Abstract
Recently, the development of an IgE receptor (FcεRI)-targeted, pH sensitive, surface-enhanced Raman spectroscopy (SERS) nanosensor has been demonstrated [1]. The targeted nanosensor enables spatial and temporal pH measurements as internalized receptors progress through endosomal compartments in live cells. Trafficking of receptor-bound nanosensors was compared at physiological temperature (37°C) versus room temperature (25°C). As expected, we observed markedly slower progression of receptors through low pH endocytic compartments at the lower temperature. We also demonstrate the utility of the nanosensors to measure directly changes in the pH of intracellular compartments after treatment with bafilomycin or amiloride. We report an increase in endosome compartment pH after treatment with bafilomycin, an H+ ATPase pump inhibitor. Decreased endosomal luminal pH was measured in cells treated with amiloride, an inhibitor of Na+/H+ exchange. The decrease in amiloride-treated cells was transient, followed by a recovery period of approximately 15–20 minutes to restore endosomal pH. These experiments demonstrate the novel application of Raman spectroscopy to monitor local pH environment in live cells with the use of targeted SERS nanosensors.
Keywords: SERS, endosome, amiloride, bafilomycin, Raman, pH
Introduction
Chemical concentrations within the lumens of cellular organelles, such as endosomes, lysosomes, mitochondria, the Golgi apparatus and the endoplasmic reticulum, are frequently different from the cytoplasm. The differences in these cellular compartments are now being individually interrogated with an ever-expanding suite of techniques. Techniques commonly used to measure metabolite concentrations, including Ca2+, NO, Na+, and pH, are often based on fluorescent sensors. In this work, we focus on the cell’s dynamic regulation of acidic pH within the endosome-lysosome system, which is necessary to support degradative enzymes as well as the proper sorting of receptors and ligands following endocytosis [2,3,4]. Endosomal sequestration of weakly basic compounds such as daunorubicin, possibly through a pH partitioning mechanism, may be a factor in drug resistance [5]. Low pH within the endosomal-lysosomal system can also play a role in development of disease states. For example, beta-amyloid plaques are believed to aggregate due to the highly acidic environment in the lysosome [6,7,8]. MCF-7 breast and Caco-2 colorectal cancer cell lines are defective in endosomal acidification, leading to aberrant secretion of (pro) cathepsin D. Increased levels of cathepsin D secretion are in turn linked to the invasive and metastatic properties of these cancer cells [9].
The ability to monitor changes in intralumenal pH, in the context of cargo moving through the endosomal-lysosomal system or in response to external stimuli, could lead to novel strategies toward drug development and understanding of pathogen survival and other disease states. There are recent reports where the cellular endosomal pH has been statically measured using fluorescence-based techniques. For example, fluorescence lifetime imaging (FLIM) techniques with fluorescent acid-sensitive molecules such as Lysosensor DND-160, OG-514 carboxylic acid dextrans, and DM-NERF dextrans have been used to measure the average intracellular endosomal pH residing near the nucleus of 3T3 fibroblasts [10]. OG-514 dextran was used to stain endosomes within resting 3T3 fibroblast cells and demonstrated a pH range of 3.5 to 5.5. Using pH-sensitive fluorophores, a localized average endosomal pH is obtained, without the ability to resolve individual endosomes. The organic fluorophores accumulate in the endosomes and become trapped, rather than being specifically targeted. To target fluorescent molecules specifically to the endosomes for individual pH measurement, sensing molecules are conjugated to a targeting molecule [11,12]. Lee, et al. reported measuring folate receptor mediated endocytosis ratiometrically, through conjugation of folate to pH-insensitive Texas-Red dextran and to pH sensitive DM-NERF dextran [13]. Drawbacks to this technique include 1) photobleaching, 2) marked differences in fluorescence intensity from molecules in alkaline and acidic pH environments, 3) unequal proportions of the two sensors in endocytic compartments, 4) temperature effects on spectrum and conjugation [13] and 5) autofluorescence from cellular proteins. Another fluorescence technique to measure endosomal pH uses genetically-encoded fluorescent proteins conjugated to a target molecule. This technique was demonstrated through conjugation of pH sensitive E1GFP to HIV-Tat protein. The HIV-Tat protein enabled localization of the molecule to the endocytic pathway while the E1GFP portion of the fusion protein was sensitive to H+ concentration for pH monitoring. The technique was valid for pH values from 5.5–7.5, with protein unfolding at pH below 5.5 and no time-lapse images [14].
A technique that overcomes the background problems and photobleaching of fluorescence dyes, while maintaining stability during temperature fluctuations is surface-enhanced Raman scattering (SERS) [15,16,17,18]. SERS provides chemically-specific information based upon unique vibrational signatures of the species adsorbed or co-localized to a roughened metal surface. SERS provides detailed information of the chemical species present, with good signal to noise, low detection limits, fast acquisition speeds and elimination of fluorescent background. When compared to fluorescence microscopy, SERS microscopy typically require more complex microscopy instruments. In addition, to enjoy high signal to noise ratios for detection increased attention to nanoparticle synthesis as the Raman scattering enhancement factors in some systems can vary over a million fold, and a minute proportion of the most intense sites contribute significantly to the overall average signal [19]. In addition, the nanoparticles themselves have unknown potential cyto-toxicity in live cells [20].Nevertheless, recent reports show an increasing trend in applying SERS-based labels to cellular and tissue studies. For example, Tkachenko et al., showed that gold nanoparticles modified with nuclear localization peptides were translocated through cell and nuclear membranes in two different cell lines [21]. Nie and coworkers showed the effectiveness of using single-chain variable fragment (scFv) antibodies to target pegylated gold nanoparticles to specific tumor cells [22]. Cellular targeting has also been accomplished with dye-embedded core/shell nanoparticles and self-assembled Au-imidazole nanoparticles [23,24].
Nanosensors are created based upon choices of a colloidal particle platform, reporter molecule and target analyte. An appropriate particle platform enables a surface-enhanced Raman response of reporter molecule’s vibrational signatures as they respond to changing analyte concentration. The nanoparticle platforms generally include noble metal systems, particularly of Ag, [25] and Au [26,27], as well as a variety of geometries including hollow shell [28] and core/shell [29,30] particles. The analytes chosen have included the identification of intracellular chemicals (e.g. DNA and phenylalanine) non-specifically bonded to the metal surface [26,31,32], folic acid in serum, [33] as well as in vivo glucose measurements [34]. These methods rely on the ability of coatings on the metal surface to exhibit unique Raman vibrational signatures, as well as sensitivity and specificity to the analyte of interest.
The most demonstrated analyte concentration studied to date with SERS nanosensors is pH. For pH nanosensors in the neutral régime, 4-mercaptobenzoic acid has been used as the pH reporter molecule on solid silver colloids [25] or hollow gold shell colloids, [28] and delivered into Chinese Hamster Ovary cells. 4-Mercaptobenzoic based nanosensors have been shown to maintain their ability to record the localized pH, despite the complex background species in a live cell. Kneipp et al., have also used 4-mercaptobenzoic acid as a pH reporter molecule on gold nanoaggregates. The pH measurements were recorded in NIH/3T3 cells, where 4-mercaptobenzoic acid was found to be sensitive over the range of pH 2–8 through exploitation of one or two photon surface-enhanced hyper-Raman scattering [35,36,37]. In contrast, 4-ethynylpyridine is sensitive to pH in the alkaline region (pH > 11) although these particles were not demonstrated inside cells [38]. Further, for measurements of acidic concentrations (3<pH<7), it has been shown that use of 4-mercaptopyridine (4-MPy), which has a Raman scattering cross section of 10−26 cm2 [39], is an effective strategy [29,40]. Prasad and colleagues effectively used 2-aminothiophenol to acquire pH-dependant SERS spectra inside buffered cells, though whole hyperspectral image cubes, and, thus, whole cell pH-maps, were not demonstrated [41].
Recently, we have shown that a targeted SERS nanosensor is capable of measuring the pH of intracellular compartments harboring internalized IgE receptors, using the RBL-2H3 mast cell line as a model system. This sensor is based upon a Au@Ag core shell particle with 4-mercaptopyridine (4MPy) as a reporter molecule and 2,4 ε-dinitrophenol-L-lysine (DNP) as a targeting molecule for DNP-specific IgE bound to the high affinity IgE receptor, FcεRI [1,42]. Here, we report the application of this nanosensor to measure the pH of endosomal compartments as sensor-bound receptors traffic through the endosome-lysosome system while the cell is exposed to dynamic external stimuli. We demonstrate that the nanosensors are resilient to changes in temperature, by lowering the temperature of cultured cells from the physiologically relevant temperature of 37°C to 25°C; this procedure also slows the transit of endocytosed receptors. More importantly, we also demonstrate that the targeted nanosensors report changes in luminal pH after treatment of cells with the H+ flux inhibiting drugs, amiloride and bafilomycin dissolved in media. Importantly, the FcεRI-targeted pH SERS nanosensors enable the direct localization of vesicles bearing internalized receptors and also report the dynamic changes in endosomal vesicle pH in response to these drugs. We show that compartments recover their normal pH after removal of these drugs from the media on the timescale of 10s of min. To our knowledge, this is the first report of the use of a targeted SERS nanosensor under external stimuli. This technology has the potential to create new techniques useful in the fields of medicine, pharmacology, and biotechnology.
Materials and Methods
Cell culture and reagents
RBL-2H3 cells were cultured in MEM media supplemented with 10% fetal clone III (p/n SH3010902, Thermo Scientific Hyclone, Logan, UT), 90 I.U./mL of penicillin (ATCC) and 90 µg/mL streptomycin (ATCC), with an additional 2 µM of L-glutamine (Sigma Aldrich, St. Louis, MO). Cells were primed with anti-DNP specific IgE (1 µg/mL) 18–24 hrs prior to use. Bafilomycin (Sigma) was dissolved in EtOH and used at a concentration 200 nM [43]. Amiloride (MP Biomedical, Inc.) was dissolved in dH2O and used at a concentration of 1 mM [44].
Preparation of targeted SERS nanosensors
Au colloids at 60 nm diameter were purchased from Ted Pella Inc. (p/n 15708-6, Redding, CA) at 2.6 × 1010 particles per mL. The Au colloids were enhanced with a Ag coating by addition of LI silver enhancing and initiator agents from Nanoprobes, Inc. (p/n 2013, Yaphank, NY). Silver enhancement was performed at a 1:1 ratio of both initiator and enhancer. The enhancement solution was added to the colloids for two minutes, centrifuged at 13,000 rpm for five minutes, and then washed with deionized H2O. 4-Mercaptopyridine (4MPy) (p/n 148202, Sigma Aldrich Chemicals, St. Louis, MO) and 2,4 ε-dinitrophenol-L-lysine (DNP) (MP Biomedical, Solon, OH), were used without further purification. Nigericin, KCl, MgCl2, CaCl2, and glucose were also from Sigma Aldrich. The nanosensors were calibrated within the cell by using pH calibrated phosphate buffers accompanied with nigericin, that equilibrates the intracellular pH with the extracellular pH [1].
Imaging experiments with the targeted nanosensors employ sonication of colloids for five minutes, filtering through a 0.2 µm filter and addition at 100× particles per cell. Cells were washed two times with Hanks buffered solution, followed by imaging in fresh Hanks buffer. In some experiments, a perfusion chamber was used (Carl Zeiss, Inc., Thornwood, NY) to introduce nanosensors to drug treated cells. The cover slips with adherent cells are then added to the environmental chamber vessel (Carl Zeiss, Inc., Thornwood, NY) equilibrated at relevant temperature and 5% CO2 for imaging.
Imaging
The microscope imaging system was based upon a Carl Zeiss Axiovert 135TV inverted microscope with an Epiplan 10× (N.A. 0.20), a LD Plan-achroplan 40× (N.A. 60), and a C-apochromat 63× (N.A. 1.2) water immersion objective. The last two objectives were primarily used for the SERS experiments. A xenon arc lamp (XBO 75, Carl Zeiss, Thornwood, NY) was used to illuminate the sample for bright field visualization in transmission mode, by video camera, using transfer optics in the trinocular head. (Optem 70XL, Labtek, Campbell, CA) The total zoom factor of the transfer optics was 0.5× in addition to the objective used. An Infinity 2-2 monochrome (Lumenera Corp., Ottawa, ON) camera was used to acquire the visible image using an acquisition rate of approximately 200 ms/frame and no detector gain. The visible image resolution was 1616×1216 pixels. Raman experiments were performed using 514.5 nm light from a Spectra Physics 177-G01 air-cooled argon ion laser. The laser light was focused to a line which is approximately 0.5 µm wide and 50 µm tall and a total power at the sample of 10 mW. The epi-directed signal was transferred using two additional 150 mm spherical lenses and reimaged onto the 100 µm wide slit of a 0.25 m f/2.2 imaging spectrograph (Holospec 2.2, Kaiser Optical Systems, Inc, Ann Arbor, MI). The signal was dispersed by a volume holographic grating (HSG-514.4-LF, Kaiser) and imaged with a LN2-cooled CCD array detector (LN/CCD-1024E, Princeton Instruments, Trenton, NJ). Each image of the CCD recorded spectral and "Y" spatial information of the sample with spectral resolution of the spectrograph at ~2 cm−1. In order to obtain spectral image cubes, the frequency and "Y" spatial axis were acquired and saved, and the sample was moved across the excitation source by stepping the computer controlled microscope stage (MS-2000, Applied Scientific Instrumentation, Eugene, OR), to sequentially build up the "X" spatial access of the cube. The schematic of this set-up was recently described [1].
For transmission electron microscopy (TEM), cells were fixed 2% glutaraldehyde in cacodylate buffer (pH 7.4), post-fixed with osmium tetroxide, dehydrated in ascending alcohols and embedded in Epon. Due to the size of the Au@Ag nanosensors, samples were sectioned with a 100–300 nm thickness on a Leica Ultracut UCT. Samples were stained with uranyl acetate and lead citrate and observed on a Hitachi 7500 electron microscope.
Surface-Enhanced Raman Scattering pH Calibration
In vivo calibration experiments employed the addition of 10 µm nigericin, 140 mM KCl, 1 mM MgCl2, 2 mM CaCl2, 5 mM glucose, 20 mM phosphate-citrate buffers of varying pH (4–8). Once the calibration solution was added, cells were returned to the incubator for ten minutes. After pH equilibration, targeted nanosensors were added to the cells and whole images were taken of internalized particles. Cellular images representing approximately 100 (0.5 µm × 0.5 µm) sized pixels with nanosensors. These images were processed to obtain individual spectra as indicated in the data analysis section. Once individual data points were collected for each pH value, the data were fit to a sigmoidal curve. This sigmoidal function was used to calculate pH from SERS signal.
Data analysis
The data analysis procedures were performed using custom written Labview code. All pixels of the hyper-spectral image cube were subject to the same correction procedures including wavelength calibration, flat field correction, background subtraction, CCD bias voltage subtraction, and spherical aberration removal. The images were thresholded by eliminating any spectral pixel information with a signal to noise ratio of less than 2.5. The signal to noise ratio was calculated by taking the highest intensity count from the integrated band at 1572 cm−1 to 1599 cm−1 subtracted from the baseline value at that point, divided by the highest intensity count from the noise at 1800 cm−1 to 1827 cm−1 subtracted from the baseline value at that point. Any pixel with a signal to noise ratio below 2.5 was considered to have no useful information and was removed from the image. The two spectral bands of interest were then integrated from 1572 cm−1 to 1599 cm−1 for the non-protonated band and 1599 cm−1 to 1626 cm−1 for the protonated band. Because the Raman spectra often occur on a weak fluorescence background, the integrated limits were evaluated against a linearly sloping background defined as the spectral intensity at 1000 cm−1 and 2000 cm−1. Since bands of interest are so close spectrally, variably sloping baselines due to cellular autofluorescence were effectively removed without altering the intrinsic ratio of the band. Unlike other pH sensors used, the 4-MPy sensor has the additional benefit of having all the reporter molecules in either one state or the other. Thus, the bands are plotted not as a ratio to a standard band, but rather as a percentage of molecules in the un-protonated state.
Results and Discussion
Microscopy
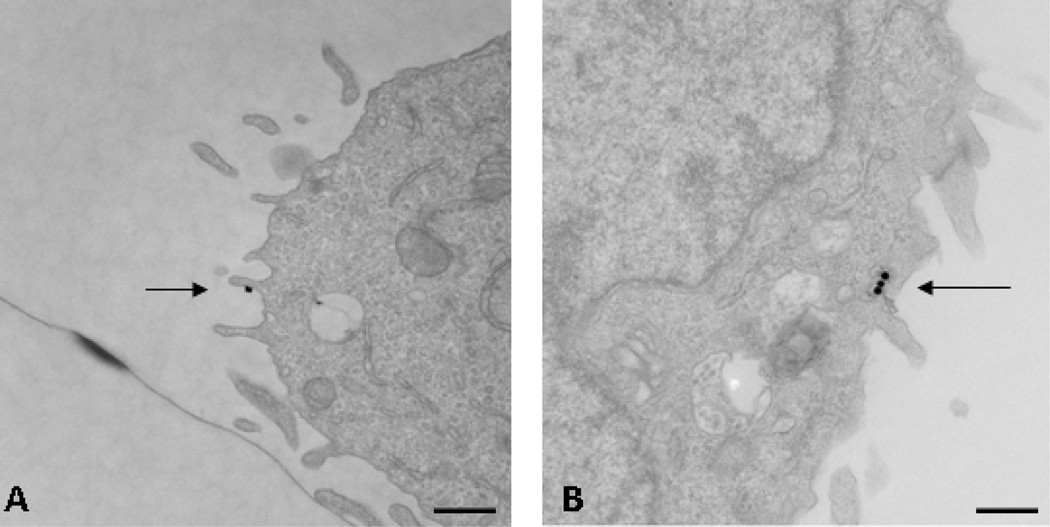
TEM images are depicted in Figure 1. Figure 1a shows a high resolution image of a RBL-2H3 cell after 5 min exposure to the nanosensors. The arrow points to the location of an individual nanosensor, seen as a black (electron-dense) dot bound to the surface of the cell, due to specific binding of DNP portion of the nanosensor to at least one IgE-bound receptor. Figure 1b illustrates a similar image of a cell exposed to a five minute pulse of nanosensors, followed by a sixty minute chase to allow for internalization of sensor-bound receptors prior to fixation and processing for TEM. In this photo, the arrow points to 3 internalized nanosensors within an endocytic compartment just under the membrane surface.
Figure 1.
Transmission electron microscopy images of RBL-2H3 cells (a) five minutes after incubation of nanosensors and (b) sixty minutes after incubation with nanosensors. Arrows are added to pinpoint locations of nanosensors within the cells. Bars = 500nm.
The pH sensitivity of the targeted nanosensors has been demonstrated previously with in vivo pH calibration over a range from pH 4.0–8.0[1]. Briefly, the calibration curve was generated by taking the integrated SERS signal from 1572 cm−1 to 1599 cm−1 divided by the total integrated SERS signal from 1572 cm−1 to 1626 cm−1, with a baseline subtraction from 900 cm−1 to 2200 cm−1. The ratiometric measurement corresponds to the percentage of the pH sensor ligand 4-mercaptopyridine (4MPy) in the deprotonated state versus the protonated state [40]. Figure 2a demonstrates how whole cell pH imaging maps are configured. The RBL-2H3 cells seen in the bright field image were incubated with IgE targeted SERS nanosensors. The brightfield image is overlaid with a Raman-based image demonstrating the individual pixels within the cell that contain targeted nanosensors. As previously discovered, there is a nearly linear correspondence of pixel number to SERS signal. This ratio is indicative of at least one nanosensor present per pixel and no evidence of nanosensor clustering [1,42].
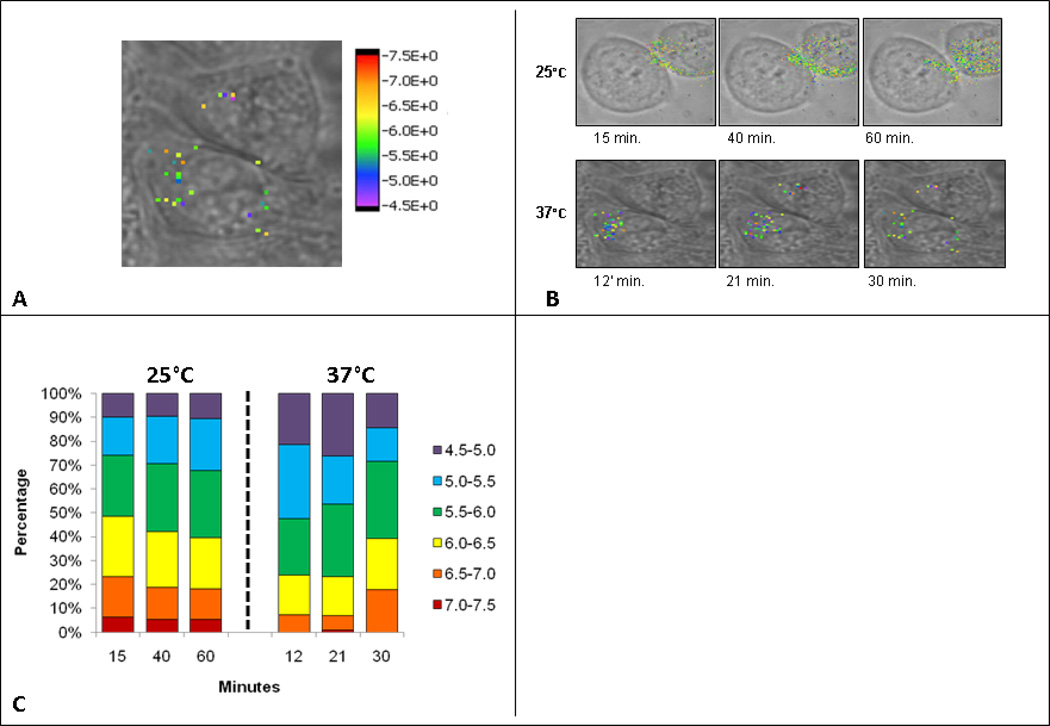
Figure 2.
Temperature effects on nanosensors at 37°C vs. 25°C. (a) Demonstration of whole cell pH imaging maps. (b) Whole cell pH image maps constructed same as in figure 2a. Top series are cells incubated at 25°C with 10 minutes incubation of nanosensors, followed by imaging at 15’, 40’, and 60’ after the initial nanosensor addition. Bottom series are cells incubated at 37°C with 10 minute incubation with nanosensors followed by imaging at 12’, 21’, and 30’ after initial nanosensor addition. (c) Bar graphs demonstrating percentage of nanosensors from whole, in each pH grouping, at specified time points. Cells were incubated for ten minutes with nanosensors and then imaged at specified time points.
The nanosensors shown in Figure 2a are colored to represent the pH of their cellular environment and are made transparent when no SERS information is present. Using the calibration curve, the Raman signature corresponding to the nanosensor within each pixel reports a specific pH value based off the calibration curve. Once the pH of the nanosensors is established, the pixel is colored according to the rainbow-colored calibration bar at the right of the image. Each image is approximately 30 × 30 µm and took approximately three minutes to acquire with the generation of individual SERS spectrum at each individual pixel.
Temperature effects
SERS targeted nanosensors were added to cells held at either 37°C or 25°C. Figure 2b depicts brightfield cellular images overlaid with color calibrated SERS images and cells incubated at the two temperatures. The brightfield images are all recorded at initial time points before the first corresponding SERS image; minor alignment issues in the overlays correct for modest movements of the adherent cells. At the physiological temperature of 37°C, FcεRI cross-linking leads to marked endocytosis of the receptors over a 2–15 minute time period; of the endocytosed receptors, most will be trafficked to lysosomes within 30 minutes [45,46]. Cells incubated at 37°C demonstrate internalization on a rapid time scale with a large portion of nanosensors displaying pH values from 4.5 to 5.5.
As expected, there was a considerable delay in receptor internalization in cells incubated at 25°C (Figure 2b). Accordingly, we extended the imaging time to over sixty minutes. We show that, at 25°C, nanosensors remained near or on the plasma membrane for at least fifteen minutes, moving towards the center by 40–60 min. At 60 min, a large proportion of the nanosensors report a pH environment comparable to the late stage endosomes with pH in the 5.0–6.0 range. At each time point, the targeted nanosensors report the local pH of their vesicular environment. To better present these data, pH groupings are assigned which reflect the specific pH environment of specific endocytic compartments. A vesicle that has just invaginated from the cellular membrane, referred to as a pre-endosomal vesicle, [47] will be within the 7.0–7.5 pH range. Recycling endosomes are expected to have a lumenal pH of 6.5–7.0 pH., while early sorting endosome are expected to have luminal pH in the range of 6.0–6.5. The late endosome is more acidic, with a pH of 5.0–6.0. Lysosomal vesicles are very acidic, expected to fall within the range of 4.5–5.5. [48]. The images in Figure 2b give a qualitative picture of the nanosensors and the pH of their localized environment. Figure 2c averages data from three cells, with the goal of reporting the total nanosensor numbers in each pH grouping as a percentage of the whole.
Figure 2c also illustrates the expeditious movement of nanosensors through the endocytic pathway when incubated at 37°C. The nanosensor groupings devised from the images show that, at twelve minutes, 80% of the nanosensors are in pH groupings indicative of late endosomal/lysosomal vesicles. At later times, the overall number of sensors is reduced, consistent with trafficking of nanosensors to the cell surface via both the recycling compartments and by exocytic release of secretory lysosomes, followed by dissociation into the medium. A pool of nanosensors is detected with pH 6.0–7.0 compartments at these later times, suggestive of their retention within early sorting and recycling endosomes.
Experiments performed at 25°C demonstrate the temperature dependence of both initial receptor-mediated internalization of sensors and their slower progress through the endocytic pathway. When compared to cells incubated at 37°C, it is notable that nanosensors tend to accumulate in the pre-endosomal/recycling endosomes at the lower temperature. There is also a notable retention of the total number of nanosensors within the lowest pH compartments, as expected if the low temperature slows the arrival of receptor-nanosensor complexes into secretory lysosomes and their subsequent return to the plasma membrane by exocytosis.
Pharmacologic effects
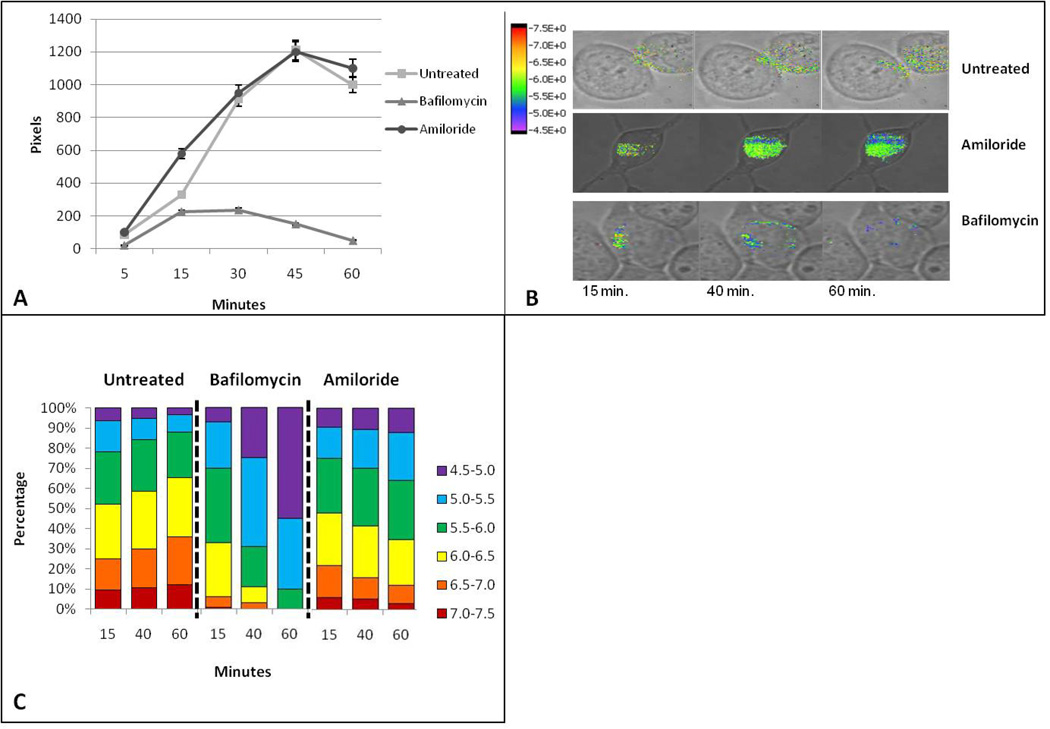
We next validated the functionality of the targeted nanosensors through a pharmacologic approach. First, we tested the ability of the nanosensor to detect increases in pH within endosomes of cells treated with bafilomycin, an inhibitor of the H+ ATPase pump that pumps H+ into the endosomal vesicle [49,50,51]. We compared these results with those in cells treated with amiloride, a known inhibitor of the Na+/H+ exchanger. This results in impairment of H+ exit from the endosomal vesicle and a decrease in endosomal pH [52,53,54]. In both cases, cells were incubated at 37°C with or without drugs for two hours prior to ten minute exposure of cells to nanosensors and imaging in the continued presence of drugs. Results are reported in Figure 3.
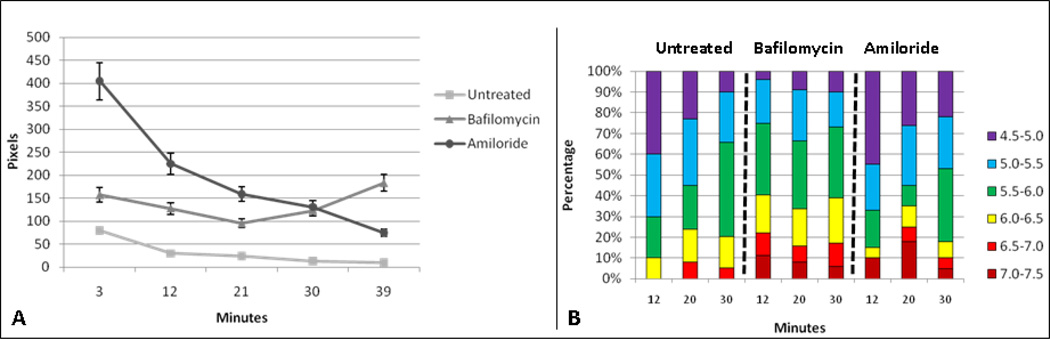
Figure 3.
Pharmacologic effects on nanosensors at 37°C. Cells incubated at 37°C with addition of bafilomycin or amiloride for two hours, followed by addition of nanosensors. Figure 3a is a demonstration of total pixel/nanosensor number averaged from three individual cells. Cells are incubated at 37°C untreated or treated for two hours with drug, followed by addition of nanosensors for ten minutes, then imaged. Figure 3b is a demonstration of the percentage of nanosensors averaged from three cells, from the whole, within the assigned pH groupings.
Figure 3A shows the total uptake of nanosensors under each condition, reported as average number of pixels at each time point for at least three different cells. Untreated cells accumulated approximately 100 nanosensors per cell during a 10 min exposure. This value slowly decreased in number after washout, likely due to a combination of 1) recycling back to the surface, 2) directed trafficking to secretory lysosomes followed by egress of receptor-bound nanosensors through exocytosis and 3) lysosomal degradation. Although the relative contributions of these two pathways are not distinguishable under these experimental conditions, it is possible to infer the locations of nanosensors from their distribution within compartments with specific pH at intervals during the chase (Figure 3B). At 12 minutes of chase, 70% of nanosensors were in compartments with pH of 4.5–5 (40%) or 5.0–5.5 (30%). Again, selective loss of this pool is consistent with arrival in the secretory lysosome, where they may be degraded or routed to the surface by exocytosis.
Remarkably, amiloride-treated cells accumulated 4-fold more nanosensors (400/cell) within the same time of exposure (Figure 3A). We speculate that this large increase in number may be attributed to increased acidification of early and recycling endosomes, shifting the balance of receptors from the recycling pathway to the lysosome-directed pathway. Disruption of the Na+/H+ exchanger may also partially block the internalization process, leaving sensors bound to receptors at the cell surface where the medium is buffered at pH 7.2. This is illustrated in Figure 3B, where 10–18% of sensors register a pH of 7.0–7.5 for up to 20 min of chase. Other notable observations in amiloride-treated cells include a detectable increase in nanosensors within very low pH compartments (pH 4.5–5) [52,53,54] and marked overall decline in number of nanosensors over the 40 min chase period. This is consistent with degradation with low pH compartments and/or exocytosis.
Bafilomycin-treated cells took up an average of 150 nanosensors during the 10 min exposure, with no obvious decline over the 40 min chase period (Figure 3A). As shown in Figure 3B, these nanosensors are trapped in compartments with significantly higher pH, consistent with blockade of the proton pump. We note that approximately 40% of the sensors reside in pH environments of 6.0–7.5 (red-orange-yellow bars in Figure 3B) for the duration of the experiment. This may reflect occupancy in endosomal and recycling compartments with aberrant pH or retention of some receptors at the cell surface [55,56]. Others have suggested that bafilomycin affects endosomal acidification primarily from the late endosomal to lysosomal stage [57,58,59]. Accordingly, less than 10% of sensors sense typical lysosomal pH in the range of pH 4.5–5. It seems likely that the sustained levels of nanosensors are due to either poor delivery to lysosomes and/or protection from degradation because of the pH dependency for lysosomal enzyme activity.
We next varied the protocol in two ways, by 1) uptake of nanosensors by untreated cells at 37°C, followed by addition of drugs and imaging or 2) treatment with drugs and nanosensors, followed by removal of the drugs and imaging to follow recovery of compartments.
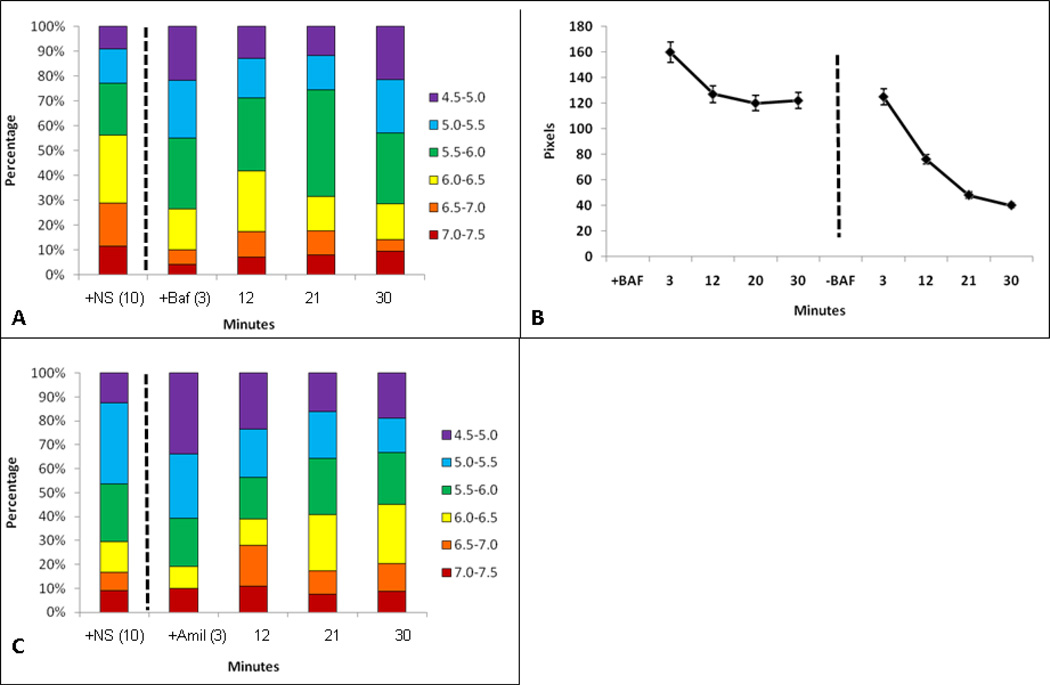
Figure 4A shows the distribution of nanosensors immediately after bafilomycin was added to the cells exposed to ten minute incubation with nanosensors. Graphs in this figure show that the effects of bafilomycin are apparent within minutes. This is most dramatic when comparisons are made at the 12 minute time point where only 25% of sensors are in lowest pH (4.5–5.5) environments compared to 70% in normal cells (see Figure 3B). Importantly, acute exposure to bafilomycin after uptake prevents loss of nanosensors. This is demonstrated in Figure 4B, where the total number of nanosensors is stabilized within 12 minutes of bafilomycin treatment (left hand plot). This effect is reversible, as shown in the right hand plot in Figure 4B, where cells were treated for 30 min with bafilomycin followed by exchange with fresh media without drug.
Figure 4.
Nanosensor recovery from pharmacological treatment. Figure 4a is representative to cells incubated at 37°C with nanosensor addition and incubation for ten minutes. The first bar of the graph is representative of nanosensors inside cells without chemical addition. After the dashed line, the bar graph is representative to nanosensor ph after addition of Bafilomycin to the cells. Figure 4b is the total pixel/nanosensor count within cells. Time points after dashed line are representative to removal of bafilomycin and addition of fresh media. Figure 4c is representative to nanosensors pH in cells incubated at 37°C with nanosensor incubation for ten minutes followed by addition of amiloride. The first bar of the graph is cells incubated with nanosensors for ten minutes. The data after the dashed line is the addition of amiloride to the media followed by imaging at respective time points.
Acute effects of amiloride treatment on cells were also examined in Figure 4C. Cells were again incubated with nanosensors for ten minutes at 37°C, followed by wash and addition of amiloride to the media (dashed line). Although 30% of nanosensors reach a low pH (4.5–5.0) environment by 3 min, there is a significant shift to higher pH environments within 12 minutes. This change appears to be complete within 20 minutes, since the range of pH compartments is similar for both 20 min and 30 min of amiloride treatment.
Room temperature experiments are consistent
The next set of experiments compared the uptake and transiting of nanosensors in cells pretreated with amiloride or bafilomycin but held at room temperature for all treatments. Under these conditions, untreated cells and amiloride-treated cells show a slow uptake of nanosensors, reaching similar plateau levels at approximately 45 min (Figure 5A). In contrast, bafilomycin-treated cells accumulate markedly fewer nanosensors per cell (Figure 5A) when held at room temperature. Representative 60 min time-lapse SERS movies showing targeted SERS nanosensors in media (a) in bafilomycin/media (b) and amiloride/media (c) are provided as supplemental information.
Figure 5.
Pharmacologic effects on nanosensors at 25°C. Cells incubated at 25°C with the addition of bafilomycin or amiloride. Cells are incubated at 25°C with simultaneous addition of nanosensors and respective drug. (a) Is a demonstration of total nanosensor number displayed as pixels, averaged from three individual cells. (b) Whole cell pH image maps of cells incubated at 25°C with the addition of nanosensors alone (untreated), with bafilomycin, or amiloride, followed by imaging at the corresponding time points below. (c) Bar graph demonstration of the percentage of nanosensors averaged from three cells, from the whole, that fall into each pH grouping in cells that are either untreated and incubated at 25°C, or treated with drug and incubated at 25°C.
Analysis of the pH environment for internalized receptors under each condition suggests complex effects of low temperature on receptor trafficking through the endosome-lysosome systems. A spatial view of nanosensor localization within the untreated and treated cells at 15, 40 and 60 minutes is demonstrated (Figure 5B). In untreated cells, the slow uptake is accompanied by marked lack of delivery to low pH (4.5–5.5) compartments. Even after 60 minutes, less than 10% of receptors would appear to have reached late endosomes or lysosomes (Figure 5C). Interpretation of receptor progression in bafilomycin-treated cells is complicated by the poor overall uptake. However, it is notable that over 35% of nanosensors taken up by amiloride-treated cells reside in a pH 4.5–5.5 environment within 60 min of uptake at 25°C. We cannot distinguish whether these nanosensors have reached lysosomal compartments at this stage or whether they reside in endosomal compartments that have dramatically acidified after amiloride blockage of the Na+/H+ exchanger on their membranes.
Conclusions
These experiments demonstrate that targeted nanosensors can be taken up by receptor-mediated internalization, providing a mechanism to simultaneously measure endocytic vesicle pH and follow the temporal and spatial progression of receptors as they traffic through the endosome-lysosome system. The targeted nanosensors show the markedly slow progression of receptors through the endocytic pathway when cells were held at 25°C (Figure 5A). The targeted nanosensors were also able to report the effects of H+ flux inhibiting drugs amiloride and bafilomycin on the receptor residing in endocytic compartments, providing reliable measurements of changes in lumenal pH. These versatile probes show promise for future applications relevant to intracellular trafficking of receptors and intelligent drug design.
Acknowledgments
This manuscript has been authored by Los Alamos National Security under Contract No. DE-RP52-05NA25396 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The authors acknowledge Los Alamos National Laboratory Laboratory Directed Research and Development grant 20080001DR (to KR) and NIH R01 AI051575 (to BW) for support of this project. We acknowledge use of the UNM-HSC Electron Microscopy Facility and the UNM Cancer Center Fluorescent Microscopy Facility. We also thank Professors Janet Oliver, Diane Lidke and Keith Lidke (University of New Mexico) for advice and input. Finally, we thank Brian Dyer of Emory University for useful discussions.
References
- 1.Nowak-Lovato KL, Rector KD. Targeted Surface-Enhanced Raman Scattering Nanosensors for Whole-Cell pH Imagery. Applied Spectroscopy. 2009;63:387–395. doi: 10.1366/000370209787944406. [DOI] [PubMed] [Google Scholar]
- 2.Bradley M, Alexander L, Duncan K, Chennaoui M, Jones AC, et al. pH sensing in living cells using fluorescent microspheres. Bioorganic & Medicinal Chemistry Letters. 2008;18:313–317. doi: 10.1016/j.bmcl.2007.10.075. [DOI] [PubMed] [Google Scholar]
- 3.Liu J, Lu W, Reigada D, Nguyen J, Laties AM, et al. Restoration of lysosomal pH in RPE cells from cultured human and ABCA4(−/−) mice: Pharmacologic approaches and functional recovery. Investigative Opthalmology & Visual Science. 2008;49:772–780. doi: 10.1167/iovs.07-0675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Varghese B, Barriere H, Carbone CJ, Banerjee A, Swaminathan G, et al. Polyubiquitination of Prolactin Receptor Stimulates Its Internalization, Postinternalization Sorting, and Degradation via the Lysosomal Pathway. Molecular Cellular Biology. 2008;28:5275–5287. doi: 10.1128/MCB.00350-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong Y, Duvvuri M, Krise J. Separate roles for the Golgi apparatus and lysosomes in the sequestration of drugs in the multidrug-resistant human leukemic cell line HL-60. The Journal of biological chemistry. 2003;278:50234–50239. doi: 10.1074/jbc.M306606200. [DOI] [PubMed] [Google Scholar]
- 6.Haugland R. The Handbook:A guide to fluorescence probes and labeling technologies: Invitrogen Corporation. 2005. p. 582. [Google Scholar]
- 7.Su Y, Chang P. Acidic pH promotes the formation of toxic fibrils from beta-amyloid peptide. Brain research. 2001;893:287–291. doi: 10.1016/s0006-8993(00)03322-9. [DOI] [PubMed] [Google Scholar]
- 8.Su Y, Chang P, Wood S, Maleeff B, Hart T, et al. Physical, morphological and functional differences between pH 5.8 and 7.4 aggregates of the Alzheimers amyloid peptide A-beta. Journal of molecular biology. 1996;256:870–877. doi: 10.1006/jmbi.1996.0133. [DOI] [PubMed] [Google Scholar]
- 9.Kokkonen N, Rivinoja A, Kauppila A, Suokas M, Kellokumpu I, et al. Defective acidification of intracellular organelles results in aberrant secretion of cathepsin D in cancer cells. The Journal of biological chemistry. 2004;279:39982–39988. doi: 10.1074/jbc.M406698200. [DOI] [PubMed] [Google Scholar]
- 10.Lin H, Herman P, Lakowicz J. Fluorescence lifetime-resolved pH imaging of living cells. Cytometry. 2003;52A:77–89. doi: 10.1002/cyto.a.10028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohkuma S, Poole B. Fluorescence Probe Measurement of Intralysosomal pH in Living Cells and Perturbation of pH by Various Agents. Proceedings of the National Academy of Sciences of the United States of America. 1978;75:3327–3331. doi: 10.1073/pnas.75.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Thomas JA, Buchsbaum RN, Zimniak A, Racker E. Intracellular pH Measurements in Ehrlich Ascites Tumor Cells Utilizing Spectroscopic Probes Generated in Situ. Biochemistry. 1979;79:2210–2218. doi: 10.1021/bi00578a012. [DOI] [PubMed] [Google Scholar]
- 13.Lee RJ, Wang S, Low PS. Measurement of endosome pH following folate receptor-mediated endocytosis. Biochmica et Biophysica Acta. 1996;1312:237–242. doi: 10.1016/0167-4889(96)00041-9. [DOI] [PubMed] [Google Scholar]
- 14.Serresi M, Bizzarri R, Cardarelli F, Beltram F. Real-time measurement of endosomal acidification by a novel genetically encoded biosensor. Analytical and Bioanalytical Chemistry. 2009;393:1123–1133. doi: 10.1007/s00216-008-2489-7. [DOI] [PubMed] [Google Scholar]
- 15.Campion A, Kambhampati P, Link S, El-Sayed M. Spectral properties and relaxation dynamics of surface plasmon electronic oscillations in gold and silver nanodots and nanorods. The journal of physical chemistry B, Materials, surfaces, interfaces & biophysical. 1999;103:8410–8426. [Google Scholar]
- 16.Hudson S, Chumanov G. Bioanalytical applications of SERS (surface-enhanced Raman spectroscopy) Analytical and Bioanalytical Chemistry. 2009;394:679–686. doi: 10.1007/s00216-009-2756-2. [DOI] [PubMed] [Google Scholar]
- 17.Moskovits M. Surface-enhanced spectroscopy. Reviews of modern physics. 1985;57:783–826. [Google Scholar]
- 18.Nie S, Emory S. Probing single molecules and single nanoparticles by surface-enhanced Raman scattering. Science. 1997;275:1102–1106. doi: 10.1126/science.275.5303.1102. [DOI] [PubMed] [Google Scholar]
- 19.Fang Y, Seong N, Dlott D. Measurment of the distribution of site enhancements in surface-enhanced Raman scattering. Science. 2008;321:388–392. doi: 10.1126/science.1159499. [DOI] [PubMed] [Google Scholar]
- 20.Boca S, Astilean S. Detoxification of gold nanorods by conjugation with thiolated poly (ethylene glycol) and their assessment as SERS-active carriers of Raman tags. Nanotechnology. 2010;21 doi: 10.1088/0957-4484/21/23/235601. [DOI] [PubMed] [Google Scholar]
- 21.Tkachenko AG, Xie H, Coleman D, Glomm W, Ryan J, et al. Multifunctional Gold Nanoparticle-Peptide Complexes for Nuclear Targeting. Journal of American Chemical Society. 2003;125:4700–4701. doi: 10.1021/ja0296935. [DOI] [PubMed] [Google Scholar]
- 22.Qian X, Xiang-Hong P, Ansari D, Yin-Goen Q, Chen G, et al. In vivo tumor targeting and spectroscopic detection with surface-enhanced Raman nanoparticle tags. Nature Biotechnology. 2008;26:83–90. doi: 10.1038/nbt1377. [DOI] [PubMed] [Google Scholar]
- 23.Doering W, Nie S. Spectroscopic Tags Using Dye-Embedded Nanoparticles and Surface-enhanced Raman scattering. Analytical chemistry. 2003;75:6171–6176. doi: 10.1021/ac034672u. [DOI] [PubMed] [Google Scholar]
- 24.Souza GR, Levin CS, Hajitou A, Pasqualini R, Arap W, et al. In Vivo Detection of Gold-Imidazole Self-Assembly Complexes: NIR-SERS Signal Reporters. Analytical Chemistry. 2006;78:6232–6237. doi: 10.1021/ac060483a. [DOI] [PubMed] [Google Scholar]
- 25.Talley C, Jusinski L, Hollars C, Lane S, Huser T. Intracellular pH sensors based on surface-enhanced raman scattering. Analytical chemistry. 2004;76:7064–7068. doi: 10.1021/ac049093j. [DOI] [PubMed] [Google Scholar]
- 26.Jin W, Costa-Fernandez J, Pereiro R, Sanz-Medel A. Surface-modified CdSe quantum dots as luminescent probes for cyanide determination. Analytica chimica acta. 2004;522:1–8. [Google Scholar]
- 27.Joo S-W. Adsorption of Bipyridine Compounds on Gold Nanoparticle Surfaces Investigated by UV-Vis Absorbance Spectroscopy and Surface Enhanced Raman Scattering. Spectroscopy Letters. 2006;39:85–96. [Google Scholar]
- 28.Schwartzberg A, Oshiro T, Zhang J, Huser T, Talley C. Improving nanoprobes using surface-enhanced Raman scattering from 30-nm hollow gold particles. Analytical chemistry. 2006;78:4732–4736. doi: 10.1021/ac060220g. [DOI] [PubMed] [Google Scholar]
- 29.Jensen R, Sherin J, Emory S. Single Nanoparticle Based Optical pH Probe. Applied Spectroscopy. 2007;61:832–838. doi: 10.1366/000370207781540105. [DOI] [PubMed] [Google Scholar]
- 30.Wu Q, Zhang C, Li F. Preparation of spindle-shape silver core-shell particles. Materials Letters. 2005;59:3672–3677. [Google Scholar]
- 31.Fujita K, Ishitobi S, Hamada K, Smith N, Taguchi A, et al. Time-resolved observation of surface-enhanced Raman scattering from gold nanoparticles during transport through a living cell. Journal of Biomedical Optics. 2009;14 doi: 10.1117/1.3119242. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Yin H, Cooper J, Haswell S. Characterization of cellular chemical dynamics using combined microfluidic and Raman techniques. Analytical and Bioanalytical Chemistry. 2008;390:833–840. doi: 10.1007/s00216-007-1564-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stokes R, McBride E, Wilson CG, Girkin JM, Smith WE, et al. Surface-enhanced Raman Scattering Spectroscopy as a Sensitive and Selective Technique for Detection of Folic Acid in Water and Human Serum. Journal of Applied Spectroscopy. 2008;62:371–376. doi: 10.1366/000370208784046812. [DOI] [PubMed] [Google Scholar]
- 34.Stuart D, Yuen J, Lyandres N, Yonzon C, Glucksberg M, et al. In vivo glucose measurement by surface-enhanced Raman spectroscopy. Analytical chemistry. 2006;78:7211–7215. doi: 10.1021/ac061238u. [DOI] [PubMed] [Google Scholar]
- 35.Kneipp J, Kneipp H, Wittig B, Kneipp K. One-and Two-Photon Excited Optical pH Probing for Cells Using Surface-Enhanced Raman and Hyper-Raman nanosensors. Nanoletter. 2007;7:2819–2823. doi: 10.1021/nl071418z. [DOI] [PubMed] [Google Scholar]
- 36.Kneipp J, Kneipp H, Wittig B, Kneipp K. Novel optical nanosensors for probing and imaging live cells. Nanomedicine. 2010;6:214–226. doi: 10.1016/j.nano.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 37.Kneipp J, Kneipp H, Wittig B, Kneipp K. Following the Dynamics of pH in Endosomes of Live Cells with SERS Nanosensors. Journal of Physical Chemistry. 2010;114:7421–7426. [Google Scholar]
- 38.Lim JK, Joo S-W. Gold Nanoparticle-based pH sensor in Highly Alkaline Region at pH >11: Surface-enhanced Raman Scattering Study. Applied Spectroscopy. 2006;60:847–852. doi: 10.1366/000370206778062183. [DOI] [PubMed] [Google Scholar]
- 39.Shegai T, Vaskevich A, Rubinstein I, Haran G. Raman spectroelectrochemistry of Molecules within Individual Electromagnetic Hot Spots. Journal of the American Chemical Society. 2009;131:14390–14398. doi: 10.1021/ja904480r. [DOI] [PubMed] [Google Scholar]
- 40.Hu J, Zhao B, Xu W, Li B, Fan Y. Surface-enhanced Raman spectroscopy study on the structure changes of 4-mercaptopyridine adsorbed on silver substrates and silver colloids. Spectrochimica acta Part A, Molecular and biomolecular spectroscopy. 2002;58A:2827–2834. doi: 10.1016/s1386-1425(02)00074-4. [DOI] [PubMed] [Google Scholar]
- 41.Wang Z, Bonoiu A, Samoc M, Cui Y, Prasad P. Biological pH sensing based on surface enhanced Raman scattering through a 2-aminothiophenol-silver probe. Biosensors and Bioelectronics. 2008;23:886–891. doi: 10.1016/j.bios.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 42.Nowak-Lovato KL, Rector KD. Use of Targeted SERS Nanosensors in Whole Cell pH Analysis. Microscopy and Microanalysis. 2009;15:52–53. [Google Scholar]
- 43.MacGlashan DW. Endocytosis, recycling, and degradation of unoccupied fc epsilon RI in human basophils. Journal of leukocyte biology. 2007;82:1003. doi: 10.1189/jlb.0207103. [DOI] [PubMed] [Google Scholar]
- 44.Stump RF, Oliver JM, Cragoe EJ, Deanin GG. The Control of Mediator Release for RBL-2H3 Cells: Roles for Ca2+, Na+, and Protein Kinase C. The Journal of Immunology. 1987;139:881–886. [PubMed] [Google Scholar]
- 45.Barker SA, Caldwell KK, Halle A, Martinez AM, Pfeiffer JR, et al. Wortmannin Blocks Lipid and Protein Kinase Activities Associated with PI 3-Kinase and Inhibits a Subset of Responses Induced by Fc-epsilon-R1 cross-linking. Molecular Biology of the Cell. 1995;6:1145–1158. doi: 10.1091/mbc.6.9.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rivera J, Mullins JM, Furuichi K, Isersky C. Endocytosis of Aggregated Immunoglobulin G by Rat Basophilic Leukemia cells Rate Extend and Effects on the Endocytosis of Immunoglobulin E. Journal of Immunology. 1986;136:623–627. [PubMed] [Google Scholar]
- 47.Hansen SH, Sandvig K, Van Deurs B. The Preendosomal Comartment Comprises Distinct Coated and Noncoated Endocytic Vesicle Populations. Journal of cell biology. 1991;113:731–742. doi: 10.1083/jcb.113.4.731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mukherjee S, Ghosh RN, Maxfield FR. Endocytosis. Physiological Reviews. 1997;77:759–803. doi: 10.1152/physrev.1997.77.3.759. [DOI] [PubMed] [Google Scholar]
- 49.Barth H, Pfeifer G, Hofmann F, Maier E, Benz R, et al. Low pH-induced Formation of Ion Channels by Clostridium difficile Toxin B in Target Cells. Journal of Biological Chemistry. 2001;276:10670–10676. doi: 10.1074/jbc.M009445200. [DOI] [PubMed] [Google Scholar]
- 50.Bidani A, Heming T. Effects of bafilomycin A-1 on functional capabilities of LPS-activated alveolar macrophages. Journal of leukocyte biology. 1995;57:275–281. doi: 10.1002/jlb.57.2.275. [DOI] [PubMed] [Google Scholar]
- 51.Tanigaki K, Sasaki S, Ohkuma S. in bafilomycin A1-resistant cells, bafilomycin A1 raised lysosomal pH and both prodigiosins and concanmycin A inhibited growth through apoptosis. FEBS Letters. 2003;537:79–84. doi: 10.1016/s0014-5793(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 52.Cabado AG, Vieytes MR, Botana LM. Amiloride-dependent transport is the main mechanism implicated in sodium iinflux regulation in rat mast cells. Journal of Cellular Physiology. 1993;156:567–570. doi: 10.1002/jcp.1041560316. [DOI] [PubMed] [Google Scholar]
- 53.Kleyman T, Cragoe E. Amiloride and Its Analogs as Tools in the Study of Ion Transport. The Journal of membrane biology. 1988;105:1–22. doi: 10.1007/BF01871102. [DOI] [PubMed] [Google Scholar]
- 54.Taouil K, Feray J, Brunet J, Christen M, Garay RP, et al. Inhibition by xipamide of amiloride-induced acidification in cultured rat cardiocytes. European Journal of Pharmacology. 1997;324:289–294. doi: 10.1016/s0014-2999(97)00087-3. [DOI] [PubMed] [Google Scholar]
- 55.Clague MJ, Urbe S, Aniento F, Gruenberg J. Vacuolar ATPase is required for edosomal carrier vesicle formation. Journal of Biological Chemistry. 1994;269:21–24. [PubMed] [Google Scholar]
- 56.van Weert AWM, Dunn KW, Geuze HJ, Maxfield fR, Stoorvogel W. Transport from Late Endosomes to Lysosomes, but Not Sorting of Integral Membrane Proteins in Endosomes, Depends on the Vacuolar Proton Pump. Journal of Cellular Biology. 1995;130:821–834. doi: 10.1083/jcb.130.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Klausner RD, Van RJ, Kempf C, Rao K, Bateman JL, et al. Failure to Release Iron from Transferring in a Chinese Hamster Ovary Cell Mutant Pleiotropically Defective in Endocytosis. Journal of Cell Biology. 1984;98:1098–1101. doi: 10.1083/jcb.98.3.1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Klausner RD, Van Renswoude J, Ashell G, Kempf C, Schechter AN, et al. Receptor Mediated Endocytosis of Transferring in K-562 Cells. Journal of Biological Chemistry. 1983;258:4715–4724. [PubMed] [Google Scholar]
- 59.Johnson LS, Dunn KW, Pytowski B, McGraw TE. Endosome Acidification and Receptor Trafficking: Bafilomycin A1 Slows Receptor Exit by a Mechanism Involving the Receptor's Internalization Motif. Molecular Biology of the Cell. 1993;4:1251–1266. doi: 10.1091/mbc.4.12.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]