Abstract
Proton transfer to and from water is critical to the function of water in many settings. However, it has been challenging to model. Here, we present proof-of-principle for an efficient yet robust model based on Lewis-inspired submolecular particles with interactions that deviate from Coulombic at short distances to take quantum effects into account. This “LEWIS” model provides excellent correspondence with experimental structures for water molecules and water clusters in their neutral, protonated and deprotonated forms; reasonable values for the proton affinities of water and hydroxide; a good value for the strength of the hydrogen bond in the water dimer; the correct order of magnitude for the stretch and bend force constants of water; and the expected time course for Grotthuss transport in water chains.
Keywords: Dissociable, Polarizable, Proton transfer, Hydrogen bonding, Water model, Eigen, Zundel, Hydronium, Hydroxide, Grotthuss mechanism
Introduction
A functional description of water is not complete without proton transfer. In the abiotic world, the amphiprotocity of water molecules provides the basis for proton diffusion that is much faster than for any other cation [1]. In many proteins, H-bonded chains of water molecules provide “wires” [2] that transfer protons to and from the active site. On the surfaces of globular proteins, protons migrating in the surrounding H-bond network alter dielectric properties and ionization states, with consequences for protein function [3]. And in cell membranes, proton transfer through water chains is critical for establishing and utilizing the proton gradients [4].
Unfortunately, simulations of these processes remain daunting. Certainly, quantum mechanics (QM) is the gold standard for modeling changes in chemical bonding. However, even with the most frugal basis sets and with the use of density functional theory, the computational demands for QM grow at a forbidding rate with the size of the system. Empirical models have the potential to provide a practical compromise between accuracy and tractability. However, fully dissociable water models are few [5–8] and less straightforward to implement than their non-reactive counterparts [9–13].
An approach that has received considerable attention in recent years involves mapping proton defects into a low-dimensional space of nuclear coordinates, a.k.a. empirical states. The MS-EVB approach, based on Warshel’s earlier empirical valence bond (EVB) theory [14], is now widely used to simulate the dynamics of excess protons in liquid water [15], through confined single-file water chains [16] and in biological systems [17]. The current limitations of this method involve the lack of an accurate, reactive hydroxide and the non-trivial setup for novice users.
In the present work, we demonstrate that full amphiproticity and polarizability can be described in a pseudo-classical manner, with intuitive constructs, useful accuracy, high efficiency, and straightforward implementation. Our approach is similar in spirit to Stillinger’s Central Force (CF) [18, 19] and Polarization (PM) [5] models. The CF model comprises a set of ingeniously designed, strictly pair-wise potentials that offer a dissociable, three-site water molecule. PM, later improved to PM6 [20], converts the partially charged CF atoms into fully charged oxygen (O2 − ) and hydrogen (H + ) ions, and provides a polarization correction through an iterative electric field calculation. PM6 has been used to study proton conduction through small water chains [21], and a TIP3P/PM6 interface has been reported [22].
In our model, we unpack each O2 − particle into an O6 + core (i.e., a combination of the oxygen nucleus and the 1s electron pair) and four explicit valence electron pairs that are free to move independent of each other and the H + particles. We name this model LEWIS, after the simple chemical bonding theory that inspires the use of electron pairs. The question is whether it is possible to construct pseudo-classical potentials between these Lewis-inspired particles that adequately mimic quantum effects. Since no predefined connectivity or geometric constraints are applied, such potentials would allow all the chemical bonds to be naturally flexible, polarizable, and dissociable [23]. It is this lifting of bonding constraints through heuristically obtained interactions that distinguishes LEWIS from EPEN [24–26] which employs similarly explicit valency in a rigid model of water.
Methods
Potentials
The LEWIS model is defined by six pairwise interactions between three types of particles, valence pairs (V), hydrogen nuclei (H), and oxygen cores (O). For computational efficiency, we are specifically interested in interactions that are smooth, isotropic, analytical and strictly pairwise. In addition, we require that
All six potentials obey the long-range electrostatic limits that correspond to the full ionic charges of the particles, i.e., qO = 6, qH = 1, and qV = −2 in elementary units,
Potentials that involve an electron pair, i.e. UVV, UVH, and UOV, are attenuated at short range to reflect the diffuse nature of the actual electron distribution,
UOV have a short-range repulsive wall that corresponds to Pauli exclusion between valence electron pairs and 1s electrons, and
The repulsions involving O cores, i.e., UOO, UOH, are somewhat attenuated by dispersive attractions [27], and
UHH is a simple monopole–monopole interaction.
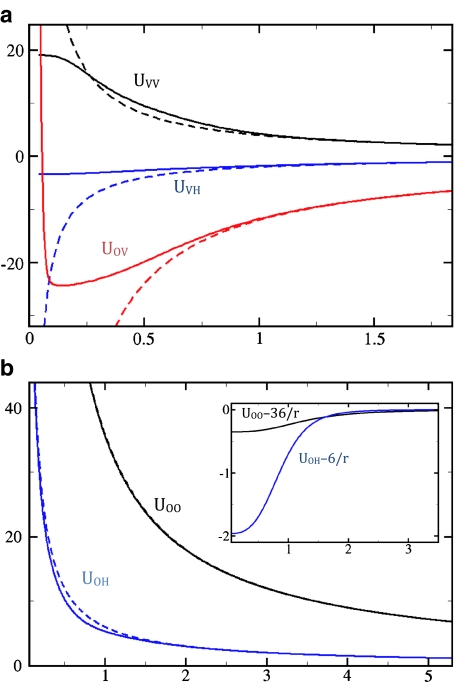
Within this vast functional space, we search for potentials that maximize agreement with structural and thermodynamic data for water monomers and dimers, with minor adjustments to accommodate bulk water properties. The potentials that give the results reported here are shown in Fig. 1.
Fig. 1.
Model potentials (in a.u. = 1,389 kJ/mol) as a function of interparticle distance (in Å): aUVV, UVH, and UOV; bUOO and UOH. For comparison, the corresponding purely Coulombic potentials are shown as dashed lines. The inset in (b) shows the difference between the model potentials and the corresponding pure Coulomb potentials. UHH (not shown) is purely Coulombic
Monte Carlo simulations
Monte Carlo (MC) simulations were performed to locate global energy minima of water clusters. The traditional Metropolis criterion [28, 29] was applied as described elsewhere [30]. Two types of movements were attempted in every step with equal likelihood: single particle displacements, and intact monomer rotations or translations. A monomer is defined as the set of particles including and surrounding an oxygen nucleus within 1.2 Å. Spatial steps are adjusted to obtain acceptance probabilities of ~40% as recommended elsewhere [31]. Low temperatures (T = 1 K) are chosen to accelerate annealing and multiple starting configurations and higher temperatures are attempted in larger clusters.
Molecular dynamics simulations
Newton’s equations of motion are solved using the velocity Verlet algorithm [32] with a time step of 0.2 fs and a fictitious electron pair mass of 1 a.m.u. Temperature is maintained at 300 K by randomly reassigning velocities every 100 steps.
Results
Monomers: water, hydronium, and hydroxide
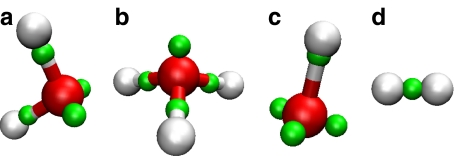
The neutral, protonated, and deprotonated forms of water are central to our training set. All bond lengths and bond angles are included, along with the two associated proton affinities (PAs), PA(H2O) and PA(OH − ). Also included in the training set, albeit with lesser weights, are PA(H − ) and the bond length for dihydrogen (H2). The results of the fit are illustrated in Fig. 2 and summarized in Table 1. Symmetric bending and stretching force constants have the proper order of magnitude but indicate a somewhat stiff water monomer. This stiffness, however, still permits strained configurations, such as in cubic water octamers (see Section 3.3).
Fig. 2.
Monomers: a Water, b hydronium, c hydroxide, and d dihydrogen. Oxygens are rendered red (dark gray in print), protons white, and electron pairs green (light gray in print)
Table 1.
Fit to features in the training set
| Molecule | Feature | Unit | Model | Reference |
|---|---|---|---|---|
| H2O | rOH | Å | 0.95 | 0.958 [33] |
| θHOH | deg | 104.7 | 104.4 [33] | |
| PA | kJ/mol | 643.2 | 691.0 [34] | |
| H3O + | rOH | Å | 0.97 | 0.976 [35] |
| θHOH | deg | 108.9 | 111.3 [35] | |
| OH − | rOH | Å | 0.99 | 0.964 [36] |
| PA | kJ/mol | 1693.6 | 1635.1 [37] | |
| H2 | rHH | Å | 0.66 | 0.741 [38] |
| H − | PA | kJ/mol | 1567.8 | 1675.3 [39] |
| (H2O)2 | rOO | Å | 2.88 | 2.976 [40] |
| θdonor | deg | −46.6 | −51.0 [40] | |
| θacceptor | deg | 32.2 | 57.0 [40] | |
| H5O2+ | rOO | Å | 2.42 | 2.40 [41] |
| H3O2- | rOO | Å | 2.45 | 2.47 [42] |
Features include rOH and rHH, covalent bond lengths, θHOH, HOH bond angle, and PA, proton affinity
The reference values are all from experiment, except for the O–O distance of the deprotonated dimer, which is from a QM calculation
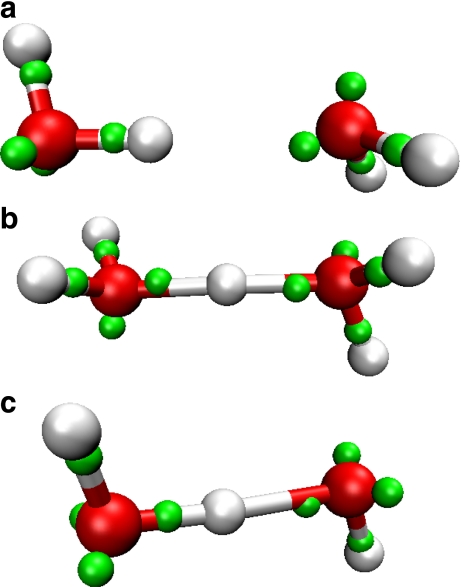
Dimers of water
All three dimers, i.e., H2O..H2O, H2O..H3O + , and H2O..OH − , are included in the training set (Table 1) to determine the UOO potential. In the fit for the neutral dimer, the oxygen–oxygen separation (rOO) is 2.88 Å, and the acceptor and donor are tilted by ~32.2° and ~46.6°, respectively (Fig. 3a). This rOO is closer to the high level QM result (2.91 Å [43]) than to the slightly longer spectroscopic value of 2.98 Å [40]. The H-bond enthalpy of the dimer is 25.9 kJ/mol, and about a fifth of this energy is due to polarization. High-level QM calculations indicate a smaller bond enthalpy (20.50 kJ/mol [43]) which is typically overestimated by empirical models (Table 2).
Fig. 3.
a Neutral, b protonated, and c deprotonated dimers, using the same color scheme is as in Fig. 2
Table 2.
Comparison of water dimers
| Model | rOO (Å) | –ΔE (kJ/mol) |
|---|---|---|
| TIP3P [10] | 2.76 | 27.2 |
| TIP4P [10] | 2.75 | 26.1 |
| TIP5P [10] | 2.68 | 28.4 |
| POL5/TZ [44] | 2.90 | 20.8 |
| TIP4P/FQ [45] | 2.92 | 18.8 |
| Amoeba [46] | 2.89 | 20.8 |
| LEWIS | 2.88 | 25.9 |
| MP2 [43] | 2.91 | 20.5 |
| Experiment | 2.98 [40] | 22.8 [47] |
non-polarizable models tend have shorter dimers to improve liquid state properties [48]
The fit for the protonated dimer (Zundel) has an rOO of 2.42 Å, and the excess proton is approximately equidistant from the two oxygens (Fig. 3b), in agreement with experimental [41, 49] and theoretical findings [50]. The deprotonated dimer (Fig. 3c) has an asymmetrically shared proton and the dangling OHs face in opposite directions, forming an approximately planar configuration. The rOO value of 2.45 Å agrees well with high-level QM calculations (2.47 Å [42]). The association enthalpies of the cationic and anionic Lewis dimers are −159.8 and −138.2 kJ/mol, respectively.
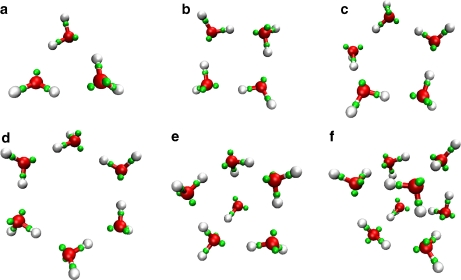
Neutral water clusters
Structures with more than two monomers are beyond the scope of our training set and provide initial tests of the prediction capabilities of LEWIS. The association enthalpies and O–O distances of small water clusters are summarized in Table 3. Among the small neutral clusters, (H2O)3, (H2O)4, and (H2O)5 form closed rings (Fig. 4a–c) whereas the lowest (H2O)6 minimum is a twisted prism, favored by by ~1.8 kJ/mol over the cyclic form (Fig. 4d, e). The water hexamer is suggested to have four low-lying minima [51] and the lowest energy configuration differs even between high-level ab initio calculations [52]. LEWIS predicts two low-lying minima for the water octamer, (H2O)8, S4 and D2d symmetric cubes (Fig. 4f), in agreement with high-level theory [53].
Table 3.
LEWIS predictions for energies of small neutral clusters
| Molecule | Number | nh | –ΔE (kJ/mol) | –ΔEbond (kJ/mol) | rOO (Å) |
|---|---|---|---|---|---|
| (H2O)4 | 4 | 4 | 142.1 | 35.53 | 2.71–2.72 |
| (H2O)5 | 5 | 5 | 191.4 | 38.28 | 2.62–2.71 |
| (H2O)6(cyclic) | 6 | 6 | 241.2 | 40.20 | 2.66–2.67 |
| (H2O)6(twistedprism) | 6 | 7 | 243.0 | 34.72 | 2.63–2.87 |
| (H2O)8(D2d) | 8 | 12 | 374.9 | 31.24 | 2.71–2.88 |
| (H2O)8(S4) | 8 | 12 | 374.9 | 31.24 | 2.71–2.90 |
nh = number of hydrogen bonds
The association enthalpy is given by ΔE = Ecluster - nEH2O, and the energy per hydrogen bond is given by ΔEbond = ΔE/nh. For O–O distances (rOO), ranges are given
Fig. 4.
Small neutral clusters of water: a trimer, b tetramer, c pentamer, d, e cyclic and twisted prism hexamers, and f D2d cubic octamer, using the same color scheme is as in Fig. 2
Small ionized water clusters
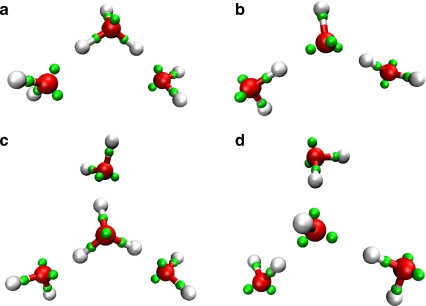
Protonated and deprotonated clusters of water are known to exhibit a wide variety of isomers with increasing size [54]. For brevity, we consider here only clusters of three and four monomers. The DFT prediction is maximum coordination of the ion: both the trimers and the tetramers have their water ion at the center such that the former structure exhibits a pseudo-2-fold- and the latter a pseudo-3-fold symmetry (Eigen-like). LEWIS predicts the same configurations (Fig. 5) with OO separations in agreement to within ∼2%. Less branched ionic tetramer configurations are unfavorable (by ∼15 kJ/mol), consistent with the greater strength of ionic H-bonds over neutral ones.
Fig. 5.
Minimum energy configurations of the protonated and deprotonated trimers (a, b) and tetramers (c, d), using the same color scheme is as in Fig. 2
Long-range proton transfer through a water chain
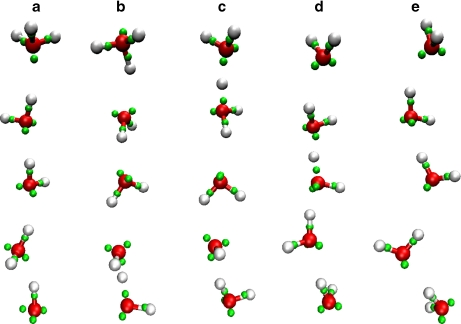
Water chains, a.k.a. “proton wires” are well-known mediators of long-range proton transfers [2, 55–57]. Transport is thought to occur via the “Grotthuss” mechanism [58, 59], which involves alternating “hop-and-turn” steps, i.e. protons hop across H-bonds between molecules and then the molecules reorient for the next passage. To mimic such a process, we set up a five-membered single file chain with water oxygens initially spaced by ∼3 Å and with one terminus protonated and the other deprotonated such that the molecules were initially oriented dysfunctionally, i.e., with their H-bonds donating toward the hydronium end of the chain (Fig. 6a). In constant temperature molecular dynamics simulations (T = 300 K), we observed initial reorientation of monomers to form H-bonds donating toward the hydroxyl end of the chain (Fig. 6b), followed by proton hops that result in migration of the excess proton and the proton hole from the ends (Fig. 6c, d) until they meet and annihilate (Fig. 6e). This series of events is completed within fractions of a picosecond, in agreement with reported estimates of water reorientation times and proton hopping rates [59].
Fig. 6.
Snapshots of Grotthuss-like proton transfer through a five-member chain: a initial arrangement of molecules at t = 0, bt = 50 fs, ct = 100 fs, dt = 140 fs, and et = 320 fs, using the same color scheme is as in Fig. 2. See text for details
Discussion
We were able to arrive at functional forms for pair-wise potentials between Lewis-inspired subatomic particles that produce an energy landscape for oxygen hydrides in which the minima have appropriate locations, depths, and shapes, and are separated by appropriate energy barriers, as judged, respectively, by the structures, proton affinities and H-bond strengths, flexibilities, and proton dynamics in predicted water clusters. The results demonstrate that, via an explicit account of valency, a pseudo-classical model can capture two related effects, polarization and bond rearrangements, in a natural and chemically consistent way, without resorting to computationally demanding multi-body interactions or iterative adjustments. The greatest possible transferability is accomplished by eschewing any reference to connectivity, such that all particles of a given type are truly interchangeable. This is an expected feature of a reactive force field.
Ongoing work is focused on an extension of this approach to carbon and nitrogen hydrides as a prelude to simulating reactions involving small organic molecules. We are also working on a hybrid interface between LEWIS and other commonly used models. We expect that the LEWIS model for water, together with its further generalizations and extensions, will be a valuable tool in various areas, including functionally important water molecules in proteins, proton conductivity in various media, water behavior at surfaces and around solutes, and acid–base catalysis of organic reactions.
Acknowledgements
We thank Peter Jordan and Michael Francis Hagan for helpful discussions, Liliya Vugmeyster for guidance in Monte Carlo simulation, and Ercan Kamber for his advice regarding high performance computation. This work was supported by NIH grant R01EB001035.
References
- 1.Agmon N. Proton solvation and proton mobility. Isr. J. Chem. 1999;39:493. [Google Scholar]
- 2.Nagle JF, Morowitz HJ.Molecular mechanisms for proton transport in membranes Proc. Natl. Acad. Sci. U. S. A. 1978752981978PNAS...75..298N 10.1073/pnas.75.1.298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Careri G. Cooperative charge fluctuations by migrating protons in globular proteins. Prog. Biophys. Mol. Biol. 1998;70:223. doi: 10.1016/S0079-6107(98)00030-3. [DOI] [PubMed] [Google Scholar]
- 4.Decoursey TE. Voltage-gated proton channels and other proton transfer pathways. Physiol. Rev. 2003;83:475. doi: 10.1152/physrev.00028.2002. [DOI] [PubMed] [Google Scholar]
- 5.Stillinger FH, David CW.Polarization model for water and its ionic dissociation products J. Chem. Phys. 19786914731978JChPh..69.1473S 10.1063/1.436773 [DOI] [Google Scholar]
- 6.Halley JW, Rustad JR, Rahman A.A polarizable, dissociating molecular-dynamics model for liquid water J. Chem. Phys. 19939841101993JChPh..98.4110H 10.1063/1.465046 [DOI] [Google Scholar]
- 7.David CW.A variable charge central force model for water and its ionic dissociation products J. Chem. Phys. 199610472551996JChPh.104.7255D 10.1063/1.471438 [DOI] [Google Scholar]
- 8.Lussetti E, Pastore G, Smargiassi E.A fully polarizable and dissociable potential for water Chem. Phys. Lett. 20033812872003CPL...381..287L 10.1016/j.cplett.2003.09.137 [DOI] [Google Scholar]
- 9.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML.Comparison of Simple Potential Functions for simulating liquid water J. Chem. Phys. 1983799261983JChPh..79..926J 10.1063/1.445869 [DOI] [Google Scholar]
- 10.Mahoney MW, Jorgensen WL.A five-site model for liquid water and the reproduction of the density anomaly by rigid, nonpolarizable potential functions J. Chem. Phys. 200011289102000JChPh.112.8910M 10.1063/1.481505 [DOI] [Google Scholar]
- 11.Berendsen, H.J.C., Postma, J.P.M., van Gusteren, W.F., Hermans, J.: Interaction models for water in relation to protein hydration. In: Intermolecular Forces, pp. 331–342. Reidel, Dordrecht (1981)
- 12.Berendsen HJC, Grigera JR, Straatsma TP. The missing term in effective pair potentials. J. Phys. Chem. 1987;91:6269. doi: 10.1021/j100308a038. [DOI] [Google Scholar]
- 13.Guillot B. A reappraisal of what we have learnt during three decades of computer simulations on water. J. Mol. Liq. 2002;101:219. doi: 10.1016/S0167-7322(02)00094-6. [DOI] [Google Scholar]
- 14.Warshel A, Weiss RM. An empirical valence bond approach for comparing reactions in solutions and in enzymes. J. Am. Chem. Soc. 1980;102:6218. doi: 10.1021/ja00540a008. [DOI] [Google Scholar]
- 15.Cuma M, Schmitt UW, Voth GA. A multi-state empirical valence bond model for acid-base chemistry in aqueous solution. Chem. Phys. 2000;258:187. doi: 10.1016/S0301-0104(00)00071-9. [DOI] [Google Scholar]
- 16.Dellago C, Hummer G.Kinetics and mechanism of proton transport across membrane nanopores Phys. Rev. Lett. 2006972459012006PhRvL..97x5901D 10.1103/PhysRevLett.97.245901 [DOI] [PubMed] [Google Scholar]
- 17.Xu JC, Voth GA.Computer simulation of explicit proton translocation in cytochrome c oxidase: the D-pathway Proc. Natl. Acad. Sci. U. S. A. 200510267952005PNAS..102.6795X 10.1073/pnas.0408117102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemberg HL, Stillinger FH.Central-force model for liquid water J. Chem. Phys. 19756216771975JChPh..62.1677L 10.1063/1.430718 [DOI] [Google Scholar]
- 19.Stillinger FH, Rahman A.Revised central force potentials for water J. Chem. Phys. 1978686661978JChPh..68..666S 10.1063/1.435738 [DOI] [Google Scholar]
- 20.Weber TA, Stillinger FH. Reactive collisions of H3O+ and OH- Studied with the polarization model. J. Phys. Chem. 1982;86:1314. doi: 10.1021/j100397a020. [DOI] [Google Scholar]
- 21.Pomes R, Roux B. Theoretical study of H+ translocation along a model proton wire. J. Phys. Chem. 1996;100:2519. doi: 10.1021/jp9525752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pomes R, Roux B.Free energy profiles for H+ conduction along hydrogen-bonded chains of water molecules Biophys. J. 199875331998BpJ....75...33P 10.1016/S0006-3495(98)77492-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lewis GN. The atom and the molecule. J. Am. Chem. Soc. 1916;38:762. doi: 10.1021/ja02261a002. [DOI] [Google Scholar]
- 24.Owicki JC, Shipman LL, Scheraga HA. Structure, energetics, and dynamics of small water clusters. J. Phys. Chem. 1975;79:1794. doi: 10.1021/j100584a010. [DOI] [Google Scholar]
- 25.Snir J, Nemenoff RA, Scheraga HA. Revised empirical potential for conformational, intermolecular, and solvation studies .1. Evaluation of problem and description of model. J. Phys. Chem. 1978;82:2497. doi: 10.1021/j100512a009. [DOI] [Google Scholar]
- 26.Nemenoff RA, Snir J, Scheraga HA. Revised empirical potential for conformational, intermolecular, and solvation studies. 2. Parameterization and testing for water and saturated organic-molecules. J. Phys. Chem. 1978;82:2504. doi: 10.1021/j100512a010. [DOI] [Google Scholar]
- 27.Koide A.A new expansion for dispersion forces and its application J. Phys. B: At. Mol. Phys. 1976931731976JPhB....9.3173K 10.1088/0022-3700/9/18/009 [DOI] [Google Scholar]
- 28.Metropolis N, Ulam S.The Monte Carlo method J. Am. Stat. Assoc. 194944335313410033.28807 10.2307/2280232 [DOI] [PubMed] [Google Scholar]
- 29.Metropolis N, Rosenbluth AW, Rosenbluth MN, Teller AH, Teller E.Equation of state calculations by fast computing machines J. Chem. Phys. 19532110871953JChPh..21.1087M 10.1063/1.1699114 [DOI] [Google Scholar]
- 30.Frenkel D, Smit B. Understanding Molecular Simulation from Algorithms to Applications. San Diego: Academic Press; 2002. [Google Scholar]
- 31.Jorgensen WL, Tirado-Rives J. Molecular modeling of organic and biomolecular systems using BOSS and MCPRO. J. Comput. Chem. 2005;26:1689. doi: 10.1002/jcc.20297. [DOI] [PubMed] [Google Scholar]
- 32.Swope WC, Andersen HC, Berens PH, Wilson KR.A computer-simulation method for the calculation of equilibrium-constants for the formation of physical clusters of molecules - application to small water clusters J. Chem. Phys. 1982766371982JChPh..76..637S 10.1063/1.442716 [DOI] [Google Scholar]
- 33.Jensen P.The potential-energy surface for the electronic ground-state of the water molecule determined from experimental-data using a variational approach J. Mol. Spectrosc. 19891334381989JMoSp.133..438J 10.1016/0022-2852(89)90203-8 [DOI] [Google Scholar]
- 34.Hunter EPL, Lias SG.Evaluated gas phase basicities and proton affinities of molecules: an update J. Phys. Chem. Ref. Data 1998274131998JPCRD..27..413H 10.1063/1.556018 [DOI] [Google Scholar]
- 35.Sears TJ, Bunker PR, Davies PB, Johnson SA, Spirko V.Diode-laser absorption-spectroscopy of D3O+ - determination of the equilibrium structure and potential function of the oxonium ion J. Chem. Phys. 19858326761985JChPh..83.2676S 10.1063/1.449270 [DOI] [Google Scholar]
- 36.Owrutsky JC, Rosenbaum NH, Tack LM, Saykally RJ.The vibration-rotation spectrum of the hydroxide anion (OH-) J. Chem. Phys. 19858353381985JChPh..83.5338O 10.1063/1.449696 [DOI] [Google Scholar]
- 37.Dewar MJS, Dieter KM. Evaluation of Am1 calculated proton affinities and deprotonation enthalpies. J. Am. Chem. Soc. 1986;108:8075. doi: 10.1021/ja00285a033. [DOI] [Google Scholar]
- 38.Lide DR. CRC Handbook of Chemistry and Physics. 76. Boca Raton: CRC Press; 1995. [Google Scholar]
- 39.Bartmess JE, Scott JA, Mciver RT. Scale of acidities in the gas-phase from methanol to phenol. J. Am. Chem. Soc. 1979;101:6046. doi: 10.1021/ja00514a030. [DOI] [Google Scholar]
- 40.Odutola JA, Dyke TR.Partially deuterated water dimers - microwave-spectra and structure J. Chem. Phys. 19807250621980JChPh..72.5062O 10.1063/1.439795 [DOI] [Google Scholar]
- 41.Fridgen TD, McMahon TB, MacAleese L, Lemaire J, Maitre P. Infrared spectrum of the protonated water dimer in the gas phase. J. Phys. Chem., A. 2004;108:9008. doi: 10.1021/jp040486w. [DOI] [Google Scholar]
- 42.Samson CCM, Klopper W. Ab initio calculation of proton barrier and binding energy of the (H2O)OH- complex. J. Mol. Struct., THEOCHEM. 2002;586:201. doi: 10.1016/S0166-1280(02)00066-0. [DOI] [Google Scholar]
- 43.Xantheas SS.On the importance of the fragment relaxation energy terms in the estimation of the basis set superposition error correction to the intermolecular interaction energy J. Chem. Phys. 199610488211996JChPh.104.8821X 10.1063/1.471605 [DOI] [Google Scholar]
- 44.Stern HA, Rittner F, Berne BJ, Friesner RA.Combined fluctuating charge and polarizable dipole models: application to a five-site water potential function J. Chem. Phys. 200111522372001JChPh.115.2237S 10.1063/1.1376165 [DOI] [Google Scholar]
- 45.Rick SW, Stuart SJ, Berne BJ.Dynamical fluctuating charge force-fields - application to liquid water J. Chem. Phys. 199410161411994JChPh.101.6141R 10.1063/1.468398 [DOI] [Google Scholar]
- 46.Ren PY, Ponder JW. Polarizable atomic multipole water model for molecular mechanics simulation. J. Phys. Chem., B. 2003;107:5933. doi: 10.1021/jp027815+. [DOI] [Google Scholar]
- 47.Curtiss LA, Frurip DJ, Blander M.Studies of molecular association in H2O and D2O vapors by measurement of thermal-conductivity J. Chem. Phys. 19797127031979JChPh..71.2703C 10.1063/1.438628 [DOI] [Google Scholar]
- 48.Ponder JW, Case DA. Force fields for protein simulations. Adv. Protein Chem. 2003;66:27. doi: 10.1016/S0065-3233(03)66002-X. [DOI] [PubMed] [Google Scholar]
- 49.Headrick JM, Bopp JC, Johnson MA.Predissociation spectroscopy of the argon-solvated H5O2+ “Zundel” cation in the 1000-1900 cm(-1) region J. Chem. Phys. 2004121115232004JChPh.12111523H 10.1063/1.1834566 [DOI] [PubMed] [Google Scholar]
- 50.Schmitt UW, Voth GA. Multistate empirical valence bond model for proton transport in water. J. Phys. Chem., B. 1998;102:5547. doi: 10.1021/jp9818131. [DOI] [Google Scholar]
- 51.Xantheas SS.Ab-Initio studies of cyclic water clusters (H2O)(n), n = 1-6 .3. Comparison of density-functional with MP2 results J. Chem. Phys. 199510245051995JChPh.102.4505X 10.1063/1.469499 [DOI] [Google Scholar]
- 52.Pedulla JM, Kim K, Jordan KD.Theoretical study of the n-body interaction energies of the ring, cage and prism forms of (H2O)(6) Chem. Phys. Lett. 1998291781998CPL...291...78P 10.1016/S0009-2614(98)00582-X [DOI] [Google Scholar]
- 53.Xantheas SS, Apra E.The binding energies of the D-2d and S-4 water octamer isomers: High-level electronic structure and empirical potential results J. Chem. Phys. 20041208232004JChPh.120..823X 10.1063/1.1626624 [DOI] [PubMed] [Google Scholar]
- 54.Chaudhuri C, Wang YS, Jiang JC, Lee YT, Chang HC, Niedner-Schatteburg G.Infrared spectra and isomeric structures of hydroxide ion-water clusters OH- (H2O)(1-5): a comparison with H3O+ (H2O)(1-5) Mol. Phys. 20019911612001MolPh..99.1161C 10.1080/00268970110046312 [DOI] [Google Scholar]
- 55.Sagnella DE, Laasonen K, Klein ML.Ab inito molecular dynamics study of proton transfer in a polyglycine analog of the ion channel Gramicidin A Biophys. J. 19967111721996BpJ....71.1172S 10.1016/S0006-3495(96)79321-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mei HS, Tuckerman ME, Sagnella DE, Klein ML. Quantum nuclear ab initio molecular dynamics study of water wires. J. Phys. Chem., B. 1998;102:10446. doi: 10.1021/jp982623t. [DOI] [Google Scholar]
- 57.Brewer ML, Schmitt UW, Voth GA. The formation and dynamics of proton wires in channel environments. Biophys. J. 2001;80:1691. doi: 10.1016/S0006-3495(01)76140-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grotthuss CJD. Sur la decomposition de l’eau et des corps qu’elle tient en dissolution a l’aide de l’electricite galvanique. Ann. Chim. 1806;LVIII:54. [Google Scholar]
- 59.Agmon N.The grotthuss mechanism Chem. Phys. Lett. 19952444561995CPL...244..456A 10.1016/0009-2614(95)00905-J [DOI] [Google Scholar]