Abstract
Background
While some recently transmitted HIV clade C (HIV-C) strains exhibited tier 1 neutralization phenotypes, most were tier 2 strains (J Virol 2010; 84:1439). Because induction of neutralizing antibodies (nAbs) through vaccination against tier 2 viruses has proven difficult, we have generated a tier 1, clade C simian–human immunodeficiency virus (SHIV-C) to permit efficacy testing of candidate AIDS vaccines against tier 1 viruses.
Methods
SHIV-1157ipEL was created by swapping env of a late-stage virus with that of a tier 1, early form.
Results
After adaptation to rhesus macaques (RM), passaged SHIV-1157ipEL-p replicated vigorously in vitro and in vivo while maintaining R5 tropism. The virus was reproducibly transmissible intrarectally. Phylogenetically, SHIV-1157ipEL-p Env clustered with HIV-C sequences. All RM chronically infected with SHIV-1157ipEL-p developed high nAb titers against autologous as well as heterologous tier 1 strains.
Conclusions
SHIV-1157ipEL-p was reproducibly transmitted in RM, induced cross-clade nAbs, and represents a tool to evaluate anti-HIV-C nAb responses in primates.
Keywords: clade C SHIV, neutralization sensitive, R5 SHIV, tier 1, vaccine development
Introduction
Human immunodeficiency virus type 1 (HIV-1) is classified into several clades based upon genetic diversity. HIV-1 clade C (HIV-C) is the major subtype and accounts for 56% of all HIV-1/AIDS cases worldwide. It is prevalent in Sub-Saharan Africa, Asia, including India and China, and Brazil (http://www.unaids.org). The epidemiological data suggest a critical need for vaccines to stop the continued spread of HIV-C across the globe.
More than 90% of all HIV-1 transmissions occur mucosally, and almost all of these are initiated by R5 viruses, even when the source persons have mixed infections [16]. Therefore, the availability of a primate model that reflects the prominent biologic features of HIV-1 transmission among humans will enhance our understanding of HIV-1 pathogenesis and facilitate the development of an effective vaccine.
We have generated a series of R5-tropic clade C simian–human immunodeficiency viruses (SHIV-Cs), among them SHIV-1157ipd3N4 [21] and SHIV-2873Nip [19]. The neutralization sensitivity of the latter viruses is typical of primary isolates (tier 2 profiles, indicating only moderate neutralization sensitivity) [18]. Both viruses have caused disease progression to AIDS in several monkeys while maintaining strict R5 tropism [4, 12].
Here, we report the adaptation and viral characteristics of SHIV-1157ipEL-p, a typical tier 1 virus that not only induced high levels of neutralizing antibodies (nAbs) against the autologous virus, but also against other tier 1 strains, including those of different clades.
Materials and methods
Animals
Colony-derived rhesus macaques (RM) of Indian origin were housed at the Yerkes National Primate Research Center (YNPRC, Emory University, Atlanta, GA) according to National Institutes of Health guidelines on the care and use of laboratory animals. YNPRC facilities are fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International. Animal experiments were approved by the Animal Care and Use Committees of Emory University and the Dana-Farber Cancer Institute.
Generation of the infectious molecular clone, SHIV-1157ipEL
SHIV-1157ipEL was constructed using the late-stage virus, SHIV-1157ipd3N4 [21] as the backbone. The env gene of the late virus was exchanged with the early form, which was excised from the R5 SHIV-1157ip [12]. Details of the SHIV-1157ipEL construction and in vitro evaluation are described elsewhere [20].
The initial virus stock of the parental infectious molecular clone, SHIV-1157ipEL, was generated by transfection of 293T cells and harvesting of cell-free supernatant, which was used to infect RM peripheral blood mononuclear cells (PBMC). This initial PBMC-derived stock was used to determine coreceptor usage, ability to replicate in PBMC of randomly selected RM blood donors, and neutralization sensitivity as was described elsewhere [20].
Intravenous and intrarectal challenges
The parental SHIV-1157ipEL stock was inoculated intravenously (i.v.) into RM REk-11. After this animal was confirmed virus positive by reverse-transcriptase polymerase chain reaction (RT-PCR), 10 ml of blood from REk-11 was transferred i.v. to the next recipient RM (RIj-11) at week 2 post-inoculation. Virus was passaged at the time of peak viremia (week 2) according to previously published protocols [12, 19] through a total of four RM. All animals were monitored for viral loads, T-cell subsets, and antibody responses. A stock of the passaged virus, SHIV-1157ipEL-p, was generated by isolating virus from the fourth recipient and expansion in RM PBMC.
To test the mucosal transmissibility of SHIV-1157ipEL-p and to establish a 5× low-dose challenge dose, six animals received repeated weekly low-dose intrarectal (i.r.) inoculations (up to a maximum of five). For intrarectal inoculations, a previously published protocol was used [4]. Monkeys that remained either aviremic or had only transient, low-level viremia (< 104 copies/ml) at the 2-week time point after the fifth low-dose SHIV-1157ipEL-p exposure were given a single high-dose i.r. challenge [approximately nine 50% animal infectious doses (AID50)]. Blood was collected at 0, 1, 2, 4, 8, and 12 weeks post-inoculation and monthly thereafter to determine viral RNA (vRNA) loads and T-cell subsets.
Measurement of plasma vRNA levels
Plasma vRNA was isolated using the QiaAmp Viral RNA Mini-kit (Qiagen, Valencia, CA, USA) and vRNA levels were measured by quantitative RT-PCR for SIV gag sequences [11]. Assay sensitivity was determined to be 50 vRNA copies/ml. We also used primers/probes for SIV gag according to Cline et al. [5].
Sequencing and phylogenetic analysis
Chromosomal DNA was extracted from PBMC of the last RM during the adaptation process using a DNA-zol genomic DNA isolation kit (Molecular Research Center Inc., Cincinnati, OH, USA). Using the following pair of primers, PCR was carried out under endpoint dilution: forward (5′-AGTCTATTATGGGGTACCTGTATGGAAAGAAGCA-3′) and reverse (5′-TCCCAGATAAGTGCCAAGGATCCGTTCACTAATC-3′); the amplified fragment was cloned into the KpnI and BamHI sites of a pcDNA6/myc-His B vector for sequencing. DNA sequencing was performed for five randomly selected clones encoding an env gene. The evolutionary history was deduced by use of the neighbor-joining method [17]. The optimal tree with the sum of branch length = 1.03331016 is depicted. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (500 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the maximum composite likelihood method [23] and are in the units of the number of base substitutions per site. The analysis involved 34 amino acid sequences. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 2063 positions in the final dataset. Phylogenetic analyses were conducted in mega4 [22].
Neutralization assays
Neutralization titers of sera obtained from RM with prolonged chronic SHIV-1157ipEL-p infection were measured using the TZM-bl reporter cell line-based neutralization assay as described previously [13, 14]. TZM-bl cells (also called JC53-bl [6] cells obtained from NIH AIDS Research and Reference Reagent Program (ARRRP) stably express CD4 and CCR5 as well as luciferase and β-galactosidase under the control of the HIV-1 long terminal repeat.
PBMC-based neutralization assays were performed using polymyxin B (15 µg/ml) throughout the assay period to rule out blocking of virus replication via chemokines induced by lipopolysaccharide [7, 25]. Human PBMC were stimulated overnight with phytohemagglutinin (5 µg/ml), followed by the addition of interleukin-2 (IL-2) (20 U/ml) for 48 hours. PBMC were washed twice and added to wells at 5 × 105/well in 96-well plates. Heat-inactivated sera at various dilutions were incubated with a pre-determined 50% tissue culture infectious dose (TCID50) of the virus for 1 hour and added to cells to yield a final volume of 250 µl and left for 3 days. As controls, virus was incubated with medium alone (negative control) or with cells alone (positive control to determine 100% virus production). Starting from day 3 after virus addition, half of the medium was replaced daily with fresh medium (containing IL-2 and polymyxin B) up to day 10. Supernatants were harvested daily, assayed for p27 levels, and the percent neutralization was measured on the culture day showing a linear phase of increase with p27 levels ranging between 8 and 15 ng/ml in the virus + cells control wells.
Results
Construction of SHIV-1157ipEL encoding a tier 1, neutralization-sensitive env
The env gene of the tier 1 neutralization-sensitive SHIV-1157ip was PCR-amplified from DNA isolated from cocultured PBMC of a SHIV-1157ip-infected RM [12]. SHIV-1157ipEL was constructed by inserting the early env gene from SHIV-1157ip into the backbone of the late-stage virus, SHIV-1157ipd3N4 [21]. Cell-free virus for initial characterization was generated by transfecting the molecular clone into 293T cells followed by a brief expansion in RM PBMC and tested for replication kinetics as described in the following.
Replication of SHIV-1157ipEL and SHIV-1157ipEL-p in RM PBMC and coreceptor usage
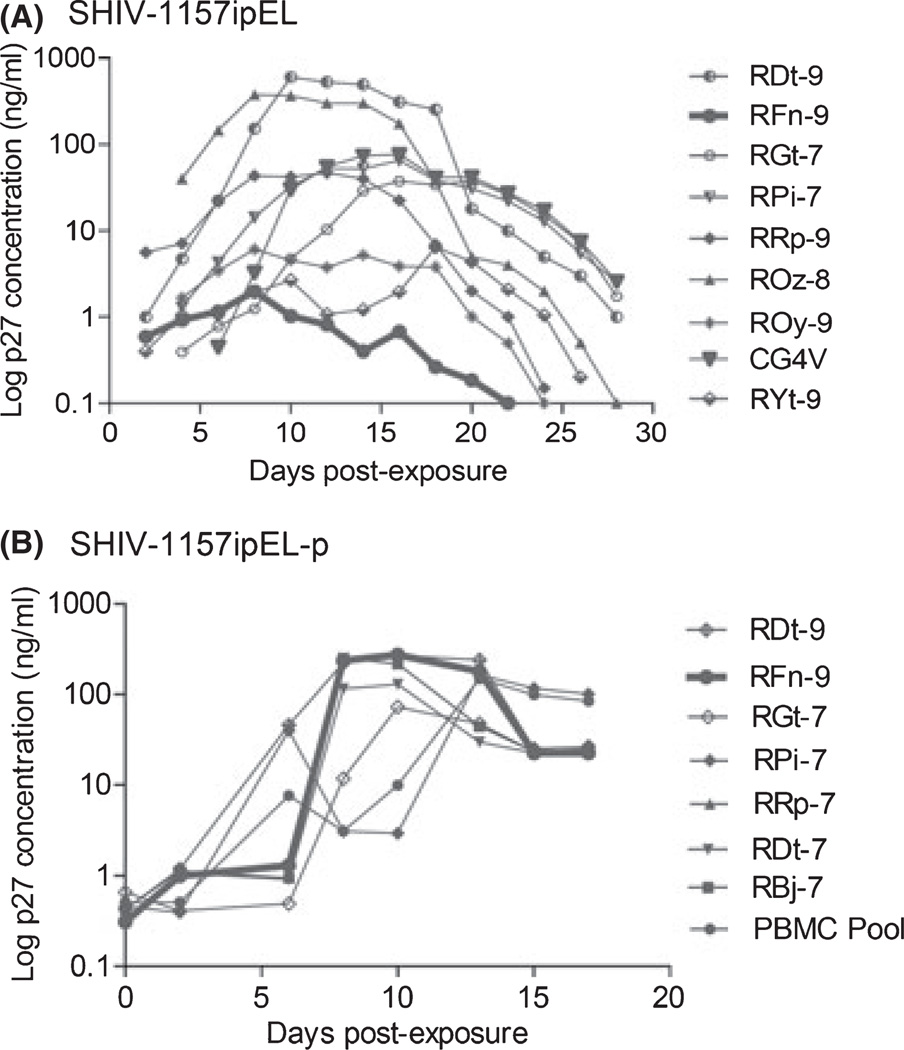
To test the replication competence of the new parental SHIV construct, SHIV-1157ipEL, we randomly selected nine naïve RM blood donors. The new virus was found to replicate in PBMC of only eight of the nine donors tested (Fig. 1A), with peak p27 production detected in the tissue culture supernatants between days 10 and 12. These data implied that the new SHIV strain needed to undergo further adaptation for optimal replication in unselected RM PBMC.
Fig. 1.
Replication of SHIV-1157ipEL and SHIV-1157ipEL-p in rhesus macaque (RM) PBMC. (A) PBMC from nine randomly selected naïve RM donors were stimulated with concanavalin A (ConA) and exposed to SHIV-1157ipEL-containing supernatant (Methods). The PBMC of RM RFn-9 did not support the replication of parental SHIV-1157ipEL (thick line). (B) PBMC from random donors were stimulated with Con A and exposed to passaged virus, SHIV-1157ipEL-p. Supernatants were harvested and p27 levels were measured at the time points indicated. Thick line, PBMC of RM RFn-9 (previously unable to support replication of parental virus, see under A) now yielded high levels of p27 production, indicating viral adaptation to the new host species. SHIV, simian–human immunodeficiency virus; PBMC, peripheral blood mononuclear cells.
After rapid passage through 4 RM, we re-isolated the adapted virus, termed SHIV-1157ipEL-p. Next, we retested its ability to replicate in PBMC of those RM, which previously had failed to replicate SHIV-1157ipEL, as well as in PBMC of other randomly selected donors. The passaged virus clearly replicated vigorously in all PBMC cultures tested (Fig. 1B), indicating its adaptation to the new host species.
We also assessed the coreceptor usage of SHIV-1157ipEL and SHIV-1157ipEL-p. As described previously [20], we observed productive infection only in U87.CD4.CCR5 cells and none in any cell line lacking CCR5, including CEMx174-GFP, U87.CD4, U87.CD4.CCR1, U87.CD4.CCR2, U87.CD4.CCR3, U87.CD4.CXCR4, GHOST-BOB, and GHOST-BONZO cells, suggesting that both viruses exclusively use CCR5 as a coreceptor for entry.
Neutralization tiers of SHIV-1157ipEL and SHIV-1157ipEL-p
Next, we tested the neutralization tiers of the parental infectious molecular clone and the passaged progeny virus, a biologic isolate, using the TZM-bl assay (Table 1). The rapid passage through RM at peak viremia had not appreciably altered the high neutralization sensitivity of the passaged virus. Thus, our adaptation strategy had avoided selection of a neutralization escape virus. In contrast, the late form, SHIV-1157ipd3N4, was considerably less sensitive to neutralization and exhibited a typical tier 2 profile.
Table 1.
Sensitivity of SHIV-C strains to soluble CD4, human nmAbs, and serum samples
| IC50 (µg/ml) in TZM-bl cells1 | IC50 (reciprocal serum dilution) in TZM-bl cells1 | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| SHIV-C strain | sCD4 | IgG1b12 | 2G12 | 2F5 | 4E10 | HIVIG | BB47 | BB55 | BB68 | BB75 | BB80 | BB81 | BB87 | Tier |
| SHIV-1157ipEL (parental infectious mol. clone with early env) | 1.7 | 0.7 | >25 | >25 | >25 | 88.6 | 4274 | 3286 | 666 | 1245 | 1822 | 691 | 3278 | 1 |
| SHIV-1157ipEL-p (passaged biologic isolate with early env) | 0.5 | 1.4 | >25 | >25 | >25 | 150.8 | 1193 | 1792 | 653 | 824 | 1135 | 427 | 2707 | 1 |
| SHIV-1157ipd3N42 (infectious mol. clone with late env) | 0.4 | 7.0 | >25 | >25 | >25 | 1160 | 105 | 131 | 86 | 79 | 72 | 47 | 260 | 2 |
SHIV, simian–human immunodeficiency virus.
Spectrum of neutralization sensitivity of R5 SHIV strains encoding HIV clade B or C env.
Values represent the concentration (µg/ml for soluble CD4 (sCD4) and human nmAbs IgG1b12, 2G12, 2F5, 4E10, or HIVIG) or the dilution (for serum samples) at which relative luciferase units (RLU) were reduced 50% compared to virus control wells. BB47, BB55, BB68, BB75, BB80, BB81, and BB87 are serum samples from individuals infected with HIV-1 clade C. HIVIG, polyclonal high-titer anti-HIV Ig preparation.
Data previously published and shown here as reference [19]; given as basis of comparison.
Phylogenetic analysis
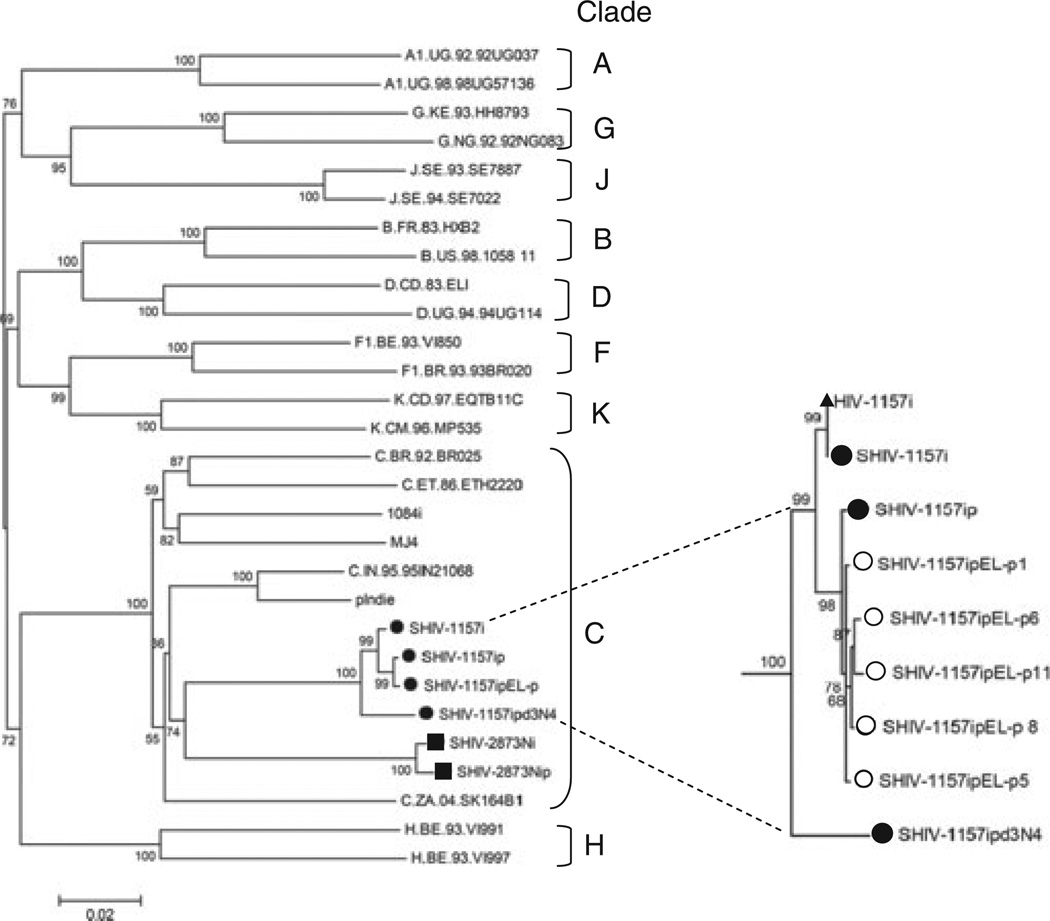
All envelopes of the 1157i series of SHIVs clustered with HIV-C (Fig. 2). The close proximity of the SHIV-1157i, SHIV-1157ip, and SHIV-1157ipEL-p as well as SHIV-1157ipd3N4 env genes on the phylogenetic tree reflects the same ancestral origin for this series of SHIVs (Fig. 2). All the newly generated clones of SHIV-1157ipEL-p were clustered together with minimal diversity (Fig. 2). Interestingly, the tier 2 envelope of the late virus, SHIV-1157ipd3N4, had a greater distance from the closely clustered tier 1 envelopes of SHIV-1157i, SHIV-1157ip, and SHIV-1157ipEL-p. We also found that the HIV-C envelopes contained in our two different sets of R5 SHIV-Cs (the 1157i and 2873i series) were more closely related to each other than to the other HIV-C Env sequences. This is probably due to the fact that the pediatric HIV-C strains (HIV1157i and HIV2873i) used to isolate the env genes for SHIV construction were derived from the same cohort of HIV-C-infected infants in Lusaka, Zambia. The divergence of two pediatric HIV-C envelopes thus reflects envelope sequence divergence in a local community.
Fig. 2.
Phylogenetic tree construction. Phylogenetic tree showing the relationship between SHIV-1157i, SHIV-1157ip, SHIV-1157ipEL-p, and SHIV-1157ipd3N4 Env sequences and those of other primary strains of HIV. Phylogenetic trees were constructed from full-length Env sequences by using MEGA4. Major clades of HIV group M were used as reference sequences; Env sequences from SHIV-2873Nip and HIV1084i were also included, as SHIV-2873Nip and HIV1084i env were genes derived from the same cohort of HIV-C-infected infants in Lusaka, Zambia. The scale bar indicates the genetic distance along the horizontal branches, and the numbers at the nodes are bootstrap values. The insert represents the phylogenetic relationship between HIV-1157i and the SHIV series related to the env gene of the latter. The open circles denote five individual SHIV-1157ipEL-p sequences obtained through end-point PCR cloning that were used to arrive at the consensus sequence of SHIV-1157ipEL-p. Closed circles represent the consensus sequence of each of the viruses as noted in the figure. The closed triangle represents the parental sequence of HIV-1157i that was used to generate the initial SHIV clone. SHIV, simian–human immunodeficiency virus; HIV-C, HIV clade C.
SHIV-1157ipEL-p inoculations in RM
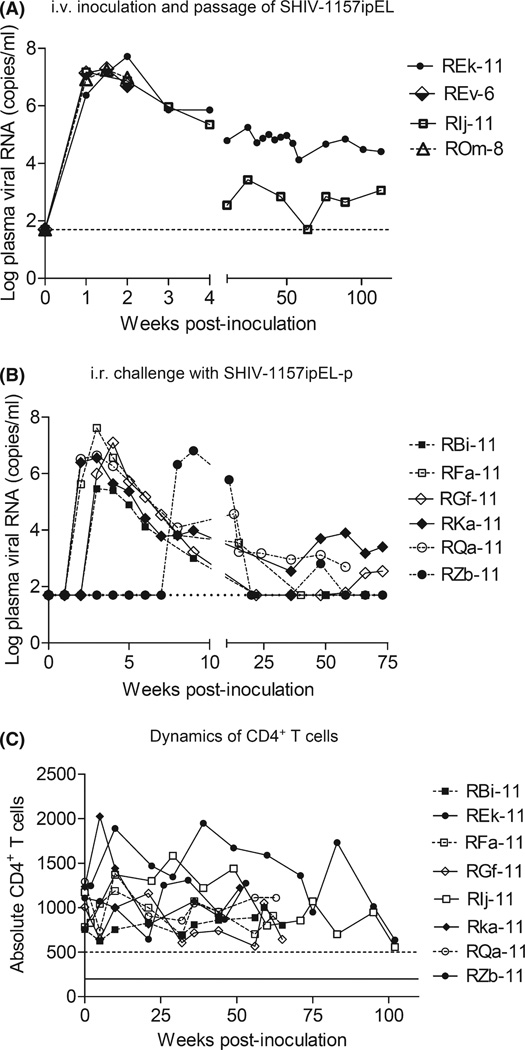
To demonstrate reliable mucosal transmissibility of SHIV-1157ipEL-p, we summarized the data from a total of 31 RM inoculated by the i.r. route (Table 2). All RM were of Indian origin. Twenty juvenile/adult RM received weekly low-dose challenges (up to a maximum of five inoculations) with virus doses ranging between 5000 and 10,000 TCID50. Another five juvenile/adult RM as well as six infants were given a single high-dose i.r. challenge. SHIV-1157ipEL-p replicated vigorously in all i.r. challenged RM with high peak viremia levels ranging from 2 × 105 to 4.1 × 107 RNA copies/ml (Fig. 3B; Table 2). We have also listed the peak vRNA levels of the four RM used in the initial adaptation of the parental SHIV-1157ipEL (Fig. 3A; Table 2). These animals were inoculated i.v. with infected blood. Chronically infected RM are currently being followed prospectively for signs of disease progression.
Table 2.
SHIV-1157ipEL-p route of challenge and mean peak viral RNA (vRNA) loads
| Age | No. of macaques | Route of exposure | Dose | Mean peak vRNA (copies/ml) × 106 |
|---|---|---|---|---|
| Juvenile/adult | 4 | i.v. (infected blood in 3 out of 4 RM) | High | 26.2 ± 17.5 |
| 6 | i.r. | High | 3.0 ± 2.1 | |
| 19 | i.r. | 5× low | 5.4 ± 9.5 | |
| Infants | 6 | i.r. | High | 6.8 ± 3.9 |
SHIV, simian–human immunodeficiency virus.
Fig. 3.
(A) Intravenous inoculation of SHIV-1157ipEL: monkey REk-11 was inoculated i.v. with 10 ml of PBMC-grown virus. This was followed by rapid passage at peak viremia (week 2 post-inoculation) through three additional RM using serial blood transfer (10 ml each). Viral RNA (vRNA) levels were measured at time points indicated. (B) Intrarectal inoculation of SHIV-1157ipEL-p in six monkeys, using limited weekly low-dose inoculations (maximally five). RM RZb-11 remained uninfected at week 2 after the fifth exposure and was given a single high-dose of SHIV-1157ipEL-p. vRNA levels were measured at the time points indicated. The horizontal dotted line indicates the lower limit of detection (<50 vRNA copies/ml). (C) Absolute CD4+ T-cell counts of RM infected with SHIV-1157ipEL-p. SHIV, simian–human immunodeficiency virus; RM, rhesus macaques; PBMC, peripheral blood mononuclear cells.
Depletion of CD4 T cells during acute viremia
SHIV-1157ipEL-p-infected monkeys (n = 8) were followed for the natural course of infection and disease. Approximately 60–110 weeks post-inoculation, these monkeys have maintained absolute CD4+ T cells in peripheral blood of >500 cells/µl (Fig. 3C). However, we observed persistently low (<10%) peripheral blood CD4+ memory T cells (assessed by CD4+CD29+ double staining) in all infected animals (data not shown). We also assessed the acute pathogenicity of SHIV-1157ipEL-p by evaluating the absolute cell counts or percentage of CD4+ T cells in blood as well as in rectal and lymph node lymphocyte populations (2–12 weeks post-inoculation). As shown previously [20], statistically significant depletion of CD4+ T cells was observed in blood, lymph nodes, and the gut in all infected RM when compared to uninfected RM.
Cross-clade nAbs induced during chronic SHIV-1157ipEL-p infection
Plasma or serum samples from SHIV-1157ipEL-p-infected animals with long-term infection (10 months to 2 years) were tested for their ability to neutralize autologous or heterologous SHIVs in TZM-bl and PBMC-based neutralization assays. All animals tested developed nAbs against the autologous virus as well as against heterologous tier 1 viruses (Table 3); in some cases, titers were very high. In contrast, the tier 2 virus SHIV-2873Nip was not neutralized. PBMC-based assays yielded similar results, although generally higher titers were observed (data not shown).
Table 3.
Neutralization titers of sera from SHIV-1157ipEL-p-challenged rhesus macaques
| Titers (plasma dilution at 50% neutralization in TZM-bl cells) | |||||
|---|---|---|---|---|---|
| Homologous clade C (tier 1) | Heterologous clade C | Heterologous clade B | |||
| Animal no. | SHIV-1157ipEL-p | SHIV-2873Nip (tier 2) | MW965.5 (tier 1) | SHIV-Bo159N41 | SHIVSF162P4 |
| RBi-11 | 128 | <20 | 1498 | 49 | 83 |
| REk-11 | 3690 | <20 | >43,740 | 9640 | 7615 |
| RFa-11 | 1038 | <20 | 8019 | 160 | 271 |
| RGf-11 | 695 | <20 | 4753 | 55 | 335 |
| RIj-11 | 1985 | <20 | 24,586 | 220 | 348 |
| RKa-11 | 779 | <20 | 8415 | 707 | 353 |
| RQa-11 | 403 | <20 | 3589 | 71 | 198 |
| RZb-11 | 243 | <20 | 1571 | 58 | 92 |
SHIV, simian–human immunodeficiency virus.
This tier 1 R5 SHIV strain is of clade B origin and encodes extra NF-κB sites in the long terminal repeats (Siddappa et al., unpublished data).
Discussion
We have described some of the biologic characteristics of the newly generated R5 SHIV-1157ipEL-p: (1) it carries env of an early, relatively recently transmitted pediatric HIV-C from Zambia; (2) it was highly replication competent in PBMC cultures of non-selected RM and induced high peak vRNA loads in all animals tested; (3) its envelope was shown to cluster with HIV-C Envs; and (4) it is highly sensitive to neutralization and exhibits a tier 1 profile.
SHIV-1157ipEL-p carries the tier 1 envelope isolated from a 6-month old Zambian infant [12]. Although several other SHIV strains encoding HIV-C envelopes have been created, they were unable to replicate in RM PBMC (SHIVCHN19) [3], could not be re-isolated after adaptation (SHIVMJ4 and SHIV-XJ02170) [15, 26], had dual tropism (SHIV-MCGP1.3) [2], or are relatively difficult to neutralize with tier 2 profiles (SHIV-1157ipd3N4 and SHIV-2873Nip) [19, 21]. In contrast, the newly created R5 SHIV-1157ipEL-p is highly sensitive to neutralization with a tier 1 profile. It not only replicated well in PBMC of all RM donors tested, but also showed robust replication in vivo with high peak vRNA loads after mucosal challenge.
After adaptation, genetic analysis of the SHIV-1157ipEL-p envelope showed only a few point mutations when compared to that of the parental chimera SHIV-1157ipEL, indicating that our rapid, every 2-week passage strategy resulted in only minor amino acid changes. Not surprisingly, the tier 1 neutralization profile was maintained after adaptation.
Based initially on the env gene of the recently transmitted pediatric HIV1157i, we have created a matched pair of tier 1/tier 2 R5 SHIV-Cs, namely SHIV-1157ipEL-p and SHIV-1157ipd3N4. The tier 2 envelope in our late SHIV-1157ipd3N4 strain evolved from the early, highly neutralization-sensitive form. Another pair of tier 1/tier 2 R5 SHIVs has been described in the literature, namely SHIVSF162P4/SHIVSF162P3 [1, 8, 24]. The envelopes of these clade B SHIVs are based upon HIVSF162. Surprisingly, the tier 1 SHIVSF162P4 arose in a macaque from a later passage compared to the tier 2 SHIVSF162P3 [1]. Interestingly, two different X4 viruses emerged from SHIVSF162P3N-infected animals [9, 10], and data obtained from these monkeys will provide important clues for factors associated with in vivo coreceptor switch. We have not yet observed any coreceptor switch in our SHIV-C-infected macaques – regardless of viral neutralization tier. We plan to follow monkeys with chronic SHIV-C viremia prospectively for evidence of expanded coreceptor usage.
To date, SHIV-1157ipEL-p has not yet induced AIDS given that follow-up times have been relatively short. However, this new tier 1 virus was constructed using components derived from pathogenic viruses: the backbone originated from SHIV-1157ipd3N4 [21] and the env gene from SHIV-1157ip [12]. Both of these viruses caused AIDS in RM independently [4, 12], and we therefore expect a similar tendency in the case of SHIV-1157ipEL-p. Prolonged, prospective follow-up will reveal the pathogenic potential of SHIV-1157ipEL-p.
In summary, we showed the adaptation and replication kinetics of the tier 1 virus, SHIV-1157ipEL-p, in vitro and in vivo. Of note, sera from all animals infected for extended time periods with this virus contained cross-clade nAbs. We suggest that this new virus represents a biologically relevant tool to evaluate anti-HIV-C nAb responses in primates and Env evolution under the pressure of autologous nAb responses. Given the robust replication in Indian RM, this tier 1 virus and/or SHIV-1157ipd3N4 can be used to test the efficacy of nAb-response-based vaccine candidates. Likewise, the new SHIV-1157ipEL-p or its tier 2 counterpart – depending on in vitro neutralization results – can be used to assess the ability of novel human neutralization monoclonal antibodies to provide protection via passive immunization.
Acknowledgments
We thank Elizabeth Samit for assistance in the preparation of this manuscript and Stephanie Ehnert, Chris Souder and Kalpana Patel for coordinating sample collections. We thank Mahesh Bachu for help with phylogenetic analysis. This work was supported by National Institutes of Health grants P01 AI048240, R01 DE016013, and R37 AI034266 to R.M.R. Base grant RR-00165 provided support to the Yerkes National Primate Research Center.
References
- 1.Balfe P, Shapiro S, Hsu M, Buckner C, Harouse JM, Cheng-Mayer C. Expansion of quasispecies diversity but no evidence for adaptive evolution of SHIV during rapid serial transfers among seronegative macaques. Virology. 2004;318:267–279. doi: 10.1016/j.virol.2003.09.022. [DOI] [PubMed] [Google Scholar]
- 2.Cayabyab M, Rohne D, Pollakis G, Mische C, Messele T, Abebe A, Etemad-Moghadam B, Yang P, Henson S, Axthelm M, Goudsmit J, Letvin NL, Sodroski J. Rapid CD4+ T-lymphocyte depletion in rhesus monkeys infected with a simian-human immunodeficiency virus expressing the envelope glycoproteins of a primary dual-tropic Ethiopian Clade C HIV type 1 isolate. AIDS Res Hum Retroviruses. 2004;20:27–40. doi: 10.1089/088922204322749477. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z, Huang Y, Zhao X, Skulsky E, Lin D, Ip J, Gettie A, Ho DD. Enhanced infectivity of an R5-tropic simian/human immunodeficiency virus carrying human immunodeficiency virus type 1 subtype C envelope after serial passages in pig-tailed macaques (Macaca nemestrina) J Virol. 2000;74:6501–6510. doi: 10.1128/jvi.74.14.6501-6510.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chenine AL, Siddappa NB, Kramer VG, Sciaranghella G, Rasmussen RA, Lee SJ, Santosuosso M, Poznansky MC, Velu V, Amara RR, Souder C, Anderson DC, Villinger F, Else JG, Novembre FJ, Strobert E, O’Neil SP, Secor WE, Ruprecht RM. Relative transmissibility of an R5 clade C simian-human immunodeficiency virus across different mucosae in macaques parallels the relative risks of sexual HIV-1 transmission in humans via different routes. J Infect Dis. 2010;201:1155–1163. doi: 10.1086/651274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cline AN, Bess JW, Piatak M, Jr, Lifson JD. Highly sensitive SIV plasma viral load assay: practical considerations, realistic performance expectations, and application to reverse engineering of vaccines for AIDS. J Med Primatol. 2005;34:303–312. doi: 10.1111/j.1600-0684.2005.00128.x. [DOI] [PubMed] [Google Scholar]
- 6.Derdeyn CA, Decker JM, Sfakianos JN, Wu X, O’Brien WA, Ratner L, Kappes JC, Shaw GM, Hunter E. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J Virol. 2000;74:8358–8367. doi: 10.1128/jvi.74.18.8358-8367.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geonnotti AR, Bilska M, Yuan X, Ochsenbauer C, Edmonds TG, Kappes JC, Liao HX, Haynes BF, Montefiori DC. Differential inhibition of human immunodeficiency virus type 1 in peripheral blood mononuclear cells and TZM-bl cells by endotoxin-mediated chemokine and gamma interferon production. AIDS Res Hum Retroviruses. 2010;26:279–291. doi: 10.1089/aid.2009.0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harouse JM, Gettie A, Tan RC, Blanchard J, Cheng-Mayer C. Distinct Distinct pathogenic sequela in rhesus macaques infected with CCR5 or CXCR4 utilizing SHIVs. Science. 1999;284:816–819. doi: 10.1126/science.284.5415.816. [DOI] [PubMed] [Google Scholar]
- 9.Ho SH, Tasca S, Shek L, Li A, Gettie A, Blanchard J, Boden D, Cheng-Mayer C. Coreceptor switch in R5-tropic simian/human immunodeficiency virus-infected macaques. J Virol. 2007;81:8621–8633. doi: 10.1128/JVI.00759-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho SH, Trunova N, Gettie A, Blanchard J, Cheng-Mayer C. Different mutational pathways to CXCR4 coreceptor switch of CCR5-using simian-human immunodeficiency virus. J Virol. 2008;82:5653–5656. doi: 10.1128/JVI.00145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann-Lehmann R, Swenerton RK, Liska V, Leutenegger CM, Lutz H, McClure HM, Ruprecht RM. Sensitive and robust one-tube real-time reverse transcriptase-polymerase chain reaction to quantify SIV RNA load: comparison of one-versus two-enzyme systems. AIDS Res Hum Retroviruses. 2000;16:1247–1257. doi: 10.1089/08892220050117014. [DOI] [PubMed] [Google Scholar]
- 12.Humbert M, Rasmussen RA, Song R, Ong H, Sharma P, Chenine AL, Kramer VG, Siddappa NB, Xu W, Else JG, Novembre FJ, Strobert E, O’Neil SP, Ruprecht RM. SHIV-1157i and passaged progeny viruses encoding R5 HIV-1 clade C env cause AIDS in rhesus monkeys. Retrovirology. 2008;5:94. doi: 10.1186/1742-4690-5-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li M, Salazar-Gonzalez JF, Derdeyn CA, Morris L, Williamson C, Robinson JE, Decker JM, Li Y, Salazar MG, Polonis VR, Mlisana K, Karim SA, Hong K, Greene KM, Bilska M, Zhou J, Allen S, Chomba E, Mulenga J, Vwalika C, Gao F, Zhang M, Korber BT, Hunter E, Hahn BH, Montefiori DC. Genetic and neutralization properties of subtype C human immunodeficiency virus type 1 molecular env clones from acute and early heterosexually acquired infections in Southern Africa. J Virol. 2006;80:11776–11790. doi: 10.1128/JVI.01730-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Montefiori DC. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol. 2005;Chapter 12(Unit 12):11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 15.Ndung’u T, Lu Y, Renjifo B, Touzjian N, Kushner N, Pena-Cruz V, Novitsky VA, Lee TH, Essex M. Infectious simian/human immunodeficiency virus with human immunodeficiency virus type 1 subtype C from an African isolate: rhesus macaque model. J Virol. 2001;75:11417–11425. doi: 10.1128/JVI.75.23.11417-11425.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pope M, Haase AT. Transmission, acute HIV-1 infection and the quest for strategies to prevent infection. Nat Med. 2003;9:847–852. doi: 10.1038/nm0703-847. [DOI] [PubMed] [Google Scholar]
- 17.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 18.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol. 2010;84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Siddappa NB, Song R, Kramer VG, Chenine AL, Velu V, Ong H, Rasmussen RA, Grisson RD, Wood C, Zhang H, Kankasa C, Amara RR, Else JG, Novembre FJ, Montefiori DC, Ruprecht RM. Neutralization-sensitive R5-tropic simian-human immunodeficiency virus SHIV-2873Nip, which carries env isolated from an infant with a recent HIV clade C infection. J Virol. 2009;83:1422–1432. doi: 10.1128/JVI.02066-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Siddappa NB, Watkins JD, Wassermann KJ, Song R, Wang W, Kramer VG, Lakhashe S, Santosuosso M, Poznansky MC, Novembre FJ, Villinger F, Else JG, Montefiori DC, Rasmussen RA, Ruprecht RM. R5 clade C SHIV strains with tier 1 or 2 neutralization sensitivity: tools to dissect env evolution and to develop AIDS vaccines in primate models. PLoS ONE. 2010;5:e11689. doi: 10.1371/journal.pone.0011689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song RJ, Chenine AL, Rasmussen RA, Ruprecht CR, Mirshahidi S, Grisson RD, Xu W, Whitney JB, Goins LM, Ong H, Li PL, Shai-Kobiler E, Wang T, McCann CM, Zhang H, Wood C, Kankasa C, Secor WE, McClure HM, Strobert E, Else JG, Ruprecht RM. Molecularly cloned SHIV-1157ipd3N4: a highly replication-competent, mucosally transmissible R5 simian-human immunodeficiency virus encoding HIV clade C Env. J Virol. 2006;80:8729–8738. doi: 10.1128/JVI.00558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 23.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tan RC, Harouse JM, Gettie A, Cheng-Mayer C. In vivo adaptation of SHIV(SF162): chimeric virus expressing, a, NSI, CCR5-specific envelope protein. J Med Primatol. 1999;28:164–168. doi: 10.1111/j.1600-0684.1999.tb00265.x. [DOI] [PubMed] [Google Scholar]
- 25.Verani A, Scarlatti G, Comar M, Tresoldi E, Polo S, Giacca M, Lusso P, Siccardi AG, Vercelli D. C-C chemokines released by lipopolysaccharide (LPS)-stimulated human macrophages suppress HIV-1 infection in both macrophages and T cells. J Exp Med. 1997;185:805–816. doi: 10.1084/jem.185.5.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu Y, Hong K, Chenine AL, Whitney JB, Xu W, Chen Q, Geng Y, Ruprecht RM, Shao Y. Molecular cloning and in vitro evaluation of an infectious simian-human immunodeficiency virus containing env of a primary Chinese HIV-1 subtype C isolate. J Med Primatol. 2005;34:101–107. doi: 10.1111/j.1600-0684.2005.00098.x. [DOI] [PubMed] [Google Scholar]