Abstract
The field of genomics has entered a new era in which the ability to identify genetic variants that impact complex human traits and disease in an unbiased fashion using genome-wide approaches is widely accessible. To date, the workhorse of these efforts has been the genome-wide association study (GWAS), which has quickly moved from novel to routine, and has provided key insights into aspects of the underlying allelic architecture of complex traits. The main lesson learned from the early GWAS efforts is that though many disease-associated variants are often discovered, most have only a minor effect on disease, and in total explain only a small amount of the apparent heritability. Here we provide a brief overview of the genetic variation classes that may harbor the heritability missing from GWAS, and touch on approaches that will be leveraged in the coming years as genomics—and by extension medicine—becomes increasingly personalized.
Keywords: Genetics, genomics, GWAS, liver, complex disease
The genome-wide association study (GWAS) has rapidly become a standard weapon in the arsenal of investigators interested in the genetic origin of complex human traits and diseases. Indeed, since 2005 when the first GWAS was reported, 654 studies have been added to the Catalog of Published Genome-Wide Association Studies (www.genome.gov/gwastudies/), a number that may well reach 1000 by the time this manuscript is published. The GWAS approach is predicated on the Common Disease / Common Variants (CDCV) hypothesis, which posits that common genetic variation is likely to play a significant role in the underlying allelic architecture of genetic traits.1 Its application is facilitated by the International HapMap Project,2 which helped to document an extensive number of common single nucleotide polymorphisms (SNPs) in the genome, and determined the patterns of linkage disequilibrium (LD) and the correlation (r2) between these variant alleles. This effort makes it possible to capture a good deal of the common genetic variation across the genome using a representative group of SNPs that can be affordably assayed by means of high-throughput technologies. As this is inherently an indirect approach, variants found to be associated with a disease or trait by GWAS are assumed “innocent until proven guilty” and extensive fine mapping is generally required to identify the “causal” variant being detected. The current iteration of genotyping arrays assess ~1 million SNPs across the genome, providing coverage of ~80–90% of the HapMap SNPs with minor allele frequency (MAF) greater than 5%, at an r2 value of 0.8 or greater in European populations.3
WHAT HAVE WE LEARNED FROM GWAS?
To date, hundreds of disease-associated genetic variants have been identified by GWAS, the majority of which are SNPs with only a small effect on the trait or disease being studied, generally with odds ratios (OR) in the range of 1.1–1.4.4 Even in instances where numerous loci have been identified for particular traits or diseases, usually very little of the apparent heritability is explained. For example, more than 30 loci have been associated with type 2 diabetes to date, yet these appear to explain only ~10% of the observed familial clustering.5 As well, the biologic effects behind a majority of the associated variants remains unclear, often due to strong LD obscuring the location of specific causative loci and the inability to ascribe function, especially when the associated alleles are located far from any recognized gene. These factors have led to the speculation that further inquiry into common genetic variation is unlikely to result in many findings of significant clinical relevance.6 However, GWAS has proven to be a useful “biologic pathway detection tool”7 and fundamental changes in the understanding of complex disease pathogenesis, such as the recognition of the significant role that autophagy plays in Crohn’s disease,8 have resulted from GWAS efforts. Although the approach as thus far employed does not appear sufficient to capture all of the apparent heritability of complex traits, GWAS is likely to remain a valuable tool in the dissection of their genetic underpinnings for some time to come.
WHERE IS THE MISSING HERITABILITY?
Put simply, heritability is the fraction of phenotypic variance in a population that is explained by the underlying genetic variation, which is comprised of both additive and nonadditive (i.e., dominance and epistatic) components.9 Heritability scores are estimates derived from family studies; due to confounding and sampling error inherent in even large investigations of closely related relatives such as siblings, the nonadditive variance is difficult to define with any precision. Thus, heritability is often described as the portion of phenotypic variance in a population attributable to only the additive genetic factors (i.e., narrow-sense heritability, denoted as h2). Theory suggests that the majority of the variance in complex traits can be explained by the additive effects, and the current data shows this is indeed the case, as the additive component generally explains over half of and usually nearly 100% of the overall genetic variance.10 However, estimates of h2 can be overstated if within-family resemblance is significantly influenced by shared familial environment or nonadditive genetic effects such as dominance or genetic interaction between genes or with the environment (i.e., epistasis).11 Because a further discussion of the subtleties of heritability is outside of the scope of this article, we refer readers to an excellent recent review by Visscher et al.9
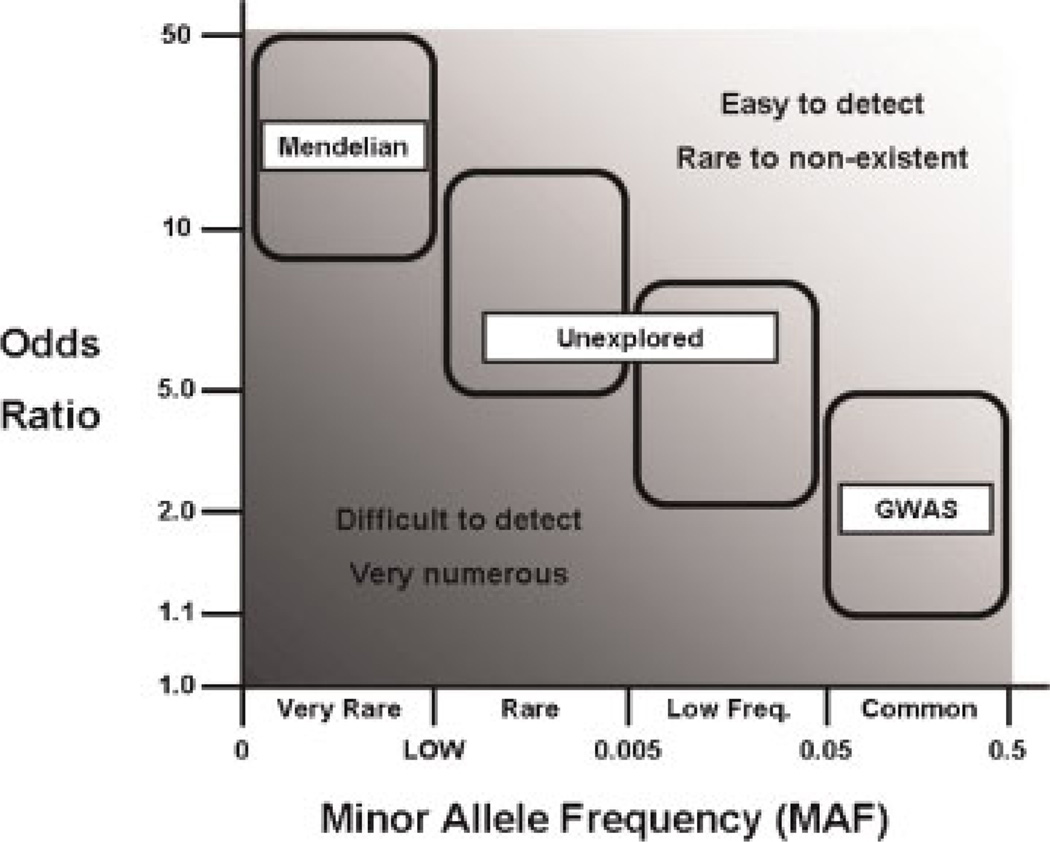
Regardless of the past conjectures and continuing debates, it is clear that our understanding of the underlying allelic architecture of complex traits and disease is limited at best. We have been long aware of the strong effects of very rare alleles on highly penetrant Mendelian disorders. Now the recent observations of GWAS confirm that for most of the studied complex diseases, common genetic variants do explain some of the risk; ranging from the relatively strong effects noted for age-related macular degeneration12 to little, if any, effect in most neuropsychiatric disorders.13 However, only minimal effort has been made to investigate the genetic variation existing in the space between these extremes of rarity and commonality in the context of complex diseases (Fig. 1).14 Moreover, non-SNP structural genetic differences such as copy number variants (CNVs, e.g., insertions and deletions) and copy neutral variants (e.g., translocations and inversions), as well as inherited epigenetic modifications (e.g., “imprinted” DNA methylation and histone modifications), may also hide some of the heritability missing in GWAS and remain understudied. Finally, nonadditive genetic effects such as dominance and epistasis, although not predicted to greatly impact heritability, could significantly influence some complex traits and have not yet been well explored. Below we briefly discuss the classes of genetic variation that are prime candidates for harboring the heritability not found by current GWAS efforts and the challenges inherent to their study.
Figure 1.
In the collective human genome, there exists many millions of genetic variants, which range in frequency among the population as well as in their contribution to the risk of developing disease. At one extreme are the polymorphisms that cause highly heritable Mendelian disorders, which are generally quite rare in the population, but are highly penetrant and thus were relatively easy to identify using linkage approaches. On the other end of the spectrum are the common variants, which are more readily identified by association analysis, and have been the focus of genome-wide association studies (GWASs). Although common variants with modest to strong effects on disease would be relatively easy to detect, we have found very few, and our experience with GWAS has shown that the majority of disease-associated common variants confer only a small risk of developing disease; most of the apparent heritability remains obscure. It is difficult to detect the likely numerous disease variants of smaller effect sizes (depending on allele frequency) using association due to power limitations. However, the low-frequency and rare variation remains relatively unexplored and could harbor detectable polymorphisms, which might explain some of the heritability missing from GWAS. Such variants are the targets of the next-generation genomics approaches.
Undetected Common Genetic Variation
Although GWAS has undoubtedly led to significant progress in the detection of disease-associated common genetic variants (defined as those with MAF ≥ 5%), by no means are these investigations complete. For instance, only a handful of the diseases so far studied by GWAS have utilized samples of sufficient size to reasonably power the identification of common associations in the frequently noted 1.1–1.4 relative risk range, especially when the variants are at the lower end of the MAF threshold. Moreover, it is very likely that many common alleles of very low effect size (i.e., risk ratios < 1.1) exist for the majority of traits, and also remain undetected due to lack of power in even the largest GWAS efforts to date.
This generalized lack of power is due to the often-implemented frequentist approach to analysis of GWAS data, which utilizes stringent restraints on the P-value to contain type I error. For example, P-values in the range of 10−6 to 10−8 are generally considered necessary evidence of association in the context of GWAS; thus, many true associations are likely to be rejected to reduce the number of false-positives represented in the resulting lists of promising candidates. This approach is well suited to identify some variants that are truly associated with the trait of interest, and therefore most likely to generate favorable findings when studied further; an important factor considering the limited availability of resources for follow-up studies. However, frequentist approaches are not practical if the goal is to identify all (or even a good portion) of the common variants associated with a particular disease.
To maximize the detection of common variant disease association significantly large patient and control populations are required, the upper limit of which will be constrained by availability and resources. For complex diseases that are rare in the population the detectable effect sizes of potentially associated genetic variants will be severely limited by the number of patients available for study, even assuming widespread cooperation between investigators and coordinated meta-analysis of all data. In contrast, diseases that are common in the population are generally well-funded and there is an abundance of patients that can potentially be observed. Thus, it is possible that many associations of very low effect could eventually be identified for these diseases; although at some point the clinical utility of cataloging an extensive list of such variants may become negligible in regards to the significant increase in cost.15
Low-Frequency Genetic Variants
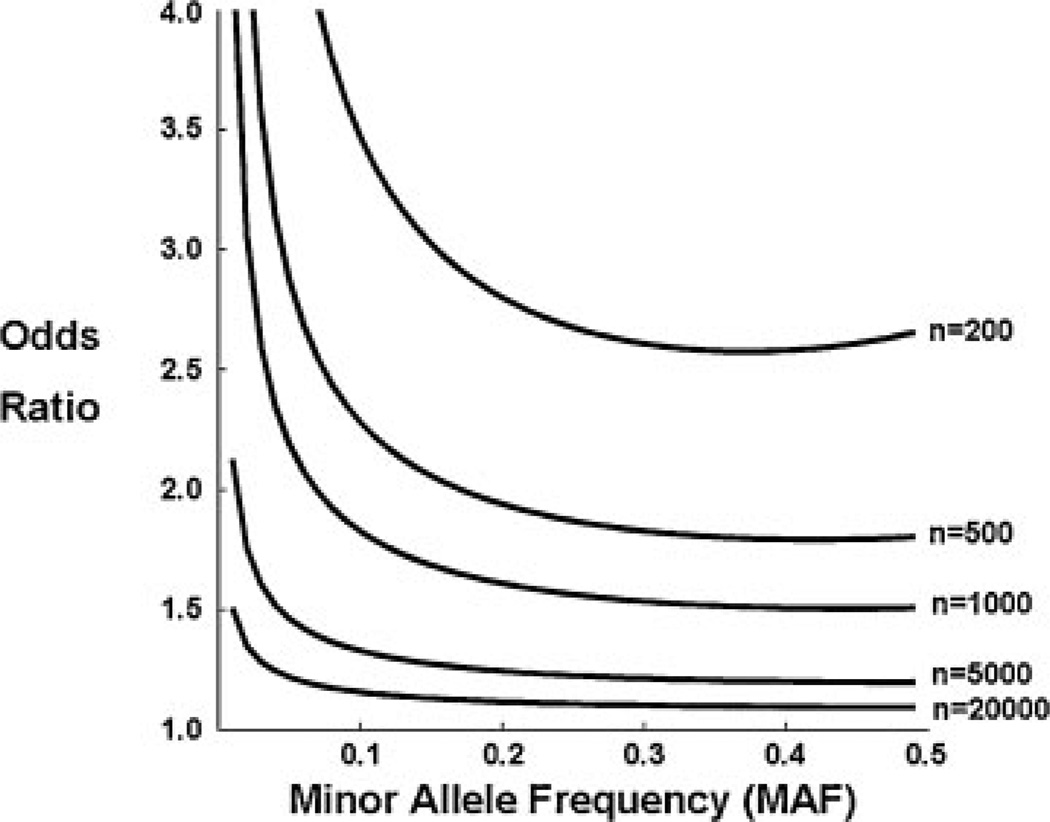
Interest in exploring the potential contribution of the so-called low-frequency genetic variants (LFGVs) has risen drastically in the past few years, and perhaps represents the middle ground between the common variant and rare variant camps. The LFGVs are loosely defined as those variants with MAF below the threshold of common variants (there is obvious overlap here), but still segregating in the population at some appreciable frequency. We will consider LFGVs to be those genetic variants that fall in the range of MAF 0.5–5.0% for sake of this discussion. As with their more common brethren, the primary challenge to the study of LFGVs in the context of association studies is power, which to maintain requires steep increases in sample size as MAF and OR decline (Fig. 2). However, it has been shown by simulations that small numbers of LFGVs contributing intermediate effects on disease would explain a large portion of the heritability missing from GWAS.16 Moreover, such variants should be detectable without requiring unrealistically large sample sizes, would provide important insights into disease mechanisms, and are not well covered by the current genome-wide16 Although the prospects for LFGVs to explain some of the missing heritability seem promising, the proposition that significant numbers will demonstrate the intermediate effects on risk required to overcome the power challenges is untested. Indeed, no evidence for a correlation between effect size and allele frequency has been revealed by the existing GWAS data.
Figure 2.
This figure illustrates the significant increases in sample size required to maintain power to detect genetic association as minor allele frequency (MAF) and odds ratio (OR) decline. For this figure, power was set to 80% and a P-value of 10−6 was assumed.
The primary obstacle to investigating LFGVs is the current lack of a complete directory of these variants upon which to build the next-generation of commercial arrays. However, the 1000 Genomes Project (www.1000genomes.org) is currently undertaking the task of building this catalog. This immense effort aims to resequence the genomes of 2500 individuals from 27 populations at low coverage (i.e., ~4 ×) with the goal of identifying most of the genetic variants with MAF of 1% in the studied populations. Although the low coverage of genotyping performed by this project is certainly not sufficient to produce “whole-genome sequences” for each individual (a task that requires 25 × –30 × coverage with current “next-gen” sequencing platforms) the data provided will allow for the detection of many copy number polymorphisms (CNPs, i.e., “common” CNVs) and other structural variants in addition to SNPs, and will provide a solid foundation for imputation17 of existing and emerging genome-wide datasets. The 1000 Genomes Project should allow us to “push-the-envelope” of utility for association study of genetic variation in human disease by providing new information to earlier GWAS efforts and facilitating the next generation of genome-wide arrays.
Rare Genetic Variation
Rare genetic variants are those with MAF below the LFGV threshold (again there is certainly some overlap, but in keeping with the above convention we’ll consider MAF < 0.5%). In the context of heritability, this would include variants of diminishing representation in the population down to those whose presence is effectively limited to a single family, but preclude de novo “private” variation in individuals. However, despite having little, if any, impact on the observed heritability, such “new” variants could certainly contribute to disease and should not be discounted. Regardless of where such variants lie within the spectrum of rarity or whether they are SNPs or CNVs, they are not very amenable to association study by GWAS due largely to their vast numbers, which would require a significant increase in array density or the use of numerous arrays to assess, as well as due to power limitations, which would necessitate the use of considerably expanded patient populations to detect associations even of large effect. For example, ~5,200 cases are required to achieve 80% power (α of 10−8) to detect an OR of 3.0 when MAF is 0.5% (bottom end of the LFGV range), but when MAF drops to 0.1%, ~26,000 cases would be required to maintain power. Taken further, the minimum detectable OR for an allele with MAF of 0.1% using the original 5,200 cases is ~7.3, which is quite high in the context of previous findings and would likely produce a compelling signal in familial linkage analysis. Of course this illustration considers only the “tip of the rare variant iceberg” so it is not hard to imagine the futility of GWAS as the alleles become more rare.
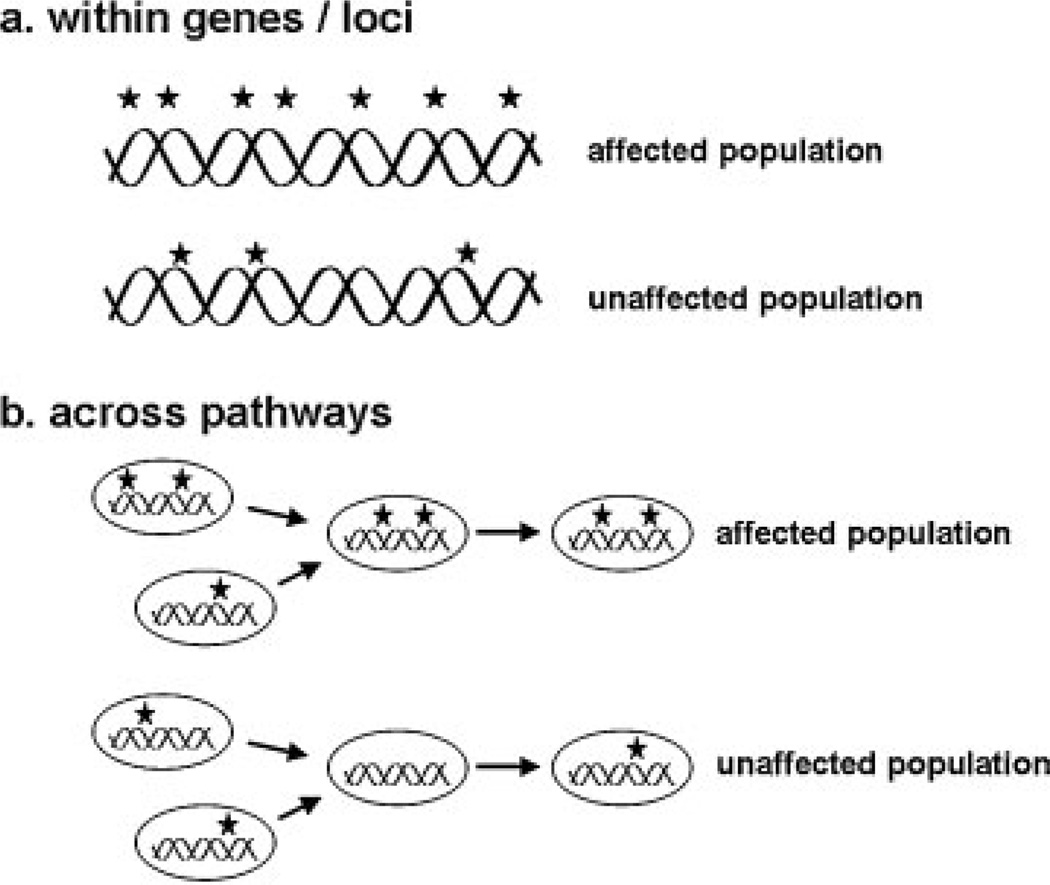
To identify and study the rare genetic variants sequencing will need to be employed, either using the current “next-gen” or promising emerging platforms. Although the price of sequencing continues to decrease dramatically it may still be many years before data handling capability and sequencing capacity reach the point that whole-genome association studies akin to the current GWAS approach in terms of patient numbers are feasible. In the meantime, more targeted approaches such as whole-exome sequencing, which focuses on the protein-coding exons and exon/intron boundaries containing splicing signals may be a more cost-effective tactic in identifying rare genetic variants that impact disease.18 This approach has already proven successful in finding causative variants for Mendelian disorders19–21 and offers promise for complex disease. Moreover, extreme-trait study designs, which focus on individuals at the far end of the phenotypic distribution, or family-based designs, which focus on families with multiple affected individuals, are currently practical using either exome or whole-genome-based approaches.6 However, initial application of these methods is not likely to reveal a large portion of the heritability missing from GWAS due simply to the limited number of individuals utilized in such studies combined with a high likelihood for extensive allelic heterogeneity within deleterious loci.22 In this regard, analytical approaches considering the “mutational load” within genes or across biological pathways may be suitable for pinning down the loci and mechanisms responsible for disease (Fig. 3). Although caution must be taken in these analyses, our current knowledge precludes any precise prediction of functional consequences resulting from most genetic variants; thus, spurious conclusions could be made.
Figure 3.
Because there is very little, if any, power to detect disease-specific effects of individual rare genetic variants using association, analytical approaches considering the “mutational load” within genes/isolated genetic loci (a) or across the genes of biological pathways (b) may be useful for identifying the genes and mechanisms effecting disease. However, caution should be taken in such analyses, as the functional consequence of all the genetic variants is not likely to be apparent, potentially leading to spurious findings.
Nonadditive Genetic Effects
Despite the evidence that additive genetic effects are the primary drivers of heritability there is still much room for nonadditive effects in the allelic architecture of many complex traits and diseases. Perhaps the most simple of these in concept are the alternative single locus genetic models such as dominant (i.e., AA vs. Aa/aa) and recessive (i.e., aa vs. AA/Aa) acting loci. In the context of GWAS, which is largely an indirect approach, these models are often not considered, as even minor deviations from perfect LD will drastically reduce the power to detect such associations.16 Moreover, the detection of recessive genetic effects inherently suffers from low power, even for relatively common variant alleles. To overcome these difficulties and improve power to detect recessive effects, especially in the context of quantitative traits, the use of multiple correlated genetic models has been shown to increase power,23 although determination of appropriate significance levels from such analysis is not straightforward and may complicate downstream efforts. More significantly, the routine consideration of nonadditive genetic models may come back into play once direct sequencing-based approaches, in contrast to indirect array-based approaches, are the norm. However, the best methods for maximizing detection while containing error in this framework are, for the large part, unexplored.
Although simple dominant and recessive models may explain some of the heritability missing from GWAS, the potential for nonadditive epistatic interaction effects between genes (gene–gene interactions) and with the environment (gene–environment interactions) has received growing interest of late. In simple terms, the interaction discussed here is defined as departure from a liner model describing the relationship between outcome and predictor variables, and thus is a statistical measure, which does not imply a physical relationship.24 As such, analysis utilizing regression models is quite fitting and easily accomplished, with various case-control and case-only approaches often employed,25–27 although other methodologies are being examined.28,29
Although the concept that genetic variation might well modify the impact of environmental effects on disease etiology and pathogenesis is widely accepted, attempting to implement the analysis of gene–environment interactions at the genome-wide scale raises several problems, not the least of which is a lack of environmental exposure data for populations studied in many of the current and previous GWAS. Defining and assessing environmental exposure is difficult as many environmental factors are multidimensional, for example, water contaminants are likely to vary substantially in their levels and overall constituency dependent on the specific source. Moreover, whether or not there are multidimensional confounders, the effect of the environmental exposure will often be, at least to some extent, dependant on timing, such as age at and/or duration of the exposure. In general, such exposures will be difficult, if not impossible, to assess with any level of accuracy for the majority of individuals available for study. Use of stratified subsamples for which more extensive exposure data are available can be of benefit30 and has been successfully utilized in identifying a genetic risk factor for smoking-related coronary heart disease.31 However, uncertainty in exposure quantification, especially when large, could generate arbitrary biases leading to spurious findings. As well, significant increases in sample size compared with GWAS (on the order of 4–5×) are required to maintain power, thus modest effects of gene–environment interaction may be difficult to detect.32 An in-depth review of the challenges of and emerging approaches to the genome-wide assessment of gene–environment interactions can be found here.33
Assessment of gene–gene interaction from genome-wide data are a bit more straightforward as the limits imposed by the esoteric environmental component are avoided. However, power and computational load place severe restraints on such analyses. For instance, an exhaustive test of all 4.5 billion two-locus interactions from 100 K chip data in 500 individuals using the fastepistasis command in PLINK is quoted as taking ~24 hours.34 Considering that the current commercial arrays assess 10 times as many SNPs and experiments often include thousands instead of hundreds of individuals, computation times in the range of weeks to months, and the utilization of multiple processing nodes is required to perform such analysis. However, new, more efficient approaches to two-locus interaction testing have been proposed.35,36 As many billions of observations are being made, concerns over multiple testing are warranted. Obviously, Bonferroni correction is far too conservative considering the abundance of correlated tests due to various levels of LD between many of the polymorphisms. Permutation testing is an obvious solution to the correlation problem, although this approach is computationally expensive. For the time being, some strict arbitrary cutoff, as generally employed in GWAS, may be the most practical approach.
Despite the multiple testing issues, exhaustive assessment of all two-locus interactions is certainly feasible. However, this approach does not scale up, as comprehensive evaluation of even three- or four-locus interactions becomes severely impractical (if not intractable) with currently available computing power, due largely to sparseness of data when spread across many genotype combinations. To get around this limitation, and perhaps more aptly approach the complexity implied by higher-order epistasis, nonlinear data mining methods such as multifactor dimensionality reduction37–39 and recursive partitioning methods,40,41 as well as Bayesian approaches42 to interaction analysis have been proposed. A good primer to these methodologies is provided in a recent review by Cordell.24
NEXT-GENERATION GENOMICS
As the study of human genomics transitions into the next generation, our ability to identify genetic variants associated with complex traits and diseases will advance dramatically, due in large part to anticipated improvements in genotyping array technology and greater access to low-cost sequencing. Nevertheless, the best approaches to leveraging these technologies remain unclear, and deciphering the functional mechanisms and determining the clinical relevance of the resulting findings will perhaps be our greatest challenge as we move into this new era. Integration of the other comprehensive “-omics” methodologies into the analysis of genetic variants will provide a global view of their potential effects, and thus, in the context of genomics are a fitting focus for the following discussion.43 However, significant advancements in traditional wet-bench approaches (both in vivo and in vitro) will also be required to quantify what are likely to be quite subtle effects of genetic variants within genes and across pathways.
The goal of genomics is not only to catalog the structure and variation present in genomes, but to understand how the information they contain is used to generate and maintain life, and by extension, to decipher how genetic variants alter biologic mechanisms and contribute to disease traits. Perhaps one of the most fundamental processes in this regard is transcriptional regulation, which has certainly been a topic of significant investigation for many years, but often focused on individual genes or gene families. However, advancements in chromatin immuno-precipitation (ChIP) coupled with genome-wide arrays (Chip-chip), or more recently with next-generation sequencing technologies (ChIP-seq), have facilitated the identification and analysis of transcription factor binding sites at genome-wide resolution. Such efforts have led to several key insights into the nature of transcription factors, such as location of binding sites, specificity of binding to consensus motifs, and functional relevance of binding site occupation.44 Nonetheless, the story is far from complete as many transcription factors remain unstudied, and ChIP-chip / ChIP-seq experiments have been traditionally focused on but one or a few cell types. Moreover, the binding of transcription factors is certainly not the only determinant of genetic transcription. Epigenetic phenomenon including histone positioning and modification, DNA methylation, and nucleosome remodeling are key aspects of transcriptional control, impacting cellular identity and potentially playing a role in complex disease. Genome-scale approaches to analyze these mechanisms are maturing (reviewed in45) and are being furthered by the NIH Roadmap Epigenomics Program (http://nihroadmap.nih.gov/epigenomics/).
Along with the greater capability to study the transcriptional control mechanisms mentioned above, the analysis of the transcriptome itself, as well as its association with inherited genetic variation, has been furthered by the recent advancements in sequencing technology coupled with GWAS. Perhaps the most notable example of this innovation is the ability to utilize the expression level of genes as quantitative traits (eQTLs) in genome-wide mapping efforts. This approach, which combines data from traditional genome-scale assessments of expression, such as commercial array-based assays (e.g., Affymetrix) or SAGE (serial analysis of gene expression) experiments, with data generated from GWAS, offers increased power to detect subtle differences in expression associated with disease,46 as well as a mechanism to narrow in on causative loci obscured by strong LD (e.g., the identification of the ORMDL3 contribution to childhood asthma47). More recently, the advent of RNA sequencing by next-generation technologies (RNA-seq), though still challenging and not without problems, provides improved sensitivity and dynamic range compared with array-based platforms, as well as single-base resolution.48 Moreover, the use of RNA-seq greatly facilitates genomic-scale examination of alternative isoform expression49 as well as allele-specific gene expression,50 and has been successfully applied using picogram quantities of mRNA, making transcriptome analysis of rare cells and tissues possible.51
PERSONALIZED GENETICS, GENOMICS, AND MEDICINE
Although substantial progress in our understanding of human genetics has occurred over the past decade and the pace of advancement continues to quicken at what seems an exponential pace, many significant challenges need to be overcome before the jump from understanding complex traits and disease at the population level to that of individuals will be possible. Indeed, despite what we have learned, our ability to classify levels of complex disease risk based on an individuals genetic background remains poor, if not essentially arbitrary, a statement that is particularly illustrated by the performance of direct-to-consumer genome-scale genetic tests.52 This is not to imply that targeted genotyping efforts, such as BRCA1/2 genotyping for breast and ovarian cancer risk, have not been beneficial. In the near term, the effort to personalize medicine is perhaps best envisioned as therapeutic selection targeted to those patients who are most likely to benefit, which will increasingly be facilitated by identifying characteristics of their genetic background or specific features identifying a subset of patients with a particular disease. For instance, the use of EGFR monoclonal antibodies for the treatment of metastatic colorectal cancer appears to only be efficacious in patients whose tumors are devoid of mutations in KRAS53 and dose selection for the anticoagulant warfarin (Coumadin) can be optimized based on the presence of gene variants in the patients CYP2C9 and VKORC1 genes.54 In this context, the immediate hurdles will be ethical, especially in cases where treatment may still offer a slight benefit to patients lacking the indicative genetic polymorphisms.
The rapidly decreasing cost of whole-genome sequencing may negate price concerns regarding genotyping as a preliminary step in treatment and dosage determination. However, such an approach raises considerable ethical, legal, and social issues (ELSI), not to mention immense technical challenges, which will need to be worked out before substantial clinical impact will be realized. For instance, our current understanding of the functional significance of individual genetic variation is severely limited, and hurried integration into clinical practice could result in inappropriate and unjustified follow-ups and procedures placing undo burden on our limited health-care resources. Moreover, the utility of whole-genome sequence as a basis for disease prevention remains untested, and it is not clear that knowledge of one’s risk of developing future illness is beneficial, particularly considering the anxiety that might result, and especially if prophylactic recommendations are lacking or potentially harmful. In this context, the use of personal genomic information in clinical management could further widen existing income-based disparities in health care when preventative measures are available, but expensive and possibly unproven. Finally, significant investments in computing infrastructure, as well as education of physicians and the public at large will be required to utilize personal genome sequences in the clinical setting, again raising the question of equity between those who will pay and those who will benefit.
A more highly personalized form of medicine, in which the characterization of genome sequence, transcriptome and proteome expression, and microbiome constituencies at global scales are routine in the context of preventative medical care, will certainly be farther off, but is no longer out of sight.
ACKNOWLEDGMENT
This work was supported by grants to Dr. K. N. Lazaridis from the NIH (RO1 DK80670 and DK84960).
ABBREVIATIONS
- ChIP
chromatin immunoprecipitation
- CNV
copy number variation
- GWAS
genome-wide association study
- LFGV
low-frequency genetic variation
- LD
linkage disequilibrium
- MAF
minor allele frequency
- OR
odds ratio
- SNP
single nucleotide polymorphism
REFERENCES
- 1.Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17(9):502–510. doi: 10.1016/s0168-9525(01)02410-6. [DOI] [PubMed] [Google Scholar]
- 2.Consortium TIH; International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437(7063):1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? J Hum Genet. 2010;55(4):195–206. doi: 10.1038/jhg.2010.19. [DOI] [PubMed] [Google Scholar]
- 4.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Voight BF, Scott LJ, Steinthorsdottir V, et al. MAGIC investigators; GIANT Consortium. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11(6):415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 7.Hirschhorn JN. Genomewide association studies—illuminating biologic pathways. N Engl J Med. 2009;360(17):1699–1701. doi: 10.1056/NEJMp0808934. [DOI] [PubMed] [Google Scholar]
- 8.Brest P, Corcelle EA, Cesaro A, et al. Autophagy and Crohn’s disease: at the crossroads of infection, inflammation, immunity, and cancer. Curr Mol Med. 2010;10(5):486–502. doi: 10.2174/156652410791608252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era—concepts and misconceptions. Nat Rev Genet. 2008;9(4):255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 10.Hill WG, Goddard ME, Visscher PM. Data and theory point to mainly additive genetic variance for complex traits. PLoS Genet. 2008;4(2):e1000008. doi: 10.1371/journal.pgen.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Katta S, Kaur I, Chakrabarti S. The molecular genetic basis of age-related macular degeneration: an overview. J Genet. 2009;88(4):425–449. doi: 10.1007/s12041-009-0064-4. [DOI] [PubMed] [Google Scholar]
- 13.Cichon S, Craddock N, Daly M, et al. Psychiatric GWAS Consortium Coordinating Committee. Genomewide association studies: history, rationale, and prospects for psychiatric disorders. Am J Psychiatry. 2009;166(5):540–556. doi: 10.1176/appi.ajp.2008.08091354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McCarthy MI. Exploring the unknown: assumptions about allelic architecture and strategies for susceptibility variant discovery. Genome Med. 2009;1(7):66. doi: 10.1186/gm66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldstein DB. Common genetic variation and human traits. N Engl J Med. 2009;360(17):1696–1698. doi: 10.1056/NEJMp0806284. [DOI] [PubMed] [Google Scholar]
- 16.McCarthy MI, Abecasis GR, Cardon LR, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 17.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 18.Teer JK, Mullikin JC. Exome sequencing: the sweet spot before whole genomes. Hum Mol Genet. 2010;19(R2):R145–R151. doi: 10.1093/hmg/ddq333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng SB, Buckingham KJ, Lee C, et al. Exome sequencing identifies the cause of a mendelian disorder. Nat Genet. 2010;42(1):30–35. doi: 10.1038/ng.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ng SB, Bigham AW, Buckingham KJ, et al. Exome sequencing identifies MLL2 mutations as a cause of Kabuki syndrome. Nat Genet. 2010;42(9):790–793. doi: 10.1038/ng.646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gilissen C, Arts HH, Hoischen A, et al. Exome sequencing identifies WDR35 variants involved in Sensenbrenner syndrome. Am J Hum Genet. 2010;87(3):418–423. doi: 10.1016/j.ajhg.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pritchard JK. Are rare variants responsible for susceptibility to complex diseases? Am J Hum Genet. 2001;69(1):124–137. doi: 10.1086/321272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lettre G, Lange C, Hirschhorn JN. Genetic model testing and statistical power in population-based association studies of quantitative traits. Genet Epidemiol. 2007;31(4):358–362. doi: 10.1002/gepi.20217. [DOI] [PubMed] [Google Scholar]
- 24.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. doi: 10.1038/nrg2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang Q, Khoury MJ, Sun F, Flanders WD. Case-only design to measure gene-gene interaction. Epidemiology. 1999;10(2):167–170. [PubMed] [Google Scholar]
- 26.Hoh J, Ott J. Mathematical multi-locus approaches to localizing complex human trait genes. Nat Rev Genet. 2003;4(9):701–709. doi: 10.1038/nrg1155. [DOI] [PubMed] [Google Scholar]
- 27.Zhao J, Jin L, Xiong M. Test for interaction between two unlinked loci. Am J Hum Genet. 2006;79(5):831–845. doi: 10.1086/508571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ruiz-Marín M, Matilla-García M, Cordoba JA, et al. An entropy test for single-locus genetic association analysis. BMC Genet. 2010;11:19. doi: 10.1186/1471-2156-11-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sucheston L, Chanda P, Zhang A, Tritchler D, Ramanathan M. Comparison of information-theoretic to statistical methods for gene-gene interactions in the presence of genetic heterogeneity. BMC Genomics. 2010;11:487. doi: 10.1186/1471-2164-11-487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas D, Stram D, Dwyer J. Exposure measurement error: influence on exposure-disease. Relationships and methods of correction. Annu Rev Public Health. 1993;14:69–93. doi: 10.1146/annurev.pu.14.050193.000441. [DOI] [PubMed] [Google Scholar]
- 31.Li R, Boerwinkle E, Olshan AF, et al. Glutathione S-transferase genotype as a susceptibility factor in smoking-related coronary heart disease. Atherosclerosis. 2000;149(2):451–462. doi: 10.1016/s0021-9150(99)00483-9. [DOI] [PubMed] [Google Scholar]
- 32.Gauderman WJ. Sample size requirements for matched case-control studies of gene-environment interaction. Stat Med. 2002;21(1):35–50. doi: 10.1002/sim.973. [DOI] [PubMed] [Google Scholar]
- 33.Thomas D. Gene-environment-wide association studies: emerging approaches. Nat Rev Genet. 2010;11(4):259–272. doi: 10.1038/nrg2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang X, Pan F, Xie Y, Zou F, Wang W. COE: a general approach for efficient genome-wide two-locus epistasis test in disease association study. J Comput Biol. 2010;17(3):401–415. doi: 10.1089/cmb.2009.0155. [DOI] [PubMed] [Google Scholar]
- 36.Brinza D, Schultz M, Tesler G, Bafna V. RAPID detection of gene-gene interactions in genome-wide association studies. Bioinformatics. 2010;26(22):2856–2862. doi: 10.1093/bioinformatics/btq529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Motsinger AA, Ritchie MD. Multifactor dimensionality reduction: an analysis strategy for modelling and detecting gene-gene interactions in human genetics and pharmacogenomics studies. Hum Genomics. 2006;2(5):318–328. doi: 10.1186/1479-7364-2-5-318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Calle ML, Urrea V, Malats N, Van Steen K. mbmdr: an R package for exploring gene-gene interactions associated with binary or quantitative traits. Bioinformatics. 2010;26(17):2198–2199. doi: 10.1093/bioinformatics/btq352. [DOI] [PubMed] [Google Scholar]
- 39.Gui J, Andrew AS, Andrews P, et al. A simple and computationally efficient sampling approach to covariate adjustment for multifactor dimensionality reduction analysis of epistasis. Hum Hered. 2010;70(3):219–225. doi: 10.1159/000319175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang R, Tang W, Wu X, Fu W. A random forest approach to the detection of epistatic interactions in case-control studies. BMC Bioinformatics. 2009;10 Suppl 1:S65. doi: 10.1186/1471-2105-10-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Günther F, Wawro N, Bammann K. Neural networks for modeling gene-gene interactions in association studies. BMC Genet. 2009;10:87. doi: 10.1186/1471-2156-10-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y. Bayesian epistasis association mapping via SNP imputation. Biostatistics. 2010 doi: 10.1093/biostatistics/kxq063. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hawkins RD, Hon GC, Ren B. Next-generation genomics: an integrative approach. Nat Rev Genet. 2010;11(7):476–486. doi: 10.1038/nrg2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farnham PJ. Insights from genomic profiling of transcription factors. Nat Rev Genet. 2009;10(9):605–616. doi: 10.1038/nrg2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Laird PW. Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet. 2010;11(3):191–203. doi: 10.1038/nrg2732. [DOI] [PubMed] [Google Scholar]
- 46.Cookson W, Liang L, Abecasis G, Moffatt M, Lathrop M. Mapping complex disease traits with global gene expression. Nat Rev Genet. 2009;10(3):184–194. doi: 10.1038/nrg2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moffatt MF, Kabesch M, Liang L, et al. Genetic variants regulating ORMDL3 expression contribute to the risk of childhood asthma. Nature. 2007;448(7152):470–473. doi: 10.1038/nature06014. [DOI] [PubMed] [Google Scholar]
- 48.Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Griffith M, Griffith OL, Mwenifumbo J, et al. Alternative expression analysis by RNA sequencing. Nat Methods. 2010;7(10):843–847. doi: 10.1038/nmeth.1503. [DOI] [PubMed] [Google Scholar]
- 50.Pastinen T. Genome-wide allele-specific analysis: insights into regulatory variation. Nat Rev Genet. 2010;11(8):533–538. doi: 10.1038/nrg2815. [DOI] [PubMed] [Google Scholar]
- 51.Ozsolak F, Goren A, Gymrek M, et al. Digital transcriptome profiling from attomole-level RNA samples. Genome Res. 2010;20(4):519–525. doi: 10.1101/gr.102129.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kuehn BM. Inconsistent results, inaccurate claims plague direct-to-consumer gene tests. JAMA. 2010;304(12):1313–1315. doi: 10.1001/jama.2010.1328. [DOI] [PubMed] [Google Scholar]
- 53.Mack GS. FDA holds court on post hoc data linking KRAS status to drug response. Nat Biotechnol. 2009;27(2):110–112. doi: 10.1038/nbt0209-110c. [DOI] [PubMed] [Google Scholar]
- 54.Klein TE, Altman RB, Eriksson N, et al. International Warfarin Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360(8):753–764. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]