Abstract
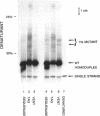
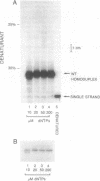
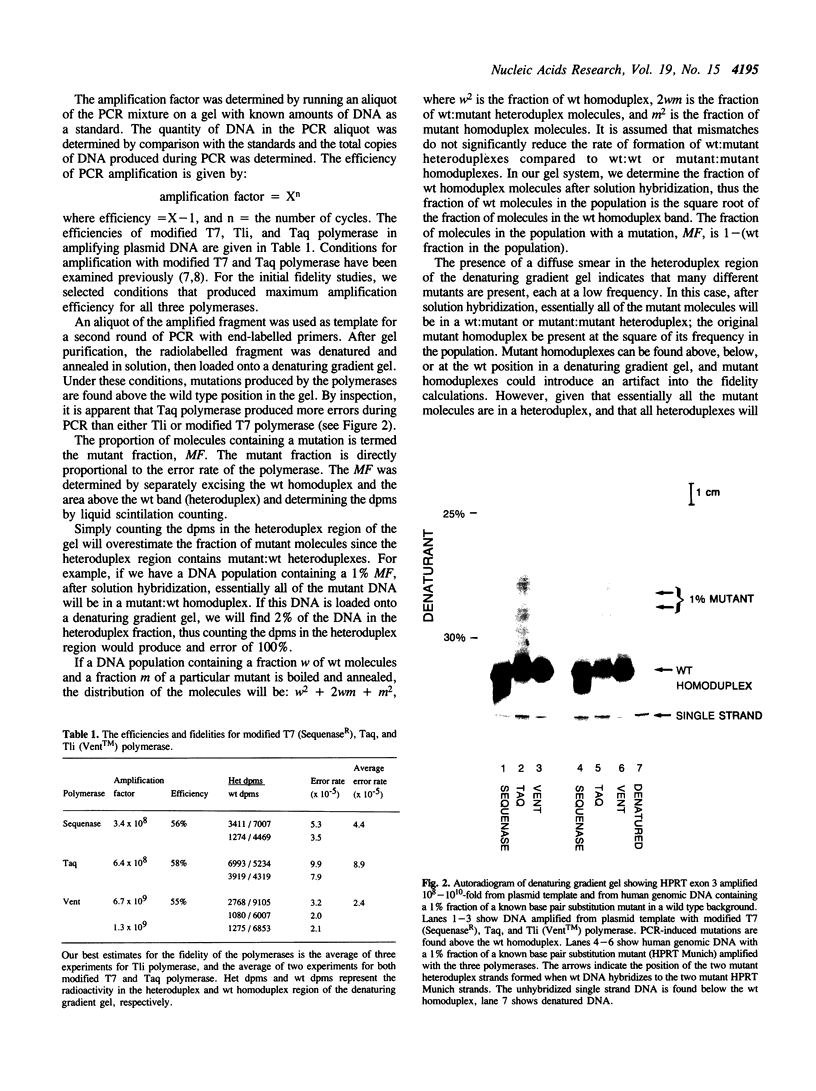
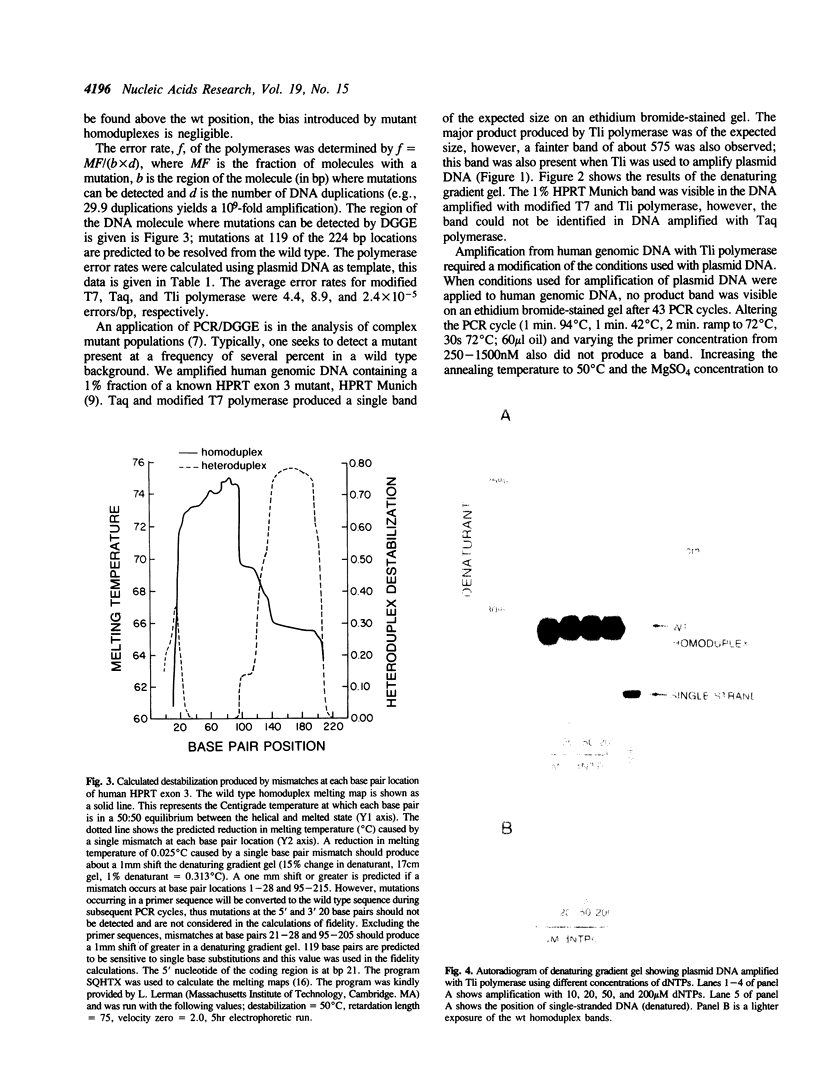
DNA synthesis fidelities of two thermostable DNA polymerases, Thermus aquaticus (Taq) and Thermococcus litoralis (Tli, also known as Vent), and a non-thermostable enzyme, a modified T7 DNA polymerase (Sequenase), were determined by analyzing polymerase chain reaction (PCR) products using denaturing gradient gel electrophoresis (DGGE). The error rates were 4.4, 8.9, and 2.4 x 10(-5) errors/bp for modified T7, Taq, and Tli polymerase, respectively. Reducing the nucleotide triphosphate concentration for Tli polymerase during PCR did not alter the fidelity. The ability of DGGE to detect a mutant present at several percent in a wild type population is related to the polymerase fidelity. To examine the sensitivity of mutant detection, human genomic DNA containing a 1% fraction of a known base pair substitution mutant was PCR-amplified with the three enzymes using primers that flank the mutant sequence. The PCR products were analyzed by DGGE. The signal from the mutant present at 1% was visible in the samples amplified with modified T7 and Tli polymerase, but the higher error rate of Taq polymerase did not permit visualization of the signal in DNA amplified with Taq polymerase.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Cariello N. F., Keohavong P., Kat A. G., Thilly W. G. Molecular analysis of complex human cell populations: mutational spectra of MNNG and ICR-191. Mutat Res. 1990 Aug;231(2):165–176. doi: 10.1016/0027-5107(90)90023-w. [DOI] [PubMed] [Google Scholar]
- Cariello N. F., Scott J. K., Kat A. G., Thilly W. G., Keohavong P. Resolution of a missense mutant in human genomic DNA by denaturing gradient gel electrophoresis and direct sequencing using in vitro DNA amplification: HPRT Munich. Am J Hum Genet. 1988 May;42(5):726–734. [PMC free article] [PubMed] [Google Scholar]
- Eckert K. A., Kunkel T. A. High fidelity DNA synthesis by the Thermus aquaticus DNA polymerase. Nucleic Acids Res. 1990 Jul 11;18(13):3739–3744. doi: 10.1093/nar/18.13.3739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ennis P. D., Zemmour J., Salter R. D., Parham P. Rapid cloning of HLA-A,B cDNA by using the polymerase chain reaction: frequency and nature of errors produced in amplification. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2833–2837. doi: 10.1073/pnas.87.7.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keohavong P., Thilly W. G. Fidelity of DNA polymerases in DNA amplification. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9253–9257. doi: 10.1073/pnas.86.23.9253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S., Silverstein K. Computational simulation of DNA melting and its application to denaturing gradient gel electrophoresis. Methods Enzymol. 1987;155:482–501. doi: 10.1016/0076-6879(87)55032-7. [DOI] [PubMed] [Google Scholar]
- Li H. H., Gyllensten U. B., Cui X. F., Saiki R. K., Erlich H. A., Arnheim N. Amplification and analysis of DNA sequences in single human sperm and diploid cells. Nature. 1988 Sep 29;335(6189):414–417. doi: 10.1038/335414a0. [DOI] [PubMed] [Google Scholar]
- Lindahl T., Nyberg B. Rate of depurination of native deoxyribonucleic acid. Biochemistry. 1972 Sep 12;11(19):3610–3618. doi: 10.1021/bi00769a018. [DOI] [PubMed] [Google Scholar]
- Loeb L. A. Apurinic sites as mutagenic intermediates. Cell. 1985 Mar;40(3):483–484. doi: 10.1016/0092-8674(85)90191-6. [DOI] [PubMed] [Google Scholar]
- Mullis K. B., Faloona F. A. Specific synthesis of DNA in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- Reiss J., Krawczak M., Schloesser M., Wagner M., Cooper D. N. The effect of replication errors on the mismatch analysis of PCR-amplified DNA. Nucleic Acids Res. 1990 Feb 25;18(4):973–978. doi: 10.1093/nar/18.4.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki R. K., Gelfand D. H., Stoffel S., Scharf S. J., Higuchi R., Horn G. T., Mullis K. B., Erlich H. A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988 Jan 29;239(4839):487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- Saiki R. K., Scharf S., Faloona F., Mullis K. B., Horn G. T., Erlich H. A., Arnheim N. Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science. 1985 Dec 20;230(4732):1350–1354. doi: 10.1126/science.2999980. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Proc Natl Acad Sci U S A. 1987 Jul;84(14):4767–4771. doi: 10.1073/pnas.84.14.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tindall K. R., Kunkel T. A. Fidelity of DNA synthesis by the Thermus aquaticus DNA polymerase. Biochemistry. 1988 Aug 9;27(16):6008–6013. doi: 10.1021/bi00416a027. [DOI] [PubMed] [Google Scholar]