Abstract
The cellular protease subtilisin kexin isozyme-1 (SKI-1)/site-1 protease (S1P) is implicated in the proteolytic processing of the viral envelope glycoprotein precursor (GPC) of arenaviruses, a step strictly required for production of infectious progeny. The small molecule SKI-1/S1P inhibitor PF-429242 was shown to have anti-viral activity against Old World arenaviruses. Here we extended these studies and show that PF-429242 also inhibits GPC processing and productive infection of New World arenaviruses, making PF-429242 a broadly active anti-arenaviral drug. In combination therapy, PF-429242 potentiated the anti-viral activity of ribavirin, indicating a synergism between the two drugs. A hallmark of arenaviruses is their ability to establish persistent infection in vitro and in vivo. Notably, PF-429242 was able to efficiently and rapidly clear persistent infection by arenaviruses. Interruption of drug treatment did not result in re-emergence of infection, indicating that PF-429242 treatment lead to virus extinction.
INTRODUCTION
Several arenaviruses cause severe viral hemorrhagic fevers (VHF) in humans and represent a serious public health problem in their endemic regions (Geisbert and Jahrling, 2004). The Old World (OW) arenavirus Lassa virus (LASV) in West Africa causes several hundred thousand infections per year (McCormick and Fisher-Hoch, 2002) and the New World (NW) arenaviruses Junin (JUNV), Machupo (MACV), Guanarito (GTOV), and Sabia virus (SABV) have emerged as etiological agents of severe VHF in the Americas (Buchmeier, de la Torre, and Peters, 2007). The worldwide distributed prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) is a neglected human pathogen of clinical significance (Barton, Mets, and Beauchamp, 2002). New arenaviruses emerge on average every three years as illustrated by the recent discoveries of Chapare virus and Lujo virus that were associated with fatal hemorrhagic fever cases in Bolivia and Southern Africa, respectively (Briese et al., 2009; Delgado et al., 2008). A predictive factor for fatality in arenavirus VHF is the viral load, suggesting competition between viral multiplication and the host’s anti-viral immune defense (McCormick and Fisher-Hoch, 2002). Drugs targeting different steps in the arenavirus life cycle may limit viral replication and spread, providing the infected host with a window of opportunity to develop an anti-viral immune response able to control, and ultimately eliminate the virus.
Arenaviruses are enveloped viruses with a bisegmented negative strand RNA genome and a non-lytic life cycle restricted to the cell cytoplasm (Buchmeier, de la Torre, and Peters, 2007). Each genomic RNA segment L (ca 7.3 kb) and S (ca 3.5 kb) contains two open reading frames that direct production of viral polypeptides in opposite orientation. The S RNA encodes the viral glycoprotein precursor, GPC, and the nucleoprotein, NP, whereas the L RNA encodes the viral RNA-dependent RNA polymerase (L), and a small RING finger protein Z. The GPC precursor is initially synthesized as a single polypeptide (ca 75 kDa) that is processed post-translationally by the cellular proprotein convertase (PC) subtilisin kexin isozyme-1 (SKI-1)/site-1 protease (S1P), yielding GP1 (40-46 kDa) and GP2 (35 kDa) (Beyer et al., 2003; Kunz et al., 2003a; Lenz et al., 2001; Rojek et al., 2008a). The GP1 portion serves as viral attachment via interaction with cellular receptors. The transmembrane GP2 resembles the fusion-active membrane-proximal parts of other enveloped viruses and is involved in fusion of the viral membrane with cellular membranes (Eschli et al., 2006).
The current therapy for treatment of human arenavirus infection is an off-labeled use of the nucleoside analogue ribavirin (Rib) (1-β-D-ribofuranosyl-1,2,4-triazole-3-carboxamide) (Parker, 2005). Early administration of Rib reduces both morbidity and mortality in humans infected with LASV (McCormick et al., 1986), and experimental infections with MACV (Kilgore et al., 1995) and JUNV (Weissenbacher, Laguens, and Coto, 1987) in animals. However, for its highest efficacy Rib needs to be administered intravenously and early infection. In addition, Rib is often associated with side effects. Therefore, it is important to develop novel anti-arenaviral drugs that could be used individually or in combination therapy to combat human pathogenic arenaviruses.
Processing of arenavirus GPC by SKI-1/S1P is crucial for infectious virus production and cell-to-cell propagation of the virus (Beyer et al., 2003; Kunz et al., 2003a; Lenz et al., 2001; Rojek et al., 2008a). In the host, SKI-1/S1P is implicated in the regulation of lipid metabolism through the processing of sterol regulatory element-binding proteins (SREBP-1 and SREBP-2), (Brown and Goldstein, 1997; Sakai et al., 1998). Another cellular substrate of SKI-1/S1P is the activating transcription 6 (ATF6), which is a key player in the cellular response to ER stress (Ye et al., 2000). In a recent studies, SKI-1/S1P-adapted serpins (Maisa et al., 2009) and peptide-based small molecule suicide inhibitors of SKI-1/S1P (Rojek et al., 2010) efficiently inhibited productive areanvirus infection. However, the inhibitors used in these proof-of-principle studies face severe limitations for their use as anti-viral drugs in vivo. Small molecule drug screens conducted by Pfizer Inc. discovered a novel specific SKI-1/S1P inhibitor, the amino-pyrrolidine amide compound PF-429242 (Hawkins et al., 2008; Hay et al., 2007). PF-429242 efficiently blocked SKI-1/S1P-mediated processing and subsequent activation of SREBP2 in cell culture and a small animal model (Hawkins et al., 2008). The toxicity profile and pharmacokinetic properties made PF-429242 a promising candidate for a novel anti-arenaviral drug. PF-429242 was able to block the biosynthesis of fusion-active mature GPC of the OW arenaviruses LCMV and LASV and showed potent anti-viral activity against LCMV and LASV in acute infection in cultured cells (Urata et al., 2011). These findings suggested the feasibility of using PF-429242 as a novel anti-arenaviral drug. Here we extended these studies and show that PF-429242 is broadly active anti-arenaviral drug that acts in synergy with ribavirin. Moreover, PF-429242 efficiently and rapidly cleared persistent virus from infected cells. Interruption of drug treatment did not result in re-emergence of infection, indicating that PF-429242 treatment resulted in virus extinction.
RESULTS
Effects of PF-429242 on SKI-1/S1P-mediated processing of SREBP and ATF6, and SKI-1/S1P autoprocessing
The SKI-1/S1P inhibitor PF-429242 was originally identified in a high-throughput small molecule screen and has been shown to efficiently block SKI-1/S1P-mediated cleavage of the cellular substrate SREBP2 in vitro and in vivo (Hawkins et al., 2008; Hay et al., 2007). To further characterize the activity of PF-429242 against cellular targets, we tested the ability of PF-429242 to process the activating transcription factor ATF6 in response to ER stress. To this end, we studied the effect of PF-429242 on the ATF6-mediated induction of the heat shock 70kDa protein 5 (HSPA5) triggered by ER stress and SREBP2-mediated upregulation of the 3-hydroxy-3-methylglutaryl-Coenzyme A synthase (HMGCS1) upon sterol depletion, respectively. To induce ER stress, we treated CHOK1 cells with tunicamycin, an inhibitor of protein N-glycosylation, for 4 hours. For sterol depletion, we treated cells with mevastatin, and inhibitor of cholesterol biosynthesis, for 18 hours. Upon ER stress induction and sterol depletion, cells were lysed, total RNA extracted, and mRNA levels for HSPA5 and HMGCS1 assessed by quantitative real-time PCR (RT-qPCR). Treatment of cells with 10 μM PF-429242 significantly blocked induction of both HSPA5 and HMGCS1, indicating efficient blocking of SKI-1/S1P-mediated cleavage of ATF6 upon ER stress and SREBP2 induced by cholesterol depletion (Fig 1A).
Figure 1. The inhibitor PF-429242 blocks SKI-1/S1P-mediated processing of SREBP and ATF6, but not SKI-1/S1P autoprocessing.
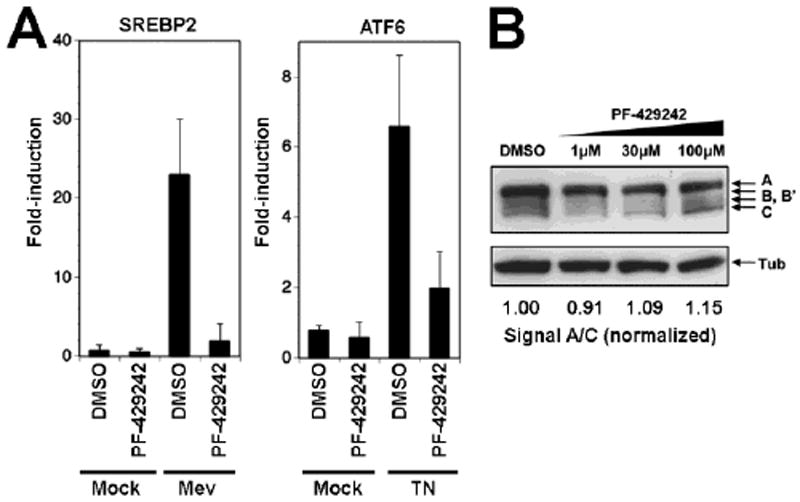
(A) Effect of PF-429242 on the ATF6-mediated induction of the heat shock 70kDa protein 5 (HSPA5) and the SREBP2-mediated upregulation of the 3-hydroxy-3-methylglutaryl-Coenzyme A synthase (HMGCS1). CHOK1 cells were seeded in a 12-well plate and cultured overnight. To induce genes downstream of ATF6 cells were treated with 5 μg/ml tunicamycin (TN) for 4 hours. Genes downstream SREBP2 were induced by treating cells with 50 μM mevastatin (Mev) for 18 hrs. At 14 hrs after addition of mevastatin and at the same time with tunicamycin treatment, PF-429242 (10μM) or DMSO vehicle were added to the cells. At 4 hours post-treatment, cells were washed twice with PBS and total RNA isolated to perform RT-qPCR analyses as described in Materials and Methods. Data were normalized using the calibrator gene hydroxymethylbilane synthase (HMBS). Data are presented as fold-induction above levels for mock (DMSO)-treated cells (means ± SD; n = 3). (B) PF-429242 has no effect on SKI-1/S1P autoprocessing. SKI-1/S1P-deficient SRD12B cells were transfected with recombinant SKI-1/S1P containing a C-terminal V5-tag. Four hours post transfection, the indicated concentrations of PF-429242 were added and left throughout the experiment. After 48 hours, cells were lysed, total protein separated by SDS-PAGE and blotted to nitrocellulose. Blots were probed with an anti-V5 antibody using a HRP-conjugated secondary antibody and ECL for detection. The positions of full-length SKI-1/S1P (A), the form processed at the B/B’ site and the C site are indicated. Tubulin was included as a loading control. To assess the degree of autoprocessing, blots were subjected to densitometric analysis (Kunz et al., 2003b) and the signal of the band corresponding to the mature enzyme (C) normalized to the precursor (A).
During biosynthesis, SKI-1/S1P undergoes maturation that involves proteolytic cleavage at three processing sites (A, B, B’, and C) to generate the active form of the enzyme (Elagoz et al., 2002; Toure et al., 2000). A previously described suicide peptide inhibitor of SKI-1/S1P derived from the C processing site, dec-RRLL-CMK, efficiently blocked processing of cellular and viral substrates (Pasquato et al., 2006; Rojek et al., 2010). Since the peptide substrate used for the small molecule screen that identified PF-429242, Ac-VFRSLK-MCA, contained the SKI-1/S1P B site consensus sequence RSLK (Hawkins et al., 2008), we assessed the effect of PF-429242 on SKI-1/S1P autoprocessing. For this purpose, we transiently expressed recombinant SKI-1/S1P bearing a C-terminal V5-tag in SKI-1/S1P deficient SRD12B cells (Rawson et al., 1998). Autoprocessing of SKI-1/S1P at the B/B’ site, followed by the C site, results in a characteristic pattern of bands that represents the uncleaved precursor, the intermediate form, and the mature protein (Fig. 1B). SKI-1/S1P autoprocessing was not affected by treatment with up to 100 μM of PF-429242 (Fig. 1B), a concentration well above the one sufficient to block processing of ATF6 and SREBP2 (Fig. 1A). Together, these data showed that PF-429242 blocks SKI-1/S1P-mediated processing of SREBP2 and ATF6, but not SKI-1/S1P autoprocessing, thus revealing important differences between SKI-1/S1P-mediated processing of the cellular substrates ATF6 and SREBP2 on the one hand and autoprocessing at the B/B’ and C site on the other hand.
Effect of PF-429242 on SKI/S1P-mediated processing of a broad range of arenavirus GPCs
PF-429242 potently inhibited processing of the GPCs of the OW arenaviruses LASV and LCMV (Urata et al., 2011). The GPC processing sites of OW arenaviruses resemble the sequence RRLL↓ of the SKI-1/S1P C autoprocessing site, whereas the recognition sequences found in NW arenaviruses are strikingly different with those of the NW Clade B viruses JUNV, TACV, and MACV resembling the B autoprocessing site RT/SLK↓ (Pasquato et al., 2011). This led us to first assess the ability of PF-429242 to block SKI-1/S1P-mediated processing of the GPCs of selected NW arenaviruses. For these studies, we chose JUNV, the most important pathogenic arenavirus in the Americas, and GTOV that is also highly pathogenic. Briefly, we transfected CHOK1 cells with expression plasmids for GPC of LASV, JUNV, and GTOV containing a C-terminal FLAG tag followed by addition of PF-429242. Because the effects of PF-429242 on cell cholesterol metabolism (Hawkins et al., 2008), we supplemented with cholesterol the media of PF-429242 treated cells (Rawson et al., 1998; Rojek et al., 2010). After 48 hours, cells were lysed and total protein probed in Western-blot with monoclonal antibody (mAb) M2 anti-FLAG. PF-429242 reduced proteolytic processing of LASV GPC as well as the GPCs of JUNV and GTOV (Fig 2A).
Figure 2. The SKI-1/S1P inhibitor PF-429242 differentially inhibits processing of a broad range of arenavirus GPCs.
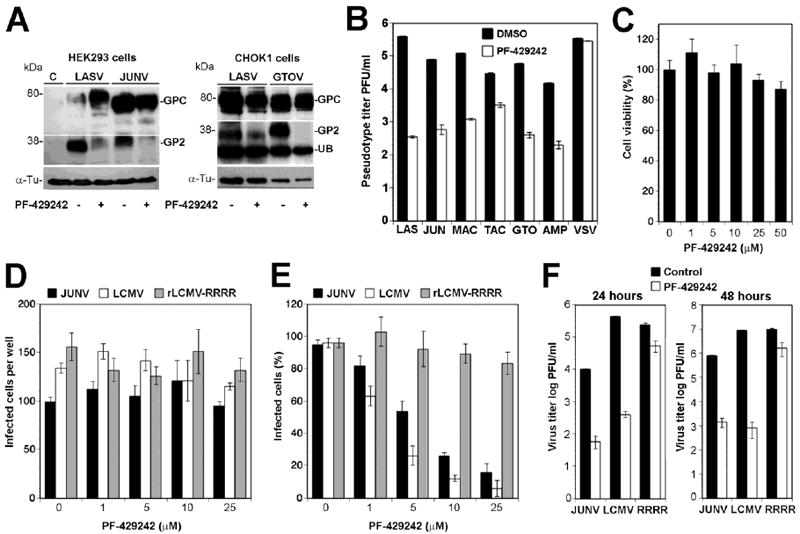
(A) PF-429242 blocks proteolytic processing of the GPCs of JUNV and GTOV. HEK293 cells and CHOK1 cells were transiently transfected with expression plasmids for LASV GPC and JUNV GPC and LASV GPC and GTOV GPC, respectively using Lipofectamine. Controls (C) were transfected with empty vector only. At 4 hours post transfection, the medium was removed and replaced by fresh medium supplemented with cholesterol and containing 10μM PF-429242 (+) or DMSO vehicle (-). After 48 hours cells were lysed and total protein probed with mAb M2 (anti-FLAG) in Western blot. After stripping of membranes tubulin (Tub) was revealed with a mAb anti-α-tubulin. The positions of GPC and GP2 are indicated. For optimal detection of GPC and processed GP2, blots have been cut and exposed at different times. UB indicates an unspecific band detected in CHOK1 cells. (B) PF-429242 blocks the expression of functional arenavirus GPCs. GP2293 packaging cell lines stable expressing MLV Gag and Pol were co-transfected with the MLV genomic plasmid pLZRS-Luc-gfp, and expression plasmids for the GPCs of LASV (LAS), JUNV (JUN), MACV (MAC), TACV (TAC), GTOV (GTO), AMPV (AMP), and VSV G protein (VSV). Four hours post-transfection, 25 μM PF-429242 or DMSO vehicle were added. After 48 hours conditioned supernatants were harvested and cleared. Serial dilutions were prepared and added to VeroE6 monolayers. Infection was detected immunofluorescence staining for GFP using a rabbit anti-GFP antibody and a FITC-labeled secondary antibody (means ± SD; n = 3). Note that the y-axis has a log scale. (C) Cytotoxicity of PF-429242 in A549 cells. A549 cells were seeded in a white 96-well clear bottom plates and cultured overnight, resulting in monolayers. Cells were treated with the indicated concentrations of PF-429242. After 48 hours, cell viability was assessed using CellTiter-GloR luminescent cell viability assay and luminescence measured using a Berthold Multimode Plate reader TriStar LB 941. Values were normalized against the controls (DMSO vehicle) (means ± SD; n = 3). (D) PF-429242 has no effect on early infection with LCMV and JUNV: Monolayers of A549 cells in LabTek tissue culture chambers were pre-treated with the indicated concentrations of PF-429242 for one hour and then infected with 200 PFU of JUNV Candid 1, LCMV ARM53b, and rLCMV-RRRR (MOI = 0.01). After 16 hours in presence of the inhibitor, cultures were fixed and foci of infected cells detected by IF using mAb BG12 to JUNV NP and mAb 113 to LCMVNP (means ± SD; n = 3). (E) The inhibitor PF-429242 reduces cell-to-cell propagation of infection: Monolayers of A549 cells were infected with JUNV, LCMV ARM53b, and rLCMV-RRRR (MOI = 0.01). One hour post-infection, the indicated concentrations of PF-429242 were added and cells cultured in presence of inhibitor. After 48 hours, cells were detached by mild trypsinization, fixed, and intracellular staining for NP performed, followed by quantification of NP positive cells by flow cytometry (see Materials and Methods). The percentage of NP positive cells are given (means ± SD; n = 3). (E) PF-429242 reduces virus production: A549 monolayers were infected with JUNV, LCMV ARM53b, and rLCMV-RRRR (MOI = 3) and PF-429242 (25 μM) added at one hour post infection. Cell culture supernatants were harvested, dialyzed against HBSS to remove the inhibitor, and infectious virus titers determined by IFA on VeroE6 cells (means ± SD; n = 3).
To complement these studies, we examined the ability of PF-429242 to inhibit the biosynthesis of functional arenavirus GP using GP-mediated cell entry as a functional readout. For this purpose, we profited from the fact that recombinant retroviruses can be pseudotyped with the GPCs of arenaviruses (Reignier et al., 2006; Rojek, Spiropoulou, and Kunz, 2006). Since host cell attachment and entry of arenaviruses is mediated exclusively by the viral envelope GP, retroviral pseudotypes are suitable to test correct processing and the biological function of arenavirus GPs. Recombinant Moloney leukemia virus (MLV) containing a green fluorescence protein (GFP) reporter gene was pseudotyped with GPCs of LASV, JUNV, MACV, Tacaribe virus (TACV), GTOV, and Amapari virus (AMPV) by co-transfection of a HEK293 packaging cell line stable expressing MLV Gag and Pol with plasmids containing the retroviral genome and the viral glycoprotein provided in trans (Rojek, Spiropoulou, and Kunz, 2006). As a control, we included MLV pseudotypes bearing the G protein of vesicular stomatitis virus (VSV), which is structurally unrelated to arenavirus GPC and does not require processing for fusion activity (Lyles and Rupprecht, 2007). To assess the impact of PF-429242 on the expression of functional viral GPs, co-transfected packaging cells were exposed to the drug at 4 hours post-transfection. After 48 hours in presence of drug, the pseudotypes were harvested and titers determined by infection of fresh monolayers of Vero E6 cells, followed by detection of GFP expressing cells in fluorescence microscopy. As expected, treatment of cells with 30 μM PF-429242 reduced the production of infectious LASV pseudotypes by circa 3 logs (Fig. 2B). Production of pseudotypes of JUNV, MACV, GTOV, and AMPV was diminished by circa 2 logs and titers of TACV pseudotypes were reduced by circa 1 log. The drug did not affect the titers of VSV pseudotypes, excluding a negative impact on the production and/or assembly of the retroviral core (Fig. 2B). Our data suggest that PF-429242 affects processing and/or incorporation of arenavirus GPCs with LASV > JUNV > TACV (Fig. 2B).
To validate the anti-viral effect of PF-429242 against the NW arenaviruses in the context of productive arenavirus infection, we used the attenuated JUNV vaccine strain Candid 1 whose GPC processing was shown to be mediated by SKI-1/S1P (Rojek et al., 2008a). As cell culture model for our study, we employed the human lung epithelial cell line A459. Our choice was based on the evidence that human pathogenic arenaviruses frequently enter the human body as infectious aerosols and that the lung is one of the major target organ of arenaviruses in humans and animal models (Kenyon et al., 1992). To exclude unspecific cell toxicity of PF-429242, A549 cells were treated with increasing concentration of PF-429242 and cell viability assessed after 48 hours by Cell Titer Glo® assay which determines cellular levels of ATP. Treatment with up to 50 μM PF-429242 did not cause detectable cytotoxicity (Fig. 2C). First, we addressed possible effects of PF-429242 on early steps of JUNV infection. For this we infected cells at low (0.01) MOI with JUNV Candid 1, LCMV-WT. As a control, we used a recombinant LCMV, rLCMV-RRRR in which the SKI-1/S1P recognition site of GPC had been substituted by a canonical furin recognition site (RRRR). As a consequence, GPC processing of rLCMV-RRRR is independent of SKI-1/S1P, but dependent on cellular furin (Rojek et al., 2010). PF-429242 was added one hour prior to infection and cultures fixed at 16 h p.i. Numbers of infected foci were determined by immunofluorescence (IF) staining for the viral nucleoproteins (NP). PF-429242 had no significant effect on the number of infected foci, making inhibition of viral entry and/or other early steps of the viral life cycle rather unlikely (Fig. 2D). To assess inhibition of cell-to-cell propagation of infection, we infected cell at low (0.01) MOI and added increasing concentrations of PF-429242 at one hour post-infection. Cells were cultured for 48 hours and the percentage of infected cells determined by intracellular staining for viral NPs. Increasing concentrations of PF-429242 resulted in a marked dose-dependent reduction in NP positive cell numbers in cultures infected with JUNV and LCMV, whereas cells infected with rLCMV-RRRR showed only a slight reduction at higher drug concentrations (Fig 2E). A similar effect of the SKI-1/1P inhibitor dec-RRLL-CMK on rLCMV-RRRR has been reported earlier (Rojek et al., 2010), likely due to a role of SKI-1/S1P in the multiplication of rLCMV-RRRR unrelated to GPC processing. Next, we assessed the ability of PF-429242 to block virus production from infected cells. For this purpose, A549 cells were infected at high (3) MOI and virus production determined in presence and absence of PF-429242. Treatment with the inhibitor at 25 μM resulted in a marked reduction of titers in cultures infected with JUNV and LCMV, whereas the titers of rLCMV-RRRR were only mildly reduced (Fig. 2F). In sum, these data showed that PF-429242 is broadly active against Old World and New World arenaviruses. However significant quantitative differences exist in the ability of PF-429242 to block GPC processing of OW versus NW arenaviruses. These findings are in line with our previous observation of a differential recognition of Old World and New World arenavirus GPCs by SKI-1/S1P (Pasquato et al., 2011).
PF-429242 and ribavirin show synergistic anti-viral effects
In vitro and in vivo studies have documented the prophylactic and therapeutic value of Rib against several arenaviruses. To investigate the combinatorial effects of PF-429242 and Rib on arenavirus multiplication we employed the prototypic arenavirus LCMV and JUNV Candid 1. To control for the target specificity of PF-429242 drug action, we included the furin-dependent rLCMV-RRRR. As an additional control, we included a recombinant LCMV expressing the G protein of vesicular stomatitis virus (VSV) (rLCMV-VSVG), which does not require proteolytic processing (Pinschewer et al., 2003). LCMV and rLCMV-RRRR exhibited similar growth curves in A549 cells, whereas, JUNV and rLCMV-VSVG grew to lower titers (Fig 3A). Both LCMV and rLCMV-VSVG showed similar sensitivities towards Rib, whereas rLCMV-RRRR and JUNV showed consistently a somewhat lower sensitivity to the drug (Fig 3B).
Figure 3. Inhibition of replication of LCMV variants by ribavirin (Rib).
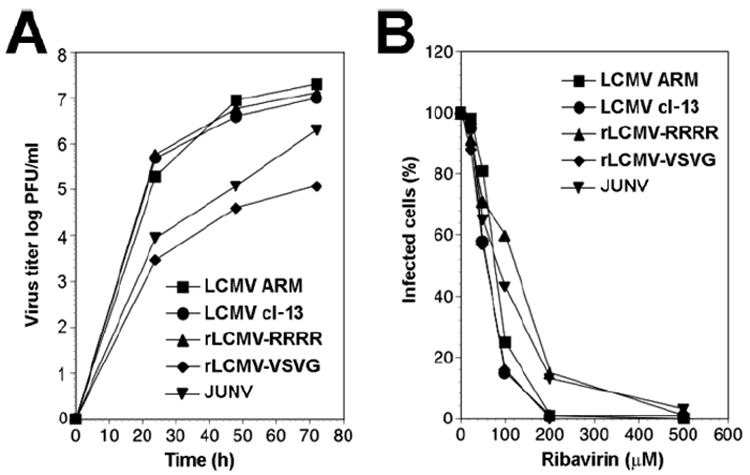
(A) Growth of LCMVs: A549 cells were infected with wild-type LCMV ARM53b, LCMV cl-13, rLCMV-RRRR, rLCMV-VSVG, and JUNV (MOI = 0.1) and virus titers determined in the supernatants after the indicated time points by plaque assay on VeroE6 cells. (B) Inhibition of replication of LCMV variants by Rib. A549 cells were seeded in a 96-well plate and cultured overnight, resulting in monolayers. Cells were infected with LCMV-ARM53b, LCMV cl-13, rLCMV-RRRR, rLCMV-VSVG, and JUNV at MOI=0.01. At 45 min post infection (p.i.), inoculums were removed and replaced by fresh medium containing increasing concentrations of Rib (25-500μM). At 48 hours p.i., cells were fixed and infection evaluated by IFA using mAb 113 to LCMV NP and mAb BG12 to JUNV NP. Cell nuclei were counterstained, 100 cells examined and NP positive cells scored. Each value is the mean of two independent experiments. Values were normalized against the controls (without drug).
Next, we determined the cytotoxicity of Rib and PF-429242 individually as well as combinations thereof in A549 cells. Cells were treated with increasing concentration of Rib (25μM-500μM) and cell viability assessed after 48 hours by Cell Titer Glo® assay. A549 cells tolerated Rib up to 500 μM without significant drop in viability (Fig. 4A). Treatment with the highest dose of Rib and PF-429242 concentrations we planned to use, alone or in combination, did not cause significant cytotoxicity during a 48 hours treatment (Fig. 4B).
Figure 4. Combined anti-viral effects of PF-429242 and Rib.
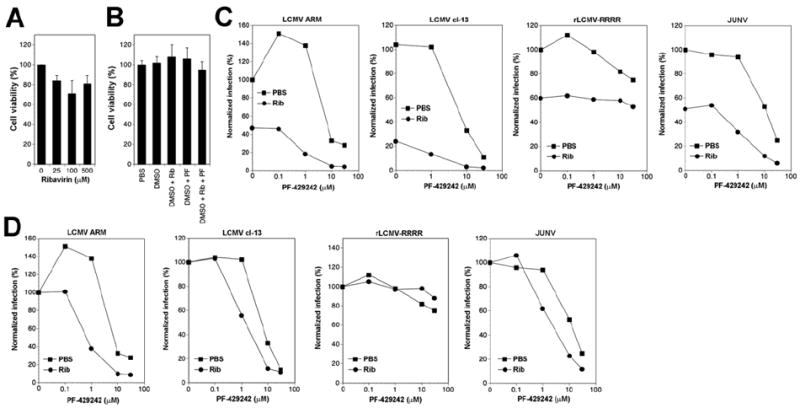
(A, B) Cytotoxicity of the combination of Rib and PF-429242 in A549 cells. Monolayers of A549 cells were treated with the indicated concentrations of Rib and PF-429242 (PF). After 48 hours, cell viability was assessed using CellTiter-Glo® cell viability assay as in Fig. 2C. Values were normalized against the controls (DMSO vehicle) (means ± SD; n = 3). (C) Treatment of infected cells with Rib and PF-429242. A549 cells were infected with the indicated viruses at MOI = 0.01. After removal of unbound virus, fresh medium was added containing the indicated concentrations of PF-429242 either alone (PBS) or in combination with 25 μM Rib (Rib). After 48 hours, cells were detached, single cell suspensions prepared, and viral NP detected by intracellular staining as in 2E. Infection was normalized setting the percentage of infection in untreated cells (PBS, PF-429242 = 0) as 100%. Each point represents the average of two independent experiments. (D) Data shown in (C) with normalization for each series: the percentage of infection in cells not treated with PF-429242 (0) of each series (Rib, PBS) was set to 100%. A simple additive effect of PF-429242 and Rib on virus infection would result in overlapping curves as e.g. seen in case of rLCMV-RRRR that is not affected by PF-429242.
To study anti-viral effects of combinations of Rib and PF-429242, A549 cells were infected with LCMV, rLCMV-RRRR, and JUNV. Infection at low MOI (0.01) was followed by treatment with increasing concentrations of PF-429242 (0-30μM), either alone or in combination with 25 μM Rib, which by itself resulted in only partial inhibition of viral infection. To assess anti-viral activity of the drug combinations, we detected viral infection at 48 hours post infection. For this purpose, we detected the viral NP in cells by intracellular staining with mAb 113 and determined the percentage of infected cells by flow cytometry. This readout allows an accurate determination of cell-to-cell propagation of virus in cells under drug treatment. Combination of PF-429242 with Rib resulted in a markedly stronger anti-viral effect (Fig 4C). To distinguish a simple additive effect of PF-429242 with Rib from a synergistic effect, or potential interference between the two drugs, we normalized the infection in Rib-treated and untreated cells (100 % infection in absence of PF-429242) and plotted the data in function of increasing concentration of PF-429242 (Fig. 4D). A simple additive effect of the drugs would result in overlapping dose-response curves for PF-429242 in presence and absence of Rib. We observed a greater than additive effect between PF-429242 and Rib in cells infected with LCMV and JUNV, but not the SKI-1/S1P-independent rLCMV-RRRR (Fig. 4D). Together the data indicated that the simultaneous targeting of viral replication (Rib) and GPC processing (PF-429242) have a synergistic inhibitory effect on arenavirus multiplication and cell-to-cell spread.
Effect of PF-429242 on arenavirus persistently infected cells
Previous studies on LCMV infection in vitro and in vivo revealed that expression of the viral NP and GPC are differently regulated during acute and persistent infections (Oldstone and Buchmeier, 1982). NP expression levels remained high and relatively constant during the acute and persistent phases of LCMV infection. In contrast, GPC was highly expressed during the first days of acute infection, followed by a marked down-regulation during the transition to persistent infection. We reasoned that low expression levels of GP in persistently infected cells could facilitate the antiviral effect of PF-429242 mediated targeting of GPC processing during arenavirus persistence. To examine this hypothesis we infected A549 cells with LCMV and the expression of GPC and NP monitored over time. Consistent with previous studies, we observed a dramatic down-regulation of GPC, but not NP after 72 hours of infection (Fig. 5A). After serial passages of LCMV-infected A549 cells for 4 weeks > 98% of cells expressed NP as assessed by intracellular immunostaining and flow cytometry, whereas GPC was barely detectable (data not shown).
Figure 5. Potent anti-viral effect of PF-429242 in persistently infected cells.
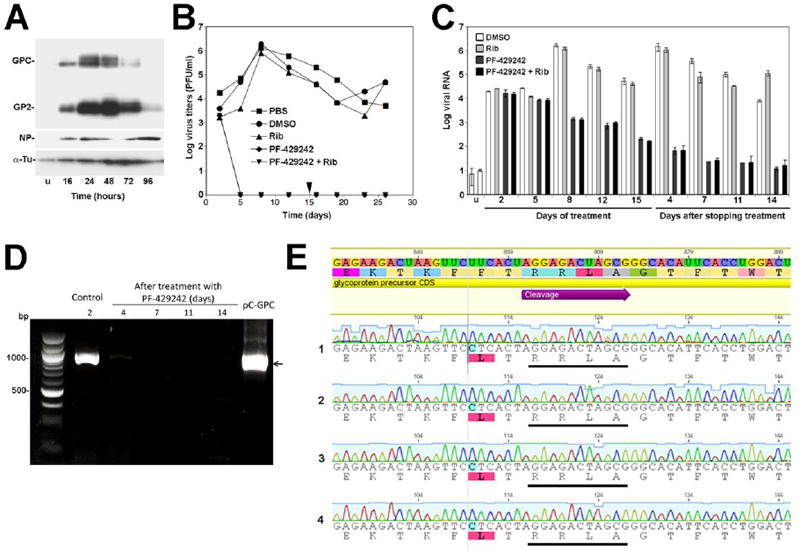
(A) Differential regulation of LCMV NP and GP expression during acute infection. A549 cells were infected with LCMV (MOI=0.1). At the indicated time points post infection (p.i.), cells were lysed in SDS-PAGE sample buffer, protein separated and probed in Western-blot using monoclonal antibody (mAb) 83.6 to LCMV GP2 (GP2), polyclonal guinea pig serum to LCMV NP (NP), and a mAb to α-tubulin (α-Tu), combined with HRP-conjugated secondary antibodies and enhanced chemiluminescence (ECL) as described (Rojek et al., 2007a). Alpha-tubulin (α-Tu) was used as loading control. The unprocessed GPC and mature GP2 are indicated. (B) PF-429242 prevents infectious virus production from persistently infected cells. A549 cells were persistently infected with LCMV cl-13 for 4 weeks. Cells were plated and either mock treated (PBS, DMSO), treated with 25 μM Rib (Rib), 10 μM PF-429242 (PF-429242), and a combination of 25 μM Rib and 10 μM PF-429242 (PF-429242 + Rib). Cells were treated with drugs for 15 days. On day 15, drugs were removed and cells cultured in drug-free medium. At the indicated time points, cell culture supernatant was removed and cells passed. Virus titers were determined in cell culture supernatants by IFA on fresh monolayers of A549 cells. Each time value represents the average of two independent experiments. (C) Detection of viral RNA in persistently infected cells treated with PF-429242 and Rib. Persistently infected A549 cells were treated with PF-429242 and Rib as in (B). At the indicated time points, cells were passed. Half of the cells were subjected to RNA extraction, followed by detection of viral RNA by RT-qPCR as described in Materials and Methods. Data were normalized using the calibrator gene GAPDH. The resulting values for viral RNA load were then normalized to the signal obtained from uninfected cells (u) (means ± SD; n = 3). (D) Detection of LCMV GPC mRNA in cells at 4, 7, 11, and 14 days after stop of PF-429242 treatment. Cellular RNA was isolated, subjected to reverse transcription, and a 1000 bp fragment of GPC containing the SKI-1/S1P recognition site amplified by PCR as detailed in Materials and Methods. As controls, RNA from control cells (DMSO treated) on day 2 of infection (Control), as well as the plasmid pC-GPC containing the LCMV GPC open reading frame were used. The position of the expected band for LCMV GPC is indicated with an arrow. (E) Representative examples of sequence analysis including chromatograms of the GPC region containing the SKI-1/S1P cleavage site in PCR products obtained in (D). The consensus sequence of LCMV GPC is given at the top and the SKI-1/S1P processing site (RRLA) is underlined. The sequences of four out of 10 PCR products are shown. Please note that only wild-type sequences were detected.
To assess the activity of PF-429242 against persistent virus, persistently infected cells were plated and after attachment exposed to PF-429242, Rib, and the combination of PF-429242 and Rib. Cells were passed every 3 days for a total of 15 days. At this time point, drugs were removed and cells passed for another 11 days in drug-free medium. The anti-viral effect of drug combinations was assessed in two ways 1) determination of infectious virus titers in cell culture supernatants from infected cells in presence and absence of drugs and 2) detection of viral RNA in treated and untreated cells. To monitor infectious virus production from persistently infected cells over time, samples of cell culture supernatant were taken at the indicated time points. Serial dilutions were added to fresh monolayers of A549 cells and virus titers determined by immunofocus assay (IFA). Treatment with Rib at 25 μM had no significant effect on infectious virus titers over time (Fig. 5B). In contrast, treatment with PF-429242 alone resulted in a dramatic reduction of infectious virus titers to undetectable levels after 5 days of treatment (Fig. 5B). In contrast to results observed during acute infection (Fig. 4), the combination of PF-429242 with Rib did not result in an enhanced anti-viral effect (Fig. 5B), indicating that PF-429242 was the major active component of the drug combination. After removal of drugs, we monitored re-emergence of infectious virus, but were unable to detect infectious particles in the supernatants of cells treated with PF-429242 or the combination of PF-429242 and Rib (Fig. 5B).
Because the limited sensitivity of the IFA used to monitor production of infectious progeny with a cut-off of circa 100 PFU/ml, we used also real-time quantitative PCR (RT-qPCR) to detect viral RNA in cells, using the house keeping gene GAPDH for normalization (Materials and Methods). Treatment of persistently infected cells with PF-429242 resulted in progressive reduction of viral RNA levels over the time of treatment by circa 3 logs. After removal of drug, cellular viral RNA levels remained 4-5 logs lower than in untreated control cells, reaching background levels on day 7 after stopping the treatment (Fig. 5C).
In order to detect possible viral escape variants, we sought to amplify mRNA of the viral GPC present in cells on day 4, 7, 11, and 14 after removal of the drug. Using classical PCR with a set of specific primers for GPC, we were able to detect low levels of GPC mRNA in cells on day 4 after removal of the drug, but not at later time points (Fig. 5D). Sequencing of the GPC open reading frame from the amplified GPC fragment from cells on day 4 after drug removal did not reveal point mutations in the SKI-1/S1P processing sites (Fig. 5E). In addition, several attempts to isolate infectious virus at this or later time points failed. Together, the data suggest that treatment of persistently infected cells with PF-429242 not only reduced infectious virus production, but led to virus extinction.
DISCUSSION
Previous studies showed efficient inhibition of LASV and LCMV GPC cleavage by PF-429242 (Urata et al., 2011), however, its activity against processing of NW arenavirus GPC was unknown. Using a panel of recombinant NW arenavirus GPCs, we found that PF-429242 differentially inhibited processing of GPCs derived from arenaviruses belonging to different phylogenetic groups. When compared to its activity against SKI-1/S1P-mediated cleavage of OW arenavirus GPCs, PF-429242 seemed overall less active against NW virus GPC processing. Anti-viral activity of PF-429242 against NW arenaviruses was validated using the JUNV vaccine strain Candid1 as a model. PF-429242 showed potent anti-viral activity preventing cell-to-cell propagation of virus and production of infectious progeny virus from infected cells. While IC50 values for LCMV were in the range of 2 μM, IC50 for JUNV was around 5 μM.
The differential inhibition of SKI-1/S1P-mediated processing of OW and NW arenavirus GPCs suggests that PF-924292 blocks SKI-1/S1P in a substrate-dependent manner. The basis of this substrate-dependence of inhibition is currently unknown, but may involve perturbation of molecular interactions between SKI-1/S1P catalytic pocket and different substrates amino acids other than those that are included in the RX(hydrophobic)X consensus motif. Indeed, previous studies showed the importance of residues N-terminal of the SKI-1/S1P recognition site for optimal processing of peptides derived from some viral substrates but not others (Pasquato et al., 2011; Pasquato et al., 2006).
Initial evaluation of PF-429242 in vivo in a small animal model raised concerns about the possible application of PF-429242 in the context of chronic diseases, such as familial hypercholesterolemia (Hawkins et al., 2008). In contrast, human pathogenic arenaviruses cause acute diseases, limiting anti-viral treatment to a time window of a few weeks. Considering these relatively short periods of treatment, the toxicological and pharmacokinetics profile of PF-429242 make it a promising anti-arenaviral drug candidate (Hawkins et al., 2008). To further evaluate the anti-viral potential of PF-429242, we tested its efficacy in combination with Rib, and found that the combination of PF-429242 with Rib revealed stronger than additive effect of the two drugs. The basis for this apparent synergism is currently unclear, but may lie in the distinct underlying anti-viral mechanisms of the two drugs. Rib inhibits arenavirus infection at the level of replication (Ruiz-Jarabo et al., 2003) and can also act as a mutagen (Moreno et al., 2011), whereas PF-429242 acts on the biosynthesis of the viral GP, blocking the formation of infectious progeny virus from infected cells. The synergistic effect of the combination of Rib with PF-4289242 suggests that the potency of the SKI-1/S1P inhibitor in infected cells may depend on the absolute expression levels of the drug target, the viral GPC.
A hallmark of arenaviruses is their ability to establish chronic persistent infections in permissive animal cells (Oldstone, 2002). The transition from acute to persistent infection is characterized by a marked down-regulation of the viral GPC in vitro and in vivo (Oldstone and Buchmeier, 1982). The resulting low expression levels of GPC likely become a limiting factor for infectious virus production and may represent an “Achilles heel” for persistent virus. Treatment of LCMV persistently infected cells with PF-429242 resulted in rapid elimination of the virus, as assessed by detection of infectious virus in the cell culture supernatant and viral RNA in cells. Interruption of drug treatment did not result in re-emergence of virus, indicating successful extinction. In contrast to acute infection, the addition of Rib did not enhance the anti-viral effect of PF-429242. A possible explanation for this discrepancy may be much lower expression levels of GPC in persistent infection, which could be less affected by Rib-mediated inhibition at the level of viral replication and transcription. Under our experimental condition, no drug resistant viral escape variants emerged. This observation is in line with previous studies from our group performed in LCMV persistently infected SKI-1/S1P-deficent cells (Rojek et al., 2010), suggesting that emergence of SKI-1/S1P-independent LCMV is a rare event. The broad anti-arenaviral activity of PF-429242, the synergistic anti-viral effect with Rib, its remarkable potency in persistent infection, together with the lack of emerging resistant viral variants, make PF-429242 an attractive drug candidate. The results of our present study warrant further evaluation of PF429242 in treatment of acute and persistent arenavirus infection in vivo, which will be investigated in follow-up studies in suitable small animal models.
MATERIALS AND METHODS
Cells lines and viruses
Human lung epithelial (A549), human embryonic kidney (HEK)293H, and African green monkey kidney (VeroE6) cells were maintained at 37°C and 5% CO2 in DMEM (Dulbecco’s modified Eagle’s medium (Invitrogen) completed with 10% fetal bovine serum (FCS) and 100units/ml penicillin and 0.1mg/ml streptomycin (P/S). Baby hamster kidney cells (BHK-21) were grown in DMEM supplemented with 5% tryptose phosphate broth solution (TPB), 10% FCS and P/S. Chinese hamster ovary (CHO)K1 cells were grown in DMEM nutrient mixture F12 Ham (GIBCO BRL, NY) supplemented with 10% FCS and P/S. SRD-12B cells, CHO knockout (KO) for SKI-1/S1P, and cells under the SKI-1/S1P inhibitor PF-429242 treatment were supplemented with 5μg/ml cholesterol (Sigma, St. Louis, MO), 1mM sodium mevalonate (Sigma), and 20μM sodium oleate (Sigma). Stocks of LCMV were prepared in BHK-21 cells and titers determined as reported (Dutko and Oldstone, 1983). JUNV Candid 1 was a kind gift from Dr. Michael Buchmeier and was produced in VeroE6 cells as described (Rojek et al., 2008a). Production and titers of retroviral pseudotypes were done as described earlier (Rojek et al., 2007b; Rojek, Spiropoulou, and Kunz, 2006). The recombinant viruses have been described elsewhere: rLCMV-LASVGP (Rojek et al., 2008b), rLCMV-RRRR (Rojek et al., 2010), and rLCMV-VSVG (Pinschewer et al., 2003).
Antibodies
Monoclonal antibodies (mAbs) 113 (anti-LCMVNP) and 83.6 (anti-LCMVGP) have been described (Buchmeier et al., 1981; Weber and Buchmeier, 1988), as has mAb SA02-BG12 anti JUNV NP (Sanchez et al., 1989). Other antibodies included: mAb anti-α-tubulin (Sigma), rabbit polyclonal anti-Flag (Sigma), rabbit polyclonal anti-GFP (Chemicon), rhodamine-X-red conjugated anti-mouse IgG, and Alexa488-conjugated anti-mouse IgG. Anti-mouse IgG and anti-rabbit IgG conjugated to horseradish peroxidase (HRP) were obtained from Pierce. Monoclonal antibody to the V5 tag and 4’-6-Diamidino-2-phenylindole (DAPI) were from Invitrogen.
Cell Viability Assay
Four times 104 A549 cells/well in a 96-well plate were treated with the compounds at various concentrations and PBS or DMSO (2%) as controls. At 48h post incubation, 100μl/well of CellTiter-Glo® luminescent cell viability assay solution (Promega, Dübendorf, Switzerland), were added and luminescence measured using a Berthold Multimode Platereader TriStar LB 941 model.
Virus infection of cells
Virus inoculums at different MOIs were added to cells and incubated at 37°C. After 45 min, inoculums were removed, fresh medium added. When indicated, media were supplemented with lipids and/or drugs at various concentrations or DMSO/PBS. For LCMV, infected cell numbers were assessed by IFA using mAb 113 and a secondary antibody coupled to rhodamine-X-red (Rojek et al., 2010). Intracellular FACS staining for LCMV NP was performed as described (Kunz et al., 2003c). Detection of JUNV NP by IFA was performed as reported (Rojek et al., 2008a). Infection of retroviral pseudotypes was detected by IF staining for GFP as reported (Rojek, Spiropoulou, and Kunz, 2006).
Pharmacological inhibitors
Ribavirin (1-β-D-ribofuranosyl-1, 2, 4-triazole-3-carboxamide) was purchased from Sigma. Once dissolved in PBS, it was kept at -80°C for long term and -20°C for short term storage. PF-429242 (4-[(diethylamino)methyl]-N-[2-(2-methoxyphenyl)ethyl]-N-[(3R)-pyrrolidin-3-yl] benzamide dihydrochloride], provided by Enamine, was dissolved in DMSO at 100mM and stored at -20°C.
Transfection
CHOK1, HEK293H or SRD-12B cells were cultured to 90% confluence in 24-well plates (2.5×105 cells/well). Plasmid DNA (800ng) was transfected using Lipofectamin2000 (Invitrogen), according to manufacturer’s instructions. Four hours post transfection, medium was removed and replaced by fresh medium supplemented with 5μg/ml cholesterol, 1mM sodium mevalonate, 20μM sodium oleate for SRD-12B cells.
Western Blot
Cells were lysed with 2× SDS polyacrylamide gel electrophoresis (PAGE) sample buffer (reducing), and samples separated through 10% SDS-PAGE. Proteins were transferred to nitrocellulose membranes and blocked with 6% (w/v) skim milk in PBS, 0.2% (wt/vol) Tween-20 (PBST). Blots were incubated with primary antibody overnight at 4°C and an appropriate HRP-conjugated secondary antibody for 1hr at RT. Either enhanced chemo luminescence (ECL) (LiteAblot®, Euroclone) or ECL AdvanceTM western blotting detection Kit (GE Healthcare) were used for revelation.
Induction of SREBP and ATF6 cleavage
To induce upregulation of genes downstream of SREPB, CHOK1 cells were treated with Ham’s F-12 DMEM 1:1, 5% LPDS, 50 μM sodium mevalonate, 50 μM mevastatin for 18 hours as described (Pullikotil et al., 2004). To induce upregulation of genes downstream of ATF6, cells were treated with tunicamycin (5 μg/ml) for 4 hours (Pasqual et al., 2011). Fourteen hours after the induction of SREPB cleavage and simultaneously with tunicamycin treatment, PF-429242(10μM) or DMSO (2%) was added to the cells. Four hours post treatment cells were washed twice with PBS and total RNA was isolated to perform RT-qPCR analyses.
Real-time quantitative PCR (RT-qPCR)
Total RNA was isolated using RNeasy Mini Kit (Qiagen, Chatsworth, CA). The lysate was homogenized with a Qiashredder kit (Qiagen). The RT reaction was performed using QuantiTect Reverse Transcription Kit by Qiagen with 1 μg of template RNA. PCR was done with SYBR® green Reagents (Applied Biosystems, Foster City, CA). Specific primers were used to determine the level of expression of HMGCS1 (5’-GCCCTTGAGATCTACTTTCC-3’, 5’-CCAGGCCAATGGTATACTTC-3’), HSPA5 (5’-CACTTGGAATGACCCTTCAG-3’, 5’-GTTTGCCCACCTCCAATATC-3’) and HMBS (5’-AGATTCTTGATACTGCACTC-3’, 5’-GAAAGACAACAGCATCACAA-3’) (Microsynth, Balgach, Switzerland). Real-time PCR analyses were performed in triplicate (real-time PCR ABI PRISM 7000 sequence detection system Applied Biosystems). The RT-qPCR on LCMV NP was performed with specific primers (5’-GAGCCTTGACAGCTTAGAAC-3’, 5’-CTTGCCGACCTCTTCAATG-3’) and a TaqMan probe (5’-CCTGCGGAAGAGCAC-3’), the TaqMan probe (GAPDH; Hs99999905_m1) (Applied Biosystems) was used as calibrator. RNA extraction, reverse transcriptase and qPCR were performed by the diagnostic facility of the Institute of Microbiology in Lausanne. The RNA was extracted using MagNA Pure 96 DNA and Viral NA Small Volume Kit (Roche) using GeneAmp ® PCR System 9700 (Applied biosystem). The RT reaction was performed with the High Capacity cDNA Reverse Transcription Kit (Applied biosystem) using GeneAmp ® PCR System 9700 (Applied biosystem). Finely the qPCR plate was prepared with Freedom EVO 150 Tecan® and the qPCR run with 7900HT Sequence Detection System (Applied biosystem). Results were analyzed using the program SDS 2.2. The analytical sensitivity of the RT-qPCR for LCMV NP was determined using an expression plasmid pCAGGS containing the cDNA LCMV NP (ARM53b) in 10-fold serial dilutions. The experiment showed a linear range between 50 and 5×104 copies and a limit of detection of ≤ 50 copies per reaction.
For the amplification of the GPC open reading frame from cellular RNA we used QuantiTect Reverse Transcription Kit (Qiagen) with 600ng of template RNA. A PCR using specific GPC primers (5’-GCC AGT GTA GAA CCT TCA GAG-3‘, 5’ CCC GGA TCC TCA GCG TCT TTT CCA GAC GGT TTT TAC ACC3’) yielded a 1000 bp PCR including the SKI-1/S1P site. PCR products were run on a 1% agarose gel. PCR products were sequenced and no mutations were found for the GPC cleavage site of the sample obtained at 4 days after the end of PF-429242 treatment.
Acknowledgments
The authors thank Dr. Nabil Seidah (Laboratory of Biochemical Neuroendocrinology, Clinical Research Institute of Montreal, Montreal, Canada) for valuable reagents. We further thank Michael S. Brown and Joseph L. Goldstein (University of Texas Southwestern Medical Center, Dallas, TX) for the S1P-deficient SRD12B cells. This research was supported by the Swiss National Science Foundation grants FN 31003A-120250 and FN 31003A-135536 (Stefan Kunz), Funds from the University of Lausanne, and NIH/NIAID grant AI079665 (Juan Carlos de la Torre).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Barton LL, Mets MB, Beauchamp CL. Lymphocytic choriomeningitis virus: emerging fetal teratogen. Am J Obstet Gynecol. 2002;187(6):1715–6. doi: 10.1067/mob.2002.126297. [DOI] [PubMed] [Google Scholar]
- Beyer WR, Popplau D, Garten W, von Laer D, Lenz O. Endoproteolytic processing of the lymphocytic choriomeningitis virus glycoprotein by the subtilase SKI-1/S1P. J Virol. 2003;77(5):2866–72. doi: 10.1128/JVI.77.5.2866-2872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briese T, Paweska JT, McMullan LK, Hutchison SK, Street C, Palacios G, Khristova ML, Weyer J, Swanepoel R, Egholm M, Nichol ST, Lipkin WI. Genetic detection and characterization of Lujo virus, a new hemorrhagic fever-associated arenavirus from southern Africa. PLoS Pathog. 2009;5(5):e1000455. doi: 10.1371/journal.ppat.1000455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89(3):331–40. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- Buchmeier MJ, de la Torre JC, Peters CJ. Arenaviridae: the viruses and their replication. In: Knipe DL, Howley PM, editors. Fields Virology. 4. Lippincott-Raven; Philadelphia: 2007. pp. 1791–1828. [Google Scholar]
- Buchmeier MJ, Lewicki HA, Tomori O, Oldstone MB. Monoclonal antibodies to lymphocytic choriomeningitis and pichinde viruses: generation, characterization, and cross-reactivity with other arenaviruses. Virology. 1981;113(1):73–85. doi: 10.1016/0042-6822(81)90137-9. [DOI] [PubMed] [Google Scholar]
- Delgado S, Erickson BR, Agudo R, Blair PJ, Vallejo E, Albarino CG, Vargas J, Comer JA, Rollin PE, Ksiazek TG, Olson JG, Nichol ST. Chapare virus, a newly discovered arenavirus isolated from a fatal hemorrhagic fever case in Bolivia. PLoS Pathog. 2008;4(4):e1000047. doi: 10.1371/journal.ppat.1000047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutko FJ, Oldstone MB. Genomic and biological variation among commonly used lymphocytic choriomeningitis virus strains. J Gen Virol. 1983;64(Pt 8):1689–98. doi: 10.1099/0022-1317-64-8-1689. [DOI] [PubMed] [Google Scholar]
- Elagoz A, Benjannet S, Mammarbassi A, Wickham L, Seidah NG. Biosynthesis and cellular trafficking of the convertase SKI-1/S1P: ectodomain shedding requires SKI-1 activity. J Biol Chem. 2002;277(13):11265–75. doi: 10.1074/jbc.M109011200. Epub 2001 Dec 26. [DOI] [PubMed] [Google Scholar]
- Eschli B, Quirin K, Wepf A, Weber J, Zinkernagel R, Hengartner H. Identification of an N-terminal trimeric coiled-coil core within arenavirus glycoprotein 2 permits assignment to class I viral fusion proteins. J Virol. 2006;80(12):5897–907. doi: 10.1128/JVI.00008-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisbert TW, Jahrling PB. Exotic emerging viral diseases: progress and challenges. Nat Med. 2004;10(12 Suppl):S110–21. doi: 10.1038/nm1142. [DOI] [PubMed] [Google Scholar]
- Hawkins JL, Robbins MD, Warren LC, Xia D, Petras SF, Valentine JJ, Varghese AH, Wang IK, Subashi TA, Shelly LD, Hay BA, Landschulz KT, Geoghegan KF, Harwood HJ., Jr Pharmacologic inhibition of site 1 protease activity inhibits sterol regulatory element-binding protein processing and reduces lipogenic enzyme gene expression and lipid synthesis in cultured cells and experimental animals. J Pharmacol Exp Ther. 2008;326(3):801–8. doi: 10.1124/jpet.108.139626. [DOI] [PubMed] [Google Scholar]
- Hay BA, Abrams B, Zumbrunn AY, Valentine JJ, Warren LC, Petras SF, Shelly LD, Xia A, Varghese AH, Hawkins JL, Van Camp JA, Robbins MD, Landschulz K, Harwood HJ., Jr Aminopyrrolidineamide inhibitors of site-1 protease. Bioorg Med Chem Lett. 2007;17(16):4411–4. doi: 10.1016/j.bmcl.2007.06.031. [DOI] [PubMed] [Google Scholar]
- Kenyon RH, McKee KT, Jr, Zack PM, Rippy MK, Vogel AP, York C, Meegan J, Crabbs C, Peters CJ. Aerosol infection of rhesus macaques with Junin virus. Intervirology. 1992;33(1):23–31. doi: 10.1159/000150227. [DOI] [PubMed] [Google Scholar]
- Kilgore PE, Peters CJ, Mills JN, Rollin PE, Armstrong L, Khan AS, Ksiazek TG. Prospects for the control of Bolivian hemorrhagic fever. Emerg Infect Dis. 1995;1(3):97–100. doi: 10.3201/eid0103.950308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz S, Edelmann KH, de la Torre J-C, Gorney R, Oldstone MBA. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology. 2003a doi: 10.1016/s0042-6822(03)00421-5. in press. [DOI] [PubMed] [Google Scholar]
- Kunz S, Edelmann KH, de la Torre J-C, Gorney R, Oldstone MBA. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology. 2003b;314:168–178. doi: 10.1016/s0042-6822(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Kunz S, Edelmann KH, de la Torre JC, Gorney R, Oldstone MB. Mechanisms for lymphocytic choriomeningitis virus glycoprotein cleavage, transport, and incorporation into virions. Virology. 2003c;314(1):168–78. doi: 10.1016/s0042-6822(03)00421-5. [DOI] [PubMed] [Google Scholar]
- Lenz O, ter Meulen J, Klenk HD, Seidah NG, Garten W. The Lassa virus glycoprotein precursor GP-C is proteolytically processed by subtilase SKI-1/S1P. Proc Natl Acad Sci U S A. 2001;98(22):12701–5. doi: 10.1073/pnas.221447598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles DS, Rupprecht CE. Rhabdoviridae. In: Knipe DM, Howley PM, editors. Fields Virology. 5. Lippincott; Philadelphia: 2007. p. 1363. [Google Scholar]
- Maisa A, Stroher U, Klenk HD, Garten W, Strecker T. Inhibition of Lassa virus glycoprotein cleavage and multicycle replication by site 1 protease-adapted alpha(1)-antitrypsin variants. PLoS Negl Trop Dis. 2009;3(6):e446. doi: 10.1371/journal.pntd.0000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormick JB, Fisher-Hoch SP. Lassa fever. Curr Top Microbiol Immunol. 2002;262:75–109. doi: 10.1007/978-3-642-56029-3_4. [DOI] [PubMed] [Google Scholar]
- McCormick JB, King IJ, Webb PA, Scribner CL, Craven RB, Johnson KM, Elliott LH, Belmont-Williams R. Lassa fever. Effective therapy with ribavirin. N Engl J Med. 1986;314(1):20–6. doi: 10.1056/NEJM198601023140104. [DOI] [PubMed] [Google Scholar]
- Moreno H, Gallego I, Sevilla N, de la Torre JC, Domingo E, Martin V. Ribavirin can be mutagenic for arenaviruses. J Virol. 2011;85(14):7246–55. doi: 10.1128/JVI.00614-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldstone MB. Biology and Pathogenesis of Lymphocytic Choriomeningitis Virus Infection. In: Oldstone MB, editor. Arenaviruses. Vol. 263. 2002. pp. 83–118. [DOI] [PubMed] [Google Scholar]
- Oldstone MB, Buchmeier MJ. Restricted expression of viral glycoprotein in cells of persistently infected mice. Nature. 1982;300(5890):360–2. doi: 10.1038/300360a0. [DOI] [PubMed] [Google Scholar]
- Parker WB. Metabolism and antiviral activity of ribavirin. Virus Res. 2005;107(2):165–71. doi: 10.1016/j.virusres.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Pasqual G, Burri DJ, Pasquato A, de la Torre JC, Kunz S. Role of the host cell’s unfolded protein response in arenavirus infection. Journal of virology. 2011;85(4):1662–70. doi: 10.1128/JVI.01782-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquato A, Burri DJ, Gomer-Ibarlucea Traba E, Hanna-El-Daher L, Seidah NG, Kunz S. Arenavirus envelope glycoproteins mimic autoprocessing sites of the cellular proprotein convertase subtilisin kexin isozyme-1/site-1 protease. Virology. 2011 doi: 10.1016/j.virol.2011.04.021. in press. [DOI] [PubMed] [Google Scholar]
- Pasquato A, Pullikotil P, Asselin MC, Vacatello M, Paolillo L, Ghezzo F, Basso F, Di Bello C, Dettin M, Seidah NG. The proprotein convertase SKI-1/S1P. In vitro analysis of Lassa virus glycoprotein-derived substrates and ex vivo validation of irreversible peptide inhibitors. J Biol Chem. 2006;281(33):23471–81. doi: 10.1074/jbc.M513675200. Epub 2006 Jun 21. [DOI] [PubMed] [Google Scholar]
- Pinschewer DD, Perez M, Sanchez AB, de la Torre JC. Recombinant lymphocytic choriomeningitis virus expressing vesicular stomatitis virus glycoprotein. Proc Natl Acad Sci U S A. 2003;100(13):7895–900. doi: 10.1073/pnas.1332709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pullikotil P, Vincent M, Nichol ST, Seidah NG. Development of protein-based inhibitors of the proprotein of convertase SKI-1/S1P: processing of SREBP-2, ATF6, and a viral glycoprotein. J Biol Chem. 2004;279(17):17338–47. doi: 10.1074/jbc.M313764200. [DOI] [PubMed] [Google Scholar]
- Rawson RB, Cheng D, Brown MS, Goldstein JL. Isolation of cholesterol-requiring mutant Chinese hamster ovary cells with defects in cleavage of sterol regulatory element-binding proteins at site 1. J Biol Chem. 1998;273(43):28261–9. doi: 10.1074/jbc.273.43.28261. [DOI] [PubMed] [Google Scholar]
- Reignier T, Oldenburg J, Noble B, Lamb E, Romanowski V, Buchmeier MJ, Cannon PM. Receptor use by pathogenic arenaviruses. Virology. 2006;353(1):111–20. doi: 10.1016/j.virol.2006.05.018. Epub 2006 Jun 21. [DOI] [PubMed] [Google Scholar]
- Rojek JM, Campbell KP, Oldstone MB, Kunz S. Old World Arenavirus Infection Interferes with the Expression of Functional {alpha}-Dystroglycan in the Host Cell. Mol Biol Cell. 2007a;29:29. doi: 10.1091/mbc.E07-04-0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek JM, Lee AM, Nguyen N, Spiropoulou CF, Kunz S. Site 1 protease is required for proteolytic processing of the glycoproteins of the South American hemorrhagic fever viruses Junin, Machupo, and Guanarito. J Virol. 2008a;82(12):6045–51. doi: 10.1128/JVI.02392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek JM, Pasqual G, Sanchez AB, Nguyen NT, de la Torre JC, Kunz S. Targeting the proteolytic processing of the viral glycoprotein precursor is a promising novel antiviral strategy against arenaviruses. J Virol. 2010;84(1):573–84. doi: 10.1128/JVI.01697-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek JM, Sanchez AB, Nguyen NT, de la Torre JC, Kunz S. Different mechanisms of cell entry by human-pathogenic Old World and New World arenaviruses. J Virol. 2008b;82(15):7677–87. doi: 10.1128/JVI.00560-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek JM, Spiropoulou CF, Campbell KP, Kunz S. Old World and Clade C New World Arenaviruses Mimic the Molecular Mechanism of Receptor Recognition Used by {alpha}-Dystroglycan’s Host-Derived Ligands. J Virol. 2007b;81(11):5685–95. doi: 10.1128/JVI.02574-06. Epub 2007 Mar 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek JM, Spiropoulou CF, Kunz S. Characterization of the cellular receptors for the South American hemorrhagic fever viruses Junin, Guanarito, and Machupo. Virology. 2006;349(2):476–91. doi: 10.1016/j.virol.2006.02.033. [DOI] [PubMed] [Google Scholar]
- Ruiz-Jarabo CM, Ly C, Domingo E, de la Torre JC. Lethal mutagenesis of the prototypic arenavirus lymphocytic choriomeningitis virus (LCMV) Virology. 2003;308(1):37–47. doi: 10.1016/s0042-6822(02)00046-6. [DOI] [PubMed] [Google Scholar]
- Sakai J, Nohturfft A, Goldstein JL, Brown MS. Cleavage of sterol regulatory element-binding proteins (SREBPs) at site- 1 requires interaction with SREBP cleavage-activating protein. Evidence from in vivo competition studies. J Biol Chem. 1998;273(10):5785–93. doi: 10.1074/jbc.273.10.5785. [DOI] [PubMed] [Google Scholar]
- Sanchez A, Pifat DY, Kenyon RH, Peters CJ, McCormick JB, Killet MP. Junin virus monoclonal antibodies: characterization and cross-reactivity with other arenaviruses. J Gen Virol. 1989;70:1125–1132. doi: 10.1099/0022-1317-70-5-1125. [DOI] [PubMed] [Google Scholar]
- Toure BB, Munzer JS, Basak A, Benjannet S, Rochemont J, Lazure C, Chretien M, Seidah NG. Biosynthesis and enzymatic characterization of human SKI-1/S1P and the processing of its inhibitory prosegment. J Biol Chem. 2000;275(4):2349–58. doi: 10.1074/jbc.275.4.2349. [DOI] [PubMed] [Google Scholar]
- Urata S, Yun N, Pasquato A, Paessler S, Kunz S, de la Torre JC. Antiviral activity of a small-molecule inhibitor of arenavirus glycoprotein processing by the cellular site 1 protease. J Virol. 2011;85(2):795–803. doi: 10.1128/JVI.02019-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber EL, Buchmeier MJ. Fine mapping of a peptide sequence containing an antigenic site conserved among arenaviruses. Virology. 1988;164(1):30–8. doi: 10.1016/0042-6822(88)90616-2. [DOI] [PubMed] [Google Scholar]
- Weissenbacher MC, Laguens RP, Coto CE. Argentine hemorrhagic fever. Curr Top Microbiol Immunol. 1987;134:79–116. doi: 10.1007/978-3-642-71726-0_4. [DOI] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6(6):1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]


