Abstract
The proteasome is the major ATP-dependent protease in eukaryotic cells, but limited structural information strongly restricts a mechanistic understanding of its activities. The proteasome regulatory particle, consisting of the lid and base subcomplexes, recognizes and processes poly-ubiquitinated substrates. We used electron microscopy and a newly-developed heterologous expression system for the lid to delineate the complete subunit architecture of the regulatory particle. Our studies reveal the spatial arrangement of ubiquitin receptors, deubiquitinating enzymes, and the protein unfolding machinery at subnanometer resolution, outlining the substrate’s path to degradation. Unexpectedly, the ATPase subunits within the base unfoldase are arranged in a spiral staircase, providing insight into potential mechanisms for substrate translocation through the central pore. Large conformational rearrangements of the lid upon holoenzyme formation suggest allosteric regulation of deubiquitination. We provide a structural basis for the ability of the proteasome to degrade a diverse set of substrates and thus regulate vital cellular processes.
Keywords: 26S proteasome, 19S regulatory particle, ubiquitin recognition, deubiquitination, AAA+ ATPase, cryoEM, UPS, PCI domain
The ubiquitin-proteasome system is the major pathway for selective protein degradation in eukaryotic cells. Covalent modification with a poly-ubiquitin chain targets damaged, misfolded, and short-lived regulatory proteins for ATP-dependent destruction by the 26S proteasome, a massive 1.5 MDa proteolytic machine. The proteasome thus controls a myriad of essential cellular processes, including the cell cycle, transcription, and protein quality control 1. Despite intensive study, however, the structural basis for substrate recognition and processing by the proteasome remains poorly understood.
The proteasome contains at least 32 different subunits that form a barrel-shaped 20S proteolytic core capped on either end by a 19S regulatory particle. The active sites of the peptidase are sequestered in an internal chamber, and access is controlled by the regulatory particle, which functions in substrate recognition, deubiquitination, unfolding, and translocation of the unfolded chains into the core 2,3,4,5.
The regulatory particle is composed of 19 subunits and can be divided into two subcomplexes, the lid and the base. The lid consists of nine non-ATPase proteins (Rpn3, 5–9, 11–12, and Sem1 in yeast), including the deubiquitinating enzyme (DUB) Rpn11, whose activity is essential for efficient substrate degradation 6,7. The base contains six distinct AAA+ ATPases, Rpt1-6, that form a hetero-hexameric ring (in the order Rpt1, 2, 6, 3, 4, 5 8) and constitute the molecular motor of the proteasome. The ATPases are predicted to use the energy of ATP binding and hydrolysis to exert a pulling force on substrate proteins, unfold them, and translocate the polypeptides through a narrow central pore into the peptidase chamber. In the presence of ATP, the C-termini of the ATPases bind dedicated sites on the α-subunit ring (α1–α7) of the 20S core, triggering the opening of a gated access channel and facilitating substrate entry 5,9,10,11. Besides Rpt1-6, the base contains four non-ATPase subunits: Rpn1, Rpn2, and the ubiquitin receptors Rpn10 and Rpn13. Additional ubiquitin shuttle receptors (Rad23, Ddi1, and Dsk2) are recruited to the base through interactions with Rpn1, which also binds a second, non-essential DUB, Ubp6 12,13,14.
While the proteolytic core has been well studied, there is only limited structural characterization of the regulatory particle 11,15,16,17. None of the 13 non-ATPase subunits, including the ubiquitin receptors and deubiquitinating enzymes, have been localized within this assembly. While it has been shown that efficient degradation depends on the length, linkage type, and placement of an ubiquitin chain, as well as the presence of an unstructured initiation site on a substrate 3,18,19, we are missing the topological information needed to explain these requirements. Thus, elucidating the architecture of the regulatory particle and the spatial arrangement of individual subunits is crucial to understanding the molecular mechanisms for substrate recognition and processing.
Here, we present the EM structure of the proteasome holoenzyme and the lid subcomplex. A newly-developed heterologous expression system for the lid facilitated the localization of all subunits within the regulatory particle, providing a complete architectural picture of the proteasome. The resulting structural understanding offers novel insight into the mechanisms of ubiquitin binding, deubiquitination, substrate unfolding, and translocation by this major eukaryotic proteolytic machine.
Recombinant expression of yeast lid in E. coli
We developed a system for the heterologous coexpression of all nine lid subunits from Saccharomyces cerevisiae in E. coli. This system allowed us to generate truncations, deletions, and fusion constructs that were used to localize individual subunits and delineate their boundaries within the lid. The recombinant, purified lid was analyzed in its subunit composition and stoichiometry by SDS PAGE (Fig. S1, S2) and tandem mass spectrometry (MS). The small, non-essential subunit Sem1 could not be detected, neither for the recombinant nor the endogenous lid that was isolated from yeast. All other subunits were present with the expected stoichiometry, and gel filtration analyses showed indistinguishable elution profiles for the heterologously expressed lid and its endogenous counterpart (data not shown). Furthermore, atomic emission spectroscopy confirmed that the essential Zn2+ ion was incorporated in Rpn11, suggesting proper folding in E. coli.
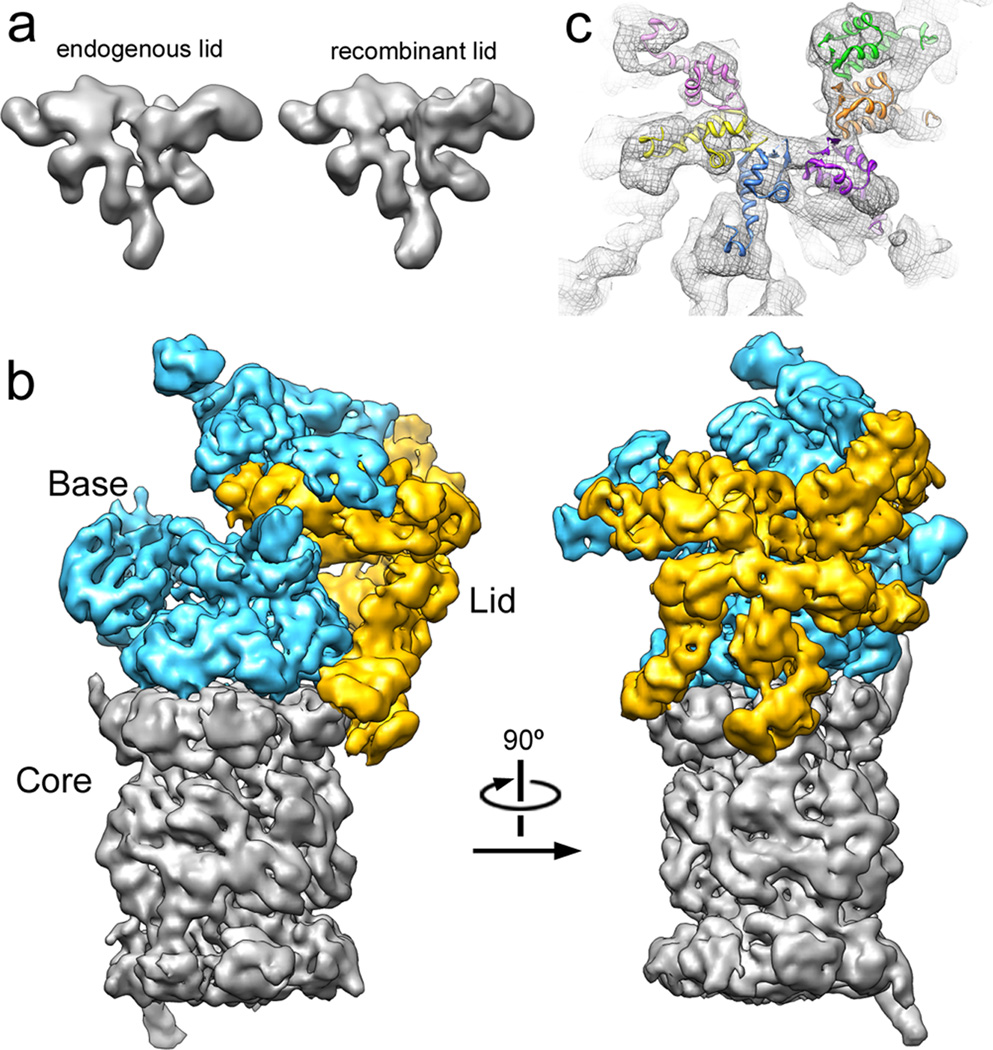
To compare the functionalities of recombinant and endogenous lid, we established conditions for their in-vitro reconstitution with base and 20S core subcomplexes from yeast to yield 26S holoenzyme. These reassembled particles were assayed for their activity in ubiquitin-dependent substrate degradation by using a poly-ubiquitinated GFP-cyclin fusion protein and following the decrease in GFP fluorescence. Proteasome reconstituted with E. coli-expressed lid supported robust substrate degradation (Fig. S3). Importantly, the 3D EM reconstructions from negative-stained samples of both lid subcomplexes are practically identical (Fig. 1a, S4), establishing this recombinant system as an ideal tool for our structural studies of the regulatory particle.
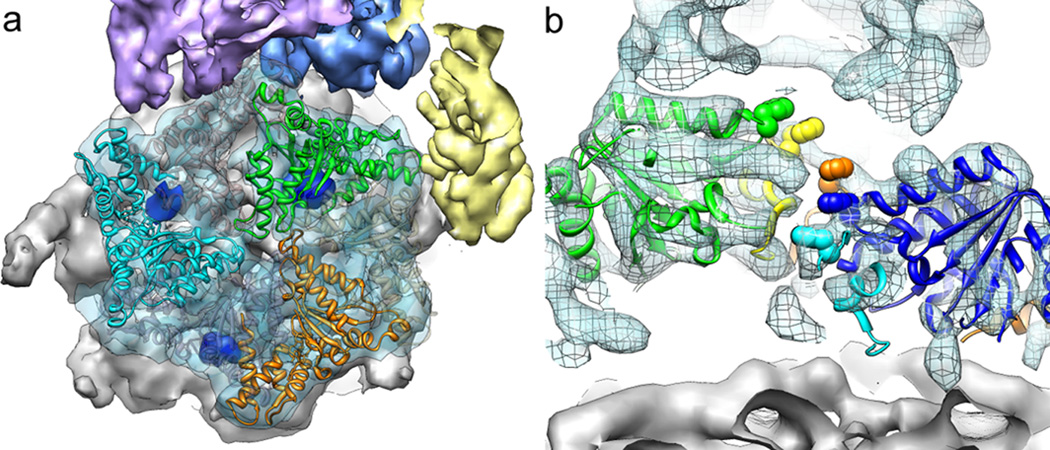
Figure 1. The lid subcomplex within the holoenzyme assembly.
a) Negative-stain 3D reconstruction at ~15 Å resolution shows resemblance between endogenous (left) and recombinant (right) lid. b) Locations of lid (yellow) and base (cyan) within the subnanometer holoenzyme reconstruction. c) Six copies of the crystal structure of a PCI domain (PDBid: 1RZ4) are docked into the lid electron density, showing a horseshoe-shaped arrangement of the winged-helix domains. Each domain is colored according to its respective lid subunit (Fig. 2).
Localization of regulatory particle subunits
As a first step in elucidating the architecture of the regulatory particle, we compared the single-particle EM reconstructions of the yeast holoenzyme and the isolated lid subcomplex obtained at 9 and 15 Å resolution, respectively (Fig. 1b, S5–7, movie S1). Docking the five-lobed, hand-shaped structure of the lid into the electron density of the holoenzyme revealed the lid’s position on one side of the regulatory particle, forming extensive interactions with the base subcomplex, but also contacting the 20S core. The lid subunits Rpn3, 5, 6, 7, 9, and 12 contain a C-terminal PCI (Proteasome-CSN-eIF3) domain that is assumed to play scaffolding roles and allow inter-subunit contacts 1. Our reconstruction provided sufficient resolution to unambiguously locate the winged-helix fold and the flanking helical segments of individual PCIs (Fig. 1c, movie S1). The C-terminal PCI domains of the six Rpn subunits thus interact laterally to form a horseshoe-shaped anchor from which the N-terminal domains radially extend. This arrangement demonstrates the scaffolding function of PCI domains in the lid, and we predict that similar interactions underlie the architecture of other PCI-containing complexes.
To determine the subunit topology of the lid, we utilized our heterologous E. coli expression system, fused maltose-binding protein (MBP) to the N- or C-terminus of individual subunits (Fig. S1), and localized the MBP within the tagged lid particles by negative-stain EM (Fig. S8a). None of the MBP fusions notably affected the lid structure, and we were able to identify the positions of all eight essential lid subunits and the relative orientation of their N- and C-termini. In combination with the PCI docking, the resolution of secondary structures in the cryo-electron density, and known molecular weights, this information allowed us to delineate approximate subunit boundaries. (Fig. 2a, movie S1)
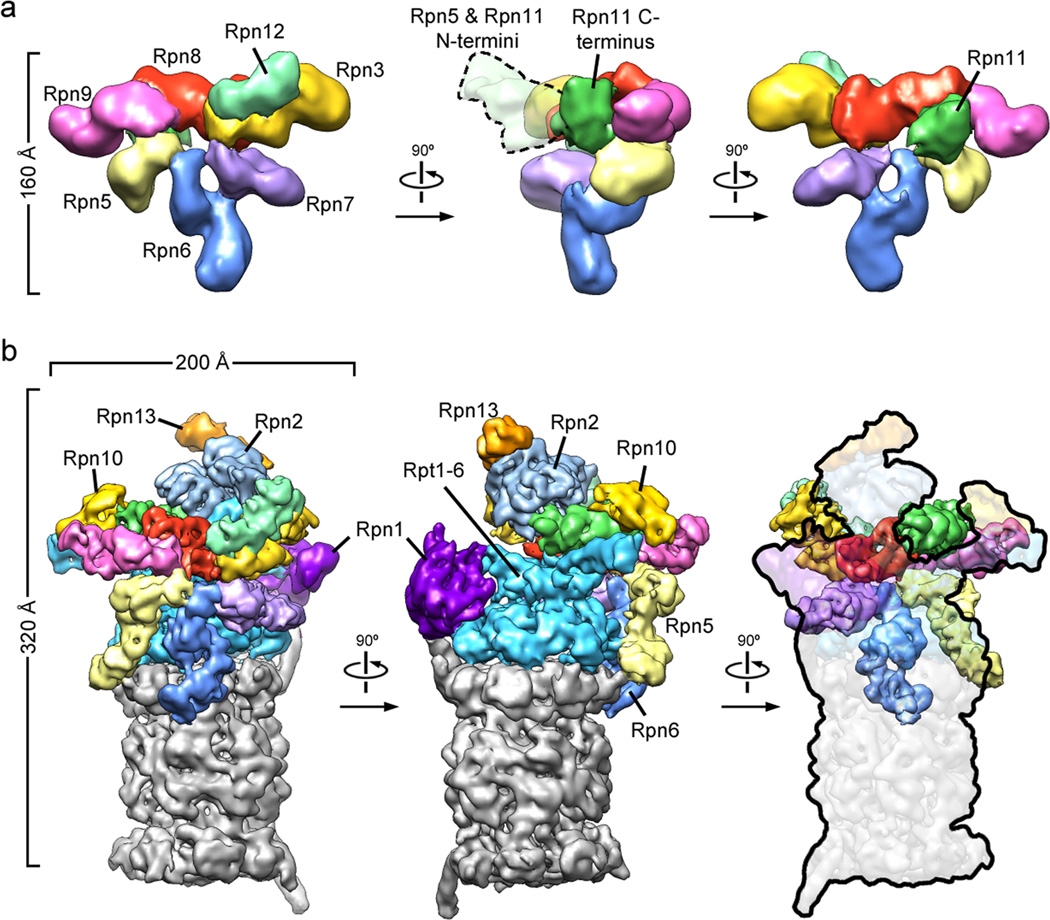
Figure 2. Three-dimensional reconstructions of the recombinant lid subcomplex and the yeast 26S proteasome.
a) Negative stain reconstruction of the isolated lid subcomplex at 15Å resolution, colored by subunit and shown from the exterior (left), the side (middle), and the interior, base-facing side (right). A dotted line (middle) indicates the highly variable electron density for the flexible N-terminal domains of Rpn5 and 11. b) Subnanometer cryoEM reconstruction of the holoenzyme, shown in three views corresponding to the isolated lid and colored as above, with the core particle in grey.
Overall, Rpn3, 7, 6, 5, and 9 form the fingers of the hand-shaped lid structure. Rpn8 shows an extended conformation that connects Rpn3 and 9, and thus closes the PCI horseshoe. In addition, it interacts with Rpn11, the only essential DUB of the proteasome, which lies in the palm of the hand and makes extensive contacts with Rpn8, 9, and 5.
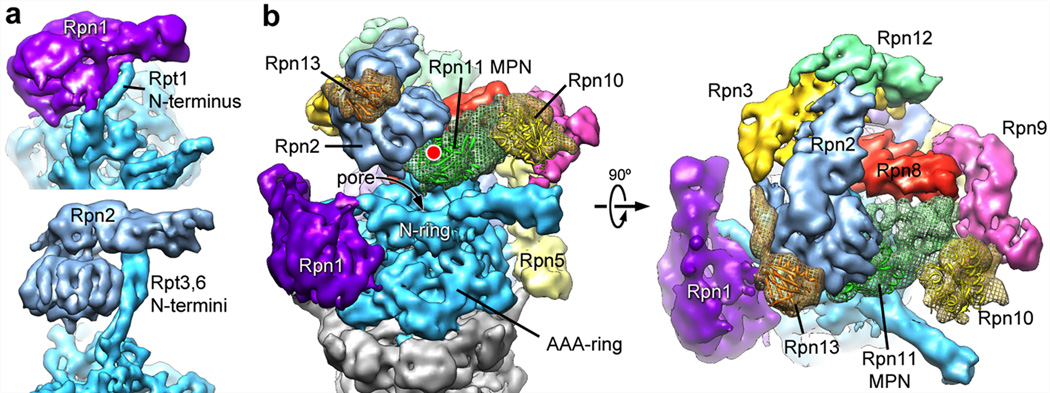
Using the topology determined for the isolated lid subcomplex, we delineated the individual lid subunits in the context of the holoenzyme (Fig. 2b). To complete the subunit assignment for the entire regulatory particle, the positions of Rpt1-6 in the base subcomplex were assigned according to established interactions with the core particle 15,20, whose crystal structure could be docked unambiguously into the EM density (Fig. S9). We localized the two large non-ATPases Rpn1 and 2 of the base subcomplex by antibody-labeling of a C-terminal FLAG tag and N-terminal fusion of gluthathione-S-transferase (GST), respectively (Fig. S2, S10a–c). Rpn1 and 2 had been predicted to contain numerous tetratricopeptide repeat (TPR)-like motifs and adopt α-solenoid structures 21. Indeed, we found a high structural resemblance between Rpn1 and 2, both consisting of a strongly curled solenoid that transitions into an extended arm towards the C-terminus (Fig. 3a). Rpn1 contacts the C-terminal helix of the 20S core subunit α4 and, based on the variability observed in our EM images, is likely to be flexible or loosely attached to the side of the base. Previous crystallography studies of the archaeal proteasome homolog PAN revealed that the N-terminal domains of the ATPases form a separate hexameric ring (N-ring) that consists of OB domains and three protruding coiled-coil segments 17,22. Each coiled coil is formed by the far N-terminal residues of two neighboring ATPases in the hexamer. Although Rpt1 and 2 do not appear to form an extended coiled coil, we find that the N-terminal helical portion of Rpt1 interacts with the solenoid and the C-terminal arm of Rpn1. Rpn2 is located above the N-ring and mounted atop the longest of the protruding coiled coils, formed by Rpt3 and 6. These interactions strongly resemble those observed between Rpt1 and Rpn1 (Fig. 3a).
Figure 3. Localization of Rpn1 and Rpn2, and ubiquitin-interacting subunits.
a) Rpn1 (top) and Rpn2 (bottom) are oriented to emphasize similarities in their domain structure and solenoid attachment to the extended N-terminal helices of Rpt1 and Rpt3/6, respectively. b) Side and top views of the regulatory particle, showing the locations of the ubiquitin receptors Rpn10 and 13, and the DUB Rpn11 relative to the central pore. Crystal structures for Rpn10 (PDBid: 2×5n), Rpn13 (PDBid: 2r2y), and an MPN domain homologous to Rpn11 (AMSH-LP, PDBid: 2znr) are shown docked into the EM density. The predicted active site of Rpn11 is indicated (red dot).
Localizing the ubiquitin receptors and DUBs within the regulatory particle is of particular interest. In addition to the DUB Rpn11 in the lid, we identified the positions of both intrinsic ubiquitin receptors, Rpn10 and 13, and of the base-associated DUB Ubp6 by imaging proteasome particles from yeast deletion strains (Fig. 3b, S10d–f). The ubiquitin receptor Rpn13 binds to Rpn2 as expected 23,24. The globular VWA domain of the second receptor Rpn10 had previously been shown to stabilize the lid-base interaction 25,26, however, we found that it does not directly contact the base. This domain bridges Rpn11 and 9, which might increase the lid-base affinity indirectly by stabilizing Rpn11 in its Rpn2-bound conformation (see below). The flexibly attached ubiquitin interacting motif (UIM) of Rpn10 likely contacts the coiled coil formed by Rpt4 and 5, stabilizing its position relative to other subunits and potentially communicating with the AAA+ motor. The DUB Ubp6 appears to be flexible and does not give rise to ordered density. Nonetheless, variance maps indicate that it interacts with the C-terminal arm of Rpn1, as suggested by immunoprecipitations 14.
Inter-subcomplex contacts
The complete localization of subunits within the holoenzyme revealed unexpected contacts between the lid and core subcomplexes. Rpn5 and Rpn6 form fingers that touch the C-termini of the core subunits α1 and α2, respectively. We confirmed the interaction between Rpn6 and α2 by in-vitro crosslinking, using an engineered cysteine in α2 and a 7-Å heterobifunctional crosslinker (Fig. S11). These previously unknown direct interactions between lid and core may stabilize the entire holoenzyme assembly, and/or be part of an allosteric network that modulates the activities of either subcomplex.
Our holoenzyme structure shows that Rpn3, 7, 8, and 11 make extensive contacts with the base. Compared to their positions in the isolated lid, Rpn8 and 11 have undergone significant conformational changes in the holoenzyme (Fig. 4). The C-terminus of Rpn8 is detached from Rpn3 to interact with the coiled-coil of Rpt3/6, while the N-terminal MPN domain of Rpn11 extends towards the center of the regulatory particle to bind the solenoid portion of Rpn2. Similarly, the N-terminal region of Rpn3 is more elongated than in the isolated lid and also contacts the Rpn2 solenoid, but from the opposite side. In turn, the extended C-terminal arm of Rpn2 interacts with Rpn3 and 12, and thus forms a direct connection between the solenoid section of Rpn2, the coiled coil of Rpt3/6, and the lid (Fig. 3b).
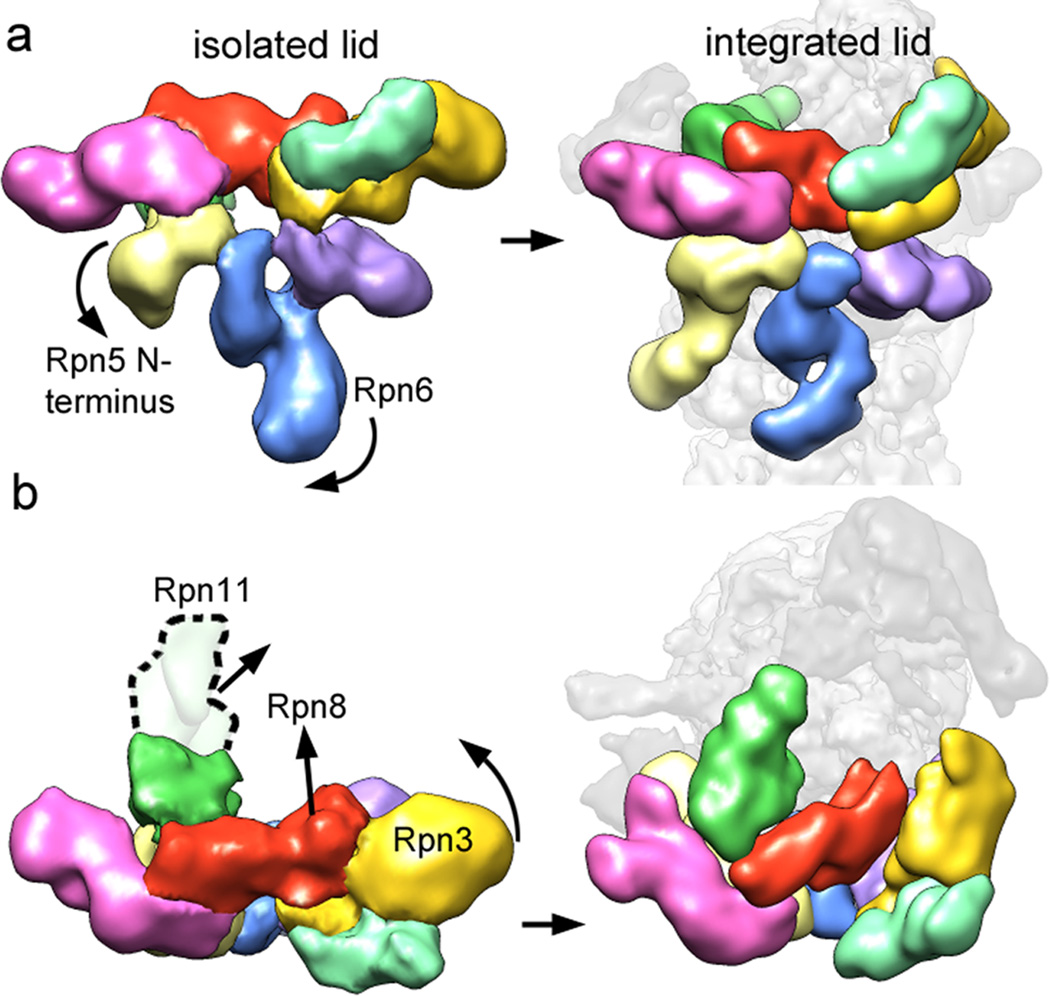
Figure 4. Conformational rearrangements of the lid subcomplex upon integration into the holoenzyme.
The lid complex in its isolated (left) and integrated (right) state is shown as viewed from the exterior (a) and top (b) of the regulatory particle. Major subunit rearrangements are depicted by arrows. The N-terminus of Rpn5 (light yellow) interacts with Rpn11 in the isolated complex, and swings down to contact the core particle upon incorporation into the holoenzyme. The N-terminal domain of Rpn6 swings to the left to interact similarly with the core particle. Rpn3, 8, and 11 undergo dramatic rearrangements, in which they move towards the center of the regulatory particle.
We speculate that Rpn2 stabilizes a lid conformation in which Rpn3, 8, and the DUB Rpn11 extend towards the base (Fig. 4b). Together, the lid, Rpn2, and the coiled coils of the N-ring appear to function as a scaffold that positions the two intrinsic ubiquitin receptors Rpn10 and 13, and the DUB Rpn11 for substrate binding, deubiquitination, and transfer to the subjacent central pore of the AAA+ motor (Fig. 3b). Interestingly, several lid subunits directly interact with AAA+ domains of the Rpts. Rpn7 contacts the AAA+ domains of Rpt2 and Rpt6, while Rpn6 and Rpn5 touch Rpt3. These interactions with contiguous motor domains are surprising, because current models for ATP-dependent unfoldases suggest significant conformational changes of individual subunits in the hexamer during ATP hydrolysis and substrate translocation 27,28,29. The observed contacts between lid and the motor domains might form only transiently; alternatively, the AAA+ ring of the proteasome may be much more static than previously assumed.
Lid conformational changes may regulate DUB activity
Comparing the structures of the lid in isolation and when bound to holoenzyme revealed major conformational changes that suggest an allosteric mechanism for the regulation of Rpn11 DUB activity (Fig. 4). In the isolated lid, the N-terminal MPN domain of Rpn11 forms extensive interactions with Rpn9 and the curled up Rpn5 finger. Upon lid binding to the holoenzyme, this Rpn5 finger swings down to contact the α1 subunit of the 20S core and thereby releases Rpn11, which then extends towards the Rpn2 solenoid. Docking the MPN domain of a related DUB (PDBid: 2znr) into the electron density of Rpn11 indicates the approximate location of the active site (Fig. 3b). The interactions of Rpn11 with Rpn9 and 5 in the free lid likely restrict access to this active site, which would prevent futile substrate deubiquitination in the absence of base and 20S core, and explain previous observations that the lid subcomplex has DUB activity only within the holoenzyme 7 (and our unpublished data).
Functional asymmetry in the AAA+ unfoldase
Our subnanometer structure of the holoenzyme provides new insights into the architecture and potential mechanisms of the base AAA+ unfoldase. As suggested by previous EM studies 15,16, the ring of the base and the 20S core are slightly offset from a coaxial alignment, with the base shifted by ~10 Å towards the lid (Fig. 5a). Despite or perhaps due to this offset, the C-terminal tails of Rpt2, Rpt3, and Rpt5 are docked into their cognate 20S binding pockets at the interfaces of the subunits α3 and α4, α1 and α2, and α5 and α6, respectively. Those three Rpt tails contain the terminal HbYX motif, which is critical for triggering gate opening in the 20S core 5,10, and indeed our structure is consistent with an open gate conformation. The tails of Rpt1, 4, and 6 lack this motif and were not observed to statically interact with 20S in our holoenzyme structure.
Figure 5. Structural features of the base ATPase subunits.
a) Positions of Rpt2 (cyan), 3 (green), and 5 (orange) within the base hexameric ring and relative to the 20S core (grey) are shown using fitted crystal structures of the homologous PAN AAA+ domain (PDBid: 3h4m). The EM density contains the molecular envelope of the C-terminal tails (dark blue), docked into their cognate binding sites on the 20S core. Corresponding densities were not found for the tails of the Rpt1, 4, and 6 (grey ribbon structure). b) Cutaway side view of the holoenzyme EM density with Rpt1-5 visible. Individually docked copies of the PAN crystal structure reveal a spiral staircase arrangement of the Rpts, emphasized by space-filling representations of the PAN pore-1 loop residues (not resolved in the Rpts).
Current mechanistic models for AAA+ unfoldases predict that ATPase subunits in the hexamer are in different nucleotide states and undergo significant conformational changes driven by coordinated ATP-hydrolysis 27,30,31. Because we determined the structure of wild-type proteasome in the presence of saturating ATP, we expected that different complexes would have any given Rpt subunit in different conformations, leading to reduced electron density or low resolution when averaging thousands of these unsynchronized motors. However, our reconstruction shows highly ordered density throughout the AAA+ domains of all six Rpts. While the C-terminal ‘small AAA+’ subdomains (except for Rpt6) arrange in one plane above the 20S core, the ‘large AAA+’ subdomains of Rpt1-5 are oriented in a spiral staircase around the hexameric ring, with Rpt3 at the highest and Rpt2 at the lowest position (Fig. 5b, movie S1). The AAA+ domain of Rpt6 adopts a tilted orientation, bridging Rpt2 and 3. Similar staircase arrangements have been previously observed for helicases of the AAA+ and RecA superfamilies 32,33. It was suggested that during ATP hydrolysis, individual subunits progress through the different conformational stages of the staircase, thereby translocating substrate through the pore. The particular staircase orientation we observed identically for all proteasome particles may represent a low-energy state of the base, adopted under our experimental conditions. Alternatively, this staircase arrangement of Rpt1-6 may be static and reflect the functional state of the base, in which substrates are translocated by local motions of the pore loops while the relative positions of the motor subunits stay fixed. Future biochemical and structural studies will be required to distinguish between these two models.
Spatial arrangement of ubiquitin receptors and DUBs
Localizing all subunits of the regulatory particle enabled us to infer the requirements and potential mechanisms for the recognition and degradation of ubiquitin-tagged substrates (Fig. 6). After a substrate binds to an ubiquitin receptor, its poly-ubiquitin chain must be removed by Rpn11 cleavage at the proximal ubiquitin to permit subsequent fast degradation 6,7. To allow cleavage without disengaging from the receptor, an ubiquitin chain must be long enough to span the distance between receptor and DUB. Based on our structure, both Rpn13 and the UIM of Rpn10 are located 70–80 Å from the predicted position of the Rpn11 MPN domain (Fig. 3b). The shuttle receptors Rad23, Ddi1, and Dsk2 are expected to reside ~80–120 Å away from Rpn11, depending on where they bind Rpn1 13. For receptor interaction, at least part of the ubiquitin chain has to be in an extended conformation with the hydrophobic patches exposed 24,34,35. Since a single ubiquitin moiety in an extended K48-linked chain contributes ~30 Å in length 36, it would take three ubiquitins to span the distance between Rpn10 or 13 and Rpn11. Moreover, both Rpn10 and Rpn13 bind between two consecutive ubiquitin moieties 24,34, such that at least a tetra-ubiquitin chain would be required on a substrate to allow interaction with a receptor and simultaneous deubiquitination by Rpn11 (Fig. 6). This model agrees with in-vitro studies that indicate a minimum of four K48-linked ubiquitins is necessary for efficient substrate degradation 3, although this number may differ for other chain types 37. Given the arrangement of Rpn10 and 13, an ubiquitin chain would have to be significantly longer to interact with both receptors. However, knockout studies have shown that ubiquitin chains are not required to bind to multiple receptors simultaneously 38.
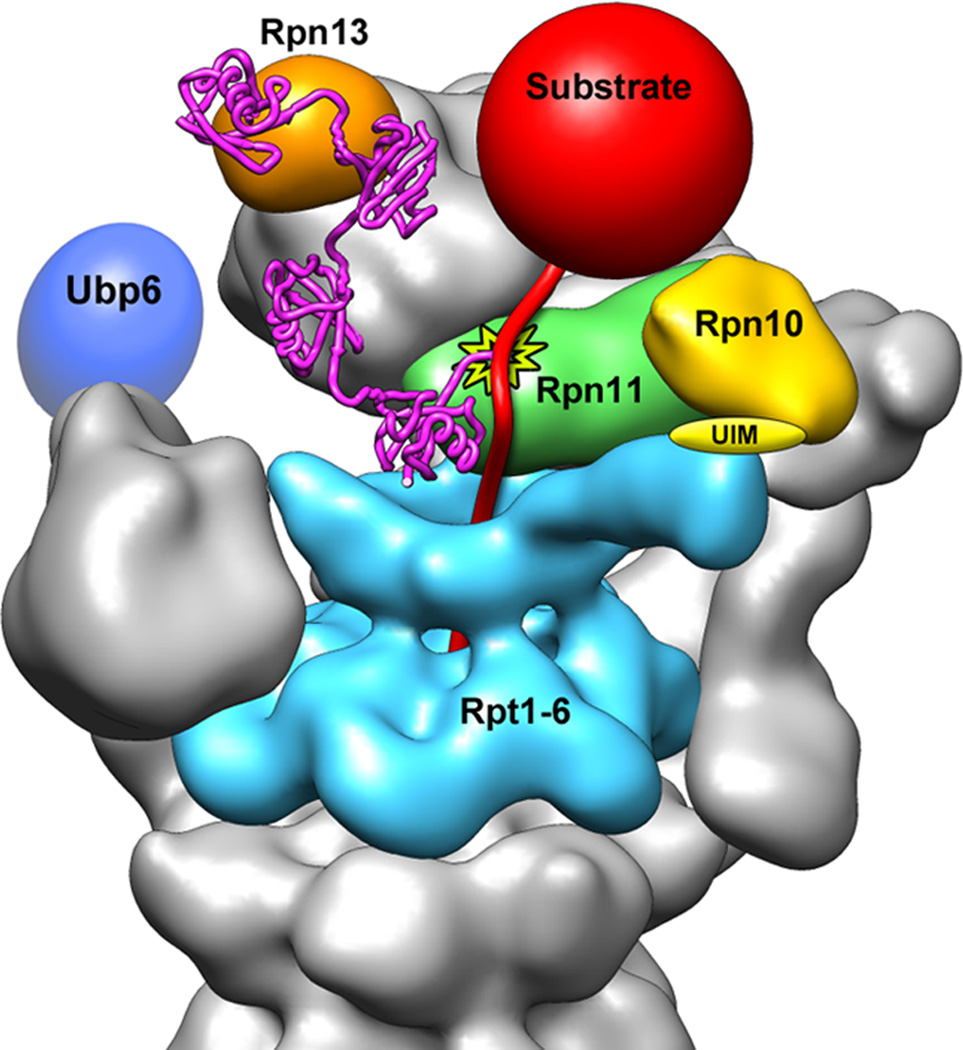
Figure 6. Model for the recognition, deubiquitination, and engagement of a poly-ubiquitinated substrate by the 26S proteasome.
A K48-linked tetra-ubiquitin chain (magenta, PDBid: 2kde) is conjugated to the unstructured initiation region of a substrate (red) and bound to the ubiquitin receptor Rpn13 (orange). The substrate is poised for deubiquitination by Rpn11 (green, active site indicated by star), and its unstructured initiation region is engaged by the translocation machinery of the base (cyan). A poly-ubiquitin chain could alternatively bind to the UIM of Rpn10 (yellow) or interact with both receptors simultaneously. The DUB Ubp6 is localized further from the central pore, in a position to trim excess ubiquitin chains.
In contrast to Rpn11, Ubp6 is known to cleave within poly-ubiquitin chains or trim them from their distal end 39. Of all the ubiquitin-interacting subunits in the regulatory particle, we found Ubp6 to be the furthest away from the entrance to the pore, which may allow it to clip extended or unnecessary ubiquitin chains from substrates. Because Ubp6 is located closer to Rad23, Dsk2, or Ddi1 than to Rpn10 or 13, it may preferentially act on substrates delivered by these shuttle receptors.
To avoid dissociation upon deubiquitination, a substrate polypeptide must be engaged with the unfolding machinery of the base before or shortly after removal of its ubiquitin chain. Engagement by the base is known to depend on an unstructured initiation site or “tail” on the substrate 40, which needs to be long enough to reach through the narrow N-ring and into the AAA+ pore (Fig. 6). In addition, this tail would have to be sufficiently spaced from the attachment point of the poly-ubiquitin chain to allow concurrent substrate engagement by the pore and deubiquitination by Rpn11. The distance between the predicted active site of Rpn11 and the AAA+ pore below the N-ring is ~60 Å, which could easily be bridged by 40–45 unstructured residues or a shorter tail combined with a folded structure.
Alternative to the above model for simultaneous receptor binding and deubiquitination, it has been proposed that commencing substrate translocation by the base might move the proximal ubiquitin from a receptor towards Rpn11 for cleavage 7. Our structure suggests for this model that efficient substrate processing would only require a mono- or di-ubiquitin for receptor binding and a 50–60 Å longer spacing between the ubiquitin and the flexible tail to reach the AAA+ pore. This length dependence of engagement is consistent with recent in-vitro degradation studies, using model substrates with different lengths and ubiquitin modifications 19. Future experiments will be required to assess whether substrates get deubiquitinated in a translocation-dependent or –independent manner.
Summary
The work presented here defines the architecture of the entire proteasome regulatory particle and provides a much-needed structural framework for the mechanistic understanding of ubiquitin-dependent protein degradation. We localized Rpn11 directly above the entrance of the pore, surrounded by the ubiquitin receptors Rpn10 and 13. This insight allows us to visualize the substrate’s path towards degradation and will be critical in elucidating how the characteristics of ubiquitin modifications affect substrate recognition and processing. Moreover, our study significantly furthers the understanding of the heterohexameric AAA+ motor of the proteasome. Individual ATPase subunits were found in a spiral staircase arrangement and may operate with more limited dynamics than previously assumed for AAA+ protein unfoldases.
Unexpectedly, the lid is bound to the side of the holoenzyme and interacts with both the base and core particle. These interactions induce major conformational changes in lid subunits that may allosterically activate the DUB Rpn11, allowing critical removal of ubiquitin chains during substrate degradation in the holoenzyme, while preventing futile deubiquitination by the isolated lid. In addition, contacts between the subcomplexes could have unexplored roles in coordinating individual substrate processing steps, for instance ubiquitin binding, deubiquitination, and the onset of translocation. The intricate architecture of the proteasome highlights the complex requirements for this proteolytic machine, which must accommodate and specifically regulate a highly diverse set of substrates in the eukaryotic cell.
Methods Summary
Protein expression and purification
Endogenous holoenzyme, core particle 41, and lid subcomplex 42 were purified from S. cerevisiae essentially as described. The base subcomplex was purified according to protocols for the holoenzyme preparation, but with minor modifications as described in the Full Methods. Details of yeast strain construction are provided in Table S1.
Yeast lid was recombinantly expressed from three plasmids in E. coli BL21-star (DE3), and purified on anti-FLAG M2 resin and by size-exclusion chromatography (see Full Methods).
Electron microscopy and image analysis
All electron microscopy data were collected using the Leginon data collection software 43 and processed in the Appion EM processing environment 44. Three-dimensional maps were calculated using libraries from the EMAN2 and SPARX software packages 45,46. UCSF Chimera was used for volume segmentation, atomic coordinate docking, and figure generation 47.
Full Methods
Recombinant lid construction and purification
Yeast Rpn5, Rpn6, Rpn8, Rpn9, and Rpn11-6xHis were cloned into pETDuet-1 (Novagen), yeast Rpn3, FLAG-Rpn7, and Rpn12 were cloned into pCOLADuet-1 (Novagen), and yeast Sem1 and Hsp90 were cloned into pACYCDuet-1 (Novagen). A T7 promoter preceded each gene and each plasmid contained a T7 terminator following the multiple cloning site. Genes for select rare tRNAs were included in the pACYCDuet-1 plasmid to account for codon-usage differences between yeast and E.coli. To ensure full-length of Rpn6 in lid particles used for biochemical experiments and the negative stain reconstruction of recombinant lid, we used a construct with the FLAG tag moved from Rpn7 to Rpn6. E. coli BL21-star (DE3) cells were co-transformed with the three plasmids mentioned above. Lid proteins and the chaperone Hsp90 were coexpressed overnight at 18 °C after inducing cells with 0.5 mM isopropyl-β-D-thiogalactopyranoside at OD600 = 0.7. Cells were harvested by centrifugation (4,000 g for 30 min), resuspended in FLAG buffer (50 mM HEPES, pH 7.6, 100 mM NaCl, 100 mM KCl and 5% glycerol) supplemented with protease inhibitors and 2 mg/mL lysozyme, and sonicated on ice for 2 min in 15-s bursts. The lysate was clarified by centrifugation (27,000 g for 30 min), and the complex was affinity-purified on anti-FLAG M2 resin (Sigma-Aldrich) using an N-terminal FLAG-tag on Rpn6 or 7. The protein was concentrated in a 30,000 MWCO concentrator (Amicon) for further purification on a Superose 6 size-exclusion column (GE Healthcare) equilibrated in FLAG buffer. Intact, assembled lid particles eluted at 13.1 mL, similar to lid purified from yeast.
His6-tagged yeast Rpn10 was expressed in E. coli and purified by Ni-NTA affinity and size-exclusion chromatography.
Yeast strain construction
Wild-type holoenzyme was purified from the strain YYS40 (MATa ade2-1 his3-11,15 leu2-3,112 trp1-1 ura3-1 can1 RPN11::RPN11-3XFLAG (HIS3))48. To generate RPN10, RPN13, and UBP6 deletion strains, the kanMX6 sequence was integrated at the respective genomic locus, replacing the gene in YYS40. To generate the strains used to purify GST-Rpn2, GFP-Rpn5 and GFP-Rpn8 holoenzyme, sequences encoding the respective tags under the control of the PGAL1 promoter were integrated 5’ of the respective genes in YYS40. To generate the strain used to purify Rpn1-FLAG holoenzyme, a sequence encoding the FLAG-tag was integrated 3’ to RPN1 in aW303 background strain (MATa ade2-1 his3-11 leu2-3,112 trp1-1 ura3-1 can1-100 bar1).
To generate the strains used to purify α2 mutant-containing core particle for the crosslinking experiments shown in Figure S11, pRS305 (LEU2) containing the mutant α2 and the genomic sequences found 500 nucleotides upstream and 100 nucleotides downstream of the gene was integrated at the LEU2 locus of RJD1144 (MATa, his3Δ200 leu2-3,112 lys2-801 trpΔ63 ura3-52 PRE1-FLAG-6xHIS::Ylplac211 (URA3)) 41, and the chromosomal copy of α2 was deleted. To generate the strain used to purify lid with Rpn6-3xHA for crosslinking, the 3xHA sequence was integrated 3’ of RPN6 in YYS40.
Expression and purification of yeast holoenzyme and subcomplexes
Endogenous holoenzyme, core particle 41, and lid subcomplex 42 were purified from S. cerevisiae essentially as described. Frozen yeast cells were lysed in a Spex SamplePrep 6870 Freezer/Mill. For holoenzyme purification, lysed cells of a strain containing a FLAG-tag on Rpn11 were resuspended in lysis buffer containing 60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.2% NP-40, and ATP regeneration mix (5 mM ATP, 0.03 mg/mL creatine kinase, 16 mM creatine phosphate). Holoenzyme was bound to anti-FLAG M2 resin and washed with wash buffer (60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 0.1% NP-40, and 500 µM ATP) before elution with 3XFLAG peptide and separation over Superose-6 in gel-filtration buffer (60 mM HEPES pH 7.6, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 10% glycerol, and 500 µM ATP). Lid, base, or core particle were purified similarly but from different yeast strains and including a salt wash to separate subcomplexes. Lid was purified from a yeast strain containing Rpn11-FLAG using a 900 mM NaCl wash. Base was purified from a yeast strain containing a C-terminal FLAG tag on Rpn2 and including a 500 mM NaCl wash, with 500 µM ATP present throughout the purification. Core particle was purified from a yeast strain containing a FLAG-6XHis tag on Pre1 and including a 500 mM NaCl wash. All subcomplexes were further purified by size-exclusion chromatography on Superose-6 in gel filtration buffer (see above).
GFP degradation assay
Proteasome holoenzyme was reconstituted from 20S core, base, Rpn10, and recombinant or endogenous yeast lid in the presence of ATP. A GFP-titin-cyclin fusion protein was modified with a K48-linked poly-ubiquitin chain 49 and degraded by reconstituted proteasome at 30°C in FLAG buffer with an ATP-regeneration system (5 mM ATP, 16 mM creatine phosphate, 6 µg ml−1 creatine phosphokinase). Degradation was monitored by the loss of fluorescence using a QuantaMaster spectrofluorimeter (PTI).
Protein crosslinking
Sulfo-MBS (Thermo Scientific) is a short (7.3Å), heterobifunctional crosslinker, whose maleimide moiety reacts primarily with sulfhydryls between pH 6.5 and 7.5, and whose NHS ester reacts with primary amines between pH 7 and 9. We purified core particle from yeast strains in which the only copy of the core α2 subunit was either WT, a D245C mutant, or an A249C mutant. Other intrinsic cysteines of the core were found largely non-reactive towards sulfhydryl-modifying agents (not shown). 10 µM reduced core particle purified from strains containing WT, A249C, and D245C α2 was incubated with 150 µM sulfo-MBS for 15 min at pH 6.5, allowing conjugation of the crosslinker to cysteines. Core particle was buffer-exchanged to remove excess crosslinker and increase the pH to 7.5, activating the amine-reactive functional group on sulfo-MBS. This core particle was added at a final concentration of 2 µM to a proteasome reconstitution mixture, containing 2 µM purified base, 10 µM purified Rpn10, 0.5 mM ATP, and 2 µM lid purified from a yeast strain in which Rpn6 was C-terminally tagged with a 3× hemagglutinin (HA) tag. Crosslinking was allowed to proceed for 15 min before reactions were stopped by the addition of 0.5 mM glycine pH 7.5 and divided equally for separation by SDS-PAGE, followed by either coomassie staining or anti-HA western blotting.
Electron Microcopy
Sample preparation for EM
Negative stain analysis of both the purified proteasome lid and holoenzyme complexes was performed using 400 mesh continuous carbon grids that had been plasma cleaned in a 75% Argon / 25% Oxygen atmosphere for 20 seconds using a Solarus plasma cleaner (Gatan, Inc). Due to the tendency for holoenzyme to adopt a preferential orientation on the carbon substrate, 5 µl of 0.1% poly L-lysine hydrobromide (Polysciences Inc. cat #09730) was placed onto the hydrophilized carbon grids and adsorbed for 90 seconds, washed twice with 5ul drops of water, and allowed to dry completely. This polylysine step was skipped when preparing grids containing the lid samples, as the lid does not adopt a preferred orientation on the carbon substrate. The remaining steps were identical for both holoenzyme and lid. A 4 µl drop of sample at a concentration of 25 µM was placed onto the grid and allowed to adsorb for one minute. The grid was blotted to near-dryness and a 4 µl drop of fresh 2% (w/v) uranyl formate was quickly placed onto the grid. In order to reduce the amount of glycerol remaining on the grids, they were subsequently floated on four successive 25 µl drops of the uranyl formate solution, waiting ten seconds on each drop. The grids were then blotted to dryness.
Preservation of both lid and holoenzyme complexes in vitreous ice was performed in the same manner. 400-mesh C-flats containing 2um holes with a spacing of 2 µm (Protochips Inc.) were plasma cleaned in a 75% Argon / 25% oxygen atmosphere for 8 s using a Solarus plasma cleaner (Gatan, Inc). The purified sample, at a concentration of 5 µM in a buffer containing 5% glycerol, was first diluted 1:5 from 60 mM HEPES, pH 7.6, 50 mM NaCl, 50 mM KCl, 5 mM MgCl2, 0.5 mM EDTA, 10% glycerol, 1 mM DTT, 0.5 mM ATP into a buffer containing 20 mM HEPES, pH 7.6, 50 mM NaCl, 50 mM KCl, 1 mM ATP, 1 mM DTT, and 0.05% NP40, and 4 µl aliquots were placed onto the grids. Grids were immediately loaded into a Vitrobot (FEI company) whose climate chamber had equilibrated to 4 °C and 100% humidity. The grids were blotted for 3 s at an offset of −1 mm, and plunged into liquid ethane. The frozen grids were transferred to a grid box and stored in liquid nitrogen until retrieved for data collection.
EM data collection
Negative stain analysis of the lid and holoenzyme samples was performed using a Tecnai T12 Bio-TWIN and a Tecnai F20 TWIN transmission electron microscope operating at 120keV. Lid samples were imaged at a nominal magnification of 68,000X (1.57 Å/pixel at the specimen level) on the T12, and 80,000X (1.45 Å/pixel) on the F20. Holoenzyme samples were imaged at a magnification of 49,000X (2.18 Å/pixel) on the T12, and 50,000X (2.16 Å/pixel) on the F20. T12 data were acquired on a F416 CMOS 4K×4K camera (TVIPS), F20 data were acquired on a Gatan 4K×4K camera, and all micrographs were collected using an electron dose of 20 e−/Å2 with a randomly set focus ranging from −0.5 to −1.2 µm. The automatic rastering application of the Leginon data collection software was used for data acquisition. Between 300–500 micrographs were collected for each of the negatively stained datasets.
For cryoEM, individual grids were loaded into a 626 single tilt cryotransfer system (Gatan, Inc) and inserted into a Tecnai F20 TWIN transmission electron microscope operating at 120 keV. Data were acquired at a nominal magnification of 100,000X (1.08 Å/pixel) using an electron dose of 20 e−/Å2 with a randomly set focus ranging from −1.2 to −2.5 µm. A total of 9,153 micrographs were collected of the holoenzyme using the MSI-T application of the Leginon software. While the holoenzyme was remained intact during the freezing process, the isolated lid specimen became completely disassembled during the freezing process. In an attempt to overcome this, the isolated lid was also frozen using grids onto which a thin carbon film was floated. Due to the elevated background noise from the addition of a carbon substrate, the resulting images lacked the sufficient signal-to-noise ratio necessary to solve a cryoEM structure of the isolated lid to a better resolution than the negative stain structure.
Image processing of negative stain data
All image pre-processing and two-dimensional classification was performed in the Appion image processing environment 44. Due to the large number of datasets acquired for both the negatively stained lid and holoenzyme complexes, a generalized schema was utilized for image analysis. This schema also minimized user bias during comparison of tagged and deletion constructs with their wild type counterparts. The contrast transfer function (CTF) of each micrograph was estimated concurrently with data collection using the ACE2 and CTFFind programs 50,51, providing a quantitative measurement of the imaging quality. Particle selection was also performed automatically concurrent with data collection. Negatively stained lid particles were selected from the micrographs using a difference of Gaussians (DoG) transform-based automated picker 52, and holoenzyme particles were selected using a template-based particle picker. Micrograph phases were corrected using ACE2, and both lid and holoenzyme particles were extracted using a 288×288-pixel box size. The data were then binned by a factor of two for processing. Each particle was normalized to remove pixels whose values were above or below 4.5 sigma of the mean pixel value using the XMIPP normalization program 53.
In order to remove aggregation, contamination, or other non-particle selections, particle stacks were decimated by a factor of 2 and subjected to five rounds of iterative multivariate statistical analysis (MSA) and multi-reference alignment (MRA) using the IMAGIC software package 54. 2D class averages depicting properly assembled complexes were manually selected, and the non-decimated particles contributing to these class averages were extracted to create a new stack for further processing. In order to include a larger range of holoenzyme views, particles contributing to doubly capped proteasome averages were removed. This stack of particles went through five rounds of MSA/MRA in IMAGIC 54, and a final correspondence analysis and classification based on Eigen images using the SPIDER software package 55 was performed to generate 2D class averages of the complexes.
Initial models for reconstructions of both the holoenzyme and lid were determined using the established “C1 startup” routines in IMAGIC. 2D class averages were manually inspected to select three images representing orthogonal views of the complex, which were in turn used to assign Eulers in a stepwise fashion to the entire dataset of reference-free class averages. The resulting low-resolution models of the lid and holoenzyme were low-pass filtered to 60Å resolution, and these densities were used as starting points for refinement of the 3D structure.
3D reconstructions were all performed using an iterative projection-matching and back-projection approach using libraries from the EMAN2 and SPARX software packages 45,46. Refinement of the starting models began using an angular increment of 25°, progressing down to 2° for the lid, and 1° for the holoenzyme. The refinement only continued to the subsequent angular increment once greater than 95% of the particles showed a pixel error of less than 1 pixel. The resolution was estimated by splitting the particle stack into two equally sized datasets, calculating the Fourier shell correlation (FSC) between the resulting back-projected volumes. The estimated resolutions for the final endogenous and recombinant lid structures based on their FSC curves at 0.5 were about 15Å.
Image processing of cryoEM holoenzyme
Processing of the holoenzyme cryo dataset proceeded in a very similar fashion to that of the negatively stained particle datasets. Only ACE2 was used to estimate CTF of the images and measure image quality, and particles were extracted using a box size of 576 pixels. Reference-free 2D classification was performed in order to remove particles that did not contribute to averages depicting a doubly capped proteasome. Three rounds of reference-free 2d classification, and particles were removed after each round. From an initial dataset of 312,483 automatically selected particles, 93,679 were kept for the 3D reconstruction. C2 symmetry was applied to one of the previously determined asymmetric negative stained reconstructions to serve as a starting model for structure refinement. The reconstruction began using an angular increment of 25°, and iterated down to 0.6°. C2 symmetry was imposed during the reconstruction. Low-resolution Fourier amplitudes of the final map were dampened matching the amplitudes of the density map to match those of an experimental GroEL SAXS curve using the SPIDER software package 55.
The estimated resolution based on the FSC of the half-volumes at 0.5 was ~9 Å, although a local resolution calculation using the “blocres” function in the Bsoft package 56 indicated a range of resolutions within the density. The majority of the core particle subunits and the AAA+ ATPases were resolved to between 7 and 8 Å resolution, while the non-ATPase subunits in the regulatory particle ranged from 8 to 12 Å resolution (Figure S7). Notably, Rpn1 and the ubiquitin receptors Rpn10 and Rpn13 were the lowest resolution features of the holoenzyme. In order to properly filter the low-resolution portions of the map, without destroying the details of the better-ordered features, a resolution-driven adaptive localized low-pass filter was applied to the final volume (G. Cardone, personal communication).
The segmentation analysis was manually performed using the “Volume Tracer” tool in the UCSF Chimera visualization software 47. This software was additionally used to perform all rigid-body fitting of crystal structures into the holoenzyme cryoEM density, as well as to generate all renderings for figure images.
Supplementary Material
Acknowledgements
We thank the members of the Martin and Nogales labs for helpful discussions, and Giovanni Cardone for help with local resolution calculations. G.C.L. acknowledges support from Damon Runyon Cancer Research Foundation, M.E.M. acknowledges support by the American Cancer Society grant 121453-PF-11-178-01-TBE, C.B. acknowledges support from the NSF Graduate Research Fellowship. This research was funded in part by the Searle Scholars Program (A.M.), start-up funds from the UC Berkeley MCB Department (A.M.), the NIH grant R01-GM094497-01A1 (A.M.), the Lawrence Berkeley National Laboratory (G.C.L.), and the Howard Hughes Medical Institute (E.N.). Some of the work presented here was conducted at the National Resource for Automated Molecular Microscopy, which is supported by the NIH through the NCRR P41 program (RR017573).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author contributions: E.E., M.E.M., and C.B. designed, expressed, and purified proteasome constructs, and performed biochemical experiments. G.C.L. performed the electron microcopy, processing, and segmentation analysis. All authors contributed to experimental design, data analysis, and manuscript preparation.
The cryoEM density map for the 26S proteasome can be found at the Electron Microscopy Data Bank under accession number EMD-1992. The negative stain reconstructions of the recombinantly expressed and yeast-purified lid have been assigned accession numbers EMD-1993 and EMD-1994, respectively.
References
- 1.Finley D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu Rev Biochem. 2009;78:477–513. doi: 10.1146/annurev.biochem.78.081507.101607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol Cell Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thrower JS, Hoffman L, Rechsteiner M, Pickart CM. Recognition of the polyubiquitin proteolytic signal. Embo J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groll M, et al. A gated channel into the proteasome core particle. Nat Struct Biol. 2000;7:1062–1067. doi: 10.1038/80992. [DOI] [PubMed] [Google Scholar]
- 5.Smith DM, et al. Docking of the proteasomal ATPases' carboxyl termini in the 20S proteasome's alpha ring opens the gate for substrate entry. Mol Cell. 2007;27:731–744. doi: 10.1016/j.molcel.2007.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yao T, Cohen RE. A cryptic protease couples deubiquitination and degradation by the proteasome. Nature. 2002;419:403–407. doi: 10.1038/nature01071. [DOI] [PubMed] [Google Scholar]
- 7.Verma R, et al. Role of Rpn11 metalloprotease in deubiquitination and degradation by the 26S proteasome. Science. 2002;298:611–615. doi: 10.1126/science.1075898. [DOI] [PubMed] [Google Scholar]
- 8.Tomko RJ, Jr., Funakoshi M, Schneider K, Wang J, Hochstrasser M. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rabl J, et al. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gillette TG, Kumar B, Thompson D, Slaughter CA, DeMartino GN. Differential roles of the COOH termini of AAA subunits of PA700 (19 S regulator) in asymmetric assembly and activation of the 26 S proteasome. J Biol Chem. 2008;283:31813–31822. doi: 10.1074/jbc.M805935200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.da Fonseca PC, Morris EP. Structure of the human 26S proteasome: subunit radial displacements open the gate into the proteolytic core. J Biol Chem. 2008;283:23305–23314. doi: 10.1074/jbc.M802716200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elsasser S, et al. Proteasome subunit Rpn1 binds ubiquitin-like protein domains. Nat Cell Biol. 2002;4:725–730. doi: 10.1038/ncb845. [DOI] [PubMed] [Google Scholar]
- 13.Gomez TA, Kolawa N, Gee M, Sweredoski MJ, Deshaies RJ. Identification of a functional docking site in the Rpn1 LRR domain for the UBA-UBL domain protein Ddi1. BMC Biol. 2011;9:33. doi: 10.1186/1741-7007-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leggett DS, et al. Multiple associated proteins regulate proteasome structure and function. Mol Cell. 2002;10:495–507. doi: 10.1016/s1097-2765(02)00638-x. [DOI] [PubMed] [Google Scholar]
- 15.Bohn S, et al. Structure of the 26S proteasome from Schizosaccharomyces pombe at subnanometer resolution. Proc Natl Acad Sci U S A. 2010;107:20992–20997. doi: 10.1073/pnas.1015530107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickell S, et al. Insights into the molecular architecture of the 26S proteasome. Proc Natl Acad Sci U S A. 2009;106:11943–11947. doi: 10.1073/pnas.0905081106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forster F, et al. An atomic model AAA-ATPase/20S core particle sub-complex of the 26S proteasome. Biochem Biophys Res Commun. 2009;388:228–233. doi: 10.1016/j.bbrc.2009.07.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xu P, et al. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 2009;137:133–145. doi: 10.1016/j.cell.2009.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Inobe T, Fishbain S, Prakash S, Matouschek A. Defining the geometry of the two-component proteasome degron. Nat Chem Biol. 2011;7:161–167. doi: 10.1038/nchembio.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tian G, et al. An asymmetric interface between the regulatory and core particles of the proteasome. Nat Struct Mol Biol. 2011;18:1259–1267. doi: 10.1038/nsmb.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Effantin G, Rosenzweig R, Glickman MH, Steven AC. Electron microscopic evidence in support of alpha-solenoid models of proteasomal subunits Rpn1 and Rpn2. J Mol Biol. 2009;386:1204–1211. doi: 10.1016/j.jmb.2009.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang F, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hamazaki J, et al. A novel proteasome interacting protein recruits the deubiquitinating enzyme UCH37 to 26S proteasomes. Embo J. 2006;25:4524–4536. doi: 10.1038/sj.emboj.7601338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schreiner P, et al. Ubiquitin docking at the proteasome through a novel pleckstrin-homology domain interaction. Nature. 2008;453:548–552. doi: 10.1038/nature06924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Glickman MH, et al. A subcomplex of the proteasome regulatory particle required for ubiquitin-conjugate degradation and related to the COP9-signalosome and eIF3. Cell. 1998;94:615–623. doi: 10.1016/s0092-8674(00)81603-7. [DOI] [PubMed] [Google Scholar]
- 26.Verma R, Oania R, Graumann J, Deshaies RJ. Multiubiquitin chain receptors define a layer of substrate selectivity in the ubiquitin-proteasome system. Cell. 2004;118:99–110. doi: 10.1016/j.cell.2004.06.014. [DOI] [PubMed] [Google Scholar]
- 27.Glynn SE, Martin A, Nager AR, Baker TA, Sauer RT. Structures of asymmetric ClpX hexamers reveal nucleotide-dependent motions in a AAA+ protein-unfolding machine. Cell. 2009;139:744–756. doi: 10.1016/j.cell.2009.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maillard RA, et al. ClpX(P) generates mechanical force to unfold and translocate its protein substrates. Cell. 2011;145:459–469. doi: 10.1016/j.cell.2011.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aubin-Tam ME, Olivares AO, Sauer RT, Baker TA, Lang MJ. Single-molecule protein unfolding and translocation by an ATP-fueled proteolytic machine. Cell. 2011;145:257–267. doi: 10.1016/j.cell.2011.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin A, Baker TA, Sauer RT. Rebuilt AAA + motors reveal operating principles for ATP-fuelled machines. Nature. 2005;437:1115–1120. doi: 10.1038/nature04031. [DOI] [PubMed] [Google Scholar]
- 31.Hersch GL, Burton RE, Bolon DN, Baker TA, Sauer RT. Asymmetric interactions of ATP with the AAA+ ClpX6 unfoldase: allosteric control of a protein machine. Cell. 2005;121:1017–1027. doi: 10.1016/j.cell.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 32.Thomsen ND, Berger JM. Running in reverse: the structural basis for translocation polarity in hexameric helicases. Cell. 2009;139:523–534. doi: 10.1016/j.cell.2009.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enemark EJ, Joshua-Tor L. Mechanism of DNA translocation in a replicative hexameric helicase. Nature. 2006;442:270–275. doi: 10.1038/nature04943. [DOI] [PubMed] [Google Scholar]
- 34.Riedinger C, et al. Structure of Rpn10 and its interactions with polyubiquitin chains and the proteasome subunit Rpn12. J Biol Chem. 2010;285:33992–34003. doi: 10.1074/jbc.M110.134510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 36.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 37.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Husnjak K, et al. Proteasome subunit Rpn13 is a novel ubiquitin receptor. Nature. 2008;453:481–488. doi: 10.1038/nature06926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hanna J, et al. Deubiquitinating enzyme Ubp6 functions noncatalytically to delay proteasomal degradation. Cell. 2006;127:99–111. doi: 10.1016/j.cell.2006.07.038. [DOI] [PubMed] [Google Scholar]
- 40.Prakash S, Tian L, Ratliff KS, Lehotzky RE, Matouschek A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat Struct Mol Biol. 2004;11:830–837. doi: 10.1038/nsmb814. [DOI] [PubMed] [Google Scholar]
- 41.Verma R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leggett DS, Glickman MH, Finley D. Purification of proteasomes, proteasome subcomplexes, and proteasome-associated proteins from budding yeast. Methods Mol Biol. 2005;301:57–70. doi: 10.1385/1-59259-895-1:057. [DOI] [PubMed] [Google Scholar]
- 43.Suloway C, et al. Automated molecular microscopy: the new Leginon system. J Struct Biol. 2005;151:41–60. doi: 10.1016/j.jsb.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 44.Lander GC, et al. Appion: an integrated, database-driven pipeline to facilitate EM image processing. J Struct Biol. 2009;166:95–102. doi: 10.1016/j.jsb.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 46.Hohn M, et al. SPARX, a new environment for Cryo-EM image processing. J Struct Biol. 2007;157:47–55. doi: 10.1016/j.jsb.2006.07.003. [DOI] [PubMed] [Google Scholar]
- 47.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Sone T, Saeki Y, Toh-e A, Yokosawa H. Sem1p is a novel subunit of the 26 S proteasome from Saccharomyces cerevisiae. J Biol Chem. 2004;279:28807–28816. doi: 10.1074/jbc.M403165200. [DOI] [PubMed] [Google Scholar]
References for full methods
- 49.Kim HC, Huibregtse JM. Polyubiquitination by HECT E3s and the determinants of chain type specificity. Mol Cell Biol. 2009;29:3307–3318. doi: 10.1128/MCB.00240-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mallick SP, Carragher B, Potter CS, Kriegman DJ. ACE: automated CTF estimation. Ultramicroscopy. 2005;104:8–29. doi: 10.1016/j.ultramic.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 51.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 52.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sorzano CO, et al. XMIPP: a new generation of an open-source image processing package for electron microscopy. J Struct Biol. 2004;148:194–204. doi: 10.1016/j.jsb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 54.van Heel M, Harauz G, Orlova EV, Schmidt R, Schatz M. A new generation of the IMAGIC image processing system. J Struct Biol. 1996;116:17–24. doi: 10.1006/jsbi.1996.0004. [DOI] [PubMed] [Google Scholar]
- 55.Frank J, et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 56.Heymann JB, Belnap DM. Bsoft: image processing and molecular modeling for electron microscopy. J Struct Biol. 2007;157:3–18. doi: 10.1016/j.jsb.2006.06.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.