Abstract
We develop fast fitting methods for generalized functional linear models. The functional predictor is projected onto a large number of smooth eigenvectors and the coefficient function is estimated using penalized spline regression; confidence intervals based on the mixed model framework are obtained. Our method can be applied to many functional data designs including functions measured with and without error, sparsely or densely sampled. The methods also extend to the case of multiple functional predictors or functional predictors with a natural multilevel structure. The approach can be implemented using standard mixed effects software and is computationally fast. The methodology is motivated by a study of white-matter demyelination via diffusion tensor imaging (DTI). The aim of this study is to analyze differences between various cerebral white-matter tract property measurements of multiple sclerosis (MS) patients and controls. While the statistical developments proposed here were motivated by the DTI study, the methodology is designed and presented in generality and is applicable to many other areas of scientific research. An online appendix provides R implementations of all simulations.
1. INTRODUCTION
Unarguably, advancements in technology and computation have led to a rapidly increasing number of applications where measurements are functions or images. These developments have been accompanied and, in some cases, anticipated by intense methodological development in regression models where some covariates are functions (James 2002; Cardot, Ferraty, and Sarda 2003; Cardot and Sarda 2005; James and Silverman 2005; Müller 2005; Ramsay and Silverman 2005; Ferraty and Vieu 2006; Reiss and Ogden 2007; Crainiceanu, Staicu, and Di 2009). In this article we develop a novel inferential approach to functional regression. Our goals are to: (1) reduce the number of tuning parameters used in functional regression; (2) increase the spectrum of models and applications where functional regression can be applied automatically; and (3) produce software that is fast and easy to generalize to more complex data and models. These goals are achieved by smoothing the covariance operators, using a large number of eigenvectors to capture the variability of the functional predictors, and modeling the functional regression parameters as penalized splines. The level of smoothing is estimated using Restricted Maximum Likelihood (REML) in an associated mixed effects model. Methods are implemented using standard mixed effects software.
Several important advantages of our penalized functional regression (PFR) approach are that (1) it provides a unified framework for functional regression in many settings, including when functions are measured with error, at equal or unequal intervals, at a dense or sparse set of points, and to multiple functional regressors observed at one or multiple levels; (2) it is computationally efficient compared to other penalized approaches and can be fit using standard software; (3) it is automated in that the smoothing parameter is estimated as a variance component in a mixed effects model, avoiding manual selection or a cross-validation procedure; and (4) confidence intervals based on mixed effects inferential machinery are readily constructed. Moreover, our methods apply to outcomes distributed in the exponential family class of models.
Briefly, functional regression seeks to quantify the relationship between a scalar outcome and a functional regressor. To illustrate the main ideas, we start with the simple example when univariate functional data are measured at a single level. More specifically, assume that for each subject, i = 1, …, I, we observe data [Yi, Xi(t), Zi], where Yi is a scalar outcome, Xi(t) ∈
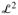 [0, 1] are random functions, and Zi is a vector of on functional covariates. We call Xi(t) “univariate” functional data because in this example we only consider one functional regressor. The case of multivariate functional regressors is considered in Section 3.1. Moreover, we call Xi (t) a “single-level” sample of random functions, because only one function, Xi (t), is sampled per subject. A multilevel or clustered case is considered in Section 3.2. The generalized functional linear model relating Yi to the covariates Xi(t), Zi is given by (McCullagh and Nelder 1989; Cardot and Sarda 2005; Müller and Stadtmüller 2005)
[0, 1] are random functions, and Zi is a vector of on functional covariates. We call Xi(t) “univariate” functional data because in this example we only consider one functional regressor. The case of multivariate functional regressors is considered in Section 3.1. Moreover, we call Xi (t) a “single-level” sample of random functions, because only one function, Xi (t), is sampled per subject. A multilevel or clustered case is considered in Section 3.2. The generalized functional linear model relating Yi to the covariates Xi(t), Zi is given by (McCullagh and Nelder 1989; Cardot and Sarda 2005; Müller and Stadtmüller 2005)
| (1.1) |
Here EF(μi, η) denotes an exponential family distribution with mean μi and dispersion parameter η, g(·) is a link function, and β(t) ∈
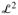 [0, 1]. The functional regression model is a powerful and practical inferential tool, in spite of the fact that observations Xi (t) are never truly functional. Rather, we observe {Xi(tij): tij ∈ [0, 1]}, with j = 1, …, Ji. Further, the regressor functions are often measured with error; that is, one often measures a proxy functional covariate, Wi(t) = Xi(t) + εi(t), where
. Thus, for subject i, data typically are of the form [Yi, {Wi (tij): tij ∈ [0, 1]}, Zi], i = 1, …, I, j = 1, …, Ji. In practice, functional data will have various sampling schemes. For example, tij, j = 1, …, Ji could be equally or unequally spaced for each subject, sparse at the subject level and dense at the population level, or dense at the subject and population level. The functions Xi (t) can be measured with no, moderate, or large measurement error.
[0, 1]. The functional regression model is a powerful and practical inferential tool, in spite of the fact that observations Xi (t) are never truly functional. Rather, we observe {Xi(tij): tij ∈ [0, 1]}, with j = 1, …, Ji. Further, the regressor functions are often measured with error; that is, one often measures a proxy functional covariate, Wi(t) = Xi(t) + εi(t), where
. Thus, for subject i, data typically are of the form [Yi, {Wi (tij): tij ∈ [0, 1]}, Zi], i = 1, …, I, j = 1, …, Ji. In practice, functional data will have various sampling schemes. For example, tij, j = 1, …, Ji could be equally or unequally spaced for each subject, sparse at the subject level and dense at the population level, or dense at the subject and population level. The functions Xi (t) can be measured with no, moderate, or large measurement error.
Of interest are all the parameters of model (1.1) including the function β(·), which characterizes the relationship between the transformed mean of Y and the covariate of interest X(·). Because of some hesitation to adopt model (1.1), on the part of both scientific collaborators and statisticians, we feel it is worth explaining the interpretation of the integral appearing there. Consider a coarse partition T = {T1, …, TG} of the interval [0, 1], and for the moment assume β(t) = βj for t ∈ Tj; that is, assume β(t) is a step function on T. Then
where X̄ij is the mean of Xi (t) on Tj. An extreme example takes β(t) = β if t ≤ 0.5 and 0 otherwise. In this case and model (1.1) becomes a standard regression model which contains the average functional covariate over the interval [0, 0.5] as a regressor. Considered this way, β(·) provides weights that are applied to all subject level functions in the same manner. Intuition for the integral in model (1.1) is enhanced by contemplating finer and finer partitions T. In practice it makes sense to consider smooth transitions in the weighting scheme, that is, a smooth β(·) function. Thus, we think of β(·) as the smooth weighting scheme which, when applied to the subject-specific predictors Xi (·), is most predictive of the outcome. Weights close to zero de-emphasize subject-level areas that are not predictive of the outcome, while large relative weights emphasize areas of the curve that are most predictive of the outcome.
Our proposed approach to estimating the coefficient function β(t) has two steps (the following uses notation from Ramsay and Silverman 2005, chap. 15). First, we estimate the random functions using a finite series expansion , where ψ = {ψ1(t), …, ψKx(t)} is the collection of the first Kx eigenfunctions of the smoothed covariance matrix ΣX(s, t) = cov[Xi (s), Xi (t)]. Second, we use a truncated power series spline basis φ(t) = {φ1(t), …, φKb(t)}for β(t), so that β(t) = φ (t)b. The truncated power series representation of β(t) imposes differentiability and allows simple control of smoothness. The tuning parameters, Kx and Kb, are considered to be very important in practice and their choice has been extensively debated in the functional and smoothing literature, respectively. In the functional regression literature, the choice of Kx is particularly important when using a low-dimensional approach that uses ψ for both the predictors and the coefficient: it must be large enough that β(t) is in the space spanned by {ψ1(t), …, ψKx(t)} but small enough to smoothly estimate β(t). In the smoothing literature, the choice of the number of knots in φ has been shown (Ruppert 2002; Li and Ruppert 2008) to be unimportant as long as it is large enough to capture the maximum complexity of the regression function: in penalized spline regression it is the explicit smoothness constraint that takes care of reducing the variability of the functional estimate and avoids the heavy computational costs associated with choosing the number and positions of knots. We emulate this principle from the smoothing literature in the current functional setting and choose Kb large; Kx is chosen large enough to satisfy the identifiability constraint Kx ≥ Kb. Choosing a large Kx avoids having to choose a “good” number of principal components (PCs). Once the bases for Xi (t) and β(t) and the parameters Kx and Kb have been selected, model (1.1) may be expressed as a generalized linear mixed effects model (GLMM); thus the GLMM inferential machinery can be applied.
Our methods are most closely related to the functional regression framework developed by Cardot, Ferraty, and Sarda (2003), Cardot and Sarda (2005), who proposed a penalized spline to estimate the functional parameter. We incorporate this idea but expand its scope to functions Xi (t) that are measured with error or are sparsely sampled; this is achieved by using a PC basis to expand Xi (t). We also greatly improve the computation time associated with fitting a penalized model by taking advantage of the link to mixed effects models and existing well-tested software. The same ideas can be extended seamlessly to functional regression when the exposure proxy has a multilevel structure (Crainiceanu, Staicu, and Di 2009; Di et al. 2009). Thus, by decoupling the estimation of the functional predictors and the coefficient function, and by exploiting the connection to mixed models, our approach leads to straightforward extensions and direct integration with other functional regression settings. Importantly, the connection to mixed models is further exploited to develop confidence intervals: while Müller and Stadtmüller (2005) derived confidence intervals for a low-dimension approach to functional regression and others (Reiss and Ogden 2007; James, Wang, and Zhu 2009) discussed empirical or bootstrap confidence intervals, we are unaware of existing explicit derivations of confidence intervals based on a penalized approach to functional regression.
It is important to distinguish our use of the PC decomposition from the widely used functional principal components regression (FPCR) techniques (Cardot, Ferraty, and Sarda 1999; Reiss and Ogden 2007). Stated shortly, FPCR regresses the vector of scalar outcomes Y on the design matrix XVKx, where X has ith row [Xi (t1), …, Xi(tT)], VKx is the truncated at Kx version of the matrix V in UDVT, the singular value decomposition of X. That is, FPCR regresses Y on the first Kx PC loadings of the functional regressors. Low-dimension FPCR techniques choose Kx either by hand or according to a rule; the resulting estimates can be very sensitive to this choice, and may be poor if β(t) is not in the space spanned by the relatively few PCs. High-dimension penalized FPCR approaches often employ an explicit penalization constraint and choose Kx via cross-validation, which is computationally expensive and does not completely alleviate the concern that β(t) is not in the space spanned by the basis. In our simulations we compare our method to both low-and high-dimensional FPCR approaches. In contrast, we use the PC decomposition only to provide estimates of the functional covariates using a small number of eigenfunctions. Indeed, using decompositions of the functional covariates in terms of other bases in the PFR method is straightforward.
The article is organized as follows. Section 2 provides the details of our approach to functional regression. Section 3 describes the seamless generalization to multiple functions, clustered functions, and to sparse functional data. Section 4 provides a detailed simulation to compare several methods in the univariate setting and explores the coverage probabilities of confidence intervals. We apply our method to the DTI data in Section 5, and conclude with a discussion in Section 6. Additional simulations for the multivariate, multilevel, and sparse functional data are found in an online appendix. To ensure reproducibility of our results we post code for all simulations at http://biostat.jhsph.edu/~jgoldsmi/Downloads/Web_Appendix_PFR.zip.
2. APPROACH
In this section we describe the PFR method for estimating the functional exposure effect β(t). We focus first on estimating the subject-specific functional effect, Xi (t), and then we describe the estimator of β(t) and its variability, respectively.
2.1 Estimation of Xi (t)
The first step in our analysis is to estimate, or predict, Xi (t) in model (1.1) using an expansion into the PC basis obtained from its covariance operator, ΣX(·, ·). As mentioned in Section 1, choosing the number of components is often viewed as both important and difficult; here we elect to use a large number of PCs and largely avoid this issue. This refocuses the problem on estimating ΣX(·, ·), which is a much simpler problem.
Assume that instead of observing Xi (t) one measures a proxy Wi (t) = Xi (t) + εi (t), where . The covariance operator for the observed data is , where ΣW (s, t) = cov{Wi (s), Wi (t)} is the covariance operator on the observed functions, ΣX(s, t) = cov{Xi (s), Xi (t)}, and δts = 1 if t = s and is 0 otherwise. This suggests the following strategy for estimating ΣX(s, t). First, construct a method of moments estimator Σ̂W (s, t) of ΣW (s, t) from the observed data. Second, smooth Σ̂W (s, t) for s ≠ t, as suggested by Staniswalis and Lee (1998), Yao et al. (2003). The only serious problem we encountered in practice occurred when the functions Xi (t) are unevenly or sparsely sampled. Consider the case when each pair of sampling locations, (tik, til), is unique. In this situation ΣW(tik, til) is estimated by {Wi (tik) − Wi (til)}2/2; the number of pairs (tik, til) can quickly explode making bivariate smoothing of the estimated covariance matrix difficult. To avoid this problem we use the ideas suggested by Di et al. (2009) to estimate ΣW (s, t):
Use a very small bandwidth smoother to obtain a rough estimate of the covariance operator.
Use a fast automatic nonparametric smoother of the undersmoothed surface obtained at the previous step.
Let be the spectral decomposition of Σ̂X(s, t), where λ1 ≥ λ2 ≥ ··· are the nonincreasing eigenvalues and ψ(·) = {ψk (·): k ∈ ℤ+} are the corresponding orthonormal eigenfunctions. An approximation for Xi (t), based on a truncated Karhunen–Loéve decomposition, is given by , where Kx is the truncation lag and . Unbiased estimators of cik are easy to obtain as the Riemann sum approximation to the integral ; for example, was proposed by Müller and Stadtmüller (2005). This method works well when data are densely sampled and each subject-specific function is sampled at many points Ji. When this is not the case a better alternative is to obtain best linear unbiased predictors (BLUP) or posterior modes in the mixed effects model (Crainiceanu, Staicu, and Di 2009; Di et al. 2009)
| (2.1) |
where cik and εij are mutually independent for every i, j, k. The subject-specific processes Xi (t) are then predicted at any t by plugging in the predictors of cij in the equality . A potential criticism of this method is that cij could be predicted with sizeable error which can lead to sizeable variability in the prediction of Xi (t). In some situations this can lead to bias in the functional regression, as discussed by Crainiceanu, Staicu, and Di (2009). When this problem is a real concern a solution is to jointly model the outcome model and model (2.1). This approach can be addressed using a fully Bayesian analysis (Crainiceanu and Goldsmith 2010; Goldsmith et al. 2010) and is not the focus of this article. Instead, we focus on the two-stage approach, which is the current state of the art in functional regression.
We emphasize that the PC decomposition in the first step of our analysis is used to estimate the Xi (t) when they are measured with error or sparsely sampled, rather than to address the ill-posed nature of the functional regression, as in FPCR. Thus, we focus on the problem of estimating β(t) using a method that does not depend on the particular choice of number of principal components, a nontrivial distinction.
2.2 Estimation of β(t)
The second step in our method is modeling β(t) and we borrow ideas from the penalized spline literature (O’Sullivan, Yandell, and Raynor 1986; Ruppert, Wand, and Carroll 2003; Wood 2006). Let φ(t) = {φ1(t), φ2(t), …, φKb(t)} be a spline basis, so that , where b = {b1, …, bKb}T. Thus, the integral in model (1.1) becomes
where and Jψφ is a Kx × Kb dimensional matrix with the (k, l)th entry equal to (Ramsay and Silverman 2005).
It would be mathematically simpler to expand β(·) in the principal component basis used for expanding the functional data, ψ1(·), …, ψKx (·). In spite of its apparent appeal, this approach is not satisfactory in many applications. The main technical reasons are that: (1) the principal component basis is typically not a parsimonious basis for the smooth parameter function; (2) the smoothing of the β(·) function is implicitly controlled by Kx, the smoothing parameter for the functional process, Xi (t), and can be very sensitive to the choice of Kx; and (3) the choice of Kx is typically undertaken either by hand, which can be subjective, or via cross-validation, which is computationally expensive. Thus, we use the truncated power series spline basis expansion where are knots. We explicitly induce smoothing by assuming that . Other choices of basis functions can be used with corresponding changes to the penalty matrix.
Denote by C the I × Kx dimensional matrix of PC loadings with ith row equal to and by Z the I × p dimensional matrix with the ith row equal to Zi. The outcome model (1.1) can be reformulated in matrix format as
| (2.2) |
which is a mixed effects model with Kb − 2 random effects, . This model can be fit robustly using standard mixed effects software (Ruppert 2002; McCulloch, Searle, and Neuhaus 2008).
Intuition for the connection between the mixed model representation of the functional regression model and the induced smoothness of β̂(t) can be built most easily when the outcome Yi is normally distributed. In this case, maximization of the likelihood of (Y, b) over the unknowns (α, b, γ) is equivalent to minimizing
This expression contains an explicit penalty on the spline terms . Moreover, the mixed model framework allows us to write the joint likelihood of all model parameters (including the smoothing parameter ) and to estimate these parameters using maximum likelihood and restricted maximum likelihood techniques.
Once the basis for β(t) is chosen, model (2.2) depends on the choice of Kb and Kx. Following Ruppert (2002) we select Kb large enough to prevent undersmoothing and, to satisfy the identifiability constraint, select Kx ≥ Kb. A rule of thumb is to choose Kb = Kx = 35; however, the specific value of Kb is unimportant as long as it is large enough to capture the maximum variability in β(t). The position of the knots in the truncated power series spline basis is typically unimportant and we place them at the quantiles of the distribution of tij.
It is important to note that the smoothing parameter is estimated as a variance component in the mixed effects model. Therefore, the only tuning parameters chosen by the user are Kb, Kx; as long as these are chosen large enough, their specific value has little impact on estimation. Thus, the procedure is highly automated and robust to changes in the selection of the tuning parameters.
We also point out that complexity of fitting model (2.2) is the same as the complexity of fitting a penalized spline model with Kb random coefficients, a well-researched problem with well-developed accompanying software (Wood 2006). In our simulations, we use the nlme package in R to fit model (2.2); this package uses first a moderate number of EM iterations to refine starting values of the variance components followed by a Newton–Raphson optimization of the restricted log-likelihood (Bates and Pinheiro 1998; Pinheiro et al. 2009; R Development Core Team 2009). Because our approach takes advantage of established mixed model theory and software, it is computationally efficient compared to penalized approaches to functional regression that employ a cross-validation step to choose the smoothing parameter.
2.3 Confidence Intervals for β(t)
Because model (2.2) is a mixed effects model the typical inferential machinery for mixed effects models can be used to obtain variance–covariance estimates of the model parameters. Variance estimators, pointwise and joint confidence intervals can be obtained following standard methods and software (Ruppert, Wand, and Carroll 2003; Wood 2006).
For illustration, consider the case when with . Take as the basis for β(t) the functions 1, t, (t − κ2)+, …, (t − κKb)+. Let β = [α bT]T; it is easy to show that β̂ = (WT R−1W+B)−1WT Y, where W = [1 CJψφ],
, and . The variance components can be estimated via REML in the corresponding mixed effects model (2.2). Recall that β(t) = φ(t)bT. One can show that
Then, at any t0 var[β̂(t0)] = var[φ(t0)b̂T] = φ(t0)Σbbφ(t0)T; we estimate , where Σ̂bb is the (Kb) × (Kb) dimensional matrix obtained by plugging the REML estimates for the variance components into the formula for var[β̂]. An approximate 95% confidence interval for E[β̂(t0)] can be constructed as .
We point out some limitations of this derivation. The first is that the confidence intervals will perform poorly in regions where β̂ (t) oversmooths the true coefficient function and is therefore biased; a possible solution would be an adaptive smoothing approach, which we do not consider here. A second limitation is that we neglect the variability in estimating the PC loadings in C; in many applications, this variability is minimal but, as pointed out by a reviewer, could be substantial when the regressors are observed sparsely at the subject level or with error. One solution would consider modeling the PC loadings and the coefficient function jointly, as in the works of Crainiceanu and Goldsmith (2010) and Goldsmith et al. (2010), and using instead the 95% posterior credible interval for β(t).
3. EXTENSIONS
As discussed earlier, a strength of the proposed method is the wide applicability that stems from treating the estimation of β(t) and the Xi (t) separately. In this section, we demonstrate this flexibility by exploring extensions to several common functional regression settings.
3.1 Multivariate Extensions
In this section, we extend our model to the case of multiple functional regressors. Suppose our observed data for subject i is of the form [Yi, Zi, {Wil (t), t ∈ [0, 1]}], where Yi is continuous or discrete, Zi is a vector of covariates, and Wil (t), 1 ≤ l ≤ L, are the observed proxies for the true functional regressors Xij (t). We emphasize that the Xil (t) are distinct functional regressors; a notationally similar—but conceptually different—setting is considered in the next section. A multivariate extension of our regression model (1.1) is given by
| (3.1) |
The approach given in Section 2 extends naturally to the multivariate functional regression setting. That is, we once again treat the estimation of the Xil (t) and of the βl (t) as separate. First, approximate each functional regressor using the truncated Karhunen–Loéve decomposition, , where ψl (·) = {ψkl (·): 1 ≤ k ≤ Kx} are the first eigenfunctions of the smoothed estimated covariance operator . Then, express each coefficient function in model (3.1) in terms of a spline basis φ (t) = {φ1(t), φ2(t), …, φKb(t)}, so that . We therefore have that
where and Jl is a Kx × Kb dimensional matrix with the (k, m)th entry equal to . As in Section 2.2, we assume the use of a truncated power series spline basis for each βl (t) and induce smoothness on the estimate of βl (t) by assuming .
The multivariate functional regression model can be expressed as the mixed effects model
| (3.2) |
with Kb − 2 random effects, , for each functional coefficient βl(t)and can be fit using standard mixed models software.
Note that we express each coefficient function in terms of the same spline basis; indeed, we typically use the truncated power series basis introduced in Section 2.2 for each βl (t). However, different bases could be used for each function. Using φl(t) = {φ1l (t), φ2l (t), …, φKbl(t)} as the basis for βl (t), the matrix Jl has (k, m)th entry equal to ; all other aspects of the multivariate regression model remain the same.
3.2 Multilevel Extensions
Here, we briefly describe an extension of our method to a multilevel setting based on the work of Crainiceanu, Staicu, and Di (2009). This extension is informative in that although the estimation of the (unobserved) Xi (t) is difficult, the same framework that we have developed applies unchanged here. That is, first we estimate the Xi (t) based on a functional principal components decomposition, then we estimate β(t) using a rich spline basis and an explicit smoothing parameter controlled via a mixed model.
Suppose for subject i we observe [Yi, Zi, {Wij (t), t ∈ [0, 1]}], where Yi is continuous or discrete, Zi is a vector of covariates, and Wij (t) is the observed functional regressor at visit j = 1, 2, …, Ji. We assume that Wij (t) is a proxy for the true underlying subject-specific function Xi (t), so that Wij (t) = μ(t) + ηj (t) + Xi (t) + Uij (t) + εij (t). Here μ(t) is the overall mean function, ηj (t) is the visit-specific deviation from the overall mean, Xi (t) is subject i’s deviation from the visit-specific mean function, Uij(t) is the remaining subject- and visit-specific deviation for the subject-specific mean, and . We further assume that Xi (t), Uij (t), and εij (t) are uncorrelated to guarantee identifiability. We construct μ̂(t) = W̄··(t) and η̂j(t) = W̄··(t) − W̄·j(t), where W̄··(t) is the mean taken over all subjects and visits and W̄·j(t) is the mean taken over all subjects at visit j. Assume these estimates have been subtracted, so that Wij(t) = Xi(t) + Uij(t) + εij(t).
We use model (1.1) as our outcome model, so that the outcome Yi depends on the subject-specific mean function Xi (t). The multilevel approach proceeds analogously to the single-level approach. First, we express the subject-specific function Xi (t) in terms of a parsimonious basis that captures most of the variability in the space spanned by the regressor functions. Second, we express the coefficient function β(t) using a truncated power series spline basis. Finally, we take advantage of the mixed models framework to construct a smooth estimate β̂(t).
We use Multilevel Functional Principal Components Analysis (MFPCA) (Crainiceanu, Staicu, and Di 2009; Di et al. 2009) to construct parsimonious bases for Xi (t), Uij (t). Following these articles, we first note that under certain assumptions the covariance operators ΣX = cov[Xi (s), Xi (t)] and ΣU = cov[Uij(s), Uij(t)] are given by cov[Wij(s, t), Wik (s, t)] and cov[Wij (s, t), Wij (s, t)] − cov[Wij(s, t), Wik(s, t)], respectively. Next, we calculate the spectral decompositions and , where and are the ordered eigenvalues and are the corresponding orthonormal eigenfunctions.
The Karhunen–Loéve decomposition is used to provide the finite series approximations and , where Kx and Lx are the truncation lags and are the PC scores with E[cik] = E[ζijk] = 0, , for every i, j, k. As in the article by Crainiceanu, Staicu, and Di (2009), we estimate cik, ζijk using the mixed model
| (3.3) |
| (3.4) |
Using the same notations as in the case of single-level regression, the functional predictor becomes
Thus, the outcome model is identical to model (2.2), with the only difference that Xi (·) are estimated using the MFPCA instead of the FPCA method. Penalized spline regression modeling is employed for modeling β(t) and mixed models software is used.
This development is related to the one proposed by Crainiceanu, Staicu, and Di (2009). Specifically, the method of Crainiceanu, Staicu, and Di (2009) uses MFPCA to construct a parsimonious basis for Xi (t) and uses a mixed model to estimate the PC loadings cik. Similarly to FPCR, the PC loadings are then treated as the regressors in a generalized linear model. In contrast, our method estimates β(t) using a truncated power series spline basis and penalized regression to construct a smooth estimate β̂(t). This method is flexible and was found to be superior in both standard simulation settings and applications.
3.3 Sparse Data
Our method also extends to the case where the functional regressor is measured sparsely at the subject level, but is dense across subjects. In this situation, we observe data of the form [Yi, {Wi (tij) : tij ∈ [0, 1]}, Zi], i = 1, …, I, j = 1, …, Ji, where Ji are small but is dense in [0, 1]. Here again, the Wi (t) are measured-with-error proxies for the true Xi(t) so that Wi(t) = Xi(t) + εi(t), where . We again point out that while some care must be taken in the estimation of the Xi (t), the same general procedure that has been used elsewhere applies here.
We use the following method, adapted from the work of Di et al. (2009), to estimate the subject-specific functional regressors based on a PC decomposition of the covariance operator ΣX(s, t). As indicated in Section 2.1, we first use a fine grid of points on [0, 1] to obtain an undersmooth of the observed covariance matrix. Call the points in this grid t1, …, ts, and for each subject let tij s be the point in this grid nearest to the observed point tij. The covariance operator can be roughly estimated using , where X̄(t) is the mean of observed functions at t, I (r, s) is the index of subjects with observed points corresponding to both tr, ts, and N(r, s) is the number of such subjects. We then smooth the off-diagonal elements of this rough covariance matrix to obtain Σ̂X(r, s), the estimated covariance operator of the sparsely observed subject-specific functional regressors on our newly defined grid t1, …, ts.
As before, the function Xi (t) is approximated by , where Kx is the truncation lag, ψ(·) = {ψk (·): 1 ≤ k ≤ Kx} are the first eigenfunctions of the smoothed estimated covariance operator, and cik are the subject-specific PC loadings. Because subject-level data are sparse, numeric integration does not yield satisfactory estimates of the cik. Instead, in this case we propose the following mixed model to describe the observed data:
where μ(t) is the mean function estimated across subjects. Here, the PC loadings are random effects and can be estimated using best linear unbiased predictions (BLUPs) or other standard inferential procedures.
Using the same notation as in other settings, the integral in the linear predictor of model (1.1) has the matrix representation . Because of the sparseness of the subject-level data, it is often necessary to reduce the number of knots used in the spline basis for β(t) and the number of PCs used to explain the variability in the Xi (t). In practice, we have found that KX = Kb = 10 typically suffices. Penalized spline regression using mixed models can be used to fit this sparsely sampled functional regression model.
4. SIMULATION
In this section, we pursue a simulation study to explore the viability of our method in the univariate functional regression setting. We compare our approach to several others, including functional principal components regression and penalized approaches, under several true coefficient functions and with varying levels of measurement error. We also include brief results regarding confidence interval performance. Additional simulations, in the multivariate, multilevel, and sparsely observed settings, are available in the online supplementary materials; the results are very similar to those in the univariate case. All code used to conduct simulations is also available online.
4.1 Univariate Simulations
We begin by investigating performance in the simplest situation—a single-level, single functional regressor model, with a continuous outcome and no nonfunctional covariates. Consider the grid { : g =0, 1, …, 100} on the interval [0, 10]. We generate scalar outcomes Yi and regressor functions Xi (t) from the following model:
| (4.1) |
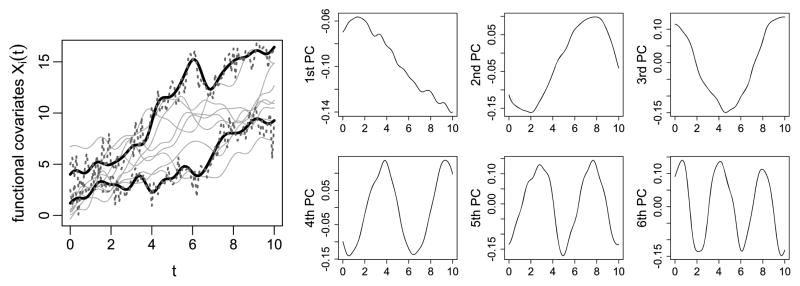
where , ui1 ~ N[0, 25], ui2 ~ N[0, 0.04], and vik1, vik2 ~ N[0, 1/k2]. For reference, Figure 1 displays a sample of 10 random functions Xi(t) as well as the first six principal components estimated from the PC decomposition of the functions. This method of generating the regressor functions Xi (t) is adapted from the work of Müller and Stadtmüller (2005). The first few PCs of the Xi (t) capture a slope on t and sine and cosine functions with one, two, and three periods on the range of t. In generating the observed functions Wi (t) we consider , and in generating the observed outcomes, we consider and three true coefficient functions β(·), yielding 12 possible parameter combinations. The choices of the coefficient functions β(·) are described below.
Figure 1.
The left panel displays a sample of 10 random functions generated from (4.1), highlighting two examples of the function measured with no error (solid) and with measurement error (dashed). The right panel displays plots of the first six estimated principal components. The online version of this figure is in color.
For each combination of the parameter values , and β(·), we simulate 1000 datasets [Yi, Wi (tg) : i = 1, …, 200]. We compare four alternative approaches to estimating β(·) to our approach as described in Section 2. Performance in estimating β(·) is compared by calculating the average mean squared error (AMSE) over the 1000 samples as
where β̂r (·) is the estimated coefficient function from the rth simulated dataset.
The first method for estimating β(·) is principal components regression (PCR). Let X be the 200 × G matrix with ith row (Xi (t1), …, Xi (tG)) and calculate the singular value decomposition UDVT of X. In PCR, the scalar outcomes Y are regressed on VA, the 200 × A matrix containing the first A columns of V, that is, the first A principal components of X. We consider two commonly used approaches for selecting A: cross-validation (PCR-CV) and percent variance explained (PCR-PVE) with a 99% threshold. For PCR-CV we implement the leave-one-out cross-validation procedure to select the number of principal components A for which the prediction sum of squares criterion is minimized. Here Ŷ−i is the predicted value for the ith data point obtained from fitting the PCR model to the data with the ith observation deleted. Though this procedure is computationally intensive, we implement a faster alternative formulation for the statistic for a linear model, namely,
where Hii is the ith diagonal element of the regression projection matrix and Ŷi is the ith fitted value. For PCR-PVE with a 99% threshold, we select the value of A satisfying
where λa is the eigenvalue corresponding to the ath principal component of X. Thus we interpret A as the minimal number of principal components needed to explain 99% of the total variation in the discretized versions of the random functions Xi (t). Further, in the presence of measurement error, we use smoothed principal components.
The third method, FPCRR (Reiss and Ogden 2007), first projects the random functions Xi(t) onto a B-spline basis and then performs a principal components analysis on the projection XB. A penalized regression model is then fit to find ξ that minimizes the criterion
where VA is the first A columns of V from the singular value decomposition XB = UDVT and P is the penalization matrix such that approximately equals the integrated squared second derivative of the coefficient function . Given a particular value of A, the smoothing parameter λ may be selected either through GCV or by representing the penalized regression in a LMM framework and using the REML estimate. The number of principal components A is selected by multi-fold cross-validation. We implement this method using code provided by the authors, which utilizes the REML estimate, a cubic B-splines basis with 40 equally spaced internal knots, and selects the number of principal components A using 8-fold cross-validation. The candidates for A are 1–10, 12, and 15–40 at intervals of 5.
Finally, we implement the method SPCR-GCV (Cardot, Ferraty, and Sarda 2003), using code provided by the authors. This approach first computes the PCR estimate using the first K principal components of the Xi (t) and then smooths the resulting function using penalized splines. In this approach, both the dimension K of the principal components basis and the smoothing parameter ρ are selected using generalized cross-validation (GCV). The number of knots of the B-spline basis and the degree of the spline functions were fixed at 20 and 4, respectively. We consider the same candidates for K as were used to select the number of principal components (A) in the implementation of FPCRR described above, and the candidates for ρ were 10−8 to 10−7 by intervals of 10−8, 10−7 to 10−6 by intervals of 10−7, and 10−6 to 10−5 by intervals of 10−6. We selected this range in order to contain the minimum GCV values for each of the three true β(·) functions. We note that without this manual tuning the method fails to work well.
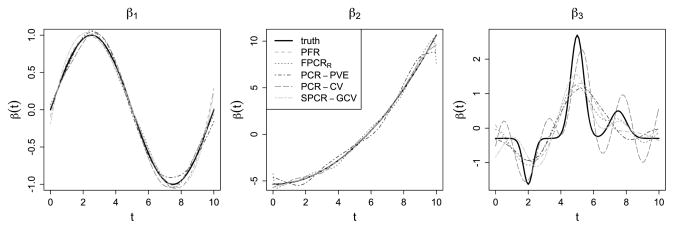
The true coefficient functions we consider in our simulations are β1(t) = sin(πt/5), β2(t) = (t/2.5)2, and β3(t) = −p(t | 2, 0.3) + 3p(t | 5, 0.4) + p(t | 7.5, 0.5), where p(· | μ, σ) is the normal density with mean μ and standard deviation σ. The function β1(t) was selected because it is one of the functions used to generate the random functions Xi (t), and is expected to favor methods that use the principal components basis for β(t). Both PCR methods (CV and PVE) and FPCRR use the principal components as a basis for the unknown β(·). The second coefficient function was chosen as an arbitrary and realistic smooth coefficient function. The third has spikes at places where the variability in the Xi (t) is low, meaning that the peaks will be very hard to detect with small sample sizes; we expect it will be difficult to estimate for all of the approaches used here.
Table 1 compares the AMSE for each set of the parameters across approaches. The function β1 was selected because it is a basis function for the Xi(t), so the methods that use the principal components as a basis for β(t) (PCR-CV, PCR-PVE, FPCRR, and SPCR-GCV) are expected to perform well. However, our method performs only slightly worse than both the FPCRR and PCR-PVE methods when and performs comparably well or slightly better when ; our method also has less than half the AMSE for β1 as the SPCR-GCV and PCR-CV approaches, with and without measurement error on the Xi (t). For the smooth β2, our approach outperformed SPCR-GCV which, in turn, performed much better than the other approaches. As expected, none of the methods performed particularly well for the third true coefficient function β3; the FPCRR and SPCR-GCV methods provide the closest estimates, with our method performing slightly worse. In Figure 2 we plot the estimated beta functions from each approach that have the median AMSE for the case where and . This plot reiterates the comparable performance across methods for β1, the superiority of our approach as well as SPCR-GCV for the smooth β2, and the relatively poor performance across all methods for β3.
Table 1.
Average MSE over 1000 repetitions for each combination of the true coefficient function β(t), the measurement error variance , and the outcome variance .
| Method |
β1(·)
|
β2(·)
|
β3(·)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| PFR | |||||||||||
| 0.0023 | 0.0037 | 2e–04 | 5e–04 | 0.19 | 0.234 | ||||||
| 0.0034 | 0.0044 | 0.0607 | 0.0607 | 0.27 | 0.282 | ||||||
| FPCRR | |||||||||||
| 0.002 | 0.0025 | 0.207 | 0.239 | 0.15 | 0.194 | ||||||
| 0.0054 | 0.0064 | 0.715 | 0.716 | 0.254 | 0.264 | ||||||
| SPCR-GCV | |||||||||||
| 0.0049 | 0.0084 | 0.0071 | 0.0114 | 0.158 | 0.178 | ||||||
| 0.0076 | 0.0103 | 0.171 | 0.173 | 0.245 | 0.256 | ||||||
| PCR-PVE | |||||||||||
| 0.002 | 0.0032 | 0.264 | 0.266 | 0.289 | 0.29 | ||||||
| 0.0035 | 0.0045 | 0.384 | 0.385 | 0.302 | 0.303 | ||||||
| PCR-CV | |||||||||||
| 0.0613 | 0.114 | 0.138 | 0.235 | 0.331 | 0.52 | ||||||
| 0.0105 | 0.0136 | 0.479 | 0.466 | 0.258 | 0.27 | ||||||
Figure 2.
For the simulation with and , we plot the estimated beta functions β̂1, β̂2, and β̂3 from each method that have the median AMSE. The online version of this figure is in color.
Another consideration in fitting functional models is computation time, particularly as the sample size n increases. To compare computation time in our approach to that in the FPCRR and SPCR-GCV approaches, we examine the case where and . To investigate how much computation time increases as sample size increases, we considered n = 100, 200, 400, and 2000. For each n, we generated a single dataset [Yi, Wi (tg) : i = 1, …, n] with true coefficient function β1 and fit each model 10 times. The average computation time for a single fit across the three methods is displayed in Table 2. The driver behind increasing computation times as sample size increases is implementation of a cross-validation or generalized cross-validation procedure. In the FPCRR method, 8-fold cross-validation selects the number of principal components A. Though generalized cross-validation reduces the computational burden of cross-validation, the SPCR-GCV approach has a nested GCV procedure, leading to a large increase in computation time as the sample size n doubles. It should not be surprising that such computational problems would snowball in more complex settings. In fact, we are unaware of competing penalized approaches that have been generalized to each of the settings considered in this article. The computational issues pointed out above are a possible reason for this.
Table 2.
Mean computation time (seconds) over 10 model fits by sample size and regression approach, for , and β(t) = β1(t)
| n | PFR | SPCR-GCV | FPCRR |
|---|---|---|---|
| 100 | 0.111 | 2.451 | 16.720 |
| 200 | 0.126 | 4.536 | 18.545 |
| 400 | 0.157 | 13.330 | 26.070 |
| 2000 | 0.390 | 231.214 | 57.469 |
4.1.1 Confidence Intervals
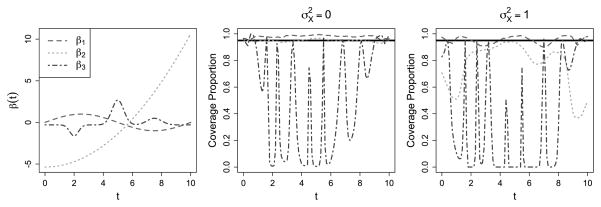
We evaluate the performance of 95% pointwise confidence intervals for β̂ (t) for our approach, using the methodology described in Section 2.3 for each of the three true β(t). For each point tg along the range [0, 10], let (lg, ug) denote the estimated 95% confidence interval about β̂(tg). We compute the proportion of times during the 1000 iterations of the simulation that the calculated interval (lg, ug) contains the truth β(tg). These proportions are displayed in Figure 3 for the cases where with and without measurement error on the Xi(t); taking yields similar confidence interval coverage probabilities.
Figure 3.
For the simulations with , a plot of the pointwise coverage probabilities for each true β(t), shown without and with measurement error in the middle and right panels, respectively. The online version of this figure is in color.
These simulations illustrate both the strengths and limitations of the confidence interval procedure. For β1(t) and β2(t), we see generally good coverage when there is no measurement error. In fact, in some situations the intervals are slightly conservative. However, for β2(t) and to some degree β1(t) the performance degrades in the presence of measurement error; we speculate that this is in part due to the treatment of C as fixed. Thus, the simulations suggest that the confidence interval procedure is useful in situations with little measurement error on the functional predictor, but may be unreliable with larger measurement error variance. For β3(t), the coverage is expectedly poor: the estimate β̂ (t) significantly oversmooths the true coefficient function, and confidence intervals based on this estimate fail to achieve nominal coverage rates.
5. APPLICATION TO DTI TRACTOGRAPHY
Our application is to a study comparing the cerebral white-matter tracts of multiple sclerosis patients to the tracts of controls. White-matter tracts consist of axons, the long projections of nerve cells that carry electrical signals, that are surrounded by a fatty insulation called myelin. The myelin sheath allows an axon in a white-matter tract to transmit signals at a much faster rate than is possible in a non-myelinated axon. Multiple sclerosis is a demyelinating autoimmune disease that causes lesions in the white-matter tracts of an affected individual and results in severe disability.
Diffusion tensor imaging (DTI) tractography is a magnetic resonance imaging (MRI) technique that allows the study of white-matter tracts by measuring the diffusivity of water in the brain: in white-matter tracts, water diffuses anisotropically in the direction of the tract, while elsewhere water diffuses isotropically. Using measurements of diffusivity along several gradients, DTI can provide relatively detailed images of white-matter anatomy in the brain (Basser, Mattiello, and LeBihan 1994,Basser, Mattiello, and LeBihan 2000; LeBihan et al. 2001; Mori and Barker 1999).
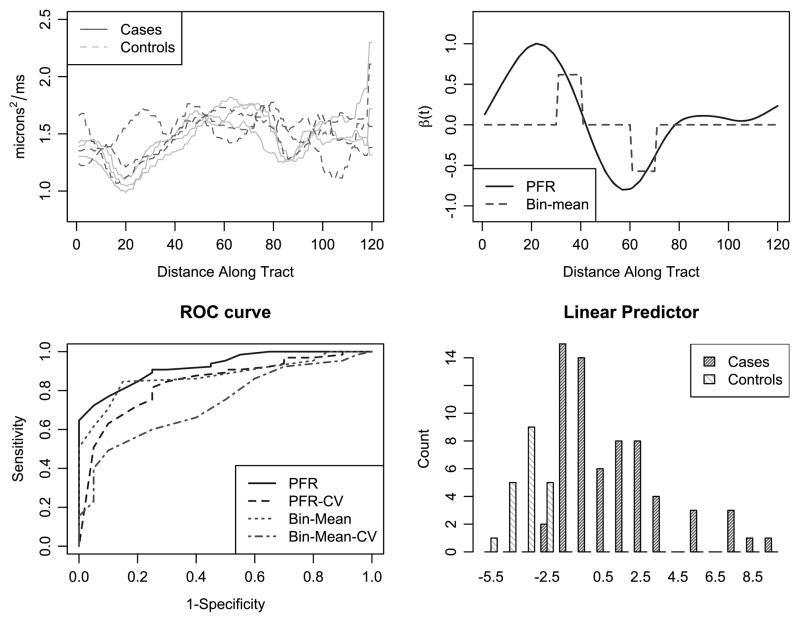
For each white-matter tract, DTI provides us several measures describing the diffusivity of water. One example of these measures is parallel diffusivity, which is the diffusivity along the principal axis of the tract. Parallel diffusivity is recorded at many locations along the tract, so that for each tract we have a continuous profile or function. The top-left panel of Figure 4 shows the parallel diffusivity profile of a single tract for three cases and three controls.
Figure 4.
The top-left panel shows a sample of functional predictors separated by case status. The top-right panel shows the estimated β(t) from the PFR and bin-mean approaches. The bottom-left panel shows the ROC curves generated by these approaches as well as leave-one-out cross-validated ROC curves. The bottom-right panel shows the distribution of the linear predictor for the PFR method. The online version of this figure is in color.
Our study consists of 20 controls and 65 cases, for whom we have a full DTI scan at baseline. Here, we focus on parallel diffusivity profiles as a way to classify subjects as cases or controls. Specifically, we take as our functional predictor the parallel diffusivity profile of the left intracranial cortico-spinal tract. Our first approach to this problem builds intuition: we bin the parallel diffusivity profiles and regress on the bin means, keeping those that are significantly related to the MS status. While straightforward, we recall that this is equivalent to constraining β(t) in a functional regression model to be a step function. We compare this to the penalized functional regression model presented in this manuscript.
The far-left panel of Figure 4 shows the estimates β̂(t) resulting from the two approaches. Both approaches emphasize the same two regions of the tract as important for distinguishing cases from controls, and give similar weights to these regions. Thus, those individuals whose parallel diffusivity profile is above average between distances 20 and 40 are more likely to be MS patients. Similarly, those individuals whose parallel diffusivity profile is above average between distances 50 and 65 are less likely to be MS patients. Moreover, the middle-left panel of this compares the predictive ability of the bin-mean and PFR methods via their respective ROC curves; also included in this plot are the leave-one-out cross-validated ROC curves for both the PFR and bin-mean methods. Note that there is a much larger drop in performance in the cross-validated curves for the bin-mean method than for PFR, perhaps indicating that the bin-mean approach is less generalizable to new datasets.
For each subject, we also compute the linear predictor ∫Xi (t)β(t) dt from the PFR method; the middle-right panel of Figure 4 shows the distribution of these quantities for 3 both cases and controls. As anticipated, ∫Xi (t)β(t) dt provides a reasonable quantity for distinguishing cases from controls based on the tract profile. The far-right panel of Figure 4 compares the tract profile resulting in the lowest three, the middle three, and the highest three linear predictors. We note that the tract profiles in this panel are Xi(t) − μ(t), where μ(t) is the overall mean profile. Thus, profiles with a low linear predictor will tend to be below zero between distances 20 and 40 and above average between distances 50 and 65, and conversely for profiles with high linear predictors.
6. DISCUSSION
By combining several well-known techniques in functional data analysis, we have developed a method for generalized functional regression with the following properties: (i) flexibly estimates β(t); (ii) treats the settings of measurement error, multilevel observations, and sparse data from the same unified framework; and (iii) compares favorably with existing methods in simulation studies. Although it builds on existing work, this method is conceptually new in that we decouple the estimation of the regressor functions Xi (t) and the coefficient function β(t). By using a PC decomposition for the Xi (t), we are able to apply our method when the Xi (t) are poorly observed (measurement error, sparse observation) or unobserved (multilevel). By using a rich spline basis for β(t) and explicitly inducing smoothness, we are able to estimate arbitrary smooth coefficient functions. Further, by expressing our method in terms of a GLMM, we take advantage of well-researched and computationally efficient machinery for fitting the model.
We tested our method in each of the settings we describe, with good results. We note that our simulation highlighted a case in which our method (as well as the others we examined) performed poorly. It is inherently difficult to detect peaks in β(t) when those peaks occur in areas of low variability in the Xi (t). Another interesting case, appearing in the online appendix, is that of sparsely observed functions in the absence of measurement error. When the measurement error variance is treated as unknown, it is estimated with bias and can result in biased estimates of the predictor processes Xt (t) and, ultimately, poor estimates of β(t); however, fixing generally resolves this issue. We note that another possible solution could be fully Bayesian treatment of the functional regression.
Several directions for future work are apparent. Handling several functional regressors, especially when those regressors are correlated, will be important as larger and larger datasets become available, as will developing new methods for multilevel functional data. More generally, examining the effectiveness of functional methods compared to less sophisticated techniques is necessary to establish the practical justification for these methods. A more robust development of confidence intervals for functional coefficients is also needed. Finally, the exploration methods in which the outcome is functional will continue to be important.
Supplementary Material
Acknowledgments
Crainiceanu’s, Goldsmith’s, and Caffo’s research was supported by Award number R01NS060910 from the National Institute of Neurological Disorders and Stroke. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Neurological Disorders and Stroke or the National Institutes of Health. Bobb’s research was supported in part by training grant 2T32ES012871, from the U.S. NIH National Institute of Environmental Health Sciences. This research was partially supported by the Intramural Research Program of the National Institute of Neurological Disorders and Stroke. We also thank the National Multiple Sclerosis Society and Peter Calabresi for the DTI tractography data.
Contributor Information
Jeff Goldsmith, Email: jgoldsmi@jhsph.edu, Johns Hopkins Bloomberg School of Public Health Biostatistics, Baltimore, MD 21205.
Jennifer Bobb, Email: jfeder@jhsph.edu, Johns Hopkins Bloomberg School of Public Health Biostatistics, Baltimore, MD 21205.
Ciprian M. Crainiceanu, Email: ccrainic@jhsph.edu, Johns Hopkins Bloomberg School of Public Health Biostatistics, Baltimore, MD 21205.
Brian Caffo, Email: bcaffo@jhsph.edu, Johns Hopkins Bloomberg School of Public Health Biostatistics, Baltimore, MD 21205.
Daniel Reich, Email: daniel.reich@nih.gov, Department of Radiology and Imaging Sciences, National Institutes of Health, Bethesda, Maryland 20892-1074.
References
- Basser P, Mattiello J, LeBihan D. MR Diffusion Tensor Spectroscopy and Imaging. Biophysical Journal. 1994;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser P, Pajevic S, Pierpaoli C, Duda J. In vivo Fiber Tractography Using DT-MRI Data. Magnetic Resonance in Medicine. 2000;44:625–632. doi: 10.1002/1522-2594(200010)44:4<625::aid-mrm17>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- Bates D, Pinheiro J. Computational Methods for Multilevel Modelling 1998 [Google Scholar]
- Cardot H, Sarda P. Estimation in Generalized Linear Model for Functional Data via Penalized Likelihood. Journal of Multivariate Analysis. 2005;92:24–41. [Google Scholar]
- Cardot H, Ferraty F, Sarda P. Functional Linear Model. Statistics and Probability Letters. 1999;45:11–22. [Google Scholar]
- Cardot H, Ferraty F, Sarda P. Spline Estimators for the Functional Linear Model. Statistica Sinica. 2003;13:571–591. [Google Scholar]
- Crainiceanu C, Goldsmith J. Bayesian Functional Data Analysis Using WinBUGS. Journal of Statistical Software. 2010;32:1–33. [PMC free article] [PubMed] [Google Scholar]
- Crainiceanu C, Staicu A, Di C. Generalized Multilevel Functional Regression. Journal of the American Statistical Association. 2009;104:1550–1561. doi: 10.1198/jasa.2009.tm08564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di C, Crainiceanu C, Caffo B, Punjabi N. Multilevel Functional Principal Component Analysis. The Annals of Applied Statistics. 2009;4:458–288. doi: 10.1214/08-AOAS206SUPP. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraty F, Vieu P. Nonparametric Functional Data Analysis: Theory and Practice. Springer; 2006. [Google Scholar]
- Goldsmith J, Crainiceanu C, Caffo B, Reich D. Longitudinal Penalized Functional Regression. 2010 to appear. [Google Scholar]
- James G. Generalized Linear Models With Functional Predictors. Journal of the Royal Statistical Society, Ser B. 2002;64:411–432. [Google Scholar]
- James G, Silverman B. Functional Adaptive Model Estimation. Journal of the American Statistical Association. 2005;100:565–576. [Google Scholar]
- James G, Wang J, Zhu J. Functional Linear Regression That’s Interpretable. The Annals of Statistics. 2009;37:2083–2108. [Google Scholar]
- LeBihan D, Mangin J, Poupon C, Clark C. Diffusion Tensor Imaging: Concepts and Applications. Journal of Magnetic Resonance Imaging. 2001;13:534–546. doi: 10.1002/jmri.1076. [DOI] [PubMed] [Google Scholar]
- Li Y, Ruppert D. On The Asymptotics Of Penalized Splines. Biometrika. 2008;95:415–436. [Google Scholar]
- McCullagh P, Nelder J. Generalized Linear Models. Chapman & Hall/CRC; 1989. [Google Scholar]
- McCulloch C, Searle S, Neuhaus J. Generalized, Linear, and Mixed Models. Wiley; 2008. [Google Scholar]
- Mori S, Barker P. Diffusion Magnetic Resonance Imaging: Its Principle and Applications. The Anatomical Record. 1999;257:102–109. doi: 10.1002/(SICI)1097-0185(19990615)257:3<102::AID-AR7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- Müller H. Functional Modelling and Classification of Longitudinal Data. Scandinavian Journal of Statistics. 2005;32:223–240. [Google Scholar]
- Müller H, Stadtmüller U. Generalized Functional Linear Models. The Annals of Statistics. 2005;33:774–805. [Google Scholar]
- O’Sullivan F, Yandell B, Raynor W. Automatic Smoothing of Regression Functions in Generalized Linear Models. Journal of the American Statistical Association. 1986;81:96–103. [Google Scholar]
- Pinheiro J, Bates D, DebRoy S, Sarkar D the R Core Team. R package version 3.1–96. 2009. nlme: Linear and Nonlinear Mixed Effects Models. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; Vienna, Austria: 2009. available at http://www.R-project.org. [8] [Google Scholar]
- Ramsay J, Silverman B. Functional Data Analysis. Springer; 2005. [Google Scholar]
- Reiss P, Ogden R. Functional Principal Component Regression and Functional Partial Least Squares. Journal of the American Statistical Association. 2007;102:984–996. [Google Scholar]
- Ruppert D. Selecting the Number of Knots for Penalized Splines. Journal of Computational and Graphical Statistics. 2002;11:735–757. [Google Scholar]
- Ruppert D, Wand M, Carroll R. Semiparametric Regression. Vol. 66. Cambridge University Press; 2003. [Google Scholar]
- Staniswalis J, Lee J. Nonparametric Regression Analysis of Longitudinal Data. Journal of the American Statistical Association. 1998;444:1403–1418. [Google Scholar]
- Wood S. Generalized Additive Models: An Introduction With R. Chapman & Hall; 2006. [Google Scholar]
- Yao F, Müller H, Clifford A, Dueker S, Follett J, Lin Y, Buchholz B, Vogel J. Shrinkage Estimation for Functional Principal Component Scores With Application to the Population. Biometrics. 2003;59:676–685. doi: 10.1111/1541-0420.00078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.