Abstract
Ileal Crohn's Disease (CD), a chronic small intestinal inflammatory disorder, is characterized by reduced levels of the antimicrobial peptides DEFA5 (HD-5) and DEFA6 (HD-6). Both of these α-defensins are exclusively produced in Paneth cells (PCs) at small intestinal crypt bases. Different ileal CD–associated genes including NOD2, ATG16L1, and recently the β-catenin–dependant Wnt transcription factor TCF7L2 have been linked to impaired PC antimicrobial function. The Wnt pathway influences gut mucosal homeostasis and PC maturation, besides directly controlling HD-5/6 gene expression. The herein reported candidate gene study focuses on another crucial Wnt factor, the co-receptor low density lipoprotein receptor-related protein 6 (LRP6). We analysed exonic single nucleotide polymorphisms (SNPs) in a large cohort (Oxford: n = 1,893) and prospectively tested 2 additional European sample sets (Leuven: n = 688, Vienna: n = 1,628). We revealed an association of a non-synonymous SNP (rs2302685; Ile1062Val) with early onset ileal CD (OR 1.8; p = 0.00034; for homozygous carriers: OR 4.1; p = 0.00004) and additionally with penetrating ileal CD behaviour (OR 1.3; p = 0.00917). In contrast, it was not linked to adult onset ileal CD, colonic CD, or ulcerative colitis. Since the rare variant is known to impair LRP6 activity, we investigated its role in patient mucosa. Overall, LRP6 mRNA was diminished in patients independently from the genotype. Analysing the mRNA levels of PC product in biopsies from genotyped individuals (15 controls, 32 ileal, and 12 exclusively colonic CD), we found particularly low defensin levels in ileal CD patients who were carrying the variant. In addition, we confirmed a direct relationship between LRP6 activity and the transcriptional expression of HD-5 using transient transfection. Taken together, we identified LRP6 as a new candidate gene in ileal CD. Impairments in Wnt signalling and Paneth cell biology seem to represent pathophysiological hallmarks in small intestinal inflammation and should therefore be considered as interesting targets for new therapeutic approaches.
Author Summary
Crohn's Disease (CD) is to date incurable and is characterized by severe, reoccurring inflammations that can affect different intestinal locations. The complicated and multifactorial pathogenesis is not completely understood but involves disturbed epithelial barriers and immune reactions against the commensal flora in genetically predisposed individuals. Some inherited disease mechanisms are specific for small intestinal CD and are often connected to impaired Paneth cells. These specialized cells are critical for epithelial defences in the small intestine, and their most abundant antimicrobials (HD-5 and HD-6) are primarily diminished in patients who suffer from CD at this location. In this context, we previously identified a primary role of the Wnt pathway factor TCF7L2, which regulates the expression of both these antimicrobial peptides. We have now studied a functional mutation in the Wnt co-receptor LRP6 that was more frequent in early onset ileal CD. It was also associated with severe penetrating behaviour and linked to especially low HD-5 expression in ileal CD patient biopsies. Independently, we found that a reduced epithelial expression likely represents an additional impairment of the Wnt co-receptor in the disorder. LRP6 is a new player in small intestinal CD and underlines the importance of Paneth cell antimicrobial defences in the disease pathogenesis.
Introduction
Ileal Crohn's disease (CD) belongs to the group of inflammatory bowel diseases characterized by chronic intestinal inflammation, ulceration and consequent diarrhoea [1]. Both, environmental and inherited factors contribute to the disease risk [2], [3] and different genetic backgrounds likely explain variability in disease severity and especially disease location. Whereas the behaviour as well as the severity might change overtime, the disease location remains stable throughout the course, arguing for different pathogenetic mechanisms in the location specific disease subgroups [4]–[6]. Supporting this, genetic associations including NOD2, ATG16L1, as well as TCF7L2 (also known as TCF4) have been specifically associated with small intestinal, but not colonic CD [7]. Since bacteria represent the main target of adaptive immune responses [8]–[10] and trigger mucosal inflammation in susceptible individuals [11], we have suggested that location specific antimicrobial immunity defects render the mucosa susceptible to microbial adhesion and invasion [12]. Such defence impairments accommodate both, the inherited and the microbial component in the pathogenesis [13] and help to explain the stability of the specific disease locations. In contrast to colonic CD and ulcerative colitis (UC), ileal CD is characterized by a specific reduction of two Paneth cell antimicrobial peptides (AMPs), the human defensins (HD) -5 and -6 [13]–[16]. These two Paneth cell α- defensins are the most abundant products of the specialized secretory cells residing at the base of small intestinal crypts of Lieberkühn and the most prominent AMPs in the ileal mucosa [17], [18]. They are secreted after activation of pattern recognition receptors with microbial products, for example with muramyldipeptide (MDP) [19], the minimal bioactive peptidoglycan motif common to all bacteria, recognized by NOD2 [20], [21]. Paneth cell antimicrobials are involved in the regulation of the luminal commensal makeup [15], [22] and protect the organism from pathogens. Different mechanisms leading to diminished PC α- defensin levels in Crohn's disease have been identified. In addition to mutations in the susceptibility gene NOD2, which explain the decrease in some patients [14], [23], alterations in the Wnt pathway seem to play an important role in the majority of patients with ileal CD [24], [25]. Given that Wnt controls Paneth cell maturation and intestinal proliferation [26], aside from directly regulating HD-5 and -6, the observed link might suggest an involvement of impaired cell differentiation in the disorder. We first identified a decrease of TCF7L2 mRNA and subsequently reduced HD-5 promoter binding activity of mucosal extracts in ileal CD patients [25]. A following genetic study in a large sample set of 3 western European cohorts uncovered an association of TCF7L2 promoter region variants specifically with ileal CD [24]. We now hypothesized that upstream Wnt factors might also be affected in ileal CD and represent interesting factors for target gene association studies. One component with a key position in canonical Wnt signalling transduction is the low density lipoprotein receptor-related protein 6 (LRP6) [27]–[29]. This Wnt co-receptor is essential for cytoplasmatic stabilization of β- catenin which upon entry into the nucleus binds to factors of the Lymphoid enhancer family (Lef)/TCF family and activates promoters of target genes including HD-5 and HD-6. In mice, impeding LRP6 receptor function leads to rapid inhibition of intestinal epithelial regeneration, loss of proliferative crypts, and eventual inflammation and architectural degeneration [30]. These findings formed the rationale to test the hypothesis that LRP6 impairment could predispose to small intestinal inflammation in human CD patients. Furthermore we aimed to understand the functional pathway and possible link to Paneth cell innate immunity.
Results
Distribution of exonic LRP6 SNPs in a western European Cohort from Oxford
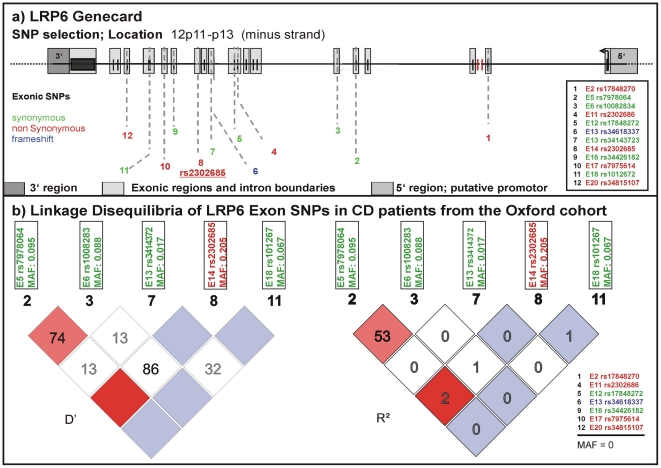
We studied frequency distributions and linkage disequilibria of all SNPs reported in the NCBI SNPdatabase in the exonic regions of LRP6 (Figure 1 upper panel). For a first analysis we used a well-defined cohort from Oxford including almost 2000 DNA samples from healthy controls and IBD patients. We determined frequencies for 5 of the 12 exonic SNPs described in the NCBI SNPdb (Figure 1 lower panel). SNPs with a minor allele frequency (MAF) of 0 in the Oxford samples were either not previously validated or so far not been found in western European cohorts. None of the tested SNPs were associated with CD or UC overall. However, in this first analysis, the coding rare allele of rs2302685 exhibited an association with a subgroup: an early disease onset phenotype in ileal CD (odds ratio (OR) 1.524, 95% confidence interval (CI) 0.988 to 2.345, p = 0.05511; for homozygous carriers OR 3.152, 95% CI 1.128 to 8.845, p = 0.02144). Since none of the other analysed SNPs showed frequency differences between controls and the different analysed disease groups we focused only on rs2302685 for additional tests. We also did not find a significant linkage between this variant and any of the other tested polymorphic SNPs and therefore did not include them in the analysis of the two additional cohorts (Figure 1 lower panel).
Figure 1. Known exonic SNPs in LRP6.
(A) LRP6 gene card with locations of exonic SNPs published in the NCBI SNPdb. Variants which lead to an amino acid exchange are marked in red. A deletion mutation causing a different protein chain is marked in blue, silent variants are marked in green. (B) Linkage disequilibria (LD) of the exonic SNPs in CD patients of the Oxford cohort. 7 of the genotyped SNPs were not present. We conclude that there is no significant LD between the sole coding variant (rs2302685) and any of the synonymous SNPs.
Association of the coding rs2302685 minor variant
After association of rs2302685 with early onset ileal CD in the Oxford patients, we prospectively tested if a higher frequency of this functional variant can also be found in other cohorts. Consistent with the first analysis in the Oxford cohort (Table 1, Oxford), subsequent analysis of two large sample sets (Table 1, Leuven and Vienna) showed the same overall result, whereas the frequency distributions among the control groups as well as the not further sub-grouped patients (IBD, CD, UC) were strikingly similar (MAFs between 18.47 and 20.59%, Table 1 and Table 2). Combining all tested samples, an association with early onset ileal CD (diagnosis at ages 17 and younger) (MAF: 29.57%; OR 1.797, 95% CI 1.298 to 2.486, p = 0.00034) and penetrating behaviour (internal fistulae; Montreal classification B3) (MAF: 23.24%; OR 1.296, 95% CI 1.066 to 1.575, p = 0.00917) suggests that Ile1062Val may influence both, disease onset and severity (Table 2, Figure 2), even though statistical significance of the latter association was lost after adjusting for multiple testing (Bonferroni adjustment for penetrating ileal CD behaviour: p = 0.10087). Gender on the other hand had no impact on the allele distribution (Table 2). The homozygous genotype of the minor allele displayed the highest risk for early onset ileal disease underlining a potential dose effect of the mutation (homozygous minor allele carriers: controls: 3,33%, early onset ileal CD: 10.75%; OR 4.093, 95% CI 1.981 to 8.455, p = 0.00004). Amongst the 237 analysed patients with exclusive colonic CD (L2) only 19 had a disease onset prior to age 18. None of these were homozygous for the risk variant and with a MAF of 13,64%, the SNP distribution showed no significant difference to controls (OR 0.803, 95% CI 0.334 to 1.926, p = 0.62172). We also compared early versus late onset in ileal CD patients and found a similar result as in the comparison with healthy controls (allele frequency: OR 1.760, 95% CI. 1.251 to 2.477, p = 0.00106; homozygous carriers: OR 4.484, 95% CI 1.995–10.077, p = 0.00009). The mean age of onset was similar between the CD patients in the different cohorts, as was the mean age of controls at the time of blood sampling for later DNA extraction (Table 3). In addition to testing for allele frequency differences and the increased risk of homozygous carriers, we also used additive, recessive and dominant models of inheritance to compare the genotype distribution between controls and early onset ileal CD as presented in Table 4.
Table 1. LRP6 rs2302685 frequency distribution in the separate cohort samples (Oxford, Leuven, Vienna).
| Overview groups | Overview ileal CD | |||||||||||
| IBD | Gender | Disease behaviour | Age at diagnosis | |||||||||
| Oxford | controls | UC | colonic CD (L2) | ileal CD (L1+L3) | male | female | B1 | B2 | B3 | >17 | </ = 17 | |
| TT | 470 | 416 | 89 | 238 | 90 | 134 | 33 | 109 | 82 | 207 | 31 | |
| Genotypes (n) | CT | 242 | 185 | 53 | 123 | 50 | 71 | 18 | 54 | 49 | 102 | 21 |
| CC | 24 | 33 | 4 | 16 | 8 | 8 | 2 | 4 | 10 | 11 | 5 | |
| samples (n) | all | 736 | 634 | 146 | 377 | 148 | 213 | 53 | 167 | 141 | 320 | 57 |
| TT | 63.86% | 65.62% | 60.96% | 63.13% | 60.81% | 62.91% | 62.26% | 65.27% | 58.16% | 64.69% | 54.39% | |
| Genotypes | CT | 32.88% | 29.18% | 36.30% | 32.63% | 33.78% | 33.33% | 33.96% | 32.34% | 34.75% | 31.88% | 36.84% |
| (frequency) | CC | 3.26% | 5.21% | 2.74% | 4.24% | 5.41% | 3.76% | 3.77% | 2.40% | 7.09% | 3.44% | 8.77% |
| Allele | T | 80.30% | 80.21% | 79.11% | 79.44% | 77.70% | 79.58% | 79.25% | 81.44% | 75.53% | 80.63% | 72.81% |
| Frequency | C | 19.70% | 19.79% | 20.89% | 20.56% | 22.30% | 20.42% | 20.75% | 18.56% | 24.47% | 19.38% | 27.19% |
The different distribution of genotypes is demonstrated for each group and subgroup: controls, ulcerative colitis (UC), Crohn's disease (CD) with solely colonic involvement (colonic CD; L2) and CD with solely ileal as well as ileal and colonic involvement (ileal CD; L1+L3). The ileal CD group was further sub-grouped according to gender, disease behaviour (inflammatory B1; stricturing B2; penetrating B3) and age at diagnosis (older than 17: >17; 17 and younger: </ = 17).
Table 2. LRP6 rs2302685 frequency distribution in the combined cohort samples.
| Overview groups | Overview ileal CD | |||||||||||
| all samples | IBD | Gender | Disease behaviour | Age at diagnosis | ||||||||
| controls | UC | colonic CD (L2) | ileal CD (L1+L3) | male | female | B1 | B2 | B3 | >17 | </ = 17 | ||
| TT | 1277 | 758 | 147 | 500 | 219 | 267 | 98 | 183 | 198 | 452 | 48 | |
| Genotypes (n) | CT | 609 | 364 | 83 | 263 | 112 | 152 | 58 | 76 | 126 | 228 | 35 |
| CC | 65 | 47 | 7 | 31 | 15 | 16 | 5 | 9 | 16 | 21 | 10 | |
| samples (n) | all | 1951 | 1169 | 237 | 794 | 346 | 435 | 161 | 268 | 340 | 701 | 93 |
| TT | 65.45% | 64.84% | 62.03% | 62.97% | 63.29% | 61.38% | 60.87% | 68.28% | 58.24% | 64.48% | 51.61% | |
| Frequencies | CT | 31.21% | 31.14% | 35.02% | 33.12% | 32.37% | 34.94% | 36.02% | 28.36% | 37.06% | 32.52% | 37.63% |
| CC | 3.33% | 4.02% | 2.95% | 3.90% | 4.34% | 3.68% | 3.11% | 3.36% | 4.71% | 3.00% | 10.75% | |
| Allele | T | 81.06% | 80.41% | 79.54% | 79.53% | 79.48% | 78.85% | 78.88% | 82.46% | 76.76% | 80.74% | 70.43% |
| Frequency | C | 18.94% | 19.59% | 20.46% | 20.47% | 20.52% | 21.15% | 21.12% | 17.54% | 23.24% | 19.26% | 29.57% |
The different distribution of genotypes is demonstrated for each group and subgroup: controls, UC, CD colonic CD and ileal CD. The ileal CD group was further sub-grouped according to gender, disease and age at diagnosis.
Figure 2. Risk for rs2302685 C-allele carriers in the different disease subgroups and the influence of detailed phenotyping.
A) Odds ratios and confidence intervals for the different comparisons are shown. The frequency distribution of the minor rs2302685 allele was analysed in different cohorts and combined samples: odds ratios and 95% confidence interval for the allele frequency are shown for patients with specific disease subtypes of ileal CD (classified as either L1 (solely ileal) or L3 (ileal and colonic involvement)) compared to healthy control individuals. The early onset as well as the penetrating behaviour subgroup displays an significantly increased Minor allele frequencies (MAF). B) An especially high risk for early onset ileal CD is found for homozygous minor C - allele carriers. This becomes apparent in the high odds ratios for such individuals in the different cohorts and combined samples (left panel) and an highly increased CC genotype frequency compared to the control samples and other disease groups (right panel). C) The MAF distribution was analysed in different cohorts and combined samples: odds ratios and 95% confidence interval for the MAF are shown for patients with ulcerative colitis (UC) (left panel) and CD patients with solely colonic involvement (right panel) compared to healthy control individuals. The lack of an increase in the odds ratios for the different comparisons allows excluding major effects of the variant in the colonic IBD subgroups.
Table 3. Age of included individuals.
| Mean age at sampling | Mean age at onset | ||
| Controls | Ileal CD (L1+L3) | Colonic CD (L2) | |
| Oxford | 42.69 (±14.62) | 27.46 (±11.79) | 34.03 (±14.71) |
| Leuven | 35.00 (±17.30) | 30.33 (±11.16) | 31.97 (±11.18) |
| Vienna | 36,73 (±11.99) | 27.89 (±9.39) | 28,07 (±10,12) |
Provided are the mean age ± standard deviation. For controls we listed the age at time of DNA sampling, for CD patients we provide the mean age at onset.
Table 4. Overall statistical analysis in controls versus early onset ileal CD.
| Controls vs early onset ileal CD | Allele positivity | Armitage's trend | |||
| allele frequency | homozygous risk | dominant model | recessive model | additive model | |
| C vs T | CC vs TT | CC vs CT+TT | TT vs CT+CC | T common odds | |
| Odds ratio | 1.797 | 4.093 | 1.776 | 0.286 | 1.877 |
| 95% CI | 1.298 to 2.487 | 1.981 to 8.455 | 1.170 to 2.696 | 0.142 to 0.577 | |
| p-value | 0.00034 | 0.00004 | 0.00632 | 0.0002 | 0.00033 |
| Bonferroni adjusted p-value | 0.00374 | 0.00044 | 0.06952 | 0.0022 | 0.00363 |
The genotypes for which the risk was calculated are marked in bold. The Bonferroni adjusted p-values were calculated with regards to all comparisons in the overall sample group (adjusted for 11 tests).
Impaired transcriptional expression of the Paneth cell antimicrobial peptide HD-5 in LRP6 mutated ileal CD patients
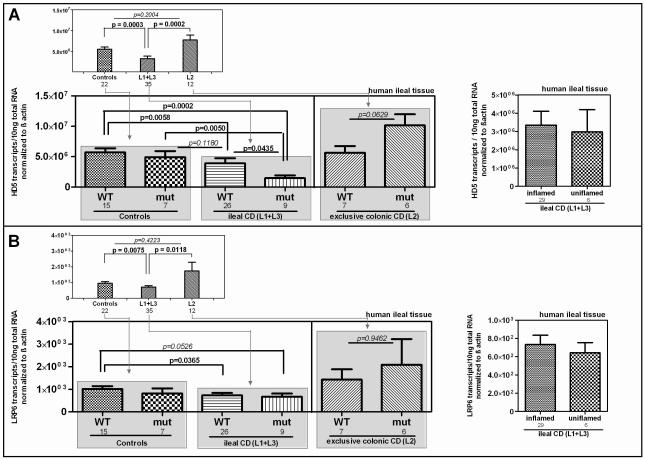
We further studied the influence of the Ile1062Val mutation on HD-5 mRNA expression in ileal mucosa obtained from a cohort endoscoped in our department. To exclude influences of NOD2 defects on the transcriptional expression of HD-5 [14], individuals with mutations in the pattern recognition receptor were excluded from the analysis. We furthermore excluded patients with a neoterminal ileum as a result to previous resection. Besides confirming the general decrease of HD-5 in ileal CD patients, we found the lowest expression of HD-5 in the ileal CD group of LRP6 Ile1062Val mutated patients (Figure 3) which suggests a specific relevance of the polymorphism in ileal CD. Confirming previous data [15], [16] inflammation per se did not seem to influence HD-5 expression (Figure 3A right panel).
Figure 3. mRNA expression in ileal CD: Effect of the coding rare rs2302685 variant on the expression levels of HD-5 and LRP6.
We excluded individuals with mutations in the pattern recognition receptor NOD2 as well as patients with a neoterminal ileum. All cohort samples were sub-grouped according to the disease state and analysed with respect to the LRP6 genotype. A) As previously reported, HD-5 mRNA levels are diminished in ileal CD as compared to controls and colonic CD. We saw no difference between ileal CD samples that came from inflamed tissue at the time of biopsy taking and material from 6 uninflamed biopsy specimens (left panel). The mRNA expression of HD-5 seems to be influenced by the coding rare LRP6 rs2302685 allele (C- allele) in ileal CD patients as carriers of the variant show the lowest HD-5 levels. B) Similar to the diminished HD-5, we found significantly decreased LRP6 transcriptional expression in ileal CD patients as compared to controls as well as to patients with exclusive colonic CD. Again similar to HD-5, we found no difference between inflamed compared to uninflamed ileal CD samples. The presence of the functional LRP6 mutation however, seems to have, as expected, no effect on LRP6 mRNA level.
Diminished LRP6 mRNA levels represent an additional impairment in ileal CD
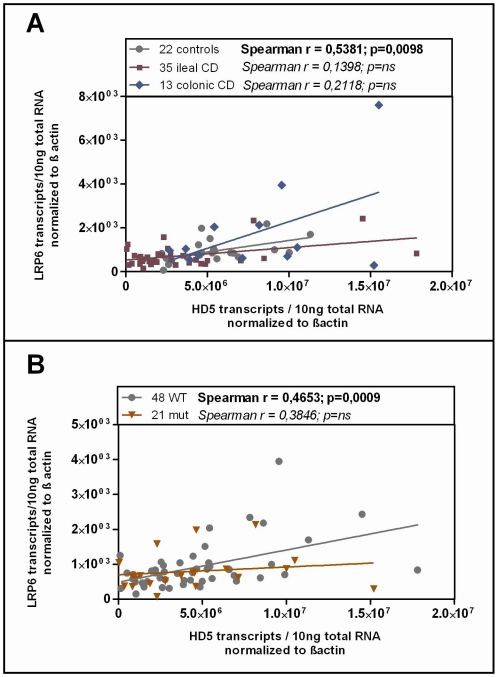
Studying the transcriptional expression level of LRP6 in our cohort mucosal biopsy samples, we found generally diminished LRP6 mRNA levels in ileal CD (Figure 3B). As expected, according to the known effect on signalling impairment -but not expression level- [30] the reduction of LRP6 was independent from the functional mutation. The reduced transcriptional expression of LRP6 might be an additional factor which contributes to the especially low levels seen in LRP6 mutated ileal CD patients. Consistent with the latter interpretation, levels of LRP6 showed a significant correlation with the Paneth cell antimicrobial HD-5 in healthy controls (Figure 4A). When analysing all samples according to the genotype we found a significant correlation in all LRP6 wild type individuals (Figure 4B). Interestingly, in carriers of the rare mutant allele (Figure 4B) the correlation was absent supporting a possible direct effect of rs2302685. Similar to HD-5 (Figure 4A), inflammation per se did not seem to affect LRP6 in our sample set, clearly it did not account for the described reduction (Figure 4B). Taken together the data suggest that both, the mutation, as well as the diminished expression level of LRP6 contribute to the reduced levels of HD-5 in ileal CD patients.
Figure 4. Correlation of HD-5 and LRP6.
Linear regression and Spearman rank analysis were used on all samples from the mRNA analysis. The samples were grouped according to A) their disease state (diseased mucosa or non-inflamed mucosa of patients) as well as in all samples B) according to the LRP6 genotype. Interestingly the two factors exhibit a correlating pattern in controls (independent of the genotype) as well as in all wild type samples but not in the C-allele carrying individuals.
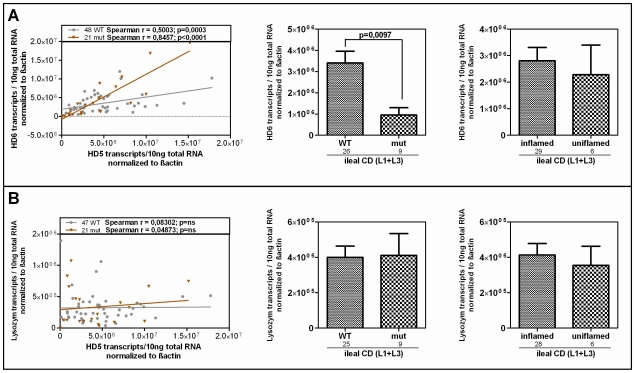
Since HD-6, the second most abundant Paneth cell product is also reduced in ileal CD, we additionally analysed its expression according to the LRP6 genotype in our patients. It is known that both Paneth cell α-defensins are regulated by the Wnt pathway, so we expected a similar effect. As hypothesized, the two factors showed a correlating pattern in wild type as well as in mutated individuals in our cohort and HD-6 exhibited the same dependence on the LRP6 genotype which was seen for HD-5 in ileal CD (Figure 5A). We also measured lysozyme, another antimicrobial found in Paneth cells which is not decreased in ileal CD and also not known to be dependent on canonical Wnt. As expected, there was no change in lysozyme mRNA levels in ileal CD carriers of the rare LRP6 SNP compared to the wild type ileal CD subgroup and also no correlation with HD-5 in either subgroup.
Figure 5. HD-6 but not lysozyme is reduced in patients who carry the rare LRP6 variant.
Linear regression and Spearman rank analysis were used on all samples from the mRNA analysis according to the LRP6 genotype. (A) The two Paneth cell α-defensins correlate in both subgroups fitting the also found genotype dependent decrease of HD-6 in ileal CD patients. This, as well as the independence from inflammation matches the pattern seen for HD-5. (B) Lysozyme on the other hand does not correlate with HD-5 and seems to be uninfluenced by the LRP6 genotype in ileal CD.
LRP6 overexpression induces β-catenin–dependent induction of HD-5 promoter activity
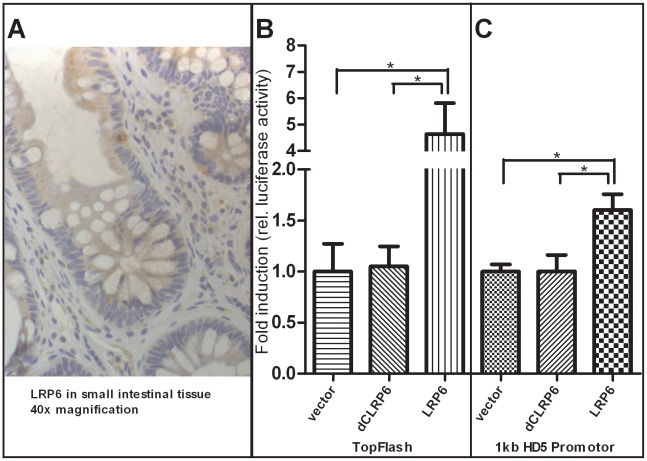
As mentioned above it is known that β-catenin dependent Wnt signaling activity is crucial for Paneth cell maturation and that disruption of the pathway precedes impaired Paneth cell gene expression and function. Since we found diminished levels of Paneth cell α-defensins in ileal CD patients carrying a coding SNP variant in LRP6, we wanted to further analyse the Wnt co-receptor's role in this context. To confirm the expression of LRP6 in small intestinal epithelia, we performed immunohistochemistry staining of the co-receptor on ileal tissue slices from healthy controls as well as ileal CD patients. LRP6 was found to be generally expressed in epithelial cells of the small intestine, and also present at the very bottom of intestinal crypts at the sites where Paneth cells as well as stem cells reside. Interestingly, LRP6 was also sporadically detected in infiltrating immune cells. A 40× magnification of a representative section is included in Figure 6A. To directly study the relationship between LRP6 activity and Paneth cell HD-5, we used an in vitro model of transiently transfected HEK293 cells. From previous work it is known that overexpression of LRP6 is sufficient to induce β-catenin dependent Wnt signalling activity, even without additional stimulation with Wnt ligands or other pathway activating compounds. As a positive control we used the Wnt responsive TopFlash promoter (Figure 6B). Overexpression of LRP6 led to an increase of transcriptional activity of an 1 kb HD-5 promoter construct (Figure 6C). As expected this was not seen using a non-functional (dominant negative) version of the co-receptor lacking important intracellular domains which are necessary for Wnt signal transduction.
Figure 6. Increased activity of β-catenin–dependent Wnt via overexpression of LRP6 transactivates human α-defensin 5 promoter activity in HEK-293 cells.
As demonstrated with immunohistochemistry staining A), the Wnt co-receptor LRP6 is widely expressed in the cells of the small intestinal epithelium and also found at the base of the crypts where Paneth and stem cells are located. Additionally, LRP6 is sporadically found in infiltrating immune cells. As previously demonstrated, overexpression of LRP6 in HEK293 cells leads to an activation of β-catenin dependent Wnt signalling B) as monitored by TopFlash activity. The transcriptional activity of a luciferase reporter under the control of an 1 kb HD-5 promoter was also increased after LRP6 mediated activation of Wnt C). Both effects were not seen using a non-functional version of the LRP6 Co-receptor which is lacking cytoplasmatic parts that are necessary for signal transduction. Values are the average of triple determinations with the SEM indicated by error bars.
Discussion
Complementing the previously described involvement of Wnt TCF7L2 [24], [25] in ileal Crohn's Disease, we now report an association of the rare coding SNP variant of rs2302685 (Ile1062Val) in the Wnt co- receptor LRP6 with early-onset ileal CD. In addition, we found a genotype independent mucosal reduction of LRP6 level in ileal CD tissue. From mouse models, it is known that specific intestinal blockage of LRP6 has devastating consequences on the regenerative and proliferative potential of intestinal epithelia which can result in inflammation as well as mucosal degeneration [30]. So far for LRP6, nothing has been known about an involvement in the development of human inflammatory bowel disorders. However, human studies could establish that the presence of the rs2302685 rare coding variant (C-allele) results in generally diminished signalling activity of LRP6 [31]. Since the Wnt signalling transcription factor TCF4 (TCF7L2) directly affects the transcription of HD-5 and HD-6 [25] and is genetically associated with ileal disease [24] we hypothesized that the LRP6 mutation also affects innate antimicrobial Paneth cell function. Ileal CD patients who feature the C-allele showed significantly further reduced Paneth cell defensin expression levels in mucosal ileal tissue but unchanged levels of lysozyme, an additional but canonical Wnt independent Paneth cell antimicrobial. To corroborate a potential direct relationship between LRP6 function and the transcriptional expression of HD-5, we performed transient transfection experiments in HEK293 cells and found an induction of a 1 kb HD-5 promoter fragment upon overexpression of LRP6 which precedes activation of β-catenin dependent Wnt signalling.
A possible limitation of the association study could be the early onset sample size. The overall studied cohort numbers are very high, but only a limited fraction (∼12%) of analysed ileal CD patients shared the phenotype of early onset. However it should be noted that the first found result could be prospectively tested and confirmed in two additional cohorts. Other limitations might apply to the mucosal tissue expression studies. For assessing genotype-depending expression levels we had no intestinal tissue from homozygous C-allele carriers and we only could test tissue from adult patients. Both factors are due to restricted biopsy material resources. A potentially even stronger effect in homozygous C-allele carriers or a cause-effect-relationship in paediatric ileal CD remains therefore to be investigated. Since healthy controls featuring the C-allele heterozygously showed almost normal levels of HD-5, it is clear that other factors must be involved, especially to explain the specific effect of the mutation in ileal CD patients. Patients who carry the rare variant might only in combination with other underlying disturbances be prone to an early symptom development. It is quite conceivable that these could be the same mechanisms which explain diminished HD-5 expression in ileal CD in general. The combination of different defects, including the mutation in LRP6, might add up to the strong decrease of HD-5 and HD-6 which is specifically seen in this subgroup. One potential additive mechanism could be the diminished transcriptional expression of LRP6, which was seen independently from the patient's genotype. However, the mechanisms explaining reduced LRP6 expression still needs to be determined. In addition to LRP6, the described decrease of TCF4 (TCF7L2) and this Wnt transcription factor is likely another additive mechanism [15], [24], [32]. To analyse such potentially additive functions of the reported impairments in canonical Wnt should be the aim of future investigations. Most previously identified Crohn's disease loci [33], [34] are common to both early and later disease onset. Exclusive early onset disease associated variants are extremely rare and none are known for the ileal version specifically. An IL-6 promoter SNP was recently associated with early onset CD in general but confined solely to male patients [35]. Five novel genomic regions were additionally associated with general early-onset IBD in a genome wide association study, including 16p11 near IL27 [34]. The identification of rs2302685 as an ileal early onset and also penetrating CD risk variant potentially provides a first location specific diagnostic marker for such high risk individuals.
Other studies on LRP6 genetic variances have so far been done in the context of variability in bone mass density, bone disorders [36], late-onset Alzheimer's disease [31], macular degeneration [37] and cardiovascular diseases [38], [39]. Complications affecting the bone are frequent in IBD with disease-inherent factors appearing to confer a risk irrespective of corticosteroid treatment. The newly reported genetic association of LRP6 might contribute to the occurrence of such extraintestinal manifestations but further studies are required to analyse a potential role in detail. Finally, it is important to acknowledge the context of the LRP6 association as it provides further evidence for the significance of the Paneth cell in the development of small intestinal CD. Multiple genetic CD variants have already been specifically associated with small intestinal involvement and most of the involved genes are important in the specialized cell's biology [7]: the Wnt transcription factor TCF7L2 [24], NOD2 [23], [40], ATG16L1 [41], [42], XBP1 [43] and very recently the potassium intermediate/small conductance calcium-activated channel KCNN4 [44]. The co-receptor LRP6 may now be added as a novel player in early onset and penetrating behaviour in ileal CD. The multiple genes linked to ileal CD support the concept that impaired Paneth cell antimicrobial function represents a primary and a not secondary defect [45], [46]. Most importantly, it provides an attractive and direct therapeutic target [47] as an alternative to the current merely anti-inflammatory approaches in Crohn's disease therapy.
Methods
Patients
All patients and healthy controls included in the present studies gave their written and informed consent after the study purpose, sample procedure, and potential adjunctive risks were clarified. All studies were approved by the ethics committees of the Medical University Vienna, Austria, the University Hospital Tübingen, Germany, the University of Leuven, Belgium and the Oxford Radcliffe Hospital Trust, Great Britain. Sub-grouping of included patients was done according to phenotype data which was based on clinical, radiological, endoscopic and histopathological diagnoses at the respective IBD centres (Figure S1). For genetic analysis, we evaluated 3 DNA cohorts (all of Western European descent) of CD and UC patients as well as healthy unrelated controls [24]. For the genetic study we included all definitely phenotyped patients, the cohorts also include patients who underwent surgery (colectomy in UC, or ileocolonic resection in CD), those with an additional involvement of the upper gastrointestinal tract (Montreal +L4) as well as patients with perianal disease. We did not further subgroup the cohorts according to these criteria as we focussed our analysis on disease location, behaviour and age of onset. Biopsies and blood were additionally collected from patients and controls in Stuttgart; in this cohort, NOD2 mutant individuals as well as patients with a neoterminal ileum (after ileocecal resection) were excluded to avoid an mRNA effect bias. We included biopsies of healthy controls and patients with and without active inflammation and also stratified the cohort according to this criterion. In line with the Montreal classification three CD subgroups were defined to accommodate the different disease locations: ileal disease only (L1), colonic disease only (L2) and ileocolonic disease (L3).
Candidate gene approach and SNP selection
We included all exonic LRP6 SNPs which were documented in the NCBI SNPdb (Genotype and allele frequency build 129). DNA was isolated by standard procedures. Multiplex genotyping was performed with the MassARRAY Compact System from Sequenom (San Diego, USA) [24].
LRP6 genotyping
All primers were designed using reference sequences as denoted by the NCBI SNPdb and Sequenom software (San Diego, USA) and are provided in Table S1. All materials originated from the Sequenom iPLEX Gold Kit and were used according to the manufacturer's protocol (Sequenom). The applied MALDI-TOF MS based SNP genotyping method measures the time of flight of ionized molecules to determine their masses. Respective fragments (∼100 bp including the SNP) for our analysis were assembled via Multiplex-PCR and verified using gelelectrophoresis. After a Shrimp Alkaline Phosphatase (SAP) clean- up procedure, a specific linear primer extension (PEX) reaction was performed creating genotype specific products with distinguishable masses. A cleanup step with Resin (exchange of cations) for optimizing mass spectrometry analysis of the extended reaction products was performed before the samples were loaded and analysed.
NOD2 mutation analysis
Genotyping for the relevant NOD2 mutations was performed in patient samples using TaqMan technology (Applied Biosystems, Foster City, California, USA), as previously described [14].
Real-time PCR
1 µl of total RNA isolated from snap frozen ileal tissue biopsies were checked for quality before transcribed into cDNA using oligo (dT) primers and the AMV- reverse transcriptase (RT) kit according to the manufacturer's protocol (Promega). Real-time PCR with cDNA corresponding to 10 ng total RNA was subsequently performed with a LightCycler 480 (Roche Diagnostics, Mannheim, Germany) using materials from the LightCycler 480 SYBR Green I Master kit according to the manufacturer's protocol (Roche). Specific plasmid standards for the selected products were utilized to calculate exact copy numbers and primers are provided in Table S2.
Cell culture
HD-5 luciferase reporter constructs have been kindly provided by Béatrice Romagnolo and Pauline Andreu (previously described by [48]). LRP6 expression plasmids were gratefully received from Xi He and Mikhail V. Semenov (previously described by [49], [50]). Vladimir Korinek kindly provided us with the Wnt responsive TopFlash luciferase reporter construct [51]. HEK293 cells in 24-well plates were transfected with 200 ng of the full-length LRP6 expressing vector, a non-functioning dnLRP6 expressing vector, or an empty vector, together with 200 ng of a TopFlash luciferase or HD-5 promoter construct and 50 ng of a Renilla luciferase expressing vector in each well using the FuGENE 6 reagent (Roche) according to the manufacturer's protocol. The luciferase activity was measured after 48 hours via the Dual Luciferase Reporter Assay System (Promega). Firefly luciferase activity corresponding to the studied promoter constructs was normalized with respect to transfection efficiencies using the respective activity of the co-transfected Renilla luciferase. Transfections were carried out in triplicates and 4 independent experiments were performed.
Immunohistochemistry
Immunostaining for LRP6 was performed using a two-step immunoperoxidase technique (EnVisionTM, Dako, Glostrup, Denmark) as described previously [52]. Slides were heated for 30 minutes in a steamer for antigen retrieval (pH 9) and incubated for 1 hour with the primary anti-LRP6 antibody (ABGENT, San Diego, USA) diluted 1∶100 in TBST (20 mM Tris-Base (pH 7.4), 0.14 M NaCl, 0.1% Tween 20). LPR6 protein was visualized by a horse-radish-peroxidase (HRP)-labelled secondary antibody (Dako) which was detected with 3′-diaminobenzidine tetrahydrochloride (Dako). Slides were counterstained with hematoxylin.
Computer analysis and statistics
mRNA levels were normalized to β- actin and evaluated by GraphPad Prism Ver. 5.0. To analyse the effect of Ile1062Val between the groups, as well as the cell culture experiments, we performed the non- parametric statistical Wilcoxon-Mann-Whitney-Test. Spearmen rank analysis was used to test for correlation. For genetic analysis we used Finetti specialized software (http://ihg2.helmholtz-muenchen.de/cgi-bin/hw/hwa1.pl). Linkage disequilibria and haplotype blocks were calculated with Haploview. To avoid statistical bias due to multiple testing between different subgroup in the overall association analysis, we calculated Bonferroni adjusted p-values for the comparison between early onset ileal CD and controls.
Supporting Information
Overview of the work flow. The association study was carried out within 3 previously assembled IBD DNA cohorts in a retrospective way. DNA and biopsies from patients and controls for the mRNA study and immunohistochemistry were additionally collected at the Robert Bosch hospital in Stuttgart.
(TIF)
Genotyping Primers: Oligonucleotides used for multiplex PCR approaches are termed 1st- respectively 2nd-PCRP. Primers used for genotype specific elongation are termed PEXP.
(DOCX)
Real-time PCR Primers: Oligonucleotides which were used for the creation of standards and mRNA quantification.
(DOCX)
Acknowledgments
We want to thank Kathleen Siegel, Jutta Bader, Andrea Schneider, Andrea Jarmuth, and Marion Schiffmann for excellent technical assistance. We furthermore want to acknowledge Dr. Stefan Winter who kindly advised us on our statistical analysis. Additionally we want to acknowledge Béatrice Romagnolo, Pauline Andreu, Xi He, and Mikhail V. Semenov, as well as Vladimir Korinek for kindly providing the plasmids which were used in the cell culture experiments.
Footnotes
Patent application (No: PCT/EP2009/009176, Publication Number: WO2010/072389): Method for determining the predisposition for Crohn's Disease.
The study was supported by the Robert Bosch Foundation, the Deutsche Forschungsgemeinschaft Emmy Noether program (WE 436/1-1), and Sonderforschungsbereich 685 (immunotherapy; Project A9). MS and ES are supported by the Federal Ministry of Science and Technology (Berlin, Germany) grant 03IS2061C. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 2.Halfvarson J, Jess T, Magnuson A, Montgomery SM, Orholm M, et al. Environmental factors in inflammatory bowel disease: a co-twin control study of a Swedish-Danish twin population. Inflamm Bowel Dis. 2006;12:925–933. doi: 10.1097/01.mib.0000228998.29466.ac. [DOI] [PubMed] [Google Scholar]
- 3.Schreiber S, Rosenstiel P, Albrecht M, Hampe J, Krawczak M. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 4.Louis E, Collard A, Oger AF, Degroote E, Aboul Nasr El Yafi FA, et al. Behaviour of Crohn's disease according to the Vienna classification: changing pattern over the course of the disease. Gut. 2001;49:777–782. doi: 10.1136/gut.49.6.777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gasche C, Grundtner P. Genotypes and phenotypes in Crohn's disease: do they help in clinical management? Gut. 2005;54:162–167. doi: 10.1136/gut.2003.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silverberg MS, Satsangi J, Ahmad T, Arnott ID, Bernstein CN, et al. Toward an integrated clinical, molecular and serological classification of inflammatory bowel disease: Report of a Working Party of the 2005 Montreal World Congress of Gastroenterology. Can J Gastroenterol. 2005;19(Suppl A):5–36. doi: 10.1155/2005/269076. [DOI] [PubMed] [Google Scholar]
- 7.Koslowski MJ, Beisner J, Stange EF, Wehkamp J. Innate antimicrobial host defense in small intestinal Crohn's disease. Int J Med Microbiol. 2009 doi: 10.1016/j.ijmm.2009.08.011. [DOI] [PubMed] [Google Scholar]
- 8.Duchmann R, May E, Heike M, Knolle P, Neurath M, et al. T cell specifity and cross reactivity towards enterobacteria, Bacteroides, Bifidobacterium, and antigens from resident intestinal flora in humans. Gut. 1999;44:812–818. doi: 10.1136/gut.44.6.812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sartor RB. Intestinal microflora in human and experimental inflammatory bowel disease. Curr Opin Gastroenterol. 2001;17:324–330. doi: 10.1097/00001574-200107000-00005. [DOI] [PubMed] [Google Scholar]
- 10.MacPherson A, Khoo UY, Forgacs I, Philpott-Howard J, Bjarnason I. Mucosal antibodies in inflammatory bowel disease are directed against intestinal bacteria. Gut. 1996;38:365–375. doi: 10.1136/gut.38.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sartor RB. Current concepts of the etiology and pathogenesis of ulcerative co and Crohn's disease. Gastroenterol Clin North Am. 1995;24:475–507. [PubMed] [Google Scholar]
- 12.Wehkamp J, Koslowski M, Wang G, Stange EF. Barrier dysfunction due to distinct defensin deficiencies in small intestinal and colonic Crohn's disease. Mucosal Immunol. 2008;1(Suppl 1):S67–S74. doi: 10.1038/mi.2008.48. [DOI] [PubMed] [Google Scholar]
- 13.Wehkamp J, Fellermann K, Herrlinger KR, Bevins CL, Stange EF. Mechanisms of disease: defensins in gastrointestinal diseases. Nat Clin Pract Gastroenterol Hepatol. 2005;2:406–415. doi: 10.1038/ncpgasthep0265. [DOI] [PubMed] [Google Scholar]
- 14.Wehkamp J, Harder J, Weichenthal M, Schwab M, Schaffeler E, et al. NOD2 (CARD15) mutations in Crohn's disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wehkamp J, Salzman NH, Porter E, Nuding S, Weichenthal M, et al. Reduced Paneth cell alpha-defensins in ileal Crohn's disease. Proc Natl Acad Sci U S A. 2005;102:18129–18134. doi: 10.1073/pnas.0505256102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kubler I, Koslowski MJ, Gersemann M, Fellermann K, Beisner J, et al. Influence of standard treatment on ileal and colonic antimicrobial defensin expression in active Crohn's disease. Aliment Pharmacol Ther. 2009 doi: 10.1111/j.1365-2036.2009.04070.x. [DOI] [PubMed] [Google Scholar]
- 17.Bevins C, Porter EM, Ganz T. Defensins and innate host defence of the gastrointestinal tract. Gut. 1999;45:911–915. doi: 10.1136/gut.45.6.911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.George MD, Wehkamp J, Kays RJ, Leutenegger CM, Sabir S, et al. In vivo gene expression profiling of human intestinal epithelial cells: analysis by laser microdissection of formalin fixed tissues. BMC Genomics. 2008;9:209. doi: 10.1186/1471-2164-9-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, et al. Secretion of microbicidal alpha-defensins by intestinal Paneth cel response to bacteria. Nat Immunol. 2000;1:113–118. doi: 10.1038/77783. [DOI] [PubMed] [Google Scholar]
- 20.Ogura Y, Inohara N, Benito A, Chen FF, Yamaoka S, et al. Nod2, a Nod1/Apaf-1 family member that is restricted to monocytes and activates NF-kappaB. J Biol Chem. 2001;276:4812–4818. doi: 10.1074/jbc.M008072200. [DOI] [PubMed] [Google Scholar]
- 21.Ouellette AJ, Bevins CL. Paneth cell defensins and innate immunity of the small bowel. Inflamm Bowel Dis. 2001;7:43–50. doi: 10.1097/00054725-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 22.Salzman NH, Hung K, Haribhai D, Chu H, Karlsson-Sjoberg J, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2009 doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ogura Y, Bonen DK, Inohara N, Nicolae DL, Chen FF, et al. A frameshift mutation in NOD2 associated with susceptibility to Crohn's diease. Nature. 2001;411:603–606. doi: 10.1038/35079114. [DOI] [PubMed] [Google Scholar]
- 24.Koslowski MJ, Kubler I, Chamaillard M, Schaeffeler E, Reinisch W, et al. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn's disease. PLoS ONE. 2009;4:e4496. doi: 10.1371/journal.pone.0004496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wehkamp J, Wang G, Kubler I, Nuding S, Gregorieff A, et al. The Paneth cell alpha-defensin deficiency of ileal Crohn's disease is linked to Wnt/Tcf-4. J Immunol. 2007;179:3109–3118. doi: 10.4049/jimmunol.179.5.3109. [DOI] [PubMed] [Google Scholar]
- 26.Gregorieff A, Clevers H. Wnt signaling in the intestinal epithelium: from endoderm to cancer. Genes Dev. 2005;19:877–890. doi: 10.1101/gad.1295405. [DOI] [PubMed] [Google Scholar]
- 27.Liu G, Bafico A, Harris VK, Aaronson SA. A novel mechanism for Wnt activation of canonical signaling through the LRP6 receptor. Mol Cell Biol. 2003;23:5825–5835. doi: 10.1128/MCB.23.16.5825-5835.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Semenov MV, Tamai K, Brott BK, Kuhl M, Sokol S, et al. Head inducer Dickkopf-1 is a ligand for Wnt coreceptor LRP6. Curr Biol. 2001;11:951–961. doi: 10.1016/s0960-9822(01)00290-1. [DOI] [PubMed] [Google Scholar]
- 29.Mao B, Wu W, Li Y, Hoppe D, Stannek P, et al. LDL-receptor-related protein 6 is a receptor for Dickkopf proteins. Nature. 2001;411:321–325. doi: 10.1038/35077108. [DOI] [PubMed] [Google Scholar]
- 30.Hoffman J, Kuhnert F, Davis CR, Kuo CJ. Wnts as essential growth factors for the adult small intestine and colon. Cell Cycle. 2004;3:554–557. [PubMed] [Google Scholar]
- 31.De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perminow G, Beisner J, Koslowski M, Lyckander LG, Stange E, et al. Defective Paneth Cell-Mediated Host Defense in Pediatric Ileal Crohn's Disease. Am J Gastroenterol. 2009;105:452–459. doi: 10.1038/ajg.2009.643. [DOI] [PubMed] [Google Scholar]
- 33.Scherr R, Essers J, Hakonarson H, Kugathasan S. Genetic determinants of pediatric inflammatory bowel disease: is age of onset genetically determined? Dig Dis. 2009;27:236–239. doi: 10.1159/000228555. [DOI] [PubMed] [Google Scholar]
- 34.Imielinski M, Baldassano RN, Griffiths A, Russell RK, Annese V, et al. Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sagiv-Friedgut K, Karban A, Weiss B, Shaoul R, Shamir R, et al. Early-onset Crohn disease is associated with male sex and a polymorphism in the IL-6 promoter. J Pediatr Gastroenterol Nutr. 2010;50:22–26. doi: 10.1097/MPG.0b013e3181b7a6a4. [DOI] [PubMed] [Google Scholar]
- 36.van Meurs JB, Trikalinos TA, Ralston SH, Balcells S, Brandi ML, et al. Large-scale analysis of association between LRP5 and LRP6 variants and osteoporosis. JAMA. 2008;299:1277–1290. doi: 10.1001/jama.299.11.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haines JL, Schnetz-Boutaud N, Schmidt S, Scott WK, Agarwal A, et al. Functional candidate genes in age-related macular degeneration: significant association with VEGF, VLDLR, and LRP6. Invest Ophthalmol Vis Sci. 2006;47:329–335. doi: 10.1167/iovs.05-0116. [DOI] [PubMed] [Google Scholar]
- 38.Sarzani R, Salvi F, Bordicchia M, Guerra F, Battistoni I, et al. Carotid artery atherosclerosis in hypertensive patients with a functional LDL receptor-related protein 6 gene variant. Nutr Metab Cardiovasc Dis. 2009 doi: 10.1016/j.numecd.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 39.Mani A, Radhakrishnan J, Wang H, Mani A, Mani MA, et al. LRP6 mutation in a family with early coronary disease and metabolic risk factors. Science. 2007;315:1278–1282. doi: 10.1126/science.1136370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hugot J-P, Chamaillard C, Zouali H, Lesage S, Cezard J-P, et al. Association of NOD2 leucine-rich repeat variants with susceptibility to Crohn's disease. Nature. 2001;411:599–603. doi: 10.1038/35079107. [DOI] [PubMed] [Google Scholar]
- 41.Hampe J, Franke A, Rosenstiel P, Till A, Teuber M, et al. A genome-wide association scan of nonsynonymous SNPs identifies a susceptibility variant for Crohn disease in ATG16L1. Nat Genet. 2007;39:207–211. doi: 10.1038/ng1954. [DOI] [PubMed] [Google Scholar]
- 42.Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaser A, Lee AH, Franke A, Glickman JN, Zeissig S, et al. XBP1 links ER stress to intestinal inflammation and confers genetic risk for human inflammatory bowel disease. Cell. 2008;134:743–756. doi: 10.1016/j.cell.2008.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Simms LA, Doecke JD, Roberts RL, Fowler EV, Zhao ZZ, et al. KCNN4 Gene Variant Is Associated With Ileal Crohn's Disease in the Australian and New Zealand Population. Am J Gastroenterol. 2010 doi: 10.1038/ajg.2010.161. [DOI] [PubMed] [Google Scholar]
- 45.Simms LA, Doecke JD, Walsh MD, Huang N, Fowler EV, et al. Reduced alpha-defensin expression is associated with inflammation and not NOD2 mutation status in ileal Crohn's disease. Gut. 2008;57:903–910. doi: 10.1136/gut.2007.142588. [DOI] [PubMed] [Google Scholar]
- 46.Bevins CL, Stange EF, Wehkamp J. Decreased Paneth cell defensin expression in ileal Crohn's disease is independent of inflammation, but linked to the NOD2 1007fs genotype. Gut. 2009;58:882–883. [PubMed] [Google Scholar]
- 47.Wang G, Stange EF, Wehkamp J. Host-microbe interaction: mechanisms of defensin deficiency in Crohn's disease. Expert Rev Anti Infect Ther. 2007;5:1049–1057. doi: 10.1586/14787210.5.6.1049. [DOI] [PubMed] [Google Scholar]
- 48.Andreu P, Colnot S, Godard C, Gad S, Chafey P, et al. Crypt-restricted proliferation and commitment to the Paneth cell lineage following Apc loss in the mouse intestine. Development. 2005;132:1443–1451. doi: 10.1242/dev.01700. [DOI] [PubMed] [Google Scholar]
- 49.Tamai K, Zeng X, Liu C, Zhang X, Harada Y, et al. A mechanism for Wnt coreceptor activation. Mol Cell. 2004;13:149–156. doi: 10.1016/s1097-2765(03)00484-2. [DOI] [PubMed] [Google Scholar]
- 50.Zeng X, Tamai K, Doble B, Li S, Huang H, et al. A dual-kinase mechanism for Wnt co-receptor phosphorylation and activation. Nature. 2005;438:873–877. doi: 10.1038/nature04185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Korinek V, Barker N, Morin PJ, van WD, de WR, et al. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science. 1997;275:1784–1787. doi: 10.1126/science.275.5307.1784. [DOI] [PubMed] [Google Scholar]
- 52.Gersemann M, Becker S, Kubler I, Koslowski M, Wang G, et al. Differences in goblet cell differentiation between Crohn's disease and ulcerative colitis. Differentiation. 2009;77:84–94. doi: 10.1016/j.diff.2008.09.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Overview of the work flow. The association study was carried out within 3 previously assembled IBD DNA cohorts in a retrospective way. DNA and biopsies from patients and controls for the mRNA study and immunohistochemistry were additionally collected at the Robert Bosch hospital in Stuttgart.
(TIF)
Genotyping Primers: Oligonucleotides used for multiplex PCR approaches are termed 1st- respectively 2nd-PCRP. Primers used for genotype specific elongation are termed PEXP.
(DOCX)
Real-time PCR Primers: Oligonucleotides which were used for the creation of standards and mRNA quantification.
(DOCX)