Abstract
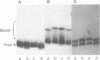
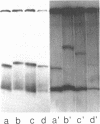
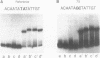
The extent of DNA bending induced by 434 repressor, its amino terminal DNA binding domain (R1-69), and 434 Cro was studied by gel shift assay. The results show that 434 repressor and R1-69 bend DNA to the same extent. 434 Cro-induced DNA bends are similar to those seen with the 434 repressor proteins. On approximately 265 base pair fragments, the cyclic AMP receptor protein of Escherichia coli (CRP) produces larger mobility shifts than does 434 repressor. This indicates that the 434 proteins bend DNA to a much smaller extent than does CRP. The effects of central operator sequence on intrinsic and 434 protein-induced DNA bending was also examined by gel shift assay. Two 434 operators having different central sequences and affinities for 434 proteins display no static bending. The amount of gel shift induced by 434 repressor on these operators is identical, showing that the 434 repressor bends operators with different central sequences to the same extent. Hence, mutations in the central region of the operator do not influence the bent structure of the unbound or bound operator.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aggarwal A. K., Rodgers D. W., Drottar M., Ptashne M., Harrison S. C. Recognition of a DNA operator by the repressor of phage 434: a view at high resolution. Science. 1988 Nov 11;242(4880):899–907. doi: 10.1126/science.3187531. [DOI] [PubMed] [Google Scholar]
- Aiba H., Fujimoto S., Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982 Feb 25;10(4):1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drak J., Crothers D. M. Helical repeat and chirality effects on DNA gel electrophoretic mobility. Proc Natl Acad Sci U S A. 1991 Apr 15;88(8):3074–3078. doi: 10.1073/pnas.88.8.3074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drew H. R., Travers A. A. DNA bending and its relation to nucleosome positioning. J Mol Biol. 1985 Dec 20;186(4):773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- Eilen E., Pampeno C., Krakow J. S. Production and properties of the alpha core derived from the cyclic adenosine monophosphate receptor protein of Escherichia coli. Biochemistry. 1978 Jun 27;17(13):2469–2473. doi: 10.1021/bi00606a001. [DOI] [PubMed] [Google Scholar]
- Fujimoto B. S., Schurr J. M. Dependence of the torsional rigidity of DNA on base composition. Nature. 1990 Mar 8;344(6262):175–177. doi: 10.1038/344175a0. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Hagerman P. J. Flexibility of DNA. Annu Rev Biophys Biophys Chem. 1988;17:265–286. doi: 10.1146/annurev.bb.17.060188.001405. [DOI] [PubMed] [Google Scholar]
- Kotlarz D., Fritsch A., Buc H. Variations of intramolecular ligation rates allow the detection of protein-induced bends in DNA. EMBO J. 1986 Apr;5(4):799–803. doi: 10.1002/j.1460-2075.1986.tb04284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Harbury P., Harrison S. C., Ptashne M. DNA twisting and the affinity of bacteriophage 434 operator for bacteriophage 434 repressor. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4633–4637. doi: 10.1073/pnas.85.13.4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koudelka G. B., Harrison S. C., Ptashne M. Effect of non-contacted bases on the affinity of 434 operator for 434 repressor and Cro. 1987 Apr 30-May 6Nature. 326(6116):886–888. doi: 10.1038/326886a0. [DOI] [PubMed] [Google Scholar]
- Liu-Johnson H. N., Gartenberg M. R., Crothers D. M. The DNA binding domain and bending angle of E. coli CAP protein. Cell. 1986 Dec 26;47(6):995–1005. doi: 10.1016/0092-8674(86)90814-7. [DOI] [PubMed] [Google Scholar]
- Messing J. New M13 vectors for cloning. Methods Enzymol. 1983;101:20–78. doi: 10.1016/0076-6879(83)01005-8. [DOI] [PubMed] [Google Scholar]
- Mondragón A., Harrison S. C. The phage 434 Cro/OR1 complex at 2.5 A resolution. J Mol Biol. 1991 May 20;219(2):321–334. doi: 10.1016/0022-2836(91)90568-q. [DOI] [PubMed] [Google Scholar]
- Porschke D., Hillen W., Takahashi M. The change of DNA structure by specific binding of the cAMP receptor protein from rotation diffusion and dichroism measurements. EMBO J. 1984 Dec 1;3(12):2873–2878. doi: 10.1002/j.1460-2075.1984.tb02223.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satchwell S. C., Drew H. R., Travers A. A. Sequence periodicities in chicken nucleosome core DNA. J Mol Biol. 1986 Oct 20;191(4):659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers A. A. DNA conformation and protein binding. Annu Rev Biochem. 1989;58:427–452. doi: 10.1146/annurev.bi.58.070189.002235. [DOI] [PubMed] [Google Scholar]
- Warwicker J., Engelman B. P., Steitz T. A. Electrostatic calculations and model-building suggest that DNA bound to CAP is sharply bent. Proteins. 1987;2(4):283–289. doi: 10.1002/prot.340020404. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Brown E. L., Ptashne M. Substituting an alpha-helix switches the sequence-specific DNA interactions of a repressor. Cell. 1984 Sep;38(2):361–369. doi: 10.1016/0092-8674(84)90491-4. [DOI] [PubMed] [Google Scholar]
- Wharton R. P., Ptashne M. A new-specificity mutant of 434 repressor that defines an amino acid-base pair contact. 1987 Apr 30-May 6Nature. 326(6116):888–891. doi: 10.1038/326888a0. [DOI] [PubMed] [Google Scholar]
- Wolberger C., Dong Y. C., Ptashne M., Harrison S. C. Structure of a phage 434 Cro/DNA complex. Nature. 1988 Oct 27;335(6193):789–795. doi: 10.1038/335789a0. [DOI] [PubMed] [Google Scholar]
- Wu H. M., Crothers D. M. The locus of sequence-directed and protein-induced DNA bending. Nature. 1984 Apr 5;308(5959):509–513. doi: 10.1038/308509a0. [DOI] [PubMed] [Google Scholar]
- Zinkel S. S., Crothers D. M. DNA bend direction by phase sensitive detection. Nature. 1987 Jul 9;328(6126):178–181. doi: 10.1038/328178a0. [DOI] [PubMed] [Google Scholar]