding the rules governing binding specificity within a protein family would benefit from comparisons of structures of multiple family members, each bound to specific and non-specific binding sites.
Acknowledgements
This work was supported by National Institutes of Health (NIH) grants GM54510 (R.S.M.) and U54 CA121852 (B.H. and R.S.M.). The authors thank Z. Shakked and T. Tullius for helpful conversations.
References
- 1.Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- 2.Berger MF, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badis G, et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol Cell. 2008;32:878–887. doi: 10.1016/j.molcel.2008.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu C, et al. High-resolution DNA-binding specificity analysis of yeast transcription factors. Genome Res. 2009;19:556–566. doi: 10.1101/gr.090233.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parker SC, Hansen L, Abaan HO, Tullius TD, Margulies EH. Local DNA topography correlates with functional noncoding regions of the human genome. Science. 2009;324:389–392. doi: 10.1126/science.1169050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greenbaum JA, Pang B, Tullius TD. Construction of a genome-scale structural map at single-nucleotide resolution. Genome Res. 2007;17:947–953. doi: 10.1101/gr.6073107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rosenberg JM, Seeman NC, Kim JJ, Suddath FL, Nicholas HB, Rich A. Double helix at atomic resolution. Nature. 1973;243:150–154. doi: 10.1038/243150a0. [DOI] [PubMed] [Google Scholar]
- 10.Seeman NC, Rosenberg JM, Rich A. Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci U S A. 1976;73:804–808. doi: 10.1073/pnas.73.3.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Viswamitra MA, Kennard O, Jones PG, Sheldrick GM, Salisbury S, Favello L, Shakked Z. DNA double helical fragment at atomic resolution. Nature. 1978;273:687–688. doi: 10.1038/273687a0. [DOI] [PubMed] [Google Scholar]
- 12.Otwinowski Z, Schevitz RW, Zhang RG, Lawson CL, Joachimiak A, Marmorstein RQ, Luisi BF, Sigler PB. Crystal structure of trp repressor/operator complex at atomic resolution. Nature. 1988;335:321–329. doi: 10.1038/335321a0. [DOI] [PubMed] [Google Scholar]
- 13.Hegde RS, Grossman SR, Laimins LA, Sigler PB. Crystal structure at 1.7 A of the bovine papillomavirus-1 E2 DNA-binding domain bound to its DNA target. Nature. 1992;359:505–512. doi: 10.1038/359505a0. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 15.Kim JL, Nikolov DB, Burley SK. Co-crystal structure of TBP recognizing the minor groove of a TATA element. Nature. 1993;365:520–527. doi: 10.1038/365520a0. [DOI] [PubMed] [Google Scholar]
- 16.Lawson CL, Berman HM. Indirect Readout of DNA Sequence by Proteins. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions: Structural Biology. Royal Society of Chemistry; 2008. [Google Scholar]
- 17.Garvie CW, Wolberger C. Recognition of specific DNA sequences. Mol Cell. 2001;8:937–946. doi: 10.1016/s1097-2765(01)00392-6. [DOI] [PubMed] [Google Scholar]
- 18.Luscombe NM, Austin SE, Berman HM, Thornton JM. An overview of the structures of protein-DNA complexes. Genome Biol. 2000;1 doi: 10.1186/gb-2000-1-1-reviews001. REVIEWS001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong M, Marmorstein R. Structural Basis for Sequence-Specific DNA Recognition by Transcription Factors and their Complexes. In: Rice PA, Correll CC, editors. Protein-Nucleic Acid Interactions: Structural Biology. Royal Society of Chemistry; 2008. [Google Scholar]
- 20.Rohs R, West SM, Liu P, Honig B. Nuance in the double-helix and its role in protein-DNA recognition. Curr Opin Struct Biol. 2009;19:171–177. doi: 10.1016/j.sbi.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McKay DB, Steitz TA. Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature. 1981;290:744–749. doi: 10.1038/290744a0. [DOI] [PubMed] [Google Scholar]
- 22.Anderson WF, Ohlendorf DH, Takeda Y, Matthews BW. Structure of the cro repressor from bacteriophage lambda and its interaction with DNA. Nature. 1981;290:754–758. doi: 10.1038/290754a0. [DOI] [PubMed] [Google Scholar]
- 23.Pabo CO, Lewis M. The operator-binding domain of lambda repressor: structure and DNA recognition. Nature. 1982;298:443–447. doi: 10.1038/298443a0. [DOI] [PubMed] [Google Scholar]
- 24.Jordan SR, Pabo CO. Structure of the lambda complex at 2.5 A resolution: details of the repressor-operator interactions. Science. 1988;242:893–899. doi: 10.1126/science.3187530. [DOI] [PubMed] [Google Scholar]
- 25.Badia D, Camacho A, Perez-Lago L, Escandon C, Salas M, Coll M. The structure of phage phi29 transcription regulator p4-DNA complex reveals an N-hook motif for DNA. Mol Cell. 2006;22:73–81. doi: 10.1016/j.molcel.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 26.Kissinger CR, Liu BS, Martin-Blanco E, Kornberg TB, Pabo CO. Crystal structure of an engrailed homeodomain-DNA complex at 2.8 A resolution: a framework for understanding homeodomain-DNA interactions. Cell. 1990;63:579–590. doi: 10.1016/0092-8674(90)90453-l. [DOI] [PubMed] [Google Scholar]
- 27.Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO. Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell. 1991;67:517–528. doi: 10.1016/0092-8674(91)90526-5. [DOI] [PubMed] [Google Scholar]
- 28.Passner JM, Ryoo HD, Shen L, Mann RS, Aggarwal AK. Structure of a DNA-bound Ultrabithorax-Extradenticle homeodomain complex. Nature. 1999;397:714–719. doi: 10.1038/17833. [DOI] [PubMed] [Google Scholar]
- 29.Joshi R, Passner JM, Rohs R, Jain R, Sosinsky A, Crickmore MA, Jacob V, Aggarwal AK, Honig B, Mann RS. Functional specificity of a Hox protein mediated by the recognition of minor groove structure. Cell. 2007;131:530–543. doi: 10.1016/j.cell.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J Mol Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 31.Schumacher MA, Choi KY, Zalkin H, Brennan RG. Crystal structure of LacI member, PurR, bound to DNA: minor groove binding by alpha helices. Science. 1994;266:763–770. doi: 10.1126/science.7973627. [DOI] [PubMed] [Google Scholar]
- 32.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 33.Van Roey P, Waddling CA, Fox KM, Belfort M, Derbyshire V. Intertwined structure of the DNA-binding domain of intron endonuclease I-TevI with its substrate. Embo J. 2001;20:3631–3637. doi: 10.1093/emboj/20.14.3631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Edgell DR, Derbyshire V, Van Roey P, LaBonne S, Stanger MJ, Li Z, Boyd TM, Shub DA, Belfort M. Intron-encoded homing endonuclease I-TevI also functions as a transcriptional autorepressor. Nat Struct Mol Biol. 2004;11:936–944. doi: 10.1038/nsmb823. [DOI] [PubMed] [Google Scholar]
- 35.Shen BW, Landthaler M, Shub DA, Stoddard BL. DNA binding and cleavage by the HNH homing endonuclease I-HmuI. J Mol Biol. 2004;342:43–56. doi: 10.1016/j.jmb.2004.07.032. [DOI] [PubMed] [Google Scholar]
- 36.Shen A, Higgins DE, Panne D. Recognition of AT-rich DNA binding sites by the MogR repressor. Structure. 2009;17:769–777. doi: 10.1016/j.str.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daniels DS, Woo TT, Luu KX, Noll DM, Clarke ND, Pegg AE, Tainer JA. DNA binding and nucleotide flipping by the human DNA repair protein AGT. Nat Struct Mol Biol. 2004;11:714–720. doi: 10.1038/nsmb791. [DOI] [PubMed] [Google Scholar]
- 38.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 39.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 40.Kodandapani R, Pio F, Ni CZ, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- 41.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR-ohrA operator complex reveals the DNA binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 42.Gajiwala KS, Chen H, Cornille F, Roques BP, Reith W, Mach B, Burley SK. Structure of the winged-helix protein hRFX1 reveals a new mode of DNA binding. Nature. 2000;403:916–921. doi: 10.1038/35002634. [DOI] [PubMed] [Google Scholar]
- 43.Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. Embo J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ma PC, Rould MA, Weintraub H, Pabo CO. Crystal structure of MyoD bHLH domain-DNA complex: perspectives on DNA recognition and implications for transcriptional activation. Cell. 1994;77:451–459. doi: 10.1016/0092-8674(94)90159-7. [DOI] [PubMed] [Google Scholar]
- 45.Nair SK, Burley SK. X-ray structures of Myc-Max and Mad-Max recognizing DNA. Molecular bases of regulation by proto-oncogenic transcription factors. Cell. 2003;112:193–205. doi: 10.1016/s0092-8674(02)01284-9. [DOI] [PubMed] [Google Scholar]
- 46.Parraga A, Bellsolell L, Ferre-D'Amare AR, Burley SK. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 1998;6:661–672. doi: 10.1016/s0969-2126(98)00067-7. [DOI] [PubMed] [Google Scholar]
- 47.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 48.Ghosh G, van Duyne G, Ghosh S, Sigler PB. Structure of NF-kappa B p50 homodimer bound to a kappa B site. Nature. 1995;373:303–310. doi: 10.1038/373303a0. [DOI] [PubMed] [Google Scholar]
- 49.Tahirov TH, et al. Structural analyses of DNA recognition by the AML1/Runx-1 Runt domain and its allosteric control by CBFbeta. Cell. 2001;104:755–767. doi: 10.1016/s0092-8674(01)00271-9. [DOI] [PubMed] [Google Scholar]
- 50.Kovall RA, Hendrickson WA. Crystal structure of the nuclear effector of Notch signaling, CSL, bound to DNA. Embo J. 2004;23:3441–3451. doi: 10.1038/sj.emboj.7600349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sidote DJ, Barbieri CM, Wu T, Stock AM. Structure of the Staphylococcus aureus AgrA LytTR domain bound to DNA reveals a beta fold with an unusual mode of binding. Structure. 2008;16:727–735. doi: 10.1016/j.str.2008.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pavletich NP, Pabo CO. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science. 1991;252:809–817. doi: 10.1126/science.2028256. [DOI] [PubMed] [Google Scholar]
- 53.Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat Rev Microbiol. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 54.Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 55.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 56.Pingoud V, Geyer H, Geyer R, Kubareva E, Bujnicki JM, Pingoud A. Identification of base-specific contacts in protein-DNA complexes by photocrosslinking and mass spectrometry: a case study using the restriction endonuclease SsoII. Mol Biosyst. 2005;1:135–141. doi: 10.1039/b503091a. [DOI] [PubMed] [Google Scholar]
- 57.Klemm JD, Rould MA, Aurora R, Herr W, Pabo CO. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 58.Pereira JH, Kim SH. Structure of human Brn-5 transcription factor in complex with CRH gene promoter. J Struct Biol. 2009;167:159–165. doi: 10.1016/j.jsb.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 59.Rhee S, Martin RG, Rosner JL, Davies DR. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc Natl Acad Sci U S A. 1998;95:10413–10418. doi: 10.1073/pnas.95.18.10413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen FE, Huang DB, Chen YQ, Ghosh G. Crystal structure of p50/p65 heterodimer of transcription factor NF-kappaB bound to DNA. Nature. 1998;391:410–413. doi: 10.1038/34956. [DOI] [PubMed] [Google Scholar]
- 61.Kwon T, Chang JH, Kwak E, Lee CW, Joachimiak A, Kim YC, Lee J, Cho Y. Mechanism of histone lysine methyl transfer revealed by the structure of SET7/9-AdoMet. Embo J. 2003;22:292–303. doi: 10.1093/emboj/cdg025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Muller CW. Transcription factors: global and detailed views. Curr Opin Struct Biol. 2001;11:26–32. doi: 10.1016/s0959-440x(00)00163-9. [DOI] [PubMed] [Google Scholar]
- 63.Chang MV, Chang JL, Gangopadhyay A, Shearer A, Cadigan KM. Activation of wingless targets requires bipartite recognition of DNA by TCF. Curr Biol. 2008;18:1877–1881. doi: 10.1016/j.cub.2008.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shakked Z, Rabinovich D. The effect of the base sequence on the fine structure of the DNA double helix. Prog Biophys Mol Biol. 1986;47:159–195. doi: 10.1016/0079-6107(86)90013-1. [DOI] [PubMed] [Google Scholar]
- 65.Rohs R, West SM, Sosinsky A, Liu P, Mann RS, Honig B. The role of DNA shape in protein-DNA recognition. Nature. 2009 doi: 10.1038/nature08473. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Honig B, Nicholls A. Classical electrostatics in biology and chemistry. Science. 1995;268:1144–1149. doi: 10.1126/science.7761829. [DOI] [PubMed] [Google Scholar]
- 67.Klapper I, Hagstrom R, Fine R, Sharp K, Honig B. Focusing of electric fields in the active site of Cu-Zn superoxide dismutase: effects of ionic strength and amino-acid modification. Proteins. 1986;1:47–59. doi: 10.1002/prot.340010109. [DOI] [PubMed] [Google Scholar]
- 68.Sharp KA, Honig B, Harvey SC. Electrical potential of transfer RNAs: codon-anticodon recognition. Biochemistry. 1990;29:340–346. doi: 10.1021/bi00454a006. [DOI] [PubMed] [Google Scholar]
- 69.Chin K, Sharp KA, Honig B, Pyle AM. Calculating the electrostatic properties of RNA provides new insights into molecular interactions and function. Nat Struct Biol. 1999;6:1055–1061. doi: 10.1038/14940. [DOI] [PubMed] [Google Scholar]
- 70.Tang CL, Alexov E, Pyle AM, Honig B. Calculation of pKas in RNA: on the structural origins and functional roles of protonated nucleotides. J Mol Biol. 2007;366:1475–1496. doi: 10.1016/j.jmb.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 71.Leslie AG, Arnott S, Chandrasekaran R, Ratliff RL. Polymorphism of DNA double helices. J Mol Biol. 1980;143:49–72. doi: 10.1016/0022-2836(80)90124-2. [DOI] [PubMed] [Google Scholar]
- 72.Jayaram B, Sharp KA, Honig B. The electrostatic potential of B-DNA. Biopolymers. 1989;28:975–993. doi: 10.1002/bip.360280506. [DOI] [PubMed] [Google Scholar]
- 73.Lavery R, Pullman B. The molecular electrostatic potential and steric accessibility of poly (dI.dC). Comparison with poly (dG.dC) Nucleic Acids Res. 1981;9:7041–7051. doi: 10.1093/nar/9.24.7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu XJ, Shakked Z, Olson WK. A-form conformational motifs in ligand-bound DNA structures. J Mol Biol. 2000;300:819–840. doi: 10.1006/jmbi.2000.3690. [DOI] [PubMed] [Google Scholar]
- 75.Shakked Z, Guerstein-Guzikevich G, Eisenstein M, Frolow F, Rabinovich D. The conformation of the DNA double helix in the crystal is dependent on its environment. Nature. 1989;342:456–460. doi: 10.1038/342456a0. [DOI] [PubMed] [Google Scholar]
- 76.Ng HL, Kopka ML, Dickerson RE. The structure of a stable intermediate in the A<-->B DNA helix transition. Proc Natl Acad Sci U S A. 2000;97:2035–2039. doi: 10.1073/pnas.040571197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Guzikevich-Guerstein G, Shakked Z. A novel form of the DNA double helix imposed on the TATA-box by the TATA-binding protein. Nat Struct Biol. 1996;3:32–37. doi: 10.1038/nsb0196-32. [DOI] [PubMed] [Google Scholar]
- 78.Wang AH, Quigley GJ, Kolpak FJ, Crawford JL, van Boom JH, van der Marel G, Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979;282:680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- 79.Arnott S, Chandrasekaran R, Birdsall DL, Leslie AG, Ratliff RL. Left-handed DNA helices. Nature. 1980;283:743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- 80.Nelson HC, Finch JT, Luisi BF, Klug A. The structure of an oligo(dA).oligo(dT) tract and its biological implications. Nature. 1987;330:221–226. doi: 10.1038/330221a0. [DOI] [PubMed] [Google Scholar]
- 81.Hizver J, Rozenberg H, Frolow F, Rabinovich D, Shakked Z. DNA bending by an adenine--thymine tract and its role in gene regulation. Proc Natl Acad Sci U S A. 2001;98:8490–8495. doi: 10.1073/pnas.151247298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Haran TE, Mohanty U. The unique structure of A-tracts and intrinsic DNA bending. Q Rev Biophys. 2009;42:41–81. doi: 10.1017/S0033583509004752. [DOI] [PubMed] [Google Scholar]
- 83.Zhurkin VB, Tolstorukov MY, Xu F, Colasanti AV, Olson WK. Sequence-dependent variality of B-DNA: an update on bending and curvature. In: Ohyama T, editor. DNA conformation and transcription. Georgetown, Tex.: Landes Bioscience; New York, NY.: Springer Science Business Media; 2005. [Google Scholar]
- 84.Goodsell DS, Kaczor-Grzeskowiak M, Dickerson RE. The crystal structure of C-C-A-T-T-A-A-T-G-G. Implications for bending of B-DNA at T-A steps. J Mol Biol. 1994;239:79–96. doi: 10.1006/jmbi.1994.1352. [DOI] [PubMed] [Google Scholar]
- 85.Crothers DM, Shakked Z. DNA bending by adenine-thymine tracts. In: Neidle S, editor. Oxford Handbook of Nucleic Acid Structures. London: Oxford University Press; 1999. pp. 455–470. [Google Scholar]
- 86.Rohs R, Sklenar H, Shakked Z. Structural and energetic origins of sequence-specific DNA bending: Monte Carlo simulations of papillomavirus E2-DNA binding sites. Structure. 2005;13:1499–1509. doi: 10.1016/j.str.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 87.Gorin AA, Zhurkin VB, Olson WK. B-DNA twisting correlates with base-pair morphology. J Mol Biol. 1995;247:34–48. doi: 10.1006/jmbi.1994.0120. [DOI] [PubMed] [Google Scholar]
- 88.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc Natl Acad Sci U S A. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mack DR, Chiu TK, Dickerson RE. Intrinsic bending and deformability at the T-A step of CCTTTAAAGG: a comparative analysis of T-A and A-T steps within A-tracts. J Mol Biol. 2001;312:1037–1049. doi: 10.1006/jmbi.2001.4994. [DOI] [PubMed] [Google Scholar]
- 90.Tolstorukov MY, Colasanti AV, McCandlish DM, Olson WK, Zhurkin VB. A novel roll-and-slide mechanism of DNA folding in chromatin: implications for nucleosome positioning. J Mol Biol. 2007;371:725–738. doi: 10.1016/j.jmb.2007.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lu XJ, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lavery R, Moakher M, Maddocks JH, Petkeviciute D, Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Janin J, Rodier F, Chakrabarti P, Bahadur RP. Macromolecular recognition in the Protein Data Bank. Acta Crystallogr D Biol Crystallogr. 2007;63:1–8. doi: 10.1107/S090744490603575X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Billeter M, Qian YQ, Otting G, Muller M, Gehring W, Wuthrich K. Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex. J Mol Biol. 1993;234:1084–1093. doi: 10.1006/jmbi.1993.1661. [DOI] [PubMed] [Google Scholar]
- 95.Ades SE, Sauer RT. Specificity of minor-groove and major-groove interactions in a homeodomain-DNA complex. Biochemistry. 1995;34:14601–14608. doi: 10.1021/bi00044a040. [DOI] [PubMed] [Google Scholar]
- 96.Tucker-Kellogg L, Rould MA, Chambers KA, Ades SE, Sauer RT, Pabo CO. Engrailed (Gln50-->Lys) homeodomain-DNA complex at 1.9 A resolution: structural basis for enhanced affinity and altered specificity. Structure. 1997;5:1047–1054. doi: 10.1016/s0969-2126(97)00256-6. [DOI] [PubMed] [Google Scholar]
- 97.Grant RA, Rould MA, Klemm JD, Pabo CO. Exploring the role of glutamine 50 in the homeodomain-DNA interface: crystal structure of engrailed (Gln50 -->ala) complex at 2.0 A. Biochemistry. 2000;39:8187–8192. doi: 10.1021/bi000071a. [DOI] [PubMed] [Google Scholar]
- 98.Hanes SD, Brent R. DNA specificity of the bicoid activator protein is determined by homeodomain recognition helix residue 9. Cell. 1989;57:1275–1283. doi: 10.1016/0092-8674(89)90063-9. [DOI] [PubMed] [Google Scholar]
- 99.Treisman J, Gonczy P, Vashishtha M, Harris E, Desplan C. A single amino acid can determine the DNA binding specificity of homeodomain proteins. Cell. 1989;59:553–562. doi: 10.1016/0092-8674(89)90038-x. [DOI] [PubMed] [Google Scholar]
- 100.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 101.Luscombe NM, Thornton JM. Protein-DNA interactions: amino acid conservation and the effects of mutations on binding specificity. J Mol Biol. 2002;320:991–1009. doi: 10.1016/s0022-2836(02)00571-5. [DOI] [PubMed] [Google Scholar]
- 102.Siggers TW, Silkov A, Honig B. Structural alignment of protein--DNA interfaces: insights into the determinants of binding specificity. J Mol Biol. 2005;345:1027–1045. doi: 10.1016/j.jmb.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 103.Cherney LT, Cherney MM, Garen CR, James MN. The structure of the arginine repressor from Mycobacterium tuberculosis bound with its DNA operator and Co-repressor, L-arginine. J Mol Biol. 2009;388:85–97. doi: 10.1016/j.jmb.2009.02.053. [DOI] [PubMed] [Google Scholar]
- 104.Ellenberger TE, Brandl CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha helices: crystal structure of the protein-DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- 105.Travers A. Transcription: activation by cooperating conformations. Curr Biol. 1998;8:R616–R618. doi: 10.1016/s0960-9822(98)70390-2. [DOI] [PubMed] [Google Scholar]
- 106.Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- 107.Love JJ, Li X, Case DA, Giese K, Grosschedl R, Wright PE. Structural basis for DNA bending by the architectural transcription factor LEF-1. Nature. 1995;376:791–795. doi: 10.1038/376791a0. [DOI] [PubMed] [Google Scholar]
- 108.Laity JH, Dyson HJ, Wright PE. DNA-induced alpha-helix capping in conserved linker sequences is a determinant of binding affinity in Cys(2)-His(2) zinc fingers. J Mol Biol. 2000;295:719–727. doi: 10.1006/jmbi.1999.3406. [DOI] [PubMed] [Google Scholar]
- 109.Holmbeck SM, Dyson HJ, Wright PE. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J Mol Biol. 1998;284:533–539. doi: 10.1006/jmbi.1998.2207. [DOI] [PubMed] [Google Scholar]
- 110.Lefstin JA, Yamamoto KR. Allosteric effects of DNA on transcriptional regulators. Nature. 1998;392:885–888. doi: 10.1038/31860. [DOI] [PubMed] [Google Scholar]
- 111.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–410. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;352:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- 113.McClarin JA, Frederick CA, Wang BC, Greene P, Boyer HW, Grable J, Rosenberg JM. Structure of the DNA-Eco RI endonuclease recognition complex at 3 A resolution. Science. 1986;234:1526–1541. doi: 10.1126/science.3024321. [DOI] [PubMed] [Google Scholar]
- 114.Drew HR, Wing RM, Takano T, Broka C, Tanaka S, Itakura K, Dickerson RE. Structure of a B-DNA dodecamer: conformation and dynamics. Proc Natl Acad Sci U S A. 1981;78:2179–2183. doi: 10.1073/pnas.78.4.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shakked Z, Guzikevich-Guerstein G, Frolow F, Rabinovich D, Joachimiak A, Sigler PB. Determinants of repressor/operator recognition from the structure of the trp operator binding site. Nature. 1994;368:469–473. doi: 10.1038/368469a0. [DOI] [PubMed] [Google Scholar]
- 116.Locasale JW, Napoli AA, Chen S, Berman HM, Lawson CL. Signatures of protein-DNA recognition in free DNA binding sites. J Mol Biol. 2009;386:1054–1065. doi: 10.1016/j.jmb.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Huang DB, Phelps CB, Fusco AJ, Ghosh G. Crystal structure of a free kappaB DNA: insights into DNA recognition by transcription factor NF-kappaB. J Mol Biol. 2005;346:147–160. doi: 10.1016/j.jmb.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 118.Elrod-Erickson M, Rould MA, Nekludova L, Pabo CO. Zif268 protein-DNA complex refined at 1.6 A: a model system for understanding zinc finger-DNA interactions. Structure. 1996;4:1171–1180. doi: 10.1016/s0969-2126(96)00125-6. [DOI] [PubMed] [Google Scholar]
- 119.Elrod-Erickson M, Benson TE, Pabo CO. High-resolution structures of variant Zif268-DNA complexes: implications for understanding zinc finger-DNA recognition. Structure. 1998;6:451–464. doi: 10.1016/s0969-2126(98)00047-1. [DOI] [PubMed] [Google Scholar]
- 120.Rozenberg H, Rabinovich D, Frolow F, Hegde RS, Shakked Z. Structural code for DNA recognition revealed in crystal structures of papillomavirus E2-DNA targets. Proc Natl Acad Sci U S A. 1998;95:15194–15199. doi: 10.1073/pnas.95.26.15194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bartfeld D, Shimon L, Couture GC, Rabinovich D, Frolow F, Levanon D, Groner Y, Shakked Z. DNA recognition by the RUNX1 transcription factor is mediated by an allosteric transition in the RUNT domain and by DNA bending. Structure. 2002;10:1395–1407. doi: 10.1016/s0969-2126(02)00853-5. [DOI] [PubMed] [Google Scholar]
- 122.Kitayner M, Rozenberg H, Kessler N, Rabinovich D, Shaulov L, Haran TE, Shakked Z. Structural basis of DNA recognition by p53 tetramers. Mol Cell. 2006;22:741–753. doi: 10.1016/j.molcel.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 123.Paillard G, Lavery R. Analyzing protein-DNA recognition mechanisms. Structure. 2004;12:113–122. doi: 10.1016/j.str.2003.11.022. [DOI] [PubMed] [Google Scholar]
- 124.Lavery R. Recognizing DNA. Q Rev Biophys. 2005;38:339–344. doi: 10.1017/S0033583505004105. [DOI] [PubMed] [Google Scholar]
- 125.Harrison SC, Aggarwal AK. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 126.Billeter M. Homeodomain-type DNA recognition. Prog Biophys Mol Biol. 1996;66:211–225. doi: 10.1016/s0079-6107(97)00006-0. [DOI] [PubMed] [Google Scholar]
- 127.Konig B, Muller JJ, Lanka E, Heinemann U. Crystal structure of KorA bound to operator DNA: insight into repressor cooperation in RP4 gene regulation. Nucleic Acids Res. 2009;37:1915–1924. doi: 10.1093/nar/gkp044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Tateno M, Yamasaki K, Amano N, Kakinuma J, Koike H, Allen MD, Suzuki M. DNA recognition by beta-sheets. Biopolymers. 1997;44:335–359. doi: 10.1002/(SICI)1097-0282(1997)44:4<335::AID-BIP3>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 129.Coulocheri SA, Pigis DG, Papavassiliou KA, Papavassiliou AG. Hydrogen bonds in protein-DNA complexes: where geometry meets plasticity. Biochimie. 2007;89:1291–1303. doi: 10.1016/j.biochi.2007.07.020. [DOI] [PubMed] [Google Scholar]
- 130.Hoogsteen K. Crystal and Molecular Structure of a Hydrogen-Bonded Complex between 1-Methylthymine and 9-Methyladenine. Acta Crystallographica. 1963;16:907–916. [Google Scholar]
- 131.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Aishima J, Gitti RK, Noah JE, Gan HH, Schlick T, Wolberger C. A Hoogsteen base pair embedded in undistorted B-DNA. Nucleic Acids Res. 2002;30:5244–5252. doi: 10.1093/nar/gkf661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Tainer JA, Cunningham RP. Molecular recognition in DNA-binding proteins and enzymes. Curr Opin Biotechnol. 1993;4:474–483. doi: 10.1016/0958-1669(93)90015-o. [DOI] [PubMed] [Google Scholar]
- 134.Joachimiak A, Haran TE, Sigler PB. Mutagenesis supports water mediated recognition in the trp repressor-operator system. EMBO J. 1994;13:367–372. doi: 10.1002/j.1460-2075.1994.tb06270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rastinejad F, Wagner T, Zhao Q, Khorasanizadeh S. Structure of the RXR-RAR DNA-binding complex on the retinoic acid response element DR1. EMBO J. 2000;19:1045–1054. doi: 10.1093/emboj/19.5.1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Kalodimos CG, Biris N, Bonvin AM, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 137.Aggarwal AK, Rodgers DW, Drottar M, Ptashne M, Harrison SC. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988;242:899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- 138.Wolberger C, Dong YC, Ptashne M, Harrison SC. Structure of a phage 434 Cro/DNA complex. Nature. 1988;335:789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- 139.Watkins D, Hsiao C, Woods KK, Koudelka GB, Williams LD. P22 c2 repressor-operator complex: mechanisms of direct and indirect readout. Biochemistry. 2008;47:2325–2338. doi: 10.1021/bi701826f. [DOI] [PubMed] [Google Scholar]
- 140.Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U. Common mode of DNA binding to cold shock domains. Crystal structure of hexathymidine bound to the domain-swapped form of a major cold shock protein from Bacillus caldolyticus. FEBS J. 2007;274:1265–1279. doi: 10.1111/j.1742-4658.2007.05672.x. [DOI] [PubMed] [Google Scholar]
- 141.Max KE, Zeeb M, Bienert R, Balbach J, Heinemann U. T-rich DNA single strands bind to a preformed site on the bacterial cold shock protein Bs-CspB. J Mol Biol. 2006;360:702–714. doi: 10.1016/j.jmb.2006.05.044. [DOI] [PubMed] [Google Scholar]
- 142.Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: structure, function, and DNA recognition. Annu Rev Biophys Biomol Struct. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Panne D, Maniatis T, Harrison SC. An atomic model of the interferon-beta enhanceosome. Cell. 2007;129:1111–1123. doi: 10.1016/j.cell.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li T, Jin Y, Vershon AK, Wolberger C. Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract. Nucleic Acids Res. 1998;26:5707–5718. doi: 10.1093/nar/26.24.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Crane-Robinson C, Dragan AI, Privalov PL. The extended arms of DNA-binding domains: a tale of tails. Trends Biochem Sci. 2006;31:547–552. doi: 10.1016/j.tibs.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 146.Privalov PL, Dragan AI, Crane-Robinson C, Breslauer KJ, Remeta DP, Minetti CA. What drives proteins into the major or minor grooves of DNA? J Mol Biol. 2007;365:1–9. doi: 10.1016/j.jmb.2006.09.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Privalov PL, Dragan AI, Crane-Robinson C. The cost of DNA bending. Trends Biochem Sci. 2009;34:464–470. doi: 10.1016/j.tibs.2009.05.005. [DOI] [PubMed] [Google Scholar]
- 148.Patikoglou G, Burley SK. Eukaryotic transcription factor-DNA complexes. Annu Rev Biophys Biomol Struct. 1997;26:289–325. doi: 10.1146/annurev.biophys.26.1.289. [DOI] [PubMed] [Google Scholar]
- 149.Travers AA. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- 150.Mann RS, Lelli KM, Joshi R. Hox specificity unique roles for cofactors and collaborators. Curr Top Dev Biol. 2009;88:63–101. doi: 10.1016/S0070-2153(09)88003-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Liu Y, Matthews KS, Bondos SE. Internal regulatory interactions determine DNA binding specificity by a Hox transcription factor. J Mol Biol. 2009;390:760–774. doi: 10.1016/j.jmb.2009.05.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu Y, Matthews KS, Bondos SE. Multiple intrinsically disordered sequences alter DNA binding by the homeodomain of the Drosophila hox protein ultrabithorax. J Biol Chem. 2008;283:20874–20887. doi: 10.1074/jbc.M800375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Phillips K, Luisi B. The virtuoso of versatility: POU proteins that flex to fit. J Mol Biol. 2000;302:1023–1039. doi: 10.1006/jmbi.2000.4107. [DOI] [PubMed] [Google Scholar]
- 154.Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 2003;17:2048–2059. doi: 10.1101/gad.269303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang W, Steitz TA. Crystal structure of the site-specific recombinase gamma delta resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 156.Santelli E, Richmond TJ. Crystal structure of MEF2A core bound to DNA at 1.5 A resolution. J Mol Biol. 2000;297:437–449. doi: 10.1006/jmbi.2000.3568. [DOI] [PubMed] [Google Scholar]
- 157.Remenyi A, Tomilin A, Pohl E, Lins K, Philippsen A, Reinbold R, Scholer HR, Wilmanns M. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol Cell. 2001;8:569–580. doi: 10.1016/s1097-2765(01)00336-7. [DOI] [PubMed] [Google Scholar]
- 158.Churchill ME, Travers AA. Protein motifs that recognize structural features of DNA. Trends Biochem Sci. 1991;16:92–97. doi: 10.1016/0968-0004(91)90040-3. [DOI] [PubMed] [Google Scholar]
- 159.Panne D. The enhanceosome. Curr Opin Struct Biol. 2008;18:236–242. doi: 10.1016/j.sbi.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 160.Escalante CR, Yie J, Thanos D, Aggarwal AK. Structure of IRF-1 with bound DNA reveals determinants of interferon regulation. Nature. 1998;391:103–106. doi: 10.1038/34224. [DOI] [PubMed] [Google Scholar]
- 161.Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO J. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Boutonnet N, Hui X, Zakrzewska K. Looking into the grooves of DNA. Biopolymers. 1993;33:479–490. doi: 10.1002/bip.360330314. [DOI] [PubMed] [Google Scholar]
- 163.Lane WJ, Darst SA. The structural basis for promoter −35 element recognition by the group IV sigma factors. PLoS Biol. 2006;4:e269. doi: 10.1371/journal.pbio.0040269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Miticka H, Rezuchova B, Homerova D, Roberts M, Kormanec J. Identification of nucleotides critical for activity of the sigmaE-dependent rpoEp3 promoter in Salmonella enterica serovar Typhimurium. FEMS Microbiol Lett. 2004;238:227–233. doi: 10.1016/j.femsle.2004.07.039. [DOI] [PubMed] [Google Scholar]
- 165.Fairall L, Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of a two zinc-finger peptide reveals an extension to the rules for zinc-finger/DNA recognition. Nature. 1993;366:483–487. doi: 10.1038/366483a0. [DOI] [PubMed] [Google Scholar]
- 166.Horton NC, Dorner LF, Perona JJ. Sequence selectivity and degeneracy of a restriction endonuclease mediated by DNA intercalation. Nat Struct Biol. 2002;9:42–47. doi: 10.1038/nsb741. [DOI] [PubMed] [Google Scholar]
- 167.Sierk ML, Zhao Q, Rastinejad F. DNA deformability as a recognition feature in the reverb response element. Biochemistry. 2001;40:12833–12843. doi: 10.1021/bi011086r. [DOI] [PubMed] [Google Scholar]
- 168.Lawson CL, Swigon D, Murakami KS, Darst SA, Berman HM, Ebright RH. Catabolite activator protein: DNA binding and transcription activation. Curr Opin Struct Biol. 2004;14:10–20. doi: 10.1016/j.sbi.2004.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schultz SC, Shields GC, Steitz TA. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991;253:1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- 170.Parkinson G, Wilson C, Gunasekera A, Ebright YW, Ebright RE, Berman HM. Structure of the CAP-DNA complex at 2.5 angstroms resolution: a complete picture of the protein-DNA interface. J Mol Biol. 1996;260:395–408. doi: 10.1006/jmbi.1996.0409. [DOI] [PubMed] [Google Scholar]
- 171.Little EJ, Babic AC, Horton NC. Early interrogation and recognition of DNA sequence by indirect readout. Structure. 2008;16:1828–1837. doi: 10.1016/j.str.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Kim YC, Grable JC, Love R, Greene PJ, Rosenberg JM. Refinement of Eco RI endonuclease crystal structure: a revised protein chain tracing. Science. 1990;249:1307–1309. doi: 10.1126/science.2399465. [DOI] [PubMed] [Google Scholar]
- 173.Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol Carcinog. 2009;48:273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 174.Kalodimos CG, Boelens R, Kaptein R. Toward an integrated model of protein-DNA recognition as inferred from NMR studies on the Lac repressor system. Chem Rev. 2004;104:3567–3586. doi: 10.1021/cr0304065. [DOI] [PubMed] [Google Scholar]
- 175.Travers A. Recognition of distorted DNA structures by HMG domains. Curr Opin Struct Biol. 2000;10:102–109. doi: 10.1016/s0959-440x(99)00056-1. [DOI] [PubMed] [Google Scholar]
- 176.Weiss MA. Floppy SOX: mutual induced fit in hmg (high-mobility group) box-DNA recognition. Mol Endocrinol. 2001;15:353–362. doi: 10.1210/mend.15.3.0617. [DOI] [PubMed] [Google Scholar]
- 177.Palasingam P, Jauch R, Ng CK, Kolatkar PR. The structure of Sox17 bound to DNA reveals a conserved bending topology but selective protein interaction platforms. J Mol Biol. 2009;388:619–630. doi: 10.1016/j.jmb.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 178.Hegde RS. The papillomavirus E2 proteins: structure, function, and biology. Annu Rev Biophys Biomol Struct. 2002;31:343–360. doi: 10.1146/annurev.biophys.31.100901.142129. [DOI] [PubMed] [Google Scholar]
- 179.Kim SS, Tam JK, Wang AF, Hegde RS. The structural basis of DNA target discrimination by papillomavirus E2 proteins. J Biol Chem. 2000;275:31245–31254. doi: 10.1074/jbc.M004541200. [DOI] [PubMed] [Google Scholar]
- 180.Hines CS, Meghoo C, Shetty S, Biburger M, Brenowitz M, Hegde RS. DNA structure and flexibility in the sequence-specific binding of papillomavirus E2 proteins. J Mol Biol. 1998;276:809–818. doi: 10.1006/jmbi.1997.1578. [DOI] [PubMed] [Google Scholar]
- 181.Nelson HB, Laughon A. The DNA binding specificity of the Drosophila fushi tarazu protein: a possible role for DNA bending in homeodomain recognition. New Biol. 1990;2:171–178. [PubMed] [Google Scholar]
- 182.Koudelka GB, Carlson P. DNA twisting and the effects of non-contacted bases on affinity of 434 operator for 434 repressor. Nature. 1992;355:89–91. doi: 10.1038/355089a0. [DOI] [PubMed] [Google Scholar]
- 183.Edwards KJ, Brown DG, Spink N, Skelly JV, Neidle S. Molecular structure of the B-DNA dodecamer d(CGCAAATTTGCG)2. An examination of propeller twist and minor-groove water structure at 2.2 A resolution. J Mol Biol. 1992;226:1161–1173. doi: 10.1016/0022-2836(92)91059-x. [DOI] [PubMed] [Google Scholar]
- 184.Lukacs CM, Aggarwal AK. BglII and MunI: what a difference a base makes. Curr Opin Struct Biol. 2001;11:14–18. doi: 10.1016/s0959-440x(00)00174-3. [DOI] [PubMed] [Google Scholar]
- 185.Tolstorukov MY, Jernigan RL, Zhurkin VB. Protein-DNA hydrophobic recognition in the minor groove is facilitated by sugar switching. J Mol Biol. 2004;337:65–76. doi: 10.1016/j.jmb.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 186.Eisenstein M, Shakked Z. Hydration patterns and intermolecular interactions in A-DNA crystal structures. Implications for DNA recognition. J Mol Biol. 1995;248:662–678. doi: 10.1006/jmbi.1995.0250. [DOI] [PubMed] [Google Scholar]
- 187.Shakked Z, Rabinovich D, Kennard O, Cruse WB, Salisbury SA, Viswamitra MA. Sequence-dependent conformation of an A-DNA double helix. The crystal structure of the octamer d(G-G-T-A-T-A-C-C) J Mol Biol. 1983;166:183–201. doi: 10.1016/s0022-2836(83)80005-9. [DOI] [PubMed] [Google Scholar]
- 188.Travers AA. Reading the minor groove. Nat Struct Biol. 1995;2:615–618. doi: 10.1038/nsb0895-615. [DOI] [PubMed] [Google Scholar]
- 189.Choo Y, Klug A. Physical basis of a protein-DNA recognition code. Curr Opin Struct Biol. 1997;7:117–125. doi: 10.1016/s0959-440x(97)80015-2. [DOI] [PubMed] [Google Scholar]
- 190.Nekludova L, Pabo CO. Distinctive DNA conformation with enlarged major groove is found in Zn-finger-DNA and other protein-DNA complexes. Proc Natl Acad Sci U S A. 1994;91:6948–6952. doi: 10.1073/pnas.91.15.6948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Pavletich NP, Pabo CO. Crystal structure of a five-finger GLI-DNA complex: new perspectives on zinc fingers. Science. 1993;261:1701–1707. doi: 10.1126/science.8378770. [DOI] [PubMed] [Google Scholar]
- 192.Kitayner M, Rozenberg H, Rabinovich D, Shakked Z. Structures of the DNA-binding site of Runt-domain transcription regulators. Acta Crystallogr D Biol Crystallogr. 2005;61:236–246. doi: 10.1107/S0907444904032378. [DOI] [PubMed] [Google Scholar]
- 193.Cassiday LA, Maher LJ., 3rd Having it both ways: transcription factors that bind DNA and RNA. Nucleic Acids Res. 2002;30:4118–4126. doi: 10.1093/nar/gkf512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Schwartz T, Rould MA, Lowenhaupt K, Herbert A, Rich A. Crystal structure of the Zalpha domain of the human editing enzyme ADAR1 bound to left-handed Z-DNA. Science. 1999;284:1841–1845. doi: 10.1126/science.284.5421.1841. [DOI] [PubMed] [Google Scholar]
- 195.Herbert A, Rich A. Left-handed Z-DNA: structure and function. Genetica. 1999;106:37–47. doi: 10.1023/a:1003768526018. [DOI] [PubMed] [Google Scholar]
- 196.Schwartz T, Behlke J, Lowenhaupt K, Heinemann U, Rich A. Structure of the DLM-1-Z-DNA complex reveals a conserved family of Z-DNA-binding proteins. Nat Struct Biol. 2001;8:761–765. doi: 10.1038/nsb0901-761. [DOI] [PubMed] [Google Scholar]
- 197.Belotserkovskaya R, Saunders A, Lis JT, Reinberg D. Transcription through chromatin: understanding a complex FACT. Biochim Biophys Acta. 2004;1677:87–99. doi: 10.1016/j.bbaexp.2003.09.017. [DOI] [PubMed] [Google Scholar]
- 198.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 199.Teytelman L, Ozaydin B, Zill O, Lefrancois P, Snyder M, Rine J, Eisen MB. Impact of chromatin structures on DNA processing for genomic analyses. PLoS One. 2009;4:e6700. doi: 10.1371/journal.pone.0006700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Segal E, Widom J. From DNA sequence to transcriptional behaviour: a quantitative approach. Nat Rev Genet. 2009;10:443–456. doi: 10.1038/nrg2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4 doi: 10.1371/journal.pcbi.1000216. e1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kaplan N, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 206.Trifonov EN, Sussman JL. The pitch of chromatin DNA is reflected in its nucleotide sequence. Proc Natl Acad Sci U S A. 1980;77:3816–3820. doi: 10.1073/pnas.77.7.3816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Chung HR, Vingron M. Sequence-dependent nucleosome positioning. J Mol Biol. 2009;386:1411–1422. doi: 10.1016/j.jmb.2008.11.049. [DOI] [PubMed] [Google Scholar]
- 209.Swinger KK, Rice PA. IHF and HU: flexible architects of bent DNA. Curr Opin Struct Biol. 2004;14:28–35. doi: 10.1016/j.sbi.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 210.Ellenberger T, Landy A. A good turn for DNA: the structure of integration host factor bound to DNA. Structure. 1997;5:153–157. doi: 10.1016/s0969-2126(97)00174-3. [DOI] [PubMed] [Google Scholar]
- 211.Rice PA, Yang S, Mizuuchi K, Nash HA. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell. 1996;87:1295–1306. doi: 10.1016/s0092-8674(00)81824-3. [DOI] [PubMed] [Google Scholar]
- 212.Lynch TW, Read EK, Mattis AN, Gardner JF, Rice PA. Integration host factor: putting a twist on protein-DNA recognition. J Mol Biol. 2003;330:493–502. doi: 10.1016/s0022-2836(03)00529-1. [DOI] [PubMed] [Google Scholar]
- 213.Piper DE, Batchelor AH, Chang CP, Cleary ML, Wolberger C. Structure of a HoxB1-Pbx1 heterodimer bound to DNA: role of the hexapeptide and a fourth homeodomain helix in complex formation. Cell. 1999;96:587–597. doi: 10.1016/s0092-8674(00)80662-5. [DOI] [PubMed] [Google Scholar]
- 214.Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- 215.Yie J, Liang S, Merika M, Thanos D. Intra- and intermolecular cooperative binding of high-mobility-group protein I(Y) to the beta-interferon promoter. Mol Cell Biol. 1997;17:3649–3662. doi: 10.1128/mcb.17.7.3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Stefl R, Wu H, Ravindranathan S, Sklenar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc Natl Acad Sci U S A. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Faiger H, Ivanchenko M, Haran TE. Nearest-neighbor non-additivity versus long-range non-additivity in TATA-box structure and its implications for TBP-binding mechanism. Nucleic Acids Res. 2007;35:4409–4419. doi: 10.1093/nar/gkm451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Ellison MJ, Feigon J, Kelleher RJ, 3rd, Wang AH, Habener JF, Rich A. An assessment of the Z-DNA forming potential of alternating dA-dT stretches in supercoiled plasmids. Biochemistry. 1986;25:3648–3655. doi: 10.1021/bi00360a026. [DOI] [PubMed] [Google Scholar]
- 219.Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 220.Lavery R, Sklenar H. Defining the structure of irregular nucleic acids: conventions and principles. J Biomol Struct Dyn. 1989;6:655–667. doi: 10.1080/07391102.1989.10507728. [DOI] [PubMed] [Google Scholar]
- 221.Stofer E, Lavery R. Measuring the geometry of DNA grooves. Biopolymers. 1994;34:337–346. doi: 10.1002/bip.360340305. [DOI] [PubMed] [Google Scholar]
- 222.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J Comput Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]