Abstract
Background
The pine shoot beetle Tomicus yunnanensis (Coleoptera: Scolytinae) is an economically important pest of Pinus yunnanensis in southwestern China. Developed resistance to insecticides due to chemical pesticides being used for a long time is a factor involved in its serious damage, which poses a challenge for management. In addition, highly efficient adaptation to divergent environmental ecologies results in this pest posing great potential threat to pine forests. However, the molecular mechanisms remain unknown as only limited nucleotide sequence data for this species is available.
Methodology/Principal Findings
In this study, we applied next generation sequencing (Illumina sequencing) to sequence the adult transcriptome of T. yunnanensis. A total of 51,822,230 reads were obtained. They were assembled into 140,702 scaffolds, and 60,031 unigenes. The unigenes were further functionally annotated with gene descriptions, Gene Ontology (GO), Clusters of Orthologous Groups (COG), and Kyoto Encyclopedia of Genes and Genome (KEGG). In total, 80,932 unigenes were classified into GO, 13,599 unigenes were assigned to COG, and 33,875 unigenes were found in KO categories. A biochemical pathway database containing 219 predicted pathways was also created based on the annotations. In depth analysis of the data revealed a large number of genes related to insecticides resistance and heat shock protein genes associated with environmental stress.
Conclusions/Significance
The results facilitate the investigations of molecular resistance mechanisms to insecticides and environmental stress. This study lays the foundation for future functional genomics studies of important biological questions of this pest.
Introduction
The bark beetle genus Tomicus (Coleoptera: Curculionidae: Scolytinae) comprises seven described species [1]. Among them, T. piniperda and T. minor have a Palearctic distribution from Europe to Japan and China and repeatedly introduced to North America, T. destruens is found only around the Mediterranean Basin, while T. yunnanensis, T. brevipilosus, T. pilifer and T. puellus are restricted to southwestern, eastern, and central China [2]. Tomicus damages various pine species mainly due to its shoot feeding behavior and trunk stem attacking, which is among the most damaging pests of pine forests in the outbreak countries. Especially, some species of Tomicus have evolved towards different ecological strategies of host use, wider range distribution, and high levels of resistance to major classes of insecticides owing to biological and genetic factors. They are as exotic pests frequently invaded into new areas. Tomicus imposes a potentially serious damage upon pine forest.
Having a globally economical importance and ecological threat, it has been studied well from general biology and ecological viewpoints [3]–[5]. But it has remained poorly investigated at the molecular level. Only few studies have been developed, leading to elucidate their population genetic and species evolution [2], [6]. Up to September 26, 2011, there are only 719 nucleotide sequences deposited on National Center for Biotechnology Information (NCBI) for Tomicus. Due to insufficient genetic background, the molecular mechanisms of Tomicus in forest ecological systems are poorly understood.
The next generation sequencing (high-throughput deep sequencing) technology recently enables efficient approach on large-scale and genome-wide for gene discovery and expression profiling, and studies in functional, comparative and evolutionary genomics in non-model organisms with little or no previous genomic information exists [7], [8]. In the past few years, several studies based on this technology have, indeed, allowed the efficient, massive and successful molecular mechanisms investigation of some insect species lacking genome information, such as Bemisia tabaci, Nilaparvata lugens, and Trialeurodes vaporariorum [9]–[11].
T. yunnanensis is a newly described pine shoot beetle only found in Yunnan, Sichuan, and Guizhou Provinces in southwestern China, and has affected 200,000 ha. of Pinus yunnanensis forests over the past 30 years [12]. Control of this pest until now depended largely on chemical insecticides. It has evolved high level of resistance to insecticides. This might be due to the increasing of metabolic capability of detoxificative systems and/or reducing target site sensitivity. In addition, this pest has been found distributed in divergent environmental ecologies. The distributional areas of T. yunnanensis are with high mounts and deep valley as geographical barriers, and its ecological environments are in large difference. It suggested that T. yunnanensis adapted efficiently to environmental stress for surviving in diversifiable ecologies during the past years. Even resistance of T. yunnanensis to insecticides and severe environmental stress is an ongoing challenge for pest management, there is no information available for uncovering the molecular mechanisms under these.
In the present work, we characterized the first global transcriptome using Illumina technology of that Tomicus species T. yunnanensis. A systematic bioinformatics strategy was engaged to functional annotation of the transcriptome data. Additionally, important homologues of genes involved in insecticide resistance and heat shock protein (HSP) genes associated with environmental stress were identified. This study dramatically provides a foundation and increases the significant promise for further functional genomics studies of T. yunnanensis and other Tomicus species.
Materials and Methods
Ethics statement
Regarding the field study, no specific permits were required. The location is not privately-owned or protected in any way. The field studies did not involve endangered or protected species.
Insects
Adult beetles identified based on morphological characters [1] as T. yunnanensis by their trunk attacking phase on P. yunnanensis were collected from Qujing city, Yunnan province, China. The samples were frozen at −80°C until use.
cDNA library and Illumina sequencing
Total RNA was extracted from each 20 female and male adult beetles using TRIzol® Reagent (Invitrogen) according to the manufacturer's instructions. RNA quality and yield were assessed by 2100 Bioanalyzer (Agilent Technologies) with a minimum RNA integrated number value of 8. The cDNA library was prepared using Illumina's kit following manufacturer's recommendations. Briefly, messenger RNA was isolated from 20 µg total RNA (pooled RNA of female and male adults) using oligo(dT) magnetic beads and fragmented into short sequences using divalent cations under elevated temperature. First and second strand cDNA were synthesized from cleaved RNA fragments. After the end repair and ligation of adaptors, the products were cleaned up with a QIAquick PCR Purification Kit to create the final cDNA library. The library was sequenced on the Illumina sequencing platform (GAII) to obtain short sequences from both ends.
Bioinformatic analysis
The raw reads from the images and quality value calculation were performed by the Illumina data processing pipeline (version 1.6). Before the assembly, the raw reads were cleaned by adaptor sequences, empty reads and low quality sequences (reads with unknown sequences ‘N’) to obtain the high-quality clean reads. Raw reads were then assembled into sequence contigs, scaffolds, and unigenes using SOAPdenovo software [13], and clustered using TGI Clustering tools [14]. All Illumina assembled unigenes were searched against nr database in NCBI, Swiss-Prot, Kyoto Encyclopedia of Genes and Genome (KEGG), and Cluster of Orthologous Groups (COG) with the BLASTX algorithm. The E-value cut-off was set at 10−5. Genes were tentatively identified according to the best hits against known sequences. Blast2GO [15] was used to predict the functions of the sequences, assign Gene Ontology (GO) terms, and predict the metabolic pathways in COG and KEGG databases. Amino acid sequence comparisons were conducted with Clustal X (v1.83) program [16]. Phylogenetic tree was constructed by MEGA 5.0 package [17] using the neighbour-joining method with the Poisson correction model after sequence alignment performed by Clustalx. Bootstrap analysis of 1,000 replication trees was performed in order to evaluate the branch strength of each tree.
Data availability
The Illumina reads of T. yunnanensis have been submitted to NCBI Short Read Archive under the accession number of SRA047283.
Results and Discussion
Sequencing and de novo assembly
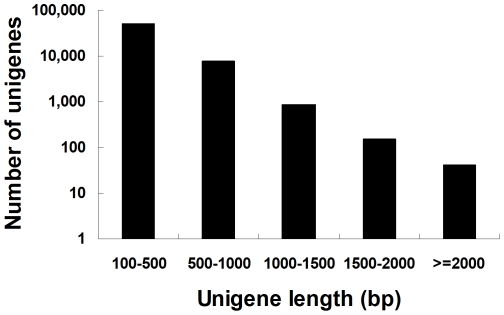
Illumina sequencing resulted in 51,822,230 raw reads, corresponding to an accumulated length of 4,664,000,700 bp (Table 1). The average raw reads length is 90 bp, which is consistent with the Illumina sequencing capacity. Using SOAPdenovo software, the raw reads were assembled into contigs after adaptor sequences, empty reads and low quality sequences filtered out. The raw reads were assembled into 642,521 contigs with a mean length of 117 bp. The range length of contigs is from 50 to 2953 bp. Although the majority of the contigs are between 50 to 400 bp, we obtained 9,331 contigs which were greater than 400 bp in length. The size distribution of these contigs is shown in Figure S1. Using paired end-joining and gap-filling, these contigs were further assembled into 140,702 scaffolds with a mean length of 233 bp, and range length of 100–2953 bp. 2,293 scaffolds were longer than 800 bp (Figure S2). Using TGI software, scaffold sequences were assembled into clusters. We obtained 60,031 unigenes with a mean length of 355 bp. The lengths of the 8,953 and 1,055 unigenes were ≥500 bp and 1000 bp, respectively, revealing that 85% of them fell between 100 and 500 bp in length (Figure 1). The result demonstrated that the scaffold and unigene length distribution followed the contig length distribution closely, with the majority being shorter sequences with relatively little redundancy, which was in a similar to other insect transcriptome projects using this technology [9]–[11], [18], [19]. The majority of scaffolds and unigenes after assembly were still less than 500 bp, which might be due to the short length sequencing capacity of Illumina sequencing and/or the low coverage of the transcriptome represented in this dataset [20]. The assembled abundant sequence data provided a rich source of information for further investigation, thus allowing for rapid characterization of a large portion of the transcriptome and better reference of interesting genes.
Table 1. Sequence statistics of the Illumina sequencing assembly.
| Reads | Contigs | scaffolds | Unigenes | |
| Number of sequences | 51,822,230 | 642,521 | 140,702 | 60,031 |
| Mean length (bp) | 90 | 117 | 233 | 355 |
| Total length (bp) | 4,664,000,700 | 74,951,788 | 32,749,102 | 21,338,135 |
Figure 1. Length distribution of unigenes.
The number of y-axis has been transfer into logarithmic scale.
Annotation of predicted proteins
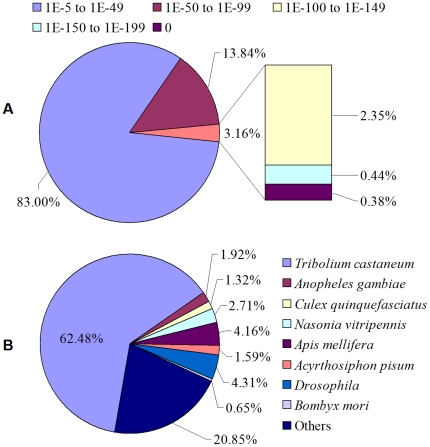
The assembled unigenes were used as a query for Blastx searches in the NCBI nr protein database with a cut-off E-value of 10−5. The search produced 34,702 hits, which comprised 57.81% of all the unigenes (Table S1). A large proportion of them (about 40%) apparently have no significant match in any of the existing databases, indicating many of them may contain novel sequences and a high number of Coleoptera or species-specific transcripts or transcript parts (orphan UTRs). This is expected, as there is very little sequence information from closely related species. The E-value distribution of the top hits in the nr database showed that 17% of the mapped sequences have strong homology (smaller than 1.0E−49), whereas 83% of the homolog sequences ranged between 1.0E−5 to 1.0E−49 (Figure 2A). The species distribution of the best match result for each sequence showed that the T. yunnanensis sequences have 62.48% matches with sequences from the Coleoptera species (Tribolium castaneum), while very low proportion (<5%) of them have matches to other insects (Figure 2B). It demonstrated that T. yunnanensis have near evolution distance with T. castaneum.
Figure 2. Characteristics of homology search of Illumina sequences against the nr database.
(A) E-value distribution of BLAST hits for each unique sequence with a cut-off E-value of 1.0E-5. (B) Species distribution of the BLASTX results. We used the first hit of each sequence for analysis.
GO assignments
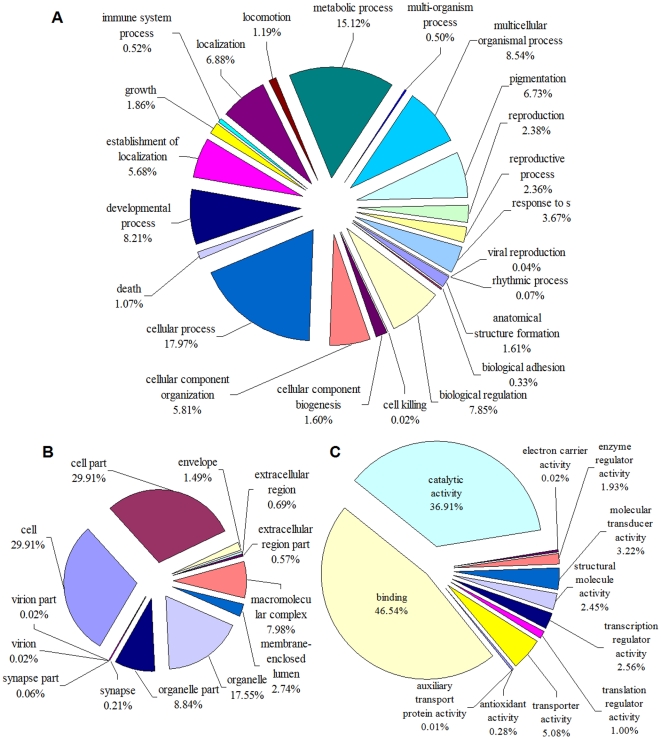
Gene ontology is widely used to standardize representation of genes across species and provide a controlled vocabulary of terms for describing gene products [21]. In total, 80,932 unigenes were assigned for GO terms based on BlastX matches with sequences whose function is previously known (Figure 3, Table S2). These GO terms were summarized into the 3 main GO categories (biological process, cellular component, and molecular function) according to the standard GO terms and 47 sub-categories. Compared to the GO annotations of Drosophila melanogaster genome [22], our sequence data do not contain any notable biases towards particular categories of genes. Biological process made up the majority of the GO annotations (38,578, 47.67% of the total), followed by cellular component (26,119, 32.27%), and molecular function (16,235, 20.06%). Among biological process category, cellular process (17.97%), and metabolic process (15.12%) were the most dominant subcategories, reflecting that the analyzed tissues were undergoing rapid growth and extensive metabolic activities. The following subcategories were multicellular organismal process (8.54%), developmental process (8.21%), biological regulation (7.85%), localization (6.88%), and cellular component organization (5.81%). The biological process illustrated all of the major cellular processes from transport and cellular organization to transcription, translation, and metabolism. Under the category of cellular component, cell (29.91%), cell part (29.91%), and organelle (17.75%) were among the most highly represented subcategories. Most of the unigenes annotated with a cellular component are localized to plastids or mitochondria. The molecular function category was mainly comprised of proteins involved in binding (46.54%), predominantly heat shock proteins (Hsp), and catalytic activities (36.91%) including hydrolases, kinases, and transferases, allowing for the identification of genes involved in the secondary metabolite synthesis pathways. Similar observations for metabolic processes were reported in transcriptomic studies of other insects [18]. These GO annotations represent a general gene expression profile signature for T. yunnanensis adults, which demonstrates that the expressed genes in this species encode diverse structural, regulatory and stress proteins.
Figure 3. Distribution of second level GO of Tomicus yunnanensis transcriptome.
(A) biological process, (B) cellular component and (C) molecular function. The percentage of a specific category of genes in that main category is shown.
COG classification
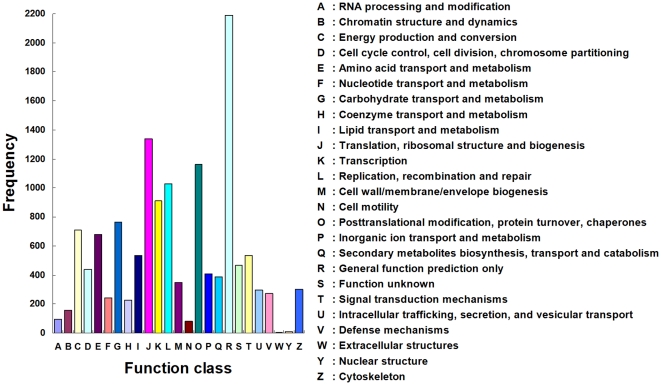
A total of 13,599 unigenes were assigned to the appropriate COG clusters (Figure 4). These COG classifications were grouped into 25 function categories that correspond to the categories observed in GO analysis. The cluster for general function prediction only (16.10%) represents the largest group. The other three largest categories include: (1) translation, ribosomal structure and biogenesis (9.85%), (2) posttranslational modification, protein turnover, chaperones (8.54%), and (3) replication, recombination and repair (7.54%). The category of secondary metabolites biosynthesis, transport and catabolism was highlighted with 2.86%, because of the importance of secondary metabolites to the insecticides in insects. The most abundant sequences in this category are cytochrome P450 monooxygenases. To some extent, the COG classifications shed light on specific responses and functions involved in the molecular processes of T. yunnanensis.
Figure 4. COG function classification of the Tomicus yunnanensis transcriptome.
All putative proteins were aligned to the COG database and can be classified functionally into at least 25 molecular families.
KEGG analysis
To identify the biological pathways that are active in T. yunnanensis, we mapped the 34,702 annotated sequences to the referential canonical pathways in KEGG. A total of 16,727 unigenes were assigned to 219 KEGG pathways. All the pathways are summarized in Table S3. The top 10 pathways are metabolic pathways (3339 members), spliceosome (665 members), huntington's disease (663 members), pathways in cancer (586 members), lysosome (527 members), purine metabolism (509), regulation of actin cytoskeleton (472 members), alzheimer's disease (471 members), ubiquitin mediated proteolysis (467 members), and focal adhesion (466 members). These annotations provide a valuable resource for investigating specific processes, functions and pathways in T. yunnanensis research.
Putative insecticides resistance related genes
Cytochrome P450 (P450)
Because of the genetic diversity, broad substrate specificity, and catalytic versatility, P450s can mediate resistance to all classes of insecticides [23]. Approximately a total of 146 P450 related unigenes were identified in the transcriptome. Even 23 T. yunnanensis P450 sequences were identified in the dataset as length longer than 600 bp (Table 2), and the majority of them were as short fragments listed in Table S4. In general, insect genomes harbor ∼100 different P450s, which can be divided into four clades (CYP2, CYP3, CYP4 and mito) [24], [25]. The number of unigenes in the T. yunnanensis transcriptome belongs to the range of P450s identified in other insect species, while the accurate gene number still remains to be identified by gene cloning based on the fragments obtained here. Of the 23 unigenes which contained longer length, it was possible to differentiate them into four clades by phylogenetic analysis. The majority of these P450s belonged to the CYP3 and CYP4 clades compared with CYP2 and mito clades, which is in agreement with Tribolium castaneum and other insect systems [25], [26]. Because some functionality of P450s can be assigned to each of the four clades based on known functionalities characterized in other insects, we can select some interesting candidates for a further investigation according to the phylogenetic results. Until recently, the increased production of P450s in resistant insects has shown to occur almost exclusively through up regulation via changes in cis-or trans-acting regulatory loci, but gene duplication or amplification of P450s has now been implicated in the resistance of four insect species [27]. From phylogeny, gene duplication events specific to T. yunnanensis are apparent, with the best example being the two CYP3-type sequences Unigene58724 and Unigene58733, and two CYP4-type sequences Unigene59997 and Unigene8705, if they were paired in the phylogeny with bootstrap support greater than 70%. However, the relevance of duplication resistance of T. yunnanensis requires a further investigation.
Table 2. Putatively identified P450 genes in Tomicus yunnanensis.
| Unigene ID | Putative assignment | Length (bp) | First hit | Identity (%) | E_value | Blast annotation/Organism |
| Unigene12883 | CYP3 | 612 | XP_975568 | 44 | 2.00E-28 | PREDICTED: similar to cytochrome P450 [Tribolium castaneum] |
| Unigene15066 | CYP4 | 819 | EFA01330 | 58 | 9.00E-59 | cytochrome P450-like protein [Tribolium castaneum] |
| Unigene1549 | CYP4 | 679 | AAT38513 | 42 | 8.00E-43 | ubiquitous cytochrome P450 [Phyllopertha diversa] |
| Unigene1697 | CYP3 | 902 | EFA04564 | 39 | 5.00E-56 | cytochrome P450 347A1 [Tribolium castaneum] |
| Unigene21111 | CYP3 | 771 | XP_972794 | 56 | 9.00E-34 | PREDICTED: similar to Probable cytochrome P450 9f2 (CYPIXF2) [Tribolium castaneum] |
| Unigene22371 | CYP4 | 694 | XP_001814854 | 45 | 1.00E-45 | PREDICTED: similar to cytochrome P450 [Tribolium castaneum] |
| Unigene23650 | mito | 645 | XP_974252 | 60 | 7.00E-65 | PREDICTED: similar to CYP302a1 [Tribolium castaneum] |
| Unigene23993 | CYP2 | 1092 | NP_503303 | 38 | 2.00E-57 | CYtochrome P450 family member (cyp-33C11) [Caenorhabditis elegans] |
| Unigene24457 | CYP3 | 1192 | EEZ99338 | 62 | 2.00E-63 | cytochrome P450 6BQ13 [Tribolium castaneum] |
| Unigene24875 | CYP2 | 1173 | XP_968477 | 53 | 1.00E-98 | PREDICTED: similar to cytochrome P450, partial [Tribolium castaneum] |
| Unigene551 | mito | 640 | NP_001123894 | 57 | 2.00E-54 | cytochrome P450 CYP314A1 [Tribolium castaneum] |
| Unigene58533 | CYP4 | 603 | ABF06546 | 67 | 2.00E-44 | CYP4BD1 [Ips paraconfusus] |
| Unigene58724 | CYP3 | 627 | NP_001164248 | 50 | 5.00E-50 | cytochrome P450 9Z4 [Tribolium castaneum] |
| Unigene58733 | CYP3 | 628 | NP_001164248 | 47 | 4.00E-51 | cytochrome P450 9Z4 [Tribolium castaneum] |
| Unigene59248 | CYP4 | 720 | ABF06544 | 46 | 5.00E-59 | CYP4AY1 [Ips paraconfusus] |
| Unigene59398 | CYP4 | 760 | XP_973153 | 45 | 1.00E-44 | PREDICTED: similar to cytochrome P450 [Tribolium castaneum] |
| Unigene59490 | CYP4 | 788 | XP_001602395 | 36 | 7.00E-41 | PREDICTED: similar to cytochrome P450 [Nasonia vitripennis] |
| Unigene59572 | CYP3 | 818 | EFA02819 | 45 | 6.00E-67 | cytochrome P450 6BQ5 [Tribolium castaneum] |
| Unigene59684 | CYP3 | 871 | NP_496108 | 42 | 4.00E-47 | CYtochrome P450 family member (cyp-13A1) [Caenorhabditis elegans] |
| Unigene59753 | CYP2 | 916 | XP_969587 | 69 | 1.00E-115 | cytochrome P450 307A1 [Tribolium castaneum] |
| Unigene59997 | CYP4 | 1443 | EEZ99364 | 45 | 1.00E-100 | cytochrome P450-like protein [Tribolium castaneum] |
| Unigene8212 | CYP4 | 650 | ABF06550 | 60 | 6.00E-71 | CYP4BG1 [Ips paraconfusus] |
| Unigene8705 | CYP4 | 686 | EEZ99364 | 32 | 4.00E-32 | cytochrome P450-like protein [Tribolium castaneum] |
Fragments less than 600 bp listed in Table S4.
Glutathione S-transferase (GST)
GSTs catalyse the glutathione conjugation reaction with reduced glutathione (GSH) to convert them, resulting in less toxic water-soluble products that can eventually be excreted, which are widespread in both prokaryotes and eukaryotes [28]. The increased expression and activity of GSTs, and amplification of the structural GST genes has been documented as a mechanism of insect resistance [27]. Seventeen putative GST unigenes were identified in T. yunnanensis transcriptome (Table 3). Based on the closest BLAST hits in the NCBI nr database and, when possible, through applying a phylogenetic analysis 14, 2, and 1 unnigenes were assigned to the Epsilon, Sigma, and Delta classes, respectively. In insects, there are two ubiquitously distributed distantly related groups of GSTs, classified according to their location within the cell: microsomal and cytosolic [29]. The microsomal class contains few gene duplicates, while the cytosolic class contains highly diverse larger gene family divided into six major subclasses: Delta, Epsilon, Sigma, Omega, Theta, and Zeta [30]. In regard to microsomal class, it may await discovery due to its absence from the current transcriptomic dataset, but this group has not been implicated in the metabolism of insecticides [29]. In microsomal class, only three subclasses have been identified from the current database, which is similar to some insect species that do not have six subclasses like Drosophila melanogaster [31]. For instance, only Sigma, Delta, and Theta type GSTs were found in Nasonia vitripennis, and Epsilon, Sigma, and Omega type GSTs were found in T. castaneum based on genomic analysis [30]. Although the Delta subclasse is absent in T. castaneum, one unnigene assigned to Delta subclasse was identified in T. yunnanensis. In addition, the majority of T. yunnanensis GSTs were assigned to Epsilon subclasse, which is in accordance with studied insect GSTs with extensive cases of gene expansions in Epsilon and Delta types [32].
Table 3. Putatively identified GST genes in Tomicus yunnanensis.
| Unigene ID | Putative assignment | Length (bp) | First hit | Identity (%) | E_value | Blast annotation/Organism |
| Unigene22819 | Epsilon | 521 | XP_971136 | 49 | 2.00E-25 | PREDICTED: similar to Glutathione S transferase E8 CG17533-PA [Tribolium castaneum] |
| Unigene23097 | Epsilon | 542 | ACU09495 | 41 | 9.00E-23 | glutathione S-transferase 16 [Helicoverpa armigera] |
| Unigene25020 | Sigma | 693 | XP_002633726 | 35 | 2.00E-16 | C. briggsae CBR-GST-3 protein [Caenorhabditis briggsae] |
| Unigene25058 | Epsilon | 715 | XP_966966 | 41 | 1.00E-37 | PREDICTED: similar to Glutathione S transferase E6 CG17530-PA [Tribolium castaneum] |
| Unigene28476 | Epsilon | 209 | XP_971389 | 65 | 4.00E-15 | PREDICTED: similar to Glutathione S transferase E5 CG17527-PA [Tribolium castaneum] |
| Unigene31256 | Epsilon | 218 | XP_966787 | 42 | 7.00E-10 | PREDICTED: similar to Glutathione S transferase E7 CG17531-PA [Tribolium castaneum] |
| Unigene34958 | Epsilon | 230 | XP_971268 | 37 | 2.00E-07 | PREDICTED: similar to Glutathione S transferase E7 CG17531-PA [Tribolium castaneum] |
| Unigene35169 | Epsilon | 231 | XP_971449 | 54 | 2.00E-11 | PREDICTED: similar to Glutathione S transferase E7 CG17531-PA [Tribolium castaneum] |
| Unigene37693 | Sigma | 241 | NP_001165920 | 52 | 6.00E-14 | glutathione S-transferase S3 [Nasonia vitripennis] |
| Unigene40017 | Epsilon | 252 | NP_611325 | 56 | 3.00E-19 | glutathione S transferase E3 [Drosophila melanogaster] |
| Unigene44809 | Epsilon | 279 | ADD19697 | 62 | 2.00E-27 | glutathione S-transferase [Glossina morsitans morsitans] |
| Unigene44817 | Epsilon | 279 | ADD19697 | 61 | 1.00E-27 | glutathione S-transferase [Glossina morsitans morsitans] |
| Unigene49693 | Epsilon | 323 | XP_967234 | 43 | 3.00E-19 | PREDICTED: similar to glutathione S-transferase 6A [Tribolium castaneum] |
| Unigene50873 | Delta | 339 | XP_974204 | 76 | 4.00E-45 | PREDICTED: similar to GST [Tribolium castaneum] |
| Unigene54951 | Epsilon | 415 | BAE80117 | 45 | 4.00E-26 | glutathione S-transferase [Plutella xylostella] |
| Unigene5052 | Epsilon | 443 | XP_971136 | 45 | 2.00E-31 | PREDICTED: similar to Glutathione S transferase E8 CG17533-PA [Tribolium castaneum] |
| Unigene8150 | Epsilon | 416 | NP_611328 | 44 | 2.00E-14 | glutathione S transferase E6 [Drosophila melanogaster] |
Other candidates
In addition to the detailed analyses above, further unigenes were identified with a high sequence similarity to important genes related to insecticide metabolism and targets. As shown in Table 4, a number of unigenes annotated as enzymes related to insecticide metabolic resistance, such as carboxylesterase, and superoxide dismutase; and insecticide targets, such as acetyl-CoA carboxylase, acetylcholinesterase, and γ-aminobutyric acid (GABA) receptor, were present in T. yunnanensis transcriptome. Although most of these unigenes are not full length, they will nevertheless facilitate a further characterisation of these targets by RACE to retrieve the full length cDNAs. The abundance of these transcripts demonstrates the quality of our sequencing data. It provided new leads for functional studies of dissecting the potential insecticide resistance role each these genes plays.
Table 4. Putative genes of interest related to insecticide resistance.
| Gene name | Uumber of unigenes had a hit with nr database |
| Cytochrome P450 | 146 |
| Glutathione S-transferase | 17 |
| Carboxylesterase | 128 |
| Superoxide dismutase | 20 |
| Acetyl-CoA carboxylase | 25 |
| Acetylcholinesterase | 6 |
| γ-aminobutyric acid (GABA) receptor | 8 |
| Nicotinic acetylcholine receptor | 17 |
| Sodium channel | 5 |
| Chloride channel | 63 |
| Ryanodine receptor | 36 |
Detection of HSP genes
Heat shock protein 10 (HSP10)
HSPs are highly conserved molecules that play vital roles in all cells. Among them, HSP10 is a near 10 kDa, highly conserved protein. In eukaryotes, HSP10, originally identified as a mitochondrial chaperone, now is also known to be present in other places such as cytosol, cell surface, and extracellular space [33]. Here, six unigenes were identified putatively encoded HSP10 (Table S5). Two of them (Unigene4430 and Unigene52967) appeared to be complete. Amino acid alignment revealed a low homology of these four HSP10s to HSP10s from the Acyrthosiphon pisum (46%), Glossina morsitans morsitans (49%), Nasonia vitripennis (51%), and Tribolium castaneum (41%) (Figure 5). HSP10 is known as a co-chaperone for heat shock protein 60 (HSP60), and exerts immunosuppressive activity in mammals [34]. To our best knowledge, HSP10 in insects has not been structurally and functionally studied in detail.
Figure 5. Alignment of the predicted amino acid sequences of Tomicus yunnanensis HSP10s with that of other insects.
Identical residues are shaded black, conserved substitutions are shaded grey. Dash (–) indicates insertion or deletion. Api, Acyrthosiphon pisum (NP_001119666); Gmo, Glossina morsitans morsitans (ADD19718); Nvi, Nasonia vitripennis (XP_001599992); Tca, Tribolium castaneum (XP_969732).
Small heat shock protein (sHSP)
sHSP are a family of molecular chaperones, with molecular mass ranging from 12 kDa to 43 kDa, usually below 30 kDa, which is involved in cellular defense under environmental stress conditions [35], [36]. Up to data, knowledgment of insect sHsps is much less than of sHsps in plants and vertebrates. Twenty three unigenes were found to be with similarities to sHSP (Table S5). Among of them, five (Unigene24062, Unigene56764, and Unigene56997, Unigene58448 and Unigene59017) appeared to be complete. These five sHSPs showed variable sequence identity, ranged from 29% to 82%, to the first hits under blast search, suggesting that sHSPs are diverse. An alignment of the predicted proteins deduced from the complete unigenes is shown in Figure 6. Interestingly, the alignment indicated that they were with very low similarity between each other, revealing that they do not display a highly conservation. Previous studies have demonstrated that sHSPs are abundant and ubiquitous in almost all organisms with different numbers, from bacteria to algae single celled and even to the higher organisms including human [37]. Representative sequences of the many demonstrated sHSPs available share a conserved sequence of approximately 90 amino acid residues, termed α-crystallin domain responsible for dimer formation and form large multimeric complexes that are known to be crucial to their chaperone activities [38]–[40]. There is α-crystalling domain in the resided C-terminus of T. yunnanensis predicted sHSPs. The N-terminal coding sequences and N-terminal ends of sHSPs are more variable than C-terminal with the α-crystalling domain in different species [41]. As expected, the deduced amino acid sequence of T. yunnanensis HSP20s showed high conservation in α-crystalling domain, and high divergence in N-terminal region and variable N-terminal end. However, there are still some conserved amino acid residues in N-terminal.
Figure 6. Alignment of the predicted amino acid sequences of Tomicus yunnanensis sHSPs. The conserved α-crystallin domain is underlined.
Identical residues are shaded black, conserved substitutions are shaded grey. Dash (–) indicates insertion or deletion.
Heat shock protein 40 (HSP40)
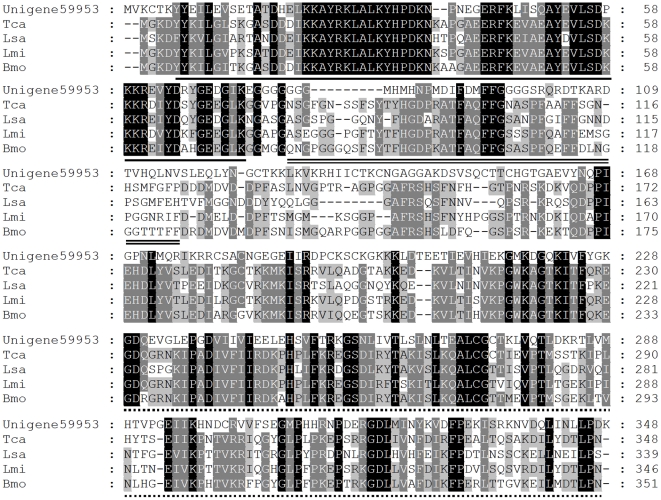
HSP40 family is one of the HSPs containing the DNAJ homologous region of Escherichia coli, J-domain of DNAJ [42]. HSP40 performs an essential molecular chaperone function in protein translation, folding, unfolding, translocation and degradation, in protein translocation across membranes, and in protecting cells from the effects of heat and other stress factors, primarily acting as cofactors or regulators of heat shock protein 70 (HSP70) [43]. Thirty seven unigenes with similarity to HSP40 were present in T. yunnanensis transcriptome. Among them, Unigene59953 is with full open reading frame. The amino acid identity is about 30% compared with those of T. castaneum, Liriomyza sativae, Locusta migratoria, and Bombyx mori (Figure 7). Typically, HSP40 proteins have three distinct domains: (1) the J domain of 70 amino acid residues in length which constitutes the most conserved region of these proteins and interacts with HSP70 and stimulates its ATPase activity; (2) a glycine and phenylalanine-rich region (G/F domain), postulated to act as a flexible hinge needed to activate the substrate binding properties of Hsp70 when it interacts with Hsp40; and (3) a cysteine-rich region (C domain) resembling a zinc-finger like structure, suggested to mediate dimer formation and molecular chaperone–peptide interactions [44]–[47]. These three domains were found in the predicted amino acid sequence of T. yunnanensis HSP40. However, not all HSP40 necessarily contain all of these 3 domains. Based on the differences in these regions, HSP40 proteins can be categorized into three groups: Type I homologs have all 3 domains (J, G/F, and C), Type II have the J and G/F but not the C domain, and Type III have the J domain alone [48], [49]. According to this, T. Yunnanensis HSP40 obtained here belongs to Type I.
Figure 7. Alignment of the Tomicus yunnanensis HSP40 amino acid sequence with other insects HSP40 amino acid sequences.
The conserved J domain is underlined. The Gly/Phe-rich domain is double underlined. The cysteine-rich domain is indicated as dotted line. Identical residues are shaded black, conserved substitutions are shaded grey. Dash (–) indicates insertion or deletion. Tca, Tribolium castaneum (EFA11191); Lsa, Liriomyza sativae (ABE57132); Lmi, Locusta migratoria (ABC84495); Bmo, Bombyx mori (BAD90846).
Heat shock protein 60 (HSP60)
HSP60, a major group of the heat shock proteins, includes stress inducible and constitutively expressed members. It is believed to be predominately mitochondrial, although some are also reported in cytosol and in extracellular compartments [50]. Eighteen unigenes coding for putative HSP60 were identified in the database (Table S5). Compared with orthologues from other organisms, the percentage identity of the deduced amino acid sequence of these unnigenes is high, between 45–93%, confirming the remarkable conservation of this family. Among them, only Unigene59944 was with the length above 1 kb, the others were shorter than 600 bp. Alignment of the predicted amino acid sequence of Unigene59944 with that of other insects revealed it contained the signature peptide known as conserved ATP-binding motif [51] (Figure 8). The classical mitochondrial HSP60 signature motif was found in the deduced amino acid sequence [52], suggesting the HSP60 coded by Unigene59944 presented here is a member of the mitochondrial HSP60 chaperone family. As Unigene59944 was 3′ and 5′-truncted, a fragment of about 30 amino acids at the N terminus required for import into the mitochondriaa and a typical GGM repeat motif for HSP60 at the C terminus were not observed.
Figure 8. Alignment of the partial amino acid sequence of Tomicus yunnanensis HSP60 with that of other insects.
(A) the ATP-binding motif. (B) The classical mt-HSP60 signature (underlined) motif. Identical amino acids are shaded black, and conserved residues are shaded grey. Tca, Tribolium castaneum (XP_971630), Ame, Apis mellifera (XP_392899), Csu, Chilo suppressalis (ACT52824); Lsa, Liriomyza sativae (AAW49251).
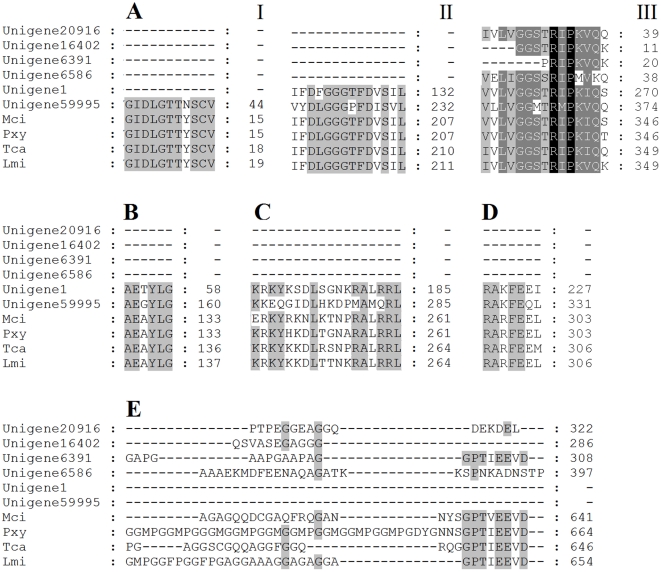
Heat shock protein 70 (HSP70)
In the HSP family, the most studied are HSP70s. There are two main forms of these 70 kD proteins, the heat shock cognate (HSC70) which is expressed constitutively and an inducible form (HSP70) which is normally expressed in response to external stimuli [53]. Seventeen four unigenes produced the best sequence matches to HSP70 (Table S5). The majority of them showed highly conserved identity above 70% to the classic inducible form of HSP70 from other organisms. All these sequences were not in completeness, and only six sequences (Unigene1, Unigene16402, Unigene20916, Unigene59995, Unigene6391, and Unigene6586) were in length longer than 1 kb. After alignment of these 6 non-redundant sequences to that of other insects (Figure 9), three signature motifs of the HSP70 family [54] were conserved. Putative ATP-GTP binding site [55] and nonorganellar consensus motif were located at the amino acid sequences. Putative bipartite nuclear localization signals, which are needed for the selective translocation of HSC70 into the nucleus [56] were identified. The terminal sequence motif of the HSP70 family identifies their cellular location, with the EEDV sequence motif attesting to its cytoplasmic localisation, whereas K/HDEL and PEAEYEEAKK characterise members localised to the endoplasmic reticulum and mitochondria respectively [53], [57]. With regard to this, the terminal sequence motif of Unigene20916 and Unigene6391 were K/HDEL and EEDV, indicating they belonged to cytoplasmic localization and endoplasmic reticulum member, respectively.
Figure 9. Alignment of the partial amino acid sequence of Tomicus yunnanensis HSP70s with that from other insects.
(A) Three highly conserved HSP70 family signatures labeled I, II, III. (B) The ATP-GTP binding site. (C) Putative bipartite nuclear localization signals. (D) The nonorganellar consensus motif. (E) The C-terminus. Identical amino acids are shaded black, and conserved residues are shaded grey. Dash (–) indicates insertion or deletion. Mci, Macrocentrus cingulum (ACD84944); Pxy, Plutella xylostella (BAF95560); Tca, Tribolium castaneum (XP_974442); Lmi, Locusta migratoria (AAO21473).
Heat shock protein 90 (HSP90)
In normal physiological conditions, HSP90 is abundant, accounting for 1% of the total soluble cytosolic protein, which is a highly conserved molecular chaperone contributing to the folding, maintenance of structural integrity and proper regulation of a subset of cytosolic protein [54], [58], [59]. Like HSP70, HSP90s has also been extensively studied. Twenty four unigenes with short length less than 800 bp were identified coding for putative HSP90 (Table S5). Except for Unigene20269 and Unigene46492, they showed above 60% amino acid identity to HSP90s from other organisms. Other sequence information was not analyzed here because of only relatively short fragments available.
Conclusions
In this work, transcriptome database has been produced on a large scale for T. yunnanensis using Illumina sequencing. The results provided new insights into the genomics of this forest pest. It contributed significantly to the rapid discovery of a wide diversity candidate gene for this organism that lacks complete genome sequences and other genetic tools and resources. This transcriptome can be used as a reference to provide new leads for comparative studies within the family. Based on this database, the predicted repertoire of genes related to insecticide resistance and environmental stress are constructed, providing valuable information regarding further investigations of the detailed mechanisms. However, as the sequences obtained by Illumina sequencing are short, most of the interesting putative genes discovery by this comprehensive tool is need to rely on RACE PCR in order to obtain full-length sequence data. Furthermore, the database will continue to be an enormous resource for genome-wide association studies of the whole picture of other important biological questions in the future.
Supporting Information
Length distribution of contigs.
(TIF)
Length distribution of scaffolds.
(TIF)
Top BLAST hits from NCBI nr database. BLAST results against the NCBI nr database for all the distinct sequences with a cut-off E value above 10−5 are shown.
(XLS)
Gene Ontology of Tomicus yunnanensis unigenes.
(XLS)
KEGG summary of Tomicus yunnanensis transciptome.
(XLS)
Putative P450 genes identified in Tomicus yunnanensis transcriptome.
(XLS)
Putative heat shock protein genes identified in Tomicus yunnanensis transcriptome.
(XLS)
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the Special Fund for Forest Scientific Research in the Public Interest of China (201004067), Science Foundation for the Excellent Youth Academic and Technological Scholars of Yunnan Province of China (2007YP01-18), Science Foundation of Southwest Forestry University (111128), and the Key Subject of Forest Protection of Yunnan Province of China (XKZ200905). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kirkendall LR, Faccoli M, Ye H. Description of the Yunnan shoot borer, Tomicus yunnanensis Kirkendall & Faccoli sp. n. (Curculionidae: Scolytinae), an unusually aggressive pine shoot beetle from southern China, with a key to the species of Tomicus. Zootaxa. 2008;1819:25–39. [Google Scholar]
- 2.Duan Y, Kerdelhué C, Ye H, Lieutier F. Genetic study of the forest pest Tomicus piniperda (Col., Scolytinae) in Yunnan province (China) compared to Europe: new insights for the systematics and evolution of the genus Tomicus. Heredity. 2004;93:416–422. doi: 10.1038/sj.hdy.6800518. [DOI] [PubMed] [Google Scholar]
- 3.Haack RA, Petrice TR, Poland TM. Tomicus piniperda (Coleoptera: Scolytidae) emergence in relation to burial depth of brood logs. J Econ Entomol. 2000;93:342–346. doi: 10.1603/0022-0493-93.2.342. [DOI] [PubMed] [Google Scholar]
- 4.Eager TA, Berisford CW, Dalusky MJ, Nielsen DG, Brewer JW, et al. Suitability of some southern and western pines as hosts for the pine shoot beetle, Tomicus piniperda (Coleoptera: Scolytidae). J Econ Entomol. 2004;97:460–467. doi: 10.1603/0022-0493-97.2.460. [DOI] [PubMed] [Google Scholar]
- 5.Faccoli M, Anfora G, Tasin M. Responses of the Mediterranean pine shoot beetle Tomicus destruens (Wollaston) to pine shoot and bark volatiles. J Chem Ecol. 2008;34:1162–1169. doi: 10.1007/s10886-008-9503-6. [DOI] [PubMed] [Google Scholar]
- 6.Horn A, Stauffer C, Lieutier F, Kerdelhué C. Complex postglacial history of the temperate bark beetle Tomicus piniperda L. (Coleoptera, Scolytinae). Heredity. 2009;103:238–247. doi: 10.1038/hdy.2009.48. [DOI] [PubMed] [Google Scholar]
- 7.Hegedus Z, Zakrzewska A, Agoston VC, Ordas A, Rácz P, et al. Deep sequencing of the zebrafish transcriptome response to mycobacterium infection. Mol Immunol. 2009;46:2918–2930. doi: 10.1016/j.molimm.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 8.Der JP, Barker MS, Wickett NJ, de Pamphilis CW, Wolf PG. De novo characterization of the gametophyte transcriptome in bracken fern, Pteridium aquilinum. BMC Genomics. 2011;12:99. doi: 10.1186/1471-2164-12-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang XW, Luan JB, Li JM, Bao YY, Zhang CX, et al. De novo characterization of a whitefly transcriptome and analysis of its gene expression during development. BMC Genomics. 2010;11:400. doi: 10.1186/1471-2164-11-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xue J, Bao YY, Li BL, Cheng YB, Peng ZY, et al. Transcriptome analysis of the brown planthopper Nilaparvata lugens. PLoS One. 2010;5:e14233. doi: 10.1371/journal.pone.0014233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karatolos N, Pauchet Y, Wilkinson P, Chauhan R, Denholm I, et al. Pyrosequencing the transcriptome of the greenhouse whitefly, Trialeurodes vaporariorum reveals multiple transcripts encoding insecticide targets and detoxifying enzymes. BMC Genomics. 2011;12:56. doi: 10.1186/1471-2164-12-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Zhang Z, Ye H, Wang H, Clarke SR, et al. Response of Tomicus yunnanensis (Coleoptera: Scolytinae) to infested and uninfested Pinus yunnanensis bolts. J Econ Entomol. 2010;103:95–100. doi: 10.1603/ec09080. [DOI] [PubMed] [Google Scholar]
- 13.Li R, Zhu H, Ruan J, Qian W, Fang X, et al. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res. 2010;20:265–272. doi: 10.1101/gr.097261.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pertea G, Huang X, Liang F, Antonescu V, Sultana R, et al. TIGR Gene Indices clustering tools (TGICL): a software system for fast clustering of large EST datasets. Bioinformatics. 2003;19:651–652. doi: 10.1093/bioinformatics/btg034. [DOI] [PubMed] [Google Scholar]
- 15.Aparicio G, Gotz S, Conesa A, Segrelles D, Blanquer I, et al. Blast2GO goes grid: developing a grid-enabled prototype for functional genomics analysis. Stud Health Technol Inform. 2006;120:194–204. [PubMed] [Google Scholar]
- 16.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acid Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bai X, Zhang W, Orantes L, Jun TH, Mittapalli O, et al. Combining next-generation sequencing strategies for rapid molecular resource development from an invasive aphid species, Aphis glycines. PLoS One. 2010;5:e11370. doi: 10.1371/journal.pone.0011370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Crawford JE, Guelbeogo WM, Sanou A, Traoré A, Vernick KD, et al. De novo transcriptome sequencing in Anopheles funestus using Illumina RNA-seq technology. PLoS One. 2010;5:e14202. doi: 10.1371/journal.pone.0014202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang H, Hu L, Hurek T, Reinhold-Hurek B. Global characterization of the root transcriptome of a wild species of rice, Oryza longistaminata, by deep sequencing. BMC Genomics. 2010;11:705. doi: 10.1186/1471-2164-11-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gene Ontology Consortium. The Gene Ontology project in 2008. Nucleic Acids Res. 2008;36:D440–D444. doi: 10.1093/nar/gkm883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ewen-Campen B, Shaner N, Panfilio KA, Suzuki Y, Roth S, et al. The maternal and early embryonic transcriptome of the milkweed bug Oncopeltus fasciatus. BMC Genomics. 2011;12:61. doi: 10.1186/1471-2164-12-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52:231–253. doi: 10.1146/annurev.ento.51.110104.151104. [DOI] [PubMed] [Google Scholar]
- 24.Feyereisen R. Insect Cytochrome P450. In: Gilbert LI, Iatrou K, Gill SS, editors. Comprehensive Molecular Insect Science Volume 4. Elsevier; 2005. pp. 1–77. [Google Scholar]
- 25.Feyereisen R. Evolution of insect P450. Biochem Soc Trans. 2006;34:1252–1255. doi: 10.1042/BST0341252. [DOI] [PubMed] [Google Scholar]
- 26.Tribolium Genome Sequencing Consortium. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 27.Bass C, Field LM. Gene amplification and insecticide resistance. Pest Manag Sci. 2011;67:886–890. doi: 10.1002/ps.2189. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto K, Teshiba S, Shigeoka Y, Aso Y, Banno Y, et al. Characterization of an omega-class glutathione S-transferase in the stress response of the silkmoth. Insect Mol Biol. 2011;20:379–386. doi: 10.1111/j.1365-2583.2011.01073.x. [DOI] [PubMed] [Google Scholar]
- 29.Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14:3–8. doi: 10.1111/j.1365-2583.2004.00529.x. [DOI] [PubMed] [Google Scholar]
- 30.Friedman R. Genomic organization of the glutathione S-transferase family in insects. Mol Phylogenet Evol. 2011;61:924–932. doi: 10.1016/j.ympev.2011.08.027. [DOI] [PubMed] [Google Scholar]
- 31.Ding Y, Ortelli F, Rossiter LC, Hemingway J, Ranson H. The Anopheles gambiae glutathione transferase supergene family: annotation, phylogeny and expression profiles. BMC Genomics. 2003;4:35. doi: 10.1186/1471-2164-4-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gullipalli D, Arif A, Aparoy P, Svenson GJ, Whiting MF, et al. Identification of a developmentally and hormonally regulated Delta-Class glutathione S-transferase in rice moth Corcyra cephalonica. Comp Biochem Physiol B. 2010;156:33–39. doi: 10.1016/j.cbpb.2010.01.016. [DOI] [PubMed] [Google Scholar]
- 33.Jia H, Halilou AI, Hu L, Cai W, Liu J, et al. Heat shock protein 10 (Hsp10) in immune-related diseases: one coin, two sides. Int J Biochem Mol Biol. 2011;2:47–57. [PMC free article] [PubMed] [Google Scholar]
- 34.Tazir Y, Steisslinger V, Soblik H, Younis AE, Beckmann S, et al. Molecular and functional characterisation of the heat shock protein 10 of Strongyloides ratti. Mol Biochem Parasitol. 2009;168:149–157. doi: 10.1016/j.molbiopara.2009.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Kim KK, Kim R, Kim SH. Crystal structure of a small heat-shock protein. Nature. 1998;394:595–599. doi: 10.1038/29106. [DOI] [PubMed] [Google Scholar]
- 36.Waters ER, Aevermann BD, Sanders-Reed Z. Comparative analysis of the small heat shock proteins in three angiosperm genomes. Cell Stress Chaperones. 2008;13:127–142. doi: 10.1007/s12192-008-0023-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li ZW, Li X, Yu QY, Xiang ZH, Kishino H, et al. The small heat shock protein (sHSP) genes in the silkworm, Bombyx mori, and comparative analysis with other insect sHSP genes. BMC Evol Biol. 2009;9:215. doi: 10.1186/1471-2148-9-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Montfort RLM, Basha E, Friedrich KL, Slingsby C, Vierling E. Crystal structure and assembly of a eukaryotic small heat shock protein. Nat Struct Biol. 2001;8:1025–1030. doi: 10.1038/nsb722. [DOI] [PubMed] [Google Scholar]
- 39.Chen H, Chu Z, Zhang Y, Yang S. Over-expression and characterization of the recombinant small heat shock protein from Pyrococcus furiosus. Biotechnol Lett. 2006;28:1089–1094. doi: 10.1007/s10529-006-9058-y. [DOI] [PubMed] [Google Scholar]
- 40.Aevermann BD, Waters ER. A comparative genomic analysis of the small heat shock proteins in Caenorhabditis elegans and briggsae. Genetica. 2008;133:307–319. doi: 10.1007/s10709-007-9215-9. [DOI] [PubMed] [Google Scholar]
- 41.Sun Y, MacRae TH. Small heat shock proteins: molecular structure and chaperone function. Cell Mol Life Sci. 2005;62:2460–2476. doi: 10.1007/s00018-005-5190-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hamajima F, Hasegawa T, Nakashima I, Isobe K. Genomic cloning and promoter analysis of the GAHSP40 gene. J Cell Biochem. 2002;84:401–407. doi: 10.1002/jcb.10029. [DOI] [PubMed] [Google Scholar]
- 43.Qiu XB, Shao YM, Miao S, Wang L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell Mol Life Sci. 2006;63:2560–2570. doi: 10.1007/s00018-006-6192-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michels AA, Kanon B, Bensaude O, Kampinga HH. Heat shock protein (Hsp) 40 mutants inhibit Hsp70 in mammalian cells. J Biol Chem. 1999;274:36757–36763. doi: 10.1074/jbc.274.51.36757. [DOI] [PubMed] [Google Scholar]
- 45.Hata M, Ohtsuka K. Murine cDNA encoding a novel type I HSP40/DNAJ homolog, mmDjA41. Biochim Biophys Acta. 2000;1493:208–210. doi: 10.1016/s0167-4781(00)00136-6. [DOI] [PubMed] [Google Scholar]
- 46.Wang HS, Wang XH, Zhou CS, Huang LH, Zhang SF, et al. cDNA cloning of heat shock proteins and their expression in the two phases of the migratory locust. Insect Mol Biol. 2007;16:207–219. doi: 10.1111/j.1365-2583.2006.00715.x. [DOI] [PubMed] [Google Scholar]
- 47.Huang LH, Kang L. Cloning and interspecific altered expression of heat shock protein genes in two leafminer species in response to thermal stress. Insect Mol Biol. 2007;16:491–500. doi: 10.1111/j.1365-2583.2007.00744.x. [DOI] [PubMed] [Google Scholar]
- 48.Cheetham ME, Caplan AJ. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones. 1998;3:28–36. doi: 10.1379/1466-1268(1998)003<0028:sfaeod>2.3.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohtsuka K, Hata M. Mammalian HSP40/DNAJ homologs: cloning of novel cDNAs and a proposal for their classification and nomenclature. Cell Stress Chaperones. 2000;5:98–112. doi: 10.1379/1466-1268(2000)005<0098:mhdhco>2.0.co;2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Arya R, Lakhotia SC. Hsp60D is essential for caspase-mediated induced apoptosis in Drosophila melanogaster. Cell Stress Chaperones. 2008;13:509–526. doi: 10.1007/s12192-008-0051-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choresh O, Loya Y, Müller WE, Wiedenmann J, Azem A. The mitochondrial 60-kDa heat shock protein in marine invertebrates: biochemical purification and molecular characterization. Cell Stress Chaperones. 2004;9:38–48. doi: 10.1379/469.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou J, Wang WN, He WY, Zheng Y, Wang L, et al. Expression of HSP60 and HSP70 in white shrimp, Litopenaeus vannamei in response to bacterial challenge. J Invertebr Pathol. 2010;103:170–178. doi: 10.1016/j.jip.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 53.Clark MS, Thorne MA, Toullec JY, Meng Y, Guan LL, et al. Antarctic krill 454 pyrosequencing reveals chaperone and stress transcriptome. PLoS One. 2011;6:e15919. doi: 10.1371/journal.pone.0015919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sonoda S, Fukumoto K, Izumi Y, Yoshida H, Tsumuki H. Cloning of heat shock protein genes (hsp90 and hsc70) and their expression during larval diapause and cold tolerance acquisition in the rice stemborer, Chilo suppressalis Walker. Arch Insect Biochem Physiol. 2006;63:36–47. doi: 10.1002/arch.20138. [DOI] [PubMed] [Google Scholar]
- 55.Saraste M, Sibbald PR, Wittinghofer A. The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci. 1990;11:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 56.Knowlton AA, Salfity M. Nuclear localization and the heat shock proteins. J Biosci. 1996;21:123–132. [Google Scholar]
- 57.Clark MS, Peck LS. HSP70 heat shock proteins and environmental stress in Antarctic marine organisms: A mini-review. Mar Gen. 2009;2:11–18. doi: 10.1016/j.margen.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 58.Welch WJ, Feramisco JR. Purification of the major mammalian heat shock proteins. J Biol Chem. 1982;257:14949–14959. [PubMed] [Google Scholar]
- 59.Picard D. Heat-shock protein 90, a chaperone for folding and regulation. Cell Mol Life Sci. 2002;59:1640–1648. doi: 10.1007/PL00012491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Length distribution of contigs.
(TIF)
Length distribution of scaffolds.
(TIF)
Top BLAST hits from NCBI nr database. BLAST results against the NCBI nr database for all the distinct sequences with a cut-off E value above 10−5 are shown.
(XLS)
Gene Ontology of Tomicus yunnanensis unigenes.
(XLS)
KEGG summary of Tomicus yunnanensis transciptome.
(XLS)
Putative P450 genes identified in Tomicus yunnanensis transcriptome.
(XLS)
Putative heat shock protein genes identified in Tomicus yunnanensis transcriptome.
(XLS)
Data Availability Statement
The Illumina reads of T. yunnanensis have been submitted to NCBI Short Read Archive under the accession number of SRA047283.